Abstract
Viruses encode silencing suppressor proteins to counteract RNA silencing. Because dsRNA plays a key role in silencing, a general silencing suppressor strategy is dsRNA binding. The p22 suppressor of the plant virus Tomato chlorosis virus (ToCV; genus Crinivirus, family Closteroviridae) has been described as having one of the longest lasting local suppressor activities. However, the mechanism of action of p22 has not been characterized. Here, we show that ToCV p22 binds long dsRNAs in vitro, thus interfering with their processing into small RNAs (sRNAs) by an RNase III-type Dicer homolog enzyme. Additionally, we have studied whether a putative zinc finger motif found in p22 has a role in dsRNA binding and suppressor function. The efficient ability of p22 to suppress RNA silencing, triggered by hairpin transcripts transiently expressed in planta, supports the relationship between its ability to bind dsRNA in vitro and its ability to inhibit RNA silencing in vivo.
Keywords: Closteroviridae, Crinivirus, Tomato chlorosis virus, RNA silencing suppressor, RNA binding protein
Highlights
-
•
The p22 mechanism of actionappears to be unusual among plant viruses.
-
•
p22 blockage of dsRNA cleavage could explain its long-lasting suppressor activity.
-
•
A link exists between long dsRNA binding in vitro and suppression of silencing in vivo.
Introduction
Post-transcriptional RNA silencing is a sequence-specific RNA-mediated gene regulatory mechanism that also serves as an antiviral defense (Baulcombe, 2005). RNA silencing is induced by double-stranded RNAs (dsRNAs) that, in the case of single-stranded RNA (ssRNA) viruses, can be derived from highly structured regions in the RNA genome or generated during the replication cycle (Szittya et al., 2002, Molnár et al., 2005). Subsequently, dsRNAs are processed by an RNase III-type Dicer-like enzyme (DCL) into double-stranded small RNAs (sRNAs) of 21–24 nucleotides (Hamilton and Baulcombe, 1999) with 2 nt 3׳-overhangs. Then, one strand of these sRNAs is incorporated into an RNA-induced silencing complex (RISC) that serves as a guide to initiate the sequence-specific degradation of target RNAs (Baulcombe, 2005). This process is amplified by host-encoded RNA-dependent RNA polymerases (RDR) that convert single-stranded RNA into dsRNA, which is subsequently processed by DCL into secondary sRNAs. To counteract this antiviral mechanism, plant viruses encode RNA silencing suppressor proteins (Voinnet, 2005), which are highly diverse in sequence, structure, and activity within and across virus families. Thus, while some viruses have delegated the suppressor functions to replication, structural or transport proteins, other viruses encode dedicated suppressor proteins without homology to any other viral or host protein (Dolja et al., 2006). Viral silencing suppressors can target all steps of RNA silencing, such as viral RNA recognition, dicing, RISC assembly, RNA targeting, and amplification (Burgyán and Havelda, 2011). Because dsRNAs play a key role in RNA silencing, viral suppressor proteins use dsRNA binding as a general strategy of suppression (Mérai et al., 2006). The dsRNA-binding suppressors (dsRBSs), based on their specificity for dsRNA size, can be broadly classified into two types: (i) dsRBSs that preferentially bind long hairpin-derived or inverted repeat dsRNA, preventing them from being processed into sRNAs by DCL, as shown for P14 of Pothos latent virus (Mérai et al., 2005); and (ii) dsRBSs that bind sRNAs and/or sequester them, preventing their incorporation into the RISC complex, as shown for the tombusvirus P19 (Lakatos et al., 2004).
Tomato chlorosis virus (ToCV; genus Crinivirus) belongs to the family Closteroviridae, in which the largest RNA genomes among plant viruses are found (Dolja et al., 2006). Closterovirus gene expression includes at least three different RNA expression mechanisms: i) proteolytic processing, ii) translational frameshifting, and iii) formation of 3´-co-terminal subgenomic RNAs (sgRNAs), resembling the mechanism in animal coronavirus (Karasev et al., 1997). The replication process generates high amounts of viral RNA species, such as dsRNA replicative intermediates that correspond to the genomic RNA and sgRNAs (Hilf et al., 1995). ToCV is transmitted in nature by the whitefly (Hemiptera: Aleyrodidae) Bemisia tabaci and has a bipartite single-stranded, positive-sense RNA genome (Wisler et al., 1998). As for other members of the family Closteroviridae (Lu et al., 2004), ToCV adopts the strategy of encoding multiple RNA silencing suppressors (Cañizares et al., 2008). Thus, while ToCV RNA-2 has delegated its suppressor function to the structural proteins CP and CPm, RNA-1 encodes a dedicated suppressor protein at its 3´-proximal end, p22, which exhibits no apparent homology to any other reported protein. It has been shown that p22 very efficiently suppresses local RNA silencing, induced either by sense RNA or by dsRNA, interfering with the initial stages of RNA silencing. Moreover, it has been reported that p22 has one of the longest lasting local suppression activities when assayed in Nicotiana benthamiana (Cañizares et al., 2008), although its mechanism of action has not been characterized.
Because the expression of viral proteins with suppressor activity seems to be essential for virus multiplication and effective systemic infection of the host, the study of the mechanism of action of these proteins will improve the understanding of the foundations of plant–virus interactions. Here, we demonstrate that the ToCV p22 suppressor preferentially binds long dsRNAs in vitro, preventing them from being cleaved by an RNase III-type Dicer homolog enzyme. Additionally, we have assessed whether a putative zinc finger domain located in the N-terminal part of the protein, highly similar to the one found in the p23 suppressor, a protein that is unique to Citrus tristeza virus (CTV) within closteroviruses, plays a role in both dsRNA binding and suppression activity. Finally, the demonstration that the p22 protein suppresses silencing triggered by hairpin transcripts transiently expressed in planta links the in vitro long dsRNA binding capability of the protein to its ability to inhibit RNA silencing in vivo.
Results
p22 preferentially binds long dsRNA in vitro
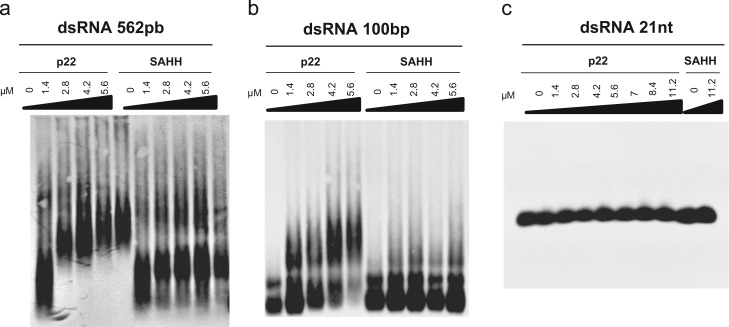
A previous report showed that the protein p22 of ToCV suppressed RNA silencing induced by either sense or double-stranded RNAs (dsRNAs), indicating that it acts downstream of the formation of dsRNA (Cañizares et al., 2008). Moreover, the drastic reduction of sRNA when p22 is present, compared to that conferred by other suppressors such as tombusviral p19 that act by specifically binding sRNAs (Takeda et al., 2002, Lakatos et al., 2006), suggested that p22 could interfere with the RNA silencing pathway upstream of sRNA synthesis. Therefore, the ability of His-tagged p22 to bind different digoxigenin (DIG)-labeled dsRNAs was analyzed by electrophoretic mobility shift assays (EMSA) using increasing amounts of protein. As a negative control in our experiments, we used a non-related protein, S-adenosylhomocysteine hydrolase (SAHH), fused to a His tag, which was expressed and purified in the same way (Cañizares et al., 2013). Long dsRNAs of 562 or 100 base pairs (bp) and small dsRNAs (sRNA) of 21 bp were used. As shown in Fig. 1a, in the case of the 562 bp dsRNA, all the RNA was shifted into a complex that gradually increased in size with increasing protein amounts, even at the lowest p22 protein concentration. No such RNA shifting was observed in the case of the control protein SAHH, confirming that the gel retardation observed was the result of an interaction of the dsRNA with p22. The gradually decreased mobility of the shifted band is likely due to the instability of the complex during electrophoresis at lower protein concentrations, which could indicate that multiple units of p22 are capable of binding to the 562 bp dsRNA, stabilizing the complex. A lower affinity of p22 to the 100 bp dsRNA was observed. In this case, although a decreased mobility of the shifted band was also shown with increasing protein amounts, some dsRNA remained unbound, even with the highest protein concentration used (Fig. 1b). No complex formation was observed for the negative SAHH control. In contrast, similarly to the negative control, no significant binding of p22 to small 21 bp dsRNAs was observed, even at the highest protein concentration used (Fig. 1c). Because in this case, the DIG molecule could be interfering with the binding of p22, the EMSA assay was also performed with 21 bp dsRNA containing a 2 nt overhang with a free OH at the 3’ end, staining the gel with ethidium bromide. As before, no evident shifting of the RNA mobility was observed (data not shown).
Fig. 1.
Affinity of Tomato chlorosis virus p22 to 562 bp, 100 bp dsRNAs, and 21 bp sRNAs. Representative EMSA assays with (a) 562 bp DIG-labeled dsRNA, (b) 100 bp DIG-labeled dsRNA, and (c) 21 nt dsRNA DIG-labeled at the 3’-end of the antisense overhang. The protein S-adenosylhomocysteine hydrolase (SAHH) was used as a negative control. The concentrations of the p22 and SAHH proteins are indicated above each lane. The RNA/proteins were transferred to nylon membranes, and dsRNAs were detected using an anti-digoxigenin antibody and a chemiluminescent substrate.
Taken together, our in vitro assays indicate that p22 is able to bind dsRNA and that it preferentially binds long dsRNAs.
p22 binds ssRNA but not ssDNA or dsDNA in vitro
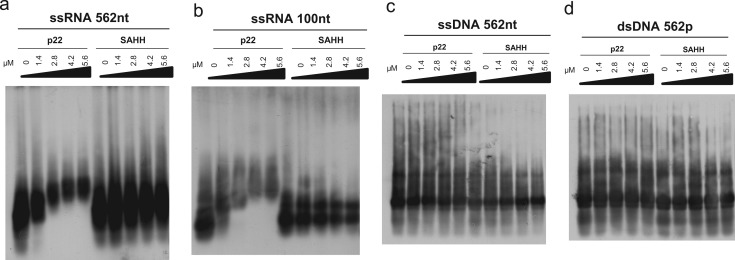
Although the described dsRNA binding activity has been shown to be an important mechanism of silencing suppression for many viral suppressors (Merai et al., 2006), we also tested whether p22 can bind other types of nucleic acids such as ssRNAs, single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA). Following the experimental approach described, the ability of His-tagged p22 to bind different DIG-labeled nucleic acid types was analyzed by EMSA. The unrelated SAHH protein purified in the same way was also used in these assays. Long ssRNAs of 562 or 100 nt and ssDNA and dsDNA of 562 nt were used. As shown in Fig. 2, while the ssRNA of both sizes was shifted into a complex of gradually decreased mobility with increasing amounts of p22 protein, no DNA shifting was observed with either ssDNA or dsDNA. No complex formation was observed with the SAHH protein.
Fig. 2.
Affinity of Tomato chlorosis virus p22 to 562 nt or 100 nt ssRNAs and to 562 nt ssDNA and dsDNA. Representative EMSA assays with (a) 562 nt DIG-labeled ssRNA, (b) 100 nt DIG-labeled ssRNA, (c) 562 nt DIG-labeled ssDNA, and (d) 562 nt DIG-labeled dsDNA. The protein S-adenosylhomocysteine hydrolase (SAHH) was used as a negative control. The concentrations of the p22 and SAHH proteins are indicated above each lane. The nucleic acids/proteins were transferred to nylon membranes, and dsRNAs were detected using an anti-digoxigenin antibody and a chemiluminescent substrate.
These results show that while p22 binds both 562 and 100 nt ssRNA with high affinity, it cannot bind either ssDNA or dsDNA.
ToCV p22 inhibits RNase III Dicer cleavage by sequestering long dsRNAs
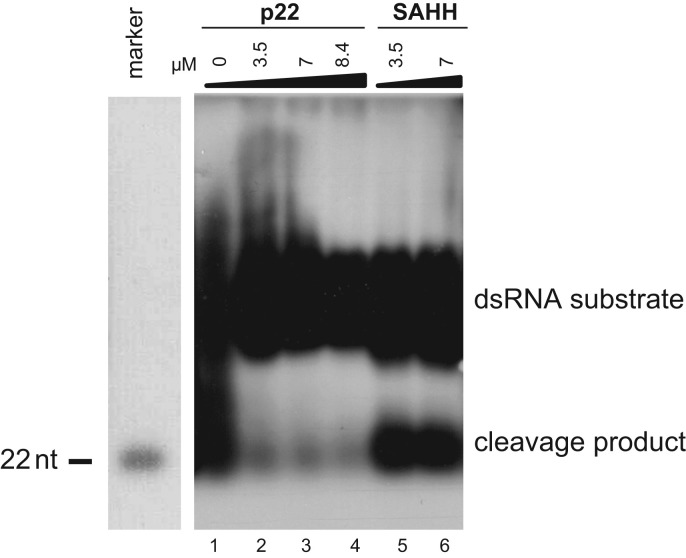
Based on the observed ability of ToCV p22 to bind long dsRNAs with high affinity in vitro, we then studied its ability to protect them from RNase III-mediated cleavage. For this purpose, we used bacterial RNase III, a Dicer homolog used for in vitro experiments as previously reported (Zhang et al., 2004, Fenner et al., 2007, Ji, 2008, Qi et al., 2011), and as a substrate we used a DIG-labeled, in vitro-transcribed 562 bp dsRNA precursor. As shown in Fig. 3, preincubation with increasing amounts of p22 resulted in a reduced amount of substrate cleaved by RNase III (see lanes 2, 3 and 4 vs. lane 1), whereas no such effect was observed for the negative control, SAHH (lanes 5 and 6).
Fig. 3.
Tomato chlorosis virus p22 protects long dsRNA from RNase III-mediated cleavage. A DIG-labeled 562 bp dsRNA was incubated with the p22 (lanes 2, 3 and 4) or S-adenosylhomocysteine hydrolase (SAHH) (lanes 5 and 6) proteins before cleavage by RNase III. The concentrations of proteins are indicated above each lane. The cleavage reaction products were then separated on 1% TBE-agarose gels. As a marker lane, an unlabeled RNA oligonucleotide 22 nt in length was run in parallel, stained with ethidium bromide before the transfer and cut (left). DIG-labeled cleavage reaction products were transferred to nylon membranes and detected using an anti-digoxigenin antibody and a chemiluminescent substrate. Positions of dsRNA precursor and cleavage product are indicated.
These results indicate that ToCV p22 blocks the in vitro RNase III cleavage activity by sequestering the dsRNA precursor.
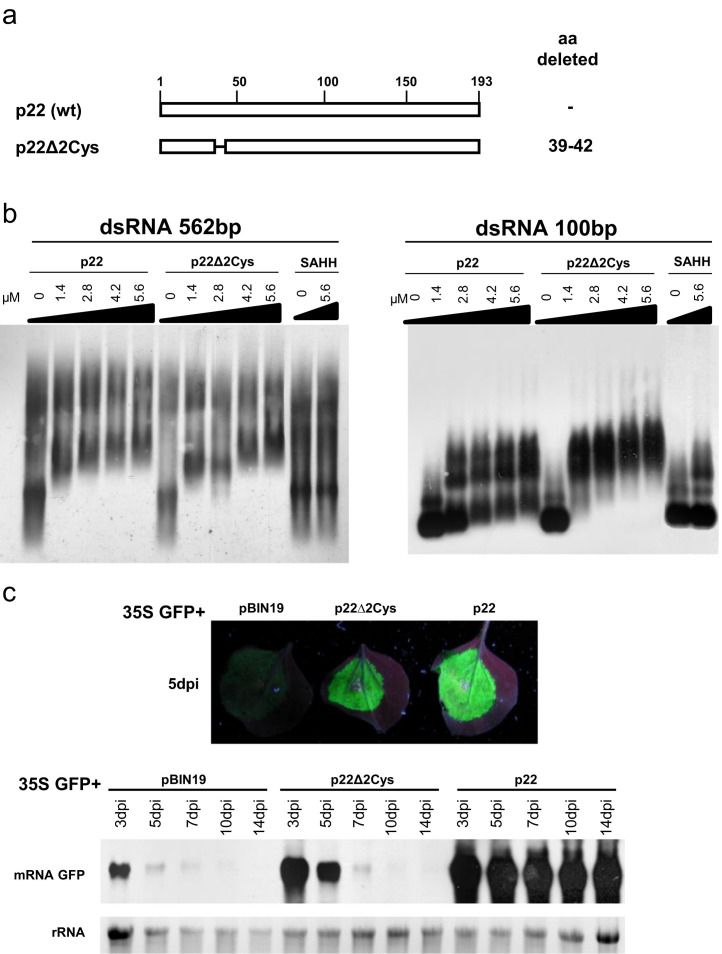
dsRNA binding and suppressor activity of the p22∆2Cys mutant
To determine whether a putative zinc-finger domain found in the N-terminal part of the p22 protein (Supplementary Fig. 1) was involved in binding to long dsRNA in vitro and suppressor activity in vivo, a deletion mutant (p22∆2Cys) in this domain was created. A similar putative zinc-finger domain, in addition to some similar basic amino acid residues, has been shown to be involved in the RNA binding of the p23 protein suppressor of another member of the family Closteroviridae, Citrus tristeza virus (CTV) (López et al., 2000). The mutant p22∆2Cys, harboring a four-amino-acid deletion ( Fig. 4a), was fused to a hexa-His tag, expressed and purified as previously described. EMSAs were performed with increasing amounts of the deletion mutant protein and DIG-labeled dsRNAs of 562 bp and 100 bp, with wild-type p22 as control. The p22∆2Cys mutant very efficiently bound both sizes of dsRNA (Fig. 4b), with an even higher affinity for the 100 bp dsRNA than wild-type p22 (Fig. 4b, right panel). No retardation was observed with the negative control SAHH.
Fig. 4.
Analysis of RNA-binding properties and silencing suppression activity of Tomato chlorosis virus -p22 deletion mutant. (a) Schematic representation of the wild-type p22 protein and the deletion mutant. The deleted amino acid (aa) residues are indicated. (b) RNA-binding properties of the p22Δ2Cys mutant detected by an EMSA assay. The increased concentration of protein is indicated above each lane. Wild-type p22 was used as a positive control, and S-adenosylhomocysteine hydrolase (SAHH) was used as a negative control. The EMSA gels were transferred to a nylon membrane, and dsRNAs were detected using an anti-digoxigenin antibody and a chemiluminescent substrate. (c) Silencing suppression assays with a 35S construct expressing the p22Δ2Cys mutant. Upper panels show photographs taken under UV light of Nicotiana benthamiana leaves at 5 days postinfiltration (dpi) with Agrobacterium tumefaciens harboring 35S GFP in combination with the empty vector pBIN19 or with a 35S construct expressing p22Δ2Cys or with p22. The lower panels show a northern blot analysis of GFP mRNA extracted from the zones infiltrated at 3, 5, 7, 10 and 14 dpi, hybridized with a probe specific to GFP mRNA. Ethidium bromide staining of rRNA was used as a loading control.
To further examine the relationship between the dsRNA binding activity in vitro and suppressor activity in vivo, we analyzed the p22∆2Cys mutant for its ability to suppress silencing using an agroinfiltration assay (Voinnet et al., 1998, Johansen and Carrington, 2001). Thus, N. benthamiana leaves were co-infiltrated with a mixture of Agrobacterium tumefaciens cultures, one expressing 35S-GFP and a second expressing the deletion construct (p22∆2Cys), also under the control of the 35S promoter, using the co-infiltration of 35S-GFP with the empty pBIN19 vector or with the wild-type suppressor p22 as negative and positive controls, respectively. At 5 days postinfiltration (dpi), tissues infiltrated with 35S-GFP plus empty vector exhibited a low level of green fluorescence under UV light (Fig. 4c, upper panels) as a consequence of RNA silencing of the GFP reporter mRNA (Brigneti et al., 1998, Voinnet et al., 2000). Consistently with this reduced fluorescence, northern blot analysis revealed that the steady-state levels of GFP mRNA were very low (Fig. 4c, lower panels). Co-infiltration with the deletion construct p22∆2Cys resulted in the suppression of GFP silencing by 5 dpi. The strong green fluorescence observed correlated with high steady-state levels of GFP mRNA (Fig. 4c). However, in contrast to co-infiltration with the wild-type p22, in which strong suppressor activity was observed at 7, 10 and 14 dpi (Fig. 4c, lower panel), the suppressor activity of the deletion construct p22∆2Cys was lost at 7 dpi. Because no p22 antibody is available, to check whether the transient expression of the deleted construct under the control of the 35S promoter was efficient, mRNA transcript levels were analyzed in agroinfiltrated patches at 4 dpi when the suppressor activity was still evident. Using a p22-specific probe, the northern blot analysis showed that in contrast to infiltration with the wild-type p22, in which mRNA transcript levels were high, the transcript levels upon agroinfiltration with the deletion construct p22∆2Cys were lower than expected (Supplementary Fig. 2). Although we do not have an explanation for this result, it is clear that even with lower levels of expression, the p22∆2Cys mutant construct exhibits suppressor activity (Fig. 4c).
Taken together, these results indicate that the putative zinc finger located at the N-terminal part of p22 is dispensable for both binding of long dsRNA in vitro and suppression activity in vivo.
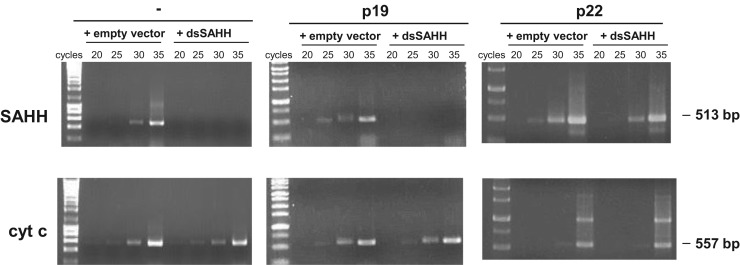
ToCV p22 differs from the sRNA binding suppressor p19 in hairpin-induced silencing assays at prolonged times post-infiltration
To assess a possible link between the in vitro long dsRNA binding capability of the p22 protein and its ability to inhibit RNA silencing in planta, we performed hairpin-transcript-induced silencing assays at longer periods of time than are routinely used in local transient assays. Thus, we compared the behavior of ToCV p22 with the behavior of the Tomato bushy stunt virus (TBSV, genus Tombusvirus, family Tombusviridae) p19 suppressor, which is known to act after sRNA generation (Vargason et al., 2003). The time for the analysis, 12 dpi, was chosen because at this time the suppressor activity of TBSV p19 in patches infiltrated with a mixture of Agrobacterium sp. expressing sense and an inverted repeat GFP construct was overcome (at 14 dpi, the infiltrated patches became deep red), and the accumulation level of GFP sRNAs was high (Takeda et al., 2002). In contrast, in co-infiltrations of p22 with the same mixture, even at 30 dpi, strong fluorescence was still observed and linked to a drastic reduction of GFP sRNAs (Cañizares et al., 2008). The hairpin-induced silencing assays were conducted by triggering the silencing of N. benthamiana SAHH through agroinfiltration with an inverted repeat SAHH construct (dsSAHH), as previously described (Cañizares et al., 2013). Agroinfiltrated patches were analyzed at 12 dpi, when high levels of sRNAs are expected to effectively silence the targeted SAHH mRNAs. Thus, no accumulation of SAHH mRNAs was observed, as shown in Fig. 5 (left panels), in which the downregulation of SAHH was determined by semiquantitative RT-PCR. An equivalent result was obtained when co-infiltration with p19 was performed (Fig. 5, central panels). In contrast, co-infiltration of the dsSAHH construct with p22 showed no reduced levels of accumulation of SAHH mRNA at this time (Fig. 5, right panels). As stated previously, because the p19 suppressor is unable to prevent sRNA accumulation in hairpin-induced silencing (Takeda et al., 2002), we could speculate that the presence of these sRNAs effectively silenced SAHH by 12 dpi. The lack of silencing of SAHH obtained in co-infiltrations with p22 indicated that this suppressor inhibits hairpin-induced silencing differently. Based on these results, we hypothesize that p22 acts at an early step of the silencing process, most likely by binding hairpin transcripts, which are the long dsRNA precursors of sRNAs.
Fig. 5.
Semiquantitative reverse-transcriptase polymerase chain reaction (RT-PCR) analysis of Nicotiana benthamiana S-adenosylhomocysteine hydrolase (NbSAHH) transcripts. RNAs extracted at 12 days postinfiltration from patches with 35S constructs expressing dsSAHH or the empty vector control, alone or in combination with constructs expressing Tomato bushy stunt virus p19 protein (p19) or Tomato chlorosis virus p22 protein (p22), were analyzed using appropriate primers. Aliquots were removed at the indicated cycles. RT-PCR of cytochrome c oxidase (cyt c) transcript was used as an internal control. Sizes of the expected RT-PCR products are indicated to the right.
Therefore, taken together, our results suggested a link between the p22 ability to bind long dsRNA and its ability to suppress gene silencing in vivo by interfering with sRNA generation.
Discussion
The expression of the complex closterovirus genomes resembling that of coronavirus (family Coronaviridae) produces high amounts of viral RNA species in the infected cell. In this set of RNAs, highly structured RNAs or dsRNAs as replicative intermediates would induce an RNA silencing response that must be efficiently counteracted by viral suppressors. In this work, the mechanism of action of the described ToCV p22 suppressor, known to suppress RNA silencing induced by both sense and double-stranded RNAs (dsRNAs) (Cañizares et al., 2008), has been studied further. The in vitro binding studies shown here indicate that p22 very efficiently binds ssRNA of both 562 and 100 nt but does not bind ssDNA or dsDNA. Moreover, p22 also possesses dsRNA binding activity, showing a high affinity for the longest dsRNA tested. The gradually decreased mobility of the shifted band observed in either ssRNA or long dsRNA binding assays might indicate that multiple units of p22 can bind to the RNA, thus retarding the complex, as the amount of RNA in all binding reactions is constant. We can speculate that both an increased number of p22 binding sites in the longest dsRNA or both ssRNAs, and a co-operative interaction might support the results obtained. Unfortunately, the lack of discrete intermediates makes it difficult to measure binding constants, thus precluding an accurate determination of cooperation.
In contrast to the described high affinity for long RNAs, no significant binding to sRNAs was observed for p22 in our conditions, either to synthetic 21 bp ds-sRNA labeled with DIG at the 3´-end or to sRNAs having a 2-nt overhang with a free OH at the 3´-end. Although we cannot rule out the binding of p22 to sRNAs, our results support the hypothesis that binding to long dsRNAs is more relevant for the p22 suppressor activity, as the co-infiltrations of p22 are always linked to reduced accumulation of sRNAs (Cañizares et al., 2008), indicating that p22 interferes with their generation. Moreover, we demonstrated that the binding of p22 to long dsRNA molecules prevented their cleavage into sRNAs by RNase III Dicer homolog, thus blocking subsequent steps of the RNA silencing process. Interestingly, although the strategy of suppressing gene silencing via binding to long dsRNA preventing Dicer cleavage has been described frequently for viruses infecting insects and mammals (Chao et al., 2005, Lingel et al., 2005, Fenner et al., 2006, van Rij et al., 2006, Kimberlin et al., 2010, Qi et al., 2011, Cui et al., 2015), it has only rarely been associated with plant viruses. In fact, it has only been reported for the protein NSs of Tomato spotted wilt virus (TSWV) (family Bunyaviridae) (Schnettler et al., 2010), and for the proteins CP and p14 of Turnip crinkle virus (TCV) and Pothos latent virus (PoLV), respectively (Mérai et al., 2005, Mérai et al., 2006), and both members of the family Tombusviridae, although in vitro cleavage assays were not performed in the latter two cases. Therefore, as proposed for the coronavirus N protein suppressor (Cui et al., 2015), we could speculate that the ability to bind either ssRNA or dsRNA shown by ToCV p22 could protect viral RNA in two ways: i) binding viral ssRNAs to prevent unnecessary intramolecular and intermolecular dsRNA formation between the positive- and negative-sense genomic or subgenomic RNAs; and ii) binding virus-derived dsRNAs to protect them from Dicer cleavage.
By analyzing the p22 amino acid sequence, we found a putative zinc finger domain in the N-terminal part of the molecule as described for the CTV p23 suppressor (López et al., 2000). Interestingly, although CTV p23 has no homologs in other closteroviruses (Lu et al., 2004), it contains a zinc-finger motif that is almost identical to the one present in ToCV p22. Thus, in CTV p23, the zinc finger-like motif has a CX2CX3HX9C signature, and in ToCV p22, the signature is CX2CX3CX9C. The zinc finger domain of p23, in addition to some basic amino acid residues, has been shown to be involved in RNA binding (López et al., 2000) and to be important for controlling asymmetrical RNA synthesis (Satyanarayana et al., 2002). In addition, a functional zinc finger domain is required for the silencing suppression activity of some suppressors (Chiba et al., 2013). With these precedents, in this study, we assessed a possible role of the putative zing finger motif of p22 in dsRNA binding and suppression activity by creating the p22∆2Cys mutant. Our results showed that, in contrast to CTV p23 (López et al., 2000), in which modifications of this domain had deleterious effects on the interaction with the RNA, the putative zinc finger motif found in p22 seemed to be dispensable for dsRNA binding. However, we found that the silencing suppressor activity of the p22∆2Cys mutant was shorter lasting than the activity of wild-type p22 and that the expression levels of the p22∆2Cys construct were lower than those for p22. Thus, a link could exist between the reduced transcript levels of this construct and the silencing suppressor activity observed. Alternatively, it can be argued that the in vivo suppression function of p22 has additional requirements for RNA binding.
The importance of the in planta p22 suppression of silencing by binding long dsRNAs is supported by the results obtained in the suppression of silencing induced by hairpin transcripts. The hairpin-induced RNA silencing pathway requires de novo processing by Dicer to produce sRNAs. Thus, the p19 silencing suppressor failed to prevent the generation of sRNAs from hairpin transcripts at long times post-infiltration (Takeda et al., 2002), and therefore, it failed to prevent silencing of the SAHH mRNA target. In contrast, the effective suppression observed for p22 in equivalent assays supported the importance of the ability of ToCV p22 to bind long dsRNA and thereby block Dicer cleavage to reduce sRNA accumulation, thus preventing silencing. In fact, the presence of p22 in GFP co-infiltrated patches can support the maintenance of a reduced level of GFP sRNAs for a long period of time (25–30 days) (Cañizares et al., 2008).
Although the use of in vitro binding assays or transient heterologous expression systems to infer the mechanism of action of certain viral suppressors has recently been questioned (Incarbone and Dunoyer, 2013), we think that the use of such indirect methodologies might help to shed light on plant–virus interactions. One of the arguments against the use of these methodologies is that the viral suppressors of RNA silencing are often multifunctional proteins that perform other essential roles in the virus life cycle that require association with viral nucleic acids, thus concluding that the binding observed in vitro could not be a feature of silencing suppression. In the case of p22, however, this argument might not apply, as p22 seems to be a dedicated suppressor protein in the ToCV genome (Cañizares et al., 2008). The property of binding dsRNA previously described for certain viral suppressors, such as TCV CP, has been questioned since the discovery of a new strategy based on the use of glycine/tryptophan (GW) mimicry to compete for and inhibit host AGOs (Azevedo et al., 2010). Recent studies, however, showed that these GW motifs are also important for binding dsRNA and, in the case of the Pelargonium line pattern virus (family Tombusviridae) p37 suppressor, an essential requirement to suppress RNA silencing (Pérez-Cañamás and Hernández, 2015). These results suggest that the RNA binding capability of suppressors containing GW motifs could have been overlooked. Thus, the property of binding dsRNA exhibited by some viral proteins that act as suppressors might not be an artifact but rather important to their function as suppressors.
In summary, we believe that the characterization of the mode of action of p22 reported in this work is another important contribution to unraveling the complex interplay occurring during plant–virus interactions. We report a mechanism of action for p22 that does not seem common for plant viruses, which could explain its long-lasting suppressor activity. The high affinity for long dsRNAs affecting the Dicer-like cleavage might block the silencing process at early steps by hindering the generation of sRNAs. The possibility that the described mechanism of action of the p22 suppressor could occur during viral infections might be supported by the results obtained using a ToCV mutant deficient in p22 (our unpublished results), which showed that systemic viral infection is impeded in wild-type N. benthamiana plants but not in rdr6 N. benthamiana plants impaired in the synthesis of dsRNA precursors of secondary sRNAs. Thus, the blockage of dsRNA cleavage through binding might be an effective way to suppress the silencing used by ToCV and sustain effective infection of host plants.
Materials and methods
Plasmid construction
For gene expression, the ToCV p22 gene was PCR-amplified from plasmid pGEM-T-p22 (Cañizares et al., 2008) using the Expand High Fidelity PCR system (Roche Diagnostics) and primers MA 1287 and MA 1288 with specific restriction sites. The PCR product was digested, purified and cloned into Escherichia coli expression vector pET-28a(+) (Novagen), resulting in the pET28a-p22 construct, harboring the p22 gene fused to a sequence coding for a hexa-His tag. To generate the p22Δ2Cys construct, which lacked four amino acid residues including two cysteines located between positions 39–42 (C-terminal region), amplification from plasmids pET28a-p22 and pBin35S-p22 (Cañizares et al., 2008) was performed. Mutations were introduced by PCR using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) and the specific primers MA 1616 and MA 1617, generating pET28a-p22Δ2Cys and pBIN35S-p22Δ2Cys.
The constructs pBIN35S-p22 and pETDuet.SAHH have been previously described (Cañizares et al., 2008, Cañizares et al., 2013). The 35S-dsSAHH construct was kindly provided by Dr. David M. Bisaro (The Ohio State University, Columbus, OH, USA), and plasmids 35S-GFP and 35S-p19 were kindly provided by Dr. David C. Baulcombe (University of Cambridge, Cambridge, United Kingdom).
Purification, analysis and quantification of p22 protein and its mutant from E. coli
Expression of His-tagged p22 and its mutant protein and of the negative control protein SAHH were performed in the E. coli strain Rosetta 2 (DE3)pLysS (Novagen). Expression was induced in the transformed bacteria with 0.4 mM isopropyl β-D-thiogalactopyranoside for 3 h at 28 °C, and the proteins were purified by chromatography on nickel-nitrilotriacetic acid (Ni-NTA) columns according to the manufacturer´s recommendations (Qiagen). The purified proteins were analyzed and quantified by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) after Coomassie brilliant blue staining.
Preparation of labeled nucleic acids
DIG-labeled dsRNAs of 562 and 100 base pairs (bp) were generated by in vitro transcription with T7 RNA polymerase (Roche Diagnostics). Primers incorporating 5′-end T7 RNA polymerase promoters into the PCR products were used to amplify two regions of 562 and 100 nucleotides (nt) of the pTOPO-GFP plasmid using either the primer pair MA 1366 and MA 1337 or the pair MA 1336 and MA 1337 (Supplementary Table 1), respectively, with the Expand High Fidelity PCR system (Roche Diagnostics). To synthesize the labeled RNA, DIG-11-UTP (Roche Diagnostics) was included in the transcription reaction. The small dsRNAs (sRNAs) were ordered as two complementary 21 nt oligonucleotides with a RNA 3’-end overhang (MA1633: 5’-ACUGGAGUUGUCCCAAUUCUU-3’ and MA 1634: 5’- GAAUUGGGACAACUCCAGUGA-3’). The antisense oligonucleotide was also prepared with a DIG molecule at the 3’-end. These small RNAs were synthesized by Sigma-Aldrich. Duplex dsRNAs of 562 bp and 100 bp were obtained by annealing the two complementary T7 transcripts produced in vitro, and small RNA (sRNA) duplexes were obtained by annealing the two complementary RNA oligonucleotides. In both cases, to anneal complementary strands, reaction mixtures were incubated for 1 min at 95 °C and cooled to room temperature. For 562 or 100 nt ssRNA synthesis, the same strategy was used but with the reverse primer MA 1337b, containing the same GFP sequence present in MA 1337 but without the T7 promoter sequence (Supplementary Table 1). The PCR products containing the T7 promoter sequence at only one of the ends were used as a template for in vitro transcription. In all transcripts, the DNA template was removed by treatment with DNase I (Invitrogen). Unincorporated nucleotides were removed using NucAway Spin Columns (Ambion). For single- and double-strand quantification, RNAs were analyzed in 5% acrylamide gel and stained with ethidium bromide, where the different mobilities of both types of molecules could be clearly observed.
DIG-labeled dsDNA of 562 nt was obtained by PCR using primers MA 1366b and MA 1337b (Supplementary Table 1), which contained the same GFP sequence present in primers MA 1366 and MA 1337 but without the T7 promoter sequence, including DIG-11-dUTP in the reaction. The ssDNA was produced by boiling the dsDNA for 5 min and quickly cooling it on ice. Labeled DNA was analyzed by electrophoresis in 1% agarose gel/TBE buffer (89 mM Tris, 89 mM boric acid, 0.5 mM EDTA) and stained with ethidium bromide.
Electrophoretic mobility shift assay
Labeled nucleic acids (~1 nM) were mixed with increasing amounts of His-tagged p22 or its mutant in a 10 µl reaction containing binding buffer [20 mM Tris/HCl pH 7.5, 1 mM MgCl2, 60 mM KCl, 100 mM NaCl, 2 mM DTT, 1% glycerol, 0.02% Tween-20, 0.1% Bovine Serum Albumin and 8 U ribonuclease inhibitor RNaseOUT (Invitrogen)]. The binding reaction was incubated at room temperature for 30 min and resolved in a 1% agarose gel run in TBE buffer. The gel was vacuum-transferred (Vacugene XL, GE Healthcare) onto positively charged nylon membranes (Roche Diagnostics) for 3 h in SSC 10× buffer (1.5 M NaCl, 0.15 M sodium citrate, pH 7). Samples were crosslinked under UV light (Ultraviolet Crosslinker RPN 2500, Amersham). The membranes were treated with anti-digoxigenin antibody (Anti-digoxigenin-AP, Fab fragments, Roche Diagnostics) and detected with the alkaline phosphatase chemiluminescent substrate (CDP-Star, Roche Diagnostics) according to the manufacturer’s protocols. As a negative control, a non-related protein involved in methylation reactions in plants, SAHH, fused to a His tag, was expressed and purified in the same manner as p22 (Cañizares et al., 2013).
RNase III-mediated cleavage assay
The RNase III-mediated cleavage assay was performed in 10 µl volumes containing ~1 nM 562 bp dsRNA, 20 mM Tris/HCl pH 7.5, 1 mM MgCl2, 60 mM KCl, 100 mM NaCl, 2 mM DTT, 1% glycerol, 0.02% Tween-20, 0.1% Bovine Serum Albumin, 1 x RNase III buffer, and different amounts of p22 or SAHH as a negative control. Following 30 min of preincubation at room temperature to allow the test proteins to bind to dsRNA, 1 U of RNase III (Ambion) was added, and the reaction mixtures were incubated at 37 °C for 1 h. The reaction products were resolved by electrophoresis in 1% agarose gel/TBE buffer. As a marker lane, an unlabeled RNA oligonucleotide 22 nt in length was run in parallel. The gel was stained with ethidium bromide before transfer, and the marker lane was cut. Nucleic acids were vacuum-transferred (Vacugene XL, GE Healthcare) to positively charged nylon membranes (Roche Diagnostics) for 3 h in SSC 10x buffer (1.5 M NaCl, 0.15 M sodium citrate, pH 7) and crosslinked under UV light (Ultraviolet Crosslinker RPN 2500, Amersham). Then, the membranes were treated with anti-digoxigenin antibody (Anti-digoxigenin-AP, Fab fragments, Roche Diagnostics) and detected with the alkaline phosphatase chemiluminescent substrate (CDP-Star, Roche Diagnostics) according to the manufacturer’s protocols.
Agroinfiltration, silencing suppression assays and fluorescence imaging
Wild-type N. benthamiana plants were grown in a chamber at 25 °C with a photoperiod of 16 h of light and 8 h of darkness. Plants at the four- to six-leaf growth stage were agroinfiltrated with A. tumefaciens strain GV3101 carrying the plasmids indicated above, as described by Voinnet et al. (1998). For co-infiltration, the A. tumefaciens cultures were adjusted to an optical density at 600 nm of 1 and mixed prior to infiltration. For the identification of suppressor activity by visual inspection of GFP fluorescence, the 35S-GFP construct was co-expressed with a construct expressing p22 and its mutant or empty vector. For the silencing assays triggered by hairpin transcripts, the 35S-dsSAHH construct or empty vector was infiltrated alone or in combination with constructs expressing the suppressor proteins p19 or p22. GFP fluorescence was observed under long-wavelength UV light (Black Ray model B 100AP, UV products). Pictures of GFP were taken using a Coolpix 8700 Nikon digital camera.
Northern blot analysis
RNA was extracted from agroinfiltrated leaf tissue as described by Noris et al. (1996). For the northern blot analysis of GFP mRNAs, total RNA aliquots (5 µg) from each sample were separated in 1% formaldehyde agarose gels, transferred to nylon membranes (Roche Diagnostics) and probed with DIG-labeled probes specific for GFP, as described previously (Cañizares et al., 2004).
Semi-quantitative RT-PCR
Total RNA was extracted from agroinfiltrated leaf tissue as described above and further treated with DNase I to eliminate genomic DNA. RNA pellets were resuspended in water and quantified in a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Total RNA (500 ng) was reverse transcribed with oligo-dT primers using AMV RT (Promega). The resulting single-stranded cDNA was used for PCR to detect SAHH transcripts using the specific primers MA 748 and MA 749. PCR with the specific primers MA 720 and MA 721 for the cytochrome c oxidase transcripts (Kadowaki et al., 1995) was used as an internal control. Aliquots were withdrawn from the PCR reaction after 20, 25, 30, and 35 cycles and analyzed by agarose gel electrophoresis.
Acknowledgments
Y. L.-R. was the recipient of an AECID (Spain) fellowship. M.C.C. was the recipient of an I3P contract (I3P-PC2004L) from the CSIC (Spain) with assistance from the European Social Fund (ESF). This work was supported by Grants AGL2010-22287-C02-01/AGL and AGL2013-48913-C2-1-R from the Ministerio de Economía y Competitividad, Spain, with assistance from the European Regional Development Fund (ERDF). J.N.C. and E.M.A. are members of the Research Group AGR-214, partially funded by the Consejería de Economía, Innovación y Ciencia, Junta de Andalucía, Spain with assistance from the ERDF and ESF. We thank F. Aparicio for providing the pET vectors and helpful discussions, and we thank M.V. Martín and R. Tovar for their technical assistance. We thank American Journal Experts (AJE) for English language editing.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2015.11.008.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- Azevedo J., Garcia D., Pontier D., Ohneshorge S., Yu A., Garcia S., Braun L., Bergdoll M., Hakimi M.A., Lagrange T., Voinnet O. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes. Dev. 2010;24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D.C. RNA silencing. Trends Biochem. Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Brigneti G., Voinnet O., Li W.X., Ji L.H., Ding S.W., Baulcombe D.C. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Burgyán J., Havelda Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011;16:265–272. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Cañizares M.C., Lozano-Durán R., Canto T., Bejarano E.R., Bisaro D.M., Navas- Castillo J., Moriones E. Effects of the crinivirus coat protein-interacting plant protein SAHH on post-transcriptional RNA silencing and its suppression. Mol. Plant Microbe Interact. 2013;26:1004–1015. doi: 10.1094/MPMI-02-13-0037-R. [DOI] [PubMed] [Google Scholar]
- Cañizares M.C., Navas-Castillo J., Moriones E. Multiple suppressors of RNA silencing encoded by both genomic RNAs of the crinivirus Tomato chlorosis virus. Virology. 2008;379:168–174. doi: 10.1016/j.virol.2008.06.020. [DOI] [PubMed] [Google Scholar]
- Cañizares M.C., Taylor K.M., Lomonossoff G.P. Surface exposed C-terminal amino acids of the small coat protein of Cowpea mosaic virus are required for suppression of silencing. J. Gen. Virol. 2004;85:3431–3435. doi: 10.1099/vir.0.80454-0. [DOI] [PubMed] [Google Scholar]
- Chao J.A., Lee J.H., Chapados B.R., Debler E.W., Schneemann A., Williamson J.R. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Chiba S., Hleibieh K., Delbianco A., Klein E., Ratti C., Ziegler-Graff V., Bouzoubaa S., Gilmer D. The benyvirus RNA silencing suppressor is essential for long distance movement, requires both zinc-finger and NoLS basic residues but not a nucleolar localization for its silencing suppression activity. Mol. Plant Microbe Interact. 2013;26:168–181. doi: 10.1094/MPMI-06-12-0142-R. [DOI] [PubMed] [Google Scholar]
- Cui L., Wang H., Ji Y., Yang J., Xu S., Huang X., Wang Z., Qin L., Tien P., Zhou X., Guo D., Chen Y. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. Virol. 2015;89:9029–9043. doi: 10.1128/JVI.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja V.V., Kreuze J.F., Valkonen J.P.T. Comparative and functional genomics of closteroviruses. Virus Res. 2006;117:38–51. doi: 10.1016/j.virusres.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner B.J., Gob W., Kwang J. Sequestration and protection of double-stranded RNA by the betanodavirus B2 protein. J. Virol. 2006;80:6822–6833. doi: 10.1128/JVI.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner B.J., Gob W., Kwang J. Dissection of double-stranded RNA binding protein B2 from betanodavirus. J. Virol. 2007;81:5449–5459. doi: 10.1128/JVI.00009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A.J., Baulcombe D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hilf M.E., Karasev A.V., Pappu H.R., Gumpf D.J., Niblett C.L., Garnsey S.M. Characterization of Citrus tristeza virus subgenomic RNAs in infected tissue. Virology. 1995;208:576–582. doi: 10.1006/viro.1995.1188. [DOI] [PubMed] [Google Scholar]
- Incarbone M., Dunoyer P. RNA silencing and its suppression: novel insights from in planta analysis. Trends Plant Sci. 2013;18:382–392. doi: 10.1016/j.tplants.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Ji X. The mechanism of RNase III action: how dicer dices. Curr. Top. Microbiol. Immunol. 2008;320:99–116. doi: 10.1007/978-3-540-75157-1_5. [DOI] [PubMed] [Google Scholar]
- Johansen L.K., Carrington J.C. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 2001;126:930–938. doi: 10.1104/pp.126.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki K., Ozawa K., Kazama S., Kubo N., Akihama T. Creation of an initiation codon by RNA editing in the coxI transcript from tomato mitochondria. Curr. Genet. 1995;28:415–422. doi: 10.1007/BF00310809. [DOI] [PubMed] [Google Scholar]
- Karasev A.V., Hilf M.E., Garnsey S.M., Dawson W.O. Transcriptional strategy of closteroviruses: mapping the 5´ termini of the Citrus tristeza virus subgenomic RNAs. J. Virol. 1997;71:6233–6236. doi: 10.1128/jvi.71.8.6233-6236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin C.R., Bornholdt Z.A., Li S., Woods V.L., MacRae I.J. Saphire, E.O., 2010. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. USA. 2010;107:314–319. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L., Csorba T., Pantaleo V., Chapman E., Carrington J.C., Liu Y.P., Dolja V.V., Calvino L.F., López-Moya J.J., Burgyán J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L., Szittya G., Silhavy D., Burgyán J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A., Simon B., Izaurralde E., Sattler M. The structure of the Flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double- stranded RNA recognition. EMBO Rep. 2005;6:1149–1155. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López C., Navas-Castillo J., Gowda S., Moreno P., Flores R. The 23-kDa protein coded by the 3׳-terminal gene of Citrus tristeza virus is an RNA-binding protein. Virology. 2000;269:462–470. doi: 10.1006/viro.2000.0235. [DOI] [PubMed] [Google Scholar]
- Lu R., Folimonov A., Shintaku M., Li W.X., Falk B.W., Dawson W.O., Ding S.W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA. 2004;101:15742–15747. doi: 10.1073/pnas.0404940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérai Z., Kerényi Z., Molnár A., Barta E., Váláczi A., Bisztray G., Havelda Z., Burgyán J., Silhavy D. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 2005;79:7217–7226. doi: 10.1128/JVI.79.11.7217-7226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérai Z., Kerényi Z., Kertész S., Magna M., Lakatos L., Silhavy D. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 2006;80:5747–5756. doi: 10.1128/JVI.01963-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár A., Csorba T., Lakatos L., Várallyay E., Lacomme C., Burgyán J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noris E., Accotto G.P., Tavazza R., Brunetti A., Crespi S., Tavazza M. Resistance to Tomato yellow leaf curl geminivirus in Nicotiana benthamiana plants transformed with a truncated viral C1 gene. Virology. 1996;224:130–138. doi: 10.1006/viro.1996.0514. [DOI] [PubMed] [Google Scholar]
- Pérez-Cañamás M., Hernández C. Key importance of small RNA binding for the activity of a glycine-tryptophan (GW) motif-containing viral suppressor of RNA silencing. J. Biol. Chem. 2015;290:3106–3120. doi: 10.1074/jbc.M114.593707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi N., Cai D., Qiu Y., Xie J., Wang Z., Si J., Zhang J., Zhou X., Hu Y. RNA binding by a novel helical fold of B2 protein from Wuhan Nodavirus mediates the suppression of RNA interference and promotes B2 dimerization. J. Virol. 2011;85:9543–9554. doi: 10.1128/JVI.00785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana T., Gowda S., Ayllón M.A., Albiach-Martí M.R., Rabindran S., Dawson W.O. The p23 protein of Citrus tristeza virus controls asymmetrical RNA accumulation. J. Virol. 2002;76:473–483. doi: 10.1128/JVI.76.2.473-483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnettler E., Hemmes H., Huismann R., Goldbach R., Prins. M., Kormelink R. Diverging affinity of tospovirus RNA silencing suppressor proteins, NSs, for various RNA duplex molecules. J. Virol. 2010;84:11542–11554. doi: 10.1128/JVI.00595-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G., Molnár A., Silhavy D., Hornyik C., Burgyán J. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell. 2002;14:359–372. doi: 10.1105/tpc.010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Sugiyama K., Nagano H., Mori M., Kaido M., Mise K., Tsuda S., Okuno T. Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 2002;532:75–79. doi: 10.1016/s0014-5793(02)03632-3. [DOI] [PubMed] [Google Scholar]
- van Rij R.P., Saleh M.C., Berry B., Foo C., Houk A., Antoniewski C., Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes. Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason J.M., Szittya G., Burgyán J., Tanaka Hall T.M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Vain P., Angell S., Baulcombe D.C. Systemic spread of sequence- specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Lederer C., Baulcombe D.C. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell. 2000;103:157–167. doi: 10.1016/s0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Wisler G.C., Li R.H., Liu H.-Y., Lowry D.S., Duffus J.E. Tomato chlorosis virus: a new whitefly-transmitted, phloem-limited bipartite closterovirus of tomato. Phytopathology. 1998;88:402–409. doi: 10.1094/PHYTO.1998.88.5.402. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material