Abstract
Non-primate hepacivirus (NPHV) has been identified in dogs, horses, bats and wild rodents. The presence of NPHV in dogs outside of the USA however is yet to be established. Here we describe for the first time the detection of NPHV in the UK dog population (described throughout the manuscript as CnNPHV). We examined tissues collected from dogs housed in a rehoming kennel where respiratory disease was endemic. CnNPHV RNA was detected in the tracheal tissues of 48/210 dogs by RT-PCR, and in the liver, lung and/or tracheal tissues of 12/20 dogs. The presence of CnNPHV RNA, and its tropism was confirmed by in situ hybridisation. Histopathological examination demonstrated a trend toward higher histopathological scores in CnNPHV RNA positive respiratory tissues, although, this was not statistically significant. Our findings broaden the geographic distribution and our understanding of CnNPHV. Further evidence of CnNPHV replication in canids warrants investigation.
Keywords: Hepaciviruses, NPHV, Canine hepacivirus, NPHV tropism, RT-PCR, ISH
Highlights
-
•
Non-primate hepacivirus (NPHV) has been detected in UK dog population.
-
•
NPHV has dual respiratory and hepatic tropism.
-
•
This is the first time NPHV RNA was detected in lower respiratory tract.
-
•
This study broaden the geographical distribution and our understanding of NPHV.
Introduction
Hepatitis C virus (HCV), the major causative agent of chronic hepatitis, has a restricted host range. Whilst higher primates are susceptible to experimental infection, natural infection has only been detected in humans (Choo et al., 1994, Choo et al., 1989, Kuo et al., 1989, Saito et al., 1990). An estimated 3% of the world population is chronically infected with HCV, which is causally linked to cirrhosis, liver failure and hepatocellular carcinoma (Shimotohno, 1995). The lack of a non-primate animal model and a limited in vitro cultivation system for HCV make vaccine and drug development for the virus challenging (Fried et al., 2002).
The hepacivirus genus, one of the four genera in the family Flaviviridae, comprises of HCV and GBV-B (Stapleton et al., 2011). GBV-B was isolated during the laboratory passage of human plasma (obtained from an individual with unexplained acute hepatitis) through Tamarins and other new world monkey species, but was never again recovered from a human sample (Stapleton et al., 2011). The natural host of GBV-B has thus remained elusive (Bukh et al., 1999, Bukh et al., 2001, Nam et al., 2004, Stapleton et al., 2011). Recently, Kapoor et al. (2011) described a novel virus with considerable genomic similarity to HCV in respiratory samples of domestic dogs and tentatively named it canine hepacivirus. More recently it has also been detected in horse serum (Burbelo et al., 2012, Lyons et al., 2012), and both viruses are currently grouped as non-primate hepacivirus (NPHV). Comparative phylogenetic analysis confirmed NPHV as the closest genetic relative of HCV to date (Burbelo et al., 2012, Kapoor et al., 2011, Lyons et al., 2012). Whether HCV has originated from animal species remains unclear. However, sustained contact between humans and other species increases the likelihood of the emergence of a virus adapted to infect and cross the species barrier.
The identification of another virus related to HCV raises the possibility that these viruses might be more widespread in mammals than previously thought. More recently, novel hepacivirus has been detected in bats and wild rodents (Drexler et al., 2013, Quan et al., 2013). However, the detection of viral sequences from single tissue or serum samples leaves unanswered questions about the prevalence and tropism of the virus, and its association with disease. In this investigation we examined tissue samples from 210 dogs for the presence of CnNPHV RNA. Due to its potential relevance for hepatitis, and its previously reported detection in respiratory tissues, respiratory and hepatic tissues from a further 20 dogs were examined for the presence of CnNPHV. Clinical respiratory scores and a detailed histological examination of these tissues were undertaken to investigate any association between the presence of CnNPHV, clinical disease and histopathological changes. This is the first study to report the detection of CnNPHV in dogs outside of the USA and the first detailed investigation into the pathogenesis of natural cases of CnNPHV infection in dogs.
Results
Viral RNA sample screening
Population [A]
Tracheal samples were screened for the presence of CnNPHV using the NS3 degenerate nested PCR. Forty eight of the 210 samples (22.9%; 95% CI: 17.4–29.1%) had amplicons between the expected size of 300–500 bp and were confirmed as CnNPHV by sequence analysis ( Table 1). The highest proportion of CnNPHV-positive dogs were detected among dogs sampled in 1999 (37.1%) compared to 21.4% in 2000 and 18.7% in 2001, but this difference was not significant (p=0.080).
Table 1.
Population A: the detection of CnNPHV RNA by measured variable.
| Measured variable |
CnNPHV PCR |
Totala | |||||
|---|---|---|---|---|---|---|---|
|
Positive |
Negative |
||||||
| Number | % | Number | % | Number | % | ||
| Year | 1999 | 13 | 27.1 | 22 | 13.6 | 35 | 16.7 |
| 2000 | 18 | 37.5 | 66 | 40.7 | 84 | 40.0 | |
| 2001 | 17 | 35.4 | 74 | 45.7 | 91 | 43.3 | |
| Length of stay | 1–7 | 5 | 10.4 | 13 | 8 | 18 | 8.6 |
| 8–14 | 25 | 52.1 | 83 | 51.2 | 108 | 51.4 | |
| 15–21 | 6 | 12.5 | 26 | 16.0 | 32 | 15.2 | |
| 22–28 | 5 | 10.4 | 17 | 10.5 | 22 | 10.5 | |
| >28 | 7 | 14.6 | 23 | 14.2 | 30 | 14.3 | |
| Clinical score | 1 | 14 | 29.2 | 60 | 37.0 | 74 | 35.2 |
| 2 | 5 | 10.4 | 32 | 19.8 | 37 | 17.6 | |
| 3 | 24 | 50.0 | 51 | 31.5 | 75 | 35.7 | |
| 4 | 3 | 6.3 | 6 | 3.7 | 9 | 4.3 | |
| 5 | 2 | 4.2 | 13 | 8.0 | 15 | 7.2 | |
| Histology score | 0 | 3 | 6.3 | 15 | 9.3 | 18 | 8.6 |
| 1 | 30 | 62.5 | 87 | 53.7 | 117 | 55.7 | |
| 2 | 10 | 20.8 | 42 | 25.9 | 52 | 24.7 | |
| 3 | 5 | 10.4 | 18 | 11.1 | 23 | 11.0 | |
| Co infection | Y | 20 | 41.7 | 61 | 37.7 | 81 | 38.6 |
| N | 28 | 58.3 | 101 | 62.3 | 129 | 61.4 | |
| Total | 48 | 100 | 162 | 100 | 210 | 100 | |
The total % represents the proportion of dogs in the measured variable out of the total 210 dogs.
Population [B]
To explore the potential of a dual respiratory and hepatic tropism of CnNPHV in UK dogs, tracheal, lung and liver tissues were screened for the presence of viral RNA by PCR using NS3-specific primers. Sequence analysis confirmed the identity of NS3 in the trachea of 7 dogs (35%), in the lung of 3 dogs (15%) and in the liver of 8 dogs (40%) from a total of 20 dogs ( Table 2). In 6 dogs, viral RNA was present in both hepatic and respiratory tissues (3 dogs: lung and liver, 3 dogs: trachea and liver). In 6 dogs viral RNA was detected in single tissues (4 dogs: trachea only, 2 dogs: liver only). In the remaining 8 dogs, all three tissues were negative for CnNPHV RNA.
Table 2.
Population B: the detection of CnNPHV RNA by measured variable.
| Measured variable |
CnNPHV PCR |
Total |
|||||
|---|---|---|---|---|---|---|---|
|
Positive |
Negative |
||||||
| Number | % | Number | % | Number | % | ||
| Clinical score | 1 | 7 | 63.6 | 6 | 85.7 | 13 | 65.0 |
| 2 | 1 | 9.1 | 0 | 0.0 | 1 | 5.0 | |
| 3 | 3 | 27.3 | 1 | 14.3 | 4 | 20.0 | |
| 4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| 5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| Missing data | 1 | 9.1 | 1 | 14.3 | 2 | 10.0 | |
| Total | 12 | 100a | 8 | 100a | 20 | 100b | |
| Co-infection | Y | 6 | 2 | 8 | 40.0 | ||
| N | 6 | 6 | 12 | 60.0 | |||
| Total | 12 | 60.0b | 8 | 40.0b | 20 | 100.0b | |
| Trachea histology score | 0 | 0 | 2 | 2 | |||
| 1 | 1 | 5 | 6 | ||||
| 2 | 2 | 0 | 2 | ||||
| 3 | 2 | 1 | 3 | ||||
| 4 | 2 | 3 | 5 | ||||
| Not examined | 0 | 2 | 2 | ||||
| Total | 7 | 35.0b | 13 | 65.0b | 20 | 100.0b | |
| Cilia (Trachea) histology score | 0 | 0 | 2 | 2 | |||
| 1 | 0 | 1 | 1 | ||||
| 2 | 2 | 3 | 5 | ||||
| 3 | 2 | 1 | 3 | ||||
| 4 | 3 | 4 | 7 | ||||
| Not examined | 0 | 2 | 2 | ||||
| Total | 7 | 35.0b | 13 | 65.0b | 20 | 100.0b | |
| Lung histology score | 0 | 0 | 3 | 3 | |||
| 1 | 1 | 5 | 6 | ||||
| 2 | 0 | 8 | 8 | ||||
| 3 | 1 | 0 | 1 | ||||
| 4 | 1 | 1 | 2 | ||||
| Not examined | 0 | 0 | 0 | ||||
| Total | 3 | 15.0b | 17 | 85.0b | 20 | 100.0b | |
| Liver histology score | 0 | 4 | 4 | 8 | |||
| 1 | 1 | 4 | 5 | ||||
| 2 | 2 | 2 | 4 | ||||
| 3 | 0 | 1 | 1 | ||||
| 4 | 0 | 0 | 0 | ||||
| Not examined | 1 | 1 | 2 | ||||
| Total | 8 | 40.0b | 12 | 60.0b | 20 | 100.0b | |
The total % represents the proportion of dogs by CnNPHV PCR results.
The total % represents the proportion of the total 20 dogs examined.
Sequence analysis
To determine the relatedness between CnNPHV variants from this study with those of previously published sequences, NS3 sequence (165 nt fragments) obtained from 4 dogs in population [A] and all CnNPHV RNA-positive tissues from the 12 dogs in population [B] (n=18) were compared to published sequences from USA, Germany, Brazil and UK. Our canine NS3 variants had overall a relatively higher nucleotide identity to canine (89.6–100% nt and 90.9–100% a.a.) and equine strains (95.7–100% nt and 89–100% a.a.) from the USA than to that of equine strains from the UK, Germany and Brazil ( Table 3).
Table 3.
The percentage of nucleotides and amino acids differences in NS3 between CnNPHV variants in UK dogs identified in this study and equine reference strains from different geographical locations.
| Species | USA | UK | Germany | Brazil | ||||
|---|---|---|---|---|---|---|---|---|
| Accession numbers |
JQ434001, JQ434003 and JQ434006, |
JX948118 and JX948121 |
KC411810 and KC411811 |
KJ469459 and KJ469466 |
||||
| Tissue types | Nucleotides | Amino acids | Nucleotides | Amino acids | Nucleotides | Amino acids | Nucleotides | Amino acids |
| Trachea | 89.6–100 | 90.9–100 | 83.6–89.6 | 89.0–98.1 | 87.8–90.9 | 89.0–98.1 | 83.6–89.6 | 89.0–100 |
| Lung | 91.5–100 | 96.3–100 | 85.4–87.8 | 96.3–98.1 | 89.6–90.9 | 96.3–98.1 | 85.4–89.6 | 96.3–100 |
| Liver | 90.9–100 | 94.5–100 | 85.4–87.8 | 94.5–98.1 | 89.0–90.9 | 94.5–98.1 | 84.8–89.6 | 94.5–98.1 |
| Overall | 89.6–100 | 90.9–100 | 83.6–89.6 | 89.0–98.1 | 87.8–90.9 | 89.0–98.1 | 83.6–89.6 | 89.0–100 |
To further assess the relatedness of our variants to published sequences in regions other than NS3, one E1/E2 tracheal variant (42-T) and two 5′UTR tracheal variants (5-T and 12-T) from population [A] were analysed. Alignment of 42-T E1/E2 (962 nt) to that of the published NPHV sequences revealed a nucleotide identity of 87.8% (92.4% a.a.) to the only published canine sequence (Accession no. JF744991) and 81.3–89.1% (88.5–94.4% a.a.) to 5 equine sequences (Accession no. JQ434001, JQ434003, JQ434004, JX948116 and KF177391). 42-T had 24 amino acid substitutions relative to that of the prototype canine isolate, and 18–38 amino acid substitutions relative to that of equine strains. Upon alignment of the 5′UTR, 5-T and 12-T shared 98.4% nucleotide identity. When 5-T and 12-T were compared to the published 5′UTR sequence, they had 94.5–100% nucleotide identity with the canine (AAK-2011) and equine NPHV sequences (Accession no. JF744991, JQ434001-JQ434008, JX948116, JX948117, JX948120, KJ472766 and KF177391).
Phylogenetic analysis of the NS3 sequence (165 nt fragment) revealed that the canine variants from this study grouped ( Fig. 1A) most closely to the USA canine strains (Accession no. JF744991-96 and JQ181558) and one USA equine isolate (Accession no. JQ434001). Similarly, the 5′UTR of 5T and 12T grouped with the only available canine sequence for this region (Accession no. AF744991) and the same USA equine isolate (Accession no. JQ434001) (Fig. 1C). In contrast, E1/E2 of 42-T grouped most closely with two different equine strains (JQ434003 and JQ434004) (Fig. 1B).
Fig. 1.
Phylogenetics analysis by the Maximum Likelihood method of (A) NS3, (B) E1/E2 and (C) 5′UTR of non-primate hepacivirus sequence amplified in this study. The evolutionary history was inferred by using the maximum Likelihood method of the MEGA 6 programme based on the Kimura- 2 Model for NS3, Hasegawa–Kishino–Yano model for E1/E2 and Tamura–Nei model for 5′UTR. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor–Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, where the topology with superior log likelihood of each tree was selected. The numbers at the nodes (bootstrap values) indicate the frequencies of occurrence for 500 replicate trees. (ο) Published canine hepacivirus sequence, (□) published equine hepacivirus sequence, (Δ) published hepacivirus sequence in bat and (⋄) M62321/HCV subtype 1a was used as an outgroup. (●) Canine hepaciviruses sequenced in this study are indicated. Tree scales represent branch length measured in the number of substitution per site.
In situ localisation of viral RNA in canine trachea, lung and liver
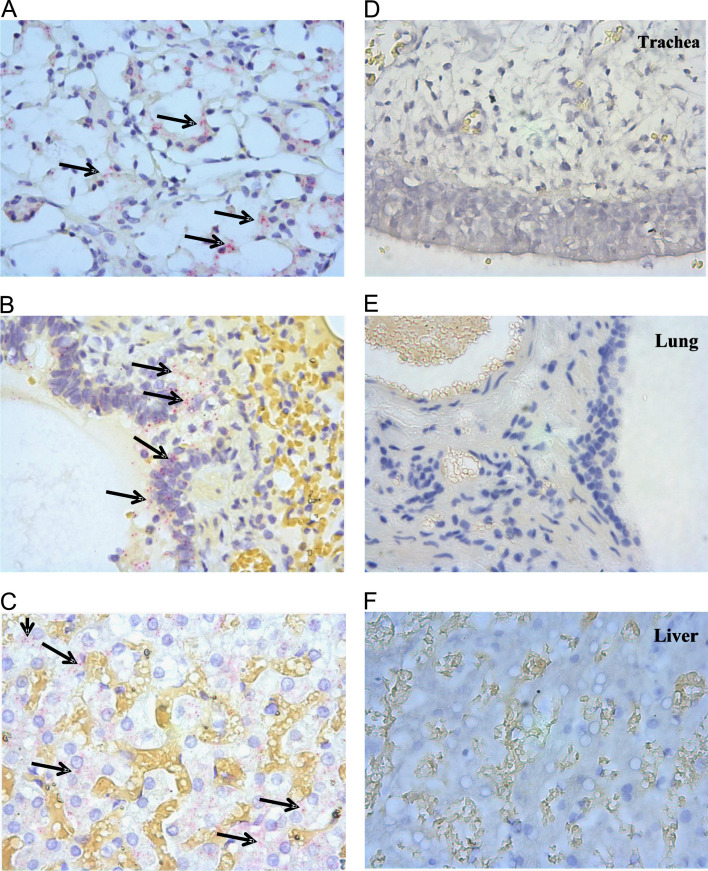
To further confirm the CnNPHV RNA positive results obtained by RT-PCR, RNA was probed by in situ hybridisation (ISH) in liver, lung and tracheal tissues of CnNPHV RNA-positive and negative dogs ( Fig. 2). Foci of viral RNA were located in the cytoplasm of ciliated epithelial cells within the trachea and alveolar epithelium of the lungs in a CnNPHV RNA-positive dog while no staining was observed in a CnNPHV RNA-negative dog. ISH in the liver revealed dispersed as well as focal infection predominantly in the cytoplasm of hepatocytes (Fig. 2). GAPDH staining was evident in all tissues tested (results not shown).
Fig. 2.
In situ hybridisation of CnNPHV RNA in canine trachea, lung and liver. Slides A, B, C are CnNPHV RNA-positive dogs and slides D, E, F are from an uninfected dog. Bright red dots indicate probe bound to CnNPHV genomic RNA as indicated by arrows. Nuclei are stained blue by haematoxylin.
Assessment of the clinical respiratory signs in CnNPHV RNA-positive dogs
To examine the association between the presence of CnNPHV RNA and respiratory disease in dogs, the clinical respiratory scores were evaluated.
Population [A]
Overall 70.8% (n=34/48; 95% CI: 55.9–83.0%) of CnNPHV RNA-positive dogs had signs of clinical respiratory disease compared to 63.0% (n=102/162; 95% CI: 54.7–70.0%) of CnNPHV RNA-negative dogs (Table 1), although this was not significantly different (p=0.118). The biggest difference was seen in dogs with a respiratory score of 3, where 50.0% (n=24/48; 95% CI: 35.2–64.8%) of CnNPHV RNA-positive dogs and 31.5% (n=51/162; 95% CI: 24.4–39.2%) of CnNPHV RNA-negative dogs had a cough and nasal discharge. Clinical respiratory scores of 4 and 5 were recorded in only a small number of CnNPHV RNA-positive dogs (n=3 and n=2).
Population [B]
Upon examination of the clinical respiratory score of the 20 dogs, one CnNPHV RNA-positive dog had a mild cough (score 2) and 3 CnNPHV RNA-positive dogs had a cough and nasal discharge (score 3). The remaining 8 CnNPHV RNA-positive dogs had no obvious respiratory disease (score 1) (one dog had no available score). None of the dogs that were CnNPHV RNA-negative had signs of respiratory disease with the exception of one dog which had a cough and nasal discharge (Table 2).
Assessment of CnNPHV RNA detection in relation to length of stay in the kennel
The association between the detection of CnNPHV RNA and length of stay in the kennel was examined.
Population [A]
Overall there was no significant association between the detection of CnNPHV RNA and the length of stay in the kennel (p=0.967). Of the 48 CnNPHV RNA-positive samples 10.4% (n=5/48, 95% CI:3.5–22.7%) were detected in dogs resident in the kennel for 1–7 days. This increased to 52.1% (n=25/48; 95% CI: 37.2–66.7%) for 8–14 days, followed by a decline to 12.5% (n=6/48; 95% CI: 4.7–25.2%) for 15–21 days and 10.4% (n=5/48; 95% CI: 3.5–22.7%) for 22–28 days (Table 1).
Population [B]
Detection of CnNPHV RNA in population [B] followed a similar trend to population [A], with regard to viral detection and length of stay in the kennel.
Co-infection with other respiratory viruses in the CnNPHV RNA-positive dogs
The incidence of co-infection with important CIRD viruses (CRCoV, CnPnV and CHV) was examined. CDV, CPIV and CAdV (1+2) were not included in this analysis as the dogs were vaccinated against these agents upon entry to the kennel.
Population [A]
Out of the 48 CnNPHV RNA-positive dogs 41.7% (n=20) had evidence of co-infection with one or more CIRD viruses (Table 1). Of these 20 dogs 14 (70%) had respiratory disease (respiratory score 2–5). Of the 28 dogs without co- infection 20 (71.4%) had respiratory disease. Overall, there was no significance difference between the respiratory score of dogs with or without co-infection (p=0.367).
Population [B]
None of the 20 dogs had evidence of co-infection with CnPnV. Of the 12 dogs screened positive for CnNPHV RNA, 5 dogs were positive for CRCoV and one dog was positive for canine herpes virus. The remaining 6 dogs had no evidence of co-infection with any of the respiratory viruses listed above. Of the six dogs with no evidence of co-infection, 50.0% (n=3) were manifested by mild cough (score 2) and cough associated with nasal discharge (score 3).
Histopathological examination of the CnNPHV RNA-positive dogs
To evaluate the relationship between CnNPHV RNA detection and histopathological changes in dogs, formalin-fixed paraffin embedded tissue sections of populations [A] and [B] were examined histologically.
Population [A]
Of the 48 CnNPHV RNA-positive dogs, 62.5% (n=30) had an overall histopathological score of 1 compared to 53.7% (n=87) of CnNPHV RNA-negative dogs (Table 1). Histopathological scores of 2 and 3 were recorded for 20.8% (n=10) and 10.4% (n=5) of CnNPHV RNA-positive dogs respectively, compared with 25.9% (n=42) and 11.1% (n=18) of CnNPHV RNA-negative dogs.
Overall there was no significant association between the histopathological score of respiratory tissues and the presence of CnNPHV RNA in this population (p=0.727). However dogs with histopathological scores of 1–3 were more likely to be positive for CnNPHV RNA than dogs with a histopathological score of 0 (OR: 2.1, 1.7 and 1.4 respectively).
Twenty eight out of the 48 dogs (58.3%) dogs were positive for CnNPHV RNA alone, with no other respiratory viruses detected. In this sub-population there was no significant association between CnNPHV RNA detection and respiratory histopathology (p=0.769).
Population [B]
Sections of trachea, lung and liver were examined.
Trachea
3/20 dogs (15.0%) were positive for only CnNPHV RNA in the trachea, with no other respiratory viruses detected. Histopathological scores for 3 dogs ranged from 2 to 4 with a mean of 3. The same dogs had a mean cilia score of 3. Histopathological changes were principally various degrees of lymphoplasmacytic tracheitis with more occasional involvement of neutrophils. In 3/20 (15.0%) CnNPHV RNA-negative dogs, also negative for other respiratory viruses, the histopathological and cilia scores were 0, 0 , 1 and 0, 1, 2 respectively.
Lung
1/20 dogs (5.0%) were positive for only CnNPHV RNA in the lung, with no other respiratory viruses detected, and had a histopathological score of 3. Histopathological changes included mild to occasionally moderate alveolar histiocytosis and mild alveolar wall thickening, suggestive of early interstitial pneumonia. In 3/20 (15.0%) CnNPHV-negative dogs, also negative for other respiratory viruses, the histopathological scores were 1, 1 and 2 with a mean of 1.33.
Liver
8/20 dogs (40.0%) were positive for CnNPHV RNA in the liver. Histopathological scores were available for 7/8 dogs and ranged from 0 to 2 with a mean of 0.71. Histopathological changes detected in these dogs were relatively non-specific and included mild to moderate sinusoidal congestion and in most cases clear to finely vacuolated zonal hepatocyte cytoplasmic swelling (hydropic degeneration), consistent with mild sublethal cell injury. In 8/20 (40.0%) CnNPHV RNA-negative dogs, histopathological scores were available for 7/8 and ranged from 0 to 3 with a mean of 1.1.
Discussion
CnNPHV was first detected in nasal swabs collected from dogs from a number of different respiratory disease outbreaks in the USA, and in the liver of dogs which died from unexplained gastroenteritis (Kapoor et al., 2011). Subsequent efforts to find CnNPHV in serum, lung, liver or spleen of dogs undergoing investigations for respiratory disease or idiopathic hepatitis were unsuccessful (Bexfield et al., 2014, Drexler et al., 2013, Lyons et al., 2012). Here, we report for the first time the detection of CnNPHV in the respiratory and hepatic tissues of UK dogs.
We screened the tracheal tissues of 210 dogs (population [A]) for CnNPHV RNA of which 48 dogs (22.9%) were positive for the virus by RT-PCR and confirmed by DNA sequencing.
To explore the previously reported dual respiratory and hepatic tropism of CnNPHV (Kapoor et al., 2011), we examined also the trachea, lung and liver of 20 dogs (population [B]). CnNPHV RNA was detected in 12 of the dogs (60%) and in all three tissue types (lung of 3 dogs, trachea of 7 dogs and liver of 8 dogs) by RT-PCR and DNA sequencing. The presence of the virus within the tissues was further confirmed by in situ hybridisation, where foci of viral RNA were detected only in the cytoplasm of ciliated epithelial cells in the trachea, alveolar epithelium in the lung and hepatocytes in CnNPHV RT-PCR positive dog.
To examine the genetic relatedness between variants detected in our study and that of published strains we aimed to sequence the 5′ UTR and E1/E2 regions of the CnNPHV genome, in addition to the NS3 domain. The conserved 5′UTR is important for virus replication/translation and has been used for HCV genotyping (Pestova et al., 1998, Simmonds, 2004). The E1/E2 envelope genes facilitate virus entry into host cells and display the most diverse sequences among HCV genomes (Hijikata et al., 1991, Kato et al., 1992, Weiner et al., 1991). Analysis of the NS3 sequence generated from some of the variants in this study, although based on a short fragment (165 nt), showed that differences existed between the strains detected in this study and with those previously published, but was most similar to the published canine strain AAK-2011 from the USA (Accession no. JF744991) and the equine strain (NZP-1). NZP-1 was previously shown to share high degree of nucleotide identity with the canine strain AAK-2011 (Burbelo et al., 2012, Kapoor et al., 2011, Lyons et al., 2014, Lyons et al., 2012). Likewise 5′UTR obtained from two tracheal variants showed substantial nucleotides similarity to canine and equines published sequences. E1/E2 sequence obtained from one tracheal variant (42-T) showed the least similarity to canine and equine published sequence compared to NS3 and 5′UTR reflecting the heterogeneity of E1/E2 genes among HCV genome (Hijikata et al., 1991, Kato et al., 1992, Weiner et al., 1991). Sequence analysis of E1/E2 and 5′UTR from more samples would be advantageous. Since the main focus of this study was to establish the presence of CnNPHV in the UK canine population further sequence analysis of E1/E2 from more samples was not perused. A standalone dedicated study focused on genomic variation is required. What this study does highlight however, even with the limited E1/E2 sequence obtained, is the high degree of diversity in E1/E2 between CnNPHV genomes, indicating the requirement for more broadly reactive primers to successfully target this genomic region from multiple variants.
The NS3, 5′UTR and E1/E2 variants identified in this study confirmed the results of RT-PCR and showed a degree of diversity from that of published sequence, particularly those of equine origin. However full genome sequences are required to accurately postulate sequence diversity.
Our study confirms the findings initially reported (Kapoor et al., 2011) by demonstrating the presence of CnNPHV-RNA in canine tissues, and extends their findings by demonstrating for the first time the presence of CnNPHV RNA in lower respiratory tract. The failure of other studies to find evidence of CnNPHV in the UK dogs (Lyons et al., 2012) may be due to differences in the dog population screened. In this study, kennelled dogs were targeted since infectious agents are often more likely to be present within a kennel environment due to crowded conditions, which facilitates transmission, and/or the often increased susceptibility of animals due to malnourishment, underlying disease or immunosuppression.
Respiratory disease in particular spreads quickly among kennelled dogs, and studies have shown that many of the associated pathogens are readily transmitted during the first two weeks of the dogs stay (Chalker et al., 2004, Erles et al., 2004, Mitchell et al., 2013b). Although not statistically significant (p=0.967), CnNPHV RNA was most frequently detected in dogs resident in the kennel for 8–14 days in population [A] (Table 1). The overall higher detection rate (60%) of CnNPHV RNA in population [B] could therefore be a result of biased sampling in which a majority of samples (41.7%) were collected from dogs resident in the kennel between 8 and 14 days.
This suggests that CnNPHV may be transmitted to dogs during the 1st and 2nd weeks of entering the kennel as has been shown for other canine viruses in this kennelled population (Erles et al., 2003, Mitchell et al., 2013b). To further investigate this would require comprehensive serological survey using paired samples, and daily swabbing of dogs following entry to the kennel for viral detection.
CnNPHV transmission within the same species and across species has been previously reported (Burbelo et al., 2012, Lyons et al., 2014). In one study the CnNPHV isolate detected in a commercial horse serum pool NZP-1 was almost identical to that from a dog, providing evidence for the ability of NPHV to cross species (Burbelo et al., 2012). Whilst in a recent study by Lyons et al. (2014), all horses that were stable mates of a CnNPHV viraemic horse seroconverted as did a dog on the same farm (Lyons et al., 2014).
Due to the presence of CnNPHV RNA in tissues from both the upper and lower airways of dogs the clinical respiratory signs were analysed against the CnNPHV RT-PCR result to explore the possibility of an association between CnNPHV and respiratory disease. Overall no significant association was detected (p=0.118). However there was a general trend toward a higher incidence of respiratory disease in CnNPHV RNA-positive dogs (70.8%) when compared to hepacivirus negative dogs (63.0%) in population [A]. The lower percentage of dogs with signs of respiratory disease in population [B] compared to population [A] may be biased by the small number of dogs.
Given this finding it is likely that the disease observed was primarily caused by other pathogens. Indeed canine infectious respiratory disease (CIRD) is a multifactorial disease involving a number of infectious viral agents (Priestnall et al., 2014). Whilst all the dogs in this study were vaccinated on the day of entry against CPIV, CDV, CAdV-2, other CIRD pathogens (CRCoV, CHV, CnPnV) have previously been associated with the onset and persistence of CIRD in these dog populations (Erles et al., 2004, Erles et al., 2003, Mitchell et al., 2013b). In examining the dogs for co-infection with these viruses, we did not observe any significant difference in the severity of respiratory disease between CnNPHV RNA-positive dogs with and without viral co-infection. Previously reported but not examined within the context of co-infections in this study is role of bacteria in the pathogenesis of CIRD. Previously Bordetella bronchiseptica, Mycoplasma cynos and Streptococcus zooepidemicus have also been associated with increased severity and persistence of disease within this cohort (Chalker et al., 2003a, Chalker et al., 2003b, Chalker et al., 2004).
Histopathological examination of the lung and tracheal tissues does not reveal a statistically significant association between the presence of CnNPHV and the histopathological changes observed. However, given the high incidence of co-infection with multiple respiratory viruses this association might be harder to prove. To address this, the number of dogs positive solely with CnNPHV RNA was assessed and, although not statistically significant, both histopathological scores in trachea and lung and cilia scores in trachea were higher in CnNPHV RNA-positive versus CnNPHV RNA-negative dogs (population [B]). The typical histopathological changes observed within these latter dogs, including mild to moderate lymphoplasmacytic tracheitis and lesions typical of early interstitial pneumonia are those that would be expected with a viral respiratory infection.
The complexity of CIRD therefore makes it difficult to tease out the contributions individual pathogens make, without much larger clinical studies, or the use of experimental challenge models. Within the context of a multifactorial disease these findings therefore cannot exclude the possibility of a role for CnNPHV in CIRD, and further work is required to fully assess this.
The high prevalence of CnNPHV RNA in 8/20 liver supports the hypothesis that CnNPHV may have a hepatic tropism like HCV. However the fact that CnNPHV RNA can also be detected from respiratory tissue indicates a wider tissue tropism. Although the liver is considered to be the primary target for HCV, extrahepatic manifestations are well recognised among patients with chronic HCV infection (Hadziyannis, 1997, Latt et al., 2012, Zignego et al., 2012). For instance, HCV RNA has been detected in saliva of infected patients (Pastore et al., 2006, Ruggieri et al., 1996). Whether or not HCV infection is associated with respiratory disease in humans remain unknown. Most clinical cases of HCV infection go unrecognised until an initial diagnosis is made during the late stages of chronic liver disease, and therefore data relating to the extrahepatic tropism of the virus during the early phase of infection is limited.
Histopathological changes in the liver were roughly equivalent in both CnNPHV RNA-positive and negative dogs and thus, although the virus is present, there is no definitive association with morphological changes in this small population [B]. The 7 available histopathological scores of the CnNPHV RNA-positive liver samples had a mean of 0.71 indicative of mild changes. Assuming these dogs were infected with CnNPHV, the liver samples would have been collected at a relatively early stage of viral infection and thus given that in HCV, hepatic lesions are most severe in late-stage chronic infections, we cannot exclude the possibility that more chronic lesions, such as fibrosis or biliary hyperplasia may have developed in these dogs. This latter theory should prompt a wider investigation into the role of CnNPHV in chronic hepatitis, a common and largely idiopathic clinical problem in dogs. Indeed morphological changes in the liver are often preceded by functional changes, such as elevations in hepatocellular enzymes which were not analysed in this retrospective study. Interestingly two recent studies in UK and Netherlands reported the absence of CnNPHV in liver samples from dogs with chronic and idiopathic hepatitis (Bexfield et al., 2014, Drexler et al., 2013). These studies however may not be representative of the global picture.
The histopathological changes observed with chronic HCV infection are characterised by portal inflammation including cholangiohepatitis and periportal injury in the form of piecemeal necrosis of hepatocytes and fibrosis (Scheuer et al., 1992) where the latter is the progressive component of the disease leading eventually to liver cirrhosis. However in HCV infection, there is a disparity between the observed numbers of patients with clinical evidence of cirrhosis as compared to the proportion of patients with chronic HCV in whom histological evidence of cirrhosis develops due to the indolent nature of HCV-induced cirrhosis (Sanchez-Tapias et al., 1990). Additionally, HCV replication occurs in the absence of obvious liver damage during the first few months after infection (Shimizu and Purcell, 1989). If CnNPHV has a similar very slow disease progression and the fact that canine species has shorter life span than human, the expected scenario would be that most CnNPHV-infected dogs would probably die before showing any evidence of liver damage. Nevertheless the lack of evidence of histological abnormalities in liver tissues may also be explained by the possibility that these dogs have recently acquired the infection whilst in the kennel. Additionally the age of these dogs ranged from 1 to 4 years old, targeting older dogs would be a useful further strategy to demonstrate any evidence of liver injury.
In conclusion, CnNPHV has been detected in two populations of dogs. Further analysis demonstrated CnNPHV RNA present within liver, lung and tracheal tissues. To our knowledge this is the first in depth investigation of CnNPHV in the UK dogs. In our study of relatively young dogs (1–4 years), we have observed the highest levels of infection in the liver of these animals but, as yet, little evident pathology. It would be valuable to examine older dogs, particularly those with liver disorders to assess whether there is any significant association between CnNPHV and hepatitis. As yet the source of CnNPHV in dogs is unknown. It is possible the virus detected in this study was introduced to these dogs from either contact with other dogs, wildlife or other sources such as vaccine contaminated with animal serum or ingestion of food. Further investigation to examine whether or not CnNPHV is replicating in these dogs is warranted, and a further evolutionary analysis of whole viral genome is required to investigate the natural history of these novel viruses.
Materials and methods
Study population
(i) Population definition
Samples used in this study were obtained from dogs residing in a well-established canine rehoming centre in London, UK with a history of endemic respiratory disease. The dogs were grouped as follows: Population [A]: 210 dogs housed in the kennel between 1999 and 2001 and population [B]: 20 dogs housed in the kennel in 2004.
All dogs were vaccinated upon entry into the kennel with KAVAK DA2 PiP69 (Fort Dodge), a live attenuated vaccine for distemper virus (CDV), canine adenovirus type 2 (CAdV-2), canine parainfluenza virus (CPIV), and canine parvovirus (CPV), and a killed leptospirosis vaccine (Fort Dodge).
(ii) Data collection
The kennel admission history, vaccination and clinical respiratory scores of the two populations were recorded as described previously (Erles et al., 2003, Mitchell et al., 2013b). The health status of each dog was assessed twice a day by a veterinary clinician and graded on the day of euthanasia as follows: (1) no respiratory signs, (2) mild cough, (3) cough and nasal discharge, (4) cough, nasal discharge, and inappetence, (5) evidence of bronchopneumonia.
(iii) Sample collection
Tissue samples were collected at post-mortem examination with informed consent where dogs were euthanised for welfare and ethical reasons unrelated to this study, ranging from behavioural problems to signs of severe respiratory disease. Multiple respiratory tissues samples were collected in duplicate where one section was frozen at −70 °C for virological analysis and the other fixed in neutral buffered formalin (NBF) prior to histological preparation.
Sample analysis
Tracheal samples from population [A] and tracheal, lung and liver samples from population [B] were analysed for the presence of CnNPHV by RT-PCR and assessed for histological changes.
(i) RNA extraction, cDNA synthesis and PCR
RNA was extracted from tracheal, lung and liver tissues using the Qiagen RNeasy tissue purification kit according to the manufacturer׳s instructions. Approximately 25–30 mg of homogenised tissue was used and the RNA was eluted in 40 µl of nuclease-free water. Total RNA was reverse transcribed into cDNA using random hexamer primers and superscript III reverse transcriptase (Life Technology) as described by the manufacturers.
Detection of GAPDH by PCR
To validate cDNA synthesis, all samples were tested for the presence of the house keeping gene glyceraldhyde-3-phosphate dehydrogenase (GAPDH) by PCR as described by Grone et al. (1996) using GoTaq® Flexi DNA Polymerase kit (Promega) following the manufacturer׳s protocol.
Detection of CnNPHV by NS3 PCR
Tracheal samples from population [A] were screened for CnNPHV by nested PCR using previously described degenerate primers targeting the CnNPHV NS3 gene (Burbelo et al., 2012, Kapoor et al., 2011). Amplicons of 300–500 bp were generated using Hotstart Taq polymerase (Qiagen). The primary round underwent an initial denaturation at 95 °C for 7 min followed by 10 cycles of [95 °C for 40 s, 60 °C (−0.5 °C/cycle) for 45 s and 72 °C for 1 min] and 30 cycles of [95 °C for 30 s, 54 °C for 40 s and 72 °C for 1 min] with final elongation step at 72 °C for 10 min. The secondary round underwent the same cycling parameters as the primary round except for the annealing temperature which was 62 °C for 10 cycles followed by 58 °C for 30 cycles. NPHV positive control plasmid DNA provided by Kapoor et al. (2011) was included in each run.
Tracheal, lung and liver samples from population [B] were screened for CnNPHV using a single round PCR targeting a 208 nt fragment of the CnNPHV NS3 gene ( Table 4). Primers sequences were deduced using Primer-Blast (Ye et al., 2012) from the initial consensus sequence for NS3 obtained from three Population [A] samples amplified using the nested PCR above. The PCR was performed using 2x PCRBIO Ultra Mix kit (PCR BIOSYSTEMS) with an initial denaturation at 95 °C for 2 min and 40 cycles of [95 °C for 15 s, 55 °C for 15 s and 72 °C for 30 s] followed by a final elongation at 72 °C for 2 min.
E1/E2 and 5′UTR amplification. A 962 bp region of the E1and E2 genes (spanning 868–1826 nt of canine hepacivirus isolate AAK-2011 (Accession number JF744991)) and the 5′UTR (301 bp) were amplified by nested PCR using Amplitaq Gold DNA polymerase (life Technologies), and primers described elsewhere (Burbelo et al., 2012, Kapoor et al., 2011). The primary round underwent initial denaturation for 8 min at 95 °C followed by 10 cycles of [95 °C for 40 s, 59 °C (−0.5 °C/cycle) for 45 s and 72 °C for 80 s] then 30 cycles of [95 °C for 30 s, 55 °C for 40 s and 72 °C for 80 s] and a final elongation at 72 °C for 5 min. The secondary round was performed as described for the primary round except the annealing temperature for the first 10 cycles was 65 °C (−0.5 °C/cycle).
Detection of co-infection with canine infectious respiratory disease (CIRD) associated viruses. Respiratory samples were screened for the presence of known CIRD viral pathogens as described previously: canine herpes virus (HCV), canine respiratory coronavirus (CRCoV) (Erles et al., 2003, Erles et al., 2004) and canine pneumovirus (CnPnV) except 2 dogs (Mitchell et al., 2013b).
Table 4.
Hepacivirus specific primer sequences for NS3 helicase gene.
| Primer name | Position | Sequence (5′-3′) |
|---|---|---|
| NS3F1 | Sense | ACTTGCTACTGCTACGCCAC |
| NS3R1 | Ani-sense | AGCATTAGCTCCGGCCTTTC |
(ii) Histopathology
Formalin-fixed paraffin-embedded tissues sections were processed and stained with haematoxylin and eosin. The histological sections were examined, blind, by board-certified veterinary pathologists (Population [A] by HB and Population [B] by SLP).
Population [A]
Multiple respiratory tissues were graded for several features (neutrophils, lymphocytes, plasma cells, macrophage, MALT reactivity, oedema, fibrin and evidence of repair) from 0 (none or normal level), through to 3 (severely affected). An overall grade (0–3) for the respiratory tract as a whole was determined based on an assessment of all the changes taken together as described previously (Mitchell et al., 2013b).
Population [B]
Tracheal, lung and liver tissues were each assigned an overall histopathological score. For the trachea, a separate score was assigned for cilia morphology as described previously (Mitchell et al., 2013a). Briefly, scores were based on the severity and distribution of lesions, ranging from 0 (no significant histological abnormality recognised) to 4 (marked, extensive/diffuse changes). Cilia were scored from 0 (dense, long cilia) to 4 (marked irregularity or diffuse cilia loss)
(iii) In situ hybridisation
Viral RNA was detected by in situ hybridisation using the QuantiGene® ViewRNA ISH Tissue 2-Plex Assay kit (Panomics/Affymetrix, Inc) according to the manufacturers׳ instruction. A mixture of three Probes sets targeting CnNPHV genomic RNA regions (nucleotides 840–2040, 3973–5010 and 7600–8634 of Accession no. JQ434001) corresponding to the core/E1 and E2 envelopes, NS3 helicase/NS4a and NS5b (RdRp) genes were used (Affymetrix). A Probe set for GAPDH RNA (Affymetrix, Accession no. NM_001003142) was employed as a positive control. Formalin-fixed, paraffin-embedded (FFPE) tissues sections (5+/−1 μm) were prepared on SuperFrost Plus slides. Sections were fixed for 2 h at 60 °C, deparaffinised, dehydrated, and permeabilized before the hybridisation steps and staining with Fast Red substrate (QG ViewRNA Chromogenic Signal Amplification kit (Panomics/Affymetrix, Inc.). The GAPDH positive control was detected with Fast Blue substrate (Panomics/Affymetrix, Inc.). Stained tissue sections were briefly treated with Meyer׳s haematoxylin (Sigma/Aldrich, UK) and dried before visualised by bright field light microscopy at 40× magnification.
(iv) Statistical analysis of data
Data were recorded in excel spread sheets and imported into IBM SPSS Statistics 22 for analysis. Associations between the presence of CnNPHV in tissues and clinical respiratory score at euthanasia, time in kennel, respiratory score, histology score and year of collection were examined using Chi-square and Fisher׳s exact tests with risk ratio calculator.
(v) DNA sequencing and phylogenetic analysis
PCR products were separated on an agarose gel and purified using the Qiaquick gel extraction purification kit (Qiagen) prior to cloning and/or DNA sequencing. PCR amplicons from the NS3 (300–500 bp or 208 bp), E1/E2 (962 bp) and 5′UTR (301 bp) were cloned into the pT7blue2 blunt vector (Novagen, UK), pGEM-T Easy vector (Promega, UK) or pCR2.1 TA cloning vector (Life Technology) and sequenced (DNA Sequencing and Services, University of Dundee, UK). A consensus sequence for each sample was generated from two or more clones, sequenced on both strands, using Clustal Omega multiple sequence alignment after the deletion of primers binding sites.
Nucleic acid similarity searches were performed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast). Amino acid (a.a.) sequences were deduced from the nt consensus sequences using EMBOSS Tools at http://www.ebi.ac.uk/Tools/st/emboss_transeq/. Multiple sequence alignments were performed and identity matrices determined using OMEGA Clustal at http://www.ebi.ac.uk/Tools/msa/clustalo/. Phylogenetic analysis was conducted using MEGA version 6 (Tamura et al., 2013).
GenBank accession numbers
CnNPHV helicase, 5′UTR and E1/E2 partial sequences used in multiple sequence alignment and phylogenetics have been assigned GenBank Accession numbers. NS3: KR349931-KR349952. The 5′UTR: KR349929 (5-T) and KR349930 (12-T), E1/E2: KR349928 (42-T).
Published CnNPHV, NPHV and HCV sequences used in this study and their accession numbers are listed below:
AK-2012 isolate NZP-1: (JQ434001), AK-2012 isolate NPHV-G1-073: (JQ434002), AK-2012 isolate NPHV-A6-006: (JQ434003), AK-2012 isolate NPHV-B10-022 (JQ434004), AK-2012 isolate NPHV-F8–068 (JQ434005), AK-2012 isolate NPHV-G5-077: (JQ434006), AK-2012 isolate NPHV-H10-094: (JQ434007), AK-2012 isolate NPHV-H3-011: (JQ434008), SMKL-2012 isolate NPHV_EF369_98: (JX948116), SMKL-2012 isolate NPHV_EF3317_98 (JX948117), SMKL isolate NPHV_EF319_98: (JX948118), SMKL-2012 isolate NPHV_EF330_97 (JX948120), SMKL isolate EF319_97: (JX948121), AAK-2011: (JF744991), AAK-2011 isolate CHV- K217: (JF744992), AAK-2011 isolate CHV-K188: (JF744993), AAK-2011 isolate CHV- K180: (JF744994), AAK-2011 isolate CHV- K154: (JF744995), AAK-2011 isolate CHV- K136: (JF744996), CHV: (JQ181558), Eq179/GER/2012 (KC411810), Eq285/GER/2012 (KC411811), AS9218 (KJ469459), AS9330 (KJ469466), horse/DH1/HUN/2013 (KF177391), Bat-PDB-491 (KC796078) HCV- subtype 1a: (M62321).
Acknowledgments
We wish to acknowledge the substantial help with providing constructive discussion and valuable reagents given by Professors Charlie Rice and Amit Kapoor. We also would like to extend our sincere acknowledgements to Dr Troels Scheel for his technical support and primers. This work is funded by the Royal Veterinary College/Internal Grant Scheme 2012 (Grant code 2578).
References
- Bexfield N.H., Watson P.J., Heaney J., Heeney J.L., Tiley L. Canine hepacivirus is not associated with chronic liver disease in dogs. J. Viral Hepat. 2014;21:223–228. doi: 10.1111/jvh.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J., Apgar C.L., Govindarajan S., Purcell R.H. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J. Med. Virol. 2001;65:694–697. doi: 10.1002/jmv.2092. [DOI] [PubMed] [Google Scholar]
- Bukh J., Apgar C.L., Yanagi M. Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology. 1999;262:470–478. doi: 10.1006/viro.1999.9941. [DOI] [PubMed] [Google Scholar]
- Burbelo P.D., Dubovi E.J., Simmonds P., Medina J.L., Henriquez J.A., Mishra N., Wagner J., Tokarz R., Cullen J.M., Iadarola M.J., Rice C.M., Lipkin W.I., Kapoor A. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 2012;86:6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker V.J., Brooks H.W., Brownlie J. The association of Streptococcus equi subsp. zooepidemicus with canine infectious respiratory disease. Vet. Microbiol. 2003;95:149–156. doi: 10.1016/S0378-1135(03)00155-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker V.J., Owen W.M., Paterson C., Barker E., Brooks H., Rycroft A.N., Brownlie J. Mycoplasmas associated with canine infectious respiratory disease. Microbiology. 2004;150:3491–3497. doi: 10.1099/mic.0.26848-0. [DOI] [PubMed] [Google Scholar]
- Chalker V.J., Toomey C., Opperman S., Brooks H.W., Ibuoye M.A., Brownlie J., Rycroft A.N. Respiratory disease in kennelled dogs: serological responses to Bordetella bronchiseptica lipopolysaccharide do not correlate with bacterial isolation or clinical respiratory symptoms. Clin. Diagn. Lab. Immunol. 2003;10:352–356. doi: 10.1128/CDLI.10.3.352-356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q.L., Kuo G., Ralston R., Weiner A., Chien D., Van Nest G., Han J., Berger K., Thudium K., Kuo C. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc. Natl. Acad. Sci. USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q.L., Kuo G., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Muller M.A., Lukashev A.N., Gmyl A., Coutard B., Adam A., Ritz D., Leijten L.M., van Riel D., Kallies R., Klose S.M., Gloza-Rausch F., Binger T., Annan A., Adu-Sarkodie Y., Oppong S., Bourgarel M., Rupp D., Hoffmann B., Schlegel M., Kummerer B.M., Kruger D.H., Schmidt-Chanasit J., Setien A.A., Cottontail V.M., Hemachudha T., Wacharapluesadee S., Osterrieder K., Bartenschlager R., Matthee S., Beer M., Kuiken T., Reusken C., Leroy E.M., Ulrich R.G., Drosten C. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013;9:e1003438. doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Dubovi E.J., Brooks H.W., Brownlie J. Longitudinal study of viruses associated with canine infectious respiratory disease. J. Clin. Microbiol. 2004;42:4524–4529. doi: 10.1128/JCM.42.10.4524-4529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Toomey C., Brooks H.W., Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003;310:216–223. doi: 10.1016/S0042-6822(03)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M.W., Shiffman M.L., Reddy K.R., Smith C., Marinos G., Goncales F.L., Jr., Haussinger D., Diago M., Carosi G., Dhumeaux D., Craxi A., Lin A., Hoffman J., Yu J. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- Grone A., Weckmann M.T., Capen C.C., Rosol T.J. Canine glyceraldehyde-3-phosphate dehydrogenase complementary DNA: polymerase chain reaction amplification, cloning, partial sequence analysis, and use as loading control in ribonuclease protection assays. Am. J. Vet. Res. 1996;57:254–257. [PubMed] [Google Scholar]
- Hadziyannis S.J. The spectrum of extrahepatic manifestations in hepatitis C virus infection. J. Viral Hepat. 1997;4:9–28. doi: 10.1046/j.1365-2893.1997.00120.x. [DOI] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Ohkoshi S., Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem. Biophys. Res. Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Gerold G., Qaisar N., Jain K., Henriquez J.A., Firth C., Hirschberg D.L., Rice C.M., Shields S., Lipkin W.I. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. USA. 2011;108:11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Ootsuyama Y., Ohkoshi S., Nakazawa T., Sekiya H., Hijikata M., Shimotohno K. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 1992;189:119–127. doi: 10.1016/0006-291x(92)91533-v. [DOI] [PubMed] [Google Scholar]
- Kuo G., Choo Q.L., Alter H.J., Gitnick G.L., Redeker A.G., Purcell R.H., Miyamura T., Dienstag J.L., Alter M.J., Stevens C.E. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- Latt N., Alachkar N., Gurakar A. Hepatitis C virus and its renal manifestations: a review and update. Gastroenterol. Hepatol. 2012;8:434–445. [PMC free article] [PubMed] [Google Scholar]
- Lyons S., Kapoor A., Schneider B.S., Wolfe N.D., Culshaw G., Corcoran B., Durham A.E., Burden F., McGorum B.C., Simmonds P. Viraemic frequencies and seroprevalence of non-primate hepacivirus and equine pegiviruses in horses and other mammalian species. J. Gen. Virol. 2014;95:1701–1711. doi: 10.1099/vir.0.065094-0. [DOI] [PubMed] [Google Scholar]
- Lyons S., Kapoor A., Sharp C., Schneider B.S., Wolfe N.D., Culshaw G., Corcoran B., McGorum B.C., Simmonds P. Nonprimate hepaciviruses in domestic horses, United kingdom. Emerg. Infect. Dis. 2012;18:1976–1982. doi: 10.3201/eid1812.120498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.A., Brooks H.W., Szladovits B., Erles K., Gibbons R., Shields S., Brownlie J. Tropism and pathological findings associated with canine respiratory coronavirus (CRCoV) Vet. Microbiol. 2013;162:582–594. doi: 10.1016/j.vetmic.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.A., Cardwell J.M., Renshaw R.W., Dubovi E.J., Brownlie J. Detection of canine pneumovirus in dogs with canine infectious respiratory disease. J. Clin. Microbiol. 2013;51:4112–4119. doi: 10.1128/JCM.02312-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J.H., Faulk K., Engle R.E., Govindarajan S., St, Claire M., Bukh J. In vivo analysis of the 3′ untranslated region of GB virus B after in vitro mutagenesis of an infectious cDNA clone: persistent infection in a transfected tamarin. J. Virol. 2004;78:9389–9399. doi: 10.1128/JVI.78.17.9389-9399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore L., Fiore J.R., Tateo M., De Benedittis M., Petruzzi M., Casalino C., Genchi C., Lo Muzio L., Angarano G., Serpico R. Detection of hepatitis C virus-RNA in saliva from chronically HCV-infected patients. Int. J. Immunopathol. Pharmacol. 2006;19:217–224. [PubMed] [Google Scholar]
- Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestnall S.L., Mitchell J.A., Walker C.A., Erles K., Brownlie J. New and emerging pathogens in canine infectious respiratory disease. Vet. Pathol. 2014;51:492–504. doi: 10.1177/0300985813511130. [DOI] [PubMed] [Google Scholar]
- Quan P.L., Firth C., Conte J.M., Williams S.H., Zambrana-Torrelio C.M., Anthony S.J., Ellison J.A., Gilbert A.T., Kuzmin I.V., Niezgoda M., Osinubi M.O., Recuenco S., Markotter W., Breiman R.F., Kalemba L., Malekani J., Lindblade K.A., Rostal M.K., Ojeda-Flores R., Suzan G., Davis L.B., Blau D.M., Ogunkoya A.B., Alvarez Castillo D.A., Moran D., Ngam S., Akaibe D., Agwanda B., Briese T., Epstein J.H., Daszak P., Rupprecht C.E., Holmes E.C., Lipkin W.I. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. USA. 2013;110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri A., Argentini C., Kouruma F., Chionne P., D‘Ugo E., Spada E., Dettori S., Sabbatani S., Rapicetta M. Heterogeneity of hepatitis C virus genotype 2 variants in West Central Africa (Guinea Conakry) J. Gen. Virol. 1996;77(Pt 9):2073–2076. doi: 10.1099/0022-1317-77-9-2073. [DOI] [PubMed] [Google Scholar]
- Saito I., Miyamura T., Ohbayashi A., Harada H., Katayama T., Kikuchi S., Watanabe Y., Koi S., Onji M., Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Tapias J.M., Barrera J.M., Costa J., Ercilla M.G., Pares A., Comalrrena L., Soley F., Bruix J., Calvet X., Gil M.P. Hepatitis C virus infection in patients with nonalcoholic chronic liver disease. Ann. Intern. Med. 1990;112:921–924. doi: 10.7326/0003-4819-112-12-921. [DOI] [PubMed] [Google Scholar]
- Scheuer P.J., Ashrafzadeh P., Sherlock S., Brown D., Dusheiko G.M. The pathology of hepatitis C. Hepatology. 1992;15:567–571. doi: 10.1002/hep.1840150402. [DOI] [PubMed] [Google Scholar]
- Shimizu Y.K., Purcell R.H. Cytoplasmic antigen in hepatocytes of chimpanzees infected with non-A, non-B hepatitis virus or hepatitis delta virus: relationship to interferon. Hepatology. 1989;10:764–768. doi: 10.1002/hep.1840100503. [DOI] [PubMed] [Google Scholar]
- Shimotohno K. Hepatitis C virus as a causative agent of hepatocellular carcinoma. Intervirology. 1995;38:162–169. doi: 10.1159/000150427. [DOI] [PubMed] [Google Scholar]
- Simmonds P. Genetic diversity and evolution of hepatitis C virus–15 years on. J. Gen. Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- Stapleton J.T., Foung S., Muerhoff A.S., Bukh J., Simmonds P. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J. Gen. Virol. 2011;92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A.J., Brauer M.J., Rosenblatt J., Richman K.H., Tung J., Crawford K., Bonino F., Saracco G., Choo Q.L., Houghton M. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zignego A.L., Gragnani L., Giannini C., Laffi G. The hepatitis C virus infection as a systemic disease. Intern. Emerg. Med. 2012;7(Suppl 3):S201–208. doi: 10.1007/s11739-012-0825-6. [DOI] [PubMed] [Google Scholar]