Abstract
The lectins DC-SIGN and DC-SIGNR augment infection by human immunodeficiency virus (HIV), Ebolavirus (EBOV) and other pathogens. The neck domain of these proteins drives multimerization, which is believed to be required for efficient recognition of multivalent ligands. The neck domain of DC-SIGN consists of seven sequence repeats with rare variations. In contrast, the DC-SIGNR neck domain is polymorphic and, in addition to the wild type (wt) allele with seven repeat units, allelic forms with five and six sequence repeats are frequently found. A potential association of the DC-SIGNR genotype and risk of HIV-1 infection is currently under debate. Therefore, we investigated if DC-SIGNR alleles with five and six repeat units exhibit defects in pathogen capture. Here, we show that wt DC-SIGNR and patient derived alleles with five and six repeats bind viral glycoproteins, augment viral infection and tetramerize with comparable efficiency. Moreover, coexpression of wt DC-SIGNR and alleles with five repeats did not decrease the interaction with pathogens compared to expression of each allele alone, suggesting that potential formation of hetero-oligomers does not appreciably reduce pathogen binding, at least under conditions of high expression. Thus, our results do not provide evidence for diminished pathogen capture by DC-SIGNR alleles with five and six repeat units. Albeit, we cannot exclude that subtle, but in vivo relevant differences remained undetected, our analysis suggests that indirect mechanisms could account for the association of polymorphisms in the DC-SIGNR neck region with reduced risk of HIV-1 infection.
Keywords: DC-SIGNR, DC-SIGN, Ebolavirus, Human immunodeficiency virus, Attachment factor, Dendritic cell, Lectin
Introduction
The C-type (i.e. calcium dependent) lectin DC-SIGN is expressed on dendritic cells (DCs) (Geijtenbeek et al., 2000b, Geijtenbeek et al., 2000c) and certain types of macrophages (Granelli-Piperno et al., 2005, Soilleux et al., 2001, Soilleux et al., 2002a). It plays a role in the establishment of cell–cell contact (Geijtenbeek et al., 2000a, Geijtenbeek et al., 2000c) and in the recognition and uptake of antigens (Engering et al., 2002, Moris et al., 2004), albeit the former function has recently been challenged (Snyder et al., 2005b). Antigens taken up by DC-SIGN can be processed for major histocompatibility complex (MHC)-mediated presentation (Engering et al., 2002, Moris et al., 2004). However, it has been proposed that a variety of viral and non-viral pathogens evade degradation upon DC-SIGN binding and employ this lectin to target DCs, thereby facilitating their dissemination in the infected host (van kooyk and geijtenbeek, 2003). Maybe most strikingly, HIV engages DC-SIGN on DCs to promote its spread to adjacent susceptible cells and possibly its transport into lymphoid tissue (Geijtenbeek et al., 2000b), a process that might be of particular relevance during the early phase of HIV infection.
The DC-SIGN related lectin DC-SIGNR (also termed L-SIGN) shares 77% amino acid identity with DC-SIGN and is expressed on endothelial cells in liver and lymph node sinusoids and in cells lining placental capillaries (Bashirova et al., 2001, Mummidi et al., 2001, Soilleux et al., 2002b, Pöhlmann et al., 2001c). DC-SIGNR interacts with the same spectrum of pathogens as DC-SIGN and might particularly promote spread of viruses that target liver and lymph nodes (Baribaud et al., 2002a). For example, binding to DC-SIGNR increases Ebolavirus (EBOV) infectivity (Alvarez et al., 2002, Simmons et al., 2003) and might augment EBOV entry into cells lining the sinusoids in lymph node and liver, which are major targets at the later stages of EBOV infection (Geisbert et al., 2003a, Geisbert et al., 2003b). DC-SIGNR might also enhance HIV infection of liver sinusoidal endothelial cells (LSECs), which are permissive (Steffan et al., 1992) and might constantly release progeny virions into the blood stream. Moreover, DC-SIGNR on LSECs might concentrate hepatitis C virus in the liver and might promote trans-infection of adjacent hepatocytes (Cormier et al., 2004, Gardner et al., 2003, Lozach et al., 2003, Lozach et al., 2004, Ludwig et al., 2004, Pöhlmann et al., 2003). Thus, DC-SIGN and DC-SIGNR (collectively referred to as DC-SIGN/R) could facilitate infection by several clinically relevant pathogens and constitute potential targets for therapeutic intervention.
DC-SIGN/R exhibit a comparable domain organization. The N-terminal domain is located in the cytoplasm and is followed by a transmembrane domain, which anchors the proteins in the cytoplasmic membrane. The extracellular domain consists of a short unique domain, a neck region formed by seven complete and one incomplete repeat of a 23 amino acid sequence and a carbohydrate recognition domain (CRD). While the CRD complexes carbohydrates of the high-mannose type (both DC-SIGN and DC-SIGNR) and fucose containing ligands (only DC-SIGN) (Feinberg et al., 2001, Guo et al., 2004), the neck region is essential for lectin tetramerization (Feinberg et al., 2005, Mitchell et al., 2001, Snyder et al., 2005a, Snyder et al., 2005b). Formation of DC-SIGN/R tetramers is believed to be important for high-affinity binding to multivalent ligands (Feinberg et al., 2005, Mitchell et al., 2001, Snyder et al., 2005a, Snyder et al., 2005b, Bernhard et al., 2004). The neck region of DC-SIGN usually comprises seven repeats, although rare exceptions have been identified (Liu et al., 2004). In contrast, the neck region of DC-SIGNR is polymorphic and can harbor between three to nine repeat units (Bashirova et al., 2001, Mummidi et al., 2001). The DC-SIGNR allele with seven repeat units is most common among Caucasians (54%) and is thus considered wild type (wt), followed by the alleles with five and six repeat units, which were detected in 29% and 12%, respectively, of the individuals analyzed in one study (Bashirova et al., 2001). Individuals can be heterozygous or homozygous for these DC-SIGNR alleles, in fact, subjects homozygous for wt DC-SIGNR or the allele harboring five repeat units are frequently found (29% and 16%, respectively, of individuals analyzed in one report; Lichterfeld et al., 2003). The number of repeat units can determine the lectin multimerization status (Feinberg et al., 2005, Snyder et al., 2005b) and the risk for HIV-1 infection (Liu et al., 2004, Liu et al., in press), albeit the latter finding is controversial (Lichterfeld et al., 2003). However, it is unknown if alleles with less than seven repeat units exhibit alterations in pathogen binding.
Here, we investigated if patient derived DC-SIGNR alleles with five and six repeat units augment infection by HIV-1 and by HIV-pseudotypes bearing the envelope glycoprotein of EBOV and the spike (S) protein of SARS-coronavirus (CoV). We report that DC-SIGNR alleles with five, six or seven repeat units bind to viral glycoproteins, enhance viral infection and form tetramers with comparable efficiency, indicating that the absence of two repeat units does not appreciably compromise DC-SIGNR interaction with pathogens. Also, coexpression of alleles with seven and five repeat units augmented viral infectivity to levels observed upon expression of each allele alone, suggesting that under conditions of robust lectin expression the potential formation of hetero-oligomers might not reduce pathogen engagement substantially.
Results
DC-SIGNR alleles with five and six repeat units are efficiently expressed at the cell surface and bind to viral glycoproteins
DC-SIGNR alleles with less than seven repeat units are frequently found in Caucasians (Bashirova et al., 2001); however, it is unclear if such alleles exhibit defects in pathogen binding. To address this question, we functionally analyzed published DC-SIGN/R constructs (Pöhlmann et al., 2001a, Pöhlmann et al., 2001c) containing seven repeat units and patient derived DC-SIGNR alleles harboring five, six and seven repeat units (Liu et al., 2005, Liu et al., in press) (Fig. 1 ). Except of the differences in the neck region, the DC-SIGNR variants analyzed exhibited identical amino acid sequences. We first investigated the importance of the number of repeat units in the DC-SIGNR neck region for lectin expression. Therefore, the lectin variants were transiently expressed in 293T cells and lectin expression was analyzed by FACS (Fig. 2 ). All lectins were expressed at the cell surface efficiently and to comparable degrees, indicating that the presence of five or six repeat units does not compromise DC-SIGNR expression.
Fig. 1.
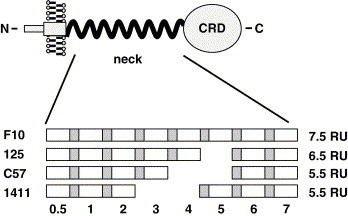
Schematic representation of the DC-SIGNR alleles analyzed. The neck domain of wt DC-SIGNR contains seven and a half repeat units (RU). Each repeat unit can be subdivided into two sequence blocks, indicated in grey and white. The allele F10 is wt, while allele 125 contains six and alleles C57 and 1411 contain five repeat units, respectively.
Fig. 2.
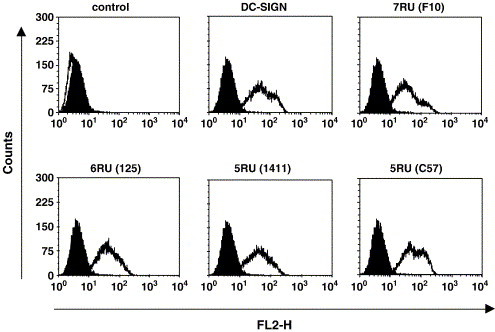
DC-SIGNR alleles with five and six repeat units are efficiently expressed at the cell surface. Published wt DC-SIGN and the indicated patient derived DC-SIGNR alleles were transiently expressed in 293T cells and surface expression analyzed by FACS using a DC-SIGN/DC-SIGNR-crossreactive monoclonal antibody. The black-filled histograms indicate staining of control cells, the white-filled histograms show staining of lectin expressing cells. Similar results were obtained in two independent experiments.
We next assessed if the DC-SIGNR variants bind to the glycoproteins of EBOV, SARS-CoV and HIV-1, all known ligands of wt DC-SIGNR. For these binding studies, chimeric proteins were employed in which the GP1 subunit of the EBOV glycoprotein, the S1 unit of the SARS-CoV-S protein or the gp120 subunit of the HIV-1 envelope protein were fused to the Fc portion of human immunoglobulin. These proteins, but not the corresponding immunoglobulin control protein, bound efficiently and to similar levels to cells expressing the DC-SIGNR variants, while no appreciable binding to control transfected cells was measured (Figs. 3A, B). Thus, the allelic DC-SIGNR forms with five and six repeat units are capable of complexing viral glycoproteins.
Fig. 3.
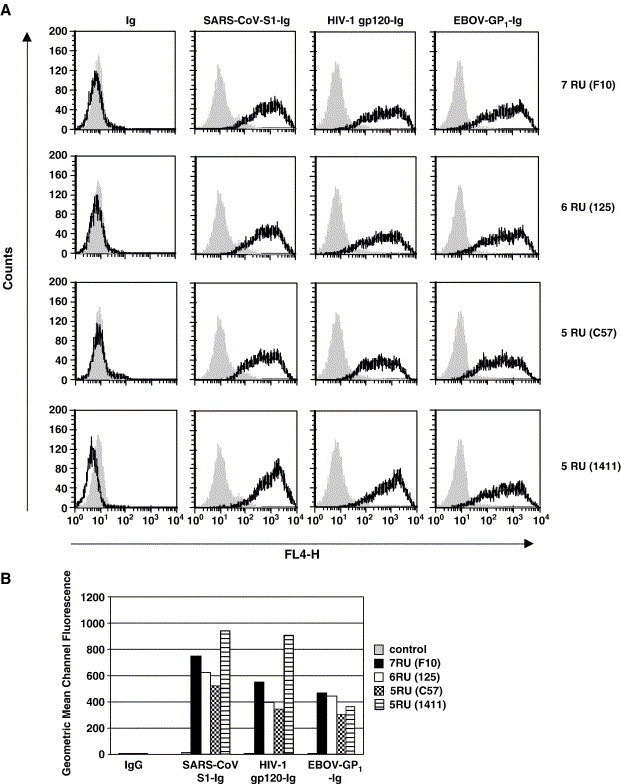
DC-SIGNR alleles with variations in the repeat region bind viral glycoproteins. (A) The indicated DC-SIGNR alleles or empty vector were transiently transfected into 293T cells and the cells incubated with concentrated supernatants containing comparable amounts of the indicated glycoproteins fused to the Fc portion of human immunoglobulin or the Fc portion of human immunoglobulin alone. Glycoprotein binding was analyzed by FACS. The grey-filled histograms indicate staining of control transfected cells, the black lines show staining of lectin expressing cells. Similar results were obtained in an independent experiment. Ig, Immunoglobulin. (B) The geometric mean channel fluorescence values measured in panel A are shown. An independent experiment yielded similar results.
Wild type DC-SIGNR and DC-SIGNR alleles with five and six repeat units show comparable augmentation of infection in cis and in trans
Since binding to viral glycoproteins must not necessarily lead to augmentation of infection (Baribaud et al., 2001, Pöhlmann et al., 2001b), we assessed if DC-SIGNR variants with variable repeat regions facilitate HIV transmission and promote infection by lentiviral reporter viruses pseudotyped with filovirus glycoproteins or SARS-CoV-S (so-called pseudotypes). These studies were again carried out with transiently transfected 293T cells. The 293T cell line lacks CD4 and is not permissive to HIV-1 infection; however, upon expression of DC-SIGN/R, these cells can bind and transfer virus to adjoining T cells (Pöhlmann et al., 2001c), a process termed infection in trans. Conversely, 293T cells are susceptible to pseudotypes bearing filovirus glycoproteins or SARS-CoV-S and infection is enhanced in the presence of DC-SIGN/R (Marzi et al., 2004), a process termed augmentation of infection in cis. To examine enhancement of infection in cis, the indicated lectins were transiently transfected into 293T cells and the cells inoculated with infectivity normalized luciferase reporter viruses bearing the glycoproteins of EBOV, Marburgvirus (MARV) or the S protein of SARS-CoV. In this experimental system, luciferase is only expressed upon successful membrane fusion, reverse transcription and integration of the proviral genome into the cellular chromosome. In agreement with previous results (Alvarez et al., 2002, Marzi et al., 2004, Simmons et al., 2003, Yang et al., 2004), expression of DC-SIGN/R enhanced filovirus glycoprotein and SARS-CoV-S dependent entry, but had no appreciable effect on infection driven by the glycoprotein of vesicular stomatitis virus (VSV-G) (Fig. 4A). Expression of all DC-SIGNR neck domain variants increased infectious entry to levels observed upon expression of the wt proteins (Fig. 4A), indicating that the presence of five repeat units in the neck region is sufficient for augmentation of infection in cis. To analyze lectin dependent infection in trans, 293T cells transiently expressing the indicated lectins were pulsed with HIV-1 NL4-3, washed and cocultivated with CEMx174 5.25 reporter T cells. Similarly to the results obtained upon infection with pseudotypes, wt DC-SIGN/R and all DC-SIGNR variants examined promoted trans infection with comparable efficiency (Fig. 4B), suggesting that the variations in the DC-SIGNR neck region analyzed do not affect the interaction with HIV-1.
Fig. 4.
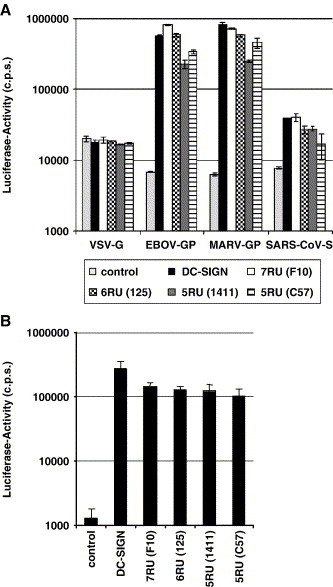
Wild type DC-SIGNR and DC-SIGNR alleles with five and six repeat units augment viral infectivity with comparable efficiency. (A) Enhancement of filovirus glycoprotein and SARS-CoV-S dependent infection. The indicated lectins were transiently expressed on 293T cells, the cells infected with lentiviral reporter viruses bearing the indicated glycoproteins and luciferase activities in cellular lysates determined. The results of a representative experiment performed in quadruplicates are shown, error bars indicate standard deviation (SD). Similar results were obtained in two independent experiments. (B) Augmentation of HIV infection. The indicated lectins were transiently expressed on 293T cells, the cells pulsed with replication competent HIV-1 reporter virus, washed, cocultivated with CEMx174 5.25 target cells and luciferase activities in the cocultures determined. The results of a representative experiment carried out in quadruplicates are shown and were confirmed in two independent experiments. Error bars indicate SD.
DC-SIGNR alleles with five and six repeat units multimerize
Considering the efficient augmentation of infection in cis and in trans, we hypothesized that the DC-SIGNR neck region variants were still capable of multimerization. To investigate this, lysates of transiently transfected 293T cells were prepared under reducing and non-reducing conditions and analyzed by SDS gel-electrophoresis followed by Western blot analysis for DC-SIGNR expression (Fig. 5 ). Only under non-reducing conditions, bands corresponding to the molecular weight of monomers, dimers, trimers and tetramers were observed for all lectins analyzed. Similar results were obtained when iodoacetamide was added before harvesting cells, indicating that multimer formation was not due to unspecific formation of disulfide bonds during preparation of cell lysates (data not shown). Thus, the loss of two repeat units does not compromise DC-SIGNR multimerization.
Fig. 5.
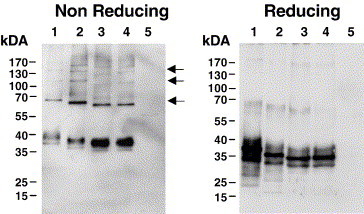
DC-SIGNR alleles with five or six repeat units multimerize. The DC-SIGNR alleles were transiently expressed in 293T cells, the cells lysed under non-reducing and reducing conditions, the lysates separated by SDS-gelelectrophoresis and lectin expression detected by Western blot. The results were confirmed in two independent experiments. Lane 1, 7RU (F10); lane 2, 6RU (125); lane 3, 5RU (C57); lane 4, 5RU (1411), lane 5, control. The arrows indicate bands of the size expected for DC-SIGNR multimers.
Coexpression of wt DC-SIGNR and allelic forms with five repeat sequences does not alter augmentation of viral infection compared to expression of each DC-SIGNR form alone
While DC-SIGNR variants with five repeat units augmented viral infection as efficiently as the wt DC-SIGNR, it is conceivable that coexpression of wt and allelic DC-SIGNR leads to formation of non-functional hetero-oligomers and thereby to reduced enhancement of viral infectivity. To address this question, we transfected 293T cells with wt DC-SIGNR, DC-SIGNR alleles with five repeat units or cotransfected cells with both wt and allelic DC-SIGNR in combination, and analyzed enhancement of EBOV glycoprotein driven infection and HIV-1 transfer to susceptible cells. Coexpression of DC-SIGNR with five and seven repeat units enhanced EBOV glycoprotein but not VSV-G dependent infection to similar levels as expression of each lectin alone (Fig. 6A). Similarly, cells coexpressing DC-SIGNR variants with five and seven repeat units transferred HIV to T cells as efficiently as cells expressing each lectin variant alone (Fig. 6B). Thus, coexpression of DC-SIGNR repeat variants might not lead to reduced interaction with pathogens, at least when cells are examined that express high levels of the lectins.
Fig. 6.
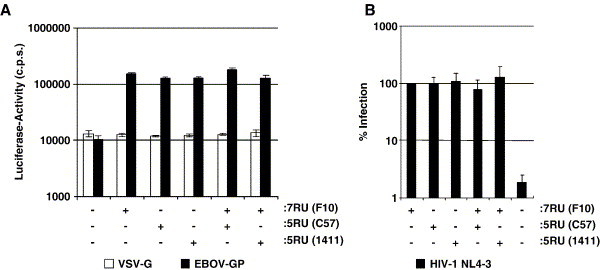
Coexpression of wt DC-SIGNR and DC-SIGNR alleles with five repeat units does not diminish augmentation of viral infectivity. (A) Augmentation of EBOV-GP dependent infection. The indicated lectins were transiently expressed on 293T cells, the cells infected with lentiviral reporter viruses bearing the indicated glycoproteins and luciferase activities in cell lysates determined. The results of a representative experiment performed in quadruplicates are shown, similar results were obtained in two independent experiments. Error bars indicate SD. (B) Augmentation of HIV-1 transfer. The indicated lectins were transiently expressed on 293T cells, the cells incubated with replication competent HIV-1 NL4-3 reporter virus, washed and cocultivated with CEMx174 5.25 target cells and luciferase activities in the cocultures determined. The average of six independent experiments performed with two separate plasmid preparations are shown. Error bars indicate standard error of the mean.
Discussion
Here, we show that DC-SIGNR alleles with five or six repeat units in the neck domain are expressed efficiently and augment viral infection similarly to wt DC-SIGNR, which contains seven repeat units. Moreover, coexpression of high levels of wt DC-SIGNR and allelic forms with five repeat units did not diminish augmentation of infection compared to cells expressing either lectin alone. While it is possible that minor but in vivo relevant differences between wt DC-SIGNR and the DC-SIGNR alleles tested were not detected in our experimental systems, our results do not hint towards a substantial defect of DC-SIGNR repeat region variants in pathogen capture. Therefore, indirect mechanisms might account, at least in part, for an association of these alleles with reduced risk of HIV-1 infection (Liu et al., 2004, Liu et al., in press).
The CRD domains of DC-SIGN/R recognize high-mannose carbohydrates on pathogens and are essential for pathogen binding (Feinberg et al., 2001, Guo et al., 2004, Pöhlmann et al., 2001a). The DC-SIGN/R neck domains form a stalk that projects the CRDs away from the cell surface (Feinberg et al., 2005, Guo et al., 2004, Snyder et al., 2005a). The neck domains are composed of repeat units, which contain helical regions (Feinberg et al., 2005). Lateral interactions between helices in the neck domains of different DC-SIGN/R molecules promote tetramerization (Feinberg et al., 2005), a process thought to be important for efficient binding to ligands displaying a high density of high-mannose glycans on the surface (Feinberg et al., 2005, Mitchell et al., 2001, Snyder et al., 2005b, Bernhard et al., 2004). It has been suggested that the N- and C-terminal repeat units of the DC-SIGN/R stalks might be particularly important for lectin multimerization (Feinberg et al., 2005). Consistent with these results, the patient derived DC-SIGNR alleles examined here exhibited deletions of internal repeats and were competent to form multimers, indicating that the deletion of the internal repeat units three and four or four and five does not interfere with DC-SIGNR oligomerization.
While variation in the DC-SIGN neck region is rare (Liu et al., 2004), DC-SIGNR alleles with five and six repeat units are frequently found, and both homozygous and heterozygous carriers have been identified (Bashirova et al., 2001, Lichterfeld et al., 2003, Liu et al., 2005, Liu et al., in press). At present, it is unclear, however, if the distribution of DC-SIGNR alleles differs between ethnic groups and if endogenous expression of these alleles on sinusoidal endothelial cells in liver and lymph node alters cellular functions or the interaction with pathogens. Our observation that the wt form of DC-SIGNR and DC-SIGNR alleles with five or six repeat units equally augment infectivity of HIV-1 and HIV-1 based pseudotypes bearing SARS-CoV-S or EBOV-GP, indicates that homozygosity for these alleles might not be beneficial against infection with viruses targeting DC-SIGNR.
It has been suggested, however, that coexpression of wt DC-SIGNR and allelic forms in cells of heterozygous carriers might result in formation of non-functional hetero-oligomers and thus in reduced expression of functional DC-SIGNR on the cell surface (Feinberg et al., 2005, Koppel et al., 2005). Since DC-SIGN dependent augmentation of infectivity strongly depends on the level of surface expression (Pöhlmann et al., 2001a), cells with reduced DC-SIGN/R expression might be less competent to augment infectivity and might confer some protection against infection with viruses that engage DC-SIGN/R. Indeed, a recent study on polymorphisms in the DC-SIGN promoter found a strong association between mutations that reduce DC-SIGN expression and protection against dengue fever (Sakuntabhai et al., 2005). Similarly, certain polymorphisms in the DC-SIGN promoter were reported to be associated with reduced risk of parenterally acquired HIV-1 infection (Martin et al., 2004), highlighting the importance of DC-SIGN expression levels for the efficient interaction with pathogens.
Under the experimental conditions employed, cells coexpressing wt DC-SIGNR and DC-SIGNR alleles with five repeat units were equally competent to augment infectivity as cells expressing each lectin alone. These observations can be explained in three ways. First, formation of hetero-oligomers might be inefficient, second, hetero-oligomers might be functional or third, under the conditions employed, expression levels were still saturating even after the formation of hetero-oligomers. Importantly, however, transfected 293T cells and monocyte derived DCs express roughly comparable amounts of DC-SIGN (Baribaud et al., 2002b, Pöhlmann et al., 2001a), and robust expression of DC-SIGNR on primary LSECs has been demonstrated (Bashirova et al., 2001). Thus, the experimental system employed to study the effects of coexpression of DC-SIGNR alleles on pathogen interactions might reflect the in vivo situation with virus exposed cells that express high amounts of DC-SIGNR. Such cells would be competent to bind pathogens in a lectin dependent manner and would not hamper dissemination of viruses that engage this lectin. However, low levels of DC-SIGNR might be expressed by cells at mucosal transmission sites (Liu et al., 2005) and might contribute to capture of sexually transmitted HIV-1. Under such circumstances, subtle differences in HIV-1 binding by mucosal cells from individuals homo- or heterozygous for wt DC-SIGNR might have profound impact on the establishment of HIV-1 infection, particularly if one assumes that HIV-1 completes several rounds of replication in the mucosal tissue during which these differences could be amplified.
It has been reported that variations in the DC-SIGNR neck domain are not associated with protection against HIV-1 infection or with slow progression to AIDS (Lichterfeld et al., 2003). In contrast, a different study found that homozygosity for wt DC-SIGNR is associated with increased risk for HIV-1 infection, while heterozygosity for DC-SIGNR (the seven/five repeat units genotype) seems to confer relative protection (Liu et al., in press). A likely explanation for this discrepancy are differences in the study populations, with the first study lacking a highly exposed seronegative cohort, a key group for such an analysis. In any case, it still needs to be determined if HIV-1 engages DC-SIGNR on endothelial cells of liver and lymph node sinusoids and if DC-SIGNR dependent binding to these cells augments viral infectivity. The observations that cultured LSECs express high levels of DC-SIGNR and are permissive to HIV-1 infection (Steffan et al., 1992) and that alternative DC-SIGNR transcripts are frequently found in genital mucosa (Liu et al., 2005) are in agreement with a role of DC-SIGNR in HIV-1 dissemination. An association between polymorphisms in the neck region and susceptibility to HIV-1 infection has also been documented for DC-SIGN (Liu et al., 2004); however, the number of individuals displaying such polymorphisms was low. Nevertheless, it will be interesting to investigate if DCs from these individuals show a reduced ability to transmit HIV-1.
In summary, we found no evidence that DC-SIGNR alleles with five or six repeat units show substantial defects in the interaction with viruses or that cells coexpressing high amounts of these alleles together with wt DC-SIGNR are less competent to augment viral infectivity compared to cells expressing wt DC-SIGNR alone. While these results do not support a beneficial role of DC-SIGNR alleles with less than seven repeat units in viral infection, it is possible that these alleles are in linkage disequilibrium with other polymorphisms, such as regulatory or structural polymorphisms in the DC-SIGNR and DC-SIGN genes, which could explain a protective effect against viral infection. Moreover, it cannot be excluded that we did not detect subtle effects that might nevertheless be relevant in vivo, particularly when being amplified during multiple rounds of viral replication. Finally, further studies need to determine whether the rarely found alleles with three, four, eight or nine repeat units are less adept in binding pathogens than the wt protein and whether these alleles are associated with relative protection against infection by HIV-1 and other viruses.
Material and methods
Cell culture
293T cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin and streptomycin. CEMx174 5.25 (Hsu et al., 2003) cells were maintained in RPMI 1640 containing 10% FBS, penicillin and streptomycin. All cells were grown at 37 °C and 5% CO2.
Plasmid construction
To generate vectors for expression of different DC-SIGNR isoforms, total RNA was extracted from monocyte derived DCs by Qiagen RNeasy Protect Kit (Qiagen, Valencia, CA) (Liu et al., 2005). Fragments spanning the entire DC-SIGNR coding region were reverse transcribed (RT) and then amplified by nested PCR as described (Liu et al., 2005). Specifically, the following primers were used: RT primer RR: 5′-GACGTTACAACATTTACCACTT-3′; first PCR primers RR and RF: 5′-AACATCTGGGGACAGCG-3′ (Soilleux et al., 2000); second PCR primers RcR: 5′-AGGCTAGCAGGGAAACAACT-3′ and RcF: 5′-TGGGGACAGCGGGAAAACAT-3′. For expression of soluble EBOV subtype Zaire glycoprotein (GP), the GP1 encoding part of the open reading frame (ORF) was fused to the Fc part of human IgG1 via PCR and cloned in frame with the amino-terminal murine IgG kappa signal peptide in the eukaryotic expression vector pAB61 (Birkmann et al., 2001). The same strategy was employed to generate an HIV-1 Env-Fc fusion construct, in which codon optimized JRFL gp120 was fused to Fc. The expression vector for the soluble S1 unit of the SARS-CoV S protein has been described previously (Hofmann et al., 2005). The glycoprotein expression plasmids employed for the generation of pseudotyped viruses have been described previously (Simmons et al., 2003). All PCR amplified sequences were confirmed by automated sequence analysis.
Expression analysis by flow cytometry
To assess cell surface expression of the DC-SIGNR variants, fluorescence activated cell sorting analysis (FACS) was performed. Transiently transfected 293T cells were harvested, washed and resuspended in ice-cold FACS buffer (PBS with 3% FBS; 0.01% NaN3). Approximately 2 × 105 cells were incubated with MAb anti-DCS 120526 (Jameson et al., 2002) at 10 μg/ml for 45 min on ice. Cells were washed and incubated with phycoerythrin-conjugated anti-mouse IgG (Vector, Burlingame, USA) at a final concentration of 10 μg/ml for 45 min on ice. Thereafter, the cells were washed, reconstituted in FACS buffer and analyzed by FACS employing a FACSCalibur flow cytometer (Becton Dickinson).
Binding of soluble viral glycoproteins to DC-SIGNR expressing cells
Soluble EBOV-GP-Ig, SARS-CoV-S-Ig or control-Ig were obtained by harvesting the supernatant of 293T cells 48 h after transient transfection with the respective expression vectors. The supernatants containing the fusion proteins were concentrated using Centricon Plus-20 centrifugal filters (Millipore, USA). To measure binding to DC-SIGNR variants, comparable amounts of soluble protein, as judged by Western blot, were incubated with transiently transfected 293T cells for 45 min on ice. After washing with FACS buffer, cells were stained with Cyan5-conjugated anti-human IgG (Jackson ImmunoResearch, USA) at a final concentration of 15 μg/ml for 45 min on ice. Cells were then washed and diluted in FACS buffer, binding was measured by flow-cytometry using a FACS Calibur flow cytometer (Becton Dickinson).
Production of lentiviral pseudotypes and analysis of DC-SIGNR-mediated enhancement of infection
EBOV-GP, SARS-CoV-S or VSV-G bearing lentiviral pseudotypes and replication competent HIV-1 NL4-3 luciferase reporter virus (NL4-3 Luc) were generated as described elsewhere (Hofmann et al., 2004, Pöhlmann et al., 2001a, Simmons et al., 2003). In brief, 293T cells were transiently cotransfected with pNL4-3 E−R− Luc (Connor et al., 1995) and an expression plasmid for EBOV-GP, SARS-CoV-S or VSV-G or transfected with NL4-3 Luc using the calcium phosphate method. The culture medium was replaced after 16 h and harvested 48 h post-transfection. The supernatants were passed through 0.4 μm filters and stored at −80 °C. To analyze the impact of DC-SIGNR expression on EBOV-GP, SARS-CoV-S or VSV-G driven infection, the indicated cell lines were seeded in 96-well plates and infected with the indicated pseudotypes. After overnight incubation, the infection medium was replenished and cells cultivated for 3 days before cells were lysed and luciferase activities determined using a commercially available kit (Promega, Madison, USA). Transfer of HIV-1 NL4-3 Luc by the different DC-SIGNR isoforms was assessed in a cocultivation assay. 293T cells were transiently transfected with the DC-SIGNR variants. After 48 h, cells were transferred in eppendorf tubes and incubated with 3 ng of luciferase reporter virus for 3 h at 37 °C. Thereafter, cells were washed with fresh RPMI medium and cocultivated with CEMx174 5.25 target cells in 96-well dishes. Three days after cocultivation, cells were lysed and luciferase activities determined as described above.
Western blot analysis of lectin multimerization
293T cells transiently expressing the indicated DC-SIGNR variants or control vector were lysed under reducing and non-reducing conditions. For reducing conditions, samples were dissolved in sodium dodecyl sulfate (SDS)-Laemmli buffer and boiled for 15 min at 95 °C, whereas under non-reducing conditions, cells were lysed in 10 mM Tris–HCl, 1 mM EDTA, 1 mM PMSF and 0.5% TritonX100 for 60 min on ice and diluted in SDS-Laemmli buffer without β-mercaptoethanol. Samples were separated via SDS gel-electrophoresis and transferred onto nitrocellulose membranes (Schleicher and Schuell, Dassel, Germany). The indicated lectins were detected by staining with the MAb DCN46 at a concentration of 1 μg/ml (Geijtenbeek et al., 2000b) and 0.3 μg/ml peroxidase-conjugated anti-mouse IgG (Jackson ImmunoResearch, USA). Chemiluminescence detection was performed according to the manufacturer's protocol (ECL Western detection kit; Amersham Pharmacia Biotech Europe, Freiburg, Germany).
Acknowledgments
We thank B. Fleckenstein und K. von der Mark for constant support. We also thank J.D. Reeves and F.H. Lee for codon optimized gp120 and N. Finze for p24-ELISA, and H. Zhu for preparation of DC-SIGNR clones. This work was supported by SFB 466 (T.G., H.H., A.M., A.W. and S.P.), Graduiertenkolleg 1071 (C.C.) and the National Institutes of Health Grants AI 56994 (T.Z.), AI 45402 (T.Z.) and AI 49109 (T.Z.).
References
- Alvarez C.P., Lasala F., Carrillo J., Muniz O., Corbi A.L., Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baribaud F., Pöhlmann S., Sparwasser T., Kimata M.T., Choi Y.K., Haggarty B.S., Ahmad N., Macfarlan T., Edwards T.G., Leslie G.J., Arnason J., Reinhart T.A., Kimata J.T., Littman D.R., Hoxie J.A., Doms R.W. Functional and antigenic characterization of human, rhesus macaque, pigtailed macaque, and murine DC-SIGN. J. Virol. 2001;75:10281–10289. doi: 10.1128/JVI.75.21.10281-10289.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baribaud F., Doms R.W., Pöhlmann S. The role of DC-SIGN and DC-SIGNR in HIV and Ebola virus infection: can potential therapeutics block virus transmission and dissemination? Expert Opin. Ther. Targets. 2002;6:423–431. doi: 10.1517/14728222.6.4.423. [DOI] [PubMed] [Google Scholar]
- Baribaud F., Pöhlmann S., Leslie G., Mortari F., Doms R.W. Quantitative expression and virus transmission analysis of DC-SIGN on monocyte-derived dendritic cells. J. Virol. 2002;76:9135–9142. doi: 10.1128/JVI.76.18.9135-9142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirova A.A., Geijtenbeek T.B., van Duijnhoven G.C., van Vliet S.J., Eilering J.B., Martin M.P., Wu L., Martin T.D., Viebig N., Knolle P.A., KewalRamani V.N., van Kooyk Y., Carrington M. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 2001;193:671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard O.K., Lai J., Wilkinson J., Sheil M.M., Cunningham A.L. Proteomic analysis of DC-SIGN on dendritic cells detects tetramers required for ligand binding but no association with CD4. J. Biol. Chem. 2004;279:51828–51835. doi: 10.1074/jbc.M402741200. [DOI] [PubMed] [Google Scholar]
- Birkmann A., Mahr K., Ensser A., Yaguboglu S., Titgemeyer F., Fleckenstein B., Neipel F. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 2001;75:11583–11593. doi: 10.1128/JVI.75.23.11583-11593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor R.I., Chen B.K., Choe S., Landau N.R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Cormier E.G., Durso R.J., Tsamis F., Boussemart L., Manix C., Olson W.C., Gardner J.P., Dragic T. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14067–14072. doi: 10.1073/pnas.0405695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engering A., Geijtenbeek T.B., van Vliet S.J., Wijers M., Van Liempt E., Demaurex N., Lanzavecchia A., Fransen J., Figdor C.G., Piguet V., van Kooyk Y. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J. Immunol. 2002;168:2118–2126. doi: 10.4049/jimmunol.168.5.2118. [DOI] [PubMed] [Google Scholar]
- Feinberg H., Mitchell D.A., Drickamer K., Weis W.I. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science. 2001;294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- Feinberg H., Guo Y., Mitchell D.A., Drickamer K., Weis W.I. Extended neck regions stabilize tetramers of the receptors DC-SIGN and DC-SIGNR. J. Biol. Chem. 2005;280:1327–1335. doi: 10.1074/jbc.M409925200. [DOI] [PubMed] [Google Scholar]
- Gardner J.P., Durso R.J., Arrigale R.R., Donovan G.P., Maddon P.J., Dragic T., Olson W.C. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4498–4503. doi: 10.1073/pnas.0831128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek T.B., Krooshoop D.J., Bleijs D.A., van Vliet S.J., van Duijnhoven G.C., Grabovsky V., Alon R., Figdor C.G., van Kooyk Y. DC-SIGN–ICAM-2 interaction mediates dendritic cell trafficking. Nat. Immunol. 2000;1:353–357. doi: 10.1038/79815. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B., Kwon D.S., Torensma R., van Vliet S.J., van Duijnhoven G.C., Middel J., Cornelissen I.L., Nottet H.S., KewalRamani V.N., Littman D.R., Figdor C.G., van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B., Torensma R., van Vliet S.J., van Duijnhoven G.C., Adema G.J., van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- Geisbert T.W., Hensley L.E., Larsen T., Young H.A., Reed D.S., Geisbert J.B., Scott D.P., Kagan E., Jahrling P.B., Davis K.J. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 2003;163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.W., Young H.A., Jahrling P.B., Davis K.J., Larsen T., Kagan E., Hensley L.E. Pathogenesis of Ebola hemorrhagic fever in primate models: evidence that hemorrhage is not a direct effect of virus-induced cytolysis of endothelial cells. Am. J. Pathol. 2003;163:2371–2382. doi: 10.1016/S0002-9440(10)63592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granelli-Piperno A., Pritsker A., Pack M., Shimeliovich I., Arrighi J.F., Park C.G., Trumpfheller C., Piguet V., Moran T.M., Steinman R.M. Dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin/CD209 is abundant on macrophages in the normal human lymph node and is not required for dendritic cell stimulation of the mixed leukocyte reaction. J. Immunol. 2005;175:4265–4273. doi: 10.4049/jimmunol.175.7.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Feinberg H., Conroy E., Mitchell D.A., Alvarez R., Blixt O., Taylor M.E., Weis W.I., Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M., Kuate S., Uberla K., Niedrig M., Pöhlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der H.L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M., Harouse J.M., Gettie A., Buckner C., Blanchard J., Cheng-Mayer C. Increased mucosal transmission but not enhanced pathogenicity of the CCR5-tropic, simian AIDS-inducing simian/human immunodeficiency virus SHIV(SF162P3) maps to envelope gp120. J. Virol. 2003;77:989–998. doi: 10.1128/JVI.77.2.989-998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson B., Baribaud F., Pöhlmann S., Ghavimi D., Mortari F., Doms R.W., Iwasaki A. Expression of DC-SIGN by dendritic cells of intestinal and genital mucosae in humans and rhesus macaques. J. Virol. 2002;76:1866–1875. doi: 10.1128/JVI.76.4.1866-1875.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel E.A., van Gisbergen K.P., Geijtenbeek T.B., van Kooyk Y. Distinct functions of DC-SIGN and its homologues L-SIGN (DC-SIGNR) and mSIGNR1 in pathogen recognition and immune regulation. Cell. Microbiol. 2005;7:157–165. doi: 10.1111/j.1462-5822.2004.00480.x. [DOI] [PubMed] [Google Scholar]
- Lichterfeld M., Nischalke H.D., van Lunzen J., Sohne J., Schmeisser N., Woitas R., Sauerbruch T., Rockstroh J.K., Spengler U. The tandem-repeat polymorphism of the DC-SIGNR gene does not affect the susceptibility to HIV infection and the progression to AIDS. Clin. Immunol. 2003;107:55–59. doi: 10.1016/s1521-6616(02)00050-5. [DOI] [PubMed] [Google Scholar]
- Liu H., Hwangbo Y., Holte S., Lee J., Wang C., Kaupp N., Zhu H., Celum C., Corey L., McElrath M.J., Zhu T. Analysis of genetic polymorphisms in CCR5, CCR2, stromal cell-derived factor-1, RANTES, and dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin in seronegative individuals repeatedly exposed to HIV-1. J. Infect. Dis. 2004;190:1055–1058. doi: 10.1086/423209. [DOI] [PubMed] [Google Scholar]
- Liu H., Hladik F., Andrus T., Sakchalathorn P., Lentz G.M., Fialkow M.F., Corey L., McElrath M.J., Zhu T. Most DC-SIGNR transcripts at mucosal HIV transmission sites are alternatively spliced isoforms. Eur. J. Hum. Genet. 2005;13:707–715. doi: 10.1038/sj.ejhg.5201409. [DOI] [PubMed] [Google Scholar]
- Liu, H., Carrington, M., Wang, C., Holte, S., Lee, J., Greene, B., Hladik, F., Koelle, D.M., Wald, A., Kurosawa, K., Rinaldo, C.R., Celum, C., Detels, R., Corey, L., McElrath, M.J., Zhu, T., in press. Effects of genetic polymorphisms in the DC-SIGNR repeat region on HIV-1 susceptibility. J. Infect. Dis. [DOI] [PubMed]
- Lozach P.Y., Lortat-Jacob H., de Lacroix d.L., Staropoli I., Foung S., Amara A., Houles C., Fieschi F., Schwartz O., Virelizier J.L., Arenzana-Seisdedos F., Altmeyer R. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 2003;278:20358–20366. doi: 10.1074/jbc.M301284200. [DOI] [PubMed] [Google Scholar]
- Lozach P.Y., Amara A., Bartosch B., Virelizier J.L., Arenzana-Seisdedos F., Cosset F.L., Altmeyer R. C-type lectins L-SIGN and DC-SIGN capture and transmit infectious hepatitis C virus pseudotype particles. J. Biol. Chem. 2004;279:32035–32045. doi: 10.1074/jbc.M402296200. [DOI] [PubMed] [Google Scholar]
- Ludwig I.S., Lekkerkerker A.N., Depla E., Bosman F., Musters R.J., Depraetere S., van Kooyk Y., Geijtenbeek T.B. Hepatitis C virus targets DC-SIGN and L-SIGN to escape lysosomal degradation. J. Virol. 2004;78:8322–8332. doi: 10.1128/JVI.78.15.8322-8332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.P., Lederman M.M., Hutcheson H.B., Goedert J.J., Nelson G.W., van Kooyk Y., Detels R., Buchbinder S., Hoots K., Vlahov D., O'Brien S.J., Carrington M. Association of DC-SIGN promoter polymorphism with increased risk for parenteral, but not mucosal, acquisition of human immunodeficiency virus type 1 infection. J. Virol. 2004;78:14053–14056. doi: 10.1128/JVI.78.24.14053-14056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., Steinkasserer A., Becker S., Bates P., Hofmann H., Pöhlmann S. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.A., Fadden A.J., Drickamer K. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 2001;276:28939–28945. doi: 10.1074/jbc.M104565200. [DOI] [PubMed] [Google Scholar]
- Moris A., Nobile C., Buseyne F., Porrot F., Abastado J.P., Schwartz O. DC-SIGN promotes exogenous MHC-I-restricted HIV-1 antigen presentation. Blood. 2004;103:2648–2654. doi: 10.1182/blood-2003-07-2532. [DOI] [PubMed] [Google Scholar]
- Mummidi S., Catano G., Lam L., Hoefle A., Telles V., Begum K., Jimenez F., Ahuja S.S., Ahuja S.K. Extensive repertoire of membrane-bound and soluble dendritic cell-specific ICAM-3-grabbing nonintegrin 1 (DC-SIGN1) and DC-SIGN2 isoforms. Inter-individual variation in expression of DC-SIGN transcripts. J. Biol. Chem. 2001;276:33196–33212. doi: 10.1074/jbc.M009807200. [DOI] [PubMed] [Google Scholar]
- Pöhlmann S., Baribaud F., Lee B., Leslie G.J., Sanchez M.D., Hiebenthal-Millow K., Munch J., Kirchhoff F., Doms R.W. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 2001;75:4664–4672. doi: 10.1128/JVI.75.10.4664-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöhlmann S., Leslie G.J., Edwards T.G., Macfarlan T., Reeves J.D., Hiebenthal-Millow K., Kirchhoff F., Baribaud F., Doms R.W. DC-SIGN interactions with human immunodeficiency virus: virus binding and transfer are dissociable functions. J. Virol. 2001;75:10523–10526. doi: 10.1128/JVI.75.21.10523-10526.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöhlmann S., Soilleux E.J., Baribaud F., Leslie G.J., Morris L.S., Trowsdale J., Lee B., Coleman N., Doms R.W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2670–2675. doi: 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöhlmann S., Zhang J., Baribaud F., Chen Z., Leslie G.J., Lin G., Granelli-Piperno A., Doms R.W., Rice C.M., McKeating J.A. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 2003;77:4070–4080. doi: 10.1128/JVI.77.7.4070-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuntabhai A., Turbpaiboon C., Casadémont I., Chuansumrit A., Lowhnoo T., Kajaste-Rudnitski A., Kalayanarooj S.M., Tangnararatchakit K., Vasanawathana S., Chaiyaratana W., Yenchitsomanus P., Suriyaphol P., Avirutnan P., Chokephaibulkit K., Matsuda F., Yoksan S., Jacob Y., Lathrop G.M., Malasit P., Desprès P., Julier C. Nat. Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Grogan C.C., Vandenberghe L.H., Baribaud F., Whitbeck J.C., Burke E., Buchmeier M.J., Soilleux E.J., Riley J.L., Doms R.W., Bates P., Pöhlmann S. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology. 2003;305:115–123. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- Snyder G.A., Colonna M., Sun P.D. The structure of DC-SIGNR with a portion of its repeat domain lends insights to modeling of the receptor tetramer. J. Mol. Biol. 2005;347:979–989. doi: 10.1016/j.jmb.2005.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder G.A., Ford J., Torabi-Parizi P., Arthos J.A., Schuck P., Colonna M., Sun P.D. Characterization of DC-SIGN/R interaction with human immunodeficiency virus type 1 gp120 and ICAM molecules favors the receptor's role as an antigen-capturing rather than an adhesion receptor. J. Virol. 2005;79:4589–4598. doi: 10.1128/JVI.79.8.4589-4598.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soilleux E.J., Barten R., Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J. Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- Soilleux E.J., Morris L.S., Lee B., Pöhlmann S., Trowsdale J., Doms R.W., Coleman N. Placental expression of DC-SIGN may mediate intrauterine vertical transmission of HIV. J. Pathol. 2001;195:586–592. doi: 10.1002/path.1026. [DOI] [PubMed] [Google Scholar]
- Soilleux E.J., Morris L.S., Leslie G., Chehimi J., Luo Q., Levroney E., Trowsdale J., Montaner L.J., Doms R.W., Weissman D., Coleman N., Lee B. Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J. Leukocyte Biol. 2002;71:445–457. [PubMed] [Google Scholar]
- Soilleux E.J., Morris L.S., Rushbrook S., Lee B., Coleman N. Expression of human immunodeficiency virus (HIV)-binding lectin DC-SIGNR: consequences for HIV infection and immunity. Hum. Pathol. 2002;33:652–659. doi: 10.1053/hupa.2002.124036. [DOI] [PubMed] [Google Scholar]
- Steffan A.M., Lafon M.E., Gendrault J.L., Schweitzer C., Royer C., Jaeck D., Arnaud J.P., Schmitt M.P., Aubertin A.M., Kirn A. Primary cultures of endothelial cells from the human liver sinusoid are permissive for human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1582–1586. doi: 10.1073/pnas.89.5.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kooyk Y., Geijtenbeek T.B. DC-SIGN: escape mechanism for pathogens. Nat. Rev., Immunol. 2003;3:697–709. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- Yang Z.Y., Huang Y., Ganesh L., Leung K., Kong W.P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


