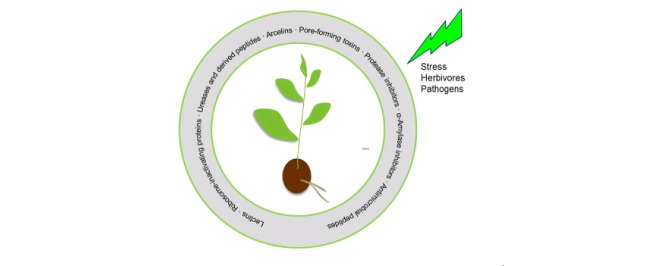
Graphical abstract
Plants produce a broad spectrum of toxic proteins that confer resistance against herbivores and pathogens. An overview of different types of toxic proteins expressed in plants is presented and their biotechnological applications are discussed.
Keywords: Plant protein, Toxicity, Biological activity, Mode of action, Plant defense, Biotechnological application
Highlights
-
•
Plants express a variety of toxic proteins.
-
•
Many toxic plant proteins confer resistance against herbivores and pathogens.
-
•
Understanding of biological activities is vital for biotechnological applications.
-
•
Applications can be envisaged in agricultural and medical fields.
Abstract
Plants have evolved to synthesize a variety of noxious compounds to cope with unfavorable circumstances, among which a large group of toxic proteins that play a critical role in plant defense against predators and microbes. Up to now, a wide range of harmful proteins have been discovered in different plants, including lectins, ribosome-inactivating proteins, protease inhibitors, ureases, arcelins, antimicrobial peptides and pore-forming toxins.
To fulfill their role in plant defense, these proteins exhibit various degrees of toxicity towards animals, insects, bacteria or fungi. Numerous studies have been carried out to investigate the toxic effects and mode of action of these plant proteins in order to explore their possible applications. Indeed, because of their biological activities, toxic plant proteins are also considered as potentially useful tools in crop protection and in biomedical applications, such as cancer treatment. Genes encoding toxic plant proteins have been introduced into crop genomes using genetic engineering technology in order to increase the plant’s resistance against pathogens and diseases. Despite the availability of ample information on toxic plant proteins, very few publications have attempted to summarize the research progress made during the last decades. This review focuses on the diversity of toxic plant proteins in view of their toxicity as well as their mode of action. Furthermore, an outlook towards the biological role(s) of these proteins and their potential applications is discussed.
1. Introduction
Being sessile organisms plants are exposed to a multitude of stress factors from their environment. In addition to the unsuitable influences from their surroundings, there is also the constant threat from predators and pathogens. To cope with a diversity of unfavorable conditions plants have undergone evolutionary adaptation such as the elaboration of sophisticated defense strategies and the synthesis of an impressive diversity of natural bioactive compounds, some of which are toxic (Maag et al., 2014). Among the different toxic compounds reported in plants is a large group of low molecular weight compounds, among which alkaloids, terpenoids, tannins and glycosides (Mithöfer and Boland, 2012). Although these small molecules do not have a primary function in plants they play an important role because of their toxicity to animals, arthropods as well as to bacteria and viruses (Cushnie et al., 2014, Mithöfer and Boland, 2012). Furthermore plants also synthesize an arsenal of proteins such as lectins and ribosome-inactivating proteins (RIPs), that help the plant in its continuous battle for survival (Lannoo and Van Damme, 2014, Virgilio et al., 2010).
Plants express a variety of toxic proteins that confer resistance against herbivores and pathogens. Some well-known families of toxic proteins include lectins, ribosome-inactivating protein, protease inhibitors, α-amylase inhibitors, ureases, arcelins, antimicrobial peptides and pore-forming toxins. Most of these proteins tend to accumulate in the vulnerable parts of the plant such as seeds and vegetative storage tissues. In fact, the first proteins classified as RIPs, arcelins, canatoxins and lectins all originated from seeds where these proteins are highly abundant (Carlini and Guimarães, 1981, Olsnes, 2004, Osborn et al., 1988). Research on toxic plant proteins has resulted in numerous data, showing evidence that these noxious proteins are involved in plant defense against phytophagous predators and pathogens, including bacteria, fungi, viruses, nematodes, and insects (Carlini and Grossi-de-Sá, 2002).
It is important to note that toxic proteins have been identified throughout the plant kingdom and have also been discovered in edible crops. For example, lectins have been reported in bean, tomato, potato, banana and garlic (Van Damme et al., 1998a). Similarly, RIPs have been identified in several edible plants, including pumpkin, cucumber, beet, and cereals (Barbieri et al., 2006). Since some of these crops are also eaten raw, knowledge about the toxic proteins in these plants is also important with respect to food safety.
Despite the availability of an enormous amount of information on toxic compounds and proteins in plants, a recent comprehensive overview of toxic proteins in the plants is lacking. This review focuses on the different classes of toxic proteins reported in plants (Table 1 ), with particular emphasis on their toxicity and mode of action. Furthermore, the potential applications of toxic plant proteins are discussed.
Table 1.
Overview of toxic plant proteins.
| Family | Source | Structural features | Biological activity | References |
|---|---|---|---|---|
| Lectin | Ubiquitous in plants | One or more CRDs | Carbohydrate-binding activity | Van Damme et al., 2008, Van Damme, 2014 |
| Ribosome-inactivating proteins | Widely distributed | N-glycosidase domain | N-glycosidase activity | Peumans et al., 2001, Shang et al., 2014 |
| Protease inhibitors/α-amylase inhibitors | Widely distributed, rich in storage tissues | N/A | Inhibition of protease/α-amylase | Leung et al., 2000, Murdock and Shade, 2002, Svensson et al., 2004 |
| Urease and canatoxin-like proteins | Mainly in legumes | A 10 kDa region, with a β-hairpin motif | Ureolytic activity Pore-forming activity |
Follmer et al., 2001, Barros et al., 2009 |
| Arcelins | Seeds of Phaseolus sp. | Legume lectin fold | N/A | Acosta-Gallegos et al., 1998, Zaugg et al., 2013 |
| Thionins | A number of monocot and dicot plants | ∼5 kDa cysteine containing proteins | Increase of cell membrane permeability | Stec (2006) |
| Cyclotides | Widely distributed | Cyclic cysteine knot | Pore-forming activity | Craik et al. (2012) |
| Pore-forming toxins | Some plants, e.g. Enterolobium contortisiliquum, wheat | Membrane-spanning region (ß-barrel/α-helical) | Pore-forming activity | Bittencourt et al., 2003, Puthoff et al., 2005 |
2. Different classes of toxic proteins in plants
2.1. Lectins
Lectins are a class of proteins endowed with carbohydrate-binding activity. They are defined as proteins with at least one non-catalytic domain that binds reversibly with specific mono- or oligosaccharides (Peumans and Van Damme, 1995). Although the majority of lectins have been characterized from plants, these proteins have also been reported in animals, insects, viruses, fungi and bacteria (Van Damme, 2014). Analysis of completed genome sequences and transcriptome data suggests that lectins are ubiquitous in the plant kingdom. Up to now, several hundreds of plant lectins have been identified, purified and at least partially characterized (Van Damme et al., 1998a, Van Damme et al., 1998b).
Lectins are globular proteins with a carbohydrate-binding site which enables them to specifically recognize and bind particular carbohydrate structures. It should be emphasized that the carbohydrate specificity of lectins is highly diverse. Although some lectins recognize and interact with monosaccharides such as mannose, glucose, galactose, fucose, most plant lectins preferentially bind to more complex oligosaccharides like N- and O-linked glycans (Ghazarian et al., 2011). The carbohydrate-binding site typically consists of five to six amino acids that bind the hydroxyls of the sugar residues mainly by hydrophobic interactions. The specific interaction between the lectin and the carbohydrate involves the formation of a network of hydrogen bonds and is often reinforced by a hydrophobic stacking of the pyranose ring of the sugar to the aromatic ring of aromatic residues (tyrosine, tryptophan or phenylalanine) located in the close vicinity of the carbohydrate binding site (del Carmen Fernández-Alonso et al., 2012).
The affinity of lectins for their substrate is usually rather weak when compared to the antigen–antibody interactions (Kd ∼ 10−8–10−12 M). The binding affinity of a lectin towards monosaccharides is typically in the order of ∼10−3 M (Duverger et al., 2003, Lis and Sharon, 1998). However it should be emphasized that most lectins preferentially recognize oligosaccharides or more complex glycans by multivalent interactions, resulting in a considerable increase of the binding affinity to Kd values of 10−6–10−8 M (Duverger et al., 2003, Liang et al., 2007).
Since the family of lectins groups all proteins that specifically interact with carbohydrate structures without altering the substrate, a large number of very diverse proteins complies with this definition. For several decades lectinologists and chemists have been trying to establish an appropriate classification system for plant lectins. A careful analysis of all available protein sequences encoding lectins or lectin domains allowed classification into 12 families based on the sequence similarities and evolutionary relationships. A detailed overview of these 12 plant lectin domains was described in several recent review papers (Lannoo and Van Damme, 2010, Lannoo and Van Damme, 2014, Van Damme et al., 2008) and therefore is beyond the scope of this review.
Lectins have a very long history. The first carbohydrate-binding protein was discovered in 1888, when Peter Herman Stillmark discovered a toxic protein in the seeds of castor bean (Ricinus communis). In the beginning of lectin research, the research was focused on lectins from seeds and later storage organs, in particular because lectins in these tissues are abundant proteins and as a consequence were rather easy to purify with the biochemical methods available at that time. It turned out that the majority of these lectins can be categorized as hololectins, being composed only of protein domains with carbohydrate-binding activity. The availability of sequence information for different lectin domains and genome sequences for different species allowed the identification of proteins containing a well-defined lectin domain or carbohydrate recognition domain (CRD) linked to other known or unknown protein domains. For example, the class of type-2 RIPs consists of chimeric proteins composed of an N-glycosidase domain linked to a lectin domain (see 2.3). In the last few years the group of chimeric lectins has been reported in many plant species. Therefore, a classification system based on the presence of particular CRDs within a sequence was introduced to cope with the increasing complexity within the whole group of plant lectins (Van Damme, 2014).
Initial studies of plant lectins started with the highly abundant lectins, which are now referred to as “the classical lectins”. Most of these lectins are synthesized on the endoplasmic reticulum (ER) and follow the secretory pathway to the vacuolar compartment. They are typically abundant proteins in seeds and vegetative storage tissues (Van Damme et al., 1998a). Several of these lectins have been studied for their toxicity towards animals, insects, fungi and also viruses (Michiels et al., 2010, Peumans and Van Damme, 1995, Yamaji et al., 2012). Due to their abundance, subcellular localization as well as their toxicity, it is generally accepted that these lectins serve a role as storage proteins and could also function in plant defense whenever the plant is attacked by a pathogen or predator (Peumans and Van Damme, 1995).
In the last decade evidence accumulated for the occurrence of a group of stress inducible lectins. In contrast to the classical lectins, these proteins are generally present at very low concentrations but transcript levels are up-regulated in specific plant tissues in response to stresses such as drought, high salt, hormone treatment, pathogen attack, or insect herbivory (Van Damme, 2008). Most inducible lectins are localized in the nucleus and/or the cytoplasm of plant cells. It is hypothesized that these lectins play an important role in stress-related pathways as signaling molecules (Lannoo and Van Damme, 2010, Lannoo and Van Damme, 2014).
In view of possible applications of lectins as plant defense proteins their biological activities and toxicity towards several organisms, including mainly bacteria, fungi, viruses and insects, have been investigated in much detail. Lectins from different sources have been described as antimicrobial proteins and are believed to be involved in the plant defense against bacteria, fungi and viruses (Peumans and Van Damme, 1995). Although plenty of lectins have been considered to have antimicrobial activities, little is known about the mode of action. Due to the existence of a cell wall in both bacteria and fungi, it is difficult to envisage a direct interaction between the lectin and the microbial membranes (Peumans and Van Damme, 1995). The lectin from Lathyrus ochrus seeds was reported to interact with components of the bacterial cell wall (Bourne et al., 1994). The antifungal activity of chitin-binding lectins has been speculated to be related to their binding with the cell wall chitin, resulting in disruption of cell wall polarity (Selitrennikoff, 2001).
A broad range of plant lectins has been tested against insects both by in vitro bioassays with artificial lectin-containing diets and in vivo experiments with transgenic plants overexpressing a lectin gene. It was shown that lectins can impose severe effects on insect fecundity, growth and development. In addition, lectins can alter insect feeding behavior as well as oviposition (Michiels et al., 2010). It is generally accepted that specific binding of lectins to particular carbohydrate structures in the insect body is essential for lectins to exert their toxicity. The best studied group of plant lectins is represented by the Galanthus nivalus agglutinin (GNA), a mannose-binding lectin, which is toxic to both hemipteran and lepidopteran insects. Feeding experiments with artificial diets and experiments with various transgenic plants have demonstrated the detrimental effects of GNA on different insects (Van Damme, 2008, Vandenborre et al., 2011a). GNA is toxic not only due to its binding to the insect gut epithelium, but can also penetrate the gut epithelium and reach the hemolymph and other tissues (Fitches et al., 2001). Since the discovery of GNA as an anti-insect protein the insecticidal activity of many mannose-binding lectins has been demonstrated. It is not surprising that especially lectins that recognize mannose structures are highly effective against insects since the glycome of insects is known to consist mainly of carbohydrate structures with terminal mannose residues (Van Damme, 2008, Vandenborre et al., 2011b). At present the exact binding sites of lectins within the insect body are still subject to further research. It is worthwhile to mention that inducible lectins can also be part of the plant defense. For instance, upon infestation with the Hessian fly wheat plants respond with the induced expression of Hessian fly-responsive proteins like Hfr-1, Hfr-2 and Hfr-3, each containing a specific lectin domain (Giovanini et al., 2007). Similarly, the Nicotiana tabacum lectin accumulates in response to chewing caterpillars (Spodoptera littoralis and Manduca sexta) and cell-content-feeding spider mites (Tetranychus urticae), while infestation with phloem-feeding herbivores such as aphids and whiteflies (Myzus nicotianae and Trialeurodes vaporariorum) did not affect lectin accumulation (Vandenborre et al., 2009a, Vandenborre et al., 2009b). Overexpression of the tobacco lectin in transgenic lines revealed a strong retardation of caterpillar development and thus confirmed the insecticidal properties of the lectin.
The wide distribution of lectins, also in edible plants and crops, makes the potential toxicity of these proteins an important issue for health of both humans and animals. The toxicity of lectins to animals can vary greatly, ranging from merely antinutritional properties to lethal effects. An important example of a highly toxic lectin is the phytohemagglutinin (PHA) from bean (Phaseolus vulgaris), which causes severe toxic effects. Overall the toxicity of lectins is mainly due to their binding to specific carbohydrate structures on the epithelial cells in the animal digestive tract. Binding of the lectins to these receptors may cause dramatic changes in the cellular morphology and metabolism of the stomach and/or the small intestine, and can activate a cascade of signals which alters the intermediary metabolism (Vasconcelos and Oliveira, 2004, Yamamoto et al., 2013). Miyake et al. (2007) reported that cell surface-bound lectins potently inhibited plasma membrane repair, and the exocytosis of mucus that normally accompanies the repair response.
In view of their toxic properties, the stability of lectins is a very critical issue. The resistance of lectins to proteolysis is a prerequisite for their toxicity. The extent of lectin resistance to degradation by gut enzymes was shown to be variable, but orally ingested lectins should be at least partially undigested to maintain their toxicity (Vasconcelos and Oliveira, 2004). It still remains a challenge to unravel the mode of action of lectins in their toxicity towards pathogens and predators and to identify the interacting partners for lectins in the tissues of the predator organism.
Lectins have drawn a lot of attention because of their possible biomedical applications, e.g., their anti-tumor activities. The anti-tumor activities of different plant lectins has been shown for several cancer cell cultures, such as human hepatocarcinoma cells (Lyu et al., 2002), human bladder cancer cells (Plattner et al., 2008), human melanoma cells (Liu et al., 2009), rat pancreatic cells (Mikkat et al., 2001). It has also been suggested that some lectins induce apoptosis and/or autophagy of cancer cells (reviewed by Liu et al., 2010).
2.2. Ribosome-inactivating proteins
Ribosome-inactivating proteins (RIPs) are a class of cytotoxic enzymes which possess highly specific rRNA N-glycosidase activity and are capable of catalytically inactivating prokaryotic or eukaryotic ribosomes (Peumans et al., 2001). Being N-glycosidases, RIPs recognize a highly conserved GAGA sequence and remove an adenine residue from the sarcin/ricin loop in the 28S rRNA of animal ribosomes or the 23S rRNA of prokaryotic ribosomes. For instance the most studied RIP ricin (from R. communis seeds) removes the adenine residue at position 4324 from the GA4324GA tetraloop motif of the sarcin/ricin loop in the 28S rRNA of rat liver ribosomes (Puri et al., 2012). Most RIPs display a rather broad N-glycosidase activity towards ribosomes from plants, bacteria, yeast and animals. Very often type-2 RIPs are more efficient for animal ribosomes (Peumans et al., 2001). As a consequence of the removal of a specific adenine residue from the large rRNA, the interaction between the elongation factor 2 and the ribosome is blocked, resulting in the arrest of protein synthesis.
At present it is generally accepted that RIPs do not exclusively act on ribosomes but display polynucleotide adenine glycosylase (PAG) activity on different nucleic acid substrates. It should be mentioned that RIPs have also been reported to possess other enzymatic activities like deoxyribonuclease, chitinase and lipase activity. However, due to lack of decisive experimental evidence and possible misconceptions resulting from sample contamination these data need to be confirmed by further investigations from independent research laboratories. Furthermore it is difficult to conceive how one protein could possess multiple binding sites to accommodate very different substrates (Peumans et al., 2001).
Sequence analyses have shown that the RIP domain is widely distributed in the plant kingdom, but is not ubiquitous. For example, bioinformatics analysis of several completed genomes provided evidence for the absence of RIP genes in at least 24 plants genomes, including the model plant Arabidopsis thaliana (Shang et al., 2014).
Based on their overall structure, RIPs are classified into two major groups. Enzymes that consist exclusively of a PAG domain are referred to as type-1 RIPs whereas type-2 RIPs are chimeric proteins where the PAG domain is linked to a C-terminal lectin domain. Besides the classical type-1 and type-2 RIPs, some special cases of RIPs are found in Poaceae. One example is the JIP60 protein (60 kDa jasmonate-induced protein) found in barley (Chaudhry et al., 1994). This is a chimeric protein where a RIP domain is linked to a domain which has similarity to the eukaryotic translation initiation factor 4E (Rustgi et al., 2014).
Most RIPs are synthesized with a signal peptide on the rough endoplasmic reticulum and follow the secretory pathway which finally guides them to storage vacuoles or the extracellular space. However, some RIPs e.g. from Poaceae lack the signal peptide and after synthesis on free ribosomes reside in the cytosol of the plant cell (Shang et al., 2014).
The biosynthesis of ricin, a typical representative of the type-2 RIPs, has been studied in great detail (Lord and Spooner, 2011). The mature ricin consists of the toxin A chain (RTA, 32 kDa) and the lectin B chain (RTB, 34 kDa) linked by a disulfide bond. Ricin is initially synthesized as a single chain precursor named preproricin, which contains the information for a 26-residue signal peptide and a 9-residue propeptide in front of the RTA sequence as well as a 12-residue linker between the RTA and RTB sequences. Because of the presence of a signal peptide preproricin is transported to the ER. During this translocation, the precursor protein undergoes several processing steps, including the cleavage of the signal peptide, the primary N-glycosylation of the protein and the formation of multiple intramolecular disulfide bonds, important for the tertiary folding of the protein. At this stage, the propeptides and linker peptides remain in the proricin polypeptide, which renders the RTA domain in an inactive state and therefore may protect the plants from any potential toxicity of the N-glycosidase domain (Lord and Spooner, 2011). Subsequently, the glycosylated proricin is translocated in vesicular carriers to the Golgi complex and eventually reaches the vacuole. During this process, the protein undergoes further modification of the glycans and the N-terminal propeptide and the linker peptide are removed by vacuolar processing enzymes, resulting in the fully active type-2 RIP (Lord, 1985, Lord and Harley, 1985).
Unlike type-2 RIPs, little is known about the biosynthesis of type-1 RIPs. Most type-1 RIPs are synthesized with a signal peptide. The mature protein consists of a single polypeptide of approximately 30 kDa that can be glycosylated. However, in some type-1 RIP sequences the signal peptide is absent, indicating that these RIPs are synthesized on free ribosomes in the cytoplasm. Furthermore the type-1 RIP polypeptide can undergo proteolytic cleavage to yield two smaller protein fragments. For example, the maize RIP b32 is synthesized in the cytoplasm as an inactive precursor which is turned into its active form only after a proteolytic activation, during which an N-terminal, a C-terminal and an internal sequence are removed (Bass et al., 2004).
It has been observed that the expression of type-1 RIPs may be toxic for the host cells when they are expressed in transgenic plants or Pichia pastoris (Lombardi et al., 2010). According to Marshall et al. (2011) the expression of saporin in tobacco protoplasts caused a significant decrease in protein synthesis, suggesting that although synthesized with a signal peptide, a small fraction of the saporin may still reach the cytosol and act upon tobacco ribosomes. Furthermore, it is proposed that the signal peptide could interfere with the catalytic activity of saporin by causing protein aggregation when the protein completely fails to be targeted to the ER (Marshall et al., 2011).
Type-2 RIPs possess an efficient strategy to enter the target cells, which makes them potent toxins, being toxic in the picomolar range (Stirpe and Battelli, 2006). The internalization of type-2 RIPs has been reviewed recently (Puri et al., 2012). In brief, type-2 RIPs such as ricin bind to glycoconjugate receptors on the cell surface with their lectin B chain which facilitates the entry of the protein in the cell through an endocytic pathway. After being transported from the endosome to the Golgi apparatus, the RIPs arrive in the ER lumen by a retrograde transport. Eventually, the type-2 RIPs exert their enzymatic activity on ribosomes after being translocated to cytosol. The carbohydrate-binding domain of most type-2 RIPs exhibits specificity towards galactosylated carbohydrate structures though a few RIPs also specifically recognize sialic acid residues (Van Damme et al., 1996). Hence galactosylated glycoconjugates, either glycoproteins or glycolipids, are the most likely targets for interaction. Because of the carbohydrate-binding activity of the B chain type-2 RIPs are also considered as lectins, and thus can also be classified as a family of lectins. Although the classical RIPs such as ricin and abrin are very toxic proteins, a few type-2 RIPs such as ebulin 1 show little or no toxicity. The cytotoxicity of ebulin 1 for HeLa cells is much lower than that of ricin, with an IC50 value of 6 × 10−8 M compared to 10−12 M for ricin (Jiménez et al., 2014).
Type-1 RIPs are generally less toxic than the type-2 RIPs due to the lack of the lectin chain. Type-1 RIPs enter the cells by endocytosis but the precise mechanism of their internalization still awaits to be elucidated. Studies using trichosanthin (TCS) and saporin-6 suggested that the endocytosis is mediated by low density lipoprotein receptors (Chan et al., 2000, Ippoliti et al., 2000) while research on saporin-S6 indicated that this process is mainly receptor-independent (Bolognesi et al., 2012). The toxicity of type-1 RIPs is limited by their low ability to reach the ribosomes in the cytosol. However, these proteins can still be very toxic if they succeed to efficiently enter the target cells, e.g., after conjugation to a lectin or antibody. This strategy has been exploited to prepare immunotoxins, which serve as a tool in cancer therapy (Polito et al., 2011; see below).
Interestingly several RIP genes are upregulated in stressed plants. For example JIP60 is produced in barley leaves treated with methyl jasmonate. It is also present in senescent plants and plays a role in the reprogramming of translation in response to stress. Currently two models for the function of JIP60 in translational reprogramming exist: (i) when JIP60 is proteolytically processed, the released RIP domain can act as an N-glycosidase thereby irreversibly inhibiting protein translation. (ii) Without processing the JIP60 protein is supposed to act as a ribosome-dissociation factor. The released eIF4E domain was shown to initiate the translation of other mRNAs encoding jasmonate-induced proteins. JIP60 thus plays a crucial role in the stress response by reprogramming the translational machinery in stressed cells (Rustgi et al., 2014). Another stress inducible RIP is PIP2 from Phytolacca insularis for which transcript levels are upregulated after both methyl jasmonate and abscisic acid treatments (Song et al., 2000). Sugar beet leaves contain a virus-inducible RIP (Iglesias et al., 2005). Jiang et al. (2008) identified 31 genes encoding RIPs in the rice (Oryza sativa) genome. Expression analysis showed that these genes are upregulated when rice is subjected to abiotic stress such as cold or salt stress. Interestingly, transgenic rice plants overexpressing one of these genes (OSRIP18) were more tolerant to salt and drought stress (Jiang et al., 2012).
Several RIPs have been studied in detail for their insecticidal, antiviral and antifungal properties. Feeding experiments with artificial diets as well as with transgenic plants suggested the insecticidal activity of several RIPs, including both type-1 and type-2 RIPs (reviewed by Vargas and Carlini, 2014). Several RIPs, mostly of the type-1 type, have also been proven to act as antifungal agents, although with less activity compared to other antifungal proteins. For instance, transgenic plants expressing PAP (from Phytolacca americana), curcin 2 (from Jatropha curcas), dianthin (from Dianthus caryophyllus) all showed enhanced resistance to Rhizoctonia solani (Qin et al., 2005, Shah and Veluthambi, 2010, Wang et al., 1998). In vitro experiments comparing the antifungal properties and the N-glycosidase activity of a type-1 RIP from Mirabilis expansa, ricin and saporin demonstrated that their antifungal activity was not necessarily linked to the depurination activity on ribosomes (Park et al., 2002). Although the mechanism of insecticidal and antifungal activity of RIPs is largely unknown, these activities are believed to be an important part of the plant defense system.
Both type-1 and type-2 RIPs also display inhibitory activity towards viral infection. For instance, pokeweed antiviral protein (PAP) has been shown to display an inhibitory effect on tobacco mosaic virus and brome mosaic virus (Karran and Hudak, 2011, Watanabe et al., 1997). The antiviral activity of RIPs towards plant viruses suggests a role in plant defense against these pathogens.
Due to their potential applications in medicine, many studies have been undertaken to investigate the toxicity of RIPs towards animal viruses, among which human immuno-deficiency virus (HIV) being the most important one. The replication of HIV can be inhibited by several RIPs, such as TCS (from Trichosanthes kirilowii), pokeweed antiviral protein (PAP) and Momordica antiviral protein (MAP30). It was reported that a mutated form for PAP, which lost its ability to depurinate ribosomes, still inhibited HIV in tobacco plants (Tumer et al., 1997). Similarly, two TCS mutants, TCSC19aa and TCSKDEL still retained N-glycosidase activity after most of their anti-HIV-1 activities were removed by site-directed mutagenesis resulting in the addition of 19 amino acids or a KDEL signal sequence to the C-terminal sequence (Wang et al., 2003). All these results suggested that the antiviral activity of RIPs is independent from their N-glycosidase activity. It has been proposed that the antiviral activity of RIPs may also be due to the direct depurination of the viral RNA/DNA. This hypothesis is supported by the observations that incubation of purified PAP and HIV-1 genomic RNA or treatment of HIV-1 long terminal repeats DNA with TCS resulted in the removal of adenine (Rajamohan et al., 1999, Zhao et al., 2010). The anti-HIV activity of TCS might be related to its ability to enhance the capabilities of chemokines to stimulate chemotaxis and G protein activation through interaction with chemokine receptors, which play important roles in HIV infection (Zhao et al., 1999). Although the inhibitory effect of RIPs towards HIV has been studied extensively and led to phase I/II clinical trials, there are still some issues that need to be resolved (Kaur et al., 2011, Puri et al., 2012).
2.3. Plant protease inhibitors and α-amylase inhibitors
Plant protease inhibitors are a vital group of proteins directed against all kinds of pathogens and invading organisms. They are widely distributed in plant tissues, especially in seeds and tubers, and their expression is often triggered by wounding or attack by pathogens or insects (Laskowski and Kato, 1980, Murdock and Shade, 2002). Due to their inhibitory activity on proteases, protease inhibitors can suppress the growth of a variety of pathogens and insects (Ryan, 1990).
Plant protease inhibitors have been reported for all four classes of proteases, including serine, cysteine, aspartyl and metalloproteinases (Christeller et al., 1998, Haq et al., 2004, Mareš et al., 1989, Otto and Schirmeister, 1997). All these protease inhibitors act similar to competitive inhibitors, which bind to the active site of the enzyme to form a complex with a very low dissociation constant (107–1014 M at neutral pH). The inhibitor directly mimics the substrate of the enzyme and thus forms an inhibitor–enzyme complex that cannot be dissociated by the normal enzyme mechanism, therefore efficiently blocking the active site and the protease activity of the enzyme (Lawrence and Koundal, 2002). Up to now, a lot of protease inhibitors have been reported from various plants and relevant information is compiled in the Plant-PIs database (http://plantpis.ba.itb.cnr.it/) and can be used for retrieval of information on plant protease inhibitors (PIs) and related genes. (De Leo et al., 2002).
Serine protease inhibitors are the largest group of protease inhibitors. The two best-characterized plant serine protease inhibitors are the Kunitz-type and the Bowman–Birk inhibitors. Kunitz-type inhibitors (18–22 kDa) usually have a low cysteine content and contain one reactive site, while Bowman–Birk type inhibitors (8–10 kDa) have a high cysteine content and possess two reactive sites. Feeding experiments with a diet supplemented with purified soybean trypsin inhibitors (the Kunitz soybean trypsin inhibitor STI and the Bowman–Birk trypsin/chymotrypsin inhibitor) caused enlargement of the pancreas in rats, chickens and mice (Birk, 1996). Furthermore, Bowman–Birk inhibitors might also be involved in the prevention of tumorigenesis and nephrotoxicity induced by the antibiotic gentamicin (Lippman and Matrisian, 2000, Smetana et al., 1992).
During the past decades, plant protease inhibitors have gained lots of attention due to their role in defense and possible applications for improvement of plant resistance to pathogens and insects (Kim et al., 2009, Valueva and Mosolov, 2004). Transgenic plants that overexpress protease inhibitors have been constructed to increase plant resistance to pathogens, insects and nematodes (Duan et al., 1996, Senthilkumar et al., 2010, Vishnudasan et al., 2005). Although the idea of using plant protease inhibitors as a pest control agent has become very attractive, some problems are of concern. During the long history of co-evolution between plants and herbivores, insects have adopted different ways to cope with protease inhibitors, such as the overexpression of proteases to maintain normal levels of enzymatic activity, the induced expression of proteases insensitive to the ingested inhibitors and the up-regulation of enzymes that degrade the protease inhibitors (Schlüter et al., 2010). The overexpression of protease inhibitors in plants not only results in the inhibition of certain insect proteases but also triggers adaptation mechanisms in some insects to minimize the effect of the protease inhibitor on food digestion (Moon et al., 2004, Dunse et al., 2010b). The use of protease inhibitors may also affect non-target organisms in the agroecosystem. Accordingly it is necessary to develop protease inhibitors with strong inhibitory activity against specific herbivores. Protein engineering methods can be used to enhance the inhibitory potency as well as broaden the activity range to improve the overall efficiency of protease inhibitors (Schlüter et al., 2010). A biotechnological approach involving transgene stacking/pyramiding can be applied to enhance the efficacy of protease inhibitors. Using a combination of potato type I and II protease inhibitors in transgenic plants, Dunse et al. (2010a) succeeded in increasing the resistance of cotton against insect damage from Helicoverpa armigera in the lab as well as in the field. Although some problems remain to be solved, it can be concluded that plant protease inhibitors show great potential for applications in pathogen control (Lawrence and Koundal, 2002).
Next to protease inhibitors plant seeds are also an important source of another group of inhibitors acting upon α-amylases. The so-called α-amylase inhibitors are present in many plants and play a role in the control of endogenous α-amylase activity as well as in defense against pathogens and pests. Since inhibitors for proteases and α-amylases function in a similar way, we refer to some review papers for more detailed information on α-amylase inhibitors and their enzymatic activity (Franco et al., 2002, Svensson et al., 2004).
2.4. Canatoxin-like proteins and ureases
Canatoxin is a toxic protein first isolated from the seeds of jack bean Canavalia ensiformis (Carlini and Guimarães, 1981). In its native form the protein exists as a non-covalently linked dimer of 95 kDa polypeptides containing zinc and nickel ions, representing up to 0.5% of the total dry weight of jack bean seeds. Based on its sequence canatoxin is considered as an isoform of the jack bean major seed urease, retaining approximately 30% of the ureolytic activity for urease (EC 3.5.1.5) (Follmer et al., 2001). Canatoxin also interacts with complex glycoconjugates and behaves like a hemilectin: erythrocytes pre-treated with canatoxin can be agglutinated by antibodies specific to canatoxin (Carlini and Guimarães, 1991).
Being a neurotoxin, canatoxin is lethal to rats and mice, with an LD50 of 2–5 μg/g upon intraperitoneal injection, but the protein is inactive when administered orally due to its instability at low pH (Carlini and Guimarães, 1981). Toxic symptoms provoked by canatoxin include respiratory distress and tonic convulsions of spinal cord origin, ultimately leading to the death of the animal. The central nervous system was identified as one of the target organs for canatoxin and certain neurotransmitters can be released dose- and time-dependently after incubation with canatoxin (Carlini et al., 1984). According to experiments using sarcoplasmic reticulum vesicles, canatoxin was deemed to disrupt the Ca2+ transport by the Ca2+ ATPase, leading to an increased cytoplasmic Ca2+ concentration, which eventually triggers exocytosis (Alves et al., 1992). It is likely that lipoxygenase pathways are somehow involved in this toxicity process since all toxic effects provoked by canatoxin known so far can be inhibited by lipoxygenase inhibitors (Barja-Fidalgo et al., 1991). Furthermore, the hemilectin activity of canatoxin mentioned above, might play a critical role in its interaction with target cell surfaces and could explain its tissue-specific toxicity (Carlini and Guimarães, 1991).
Canatoxin, together with other ureases such as jackbean major seed urease, soybean embryo-specific urease and pigeon pea urease, exhibits insecticidal and antifungal activity (Balasubramanian et al., 2013, Carlini et al., 1997, Carlini and Polacco, 2008, Postal et al., 2012). Nymphs of the hemipteran cotton stainer bug Dysdercus peruvianus are more susceptible to canatoxin compared to adults due to the distinct pattern of enzymatic activities of cathepsin-like protease in midgut homogenates depending on their developmental stages (Staniscuaski et al., 2005). Upon digestion of the native canatoxin by cathepsin-like enzymes present in the insect digestive tract, a 10 kDa internal peptide named pepcanatox is released which accounts for the insecticidal activity of this protein (Ferreira-DaSilva et al., 2000). Later, it turned out that pepcanatox is responsible for both the insecticidal and antifungal activities of urease (Postal et al., 2012). To elucidate the mechanism of action, a recombinant peptide equivalent to pepcanatox, named jaburetox-2Ec was used. Irrespective of proteolytic release, Jaburetox-2Ec exhibited similar insecticidal activity towards insects with both cathepsin-based and trypsin-based digestion. Molecular modeling showed that jaburetox-2Ec forms a large, generally exposed β-hairpin structure, which shares similar features with some pore-forming toxins and some neurotoxins (Mulinari et al., 2007). Crystal structures for jackbean major seed urease confirmed that a 10 kDa region corresponding to Jaburetox-2Ec, consists of an alpha-helix, a long loop, another short helix and a β-hairpin motif (Balasubramanian and Ponnuraj, 2010). This 10 kDa peptide was reported to have a cation-selective pore-forming activity, which explained the mechanism of jack bean urease to permeabilize phospholipid membranes (Piovesan et al., 2014). Both the insecticidal and antifungal activities of urease rely on this ability for membrane permeabilization. Surprisingly, studies with different mutants of Jaburetox (a peptide modified from Jaburetox-2Ec) showed that the N-terminal portion of Jaburetox maintained the pore-forming activity similar to the full peptide, despite of the absence of the β-hairpin motif (Martinelli et al., 2014), indicating that it is mainly the helix structure rather than β-hairpin motif that is essential for the membrane permeabilizing activity of the peptide. Taking all these facts into consideration, Jaburetox probably possesses an action mechanism similar to that of some α-pore-forming toxins, which bind to and act upon membrane K+ channels (Brogden, 2005, Degiacomi et al., 2013).
2.5. Arcelins
Arcelins are seed proteins discovered in wild accessions of common bean (P. vulgaris L.). The arcelin sequences belong to the arcelin/phytohemagglutinin/α-amylase inhibitor (APA) family, a group of sequences all encoded in a single locus, the so-called APA locus (Blair et al., 2010, Osborn et al., 1988). Although arcelins and α-amylase inhibitors exhibit high sequence similarity to lectins and have a similar three-dimensional conformation, they do not possess functional carbohydrate-binding sites (Sparvoli and Bollini, 1998). At present, eight electrophoretic variants of the arcelin proteins (named arcelin 1–8) have been reported, with molecular weights ranging from 27 to 42 kDa (Acosta-Gallegos et al., 1998, Osborn et al., 1986, Zaugg et al., 2013).
Characterization of wild P. vulgaris L. accessions showed different levels of resistance, depending on the type of arcelin present. Some arcelins were shown to have insecticidal activity on the larval development of Zabrotes subfasciatus, one of the two major bruchid species affecting beans, whereby arcelin-5 conferred the highest level of resistance to Z. subfasciatus and arcelin-3 showed the lowest activity (Acosta-Gallegos et al., 1998, Cardona et al., 1990). However, transgenic Phaseolus acutifolius seeds overexpressing the arcelin-5 isoform did not achieve adequate levels of resistance against Z. subfasciatus, indicating that arcelins may only be partially responsible for the resistance to Z. subfasciatus (Goossens et al., 2000). Furthermore, two arcelin-containing P. vulgaris genotypes containing arcelin-4 and arcelin-8 were also resistant to Acanthoscelides obtectus, the second major bruchid species (Zaugg et al., 2013).
Despite extensive studies, the mechanism of arcelin toxicity remains controversial. Being the first discovered arcelin, arcelin-1 has been studied most extensively. Native arcelin 1 is a 60 kDa dimeric glycoprotein, with non-covalent linkage of two identical monomers. Paes et al. (2000) discovered that Arc-1 altered the gut structure of Z. subfasciatus, (but not for A. obtectus) and penetrated into the hemolymph. They proposed that the severe deleterious effects of arcelins on the gut of Z. subfasciatus might be due to the recognition and interaction of arcelin with glycoproteins and other membrane constituents along the digestive tract. However, according to Minney et al. (1990) arcelins are indigestible by the gut proteases of Z. subfasciutus and thus caused starvation of Z. subfasciatus larvae. It is very likely that both factors contribute to the toxicity of arcelin for Z. subfasciutus. With respect to the insecticidal activity of Arc-4 and Arc-8 to A. obtectus, their resistance to proteolysis might be the main reason for their toxicity (Zaugg et al., 2013).
2.6. Antimicrobial peptides
Antimicrobial peptides (AMPs) are ubiquitous, low molecular weight peptides that directly target a broad spectrum of microbial pathogens. In plants, AMPs can be grouped into different classes, including cyclotides, thionins, defensins, lipid transfer proteins, snakins, hevein-like peptides, vicilin-like peptides and knottins (Goyal and Mattoo, 2014). Generally, the biological activity of these bioactive peptides relies on their binding to the target membrane followed by membrane permeabilization and disruption. Considering the high similarity between different AMPs in terms of their toxicity and antimicrobial activity, only two major groups of antimicrobial peptides, in particular thionins and cyclotides, are discussed below. For more information with respect to the antimicrobial activity of AMPs we refer to some recent review papers (Goyal and Mattoo, 2014, Salas et al., 2015).
2.6.1. Thionins
Thionins are small cysteine-containing, usually basic proteins of approximately 5 kDa, found in a number of monocot and dicot plants (Bohlmann, 1999). They consist of 45–48 amino acids bound by three or four disulfide bonds and are highly basic. Thionins can be divided into two classes: α/β-thionins and γ-thionins. All the α/β-thionins are highly homologous at the amino acid level and exhibit the same three-dimensional structure. They are classified into five different groups mainly based on their distribution in the plant kingdom (Bohlmann and Apel, 1991). Type I thionins are present in the endosperm of cereals (the family Poaceae). Type II thionins have been isolated from leaves and nuts of the parasitic plant Pyrularia pubera and from the leaves of barley Hordeum vulgare. Type III thionins have been extracted from leaves and stems of mistletoe species. Type IV thionins are found in seeds of Abyssinian cabbage (Crambe abyssinica). Type V thionins are truncated forms of regular thionins found in some cereals such as wheat. Unlike the α/β-thionins, the γ-thionins show distinct three-dimensional structures and share more similarity with another family of peptides named defensins, which have been reported in plants, but also in insects and animals (Stec, 2006, Stotz et al., 2009).
Thionins show toxicity to a wide range of biological systems, such as bacteria, fungi, cultured mammalian cells (Carrasco et al., 1981), insect larvae (Kramer et al., 1979) and Leishmania donovani (Berrocal-Lobo et al., 2009). In terms of antibacterial activity, thionins from the endosperm of wheat and barley (type I), and from barley leaves (type II) exhibited similar EC50 values around 2–3 × 10−7 M to the bacterial species Clavibacter michiganensis subsp. sepedonicus or Pseudomonas solanacearum, and an EC50 value of 1–4 × 10−6 M to fungal pathogens, such as Rosellinia necatrix, Colletotrichum lagenarium and Fusarium solani (Molina et al., 1993). For insecticidal activity, thionins from wheat, barley and rye showed an LC50 of 17–46 μg/g towards larvae of Manduca sexta upon injection. Berrocal-Lobo et al. (2009) also reported the leishmanicidal activity for a mixture of different type I thionins. Due to their toxicity, thionins have been suggested to play a role in plant defense against pathogen attack (Asano et al., 2013, Florack and Stiekema, 1994).
The primary effect of thionin toxicity is an increase of cell membrane permeability (Carrasco et al., 1981, Thevissen et al., 1999), which was inhibited by mono- or divalent metal ions (Oard et al., 2007). This change in permeability provokes several subsequent effects, including a membrane depolarization, increase in Ca2+ and K+ ion permeability and also activation of some enzymes (Stec, 2006). All these secondary events might strengthen the initial toxicity and lead to final cell destruction.
So far, there have been different hypotheses to explain the mechanism of thionin toxicity. The wide range of toxicity suggests that the permeabilization of cells by thionins relies on some universal process rather than a specific cell surface receptor. According to Hughes et al. (2000) the universal toxicity of thionins is due to the formation of ion channels in the cell membrane by binding to the lipid surface itself. However, Richard et al. (2002) proposed that thionins can partially insert into the lipid membrane through an electrostatic interaction, which subsequently rigidifies the membrane and increases the fluidity at edges of the interfacial region. Further studies suggested that the formation of negatively charged patches of phospholipid molecules is promoted by the electrostatic interaction between thionins and individual phospholipid head groups. These patches of toxins increase membrane fluidity and withdraw phospholipids from the membrane by lowering the energy penalty for the phospholipid membrane separation, which leads to additional membrane instability and ultimately irreparable lysis (Stec et al., 2004). Despite all the evidence mentioned above, more experimental work is still needed to elucidate the detailed mode of action of thionins and decipher their biological role.
2.6.2. Cyclotides
The family of cyclotides (from cyclic peptides) groups all proteins defined by a cyclic backbone and a cyclic cysteine knot motif built from six conserved cysteine residues (Craik et al., 1999). They are widespread in nature, from bacteria to animals, and many of these small globular microproteins have been studied in plants. These proteins typically consist of 28–37 amino acids with six cysteine residues that form three conserved disulfide bonds. The polypeptides possess a unique head-to-tail cyclic cysteine knot topology, in which a ring is formed by opposing peptide backbone segments. The unique structure of cyclotides results in an extraordinary stability towards thermal and chemical denaturation as well as enzymatic degradation (Gould et al., 2011).
The discovery of the first cyclotide kalata B1 dates back to the 1960s. Kalata B1 was reported as the main active component of the Rubiaceae plant Oldenlandia affinis, which was used by natives to make a tea for the purpose of accelerating childbirth (Gould et al., 2011). Since then, more than 200 cyclotide sequences have been discovered in the families Rubiaceae, Violaceae, Cucurbitaceae and Fabaceae (Nguyen et al., 2012). Meanwhile, cyclotides have been reported in diverse tissues, including leaves, stems, roots and flowers. Furthermore, extensive analysis of the distribution of cyclotides in flowering plants indicated that cyclotides represent one of the largest peptide families within the plant kingdom (Gruber et al., 2008). A database for cyclotides is available at Cybase (http://www.cybase.org.au) (Wang et al., 2008).
Judging from their activity against insects (Gruber et al., 2007), nematodes (Colgrave et al., 2008) and mollusks (Plan et al., 2008), the natural function of cyclotides in plants probably relates to host defense. Several cyclotides from the Rubiaceae family including kalata B1, kalata B2 and parigidin-br1 possess insecticidal activity towards lepidopteran larvae, causing retardation of development as well as mortality (Gruber et al., 2007, Pinto et al., 2012). Studies on the insecticidal activity of kalata B1 revealed a disruption of midgut epithelial cells in the midgut of lepidopteran larvae, which resembled the morphological changes of insect midguts induced by delta-endotoxins from Bacillus thuringiensis (Barbeta et al., 2008). Overall, due to their toxicity against insects, nematodes and mollusks, cytoclotides offer great potential as a class of pest control agents.
Besides their anti-insects properties, a diversity of activities have been ascribed to cyclotides, including uterotonic activity, anti-HIV activity, anti-tumor activity, neurotensin antagonism and hemolytic properties (Gould et al., 2011). Because most of these properties are poorly studied, only some of them will be discussed. Ever since the discovery of their anti-HIV activity in early screening studies, cyclotides have attracted lots of attention. Although several cyclotides show anti-HIV activity, the exact mode of action is still unclear. Current data suggest that cyclotides affect the binding and/or fusion of the virus to the target membrane of host cells (Ireland et al., 2008). However, cyclotides are not being considered as anti-HIV agents due to their low therapeutic index (i.e., the ratio of their therapeutic effects to toxic effects). Hemolytic activity has been tested with numerous cyclotides, indicating a very low potency with a median hemolytic dose higher than 10 μM (Craik, 2010).
The biological activities of cyclotides are most probably related to their ability to form pores in the host membrane (Huang et al., 2009). Electrophysiological experiments demonstrated that conductive pores were induced in liposome patches after incubation with kalata B1. Alanine-scanning mutagenesis of kalata B1 revealed that the residues essential for membrane disruptive activity are clustered, forming the bioactive side of cyclotides (Huang et al., 2009). Interestingly, the hemolytic and insecticidal activities both depend on a common, well-defined cluster of hydrophilic residues on one face of the cyclotides, separated from the membrane binding side of the protein (Simonsen et al., 2008).
All these fascinating properties of cyclotides make them an ideal tool for drug development. Thanks to their relatively small size, cyclotides can be produced by recombinant expression systems as well as by chemical synthesis methods. The pharmaceutical applications of cyclotides as well as details of their bioactivities have been compiled in several review papers (Craik et al., 2012, Gould et al., 2011).
2.7. Pore-forming toxins
Pore-forming toxins are widely distributed proteins that form water-filled pores in biological membranes. They are best characterized in bacteria, but they have also been identified in plants, fungi and animals (Bischofberger et al., 2012, Gilbert, 2002). Many pathogens produce pore-forming toxins to attack the host by forming holes in the target cell membrane. Pore-forming toxins usually undergo a conformational change and then assemble into an oligomeric structure, which would promote a spontaneous membrane insertion (Iacovache et al., 2010). Eventually the disruption of the membrane permeability barrier can lead to cell death (Parker and Feil, 2005). In recent years there have been several reviews about pore-forming toxins (Bischofberger et al., 2012, Iacovache et al., 2010, Leippe, 2014, Los et al., 2013, Ramarao and Sanchis, 2013). These papers focus on bacterial pore-forming toxins or pore-forming toxins in general. Here we will mainly concentrate on pore-forming toxins from plants.
The best studied pore-forming toxin from plants is Enterolobin, a 54.8 kDa cytolytic protein from the seeds of the tropical tree Enterolobium contortisiliquum (Bischofberger et al., 2012, Sousa and Morhy, 1989). Enterolobin is structurally similar to the plant cytolysin aerolysin and occurs as a dimer in solution (Bittencourt et al., 2003). Insect feeding experiments showed that enterolobin is toxic to larvae of the bruchid Callosobruchus maculatus, causing 70% mortality at a concentration of 0.01% and 100% mortality at 0.025%. In vitro proteolysis studies showed that Entrolobin is resistant to the digestion by larval gut enzymes of C. maculatus (Sousa et al., 1993). Enterolobin also induces inflammation upon injection in rats (Castro Faria Neto et al., 1991). Similar to other pore-forming toxins, the oligomerization of enterolobin is promoted by low pH and high ionic strength (Fontes et al., 1997).
Interestingly evidence for the occurrence of pore-forming toxins was also obtained for wheat. Upon infestation of susceptible wheat (Triticum aestivum) plants by larvae of the Hessian fly (Mayetiola destructor) wheat gene expression is changed. Up-regulation of gene expression was observed in particular for the Hessian fly responsive-2 (Hfr-2) gene, which encodes a protein consisting of a domain with sequence similarity to the seed-specific lectin from Amaranthus linked to a domain with sequence similarity to pore-forming toxins. Further support for the involvement of Hfr-2 in interactions with insects came from experiments with fall armyworm (Spodoptera frugiperda) and bird cherry-oat aphid (Rhopalosiphum padi). Wheat infestation with both insects resulted in enhanced transcript levels for the Hfr-2 gene (Puthoff et al., 2005). Unfortunately at present no information is available at protein level to proof the pore-forming activity of Hfr-2 and its importance for the biological activity of the protein.
Although pore-forming toxins have been reported in plants (Puthoff et al., 2005, Szczesny et al., 2011), little information is available especially with respect to their mode of action. More studies are needed to understand the distribution and biological importance of plant pore-forming proteins.
3. Potential applications
Plants express a broad range of toxic proteins encompassing toxicity to animals, bacteria, fungi and viruses, and as such these noxious compounds can play an important role in plant defense. In addition to their role in plant protection this group of toxic proteins also represents a powerful tool for agricultural and medical applications.
3.1. Agricultural applications
A lot of research has been performed to exploit the antiviral, antifungal and insecticidal properties of some toxic plant proteins. Genetic engineering technology has been applied in plants to introduce the genes encoding toxic proteins derived from another plant or regulate the transcription levels of endogenous genes in order to obtain enhanced resistance to various pathogens (Ahmad et al., 2012). Plant toxic proteins such as lectins, RIPs, protease inhibitors and thionins have been introduced in different plants including important crops to increase their resistance to biotic as well as abiotic stresses (Dias et al., 2010, Muramoto et al., 2012, Shahidi-Noghabi et al., 2009, Vila et al., 2005).
In the last few decades several strategies have emerged to enhance the resistance of transgenic plants. The general idea is ‘to use a plant protein to solve a plant problem’ which – in view of consumer acceptability is a better strategy compared to the use of non-plant proteins. On the long term co-expression of multiple genes should be used rather than the expression of a single gene with the idea to circumvent problems related to development of resistance (Dowd et al., 2012, Dunse et al., 2010b). Similarly synthetic genes comprising a combination of different toxins have been tested. For instance, Liu et al. (2015) applied a fusion protein consisting of a GNA lectin domain linked to AaIT, the insecticidal neurotoxin of the scorpion Androctonus australis venom and reported that transgenic tobacco and rice plants which overexpress this fusion protein show enhanced resistance to both chewing and sucking pests. As such these lectins can contribute to the development of integrated insect pest control strategies and provide a nice alternative to the use of classical chemical insecticides or the use of B. thuringiensis toxins that exhibit little toxicity against sap sucking insects. Similar strategies can be used to engineer plants with enhanced resistance to bacterial, viral and fungal diseases. The use of inducible or tissue specific promoters rather than constitutive promoters could also achieve a better and more sustainable protection of plants (Abdeen et al., 2005, Checker et al., 2012, Corrado et al., 2005).
Of course, there are still some concerns related to the application of genetically modified plants for crop protection purposes. For example, the variable susceptibility of different insect species, the adaptation of pathogens leading to resistance, the potential toxicity of the overexpressed toxic proteins in the transgenic plants for the consumer are major issues that need to be solved (Macedo et al., 2015, Murdock and Shade, 2002). In addition there are the biosafety concerns and the issues related to (lack of) regulation and legislation for the commercial use of genetically modified plants for field applications. Future experiments will need to focus on these problems and provide answers for these questions.
3.2. Medical applications
Plant toxins, mainly RIPs, have been used to fabricate immunotoxins for therapeutic purpose. The idea of immunotoxins, also named targeted toxins, was brought up more than four decades ago, and intended to make an A-B toxin specifically targeting cancer cells (Kreitman, 2006). The strategy is to link a toxin to another molecule that can direct the toxin in a selective way towards the cells to be eliminated. So far, there is only one successful example, an engineered protein combining Interleukin-2 and Diphtheria toxin named denileukin diftitox (Ontak), which has been approved by the U.S. Food and Drug Administration for treatment of cutaneous T-cell lymphoma (Duvic and Talpur, 2008). In recent years a wide variety of immunotoxins have been synthesized and tested against several malignancies in cell cultures, animal models and patients. A list including more than 450 RIP-based immunotoxins can be found in Gilabert-Oriol et al. (2014). Normally, type-1 RIPs or the A chain of type-2 RIPs are used to construct the targeted toxins. Whole type-2 RIPs can only be used after modification of the protein since their B chain will affect the specific delivery of the toxins (Becker and Benhar, 2012). The toxins can be fused to carriers such as monoclonal antibodies, lectins, hormones, growth factors using a chemical linkage. Alternatively, genetic engineering methods can be employed offering a more standardized composition of the resulting molecule (Stirpe, 2013). Experiments with the RIP-based toxins yielded promising results in vitro and in experimental animals (Fracasso et al., 2010). However, some adverse reactions like vascular leak syndrome and fatigue have been observed. The use of mutant RIPs could be an option to reduce these side effects (Smallshaw et al., 2003). Nevertheless, there are still some obstacles for the use of immunotoxins. For example, the immunotoxins are recognized as foreign proteins, and hence might trigger an immunological response preventing their repeated use. A possible way to reduce the immunogenicity of the proteins is to use human or humanized antibodies, make a PEGylated form or deplete the immunodominant epitopes of RIPs (Lorberboum-Galski, 2011, Meng et al., 2012). Other hurdles include the expensive production, protein instability and short biological half-life and insufficient endosomal escape, which hopefully can be solved in the future (Gilabert-Oriol et al., 2014).
The traditional Chinese medicine makes use of Trichosanthin (TCS), a RIP from T. kirilowii, to induce abortion and to treat hydatidiform moles (Ng et al., 1992). TCS was also the first RIP for which anti-HIV activity was demonstrated in vitro. The inhibitory effect of RIPs on HIV proliferation in cells led to phase I/II clinical trials with TCS, and modified PAP was tested on AIDS patients (Puri et al., 2012). Although there is no success so far, the clinical trials showed the potential of TCS and PAP for treating HIV positive patients. Besides RIPs, lectins, especially some mannose-binding lectins, have also been shown to possess anti-HIV activity. These carbohydrate-binding agents can inhibit HIV infection of susceptible cells, but can also inhibit syncytia formation between persistently HIV-infected cells and uninfected lymphocytes. The lectins presumably act through binding with the glycans present on the HIV envelope, e.g. on gp120. Furthermore long-term exposure of HIV to lectins results in the progressive deletion of N-glycans on the viral surface in an attempt of the virus to escape drug pressure. Thus lectin treatment of HIV may form the basis for a novel chemotherapeutic concept relying on (1) the direct antiviral activity of lectins by preventing virus entry and transmission to target cells, and (2) an indirect antiviral activity of lectins by forcing the virus to change the glycosylation of the viral surface proteins. Eventually this novel strategy to combat HIV might also be extended to other enveloped viruses such influenza virus and coronavirus (François and Balzarini, 2012).
Due to their specific glycan-binding activity, lectins are interesting tools for different medical applications. For example, next to being a carrier molecule in immunotoxins, lectins can also be useful for drug delivery (Lehr, 2000). The idea behind this application is simple, because different cells express different glycans on their cell surface and diseased or cancer cells often show altered glycosylation patterns compared to normal cells. Therefore these cell surface glycoconjugates can serve as binding sites for lectins. More information can be found in a review by Bies et al. (2004).
Another well-known application is the use of lectin microarrays as a powerful tool for high-throughput and high-sensitivity profiling of complex glycans without the need for glycan release. This technology allows testing a broad range of glycoconjugates from cells, tissues, body fluids as well as microbes (Hirabayashi et al., 2013, Qin et al., 2012, Qin et al., 2013), and has already been widely used in medical research for the discovery of disease-related biomarkers (Katrlik et al., 2010, Zhou et al., 2011). Lectins have also been linked to beads for the separation of glycoproteins based on their glycan binding specificity. For instance, Concanavalin A-magnetic particle conjugate-based methods have been used in the analysis of hepatocellular carcinoma related glycoprotein profile changes in human sera, which might be useful for the identification of disease-specific biomarkers (Yang et al., 2012).
In addition to lectins and RIPs, cyclotides and the α-amylase inhibitors also deserve some attention with respect to their applications for medical science. Due to their unique structure and ultra-stability, cyclotides are promising templates for drug design and discovery. Studies have shown that bioactive peptide sequences directed at cancer, cardiovascular and infectious diseases can be grafted into cyclotide frameworks and thereby are stabilized, while maintaining biological activity (Craik et al., 2012, Gould et al., 2011). The α-amylase inhibitor from white bean (P. vulgaris) is effective in reducing glycaemia in type-2 diabetic rats when orally administered (Tormo et al., 2006). A nutritional supplement product named Phase 2® Carb Controller (Pharmachem Laboratories, Kearny, NJ) has been brought to the market, and causes weight loss and reduces post-prandial spikes in blood sugar in a dose-related manner (Barrett and Udani, 2011).
4. Conclusions
Plants synthesize and store a broad range of chemical and protein based toxic compounds as part of their defense system designed to detect and respond to invading organisms. It is extremely important to obtain a detailed understanding of the protein structure and diversity, and their biological activities, since this knowledge is a prerequisite for biotechnological applications of these proteins. In addition, understanding how plants defend themselves is essential to preserve the food supply and develop disease-resistant crops. Throughout evolution plants have built up a first line of defense including constitutive as well as inducible defense mechanisms, consisting of an array of structural barriers, chemical compounds and toxic defense proteins. Although there is a large diversity in the types of proteins and their biological activities some of them are clearly evolutionary related. For example, sequence data proof the evolutionary relationships between legume lectins and arcelins. The same holds true for the ricin-related lectins and the RIPs that in addition to the lectin domain have acquired a domain with enzymatic activity. Ureases share some structural features with certain pore-forming toxins. Although these proteins also resemble each other with respect to their working mechanism there is a huge difference in the size of these proteins, ranging from small peptide proteins such as the antimicrobial peptides to high molecular weight multimeric structures as in the case of the pore-forming toxins. Irrespective of their sizes, small peptides like cyclotides can possess a similar pore-forming activity as the high molecular weight pore-forming toxins. Interestingly, several types of toxic proteins co-exist within the same plant species and can act together. This defense network of plants gets solidified not only by different plant toxic proteins that reinforce each other but also with low molecular weight chemicals such as terpenoids, phenolics, and alkaloids. Additionally, there is also a large group of non-toxic proteins such as hydrolytic enzymes, chitinases, defensins that are produced for defense purposes, and can support the action of those proteins with intrinsic toxicity. Although some toxic proteins clearly differ in their molecular structure and three-dimensional conformation they resemble each other in their mode of action. For instance, thionins, cyclotides and pore-forming toxins are active at the level of the membrane and both rely on the disruption of the membrane to execute their toxic activity.
Research of toxic plant proteins has attracted much interest mainly because of the potential applications in agriculture and medicine. Although a lot of progress has been made in our understanding of the biological activity of toxic proteins there are still a few important questions that remain. First, what is the distribution of toxic proteins? The availability of more completed sequences for different plant genomes will help to solve this question. Second, what is the physiological role of toxic proteins for plants? Although it is believed that toxic proteins are critical for plant defense, more efforts are required to understand the whole story. Finally, how could we benefit from our knowledge on toxic proteins and make use of these toxic proteins? A better understanding of the mode of action for each group of toxic proteins will facilitate manipulating and improving the use of these compounds, as part of a better strategy for the applications of plant toxic proteins. With respect to the use of toxic plant proteins for biomedical applications such as the use of proteins as therapeutic drugs some issues still remain and will require major consideration. But efforts are under way to improve the proteins and the available technologies to deliver them to specific locations in the organism or the cells.
Acknowledgements
This work was funded primarily by Ghent University and the Fund for Scientific Research-Flanders. Liuyi Dang is a recipient of a scholarship from the China Scholarship Council and receives doctoral co-funding from the Special Research Council of Ghent University.
Biographies

Prof. Els J.M. Van Damme graduated in Plant Sciences in 1986 at the Catholic University of Leuven (Belgium), and obtained her PhD in Plant Sciences in 1991 at the same university. From 1992 to 2001 she was working as a Postdoctoral Fellow of the Fund for Scientific Research – Flanders. She started her research with the isolation and characterization of novel interesting plant lectins (1986–1990). Afterwards her interest was focused on the molecular biology of plant lectins and the exploitation of the defense properties of lectins in transgenic plants. In 2002 she has been appointed as a Research Professor at Ghent University. Currently she is affiliated with the Department of Molecular Biotechnology at Faculty of Bioscience Engineering as a Senior Full Professor. The research is situated in the field of plant biochemistry, molecular biotechnology and glycobiology, and concentrates primarily on the study of plant proteins with specific biological activities. At present her interest focuses at unravelling the physiological role of different carbohydrate-binding proteins in plants. The ultimate goal of the research is to exploit the possibility of using these plant proteins to increase the stress resistance of the plant.

Liuyi Dang obtained his B.Sc. degree in Life Science and Biotechnology from Northwest University, China in 2009. At the same university, he finished his M.Sc study in Biochemistry and Molecular Biology, working on Glycan-binding proteins in Hepatocellular Carcinoma. He is currently a PhD student in the Department of Molecular Biotechnology at the Faculty of Bioscience Engineering, Ghent University, Belgium. The research focuses on the biological role of two groups of lectins in Cucumis sativus, in particular proteins with an Amaranthin domain and ribosome-inactivating proteins.
Contributor Information
Liuyi Dang, Email: Liuyi.Dang@UGent.be.
Els J.M. Van Damme, Email: Elsjm.VanDamme@UGent.be.
References
- Abdeen A., Virgos A., Olivella E., Villanueva J., Aviles X., Gabarra R., Prat S. Multiple insect resistance in transgenic tomato plants over-expressing two families of plant proteinase inhibitors. Plant Mol. Biol. 2005;57:189–202. doi: 10.1007/s11103-004-6959-9. [DOI] [PubMed] [Google Scholar]
- Acosta-Gallegos J.A., Quintero C., Vargas J., Toro O., Tohme J., Cardona C. A new variant of arcelin in wild common bean, Phaseolus vulgaris L., from southern Mexico. Genet. Resour. Crop Evol. 1998;45:235–242. [Google Scholar]
- Ahmad P., Ashraf M., Younis M., Hu X., Kumar A., Akram N.A., Al-Qurainy F. Role of transgenic plants in agriculture and biopharming. Biotechnol. Adv. 2012;30:524–540. doi: 10.1016/j.biotechadv.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Alves E.W., Ferreira A.T., Ferreira C.T., Carlini C.R. Effects of canatoxin on the Ca2+-ATPase of sarcoplasmic reticulum membranes. Toxicon. 1992;30:1411–1418. doi: 10.1016/0041-0101(92)90516-8. [DOI] [PubMed] [Google Scholar]
- Asano T., Miwa A., Maeda K., Kimura M., Nishiuchi T. The secreted antifungal protein thionin 2.4 in Arabidopsis thaliana suppresses the toxicity of a fungal fruit body lectin from Fusarium graminearum. PLoS Pathog. 2013;9:e1003581. doi: 10.1371/journal.ppat.1003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian A., Ponnuraj K. Crystal structure of the first plant urease from jack bean: 83 years of journey from its first crystal to molecular structure. J. Mol. Biol. 2010;400:274–283. doi: 10.1016/j.jmb.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Balasubramanian A., Durairajpandian V., Elumalai S., Mathivanan N., Munirajan A.K., Ponnuraj K. Structural and functional studies on urease from pigeon pea (Cajanus cajan) Int. J. Biol. Macromol. 2013;58:301–309. doi: 10.1016/j.ijbiomac.2013.04.055. [DOI] [PubMed] [Google Scholar]
- Barbeta B.L., Marshall A.T., Gillon A.D., Craik D.J., Anderson M.A. Plant cyclotides disrupt epithelial cells in the midgut of lepidopteran larvae. Proc. Natl. Acad. Sci. USA. 2008;105:1221–1225. doi: 10.1073/pnas.0710338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri L., Polito L., Bolognesi A., Ciani M., Pelosi E., Farini V., Jha A.K., Sharma N., Vivanco J.M., Chambery A., Parente A., Stirpe F. Ribosome-inactivating proteins in edible plants and purification and characterization of a new ribosome-inactivating protein from Cucurbita moschata. Biochim. Biophys. Acta. 2006;1760:783–792. doi: 10.1016/j.bbagen.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Barja-Fidalgo C., Guimarães J.A., Carlini C.R. Lipoxygenase-mediated secretory effect of canatoxin the toxic protein from Canavalia ensiformis seeds. Toxicon. 1991;29:453–459. doi: 10.1016/0041-0101(91)90019-n. [DOI] [PubMed] [Google Scholar]
- Barrett M.L., Udani J.K. A proprietary alpha-amylase inhibitor from white bean (Phaseolus vulgaris): a review of clinical studies on weight loss and glycemic control. Nutr. J. 2011;10:24. doi: 10.1186/1475-2891-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros P.R., Stassen H., Freitas M.S., Carlini C.R., Nascimento M.A., Follmer C. Membrane-disruptive properties of the bioinsecticide Jaburetox-2Ec: implications to the mechanism of the action of insecticidal peptides derived from ureases. Biochim. Biophys. Acta. 2009;1794:1848–1854. doi: 10.1016/j.bbapap.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Bass H.W., Krawetz J.E., OBrian G.R., Zinselmeier C., Habben J.E., Boston R.S. Maize ribosome-inactivating proteins (RIPs) with distinct expression patterns have similar requirements for proenzyme activation. J. Exp. Bot. 2004;55:2219–2233. doi: 10.1093/jxb/erh243. [DOI] [PubMed] [Google Scholar]
- Becker N., Benhar I. Antibody-based immunotoxins for the treatment of cancer. Antibodies. 2012;1:39–69. [Google Scholar]
- Berrocal-Lobo M., Molina A., Rodríguez-Palenzuela P., García-Olmedo F., Rivas L. Leishmania donovani: thionins, plant antimicrobial peptides with leishmanicidal activity. Exp. Parasitol. 2009;122:247–249. doi: 10.1016/j.exppara.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Bies C., Lehr C.M., Woodley J.F. Lectin-mediated drug targeting: history and applications. Adv. Drug Delivery Rev. 2004;56:425–435. doi: 10.1016/j.addr.2003.10.030. [DOI] [PubMed] [Google Scholar]
- Birk Y. Protein proteinase inhibitors in legume seeds–overview. Arch. Latinoam. Nutr. 1996;44:26S–30S. [PubMed] [Google Scholar]
- Bischofberger M., Iacovache I., Gisou van der Goot F. Pathogenic pore-forming proteins: function and host response. Cell Host Microbe. 2012;12:266–275. doi: 10.1016/j.chom.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Bittencourt S.E., Silva L.P., Azevedo R.B., Cunha R.B., Lima C.M., Ricart C.A., Sousa M.V. The plant cytolytic protein enterolobin assumes a dimeric structure in solution. FEBS Lett. 2003;549:47–51. doi: 10.1016/s0014-5793(03)00763-4. [DOI] [PubMed] [Google Scholar]
- Blair M.W., Munoz C., Buendia H.F., Flower J., Bueno J.M., Cardona C. Genetic mapping of microsatellite markers around the arcelin bruchid resistance locus in common bean. Theor. Appl. Genet. 2010;121:393–402. doi: 10.1007/s00122-010-1318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann H. The role of thionins in the resistance of plants. In: Datta S.K., Muthukrishnan S., editors. Pathogenesis-related proteins in plants. CRC Press Inc.; Florida: 1999. pp. 207–234. [Google Scholar]
- Bohlmann H., Apel K. Thionins. Annu. Rev. Plant Biol. 1991;42:227–240. [Google Scholar]
- Bolognesi A., Polito L., Scicchitano V., Orrico C., Pasquinelli G., Musiani S., Santi S., Riccio M., Bortolotti M., Battelli M.G. Endocytosis and intracellular localisation of type-1 ribosome-inactivating protein saporin-s6. J. Biol. Regul. Homeost. Agent. 2012;26:97–109. [PubMed] [Google Scholar]
- Bourne Y., Ayouba A., Rougé P., Cambillau C. Interaction of a legume lectin with two components of the bacterial cell wall. A crystallographic study. J. Biol. Chem. 1994;269:9429–9435. [PubMed] [Google Scholar]
- Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Cardona C., Kornegay J., Posso C.E., Morales F., Ramirez H. Comparative value of four arcelin variants in the development of dry bean lines resistant to the Mexican bean weevil. Entomol. Exp. Appl. 1990;56:197–206. [Google Scholar]
- Carlini C.R., Grossi-de-Sá M.F. Plant toxic proteins with insecticidal properties. A review on their potentialities as bioinsecticides. Toxicon. 2002;40:1515–1539. doi: 10.1016/s0041-0101(02)00240-4. [DOI] [PubMed] [Google Scholar]
- Carlini C.R., Guimarães J.A. Isolation and characterization of a toxic protein from Canavalia ensiformis (jack bean) seeds, distinct from concanavalin A. Toxicon. 1981;19:667–675. doi: 10.1016/0041-0101(81)90104-5. [DOI] [PubMed] [Google Scholar]
- Carlini C.R., Guimarães J.A. Plant and microbial toxic proteins as hemilectins: emphasis on canatoxin. Toxicon. 1991;29:791–806. doi: 10.1016/0041-0101(91)90216-e. [DOI] [PubMed] [Google Scholar]
- Carlini C.R., Polacco J.C. Toxic properties of urease. Crop Sci. 2008;48:1665–1672. [Google Scholar]
- Carlini C.R., Gomes C., Guimaraes J.A., Markus R.P., Sato H., Trolin G. Central nervous effects of the convulsant protein canatoxin. Acta Pharmacol. Toxicol. 1984;54:161–166. doi: 10.1111/j.1600-0773.1984.tb01912.x. [DOI] [PubMed] [Google Scholar]
- Carlini C.R., Oliveira A.E., Azambuja P., Xavier-Filho J., Wells M.A. Biological effects of canatoxin in different insect models: evidence for a proteolytic activation of the toxin by insect cathepsin like enzymes. J. Econ. Entomol. 1997;90:340–348. doi: 10.1093/jee/90.2.340. [DOI] [PubMed] [Google Scholar]
- Carrasco l., Vazquez D., Hernández-lucas C., Carbonero P., García-olmedo F. Thionins: plant peptides that modify membrane permeability in cultured mammalian cells. Eur. J. Biochem. 1981;116:185–189. doi: 10.1111/j.1432-1033.1981.tb05317.x. [DOI] [PubMed] [Google Scholar]
- Castro Faria Neto H.C., Cordeiro R.S.B., Martins M.A., Correia da Silva A.C.V., Bozza P.T., Sousa M.V., Morhy L. Enterolobin induces rat paw oedema independently of PAF-acether. Mem. Inst. Oswaldo Cruz. 1991;86:129–131. doi: 10.1590/s0074-02761991000600029. [DOI] [PubMed] [Google Scholar]
- Chan W.L., Shaw P.C., Tam S.C., Jacobsen C., Gliemann J., Nielsen M.S. Trichosanthin interacts with and enters cells via LDL receptor family members. Biochem. Biophys. Res. Commun. 2000;270:453–457. doi: 10.1006/bbrc.2000.2441. [DOI] [PubMed] [Google Scholar]
- Chaudhry B., Müller-Uri F., Cameron-Mills V., Gough S., Simpson D., Skriver K., Mundy J. The barley 60 kDa jasmonate-induced protein (JIP60) is a novel ribosome-inactivating protein. Plant J. 1994;6:815–824. doi: 10.1046/j.1365-313x.1994.6060815.x. [DOI] [PubMed] [Google Scholar]
- Checker V.G., Chhibbar A.K., Khurana P. Stress-inducible expression of barley Hva1 gene in transgenic mulberry displays enhanced tolerance against drought, salinity and cold stress. Transgenic Res. 2012;21:939–957. doi: 10.1007/s11248-011-9577-8. [DOI] [PubMed] [Google Scholar]
- Christeller J.T., Farley P.C., Ramsay R.J., Sullivan P.A., Laing W.A. Purification, characterization and cloning of an aspartic proteinase inhibitor from squash phloem exudate. Eur. J. Biochem. 1998;254:160–167. doi: 10.1046/j.1432-1327.1998.2540160.x. [DOI] [PubMed] [Google Scholar]
- Colgrave M.L., Kotze A.C., Huang Y.H., O’Grady J., Simonsen S.M., Craik D.J. Cyclotides: natural, circular plant peptides that possess significant activity against gastrointestinal nematode parasites of sheep. Biochemistry. 2008;47:5581–5589. doi: 10.1021/bi800223y. [DOI] [PubMed] [Google Scholar]
- Corrado G., Bovi P.D., Ciliento R., Gaudio L., Di Maro A., Aceto S., Lorito M., Rao R. Inducible expression of a Phytolacca heterotepala ribosome-inactivating protein leads to enhanced resistance against major fungal pathogens in tobacco. Phytopathology. 2005;95:206–215. doi: 10.1094/PHYTO-95-0206. [DOI] [PubMed] [Google Scholar]
- Craik D.J. Discovery and applications of the plant cyclotides. Toxicon. 2010;56:1092–1102. doi: 10.1016/j.toxicon.2010.02.021. [DOI] [PubMed] [Google Scholar]
- Craik D.J., Daly N.L., Bond T., Waine C. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cysteine knot structural motif. J. Mol. Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- Craik D.J., Swedberg J.E., Mylne J.S., Cemazar M. Cyclotides as a basis for drug design. Expert Opin. Drug Discov. 2012;7:179–194. doi: 10.1517/17460441.2012.661554. [DOI] [PubMed] [Google Scholar]
- Cushnie T.T., Cushnie B., Lamb A.J. Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int. J. Antimicrob. Agents. 2014;44:377–386. doi: 10.1016/j.ijantimicag.2014.06.001. [DOI] [PubMed] [Google Scholar]
- De Leo F., Volpicella M., Licciulli F., Liuni S., Gallerani R., Ceci L.R. PLANT-PIs: a database for plant protease inhibitors and their genes. Nucleic Acids Res. 2002;30:347–348. doi: 10.1093/nar/30.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degiacomi M.T., Iacovache I., Pernot L., Chami M., Kudryashev M., Stahlberg H., van der Goot F.G., Dal Peraro M. Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism. Nat. Chem. Biol. 2013;9:623–629. doi: 10.1038/nchembio.1312. [DOI] [PubMed] [Google Scholar]
- del Carmen Fernández-Alonso M., Díaz D., Berbis M.Á., Marcelo F., Cañada J., Jiménez-Barbero J. Protein-carbohydrate interactions studied by NMR: from molecular recognition to drug design. Curr. Protein Pept. Sci. 2012;13:816–830. doi: 10.2174/138920312804871175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S.C., da Silva M.C.M., Teixeira F.R., Figueira E.L.Z., de Oliveira-Neto O.B., de Lima L.A., Grossi-de-Sa M.F. Investigation of insecticidal activity of rye α-amylase inhibitor gene expressed in transgenic tobacco (Nicotiana tabacum) toward cotton boll weevil (Anthonomus grandis) Pestic. Biochem. Physiol. 2010;98:39–44. [Google Scholar]
- Dowd P.F., Johnson E.T., Price N.P. Enhanced pest resistance of maize leaves expressing monocot crop plant-derived ribosome-inactivating protein and agglutinin. J. Agric. Food Chem. 2012;60:10768–10775. doi: 10.1021/jf3041337. [DOI] [PubMed] [Google Scholar]
- Duan X., Li X., Xue Q., Abo-El-Saad M., Xu D., Wu R. Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat. Biotechnol. 1996;14:494–498. doi: 10.1038/nbt0496-494. [DOI] [PubMed] [Google Scholar]
- Dunse K.M., Stevens J.A., Lay F.T., Gaspar Y.M., Heath R.L., Anderson M.A. Coexpression of potato type I and II proteinase inhibitors gives cotton plants protection against insect damage in the field. Proc. Natl. Acad. Sci. USA. 2010;107:15011–15015. doi: 10.1073/pnas.1009241107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunse K.M., Kaas Q., Guarino R.F., Barton P.A., Craik D.J., Anderson M.A. Molecular basis for the resistance of an insect chymotrypsin to a potato type II proteinase inhibitor. Proc. Natl. Acad. Sci. USA. 2010;107:15016–15021. doi: 10.1073/pnas.1009327107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duverger E., Frison N., Roche A.C., Monsigny M. Carbohydrate-lectin interactions assessed by surface plasmon resonance. Biochimie. 2003;85:167–179. doi: 10.1016/s0300-9084(03)00060-9. [DOI] [PubMed] [Google Scholar]
- Duvic M., Talpur R. Optimizing denileukin diftitox (Ontak®) therapy. Future Oncol. 2008;2008(4):457–469. doi: 10.2217/14796694.4.4.457. [DOI] [PubMed] [Google Scholar]
- Ferreira-DaSilva C.T., Gombarovits M.E., Masuda H., Oliveira C.M., Carlini C.R. Proteolytic activation of canatoxin, a plant toxic protein, by insect cathepsin-like enzymes. Arch. Insect Biochem. Physiol. 2000;44:162–171. doi: 10.1002/1520-6327(200008)44:4<162::AID-ARCH3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Fitches E., Woodhouse S.D., Edwards J.P., Gatehouse J.A. In vitro and in vivo binding of snowdrop (Galanthus nivalis agglutinin; GNA) and jackbean (Canavalia ensiformis; Con A) lectins within tomato moth (Lacanobia oleracea) larvae: mechanisms of insecticidal action. J. Insect Physiol. 2001;47:777–787. doi: 10.1016/s0022-1910(01)00068-3. [DOI] [PubMed] [Google Scholar]
- Florack D.E.A., Stiekema W.J. Thionins: properties, possible biological roles and mechanisms of action. Plant Mol. Biol. 1994;26:25–37. doi: 10.1007/BF00039517. [DOI] [PubMed] [Google Scholar]
- Follmer C., Barcellos G.B., Zingali R.B., Machado O.L., Alves E.W., Barja-Fidalgo C., Guimaraes J.A., Carlini C.R. Canatoxin, a toxic protein from jack beans (Canavalia ensiformis), is a variant form of urease (EC 3.5.1.5): biological effects of urease independent of its ureolytic activity. Biochem. J. 2001;15:217–224. doi: 10.1042/0264-6021:3600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes W., Sousa M.V., Aragão J.B., Morhy L. Determination of the amino acid sequence of the plant cytolysin enterolobin. Arch. Biochem. Biophys. 1997;347:201–207. doi: 10.1006/abbi.1997.0358. [DOI] [PubMed] [Google Scholar]
- Fracasso G., Stirpe F., Colombatti M. Ribosome-inactivating protein-containing conjugates for therapeutic use. In: Lord J.M., Hartley M.R., editors. Toxic Plant Proteins. Springer; Berlin Heidelberg: 2010. pp. 225–263. [Google Scholar]
- Franco O.L., Rigden D.J., Melo F.R., Grossi-de-Sá M.F. Plant α-amylase inhibitors and their interaction with insect α-amylases. Eur. J. Biochem. 2002;269:397–412. doi: 10.1046/j.0014-2956.2001.02656.x. [DOI] [PubMed] [Google Scholar]
- François K.O., Balzarini J. Potential of carbohydrate-binding agents as therapeutics against enveloped viruses. Med. Res. Rev. 2012;32:349–387. doi: 10.1002/med.20216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazarian H., Idoni B., Oppenheimer S.B. A glycobiology review: carbohydrates, lectins and implications in cancer therapeutics. Acta Histochem. 2011;113:236–247. doi: 10.1016/j.acthis.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilabert-Oriol R., Weng A., von Mallinckrodt B., Melzig M.F., Fuchs H., Thakur M. Immunotoxins constructed with ribosome-inactivating proteins and their enhancers: a lethal cocktail with tumor specific efficacy. Curr. Pharm. Des. 2014;20:6584–6643. doi: 10.2174/1381612820666140826153913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert R.J.C. Pore-forming toxins. Cell. Mol. Life Sci. 2002;59:832–844. doi: 10.1007/s00018-002-8471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanini M.P., Saltzmann K.D., Puthoff D.P., Gonzalo M., Ohm H.W., Williams C.E. A novel wheat gene encoding a putative chitin-binding lectin is associated with resistance against Hessian fly. Mol. Plant Pathol. 2007;8:69–82. doi: 10.1111/j.1364-3703.2006.00371.x. [DOI] [PubMed] [Google Scholar]
- Goossens A., Quintero C., Dillen W., De Rycke R., Valor J.F., De Clercq J., Van Montagu M., Cardona C., Angenon G. Analysis of bruchid resistance in the wild common bean accession G02771: no evidence for insecticidal activity of arcelin 5. J. Exp. Bot. 2000;51:1229–1236. [PubMed] [Google Scholar]
- Gould A., Ji Y., Aboye T.L., Camarero J.A. Cyclotides, a novel ultrastable polypeptide scaffold for drug discovery. Curr. Pharm. Des. 2011;17:4294–4307. doi: 10.2174/138161211798999438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal R.K., Mattoo A.K. Multitasking antimicrobial peptides in plant development and host defense against biotic/abiotic stress. Plant Sci. 2014;228:135–149. doi: 10.1016/j.plantsci.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Gruber C.W., Anderson M.A., Craik D.J. Insecticidal plant cyclotides and related cystine knot toxins. Toxicon. 2007;49:561–575. doi: 10.1016/j.toxicon.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Gruber C.W., Elliott A.G., Ireland D.C., Delprete P.G., Dessein S., Göransson U., Trabi M., Wang C.K., Kinghorn A.B., Robbrecht E., Craik D.J. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20:2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq S.K., Atif S.M., Khan R.H. Protein proteinase inhibitor genes in combat against insects, pests, and pathogens: natural and engineered phytoprotection. Arch. Biochem. Biophys. 2004;431:145–159. doi: 10.1016/j.abb.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J., Yamada M., Kuno A., Tateno H. Lectin microarrays: concept, principle and applications. Chem. Soc. Rev. 2013;42:4443–4458. doi: 10.1039/c3cs35419a. [DOI] [PubMed] [Google Scholar]
- Huang Y.H., Colgrave M.L., Daly N.L., Keleshian A., Martinac B., Craik D.J. The biological activity of the prototypic cyclotide kalata b1 is modulated by the formation of multimeric pores. J. Biol. Chem. 2009;284:20699–20707. doi: 10.1074/jbc.M109.003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes P., Dennis E., Whitecross M., Llewellyn D., Gage P. The cytotoxic plant protein, β-purothionin, forms ion channels in lipid membranes. J. Biol. Chem. 2000;275:823–827. doi: 10.1074/jbc.275.2.823. [DOI] [PubMed] [Google Scholar]
- Iacovache I., Bischofberger M., van der Goot F.G. Structure and assembly of pore-forming proteins. Curr. Opin. Struct. Biol. 2010;20:241–246. doi: 10.1016/j.sbi.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Iglesias R., Perez Y., de Torre C., Ferreras J.M., Antolin P., Jiminez P., Rojo M.A., Mendez E., Girbes T. Molecular characterization and systemic induction of single-chain ribosome-inactivating proteins (RIPs) in sugar beet (Beta vulgaris) leaves. J. Exp. Bot. 2005;56:1675–1684. doi: 10.1093/jxb/eri164. [DOI] [PubMed] [Google Scholar]
- Ippoliti R., Lendaro E., Benedetti P.A., Torrisi M.R., Belleudi F., Carpani D., Soria M.R., Fabbrini M.S. Endocytosis of a chimera between human pro-urokinase and the plant toxin saporin: an unusual internalization mechanism. FASEB J. 2000;14:1335–1344. doi: 10.1096/fj.14.10.1335. [DOI] [PubMed] [Google Scholar]
- Ireland D.C., Wang C.K., Wilson J.A., Gustafson K.R., Craik D.J. Cyclotides as natural anti-HIV agents. Biopolymers. 2008;90:51–60. doi: 10.1002/bip.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S.Y., Ramamoorthy R., Bhalla R., Luan H.F., Venkatesh P.N., Cai M., Ramachandran S. Genome-wide survey of the RIP domain family in Oryza sativa and their expression profiles under various abiotic and biotic stresses. Plant Mol. Biol. 2008;67:603–614. doi: 10.1007/s11103-008-9342-4. [DOI] [PubMed] [Google Scholar]
- Jiang S.Y., Bhalla R., Ramamoorthy R., Luan H.F., Venkatesh P.N., Cai M., Ramachandran S. Over-expression of OSRIP18 increases drought and salt tolerance in transgenic rice plants. Transgenic Res. 2012;21:785–795. doi: 10.1007/s11248-011-9568-9. [DOI] [PubMed] [Google Scholar]
- Jiménez P., Gayoso M.J., Girbés T. Non-toxic type-2 ribosome-inactivating proteins. In: Stirpe F., Lappi D., editors. Ribosome-inactivating proteins: ricin and related proteins. Willy Blackwell; Oxford: 2014. pp. 67–82. [Google Scholar]
- Karran R.A., Hudak K.A. Depurination of brome mosaic virus RNA3 inhibits its packaging into virus particles. Nucleic Acids Res. 2011;39:7209–7222. doi: 10.1093/nar/gkr383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrlik J., Švitel J., Gemeiner P., Kožár T., Tkac J. Glycan and lectin microarrays for glycomics and medicinal applications. Med. Res. Rev. 2010;30:394–418. doi: 10.1002/med.20195. [DOI] [PubMed] [Google Scholar]
- Kaur I., Gupta R.C., Puri M. Ribosome inactivating proteins from plants inhibiting viruses. Virol. Sin. 2011;26:357–365. doi: 10.1007/s12250-011-3223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Park S.C., Hwang I., Cheong H., Nah J.W., Hahm K.S., Park Y. Protease inhibitors from plants with antimicrobial activity. Int. J. Mol. Sci. 2009;10:2860–2872. doi: 10.3390/ijms10062860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer K.J., Klassen L.W., Jones B.L., Speirs R.D., Kammer A.E. Toxicity of purothionin and its homologues to the tobacco hornworm, Manduca sexta (L.) (lepidoptera: sphingidae) Toxicol. Appl. Pharmacol. 1979;48:179–183. doi: 10.1016/s0041-008x(79)80020-4. [DOI] [PubMed] [Google Scholar]
- Kreitman R.J. Immunotoxins for targeted cancer therapy. AAPS J. 2006;8:E532–E551. doi: 10.1208/aapsj080363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannoo N., Van Damme E.J.M. Nucleocytoplasmic plant lectins. Biochim. Biophys. Acta. 2010;1800:190–201. doi: 10.1016/j.bbagen.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Lannoo N., Van Damme E.J.M. Lectin domains at the frontiers of plant defense. Front. Plant Sci. 2014;5:397. doi: 10.3389/fpls.2014.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M., Jr, Kato I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- Lawrence P.K., Koundal K.R. Plant protease inhibitors in control of phytophagous insects. Electron. J. Biotechnol. 2002;5:5–6. [Google Scholar]
- Lehr C.M. Lectin-mediated drug delivery: the second generation of bioadhesives. J. Controlled Release. 2000;65:19–29. doi: 10.1016/s0168-3659(99)00228-x. [DOI] [PubMed] [Google Scholar]
- Leippe M. Pore-forming toxins from pathogenic amoebae. Appl. Microbiol. Biotechnol. 2014;98:4347–4353. doi: 10.1007/s00253-014-5673-z. [DOI] [PubMed] [Google Scholar]
- Leung D., Abbenante G., Fairlie D.P. Protease inhibitors: current status and future prospects. J. Med. Chem. 2000;43:305–341. doi: 10.1021/jm990412m. [DOI] [PubMed] [Google Scholar]
- Liang P.H., Wang S.K., Wong C.H. Quantitative analysis of carbohydrate-protein interactions using glycan microarrays: determination of surface and solution dissociation constants. J. Am. Chem. Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
- Lippman S.M., Matrisian L.M. Protease inhibitors in oral carcinogenesis and chemoprevention. Clin. Cancer Res. 2000;6:4599–4603. [PubMed] [Google Scholar]
- Lis H., Sharon N. Lectins: carbohydrate-specific proteins that mediate cellular recognition. Chem. Rev. 1998;98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- Liu B., Cheng Y., Zhang B., Bian H.J., Bao J.K. Polygonatum cyrtonema lectin induces apoptosis and autophagy in human melanoma A375 cells through a mitochondria-mediated ROS–p38–p53 pathway. Cancer Lett. 2009;275:54–60. doi: 10.1016/j.canlet.2008.09.042. [DOI] [PubMed] [Google Scholar]
- Liu B., Bian H.J., Bao J.K. Plant lectins: potential antineoplastic drugs from bench to clinic. Cancer Lett. 2010;287:1–12. doi: 10.1016/j.canlet.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Liu S.M., Li J., Zhu J.Q., Wang X.W., Wang C.S., Liu S.S., Chen X.X., Li S. Transgenic plants expressing the AaIT/GNA fusion protein show increased resistance and toxicity to both chewing and sucking pests. Insect Sci. 2015 doi: 10.1111/1744-7917.12203. in press. [DOI] [PubMed] [Google Scholar]
- Lombardi A., Bursomanno S., Lopardo T., Traini R., Colombatti M., Ippoliti R., Flavell D.J., Flavell S.U., Ceriotti A., Fabbrini M.S. Pichia pastoris as a host for secretion of toxic saporin chimeras. FASEB J. 2010;24:253–265. doi: 10.1096/fj.08-118042. [DOI] [PubMed] [Google Scholar]
- Lorberboum-Galski H. Human toxin-based recombinant immunotoxins/chimeric proteins as a drug delivery system for targeted treatment of human diseases. Exp. Opin. Drug Delivery. 2011;8:605–621. doi: 10.1517/17425247.2011.566269. [DOI] [PubMed] [Google Scholar]
- Lord J.M. Precursors of ricin and Ricinus communis agglutinin. Glycosylation and processing during synthesis and intracellular transport. Eur. J. Biochem. 1985;146:411–416. doi: 10.1111/j.1432-1033.1985.tb08667.x. [DOI] [PubMed] [Google Scholar]
- Lord J.M., Harley S.M. Ricinus communis agglutinin B chain contains a fucosylated oligosaccharide side chain not present on ricin B chain. FEBS Lett. 1985;189:72–76. [Google Scholar]
- Lord J.M., Spooner R.A. Ricin trafficking in plant and mammalian cells. Toxins. 2011;3:787–801. doi: 10.3390/toxins3070787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los F.C., Randis T.M., Aroian R.V., Ratner A.J. Role of pore-forming toxins in bacterial infectious diseases. Microbiol. Mol. Biol. Rev. 2013;77:173–207. doi: 10.1128/MMBR.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu S.Y., Choi S.H., Park W.B. Korean mistletoe lectin-induced apoptosis in hepatocarcinoma cells is associated with inhibition of telomerase via mitochondrial controlled pathway independent of p53. Arch. Pharmacal Res. 2002;25:93–101. doi: 10.1007/BF02975269. [DOI] [PubMed] [Google Scholar]
- Maag D., Erb M., Köllner T.G., Gershenzon J. Defensive weapons and defense signals in plants: Some metabolites serve both roles. BioEssays. 2014;37:167–174. doi: 10.1002/bies.201400124. [DOI] [PubMed] [Google Scholar]
- Macedo M.L., de Oliveira C.F., Costa P.M., Castelhano E.C., Silva-Filho M.C. Adaptive mechanisms of insect pests against plant protease inhibitors and future prospects related to crop protection: a review. Protein Pept. Lett. 2015;22:149–163. doi: 10.2174/0929866521666141020111631. [DOI] [PubMed] [Google Scholar]
- Mareš M., Meloun B., Pavlik M., Kostka V., Baudyš M. Primary structure of cathepsin D inhibitor from potatoes and its structure relationship to soybean trypsin inhibitor family. FEBS Lett. 1989;251:94–98. doi: 10.1016/0014-5793(89)81435-8. [DOI] [PubMed] [Google Scholar]
- Marshall R.S., D’Avila F., Di Cola A., Traini R., Spanò L., Fabbrini M.S., Ceriotti A. Signal peptide regulated toxicity of a plant ribosome-inactivating protein during cell stress. Plant J. 2011;65:218–229. doi: 10.1111/j.1365-313X.2010.04413.x. [DOI] [PubMed] [Google Scholar]
- Martinelli A.H., Kappaun K., Ligabue-Braun R., Defferrari M.S., Piovesan A.R., Stanisçuaski F., Demartini D.R., Dal Belo C.A., Almeida C.G.M., Follmer C., Verli H., Carlini C.R., Pasquali G. Structure-function studies on jaburetox, a recombinant insecticidal peptide derived from jack bean (Canavalia ensiformis) urease. Biochim. Biophys. Acta. 2014;1840:935–944. doi: 10.1016/j.bbagen.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Meng Y., Liu S., Li J., Meng Y., Zhao X. Preparation of an antitumor and antivirus agent: chemical modification of α-MMC and MAP30 from Momordica charantia L. with covalent conjugation of polyethylene glycol. Int. J. Nanomed. 2012;7:3133–3142. doi: 10.2147/IJN.S30631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels K., Van Damme E.J.M., Smagghe G. Plant-insect interactions: what can we learn from plant lectins? Arch. Insect Biochem. Physiol. 2010;73:193–212. doi: 10.1002/arch.20351. [DOI] [PubMed] [Google Scholar]
- Mikkat U., Damm I., Kirchhoff F., Albrecht E., Nebe B., Jonas L. Effects of lectins on CCK-8-stimulated enzyme secretion and differentiation of the rat pancreatic cell line AR42J. Pancreas. 2001;23:368–374. doi: 10.1097/00006676-200111000-00006. [DOI] [PubMed] [Google Scholar]
- Minney B.H.P., Gatehouse A.M.R., Dobie P., Dendy J., Cardona C., Gatehouse J.A. Biochemical bases of seed resistance to Zabrotes subfasciatus (Bean weevil) in Phaseolus vulgaris (common bean): a mechanism for arcelin toxicity. J. Insect Physiol. 1990;36:757–767. [Google Scholar]
- Mithöfer A., Boland W. Plant defense against herbivores: chemical aspects. Annu. Rev. Plant Biol. 2012;63:431–450. doi: 10.1146/annurev-arplant-042110-103854. [DOI] [PubMed] [Google Scholar]
- Miyake K., Tanaka T., McNeil P.L. Lectin-based food poisoning: a new mechanism of protein toxicity. PLoS ONE. 2007;2:e687. doi: 10.1371/journal.pone.0000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina A., Goy P.A., Fraile A., Sánchez-Monge R., García-Olmedo F. Inhibition of bacterial and fungal plant pathogens by thionins of types I and II. Plant Sci. 1993;92:169–177. [Google Scholar]
- Moon J., Salzman R.A., Ahn J.E., Koiwa H., Zhu-Salzman K. Transcriptional regulation in cowpea bruchid guts during adaptation to a plant defence protease inhibitor. Insect Mol. Biol. 2004;13:283–291. doi: 10.1111/j.0962-1075.2004.00485.x. [DOI] [PubMed] [Google Scholar]
- Mulinari F., Stanisçuaski F., Berthodo-Vargas L.R., Postal M., Oliveira-Neto O.B., Ridgen D.J., Grossi-de-Sá M.F., Carlini C.R. Jaburetox-2Ec: An insecticidal peptide derived from an isoform of urease form the Canavalia ensiformis plant. Peptides. 2007;28:2042–2050. doi: 10.1016/j.peptides.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Muramoto N., Tanaka T., Shimamura T., Mitsukawa N., Hori E., Koda K., Otani M., Hirai M., Nakamura K., Imaeda T. Transgenic sweet potato expressing thionin from barley gives resistance to black rot disease caused by Ceratocystis fimbriata in leaves and storage roots. Plant Cell Rep. 2012;31:987–997. doi: 10.1007/s00299-011-1217-5. [DOI] [PubMed] [Google Scholar]
- Murdock L.L., Shade R.E. Lectins and protease inhibitors as plant defenses against insects. J. Agric. Food Chem. 2002;50:6605–6611. doi: 10.1021/jf020192c. [DOI] [PubMed] [Google Scholar]
- Ng T.B., Chan W.Y., Yeung H.W. Proteins with abortifacient, ribosome inactivating, immunomodulatory, antitumor and anti-AIDS activities from Cucurbitaceae plants. Gen. Pharmacol. 1992;23:575–590. doi: 10.1016/0306-3623(92)90131-3. [DOI] [PubMed] [Google Scholar]
- Nguyen G.K.T., Lim W.H., Nguyen P.Q.T., Tam J.P. Novel cyclotides and uncyclotides with highly shortened precursors from Chassalia chartacea and effects of methionine oxidation on bioactivities. J. Biol. Chem. 2012;287:17598–17607. doi: 10.1074/jbc.M111.338970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oard S., Karki B., Enright F. Is there a difference in metal ion-based inhibition between members of thionin family: molecular dynamics simulation study. Biophys. Chem. 2007;130:65–75. doi: 10.1016/j.bpc.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Olsnes S. The history of ricin, abrin and related toxins. Toxicon. 2004;44:361–370. doi: 10.1016/j.toxicon.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Osborn T.C., Blake T., Gepts P., Bliss F.A. Bean arcelin 2. Genetic variation, inheritance and linkage relationships of a novel seed protein of Phaseolus vulgaris L. Theor. Appl. Genet. 1986;71:847–855. doi: 10.1007/BF00276428. [DOI] [PubMed] [Google Scholar]
- Osborn T.C., Alexander D.C., Sun S., Cardona C., Bliss F.A. Insecticidal activity and lectin homology of arcelin seed protein. Science. 1988;240:207–210. doi: 10.1126/science.240.4849.207. [DOI] [PubMed] [Google Scholar]
- Otto H.H., Schirmeister T. Cysteine proteases and their inhibitors. Chem. Rev. 1997;97:133–172. doi: 10.1021/cr950025u. [DOI] [PubMed] [Google Scholar]
- Paes N.S., Gerhardt I.R., Coutinho M.V., Yokoyama M., Santana E., Harris N., Chrispeels M.J., Grossi-de-Sa M.F. The effect of arcelin-1 on the structure of the midgut of bruchid larvae and immune localization of the arcelin protein. J. Insect Physiol. 2000;46:393–402. doi: 10.1016/s0022-1910(99)00122-5. [DOI] [PubMed] [Google Scholar]
- Park S.W., Lawrence C.B., Linden J.C., Vivanco J.M. Isolation and characterization of a novel ribosome-inactivating protein from root cultures of pokeweed and its mechanism of secretion from roots. Plant Physiol. 2002;130:164–178. doi: 10.1104/pp.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.W., Feil S.C. Pore-forming protein toxins: from structure to function. Prog. Biophys. Mol. Biol. 2005;88:91–142. doi: 10.1016/j.pbiomolbio.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Peumans W.J., Van Damme E.J.M. Lectins as plant defense proteins. Plant Physiol. 1995;109:347–352. doi: 10.1104/pp.109.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans W.J., Hao Q., Van Damme E.J.M. Ribosome-inactivating proteins from plants: more than RNA N-glycosidases? FASEB J. 2001;15:1493–1506. doi: 10.1096/fj.00-0751rev. [DOI] [PubMed] [Google Scholar]
- Pinto M.F., Fensterseifer I.C., Migliolo L., Sousa D.A., de Capdville G., Arboleda-Valencia J.W., Colgrave M.L., Craik D.J., Magalhães B.S., Dias S.C., Franco O.L. Identification and structural characterization of novel cyclotide with activity against an insect pest of sugar cane. J. Biol. Chem. 2012;287:134–147. doi: 10.1074/jbc.M111.294009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan A.R., Martinelli A.H., Ligabue-Braun R., Schwartz J.L., Carlini C.R. Canavalia ensiformis urease, Jaburetox and derived peptides form ion channels in planar lipid bilayers. Arch. Biochem. Biophys. 2014;547:6–17. doi: 10.1016/j.abb.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Plan M.R.R., Saska I., Cagauan A.G., Craik D.J. Backbone cyclised peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail) J. Agric. Food Chem. 2008;56:5237–5241. doi: 10.1021/jf800302f. [DOI] [PubMed] [Google Scholar]
- Plattner V.E., Wagner M., Ratzinger G., Gabor F., Wirth M. Targeted drug delivery: binding and uptake of plant lectins using human 5637 bladder cancer cells. Eur. J. Pharm. Biopharm. 2008;70:572–576. doi: 10.1016/j.ejpb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Polito L., Bortolotti M., Pedrazzi M., Bolognesi A. Immunotoxins and other conjugates containing saporin-S6 for cancer therapy. Toxins. 2011;3:697–720. doi: 10.3390/toxins3060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postal M., Martinelli A.H., Becker-Ritt A.B., Ligabue-Braun R., Demartini D.R., Ribeiro S.F., Pasquali G., Gomes V.M., Carlini C.R. Antifungal properties of Canavalia ensiformis urease and derived peptides. Peptides. 2012;38:22–32. doi: 10.1016/j.peptides.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Puri M., Kaur I., Perugini M.A., Gupta R.C. Ribosome-inactivating proteins: current status and biomedical applications. Drug Discov. Today. 2012;17:774–783. doi: 10.1016/j.drudis.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Puthoff D.P., Sardesai N., Subramanyam S., Nemacheck J.A., Williams C.E. Hfr-2, a wheat cytolytic toxin-like gene, is up-regulated by virulent Hessian fly larval feeding. Mol. Plant Pathol. 2005;6:411–423. doi: 10.1111/j.1364-3703.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- Qin W., Ming-Xing H., Ying X., Xin-Shen Z., Fang C. Expression of a ribosome inactivating protein (curcin 2) in Jatropha curcas is induced by stress. J. Biosci. 2005;30:351–357. doi: 10.1007/BF02703672. [DOI] [PubMed] [Google Scholar]
- Qin Y., Zhong Y., Dang L., Zhu M., Yu H., Chen W., Cui J., Bian H., Li Z. Alteration of protein glycosylation in human hepatic stellate cells activated with transforming growth factor-β1. J. Proteomics. 2012;75:4114–4123. doi: 10.1016/j.jprot.2012.05.040. [DOI] [PubMed] [Google Scholar]
- Qin Y., Zhong Y., Zhu M., Dang L., Yu H., Chen Z., Chen W., Wang X., Zhang H., Li Z. Age- and sex-associated differences in the glycopatterns of human salivary glycoproteins and their roles against influenza A virus. J. Proteome Res. 2013;12:2742–2754. doi: 10.1021/pr400096w. [DOI] [PubMed] [Google Scholar]
- Rajamohan F., Venkatachalam T.K., Irvin J.D., Uckun F.M. Pokeweed antiviral protein isoforms PAP-I, PAP-II, and PAP-III depurinate RNA of human immunodeficiency virus (HIV)-1. Biochem. Biophys. Res. Commun. 1999;260:453–458. doi: 10.1006/bbrc.1999.0922. [DOI] [PubMed] [Google Scholar]
- Ramarao N., Sanchis V. The pore-forming haemolysins of Bacillus cereus: a review. Toxins. 2013;5:1119–1139. doi: 10.3390/toxins5061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J.A., Kelly I., Marion D., Pézolet M., Auger M. Interaction between β-purothionin and dimyristoyl phosphatidylglycerol: a 31P-NMR and infrared spectroscopic study. Biophys. J. 2002;83:2074–2083. doi: 10.1016/S0006-3495(02)73968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustgi S., Pollmann S., Buhr F., Springer A., Reinbothe C., von Wettstein D., Reinbothe S. JIP60-mediated, jasmonate- and senescence-induced molecular switch in translation toward stress and defense protein synthesis. Proc. Natl. Acad. Sci. USA. 2014;111:14181–14186. doi: 10.1073/pnas.1415690111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C.A. Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu. Rev. Phytopathol. 1990;28:425–449. [Google Scholar]
- Salas C.E., Badillo-Corona J.A., Ramírez-Sotelo G., Oliver-Salvador C. Biologically active and antimicrobial peptides from plants. Biomed Res. Int. 2015 doi: 10.1155/2015/102129. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter U., Benchabane M., Munger A., Kiggundu A., Vorster J., Goulet M.C., Cloutier C., Michaud D. Recombinant protease inhibitors for herbivore pest control: a multitrophic perspective. J. Exp. Bot. 2010;61:4169–4183. doi: 10.1093/jxb/erq166. [DOI] [PubMed] [Google Scholar]
- Selitrennikoff C.P. Antifungal proteins. Appl. Environ. Microbiol. 2001;67:2883–2894. doi: 10.1128/AEM.67.7.2883-2894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthilkumar R., Cheng C.P., Yeh K.W. Genetically pyramiding protease-inhibitor genes for dual broad-spectrum resistance against insect and phytopathogens in transgenic tobacco. Plant Biotechnol. J. 2010;8:65–75. doi: 10.1111/j.1467-7652.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- Shah J., Veluthambi K. Dianthin, a negative selection marker in tobacco, is non-toxic in transgenic rice and confers sheath blight resistance. Biol. Plant. 2010;54:443–450. [Google Scholar]
- Shahidi-Noghabi S., Van Damme E.J.M., Smagghe G. Expression of Sambucus nigra agglutinin (SNA-I) from elderberry bark in transgenic tobacco plants results in enhanced resistance to different insect species. Transgenic Res. 2009;18:249–259. doi: 10.1007/s11248-008-9215-2. [DOI] [PubMed] [Google Scholar]
- Shang C., Peumans W.J., Van Damme E.J.M. Occurrence and taxonomical distribution of ribosome inactivating proteins belonging to the Ricin/Shiga toxin superfamily. In: Stirpe F., Lappi D., editors. Ribosome-inactivating proteins: ricin and related proteins. Willy Blackwell; Oxford: 2014. pp. 11–27. [Google Scholar]
- Simonsen S.M., Sando L., Rosengren K.J., Wang C.K., Colgrave M.L., Daly N.L., Craik D.J. Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J. Biol. Chem. 2008;283:9805–9813. doi: 10.1074/jbc.M709303200. [DOI] [PubMed] [Google Scholar]
- Smallshaw J.E., Ghetie V., Rizo J., Fulmer J.R., Trahan L.L., Ghetie M.A., Vitetta E.S. Genetic engineering of an immunotoxin to eliminate pulmonary vascular leak in mice. Nat. Biotechnol. 2003;21:387–391. doi: 10.1038/nbt800. [DOI] [PubMed] [Google Scholar]
- Smetana S., Khalef S., Bar-Khayim Y., Birk Y. Antimicrobial gentamicin activity in the presence of exogenous protease inhibitor (Bowman–Birk inhibitor) in gentamicin-induced nephrotoxicity in rats. Nephron. 1992;61:68–72. doi: 10.1159/000186837. [DOI] [PubMed] [Google Scholar]
- Song S.K., Choi Y., Moon Y.H., Kim S.G., Choi Y.D., Lee J.S. Systemic induction of a Phytolacca insularis antiviral protein gene by mechanical wounding, jasmonic acid, and abscisic acid. Plant Mol. Biol. 2000;43:439–450. doi: 10.1023/a:1006444322626. [DOI] [PubMed] [Google Scholar]
- Sousa M.V., Morhy L. Enterolobin, a hemolytic protein from Enterolobium contortisiliquum seeds (Leguminosae–Mimosoideae). Purification and characterization. An. Acad. Bras. Cienc. 1989;61:405–412. [PubMed] [Google Scholar]
- Sousa M.V., Morhy L., Richardson M., Hilder V.A., Gatehouse A.M.R. Effects of the cytolytic seed protein enterolobin on coleopteran and lepidopteran insect larvae. Entomol. Exp. Appl. 1993;69:231–238. [Google Scholar]
- Sparvoli F., Bollini R. Arcelin in wild bean (Phaseolus vulgaris L.) seeds: sequence of arcelin 6 shows it is a member of the arcelins 1 and 2 subfamily. Genet. Resour. Crop Evol. 1998;45:383–388. [Google Scholar]
- Staniscuaski F., Ferreira-Da Silva C.T., Mulinari F., Pires-Alves M., Carlini C.R. Insecticidal effects of canatoxin on the cotton stainer bug Dysdercus peruvianus (Hemiptera: Pyrrhocoridae) Toxicon. 2005;45:753–760. doi: 10.1016/j.toxicon.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Stec B. Plant thionins–the structural perspective. Cell. Mol. Life Sci. 2006;63:1370–1385. doi: 10.1007/s00018-005-5574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec B., Markman O., Rao U., Heffron G., Henderson S., Vernon L.P., Teeter M.M. Proposal for molecular mechanism of thionins deduced from physico-chemical studies of plant toxins. J. Pept. Res. 2004;64:210–224. doi: 10.1111/j.1399-3011.2004.00187.x. [DOI] [PubMed] [Google Scholar]
- Stirpe F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon. 2013;67:12–16. doi: 10.1016/j.toxicon.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Stirpe F., Battelli M.G. Ribosome-inactivating proteins: progress and problems. Cell. Mol. Life Sci. 2006;63:1850–1866. doi: 10.1007/s00018-006-6078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz H.U., Thomson J.G., Wang Y. Plant defensins: defense, development and application. Plant Signal. Behav. 2009;4:1010–1012. doi: 10.4161/psb.4.11.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson B., Fukuda K., Nielsen P.K., Bønsager B.C. Proteinaceous α-amylase inhibitors. Biochim. Biophys. Acta. 2004;1696:145–156. doi: 10.1016/j.bbapap.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Szczesny P., Iacovache I., Muszewska A., Ginalski K., Van Der Goot F.G., Grynberg M. Extending the aerolysin family: from bacteria to vertebrates. PLoS ONE. 2011;6:e20349. doi: 10.1371/journal.pone.0020349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevissen K., Terras F.R., Broekaert W.F. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl. Environ. Microbiol. 1999;65:5451–5458. doi: 10.1128/aem.65.12.5451-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo M.A., Gil-Exojo I., Romero de Tejada A., Campillo J.E. White bean amylase inhibitor administered orally reduces glycaemia in type-2 diabetic rats. Br. J. Nutr. 2006;96:539–544. [PubMed] [Google Scholar]
- Tumer N.E., Hwang D.J., Bonness M. C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc. Natl. Acad. Sci. USA. 1997;94:3866–3871. doi: 10.1073/pnas.94.8.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valueva T.A., Mosolov V.V. Role of inhibitors of proteolytic enzymes in plant defense against phytopathogenic microorganisms. Biochemistry (Moscow) 2004;69:1305–1309. doi: 10.1007/s10541-005-0015-5. [DOI] [PubMed] [Google Scholar]
- Van Damme E.J.M. Plant lectins as part of the plant defense system against insects. In: Schaller A., editor. Induced plant resistance to herbivory. Springer; Netherlands: 2008. pp. 285–307. [Google Scholar]
- Van Damme E.J.M. History of plant lectin research. In: Hirabayashi J., editor. Lectins: methods and protocols. Springer; New York: 2014. pp. 3–13. [Google Scholar]
- Van Damme E.J.M., Barre A., Rougé P., Leuven F., Peumans W.J. The NeuAc (α-2,6)-Gal/GalNAc binding lectin from elderberry (Sambucus nigra) bark, a type 2 ribosome inactivating protein with an unusual specificity and structure. Eur. J. Biochem. 1996;235:128–137. doi: 10.1111/j.1432-1033.1996.00128.x. [DOI] [PubMed] [Google Scholar]
- Van Damme E.J.M., Peumans W.J., Barre A., Rougé P. Plant lectins: a composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. Crit. Rev. Plant Sci. 1998;17:575–692. [Google Scholar]
- Van Damme E.J.M., Peumans W.J., Pusztai A., Bardocz S. John Wiley & Sons; New York: 1998. Handbook of plant lectins: properties and biomedical applications. [Google Scholar]
- Van Damme E.J.M., Lannoo N., Peumans W.J. Plant lectins. Adv. Bot. Res. 2008;48:107–209. [Google Scholar]
- Vandenborre G., Van Damme E.J.M., Smagghe G. Nicotiana tabacum agglutinin expression in response to different biotic challengers. Arthropod Plant Interact. 2009;3:193–202. [Google Scholar]
- Vandenborre G., Miersch O., Hause B., Smagghe G., Wasternack C., Van Damme E.J.M. Spodoptera littoralis-induced lectin expression in tobacco. Plant Cell Physiol. 2009;50:1142–1155. doi: 10.1093/pcp/pcp065. [DOI] [PubMed] [Google Scholar]
- Vandenborre G., Smagghe G., Van Damme E.J.M. Plant lectins as defense proteins against phytophagous insects. Phytochemistry. 2011;72:1538–1550. doi: 10.1016/j.phytochem.2011.02.024. [DOI] [PubMed] [Google Scholar]
- Vandenborre G., Smagghe G., Ghesquiere B., Menschaert G., Rao R.N., Gevaert K., Van Damme E.J.M. Diversity in protein glycosylation among insect species. PLoS ONE. 2011;6:e16682. doi: 10.1371/journal.pone.0016682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas L.R.B., Carlini C.R. Insecticidal and antifungal activities of ribosome-inactivating proteins. In: Stirpe F., Lappi D., editors. Ribosome-inactivating proteins: ricin and related proteins. Willy Blackwell; Oxford: 2014. pp. 212–222. [Google Scholar]
- Vasconcelos I.M., Oliveira J.T.A. Antinutritional properties of plant lectins. Toxicon. 2004;44:385–403. doi: 10.1016/j.toxicon.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Vila L., Quilis J., Meynard D., Breitler J.C., Marfà V., Murillo I., Vassal J.M., Messeguer J., Guiderdoni E., San Segundo B. Expression of the maize proteinase inhibitor (mpi) gene in rice plants enhances resistance against the striped stem borer (Chilo suppressalis): effects on larval growth and insect gut proteinases. Plant Biotechnol. J. 2005;3:187–202. doi: 10.1111/j.1467-7652.2004.00117.x. [DOI] [PubMed] [Google Scholar]
- Virgilio M.D., Lombardi A., Caliandro R., Fabbrini M.S. Ribosome-inactivating proteins: from plant defense to tumor attack. Toxins. 2010;2:2699–2737. doi: 10.3390/toxins2112699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnudasan D., Tripathi M.N., Rao U., Khurana P. Assessment of nematode resistance in wheat transgenic plants expressing potato proteinase inhibitor (PIN2) gene. Transgenic Res. 2005;14:665–675. doi: 10.1007/s11248-005-5696-4. [DOI] [PubMed] [Google Scholar]
- Wang P., Zoubenko O., Tumer N.E. Reduced toxicity and broad spectrum resistance to viral and fungal infection in transgenic plants expressing pokeweed antiviral protein II. Plant Mol. Biol. 1998;38:957–964. doi: 10.1023/a:1006084925016. [DOI] [PubMed] [Google Scholar]
- Wang J.H., Nie H.L., Huang H., Tam S.C., Zheng Y.T. Independency of anti-HIV-1 activity from ribosome-inactivating activity of trichosanthin. Biochem. Biophys. Res. Commun. 2003;302:89–94. doi: 10.1016/s0006-291x(03)00119-0. [DOI] [PubMed] [Google Scholar]
- Wang C.K., Kaas Q., Chiche L., Craik D.J. CyBase: a database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 2008;36:D206–D210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Kawasaki T., Sako N., Funatsu G. Actions of pokeweed antiviral protein on virus-infected protoplasts. Biosci. Biotechnol. Biochem. 1997;61:994–997. doi: 10.1271/bbb.61.994. [DOI] [PubMed] [Google Scholar]
- Yamaji Y., Maejima K., Komatsu K., Shiraishi T., Okano Y., Himeno M., Sugawara K., Neriya Y., Minato N., Miura C., Hashimoto M., Namba S. Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell. 2012;24:778–793. doi: 10.1105/tpc.111.093658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Tomiyama M., Nemoto R., Naganuma T., Ogawa T., Muramoto K. Effects of food lectins on the transport system of human intestinal caco-2 cell monolayers. Biosci. Biotechnol. Biochem. 2013;77:1917–1924. doi: 10.1271/bbb.130367. [DOI] [PubMed] [Google Scholar]
- Yang G., Cui T., Chen Q., Ma T., Li Z. Isolation and identification of native membrane glycoproteins from living cell by concanavalin A–magnetic particle conjugates. Anal. Biochem. 2012;421:339–341. doi: 10.1016/j.ab.2011.10.033. [DOI] [PubMed] [Google Scholar]
- Zaugg I., Magni C., Panzeri D., Daminati M.G., Bollini R., Benrey B., Bacher S., Sparvoli F. QUES, a new Phaseolus vulgaris genotype resistant to common bean weevils, contains the Arcelin-8 allele coding for new lectin-related variants. Theor. Appl. Genet. 2013;126:647–661. doi: 10.1007/s00122-012-2008-2. [DOI] [PubMed] [Google Scholar]
- Zhao J., Ben L.H., Wu Y.L., Hu W., Ling K., Xin S.M., Nei H.L., Ma L., Pei G. Anti-HIV agent trichosanthin enhances the capabilities of chemokines to stimulate chemotaxis and G protein activation, and this is mediated through interaction of trichosanthin and chemokine receptors. J. Exp. Med. 1999;190:101–112. doi: 10.1084/jem.190.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.L., Feng D., Wu J., Sui S.F. Trichosanthin inhibits integration of human immunodeficiency virus type-1 through depurinating the long-terminal repeats. Mol. Biol. Rep. 2010;37:2093–2098. doi: 10.1007/s11033-009-9668-2. [DOI] [PubMed] [Google Scholar]
- Zhou S., Cheng L., Guo S., Zhu H., Tao S. Lectin microarrays: a powerful tool for glycan-based biomarker discovery. Comb. Chem. High T. Scr. 2011;14:711–719. doi: 10.2174/138620711796504398. [DOI] [PubMed] [Google Scholar]