Abstract
Aminopeptidase N, or CD13, is a receptor for serologically related coronaviruses of humans, pigs, and cats. A mouse line transgenic for the receptor of human coronavirus-229E (HCoV-229E) was created using human APN (hAPN) cDNA driven by a hAPN promoter. hAPN-transgenic mice expressed hAPN mRNA in the kidney, small intestine, liver, and lung. hAPN protein was specifically expressed on epithelial cells of the proximal convoluted renal tubules, bronchi, alveolar sacs, and intestinal villi. The hAPN expression pattern within transgenic mouse tissues matched that of mouse APN and was similar in mice heterozygous or homozygous for the transgene. Primary embryonic cells and bone marrow dendritic cells derived from hAPN-transgenic mice also expressed hAPN protein. Although hAPN-transgenic mice were resistant to HCoV-229E in vivo, primary embryonic cells and bone marrow dendritic cells were infected in vitro. hAPN-transgenic mice are valuable as a source of primary mouse cells expressing hAPN. This hAPN-transgenic line will also be used for crossbreeding experiments with other knockout, immune deficient, or transgenic mice to identify factors, in addition to hAPN, that are required for HCoV-229E infection.
Keywords: Coronavirus, HCoV-229E, Transgenic, Receptor, Virus, Aminopeptidase N, CD13, Animal model, Dendritic cell, HCV-229E
Introduction
Several different coronaviruses (CoV), including HCoV-229E, SARS-CoV, HCoV-OC43, HCoV-NL63 (McIntosh, 2005), and HKU1 (Woo et al., 2005), infect humans. Serological studies suggest that the prototypical strains HCoV-229E and HCoV-OC43 cause from 15% to 30% of human upper respiratory infections or colds (Holmes, 2001). Studies on the pathogenesis of HCoV-229E respiratory infections were limited to intranasal inoculation of human volunteers because there was no small animal model. HCoV-229E infection leads to coryza, headache, cough, fever, disruption of nasal epithelium, and it can occasionally cause lower respiratory tract infection (Chilvers et al., 2001). There is in vitro and in vivo evidence for infection of cells of the central nervous system (CNS) by human coronaviruses, although no neurological symptoms have been associated with HCoV-229E infection (Arbour et al., 2000).
A cell surface receptor that mediates entry of HCoV-229E into cells is human aminopeptidase N (hAPN) (Yeager et al., 1992). When the single-stranded positive-sense genomic RNA of HCoV-229E is transfected into cells derived from many species, productive virus replication ensues and progeny virions are efficiently released (Thiel et al., 2001). Thus, hAPN plays a critical role in the tissue tropism and species specificity of HCoV-229E. For example, mouse fibroblasts that are nonpermissive for HCoV-229E become susceptible after transfection with cDNA encoding hAPN (Yeager et al., 1992). Porcine and feline coronaviruses also use aminopeptidase N (APN) as their cellular receptors (Delmas et al., 1992, Tresnan et al., 1996).
APN or CD13 is a 150–160 kDa type II glycoprotein that is a metalloprotease (Noren et al., 1997). APN is expressed as a dimer on the surface of epithelial cells of the kidney, intestine, and respiratory tract. APN is also expressed by granulocytes, fibroblasts, endothelial cells, cerebral pericytes at the blood–brain barrier, synaptic membranes in the central nervous system, as well as on antigen presenting cells, such as macrophages and dendritic cells (Dong et al., 2000, Noren et al., 1997, Riemann et al., 1999). The biological activities of APN include the removal of individual amino acids from amino-termini of small peptides in the lumen of the small intestine, cleavage of peptides bound to MHC class II molecules on antigen presenting cells, angiogenic regulation, and degradation of neurotransmitters at synaptic junctions (Bhagwat et al., 2003, Noren et al., 1997, Riemann et al., 1999). Human APN is also a marker for acute myeloid leukemia and plays a role in tumor invasion (Riemann et al., 1999).
Several transgenic mouse models have proven useful for studying viruses that normally infect humans. Therefore, we developed mice transgenic for hAPN, and tested these animals, or primary cells isolated from them, for susceptibility to HCoV-229E. The hAPN transgene was designed to be expressed in the epithelial cells of respiratory and intestinal tracts where coronaviruses enter the host. The human APN operon has two promoters separated by more than 8 Kb and they direct the differential expression of hAPN mRNA in various tissues (Shapiro et al., 1991). The transgene was engineered to use the proximal human APN promoter, active in epithelial cells of the intestine, lung, liver, and kidney (Shapiro et al., 1991), to direct the transcription of hAPN cDNA and the polyadenylation signal from SV40 virus (Fig. 1 ). Eight lines of mice were produced by pronuclear microinjection and hAPN expression was analyzed. This study focuses on a single line of mice that expressed the highest levels of hAPN mRNA and protein in a variety of tissues. This hAPN transgenic line of mice, and cells derived from these animals, were tested for susceptibility to HCoV-229E.
Fig. 1.
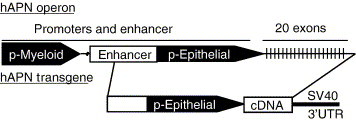
Diagram of the hAPN operon and transgene cassette. The hAPN cDNA was cloned downstream of the natural human APN epithelial promoter and upstream of the 3′ UTR of SV40 virus.
Results
Mouse cells transfected with the hAPN transgene construct were permissive to HCoV-229E and produced infectious virus
The transgene construct and a plasmid that encodes a neomycin resistance (Neo) gene driven by the SV40 promoter were co-transfected into mouse CMT93 cells to show that hAPN protein was produced from the transgene and that it functioned as a receptor for HCoV-229E. FACS analysis with anti-hAPN MAb-WM15 showed that 29% of the G418 resistant co-transfected cell population expressed hAPN on the cell surface, whereas the control cells transfected with Neo vector alone did not show any reactivity to the anti hAPN MAb (data not shown). Control cells and the hAPN co-transfected cells were inoculated with HCoV-229E and the titer of released virus (Control-S and hAPN-S) or the total cell associated plus released virus (Control-T and hAPN-T) was determined at various times post-inoculation by plaque assay (Fig. 2 ). The data show that HCoV-229E entered, replicated, and infectious virus was efficiently released from mouse cells expressing hAPN protein (Fig. 2). The hAPN transgene construct/Neo co-transfected cells were also sorted for hAPN expression by FACS, and 88% were positive for cell surface expression post-sort (data not shown). Transfected cells sorted by hAPN expression were inoculated with a low MOI (0.001) and the supernatants were harvested over 72 h (Fig. 2, hAPN sorted, solid squares). These data show that HCoV-229E completed multiple rounds of replication in mouse cells that expressed hAPN. Cell lines were generated from the hAPN-positive population and analyzed for reactivity with a panel of anti-hAPN MAbs (WM15, WM47, MY7, DW1, Y2K, and BB1) that bind to multiple epitopes (Ashmun et al., 1995). MAb-DW1, -Y2K, and -BB1 are new anti-hAPN receptor MAbs and inhibit virus infection. FACS analysis of the hAPN-positive cell lines showed they reacted similarly with all six of the anti-hAPN MAbs, indicating proper surface conformation of hAPN on the mouse CMT93 cells (data not shown).
Fig. 2.
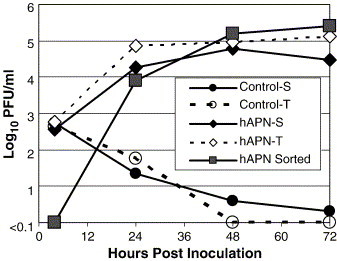
Mouse CMT93 cells transfected with the hAPN transgene construct are productively infected by HCoV-229E. The hAPN transgene was co-transfected with a neomycin-resistant plasmid (PCR 3.0) into a mouse epithelial cell line (CMT93) and selected for G418 resistance. Control cells were transfected with PCR 3.0 alone (open and solid circles), the other cells were co-transfected with the hAPN transgene plasmid and PCR 3.0 (open and solid diamonds). The cells were inoculated at an MOI of 0.6 with HCoV-229E and the titer of virus released into the supernatant (solid circle and diamond) and total virus produced (open circle and diamond) was determined by plaque assay. hAPN-expressing cells were also sorted by FACS (88% were hAPN-positive post-sort) and were inoculated with HCoV-229E at an MOI of 0.001. The titer of virus released into the supernatant from the sorted cells was determined over 72 h (solid squares).
hAPN mRNA was expressed in a tissue-specific manner by the transgene construct
The proximal hAPN promoter used in the transgene was expected to be very active in the epithelial cells of the respiratory and enteric tracts and in the proximal convoluted tubules of the kidney. Progeny mice from 8 founder animals were initially screened by a hAPN-specific RT-PCR assay designed to amplify only polyadenylated RNA from total RNA isolated from the small intestine. One of these lines expressed higher levels of hAPN mRNA and was used for subsequent studies (data not shown). Total RNA was isolated from the kidney (K), small intestine (SI), lung (L), spleen (S), liver (Li), muscle (M), and heart (H) of non-transgenic and hAPN-transgenic mice. The hAPN transcript was identified by a 712-bp amplicon visible by ethidium bromide staining in the kidney, small intestine, and liver of the transgenic mice and it was not amplified from the RNA of the non-transgenic animal tissues (Fig. 3 , RT). The hAPN transcript amplicon was shown to be specific by Southern blot using a 32P labeled RNA probe complementary to the 3′ terminus of hAPN and part of the SV40 3′UTR that is specific to the transgene (data not shown). This assay demonstrated that the 712-bp amplicons visible by ethidium bromide staining and a band amplified from the lung RNA of the hAPN-transgenic mouse were specific to hAPN mRNA transcription (data not shown). The smaller amplicons observed in the lung, brain, and liver of both the non-transgenic and the hAPN-transgenic mice did not hybridize with the hAPN-specific probe and therefore were non-specific amplification products (data not shown). When the sensitivity of hAPN mRNA detection was increased using hemi-nested PCR that maintained the specificity for a polyadenylated hAPN transcript, a 439-bp amplicon was identified in every tissue tested in the transgenic mice (Fig. 3, HN). The data showed that hAPN mRNA was expressed at qualitatively higher levels in the kidney, small intestine, liver, and to a lesser extent in the lung, which was identified by Southern blot. This expression pattern corresponded to the tissue-specific expression pattern found in previous transcriptional analysis of the proximal hAPN promoter (Shapiro et al., 1991).
Fig. 3.
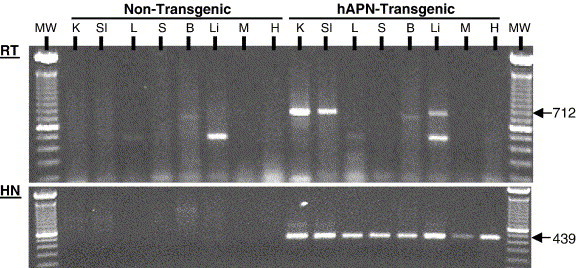
The hAPN-transgenic mice express hAPN mRNA with a tissue-specific distribution. Total RNA was isolated from the kidney (K), small intestine (Si), lung (L), spleen (S), brain (B), liver (Li), muscle (M), and heart (H) of non-transgenic and hAPN-transgenic mice. Amplicons from an RT-PCR reaction that used a forward primer specific for hAPN (hAPN-2880) and a tailed oligo dT reverse primer specific for the poly-A tail (oligodT/Xho1) were subjected to agarose gel electrophoresis and stained with ethidium bromide (RT). The expected size of the RT-PCR amplicons was 712 bp and molecular weight markers, 100-bp DNA ladder (Invitrogen), were run in the first and last lanes. Hemi-nested (HN) PCR of an aliquot of the hAPN RT-PCR reaction was also preformed using an internal hAPN forward primer and the same reverse primer used for the RT-PCR reaction. This HN-PCR reaction increases the sensitivity and specificity of the assay and produces a 439-bp DNA fragment when hAPN transgene mRNA is present.
Human APN protein was expressed with the same cellular distribution as mouse APN in the small intestine and the kidney
To identify expression of hAPN and mouse APN (mAPN) proteins in the small intestine and kidney of hAPN-transgenic and non-transgenic mice, we used immunocytochemistry with a biotinylated MAb-DW1 or a rat MAb-R3-242 against mAPN. These were detected by either streptavidin-HRP or a biotinylated rabbit anti-rat followed by streptavidin-HRP, respectively. The Vector Red substrate used stained regions bound by antibodies red and was also fluorescent. The sections were counterstained with hematoxylin. The data showed the cell type distribution expected for mAPN in the epithelial cells of the brush border along the villi of the small intestine and by cells of the proximal convoluted tubule in the kidney (Fig. 4 , anti-mAPN). Both the non-transgenic and the hAPN-transgenic mice showed similar mAPN protein distribution patterns, demonstrating that expression of the hAPN protein in the transgenic animals did not dramatically influence the level of mAPN expressed. Human APN protein was expressed in the small intestine and kidney of the transgenic mice in the same manner as mAPN (Fig. 4, anti-hAPN). The hAPN protein was expressed on the brush border membrane by epithelial cells on the intestinal villi and by cells in the proximal convoluted tubules of the kidney (Fig. 4, anti-hAPN). MAb-DW1 binding suggests that hAPN expressed in the mouse tissues was in the proper conformation for interaction with HCoV-229E.
Fig. 4.
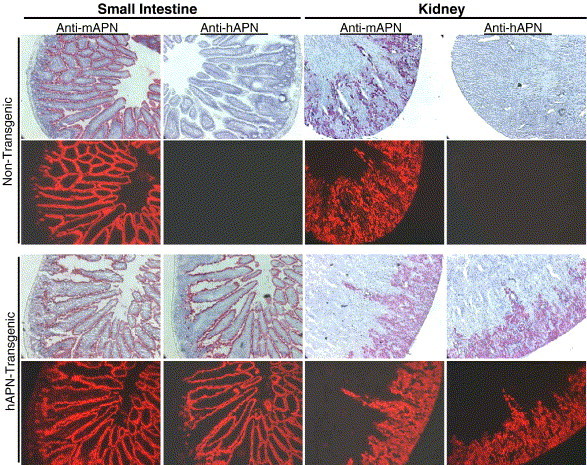
Immunohistochemistry of cryosections shows that the hAPN has the same cellular distribution as mouse APN in transgenic animals. Serial sections from the small intestine or kidney of age matched non-transgenic and hAPN-transgenic mice were tested for reactivity with a rat anti mouse APN (mAPN) MAb-R3-242 or with a mouse anti-hAPN MAb-DW1 and stained using Vector Red and counter stained with hematoxylin. The Vector Red® substrate is also fluorescent and these images are shown in the panels below the hematoxylin counter-stained images.
The hAPN protein was expressed similarly in homozygous female, heterozygous female, and hemizygous male transgenic mice
The hAPN expression corresponded with a transgene cassette that was inserted into the X chromosome of this transgenic line. The APN expression in the various genotypes was determined using cryosections of the small intestine from non-transgenic (XX, XY) and hAPN homozygous (XhXh), heterozygous (XhX), and hemizygous (XhY) transgenic mice. The same expression pattern of both hAPN and mAPN proteins was detected in the brush border membrane of the small intestine (Fig. 5A). Fluorescent Vector red staining of mAPN was observed in all mice, and hAPN protein was detected only in transgenic mice (Fig. 5A).
Fig. 5.
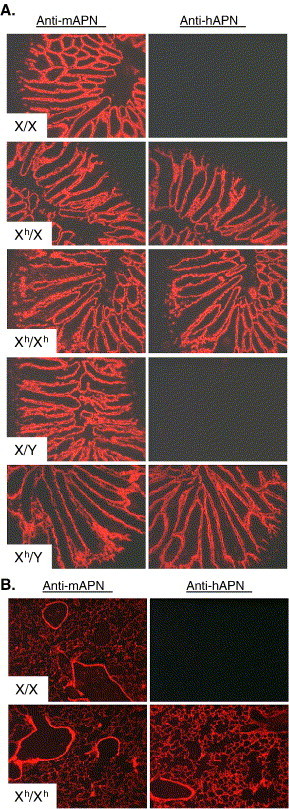
Homozygous, heterozygous, and hemizygous hAPN-transgenic mice all express hAPN with a similar expression pattern and hAPN was expressed on the epithelial surface of the lungs. (A) Serial cryosections of small intestine from age matched non-transgenic females (XX), heterozygous females (XhX), homozygous females (XhXh), non-transgenic males (XY), and hemizygous males (XhY) were analyzed for expression of mouse APN (mAPN) using rat MAb-R3-242 to mAPN or with a mouse anti-hAPN MAb-DW1 and binding was determined by a fluorescent substrate (Vector Red®). (B) Cryosections of lungs from age matched non-transgenic and homozygous hAPN-transgenic mice were analyzed with a MAb-R3-242 specific to mouse APN (mAPN) or a MAb-DW1 against hAPN. Binding of the MAbs was revealed by a fluorescent substrate (Vector Red®).
Epithelial cells of the secondary bronchi and alveolar sacs in the hAPN-transgenic mice expressed hAPN that corresponded with mouse APN expression
We were particularly interested in the localization of hAPN in respiratory tissues of the hAPN-transgenic mice, because HCoV-229E is a respiratory pathogen. Our goal was to express hAPN on the epithelial cells of the respiratory tract that are typically infected by HCoV-229E. Cryosections of lungs from non-transgenic and homozygous hAPN-transgenic mice stained with anti-mAPN-R3-242 or with anti-hAPN-DW1 were used to analyze expression of mAPN or hAPN proteins, respectively. Mouse APN was detected in the secondary bronchi and alveolar sacs of both the non-transgenic and hAPN-transgenic mice (Fig. 5B). In transgenic animals, hAPN protein was expressed with the same distribution pattern as mAPN (Fig. 5B). The data show that the proximal human APN promoter directed expression of hAPN protein in the respiratory tract of transgenic mice with the appropriate cell-specific distribution.
HCoV-229E infection of the hAPN-transgenic mice was not detected after intranasal, intracranial, or intragastric inoculation
Many trials were conducted to determine if adult transgenic animals were susceptible to HCoV-229E after intranasal inoculation, the natural route of infection. Mice of a variety of ages (10 days, 3 weeks, 3 months, and 1.5–2 years) were tested and evidence of infection of the hAPN-transgenic animals was for the most part negative. Anesthetized mice were inoculated with 1 × 107 to 1 × 108 PFU of HCoV-229E and examined for evidence of virus replication by clinical signs, gross pathology, virus titration, and sensitive RT-PCR assays specific for HCoV-229E. No clinical signs or gross pathology were observed in either the hAPN-transgenic or non-transgenic animals post inoculation (pi). In some trials, the titer of infectious virus in respiratory washes from hAPN-transgenic and non-transgenic mice was determined. This showed that the titers of both hAPN-transgenic and non-transgenic mice were very low (101–102 PFU/ml) 1 day pi and were below the limit of detection by 3 days pi (data not shown).
Very sensitive analysis of HCoV-229E genomic RNA (gRNA) was done using RT-PCR specific for the nucleocapsid gene, or multiplex RT-PCR specific for the spike and polymerase genes followed by nested PCR of the amplicons. The nested PCR procedure consistently identified gRNA, presumably from the input virus, in the RNA from the lungs of both hAPN-transgenic and non-transgenic mice (data not shown). To differentiate input virus gRNA from low level HCoV-229E replication, we used RT-PCR to detect leader-body junction containing subgenomic RNA (sgRNA) encoding the N gene. The results showed that the RNA from the lungs and/or nasal turbinates of HCoV-229E-inoculated mice that were positive for gRNA consistently tested negative for sgRNA that is produced during viral replication (data not shown). However, one hAPN-transgenic animal tested positive for N sgRNA in the lung (data not shown).
There is evidence of CNS infection by HCoV-229E in humans, so we inoculated 2-day-old hAPN-transgenic and non-transgenic littermates intracranially (Arbour et al., 2000). The mice were tested for hAPN mRNA and HCoV-229E sgRNA specific to virus entry and initiation of replication. We found that the hAPN-specific mRNA was not detectable (data not shown) by the RT-PCR, hemi-nested PCR procedure that detects hAPN transcripts in the brains of adult mice (Fig. 3), and HCoV-229E sgRNA production was not detected (data not shown). The data suggest that the expression of the hAPN transgene correlated with the natural expression of hAPN on synaptic junctions, which increases with age, and infection of CNS of the hAPN-transgenic mice needs to be examined further using older animals.
Transmissible gastroenteritis virus and porcine respiratory coronaviruses are genetically related to HCoV-229E and use porcine APN as a cellular receptor (Delmas et al., 1992). Hence, HCoV-229E has the potential to infect epithelial cells in the intestine of hAPN-transgenic mice, which expressed hAPN mRNA and protein (Fig. 3, Fig. 4). However, no evidence of HCoV-229E enteric infection was observed after intragastric inoculation of adult hAPN-transgenic and non-transgenic mice (data not shown). This may be due to the fact that the spike glycoprotein of HCoV-229E has a deletion when it is compared to the TGEV spike glycoprotein. The HCoV-229E spike glycoprotein deletion is similar to a deletion in the spike glycoprotein of some porcine respiratory coronaviruses that only infect the respiratory tract (Enjuanes et al., 1993).
Primary cells isolated from transgenic mouse embryos were infected by HCoV-229E and produced infectious virus
Cell lines from embryos derived from hAPN-transgenic and non-transgenic mice were established to determine if hAPN protein expressed by the hAPN-transgenic mice was a functional receptor for HCoV-229E. Human APN expression on cells of the embryonic cultures was analyzed by flow cytometry, and 3% of the cells derived from the hAPN-transgenic mouse expressed the HCoV-229E receptor within a range typically observed on human MRC5 cells, while hAPN expression was not detected on cells derived from non-transgenic mice (data not shown). Cells from the non-transgenic and hAPN-transgenic mice were grown on glass cover slips and inoculated with HCoV-229E at an MOI of 1. Twenty hours post-inoculation, the cells were fixed and expression of human APN or HCoV-229E proteins was detected by immunofluorescence assay (IFA) using anti-hAPN MAb-DW1 or polyclonal goat anti-HCoV-229E. The cells derived from hAPN-transgenic embryos that expressed hAPN protein were infected by HCoV-229E (Fig. 6A). Embryonic cells derived from non-transgenic mice and hAPN-transgenic mice, or human lung fibroblasts (MRC5), were inoculated at an MOI of 1, and the virus in the supernatant was quantified by TCID50 4 h and 24 h post-inoculation. The HCoV-229E titer decreased below the limit of detection in the non-transgenic embryo cells by 24 h post-inoculation (Fig. 6B). In contrast, the titer of HCoV-229E in the supernatant from hAPN-transgenic embryo cells increased to 1 × 103 TCID50/ml by 24 h post-inoculation (Fig. 6B). Only the cells derived from hAPN-transgenic embryos that expressed hAPN were infected by HCoV-229E, and this hAPN-positive population (3%) efficiently produced infectious virus.
Fig. 6.
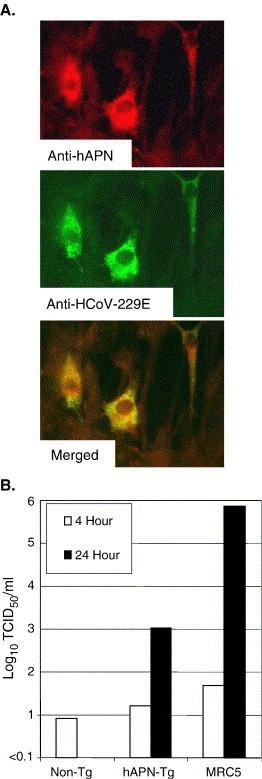
Some embryonic cells from the hAPN-transgenic mice express hAPN protein and are infected by HCoV-229E. (A) Cells derived from non-transgenic and hAPN-transgenic embryos were inoculated with HCoV-229E at an MOI of 1 and fixed 20 h post-inoculation. Cells were tested for hAPN expression (red) with a mouse anti-hAPN MAb-DW1 and for expression of HCoV-229E proteins (green) with polyclonal goat antisera to HCoV-229E. In addition, the overlay of these two images is shown (merged). (B) Embryonic cells from non-transgenic and hAPN-transgenic mice were inoculated with HCoV-229E at an MOI of 1, unbound virus was removed by washing, and the titer of the virus in the supernatant at 4 h and 24 h post-infection was determined by TCID50 in MRC5 cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Dendritic cells derived from the bone marrow of the hAPN-transgenic mice expressed hAPN and were susceptible to HCoV-229E infection
Dendritic cells (DCs) are potent antigen presenting cells that can present peptides in the context of both MHC class I and class II molecules. In addition, both mouse and human DCs are known to express APN (CD13) (Dong et al., 2000, Summers et al., 2001, Woodhead et al., 2000). To determine if dendritic cells of the hAPN-transgenic mice expressed hAPN protein and were susceptible to HCoV-229E, we derived DCs from the bone marrow (BM) of non-transgenic and hAPN-transgenic mice. The BM–DCs were prepared by isolating the BM from male littermates using magnetic separation to remove T cell, B cell, and macrophage precursors, and then culturing the remaining cells in medium containing IL4 and GMCSF for 5–7 days (Turner et al., 2004). Comparison of the fluorescence between BM–DCs treated with no primary antibody (gray line) or a hamster MAb to an irrelevant antigen (dotted line) by FACS showed there was little non-specific reactivity (Fig. 7A, a and b). The BM–DCs isolated from non-transgenic and hAPN-transgenic mice reacted similarly with a hamster MAb against the DC marker CD11c (Fig. 7A, a and b, solid line with shaded area under the curve). Fig. 7A, panels c and d, shows that the BM–DCs from non-transgenic and hAPN-transgenic mice reacted similarly with rat anti-mAPN MAb-R3-242 (solid line shaded under the curve) when compared to cells treated with no primary antibody (gray line) or nonspecific control antibody (dotted line). MHC I (dashed line) and CEACAM1a (not shown) also showed very similar levels of expression in DCs derived from both the non-transgenic and transgenic littermates (Fig. 7A, e and f). Thus, the cells isolated and cultured from both hAPN-transgenic and non-transgenic mice showed consistent expression of multiple surface antigens. In contrast to the non-transgenic mice, substantial hAPN expression (solid line with shading under the curve) was observed on BM–DCs isolated from the hAPN-transgenic mice, indicating that the DC-enriched population of cells expressed the hAPN protein from the transgene (Fig. 7A, e and f). FACS analysis using three different anti-hAPN MAbs (DW1, Y2K, and WM47) that bind to different regions of hAPN showed very similar reactivity with BM–DCs from the hAPN-transgenic animals and very little reactivity with BM–DCs from the non-transgenic mice (data not shown). This indicates that the hAPN on the DC surface was folded correctly and expressed several epitopes important in HCoV-229E receptor activity, as receptor binding MAb-DW1 and MAb-Y2K block HCoV-229E infection (data not shown).
Fig. 7.
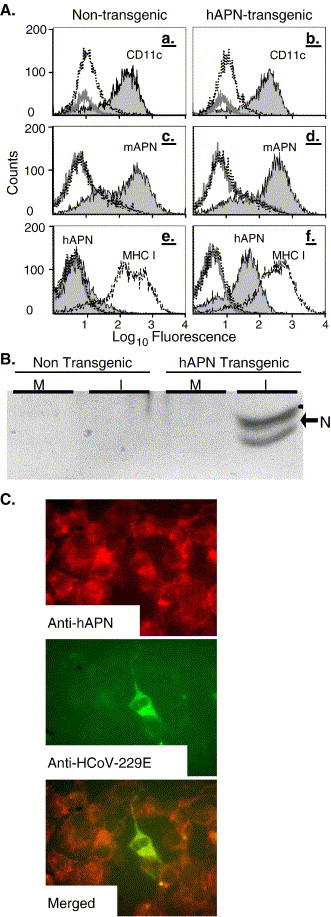
Dendritic cells derived from the bone marrow of hAPN-transgenic mice express hAPN and are infected by HCoV-229E. (A) Dendritic cells were isolated from non-transgenic (a, c, and e) and hAPN-transgenic (b, d, and f) male littermates and analyzed by cytofluorometry. The controls were cells that were not treated with primary antibody (gray line) or treated with antibodies (hamster, rat, or mouse) to an irrelevant antigen (dotted line). The surface expression of CD11c (a and b, solid line with gray shading), mouse APN (c and d, solid line with gray shading), MHC class I (e and f, dashed line), and human APN (e and f, solid line with gray shading) were identified with hamster MAb-HL3, rat MAb-R3-242, mouse MAb-KH117, and MAb-DW1, respectively. Dendritic cells isolated from non-transgenic and hAPN-transgenic mice were mock inoculated (M) or inoculated (I) with HCoV-229E at an MOI of 1.0 at 4 °C. Unbound virus was removed by washing and cells were incubated at 34 °C for 20 h. (B) Immunoblot analysis of protein lysates was performed using polyclonal goat anti-HCoV-229E that reacts strongly with nucleocapsid protein (arrow). (C) IFA of acetone fixed hAPN-transgenic dendritic cells was done using anti-hAPN MAb-DW1 (red) and with polyclonal goat anti-HCoV-229E (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To test their susceptibility to HCoV-229E, the same BM–DCs analyzed in Fig. 7A were inoculated with HCoV-229E at an MOI of 1 or were mock inoculated. Analysis of the supernatant media collected from the BM–DCs at 2 h and 20 h pi showed a two log10 increase in HCoV-229E titer from the transgenic mice (data not shown). Whereas, a decrease in virus titer was observed in the supernatant media of BM–DC derived from the non-transgenic mice (data not shown). Immunoblot analysis of proteins isolated from mock (M) or HCoV-229E inoculated (I) cells showed that BM–DCs from hAPN-transgenic mice were susceptible to HCoV-229E as indicated by the doublet at ∼45 kDa characteristic of viral nucleocapsid (N) protein (Fig. 7B, arrow). No viral antigens were detected in BM–DCs derived from the non-transgenic mice. IFA showed expression of hAPN protein (red) in most of the BM–DCs and infection of some of the BM–DCs (green) from the hAPN-transgenic mice (Fig. 7C). Thus, hAPN expression on the BM–DCs of transgenic mice permitted infection by HCoV-229E and resulted in the replication and release of virus in vitro.
Discussion
The goals of this study were to develop hAPN-transgenic mice that express hAPN protein in epithelial tissues in the hopes that the animals would prove to be beneficial for the study of HCoV-229E infection in vivo or that the response of specific cell types to HCoV-229E could be studied in vitro. APN has numerous biological activities, and many of these depend on its enzymatic activity and tissue localization. For example, APN plays important roles in digestion, cancer, angiogenesis, synaptic activity, and is responsible for cleaving peptides bound to MHC molecules of antigen presenting cells (Bhagwat et al., 2003, Ishii et al., 2001, Noren et al., 1997, Riemann et al., 1999). Thus, hAPN-transgenic mice may prove valuable for many areas of research in addition to HCoV-229E–host interactions.
The hAPN-transgene construct directed expression of hAPN mRNA and protein in vitro when it was transfected into CMT93 cells, which are derived from mouse intestinal epithelium. The hAPN protein expressed by CMT93 cells was recognized by a panel of six anti-hAPN MAbs that recognize different epitopes of hAPN and also had enzymatic activity. HCoV-229E inoculation of CMT93 cells expressing hAPN protein produced considerable titers (4.5–5 log10) of HCoV-229E through multiple replication cycles over 3 days. Therefore, hAPN protein expressed from the transgene construct was transported to the surface of mouse cells with normal epitopes, enzymatic activity, and in the proper conformation for productive HCoV-229E replication.
In vivo hAPN mRNA and protein were detected in the lung, small intestine, and kidneys, which corresponded with expression of wild-type mAPN. The anti-hAPN MAb-DW1 used to specifically identify hAPN blocks virus infection. Consequently, the region of hAPN that HCoV-229E recognizes is probably in the proper conformation for virus binding and entry in vivo as well as in vitro. The hAPN transgene that expressed mRNA and protein was inserted in the X chromosome and similar levels of hAPN protein were expressed in tissues of hemizygous male as well as heterozygous and homozygous female mice.
In vitro experiments with primary embryo cells and BM–DCs derived from hAPN-transgenic mice showed that the hAPN expressed on the cell surface by the transgene was a functional receptor for HCoV-229E. Although a small percentage of the hAPN-transgenic mouse embryo cell population expressed hAPN, these positive cells were susceptible to HCoV-229E infection and efficiently produced infectious virus. Therefore, cells isolated from hAPN-transgenic mice were productively infected by HCoV-229E.
Dendritic cells (DCs) are extremely important antigen presenting cells that are critical to the initiation of both the adaptive and innate immune response of the host, and it is clear that their interactions with pathogens can have both positive and negative effects on pathogenesis (Liu, 2001, Mellman and Steinman, 2001). Coronaviruses gain entry into the host via respiratory or intestinal epithelium where there are numerous DCs. APN/CD13 is expressed by specific DC subsets and is important in maturation of DCs, extracellular degradation of antigens, and may be involved in T-cell activation (Amoscato et al., 1998, Liu, 2001, Summers et al., 2001, Woodhead et al., 2000). Consequently, we studied mAPN and hAPN expression on dendritic cells derived from the bone marrow (BM–DCs) of non-transgenic and hAPN-transgenic mice. Expressions of surface proteins including MHC I, CEACAM1a, mouse APN, and CD11c were very similar in both the non-transgenic and the hAPN-transgenic mice. In contrast, hAPN was only expressed by the DC-enriched population derived from the hAPN-transgenic mice. BM–DCs isolated from the hAPN-transgenic mice were susceptible to HCoV-229E, whereas BM–DCs derived from non-transgenic mice were not. Our data demonstrate that DCs from hAPN-transgenic mice are infected by HCoV-229E in a receptor-dependent fashion, and Thiel et al. have shown that HCoV-229E virus-like particles transduce human DCs (Thiel et al., 2003). Therefore, DCs in the human respiratory epithelium may play a role in the initiation of HCoV-229E infection and its dissemination to other tissues. Other group 1 coronaviruses that infect cats and pigs may also infect DCs via their cognate APN receptors (Delmas et al., 1992, Tresnan et al., 1996). Mouse DCs are also infected by mouse hepatitis virus-A59, a group 2 coronavirus, in a CEACAM1a receptor-dependent manner (Turner et al., 2004). DC–coronavirus interaction may be a common evolutionary mechanism of entry, dissemination, immunomodulation, and pathogenesis observed in diverse coronaviruses, including feline infectious peritonitis virus, mouse hepatitis virus, severe acute respiratory syndrome coronavirus, and HCoV-229E (Ding et al., 2004, Holmes, 2001, Jeffers et al., 2004, Konikova et al., 1998, Thiel et al., 2003, Turner et al., 2004, Yang et al., 2004). In other viruses, such as LCMV and measles, infection of DCs leads to severe immunomodulation, impairment of antigen presentation, and cytokine expression (Borrow et al., 1995, Hahm et al., 2004, Naniche and Oldstone, 2000). The hAPN-transgenic mice developed in this study will provide an excellent reproducible source of hAPN-expressing mouse DCs to study the effect HCoV-229E infection has on DC function.
Although the hAPN transgene was expressed in respiratory epithelial cells that are the principle targets of HCoV-229E infection, our attempts to infect the hAPN-transgenic mice with HCoV-229E by intranasal inoculation were unsuccessful. HCoV-229E did not cause any clinical signs of disease such as lethargy, weight loss, ruffled fur, or rhinorrhea in the mice; however, many coronavirus infections are subclinical (Holmes, 2001). There was no increase in virus titer in respiratory washes from hAPN-transgenic mice compared to non-transgenic controls. The RT-PCR/hemi-nested PCR procedure utilized to identify genomic RNA was more sensitive than plaque assay and could detect as little as 1 × 10−5 PFU in vitro. We consistently observed that the lung RNA from both non-transgenic and hAPN-transgenic mice inoculated with HCoV-229E contained viral gRNA, most likely from the inoculum. When lung samples were tested for expression of sgRNAs that are produced during replication of HCoV-229E, all but one mouse were negative. Preliminary studies using other routes of inoculation showed no evidence of HCoV-229E infection.
Other initial attempts to generate small animal models by producing transgenic mice or rats that express specific viral receptors have also had limited success (Blixenkrone-Moller et al., 1998, Horvat et al., 1996, Hughes et al., 2003, Niewiesk et al., 1997, Ren et al., 1990, Thorley et al., 1997, Zhang and Racaniello, 1997). Subsequent modification of the transgene constructs, inoculation strategies/routes, crossbreeding with alpha/beta interferon-defective mice, identification of new receptors, and virus strain variation improved models for measles virus and poliovirus (Crotty et al., 2002, Hahm et al., 2004, Mrkic et al., 1998, Nagata et al., 2004, Oldstone et al., 1999, Rall et al., 1997, Tatsuo and Yanagi, 2002).
The reason(s) that cells from hAPN-transgenic mice were susceptible to HCoV-229E in vitro but hAPN-transgenic mice were not permissive to HCoV-229E infection remains to be identified. Many factors that can be eliminated or controlled in in vitro experiments cannot be controlled in vivo. These include adsorption time, temperature, concentration of virus at the cell surface, innate immune response, and presence of non-specific inhibitors such as mucins or pH. Some of these problems will be overcome by crossing these hAPN-transgenic mice with knockout mice (e.g., CD13, STAT1) or other mouse lines. Identification of new species-specific molecules that aid HCoV-229E binding or entry will aid in the development of a better transgenic mouse model. Angiotensin converting enzyme 2 (Ace2) is a functional receptor for SARS-CoV, and like APN it is a surface metalloprotease that is expressed in the respiratory tract, small intestine, spleen, brain, liver, and kidney (Hamming et al., 2004, Li et al., 2003, Wang et al., 2004). DC-SIGN binds SARS-CoV spike glycoprotein (Marzi et al., 2004, Yang et al., 2004) and we have shown that CD209L can act as an alternative receptor for SARS-CoV (Jeffers et al., 2004). CD209L, DC-SIGN, or other species-specific receptors may also facilitate entry of HCoV-229E and their identification will provide new tools to improve hAPN-transgenic mice as a small animal model.
Factors specific to HCoV-229E could also produce the results observed with the hAPN-transgenic mice. HCoV-229E was isolated by serial blind passage in human embryonic kidney cells in 1966 (Hamre et al., 1967, Holmes, 2001). Serial passage may have resulted in the accumulation of mutations that are advantageous in vitro, but these mutations could have adverse affects in vivo. New genomic sequence information from HCoV-229E-related viruses that are currently circulating may identify differences between natural isolates and the laboratory adapted HCoV-229E strain. Reverse genetics of HCoV-229E will allow the correction of these mutations and allow the generation of viruses that identify determinants critical to infection/pathogenesis in hAPN-transgenic mice (Thiel et al., 2001).
The natural cellular distribution of the transgene product is critical to the development of transgenic mice as models to study pathogenesis. We demonstrated the tissue-specific expression of hAPN mRNA and found that hAPN protein distribution within these tissues corresponded with natural mAPN expression. Our results show the value of the present hAPN-transgenic line as a source of different primary cell types for the elucidation of factors critical to virus entry and the host response to HCoV-229E. For example, hAPN-transgenic mouse BM–DCs can be used to gain a better understanding of DC–coronavirus interactions. A similar approach has proven very helpful for study of the interaction of wild-type measles virus with DCs isolated from CD150-transgenic mice (Hahm et al., 2004). These hAPN-transgenic mice may also be useful as models for new coronaviruses or other human pathogens that may interact with hAPN (Gredmark et al., 2004, Larsson et al., 1998, Moller et al., 1999, van der Hoek et al., 2004). Crossbreeding the present FVB/NJ hAPN-transgenic mouse line with other mouse strains, knockout, or transgenic mice will aid in the discovery of host mechanisms critical to coronavirus resistance.
Materials and methods
Cells and viruses
Cells were maintained in appropriate medium supplemented with 2% Antibiotic-Antimycotic (GIBCO BRL, Grand Island, NY) at 37 °C and 5% CO2 in Falcon tissue culture-treated flasks or plates (Becton Dickinson and Company, Franklin Lakes, NJ). Human diploid lung fibroblasts, MRC5 cells (American Type Culture Collection (ATCC), Rockville, MD), were grown from passage 19 to 27 in Eagle's minimal essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1 mM sodium pyruvate, and 1 mM non-essential amino acids (GIBCO BRL). CMT93 cells (ATCC, Rockville, MD) were grown in Dulbecco's modified Eagle's minimal essential medium (DMEM) supplemented with 10% heat-inactivated FBS (GIBCO BRL). Embryonic cell lines were established from pregnant non-transgenic and homozygous hAPN-transgenic females on the 14th day of pregnancy (Spector et al., 1998).
The prototypical strain, HCoV-229E, used in this study was isolated from a patient with an upper respiratory infection in 1966 (Hamre et al., 1967). This virus was isolated by serial blind passage in human embryonic kidney cells, and it can now be propagated in human lung fibroblasts (Hamre et al., 1967). HCoV-229E was obtained from ATCC (VR740) and a variant that was previously adapted to growth at 37 °C (RW) was propagated in MRC5 cells at 37 °C and was used for the majority of the in vivo experiments. HCoV-229E (DW6-W) was originally obtained from ATCC and was propagated in MRC5 cells at 34 °C (Wentworth and Holmes, 2001).
hAPN transgene construction and testing
The hAPN expression plasmid was produced by subcloning an hAPN cDNA fragment (BamHII–BglII, partial digestion), GenBank accession number M22324, from pBSSK+ (Stratagene, La Jolla CA) into the pGem11 plasmid (Promega CO, Madison, WI) that contained a 1789-bp fragment of the proximal promoter of hAPN from SacI to BamHI (the BamH1 site is within the coding sequence of hAPN). Part of the 3′ UTR of hAPN was replaced with the 3′ UTR and poly A signal from SV40 virus by ligating a BamHI–XbaI fragment from pRep4 (Invitrogen) into the hAPN plasmid previously digested with BglII and XbaI. This created the transgene plasmid pGem.APN.ds that could be linearized with SacI and XbaI to yield the complete transgene (Fig. 1).
Mouse CMT93 cells at 50% confluency in 100-mm dishes were transfected with 27 μg of pCR3.0 (Invitrogen, Carlsbad, CA) alone or were co-transfected with 5 μg of pCR3.0 and 22 μg of pGem.APN.ds and 45 μl of pFX2 (Invitrogen) in 1.5 ml of Optimem (Invitrogen). Two days post-transfection, the medium was replaced with media containing 600 μg/ml G418 (Invitrogen) and cells resistant to G418 were selected. These stably transfected cells were used for FACS analysis and HCoV-229E infection experiments as a mixed population. This mixed cell population was also sorted for hAPN expression using MAb WM15 as previously described (Wentworth and Holmes, 2001). To test the mixed population for receptor activity, HCoV-229E was inoculated onto replicate wells of the control cells or the hAPN transgene-transfected cells at an MOI of 0.6 in 0.3 ml for 1 h at 34 °C and 5% CO2. The inoculum was removed and the cells were washed 2 times with 2 ml of medium, and 3 ml of medium was added. The cells were incubated at 34 °C and 5% CO2, at 4 h, 24 h, 48 h, and 72 h post-inoculation, the supernatants were collected, or the cells in duplicate plates were scraped and pooled with the supernatants for virus titration. The pooled cells and supernatants were frozen and thawed 3 times, and cell debris removed by centrifugation. Virus titration was determined by plaque assay in MRC5 cells.
Transgenic mouse production and maintenance
The hAPN transgene cassette was microinjected into the pronucleus of oocytes from FVB/NJ mice (Jackson laboratories, Bar Harbor, MA). Fertilized ova bearing the new hAPN transgene cassette were introduced into the uteri of pseudopregnant FVB/NJ mice by microsurgery at the University of Colorado Health Sciences Center Transgenic Core Mouse Laboratory. DNA isolated from 8 founder lines of mice was analyzed by PCR using intron-spanning primers (DT-21 and DT-22) specific for hAPN cDNA. APN has many biological functions and is involved in a variety of cancers; however, we did not observe any consistent abnormalities such as tumors, developmental problems, or other obvious disease due to the expression of hAPN in transgenic mice. Mice were humanely euthanized consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. All work on mice was done under a protocol approved by the Institutional Animal Care and Use Committee. The animal facilities were supervised by veterinarians specializing in lab animal medicine, and the facility is operated under the NIH Guide for the Care and Use of Laboratory Animals.
Antibodies, fluorescence activated cell sorting (FACS), and immunofluorescence assay
Three new anti-hAPN MAbs (Y2K, BB1, and DW1) that block HCoV-229E infection were generated by inoculating Swiss Webster mice with NIH 3T3 cells expressing hAPN (Zip cells) (Yeager et al., 1992) and screening the supernatants of individual clones for the ability to block HCoV-229E infection of MRC5 cells. The hybridomas were cloned by limiting dilution and expanded to produce the MAbs. MAb-Y2K blocked virus infection, but had no effect on the enzymatic activity of hAPN. MAb-DW1 and MAb-BB1 also blocked virus infection, but decreased enzymatic activity by approximately 55% (data not shown). MAb DW1 was purified, biotinylated, and used to stain the transgenic mouse tissues. Other anti-hAPN MAbs, WM47 (DAKO Corporation, Carpinteria, CA), MY7 (BioGenex, San Ramon, CA), and WM15 (Biodesign, Kennebunk, ME) were purchased. Control MAb against cholera toxin was kindly provided by Dr. Randall Holmes (University of Colorado Health Sciences Center, Denver, CO). Rat anti-mouse APN MAb-R3-242, hamster anti-CD11c MAb-HL3, control MAb-G235-2356 against the hapten trinitrophenol, and mouse anti-MHC I Mab-KH117 to H-2Dq/H2Lq haplotypes were also purchased (PharMingen Int., San Diego, CA). Polyclonal goat anti-HCoV-229E was produced from NP40-disrupted, UV-inactivated HCoV-229E purified by sucrose density gradient centrifugation in collaboration with Lawrence Sturman (Wadsworth Center, New York State Health Department, Albany, NY). FACS and IFA procedures were done as previously described (Wentworth and Holmes, 2001).
RT-PCR and nested PCR
An anchored and tailed oligo-dT (oligo-dT/Xho1) primer was used for reverse transcription (RT) of 5–10 μg of total cellular RNA with Superscript reverse transcriptase (Invitrogen) in a 20-μl reaction. 2-μl of cDNA was amplified by PCR with an hAPN-specific forward primer (APN-2880) and oligo-dT/Xho1 reverse primer which maintains specificity for polyadenylated RNA. Temperature cycling parameters were 30 cycles of 94 °C for 30 s, 57 °C for 45 s, and 72 °C for 1 min. This yielded a 712-base pair (bp) product from RNA produced by the transgene construct and a 614-bp from RNA of the natural human transcript expressed in MRC5 cells. To control for RNA integrity and cDNA production, mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also amplified from 2 μl of the same cDNA using a GAPDH-specific forward primer (G3P-279) and the oligo-dT/Xho1 reverse primer. An aliquot (2 μl) from the hAPN RT-PCR reaction was used as template in another PCR reaction with a hemi-nested set of primers that amplify a 439-bp fragment. The forward primer was specific to hAPN (DT31) and the same reverse primer used in the first round amplification (oligodT/Xho1) was used to maintain the specificity for mRNA. The reaction conditions were the same as used for the first round of RT-PCR. A 20-μl aliquot of this hemi-nested (HN) reaction was electrophoresed through an agarose gel with molecular weight markers (100-bp DNA ladder, Invitrogen) and visualized by ethidium bromide staining.
A very sensitive RT-PCR-based procedure to detect HCoV-229E genomic viral RNA and subgenomic RNA that is indicative of virus entry and initiation of RNA transcription in the tissues of inoculated mice was developed. The reaction conditions and oligonucleotide primers will be provided upon request (Table 1 ).
Table 1.
Oligonucleotides used for PCR
| Forward primer | Sequence (5′–3′) | Reverse primer | Sequence (5′–3′) |
|---|---|---|---|
| DT-021 | CCCACTTCCAGAAGACCCC | DT-022 | AGTTCTCCCGGTAGGTCACC |
| DT-031 | AGCCCACGTTCTCTCTGCC | OligodT-Xhol | TCCTCGAGGTTTTTTTTTTTTTTTTV |
| Leader-1 | ACTTAAGTACCTTATCTATCTACA | N-359R | CCGTTTGCCCTTTCTAGTTCT |
| HAPN-2880 | GTTCAAGAAGGACAACGAGGAA | N-520R | TCTGGTTCTGAATTCTTGCGC |
Tissue sections
Wild type and hAPN-transgenic mice were euthanized as approved by the University of Colorado Animal Care and Use Committee. Tissues were excised, placed in Tissue-Tek O.C.T compound (VWR Int. Aurora, CO), and frozen in liquid nitrogen-cooled isopentane. Frozen tissues were sectioned 10-μm-thick with a microtome/cryostat (Leica CM 1850-3-1 Chantilly, VA) using Accu-edge low profile blades (VWR Int.). Initially, the sections were incubated in C-buffer [PBS containing 1% FBS (GIBCO BRL)] for 20 min. Human APN was detected in frozen sections with 5 ng of a biotinylated mouse anti-hAPN MAb-DW1. Mouse APN was detected with a rat MAb (R3-242, PharMingen Int., San Diego, CA) against mAPN. Sections were incubated with the primary antibodies for 1 h in a humidified chamber, then rinsed 4 times for 5 min in C-buffer. Anti-mAPN-treated sections were incubated for an additional hour with 5 ng of biotinylated rabbit anti-rat antibody (H0408, Vector Laboratories) and rinsed 4 times for 5 min in C-buffer. To reveal antibody binding, the sections were incubated in Vectastain ABC-AP (Vector Laboratories, Burlingame, CA) for 30 min and rinsed 3 times for 5 min with C-buffer. Next, the sections were incubated in Vector Red substrate (Vector Laboratories) for 1–3 min, rinsed in C-buffer, stained with Hematoxylin QS (Vector Laboratories) for 30 s, and rinsed with tap water. Sections were dehydrated by serial washes in 75%, 95%, and 100% ethanol and incubated for 2 min in xylene prior to mounting with Vectamount (Vector Laboratories).
Acknowledgments
The authors would like to thank Justin Hagee and Amy Hopkins for their excellent technical assistance, Karen Helm for FACS analysis, and Dr. Christopher Korch who sequenced the DNA samples. We also thank Drs. Bruce Zelus and Dianna Blau for their very helpful advice. This work was supported by NIH grant AI26075; Dr. Wentworth was supported by NIH Neurovirology-Molecular Biology Training Grant-T32 NS07321 and Dr. Tresnan by NIH training grant K11 AI01151. FACS analysis at the University of Colorado Cancer Center Flow Cytometry Core and DNA sequencing at the University of Colorado Cancer Center DNA Sequencing and Analysis Core were supported by the NIH/NCI Cancer Core Support Grant CA46934-09.
References
- Amoscato A.A., Prenovitz D.A., Lotze M.T. Rapid extracellular degradation of synthetic class I peptides by human dendritic cells. J. Immunol. 1998;161:4023–4032. [PubMed] [Google Scholar]
- Arbour N., Day R., Newcombe J., Talbot P.J. Neuroinvasion by human respiratory coronaviruses. J. Virol. 2000;74:8913–8921. doi: 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashmun R.A., Holmes K.V., Shapiro L.H., Favaloro E.J., Razak K., de Crom R.P.G., Howard C.J., Look A.T. M3 CD13 (aminopeptidase N) cluster workshop report. In: Schlossman S.F., Boumsell L., Gilks W., Harlan J.M., Kishimoto T., Morimoto C., Ritz J., Shaw S., Silverstein R., Springer T., Tedder T.F., Todd R.F., editors. 1(Leucocyte typing V) Oxford University Press; Oxford: 1995. pp. 771–775. White cell differentiation antigens, Proceedings of the Fifth International Workshop and Conference. 3-11-0093. [Google Scholar]
- Bhagwat S.V., Petrovic N., Okamoto Y., Shapiro L.H. The angiogenic regulator CD13/APN is a transcriptional target of Ras signaling pathways in endothelial morphogenesis. Blood. 2003:1818–1826. doi: 10.1182/blood-2002-05-1422. [DOI] [PubMed] [Google Scholar]
- Blixenkrone-Moller M., Bernard A., Bencsik A., Sixt N., Diamond L.E., Logan J.S., Wild T.F. Role of CD46 in measles virus infection in CD46 transgenic mice. Virology. 1998;249:238–248. doi: 10.1006/viro.1998.9301. [DOI] [PubMed] [Google Scholar]
- Borrow P., Evans C.F., Oldstone M.B. Virus-induced immunosuppression: immune system-mediated destruction of virus-infected dendritic cells results in generalized immune suppression. J. Virol. 1995;69:1059–1070. doi: 10.1128/jvi.69.2.1059-1070.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilvers M.A., McKean M., Rutman A., Myint B.S., Silverman M., O'Callaghan C. The effects of coronavirus on human nasal ciliated respiratory epithelium. Eur. Respir. J. 2001;18:965–970. doi: 10.1183/09031936.01.00093001. [DOI] [PubMed] [Google Scholar]
- Crotty S., Hix L., Sigal L.J., Andino R. Poliovirus pathogenesis in a new poliovirus receptor transgenic mouse model: age-dependent paralysis and a mucosal route of infection. J. Gen. Virol. 2002;83:1707–1720. doi: 10.1099/0022-1317-83-7-1707. [DOI] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L'Haridon R., Vogel L.K., Sjostrom H., Noren O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J., Wang H., Shen H., Qiu L., Li Z., Geng J., Cai J., Han H., Li X., Kang W., Weng D., Liang P., Jiang S. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., An B., Salvucci K.L., Storkus W.J., Amoscato A.A., Salter R.D. Modification of the amino terminus of a class II epitope confers resistance to degradation by CD13 on dendritic cells and enhances presentation to T cells. J. Immunol. 2000;164:129–135. doi: 10.4049/jimmunol.164.1.129. [DOI] [PubMed] [Google Scholar]
- Enjuanes L., Sanchez C., Gebauer F., Mendez A., Dopazo J., Ballesteros M.L. Evolution and tropism of transmissible gastroenteritis coronavirus. Adv. Exp. Med. Biol. 1993;342:35–42. doi: 10.1007/978-1-4615-2996-5_6. [DOI] [PubMed] [Google Scholar]
- Gredmark S., Britt W.B., Xie X., Lindbom L., Soderberg-Naucler C. Human cytomegalovirus induces inhibition of macrophage differentiation by binding to human aminopeptidase N/CD13. J. Immunol. 2004;173:4897–4907. doi: 10.4049/jimmunol.173.8.4897. [DOI] [PubMed] [Google Scholar]
- Hahm B., Arbour N., Oldstone M.B. Measles virus interacts with human SLAM receptor on dendritic cells to cause immunosuppression. Virology. 2004;323:292–302. doi: 10.1016/j.virol.2004.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M., Lely A., Navis G., Van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Kindig D.A., Mann J. Growth and intracellular development of a new respiratory virus. J. Virol. 1967;1:810–816. doi: 10.1128/jvi.1.4.810-816.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. Coronaviruses. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., editors. vol. 1. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 1187–1203. (Fields Virology). [Google Scholar]
- Horvat B., Rivailler P., Varior-Krishnan G., Cardoso A., Gerlier D., Rarourdin-Combe C. Transgenic mice expressing human measles virus (MV) receptor CD46 provide cells exhibiting different permissivities to MV infections. J. Virol. 1996;70:6673–6681. doi: 10.1128/jvi.70.10.6673-6681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S.A., Thaker H.M., Racaniello V.R. Transgenic mouse model for echovirus myocarditis and paralysis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15906–15911. doi: 10.1073/pnas.2535934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Usui S., Sugimura Y., Yoshida S., Hioki T., Tatematsu M., Yamamoto H., Hirano K. Aminopeptidase N regulated by zinc in human prostate participates in tumor cell invasion. Int. J. Cancer. 2001;92:49–54. [PubMed] [Google Scholar]
- Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., Demartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konikova E., Glasova M., Kusenda J., Babusikova O. Intracellular markers in acute myeloid leukemia diagnosis. Neoplasma. 1998;45:282–291. [PubMed] [Google Scholar]
- Larsson S., Soderberg-Naucler C., Moller E. Productive cytomegalovirus (CMV) infection exclusively in CD13-positive peripheral blood mononuclear cells from CMV-infected individuals: implications for prevention of CMV transmission. Transplantation. 1998;65:411–415. doi: 10.1097/00007890-199802150-00021. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J. Dendritic cell subsets and lineages, and their functions in innate and adaptive immunity. Cell. 2001;106:259–262. doi: 10.1016/s0092-8674(01)00456-1. [DOI] [PubMed] [Google Scholar]
- Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., Steinkasserer A., Becker S., Bates P., Hofmann H., Pohlmann S. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K. Coronaviruses in the limelight. J. Infect. Dis. 2005;191:489–491. doi: 10.1086/428510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., Steinman R.M. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- Moller E., Soderberg-Naucler C., Sumitran-Karuppan S. Role of alloimmunity in clinical transplantation. Rev. Immunogenet. 1999;1:309–322. [PubMed] [Google Scholar]
- Mrkic B., Pavlovic J., Rulicke T., Volpe P., Buchholz C.J., Hourcade D., Atkinson J.P., Aguzzi A., Cattaneo R. Measles virus spread and pathogenesis in genetically modified mice. J. Virol. 1998;72:7420–7427. doi: 10.1128/jvi.72.9.7420-7427.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N., Iwasaki T., Ami Y., Sato Y., Hatano I., Harashima A., Suzaki Y., Yoshii T., Hashikawa T., Sata T., Horiuchi Y., Koike S., Kurata T., Nomoto A. A poliomyelitis model through mucosal infection in transgenic mice bearing human poliovirus receptor, TgPVR21. Virology. 2004;321:87–100. doi: 10.1016/j.virol.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Naniche D., Oldstone M.B. Generalized immunosuppression: how viruses undermine the immune response. Cell. Mol. Life Sci. 2000;57:1399–1407. doi: 10.1007/PL00000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiesk S., Schneider-Schaulies J., Ohnimus H., Jassoy C., Schneider-Schaulies S., Diamond L., Logan J.S., ter M.V. CD46 expression does not overcome the intracellular block of measles virus replication in transgenic rats. J. Virol. 1997;71:7969–7973. doi: 10.1128/jvi.71.10.7969-7973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren O., Sjostrom H., Olsen J. Aminopeptidase N. In: Kenney A.J., Boustead C.M., editors. Cell-surface peptidases in health and disease. BIOS Scientific Publishers; Oxford: 1997. pp. 175–191. [Google Scholar]
- Oldstone M.B., Lewicki H., Thomas D., Tishon A., Dales S., Patterson J., Manchester M., Homann D., Naniche D., Holz A. Measles virus infection in a transgenic model: virus-induced immunosuppression and central nervous system disease. Cell. 1999;98:629–640. doi: 10.1016/s0092-8674(00)80050-1. [DOI] [PubMed] [Google Scholar]
- Rall G.F., Manchester M., Daniels L.R., Callahan E.M., Belman A.R., Oldstone M.B. A transgenic mouse model for measles virus infection of the brain. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4659–4663. doi: 10.1073/pnas.94.9.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R.B., Costantini F., Gorgacz E.J., Lee J.J., Racaniello V.R. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell. 1990;63:353–362. doi: 10.1016/0092-8674(90)90168-e. [DOI] [PubMed] [Google Scholar]
- Riemann D., Kehlen A., Langner J. CD13—not just a marker in leukemia typing [Review] Immunol. Today. 1999;20:83–88. doi: 10.1016/S0167-5699(98)01398-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L.H., Ashmun R.A., Roberts W.M., Look A.T. Separate promoters control transcription of the human aminopeptidase N gene in myeloid and intestinal epithelial cells. J. Biol. Chem. 1991;266:11999–12007. [PubMed] [Google Scholar]
- Spector D.L., Goldman R.D., Leinwand L.A. Cold Spring Harbor Laboratory Press; Plainview, NY: 1998. Cells a laboratory manual. 11803-2500. [Google Scholar]
- Summers K.L., Hock B.D., McKenzie J.L., Hart D.N. Phenotypic characterization of five dendritic cell subsets in human tonsils. Am. J. Pathol. 2001;159:285–295. doi: 10.1016/S0002-9440(10)61694-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuo H., Yanagi Y. The morbillivirus receptor SLAM (CD150) Microbiol. Immunol. 2002;46:135–142. doi: 10.1111/j.1348-0421.2002.tb02678.x. [DOI] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Thiel V., Karl N., Schelle B., Disterer P., Klagge I., Siddell S.G. Multigene RNA vector based on coronavirus transcription. J. Virol. 2003;77:9790–9798. doi: 10.1128/JVI.77.18.9790-9798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley B.R., Milland J., Christiansen D., Lanteri M.B., McInnes B., Moeller I., Rivailler P., Horvat B., Rabourdin-Combe C., Gerlier D., McKenzie I.F., Loveland B.E. Transgenic expression of a CD46 (membrane cofactor protein) minigene: studies of xenotransplantation and measles virus infection. Eur. J. Immunol. 1997;27:726–734. doi: 10.1002/eji.1830270322. [DOI] [PubMed] [Google Scholar]
- Tresnan D.B., Levis R., Holmes K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996;70:8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.C., Hemmila E.M., Beauchemin N., Holmes K.V. Receptor-dependent coronavirus infection of dendritic cells. J. Virol. 2004;78:5486–5490. doi: 10.1128/JVI.78.10.5486-5490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen J., Zheng A., Nie Y., Shi X., Wang W., Wang G., Luo M., Liu H., Tan L., Song X., Wang Z., Yin X., Qu X., Wang X., Qing T., Ding M., Deng H. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 2004;315:439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth D.E., Holmes K.V. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation. J. Virol. 2001;75:9741–9752. doi: 10.1128/JVI.75.20.9741-9752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead V.E., Stonehouse T.J., Binks M.H., Speidel K., Fox D.A., Gaya A., Hardie D., Henniker A.J., Horejsi V., Sagawa K., Skubitz K.M., Taskov H., Todd R.F., III, van Agthoven A., Katz D.R., Chain B.M. Novel molecular mechanisms of dendritic cell-induced T cell activation. Int. Immunol. 2000;12:1051–1061. doi: 10.1093/intimm/12.7.1051. [DOI] [PubMed] [Google Scholar]
- Yang Z.Y., Huang Y., Ganesh L., Leung K., Kong W.P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Racaniello V.R. Expression of the poliovirus receptor in intestinal epithelial cells is not sufficient to permit poliovirus replication in the mouse gut. J. Virol. 1997;71:4915–4920. doi: 10.1128/jvi.71.7.4915-4920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


