Abstract
Background:
Severe acute respiratory syndrome (SARS) is an emerging infection caused by a novel coronavirus known as SARS-CoV, characterized by an over-exuberant immune response with lung lymphomononuclear cells infiltration and proliferation that may account for tissue damage more than the direct effect of viral replication. This study is aimed at investigating the capability of SARS-CoV to activate IFN-α and -γ expression in lymphomonocytes (PBMC) from healthy donors, evaluating whether viral replication is necessary for this activation.
Results:
SARS-CoV virus is able to induce both IFN-α and -γ mRNA accumulation and protein release in a dose-dependent manner, MOI 10 being the most effective. The time course curve indicated that IFN-α mRNA induction peaked at 24 h.p.i,. whereas IFN-γ mRNA was still increasing at 48 h.p.i. Released IFN (both types) reached a plateau after 24–48 h.p.i. and remained rather stable over a 5-day period. A transient peak of negative strand viral RNA was detected after 1–2 days of infection, but neither infectious virus progeny yield nor newly produced viral genomic RNA could be evidenced in infected cultures, even after prolonged observation time (up to 13 days). Cocultivation of PBMC with fixed SARS-CoV-infected Vero cells was even more efficient than exposure to live virus in eliciting IFN-α and -γ induction. A combination of IFN-α and -γ strongly inhibited SARS-CoV replication in Vero cells, while the single cytokines were much less effective.
Conclusions:
This study provides evidence that SARS-CoV is able to induce in normal PBMC a coordinate induction of IFN-α and -γ gene expression. Virus replication is not necessary for IFN induction since efficient IFN expression could be obtained also by the cocultivation of normal PBMC with fixed SARS-CoV-infected cells. Concomitant activation of IFN-α and -γ gene expression by SARS-CoV in vivo may be relevant for the pathogenesis of the disease, both for the possible involvement in immunomediated damage of the tissues and for the strong inhibition of SARS-CoV replication as a result of combined cytokine action.
Abbreviations: IFN, interferon; SARS-CoV, severe acute respiratory syndrome coronavirus; MOI, multiplicity of infection
Keywords: SARS-CoV, PBMC, Fixed cell
Background
Severe acute respiratory syndrome (SARS) is an emerging infection caused by a novel coronavirus known as SARS-CoV (Chan et al., 2003, Drosten et al., 2003). The illness is characterized by inflammatory exudation in the alveoli and interstitial tissue accompanied by hyperplasia of fibrous tissues and fibrosis (Nicholls et al., 2003). Destruction of lung tissue is thought to result from an over-exuberant immune response rather than from the direct effects of viral replication. Particularly, it has been shown that IFN-γ may be activated in SARS patients (Wong et al., 2004; Cameron M.J. International Conference on SARS, Lubeck, May 8–11, 2004), suggesting that proinflammatory cytokines may be associated with lung infiltration and proliferation of lymphomononuclear cells (Nicholls et al., 2003, Wong et al., 2004, Bauer et al., 2000, Beijing Group of National Research Project for SARS, 2003). Another clinical feature of SARS is leukopenia (Wong et al., 2003) which may be consequent to the recruitment of lymphomonocytes from the periphery to the inflamed tissue. A short time after the onset of the disease, the presence of the replicative intermediates of SARS-CoV has been shown in peripheral blood mononuclear cells (PBMC) taken from patients (Li et al., 2003). In addition, SARS-CoV genome replication has been detected in PBMC in vitro (Ng et al., 2004), and low titers of infectious progeny production have been observed in cultures of purified monocyte/macrophage (Yilla et al., 2005). Minute and variable amounts of IFN-α have been detected in these cultures, suggesting a potential role of this cytokine in restricting SARS-CoV replication in monocytes in vitro. Recently, both macrophages (mϕ) and monocyte-derived dendritic cells (DC) have been shown to be unable to sustain productive replication of SARS-CoV, although initial phases of virus replication cycle have been detected in the latter cells (Tseng et al., 2005, Law et al., 2005).
This study was aimed at investigating the capability of SARS-CoV to activate IFN-α and -γ production in PBMC from healthy donors and to establish whether productive infection is essential for the activation of IFN response.
For the first issue, normal PBMC were exposed to infectious SARS-CoV at various multiplicity of infection (MOI), and both virus replication and activation of IFN-α and -γ gene expression were determined. For the second issue, normal PBMC were cocultivated with fixed SARS-CoV-infected cells, and the activation of IFN response was determined as well.
Results
IFN induction by live SARS-CoV in normal PBMC
To test the ability of SARS-CoV to induce IFN in normal PBMC, dose-dependence experiments were performed, exposing purified blood cells to the virus at an MOI ranging from 0.1 to 10. The levels of IFN-α and -γ mRNA in the cells and the corresponding cytokine levels in the supernatants were measured. The results of a representative experiments are shown in Fig. 1 and indicated that SARS-CoV is able to induce both IFN-α and -γ mRNA in a dose-dependent manner, being MOI 10 the most effective (Fig. 1a). In addition, IFN-α mRNA levels reached a peak after 24 h, whereas IFN-γ mRNA levels were steadily increasing at 48 h of culture (Fig. 1c). The dose-dependence and the kinetics of IFN-α and -γ release in the supernatants, shown in Figs. 1b and d, were consistent with the mRNA results. Released IFN (both types) reached a plateau after 24–48 h.p.i. (Fig. 1d) and remained rather stable up to day 4 p.i., then started to decline thereafter (not shown). The response to SARS-CoV infection was consistently found in PBMC from different donors, although with great variation in the extent of response. To give an idea of this variation, individual results from 4 different donors, referring to one representative time and dose, are shown in Figs. 1e and g (for mRNA) and Figs. 1f and h (for immunoreactive cytokines). The stimulation of IFN response to SARS-CoV was observed in all experiments, but, as can be seen, the extent of response for mRNA varied from 3 to 45 stimulation over background and was even more pronounced when measuring cytokine levels by ELISA. The overall pattern of variation was not concordant for mRNA and ELISA at individual PBMC level, suggesting that it did not merely reflect the different activation state of PBMC from different donors.
Fig. 1.
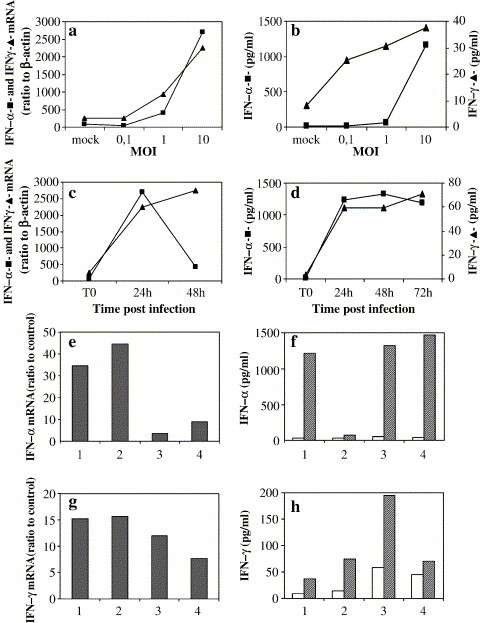
Induction of mRNA for IFN-α and -γ and of immunoreactive cytokines by live SARS-CoV. (a and b) Representative dose-dependence experiment: PBMC were infected with SARS-CoV at different MOI (0.1, 1, and 10). After overnight incubation, mRNA levels specific for IFN-α (–▲–) and -γ (–■–) were measured by limiting dilution RT-PCR and expressed as ratio to β-actin mRNA (×10−3), as described in the Methods section (a). Released cytokines were detected by ELISA (b) and expressed as pg/ml. (c and d) Representative time course experiment: PBMC were exposed for the indicated times to SARS-CoV at MOI 10. Levels of mRNA for IFN-α (–▲–) and -γ (–■–) and of released cytokines were determined as in panels a and b, respectively. (e and g) Peak levels of mRNA for IFN-α (24 h.p.i.) and IFN-γ (48 h.p.i) in PBMC from 4 different donors infected with SARS-CoV at MOI 10. Results expressed as ratio to the levels in the unstimulated cultures. (f and h) Levels of released IFN-α (48 h.p.i.) and IFN-γ (72 h.p.i) by the same PBMC cultures shown in panels e and g. Results are expressed as pg/ml.
SARS-CoV replication in normal PBMC
The ability of SARS-CoV to productively infect normal PBMC was determined in the same cultures used for the IFN induction experiments, extending the observation period to up 13 days, with periodical sampling of the cultures to measure several parameters indicative of virus replication. In Fig. 2a, the infectious virus concentration in PBMC infected at the various MOI is shown. A decreasing amount of infectious virus was observed along the whole study period, probably resulting from thermal inactivation of residual viral inoculum, suggesting the absence of newly formed progeny yield. Similar results were also obtained with mitogen (phytohemoagglutinin, PHA)-activated PBMC (not shown). To test if at least part of the virus replication cycle took place in PBMC, we measured the levels of both virus-specific minus-RNA strand and genomic RNA. A peak of minus-RNA strand could be visualized only at low MOI 0.1 due to high background in the inoculum at higher MOI, whereas genomic RNA quantification revealed a trend similar to infectious virus yield titration, with a decreasing residual inoculum detected along the whole observation period (not shown). To lower the background noise at high MOI, we repeated the infection experiments in PBMC from 3 additional donors and performed trypsinization and extensive washing of the cells after the adsorbtion phase. Under these conditions, we could clearly appreciate a peak of minus strand viral RNA in 3 out of 3 PBMC samples, ranging from 50- to 250-fold increase over background levels. In two PBMC samples, the peak was observed at day 1 p.i., while in one PBMC sample, it was observed at day 2 p.i. followed by a decline to background levels thereafter in all 3 cases. Fig. 2c shows one representative case. However, also in these conditions, no increase of either infectivity or genomic RNA could be appreciated, as shown in Figs. 2b and c. No damage of PBMC apparently occurred in such cultures, as assessed as both overall mortality and apoptotic cell death (never exceeding the background levels of 5–6%). These results suggested the occurrence of an incomplete virus replication cycle in PBMC.
Fig. 2.
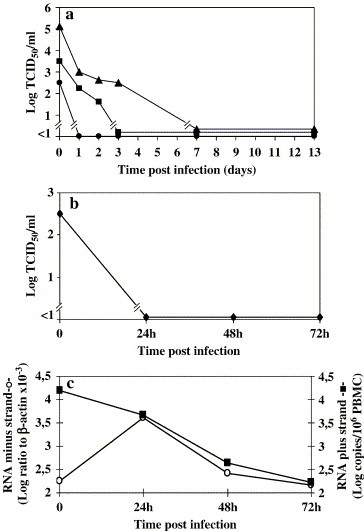
Replication kinetic of SARS-CoV on PBMC. (a) PBMC were infected at MOI 0.1 (–●–), 1 (–■–), and 10 (–▲–) with SARS-CoV. Then, the cells were extensively washed, and fresh medium was added (time 0). Sampling of the cultures was performed at the indicated time points, up to day 13. One representative experiment is shown. Results are expressed as TCID50/ml. (b and c) Infectivity (b) and viral RNA (positive and negative strand), (c) in PBMC infected at MOI 10, treated with trypsin after the adsorbtion phase. One representative experiment is shown. Infectivity is expressed as TCID50/ml, viral genomic RNA as Log copies/106 PBMC, and negative strand viral RNA as Log ratio to β-actin ×10−3.
IFN induction by fixed SARS-CoV-infected Vero cells and reduction of SARS-CoV replication by combined cytokine treatment
To verify if the occurrence of a complete or an even partial replication cycle of SARS-CoV is required for IFN induction in PBMC or whether IFN induction could be triggered also in the absence of virus replication, we used fixed SARS-CoV-infected Vero cells as IFN inducers. To this aim, normal PBMC were cocultivated with fixed SARS-CoV-infected Vero cells at different ratios (from 6:1 to 200:1). Fixed uninfected Vero cells were used as negative control. Fig. 3 shows the results of a representative dose- and time-dependence experiment. As can be seen, both IFN types were dose-dependently induced by the fixed SARS-CoV-infected cells. Furthermore, IFN-α release peaked at day 1, remaining at plateau levels thereafter, whereas IFN-γ reached plateau levels at 48 h.p.i. only with the highest concentration of the inducer, while it was still increasing at this time at lower inducer concentrations. Similar results were obtained with PBMC from different donors, although with some variability in the extent of response. In fact, range for peak IFN-α and -γ levels were 800–2500 and 200–2000, respectively (not shown). On the whole, the IFN response was higher than that observed in the experiments using live virus since average cytokine levels were about 2- and 5-fold higher for IFN-α and -γ, respectively.
Fig. 3.
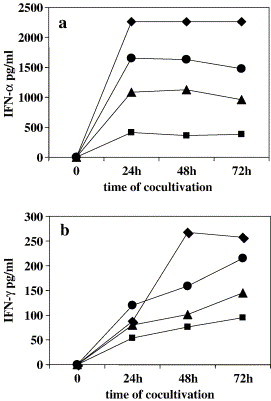
Induction of IFN by SARS-CoV-fixed-infected cells. PBMC were cocultivated with fixed SARS-CoV-Vero-infected cells at different ratios, 6:1 (–■–), 20:1 (–▲–), 60:1 (–●–), 200:1 (–♦–). At the indicated time points, supernatants were collected, and immunoreactive IFN-α (a) and -γ (b) were detected by ELISA. IFN-α and -γ levels in supernatants of cocultures of PBMC with uninfected fixed Vero cells were lower than the detection limit (5 pg/ml for IFN-α and >2 pg/ml for IFN-γ, respectively) (not shown).
In classical virus yield experiments, Vero cells were treated with either IFN-α alone (5000 IU/ml), IFN-γ alone (1000 IU/ml), or with a combination of both and then infected with SARS-CoV at an MOI of 0.01. The results from three experiments, shown in Fig. 4 , indicated a mean reduction of virus yield at day 3 p.i. of 1.72 and 0.57 Log10 for IFN-α and IFN-γ, respectively (P = 0.072 and 0.451), but a much stronger reduction (4.63 Log10) when a combination of both IFN types was used (P < 0.01).
Fig. 4.

Reduction of infectious virus yield in Vero cells by IFN-α and -γ used singularly or in combination. Vero cells were treated with either IFN-α alone (5000 IU/ml), IFN-γ alone (1000 IU/ml), or with a combination of both and then infected with SARS-CoV at MOI 0.01. After 3 days, progeny virus was harvested and titrated. Results are expressed as mean virus yield (Log TCID50) over 3 experiments. Bars indicate standard error over the mean. Statistical evaluation of the reduction versus control cultures: IFN-α P = 0.072; IFN-γ P = 0.451; IFN-α + IFN-γ P < 0.01.
Discussion
The coordinate induction of IFN-α and -γ in unprimed PBMC cultures in vitro is a well-established phenomenon common to different viruses such as influenza and parainfluenza viruses, as well as HIV-1 (Capobianchi et al., 1993). In this study, we extended the observations to the recently discovered etiologic agent of SARS, obtaining evidence that SARS-CoV has the intrinsic ability to activate both IFN-α and -γ gene expression in PBMC cultures. In fact, we observed a dose- and time-dependent induction of both mRNA and protein release for both cytokines when using live virus as IFN inducer in normal PBMC cultures. Recently, SARS-CoV antigens have been shown to be able to induce a recall response, including the production of IFN-γ, in PBMC from vaccinated mice (Takasuka et al., 2004), but, to our knowledge, there were no previous data showing IFN-γ response to SARS-CoV by unprimed human PBMC.
The kinetics of induction of IFN-α and -γ mRNA observed in SARS-CoV-exposed PBMC cultures is in line with the expected kinetics of induction by other viruses (Capobianchi et al., 1993), with more transient IFN-α mRNA activation. However, the levels of cytokines in the supernatants showed a similar pattern for both IFN types, being stable for up to 4 days. The response to SARS-CoV infection was consistently found in PBMC from different donors, although the extent of responses varied among individuals, as also observed in SARS-CoV-infected monocytes (Yilla et al., 2005).
In this study, we also obtained evidence that productive infection of PBMC is not mandatory for IFN induction by two lines of evidence: no peak of SARS-CoV infectious progeny was detected in PBMC cultures; efficient IFN induction could be achieved in PBMC by cocultivation with fixed SARS-CoV-infected cells.
Concerning the first line of evidence, the present study adds significant advancement to previous data. In fact, recent results from another group indicated that SARS-CoV is able to infect in vitro PBMC and to determine virus-specific RNA production (Ng et al., 2004). However, infectious viral yield was not tested in this paper. Here, we show that, under our experimental conditions, productive viral replication does not occur in PBMC, although initial phases of virus infection take place. In fact, both viral infectivity and genomic RNA progressively decreased along the observation time. Even after removal of background levels by trypsinization, we failed to detect any increase of both infectious progeny yield and genomic RNA. However, removal of background noise allowed us to consistently detect a peak of newly formed negative strand at day 1 or 2 p.i., declining thereafter. These findings suggest that, under these conditions, initial phases of virus replication started in PBMC, but the cycle did not progress to completion. No damage of PBMC apparently occurred in such cultures, as assessed as both overall mortality and apoptotic cell death, further supporting the absence of virus replication in bulk PBMC cultures. These results remind what has been observed in two recent studies in blood mononuclear cell cultures (Tseng et al., 2005, Law et al., 2005). Particularly, in the study by Tseng et al., the infectious virus titer progressively decreased, and viral subgenomic RNA was not produced in primary mϕ or monocyte-derived DC. The authors conclude that in these cells SARS-CoV fails to establish productive infection, although affecting some functions of both cell types (Tseng et al., 2005). Similarly, Law et al. did not observe production of infectious virus yield in DC, although initial phases of viral replication seemed to occur (Law et al., 2005).
Concerning the second line of evidence, the fact that efficient induction of IFN-α and -γ could be achieved in PBMC also by cocultivation with fixed SARS-CoV-infected cells may suggest the involvement in IFN induction of membrane mechanisms leading to activation of internal pathways of IFN gene expression. As previously shown for other viruses, including HIV, HSV, and animal coronaviruses (Capobianchi et al., 1988, Capobianchi et al., 1993, Charley and Laude, 1988), glycoproteins exposed on infected cells could be involved in IFN induction, although the mechanisms need still to be elucidated.
Conclusions
It is generally believed that the IFN system can play a pivotal role in host defense against viruses.
It is reasonable to assume that lymphomonocytes can be exposed to high concentration of SARS-CoV in the body sites where virus replication takes place and where IFN activation can occur as a result of either direct (although non productive) PBMC infection or exposure to infected cells as bystander effect.
Therefore, IFN-α and -γ production by PBMC exposed to SARS-CoV is expected to occur in vivo and to participate to the inflammatory events, taking place in the diseased tissues. Concomitant activation of both IFN-α and -γ gene expression, here shown for the first time, is relevant for the pathogenesis of the disease. In fact, SARS-CoV has been shown to be poorly sensitive to the antiviral action of IFN-α and -γ when used singularly but is strongly inhibited by the combination of these two cytokines (Stroher et al., 2004, Hensley et al., 2004, Antonelli et al., 2003 and present results), as observed with many other viruses. Therefore, the natural response to SARS-CoV exerted by lymphomononuclear cells, leading to the concomitant activation of type I and II IFN response, can result in a strong inhibition of SARS-CoV replication.
In the light of these results, the possible added value of therapeutic intervention by combined IFN administration to diseased patients should be taken into consideration, although more data are necessary to clarify the role of endogenously activated IFN system in SARS.
Methods
Cells and viral stocks
Vero E6 cells were maintained in Modified Eagle Medium (MEM) supplemented with 10% Fetal Calf Serum (FCS) at 37 °C in a humidified atmosphere. For virus stock preparation, Vero E6 cells were infected with SARS-CoV (Tor2 isolate, provided by H. Feldmann, Dept. of Medical Microbiology of Manitoba, Canada) at a MOI of 0.01 TCID50/cell. The virus was harvested when 70–80% of cell monolayer showed cytopathic effect. After freezing and thawing three times, cell lysates were clarified, aliquoted, and stored at −70 °C. Virus titration was performed on Vero E6 cells with limiting dilution assay, and the results expressed as TCID50/ml (sensitivity 1 Log TCID50/ml). The virus stock titer was 108 TCID50/ml.
For the preparation of fixed cells, Vero E6 cells were infected with a mock virus preparation or with SARS-CoV at MOI 10 TCID50/cell. Seven hours post-infection, cell monolayers were washed twice with PBS, detached with 5 mM EDTA, extensively washed, and exposed to 4% paraformaldehyde in PBS for 20 min at 4 °C. Then, the cells were extensively washed and resuspended at 3 × 106 cells/ml in RPMI containing 10% FCS and kept on ice until use.
PBMC infection and detection of SARS-CoV replication
PBMC were obtained from healthy donors by Ficoll/Hypaque (Pharmacia, Sweden) density centrifugation. Cultures were performed in RPMI 1640 medium (GIBCO, USA) containing 10% FCS and antibiotics. PBMC from 10 donors were used in the infection experiments. Peripheral blood mononuclear cells were infected for 30 min at 37 °C with different MOI, ranging from 0.1 to 10, washed, reseeded at 2 × 106/ml in RPMI 10% FCS, and incubated at 37 °C. In some experiments, PBMC were exposed to trypsin (0.5 mg/ml) for 3 min after removal of virus inoculum, washed 3 times, and then reseeded. At days 5 and 10, half amount of medium was replaced with fresh complete medium. At the indicated times, total cellular RNA was extracted by Trizol (Life Technologies, NY, USA), according to the manufacturer's protocol. In parallel, whole clarified culture lysates, obtained by three cycles of freeze–thawing, were assayed for virus titration on Vero cells, according to the method of Reed and Muench. Results were expressed as TCID50/ml. In some experiments, both PBMC unstimulated and activated with 0.5 μg/ml phytohemoagglutinin (PHA) (GIBCO, USA) were used. Cell mortality and apoptosis (hypo-diploid cells) were detected, at days 1, 2, and 6 p.i., by trypan blue and propidium iodide staining, respectively.
In order to detect virus-specific minus-RNA strand, total RNA extracted from SARS-CoV-infected PBMC was retrotranscribed in the presence of the sense primer BNIoutS2 (5′ATGAATTACCAAGTCAATGGTTAC3′) 200 nM, RT buffer (Invitrogen), 0.5 mM (each) dNTP, 20 mM DTT, and 50 U of MMLV RT (Invitrogen) for 20 min at 42 °C, after denaturation of the target at 70 °C for 10 min. Amplification was carried out with 1 U of AmpliTaq Gold (Applied Biosystems), 1× PCR buffer (Applied Biosystems), 2 mM MgCl2, 0.5 mM (each) dNTP and sense BNIoutS2 and antisense primer BNIoutAS (5′CATAACCAGTCGGTACAGCTAC3′) 500 nM each, as described (Drosten et al., 2003). The amplified products (186 bp) were detected by agarose gel electrophoresis and visualized by UV fluorescence, after staining with ethidium bromide. Semiquantitative evaluation was obtained by limiting dilution RT-PCR, and normalization for an housekeeping gene expression was performed by running parallel amplification with β-actin mRNA.
Measurement of SARS-CoV genomic RNA was performed by quantitative real time RT-PCR, using the commercial kit RealArt TM HPA-Coronavirus LC RT-PCR (Artus, Hamburg, Germany), on a LightCycler Instrument (Roche Diagnostic, Basel, Switzerland).
IFN induction
All the induction experiments were performed on freshly collected PBMC.
IFN induction in PBMC was performed by infection with live SARS-CoV or by cocultivation with fixed SARS-CoV-infected Vero cells. For the first issue, 5 × 106 (2 × 106 cells/ml) PBMC were infected with SARS-CoV virus at an MOI ranging from 0.1 to 10 and sampled at various time points. PBMC from 9 donors were used in this set of experiments. For IFN induction by fixed cells, freshly isolated PBMC (2 × 106 cells/ml) were cultivated in the presence of Vero cells, either uninfected or infected with SARS-CoV, fixed with paraformaldehyde as above described, at different ratio. PBMC from 3 donors were used in this set of experiments. As positive control for IFN induction, Newcastle Disease virus (NDV) was used in all the experiments, at 10 hemagglutination units (HU)/106 cells. In all cases, a strong response was observed in NDV-exposed PBMC, indicating a good inducibility of both IFN-α and -γ in our experimental conditions (not shown).
IFN-α and -γ expression
Total RNA was extracted by Trizol and retrotranscribed by the random examer extension method. The determination of the mRNA levels for IFN-α and -γ was performed by limiting dilution RT-PCR as previously described (Abbate et al., 2003). The amplified products were analyzed by agarose gel electrophoresis, and results were expressed as ratio to β-actin.
Released IFN-α and -γ were detected by enzyme-linked immunosorbent assays (ELISA), namely Human Interferon Alpha (Hu-IFN-α) multi-subtype ELISA kit (PBL Biomedical Laboratories), measuring all isotypes of Hu-IFN-α, and ELISA for IFN-γ, purchased from Pierce Biotechnology (Rockford, IL). Before the ELISA assay, culture supernatants of PBMC exposed to live virus were treated with 0.1% Triton X-100 to inactivate residual viral infectivity. Results were expressed as IFN pg/ml. To give an idea of the inter-donor extent of variation, individual results of PBMC from 4 different donors, referring to peak stimulation, are shown in the results section, expressed as ratio to control for mRNA and in pg/ml for immunoreactive proteins.
Statistical evaluation
Mean reduction of virus yield by single or combined IFN treatment was evaluated by Student's t test.
Acknowledgments
This work has been supported by Ricerca Corrente e Finalizzata del “Ministero della Salute” to INMI “L. Spallanzani”.
CC carried out IFN induction experiments and drafted the manuscript. LB carried out IFN induction experiments. EL, CA, and GR performed molecular analysis and immunoassays. IA, FP, and MRC participated in the study design and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Concetta Castilletti, Email: castilletti@inmi.it.
Licia Bordi, Email: colault@yahoo.it.
Eleonora Lalle, Email: eleonoralalle@interfree.it.
Gabriella Rozera, Email: gabriellarozera@virgilio.it.
Fabrizio Poccia, Email: poccia@inmi.it.
Chiara Agrati, Email: agrati@inmi.it.
Isabella Abbate, Email: abbate@inmi.it.
Maria R. Capobianchi, Email: capobianchi@inmi.it.
References
- Abbate I., Romano M., Longo R., Cappiello G., Lo Iacono O., Di Marco V., Paparella C., Spano A., Capobianchi M.R. Endogenous levels of mRNA for IFNs and IFN-related genes in hepatic biopsies of chronic HCV-infected and non alcoholic steato hepatitis patients. J. Med. Virol. 2003;70:581–587. doi: 10.1002/jmv.10433. [DOI] [PubMed] [Google Scholar]
- Antonelli G., Scagnolari C., Vincenzi E., Clementi M. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)14482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer T.T., Montòn C., Torres A., Cabello H., Fillela X., Maldonado A., Nicolas J.M., Zavala E. Comparison of systemic cytokine levels in patients with acute respiratory distress syndrome, severe pneumonia, and controls. Thorax. 2000;55:46–52. doi: 10.1136/thorax.55.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijing Group of National Research Project for SARS Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Clin. Med. J. 2003;116:1283–1287. [PubMed] [Google Scholar]
- Capobianchi M.R., Malavasi F., Di Marco P., Dianzani F. Differences in the mechanism of induction of interferon-α by herpes simplex virus and herpes simplex virus-infected cells. Arch. Virol. 1988;103:219–229. doi: 10.1007/BF01311094. [DOI] [PubMed] [Google Scholar]
- Capobianchi M.R., Ameglio F., Cordiali F.P., Castilletti C., Mercuri F., Fais S., Dianzani F. Coordinate induction of IFN-α and -γ by recombinant HIV-1 glycoprotein 120. AIDS Res. Hum. Retrovir. 1993;9:957–962. doi: 10.1089/aid.1993.9.957. [DOI] [PubMed] [Google Scholar]
- Chan P.K., Ip M., Ng K.C., Rickjason C.W., Wu A., Lee N., Rainer T.H., Joynt G.M., Sung J.J., Tam J.S. Severe acute respiratory syndrome-associated coronavirus infection. Emerg. Infect Dis. 2003;9:1453–1454. doi: 10.3201/eid0911.030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charley B., Laude H. Induction of α interferon by transmissible gastroenteritis coronavirus role of transmembrane glycoprotein E1. J. Virol. 1988;62:8–11. doi: 10.1128/jvi.62.1.8-11.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Managuerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Hensley L.E., Fritz L.E., Jahhrling P.B., Karp C.L., Huggins J.W., Geisbert T.W. Interferon-α 1a and SARS coronavirus replication. Emerg. Infect Dis. 2004;10:317–319. doi: 10.3201/eid1002.030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law H.K., Cheung C.Y., Ng H.Y., Sia S.F., Chan Y.O., Luk W., Nicholls J.M., Peiris J.S., Lau Y.L. Chemokine upregulation in SARS coronavirus infected human monocyte derived dendritic cells. Blood. 2005 doi: 10.1182/blood-2004-10-4166. (Apr. 28; [electronic publication]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wo J., Shao J., Zhu H., Wu N., Li M., Yao H., Hu M., Dennin R.H. SARS-coronavirus replicates in mononuclear cells of peripheral blood (PBMCs) from SARS patients. J. Clin. Virol. 2003;28:239–244. doi: 10.1016/S1386-6532(03)00195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.F., Hibberd M.L., Ooi E., Tang K.F., Neo S.Y., Tan J., Murthy K.R., Vega V.B., Chia J.M., Liu E.T., Ren E.C. A human in vitro model system for investigation genome-wide host responses to SARS coronavirus infection. BMC Infect. Dis. 2004;4:34. doi: 10.1186/1471-2334-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., Yan K.W., Chan K.H., Tsang N.C., Guan Y., Yuen K.Y., Peiris J.S.M. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroher U., Di Caro A., Li Y., Strong J.E., Aoki F., Plummer F., Jones S.M., Feldmann H. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon-α. J. Infect. Dis. 2004;189:1164–1167. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasuka N., Fujii H., Takahashi Y., Kasai M., Morikawa S., Itamura S., Ishii K., Sakaguchi M., Ohnishi K., Ohshima M., Hashimoto S., Odagiri T., Tashiro M., Yoshikura H., Takemori T., Tsunetsugu-Yokota Y. A subcutaneously injected UV-inactivated SARS coronavirus vaccine elicits systemic humoral immunity in mice. Int. Immunol. 2004;16:1423–1430. doi: 10.1093/intimm/dxh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.T., Perrone L.A., Zhu H., Makino S., Peters C.J. Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. J. Immunol. 2005;174:7977–7985. doi: 10.4049/jimmunol.174.12.7977. [DOI] [PubMed] [Google Scholar]
- Wong R.S., Wu A., To K.F., Lee N., Lam C.W., Wong C.K., Chan P.K., Ng M.H., Yu L.M., Hui D.S., Tam J.S., Cheng G., Sung J.J. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326:1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilla M., Harcourt B.H., Hickman C.J., McGrew M., Tamin A., Goldsmith C.S., bellini W.J., Anderson L.J. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Research. 2005;107:93–101. doi: 10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


