Abstract
The nucleocapsid (N) protein is a structural component of severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) and can induce antibody responses in SARS patients during infection. However, it is not known whether SARS-CoV N protein can induce a long persistence of memory T-cell response in human. In this study, we found that peripheral blood mononuclear cells (PBMCs) from fully recovered SARS individuals rapidly produced IFN-γ and IL-2 following stimulation with a pool of overlapping peptides that cover the entire N protein sequence. The N-specific IFN-γ+CD4+ T cells were mainly composed of CD45RA−CCR7+CD62L− cells, whereas IFN-γ+CD8+ memory T cells were mostly contained within CD45RA+CCR7−CD62L− cell population. Epitope mapping study indicated that a cluster of overlapping peptides located in the C-terminal region (amino acids [aa] 331 to 362) of N protein contained at least two different T-cell epitopes. The results indicated that human memory T-cell responses specific for SARS-CoV N protein could persist for 2 years in the absence of antigen, which would be a valuable for the design of effective vaccines against SARS-CoV and for basic studies of human T-cell memory.
Keywords: Severe acute respiratory syndrome (SARS), N protein, Memory T cells, Epitope
Introduction
At the end of 2002, severe acute respiratory syndrome (SARS) emerged as a new epidemic form of life-threatening infectious disease (Drosten et al., 2003, Peiris et al., 2003). More than 8400 individuals were infected, and 800 of them died in approximately 7 months over 30 countries. The causative agent of severe acute respiratory syndrome (SARS) was identified as a new type of coronavirus, SARS coronavirus (SARS-CoV) (Falsey and Walsh, 2003, Peiris et al., 2003, Tsang et al., 2003). Although the first SARS epidemic has been successfully controlled, SARS still remains a potential threat to humans because the pathogenesis of SARS is not well understood and no effective approaches to prevent and treat the disease are developed.
As we know, SARS-CoV is a positive-sense RNA virus and the virion consists of a nucleocapsid core surrounded by an envelope containing three membrane proteins, spike (S), membrane (M) and envelope (E) (Marra et al., 2003, Tan et al., 2004). The N protein of CoV is a structural component of the helical nucleocapsid and plays an important role in viral pathogenesis, replication and RNA packaging (Hiscox et al., 2001, Narayanan et al., 2003). Furthermore, this protein is more conserved than other proteins of the virus, such as spike and membrane glycoproteins. Among all the coronavirus proteins, the N protein is the most abundant throughout infection at both mRNA and protein levels (Hiscox et al., 2001). Previous studies have shown that the N protein is one of the immunodominant antigens in the CoV family (Boots et al., 1992, Stohlman et al., 1994, Wesseling et al., 1993). In addition, it has been proved that cellular immune response against N protein of some animal coronavirus can generate protective effects (Collisson et al., 2000, Stohlman et al., 1995). Recently, several studies in animals demonstrated that the SARS-CoV N could induce specific T-cell responses (Gao et al., 2003, Jin et al., 2005, Kim et al., 2004, Zhu et al., 2004), as have been observed with other coronaviruses. Therefore, N protein may be an important target for SARS vaccine.
Evidence suggests that the most effective prevention method against a pathogen is vaccination. Immunological memory is the basis of vaccination. At present, most studies of immune memory after SARS-CoV infection focused on memory B cells. It is clear that antibody responses specific to SARS-CoV has been observed in all of SARS patients (Li et al., 2003) and can last up to 540 days after the onset of symptoms (Mo et al., 2005). However, little is known about the memory T lymphocyte responses in human SARS-CoV infection.
To explore the persistence and quality of T-cell memory to SARS-CoV in human, we investigated SARS-CoV N-protein-specific memory T-cell response in a group of Chinese individuals who had clinical infections with SARS-CoV 2 years earlier. We were able to demonstrate that SARS-CoV N-protein-specific memory CD4+ and CD8+ T cells were existed in all SARS donors and maintained for 2 years in the absence of pathogen. Furthermore, we report the detailed mapping of T-cell epitopes using an overlapping peptide library spanning the entire N protein. These results provide critical information for the design of effective vaccines against SARS-CoV and for basic studies of human T-cell memory.
Results
Memory T-cell response specific for SARS-CoV N protein persists for 2 years after recovery
To identify memory T-cell response to N protein after SARS-CoV infection in humans, PBMCs from seven individuals who had fully recovered from SARS 2 years after infection were stimulated with a pool of 57 peptides spanning the entire amino acid sequence of the N protein in vitro for 72 h. The culture supernatants were collected for detection of IFN-γ by ELISA. As shown in Fig. 1A, although the level of IFN-γ varied from donor to donor (ranged from 54 to 230 pg/ml), all of SARS-recovered donors were capable of producing IFN-γ in response to N peptides. However, the levels of IFN-γin normal individuals were less than 25 pg/ml.
Fig. 1.
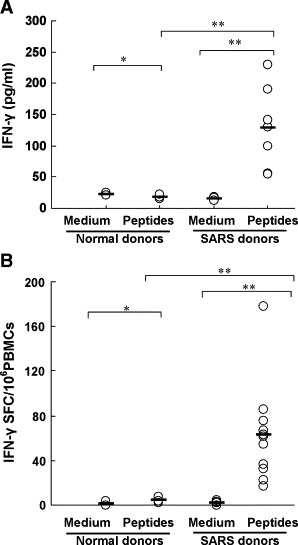
Production of IFN-γ by PBMCs from SARS-recovered individuals induced with a pool of SARS-CoV N peptides. (A) PBMCs from seven SARS-recovered donors were cultured in 96-well plates with or without a pool of N peptides for 72 h. The culture supernatants were collected and assessed for the production of IFN-γ by ELISA. All of assays were performed in triplicate. Statistical analysis used a Student's t test. Statistical significant difference was set at P < 0.05(**). *P > 0.05. Bars indicate mean values. (B) PBMCs from eleven SARS-recovered donors were stimulated with a pool of N peptides. IFN-γ-producing cells were detected by ELIspot assay. PBMCs incubated with medium alone were used as negative controls. The number of spots in the medium control wells ranged from 0 to 2. All of assays were performed in triplicate. Statistical analysis used a Student's t test. Statistical significant difference was set at P < 0.05(**). *P > 0.05. Bars represent mean values.
Next, we determined the frequency of N-protein-specific T cells by using IFN-γ ELIspot assay. Fig. 1B showed that the PBMCs from all of the 11 SARS-recovered individuals tested displayed significantly higher frequency of IFN-γ-producing cells than those from normal donors after stimulation with a pool of N peptides. The frequency of IFN-γ-producing cells from SARS-recovered individuals was at 63 SFC/million PBMCs, with a range of 10–178 SFC/million PBMCs. It is also noticed that there was no obvious difference in the frequencies of IFN-γ-producing T cells of the SARS donors and normal controls when PBMCs were stimulated with anti-CD3 and anti-CD28 mAbs (data not shown). These results demonstrated that the N protein of SARS-CoV was highly immunogenic and N-protein-specific T-cell responses could persist at least for 2 years after SARS-CoV infection.
Characterization of long-term memory T cells specific for N protein
To assess the cell populations involved in the N-specific T-cell response, PBMCs from six SARS-recovered donors were incubated with a pool of N peptides and stained for cell surface and intracellular IFN-γ expression (Fig. 2 ). The results showed that IFN-γ-producing CD8+ T cells specific to N protein were detected in all of the SARS-recovered donors with a mean frequency of 0.132% (range: 0.06%–0.23%). However, the frequency of IFN-γ-producing CD4+ T cells specific to N protein was lower than that of CD8+ T cells (mean: 0.072%, range: 0.05%–0.10%). These results demonstrated that both CD4+ and CD8+ T cells were involved in SARS-CoV N-specific immune responses and that CD8+ T cells were the major component of T-cell responses to SARS-CoV N-specific antigen.
Fig. 2.
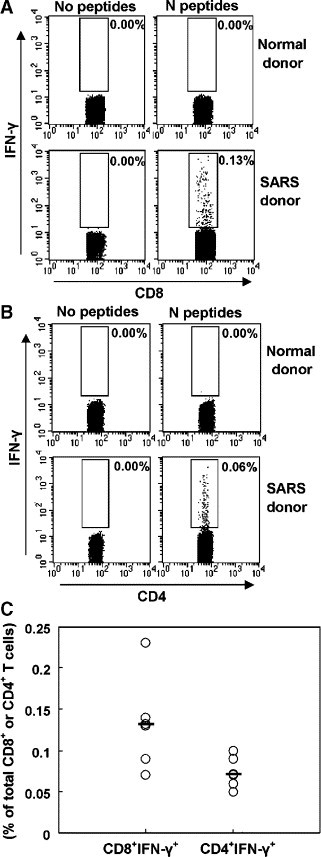
Detection of IFN-γ-producing T cells specific to SARS-CoV N peptides by intracellular staining. PBMCs from SARS-recovered donors were incubated with a pool of peptides for 6 h. Cell surface and intracellular cytokine staining for IFN-γ was performed. The cells were first gated on CD4−CD8+ T cells (A) and CD8−CD4+ T cells (B) and analyzed for IFN-γ expression. Results shown are from one experiment performed on PBMC from a SARS patient and are representative of 5 independent experiments from 5 SARS patients. C represents the expression of IFN-γ as percentage of total CD4+ or CD8+ T cells in different patients. Bars indicate mean values.
Besides studying IFN-γ, the function of memory T cells was also evaluated on the basis of their ability to secrete IL-2 by flow cytometry. In this experiment, PBMCs from SARS-recovered donors were stimulated with a pool of N peptides and were stained for intracellular IL-2 expression (Fig. 3 ). From Fig. 3A, it was seen that both of SARS-CoV N-protein-specific CD4+ and CD8+ memory T cells would produce IL-2. To establish the relationship between IL-2- and IFN-γ-producing T cells, co-staining for IFN-γ and IL-2 expression was carried out and analyzed by flow cytometry. The results indicated in Fig. 3B that T cells specific for N peptides of SARS-CoV could be divided into three subsets based on IL-2 and IFN-γ expression: single IFN-γ-secreting cells; co-expression of IL-2 and IFN-γ cells; and single IL-2-secreting cells. The majority of T cells specific for N peptides were single IFN-γ-secreting cells. Compared to CD8+ T cells, higher frequency of CD4+ T cells could express IL-2. Co-expression of IL-2 and IFN-γ cells was rare in both CD4+ and CD8+ T cells (Fig. 3C).
Fig. 3.
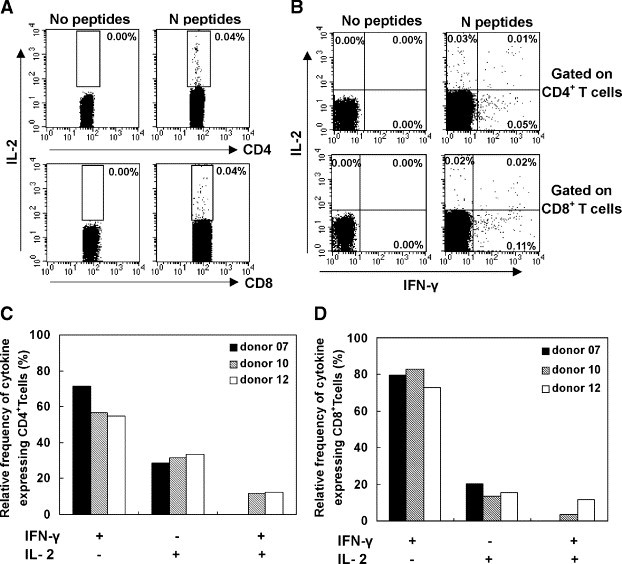
Cytokine profiles of IL-2 and IFN-γ in CD4+ and CD8+ T cells in response to N peptides. PBMCs from SARS-recovered donors were stimulated with a pool of N peptides for 6 h. The expression of IL-2 and IFN-γ was assessed by flow cytometry. (A) Dot plots show intracellular staining for IL-2 and IFN-γ in the CD8+ T cells and CD4+ T cells. Data are representative of three independent experiments with similar results. (B) The frequency of cells expressing both IL-2 and IFN-γ in PBMCs from SARS-recovered donors. Data are presented as the percentage of the total number of cells expressing at least one cytokine.
Additionally, the phenotypic characteristics of N-protein-specific CD4+ and CD8+ T cells were also investigated here. After the stimulation with N peptides, PBMCs from three SARS-recovered donors were stained with anti-CD4, CD8, IFN-γ, CD45RA, CCR7 and CD62L (Fig. 4 and Table 1 ). The results showed that most of IFN-γ+CD4+ T cells expressed CCR7 (71.4–91.3%) but did not express CD45RA (73.7–92%) and CD62L (75–76.6%). Compared with IFN-γ+CD4+ T cells, most of IFN-γ+CD8+ T cells expressed CD45RA (57.8–73%) but did not express CCR7 (61.9–87.7%) and CD62L (90.4–97.4%). These results suggested that IFN-γ+CD4+T cells should be mostly contained within the CD45RA−CCR7+CD62L− cell population, whereas IFN-γ+CD8+ T cells should be mostly contained within the CD45RA+CCR7−CD62L− cell population. Our results showed that a relatively larger number of IFN-γ+CD8+ T cells were CD45RA+CCR7− compared with IFN-γ+CD4+ T cells, suggesting the difference of phenotype between CD4+ and CD8+ memory T cells and also demonstrating the heterogeneity of memory T cells.
Fig. 4.
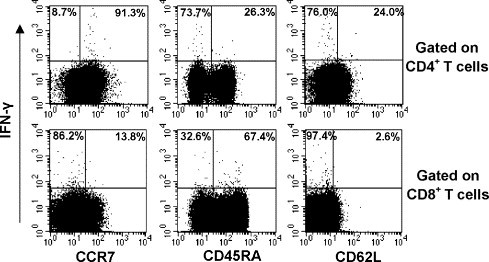
Determination of phenotype of N-protein-specific memory T cells in SARS-recovered donors. PBMCs from SARS-recovered donors were stimulated with a pool of N peptides for 6 h. The expression of CD45RA, CCR7 and CD62L in IFN-γ+CD4+ or IFN-γ+CD8+ T cells was assessed by flow cytometry. CD4 +CD8− or CD4 +CD8− cells were gated and analyzed. Results shown are from one experiment performed on PBMC from a SARS patient and are representative of 3 independent experiments from 3 SARS patients.
Table 1.
Phenotypic analysis of SARS-CoV N-specific memory T cells from donors who have recovered from SARS 2 years after infection
| Donors | IFN-γ+CD4+ T |
IFN-γ+CD8+ T |
||||
|---|---|---|---|---|---|---|
| CCR7+ | CD45RA+ | CD62L+ | CCR7+ | CD45RA+ | CD62L+ | |
| 07 | 71.4 | 18.0 | 23.4 | 12.3 | 57.8 | 5.2 |
| 12 | 76.0 | 8.0 | 25.0 | 38.1 | 73.0 | 9.6 |
| 13 | 91.3 | 26.3 | 24.0 | 13.8 | 67.4 | 2.6 |
Mapping of dominant epitopes in N protein
To identify dominant epitopes in N protein, 57 individual peptides were divided into six pools: pool I (N 1 to N 10), pool II (N 11 to N 20), pool III (N 21 to N 30), pool IV (N 31 to N 40), pool V (N 41 to N 50) and pool VI (N 51 to N 57). PBMCs from five SARS-recovered donors were stimulated with these six peptide pools individually for 72 hour, and the levels of IFN-γ in culture supernatants were measured by ELISA. The results showed that PBMCs from all of five SARS-recovered individuals had specific T-cell responses to one or more peptide pools based on the production of IFN-γ. But the hierarchy of epitope recognition varied from donor to donor. As shown in Fig. 5A, 100% and 60% of the donors responded to pool V and pool IV corresponding to the sequences of amino acid (aa) residues 293–376 and 219–302 respectively, whereas frequencies of other pools were below 50%. Similar results were also confirmed by the detection of IFN-γ-secreting cells using ELIspot assay (Fig. 5B). The results showed that all of four SARS donors responded to pool V and the frequency of IFN-γ-producing T cells in response to pool V was the highest among six peptide pools. This result suggested that the pool V contained major immunodominant epitopes corresponding to the sequences of residues 293–376 in N protein. Therefore, the pool V was chosen for further mapping the individual peptide responsible for the induction of IFN-γ expression.
Fig. 5.
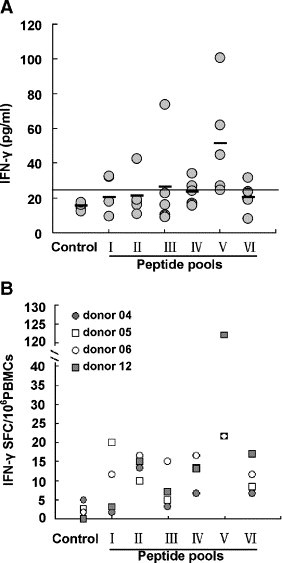
Specific T-cell responses to pooled peptides of the N protein in SARS-recovered donors by ELISA and ELIspot assay. Fifty seven overlapping individual peptides that covered the entire N protein sequence were divided into 6 pools (each pool contains 10 peptides) as described in Materials and methods. (A) PBMCs from 5 SARS-recovered donors were stimulated with a pool of 10 peptides (1 μg/ml of each peptide) for 72 h. The culture supernatants were collected for detection of IFN-γ by ELISA. Bars indicate mean values. (B) PBMCs from four SARS-recovered donors were stimulated with a pool of 10 peptides (1 μg/ml of each peptide) for detection of IFN-γ expression by ELIspot assay. PBMCs incubated with media alone were used as negative controls. The experiments were carried out in triplicate.
In the following, PBMCs from SARS-recovered donors were tested for a single peptide screening from 10 overlapping peptides in pool V by IFN-γ ELIspot assay. Although positive responses were obtained with all of the peptides in one or more donors, the highest responses (ranging from 62 to 85%) were obtained with peptides N46 (aa 331 to 347), N47 (aa 339 to 354) and N48 (aa 346 to 362), whereas the rest of peptides in pool V exhibited lower responses (20%–44%) (as shown in Table 2 ). Therefore, aa 331 to 362 peptide should be the major dominant antigenic site of N protein and contain at least two different T-cell epitopes (N46: aa 331–347 and N48: aa 346–362). Based on ELIspot assay, IFN-γ-producing T cells specific to these two epitopes were 25.7% of total N-protein-specific IFN-γ-producing T cells in donor 13, 58.4% in donor 7, 38.4% in donor 10 and 65% in donor 12, respectively (as shown in Fig. 6 ). Therefore, these peptides would be chosen for further characterization.
Table 2.
Production of IFN-γ by PBMCs from SARS-recovered donors in response to single SARS-CoV N peptide by ELIspot
| Peptides | Peptide position in protein | Number of donors tested | Positive response (%) |
|---|---|---|---|
| N41 | 293–309 | 4 | 25 |
| N42 | 300–316 | 5 | 40 |
| N43 | 307–323 | 5 | 20 |
| N44 | 315–331 | 4 | 20 |
| N45 | 323–339 | 6 | 17 |
| N46 | 331–347 | 7 | 85 |
| N47 | 339–354 | 8 | 62 |
| N48 | 346–362 | 8 | 75 |
| N49 | 354–370 | 5 | 40 |
| N50 | 362–376 | 9 | 44 |
The peptides of pool V were used to scan SARS-recovered donors by IFN-γ ELIspot. The number of spots in the negative control wells ranged from 0 to 2.
Fig. 6.
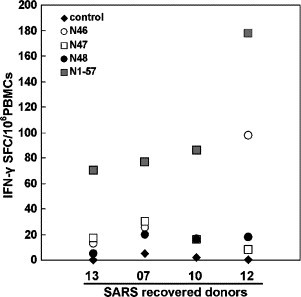
T-cell responses to individual peptide of SARS-CoV N protein by ELIspot assay. PBMCs from SARS-recovered donors were stimulated with a single peptide (1 μg/ml). The production of IFN-γ was assessed by ELIspot assay. The number of spots in the media control wells was ranged from 0 to 2. The experiments were carried out in triplicate.
IFN-γ response by CD4+ and CD8+ T cells for recognition with individual peptide
As described above, peptides of N46, N47 and N48 could effectively stimulate T-cell responses when detected with ELIspot assay. Next, we would discuss the response of CD4+ and CD8+ T subpopulations for recognition of individual peptide by flow cytometry. Freshly isolated PBMCs from SARS-CoV-recovered donors were analyzed for IFN-γ expression after stimulation for 6 h in the presence or absence of peptides N46, N47 or N48, respectively. CD8+ T and CD4+ T cells were gated and analyzed by flow cytometry (Fig. 7A). The frequency of SARS-CoV N-peptide-specific CD8+ T cells induced by the selected peptide was ranged from 0.02 to 0.4% of CD8+ T lymphocytes (Fig. 7B). However, much lower frequencies of specific IFN-γ-producing CD4+ T cells ranging from 0.02 to 0.29% of CD4+ T lymphocyte were detected (Fig. 7C). These data indicated that the individual peptides N46, N47 or N48 were able to induce both CD4+ and CD8+ T-cell subsets with a predominance of CD8+ T-cell response.
Fig. 7.
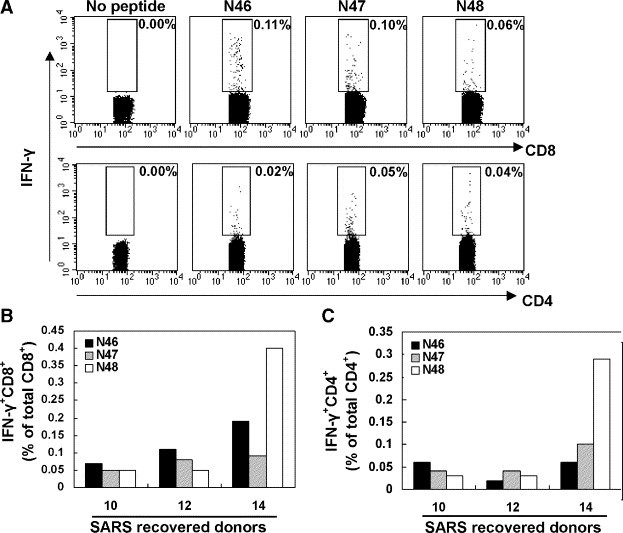
Memory T-cell responses specific to the peptides N46, N47 or N48 by intracellular staining. PBMCs from SARS-recovered donors were incubated with the indicated peptides (N46, N47 or N48) for 6 h. The intracellular staining for IFN-γ was performed. CD4+ and CD8+ T cells were gated and analyzed for IFN-γ expression by flow cytometry (A). Results shown are representative of 3 independent experiments from 3 SARS patients. Similar results were also obtained in other experiments for CD8+ (B) and CD4+ (C) T cells.
Discussion
In this study, we analyzed memory T-cell responses in a group of Chinese individuals who had clinical SARS-CoV infection between January 2003 to May 2003. We were able to demonstrate SARS-CoV N-protein-specific T-cell response in all donors, and the memory T cell specific for SARS-CoV could persist for 2 years in the absence of antigen. The N protein was chosen because it was the most conserved protein in SARS-CoV, and it was previously reported to be a major target for the CTL memory response (Collisson et al., 2000, Stohlman et al., 1995). Some studies in animals also demonstrated that the SARS-CoV N could induce specific T-cell responses (Gao et al., 2003, Jin et al., 2005, Kim et al., 2004, Zhu et al., 2004). In addition, our results demonstrated that both CD4+ and CD8+T cells were participated in SARS-CoV N-specific memory immunity with a predominance of CD8+ T-cell memory responses in humans. Therefore, the N protein of SARS-CoV would be highly immunogenic and could be an important target for SARS vaccine.
Perhaps the most important finding of this study is that N-specific memory CD4+ and CD8+ T cells can persist in individuals who had clinical infections with SARS-CoV 2 years ago, in the absence of re-exposure to this pathogen. There is no evidence of persistent SARS-CoV infection, and clinical reinfections with SARS-CoV have never been reported. Viral RNA can be easily detected within the first 14 days of symptomatic SARS-CoV infection, but convalescent samples (50 days after the onset of symptoms) are consistently PCR negative (Xu et al., 2005). Therefore, the maintenance of N-protein-specific T-cell memory in vivo appears to be antigen-independent.
There are some debates regarding the role of antigen in the maintenance of memory T cells. On one hand, periodic re-exposure to the pathogen which serves as a natural “booster” to the immune system is an effective way to maintain high levels of immunity though such reinfections are usually asymptomatic or only produce mild clinical symptoms. On the other hand, it has been already clear that memory CD4+ T cells and memory CD8+ T cells could persist in the absence of antigen in mice (Murali-Krishna et al., 1999, Lau et al., 1994). Furthermore, memory CD4+ T cells and CD8+ T cells could be maintained for decades after smallpox vaccination in human (Hammarlund et al., 2003). Since vaccinia virus does not cause a chronic infection in humans and re-exposure to the virus is unlikely, the maintenance of T-cell memory in this system also appears to be antigen-independent. Therefore, both antigen-dependent and antigen-independent mechanisms should be involved in sustaining immunological memory.
Previous studies proposed that memory T cells could be divided into two functionally distinct subsets based on expression of CCR7 (Sallusto et al., 1999, Seder and Ahmed, 2003). In these two subsets, CCR7− effector memory T cells (TEM cells) would be present in the blood, spleen and nonlymphoid tissues, whereas CCR7+ central memory T cells (TCM cells) would be present in lymphoid, spleen and blood but not in nonlymphoid tissues. Recent studies of both in mice and humans have demonstrated that both cell subsets can rapidly produce IFN-γ and TNF-α but that IL-2 production remains a property of TEM cells following stimulation with cognate antigen (Harari et al., 2005, Lier et al., 2003, Wherry and Ahmed, 2004, Wills et al., 2002). Our results demonstrated that SARS-CoV-specific memory T cells from SARS-recovered donors rapidly produced IFN-γ and IL-2 following a short-term stimulation with N peptides. IL-2-secreting CD4+ memory T cells were more than those of CD8+ memory T cells. Phenotypic analysis suggested that a higher proportion of IFN-γ+CD8+ T cells were CD45RA+CCR7− compared with those of IFN-γ+CD4+ T cells. These larger numbers of IFN-γ+ CD8+ T cells were TEM cells and produced less IL-2 compared with those of IFN-γ+CD4+ T cells. Similar observations were also obtained in CMV infection in which reactive CD8+ T cells expressed CD45RA during recovery (Wills et al., 2002) and a larger number of IFN-γ+CD8+ T cells were CD45RA+ CCR7− compared with those of IFN-γ+CD4+ T cells (Seder and Ahmed, 2003).
As the T-cell epitopes are usually 8 to 10 aa long, the N peptides used in our study were overlapped by 10 aa in each peptide to minimize the possibility of missing the T-cell epitopes of SARS N protein. The clear difference in protein recognition among SARS-CoV immune donors suggests that the nature and composition of the immune response against the virus vary between individuals possibly due to the difference in HLA types. Although the results showed that the T-cell epitopes were scattered throughout the sequence of N protein, some peptides were recognized more frequently than others. The peptides that were frequently recognized by T cells were N46 (aa 331 to 347), N47 (aa 339 to 354) and N48 (aa 346 to 362). Therefore, a cluster of peptides corresponding to aa 331 to 362 were the major dominant antigenic site on N protein and contained at least two different T-cell epitopes (N46 and N48). IFN-γ-producing T cells specific to these two epitopes were from 25.7% to 65% of total N-protein-specific IFN-γ-producing T cells in four SARS donors based on ELIspot assays.
Although SARS-CoV N protein has previously been tested for induction of T-cell responses in animals (Gao et al., 2003, Jin et al., 2005, Kim et al., 2004, Zhao et al., 2005, Zhu et al., 2004), our data provide the first evidence in which the T-cell epitopes of N protein recognized by human T cells were mapped. Our results suggest that the C-terminal end of N protein may be highly immunogenic. This had been confirmed by Kim et al. who observed a strong T-cell response to the N peptides at aa 346 to 354 when mice were immunized with N protein (Kim et al., 2004). These peptides were overlapped with the peptide N47 and peptide N48 in our study. Like other CoVs, SARS-CoV N proteins clearly contain multiple immunodominant epitopes and antigenic sites (Stohlman et al., 1994). Several groups have been extensively studied for B-cell epitopes of N protein in SARS patients (Chen et al., 2004, He et al., 2004, Liang et al., 2005, Lin et al., 2003, Zhong et al., 2005). He and his co-workers demonstrated that two major immunodominant epitopes that reacted with more than 75% of sera from SARS patients were localized at the C-terminal and middle regions, corresponding to aa 362 to 412 and 153 to 178, respectively (He et al., 2004). This suggests that B cells and T cells target to distinct regions of the N protein during the immune response to SARS-CoV. This may be an important consideration in the design of effective vaccines capable of eliciting both humoral and cellular responses.
The frequencies of SARS-CoV-specific memory T cells reported here are comparable to the frequencies of T cell specific for dominant influenza A virus epitopes (Lalvani et al., 1997). In a previous study, the highest frequencies of memory T cells specific for dominant influenza A virus epitopes on the M1 and NP proteins ranged from 9 to 286/106 PBMCs and from 15 to 67/106 PBMCs, respectively (Lalvani et al., 1997, Jameson et al., 1998). The highest frequencies of SARS-CoV N46-specific T cells measured in our study were 12–90/106 PBMCs. Influenza virus can periodically reinfect individuals with existing influenza virus-specific T-cell memory and thus boost T-cell responses. However, periodic exposures to SARS-CoV have never occurred and secondary infections have never been reported for this. Thus, the memory T-cell response of SARS-CoV is unlikely to be effectively boosted. Overall, the frequencies of SARS-CoV-specific memory T cells measured in our study are comparable to those measured in other acute virus infections such as influenza. This system could provide a model system of T-cell memory to study the maintenance of antiviral T-cell memory in humans after an acute infection in the absence of antigen.
In conclusion, our study has demonstrated that both CD4+ and CD8+ T cells are involved in SARS-CoV N-specific memory immunity and that the memory T-cell responses specific for SARS-CoV have been maintained for 2 years in the absence of antigen. We also find that peptide 331–362 is the most dominant antigen site of the N protein and contains at least two different T-cell epitopes (N46: aa 331–347 and N48: aa 346–362). The identification of specific T-cell responses in SARS patients is essential for understanding the mechanism involved in recovery from infection and the pathology of infection. This information should be useful for the design of effective vaccines against SARS-CoV and for fundamental studies of human T-cell memory.
Materials and methods
Subjects
Fourteen recovered SARS individuals (7 men and 7 women, aged 20 to 37) were obtained from the Second Affiliated Hospital of Sun Yat-sen University and Guangdong Provincial Hospital of Traditional Chinese Medicine, Guangzhou, Guangdong, China, respectively. All of the donors had been diagnosed as SARS patients based on clinical examination during the period of January to May 2003. The diagnostic criteria for SARS-CoV infection followed the World Health Organization definition of SARS (Hoey, 2003). The diagnosis of SARS-CoV infection was further confirmed by serological detection of SARS-CoV-specific antibodies (Huang et al., 2005, Wu et al., 2004). Three normal subjects without any contact history with SARS patients were used as controls.
Synthetic peptides
Fifty-seven synthetic peptides that spanned the entire sequence of the SARS-CoV N protein were kindly provided by Drs. Koup and Bailer at the Vaccine Research Center of National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), USA. The peptides were 15–20 mers overlapped by 10 amino acids and named as N1 through N57. In order to map epitopes, 57 peptides were divided into six pools: pool I, N1 to N10; pool II, N11 to N20, pool III, N21 to N30; pool IV, N31 to N40; pool V, N41 to N50; and pool VI, N51 to N57. These pools and single peptide were used in the subsequent experiments.
Isolation of PBMCs
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood by Ficoll-Hypaque density gradient centrifugation. Cells were washed twice with RPMI 1640 (GiBco) and suspended in complete culture medium (RPMI 1640 containing 10% fetal calf serum, 100 U of penicillin per ml, 100 mg of streptomycin per ml).
IFN-γ assay by ELISA
PBMCs were seeded into the wells of 96-well culture plates (Becton Dickinson) (2 × 105 cells/well) in triplicate. Peptides and costimulatory mAbs to CD28 (BD Pharmingen) and CD49d (BD Pharmingen) were added each at 1 μg/ml to the wells. In addition, PBMCs stimulated with mAbs to CD3 and CD28 were used as positive controls and PBMCs unstimulated as negative controls. The plates were incubated for 72 h at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air. Supernatants were collected, and the level of IFN-γ was measured by ELISA kit (R&D) according to the manufacturer's instruction. The detection limit of the IFN-γ assay kit was 15 pg/ml.
IFN-γ ELIspot
ELIspot assay kit for IFN-γ was purchased from BD Biosciences and performed as described (Lalvani et al., 1997). Briefly, 96-well plates (Millipore) were coated with anti-IFN-γ mAb at 4 °C overnight. The plates were washed three times before blocking with complete culture medium. Fresh PBMCs were plated at 2 × 105 cells per well in triplicate. Peptides and costimulatory mAbs to CD28 and CD49d were added each at 1 μg/ml. PBMCs stimulated with CD3 and CD28 (1 μg/ml) mAbs were used as positive controls, and PBMCs were not stimulated as negative controls. After incubation for 20 h at 37 °C, the cells were removed and incubated with biotinylated anti-human IFN-γ detection antibody for 2 h at room temperature. After washing, wells were developed for 1 h with streptavidin–HRP and incubated with substrate reagent according to the manufacturer's protocol. Spot-forming cells (SFC) were detected with ELIspot image analysis system (Sage Creation). The frequency of IFN-γ-producing cells was calculated as the number of spots/number of total PBMCs per well and was adjusted as the number of IFN-γ-producing cells/106 PBMCs. The number of spots in negative control wells was at a range of 0–2 spots.
Intracellular cytokine staining and flow cytometry
Intracellular IFN-γ and IL-2 expression was assessed as previously described (Hoffmeister et al., 2003). 1 × 106 cells (PBMCs) were placed into polystyrene tissue culture tubes (Becton Dickinson) with 1 ml of complete medium (CM) and were stimulated with peptides plus costimulatory mAbs as described above. Culture tubes were incubated at 37 °C in a humidified 5% CO2 atmosphere for 6 h. Brefeldin A (Sigma-Aldrich) was added to cells at a final concentration of 10 μg/ml to prevent secretion of cytokines. After incubation, the cells were harvested and washed twice with PBS containing 0.1% BSA plus 0.05% sodium azide. Cell phenotype was determined by cell surface staining with PerCP-labeled anti-CD4, PerCP-labeled anti-CD8, FITC-labeled anti-CD45RA, PE-labeled anti-CCR7 and PE-labeled anti-CD62L mAbs (BD Pharmingen) at 4 °C for 30 min. Intracellular IFN-γ or IL-2 detection used APC-labeled anti-IFN- and PE-labeled anti-IL-2 (BD Pharmingen). Briefly, cells were fixed with PFA 4% for 8 min, washed in PBS–BSA–0.1% saponin buffer and incubated for 1 h with 0.1% saponin buffer. Cells were washed with PBS–BSA–0.1% saponin buffer and stained with the labeled Abs at 4 °C for 30 min. Data acquisition was performed on a flow cytometer (FACS calibur). Data were analyzed using CellQuest 4.3 software (Becton Dickinson).
Acknowledgments
This study was supported by grants from National Nature Science Foundation of China (No. 30340012), Scientific Technology Program of Guangdong (No. 2003Z3-E0491), Ministry of Education, Guangdong Province of China and the National Key Basic Research Program of China (973) (No. 2001CB510007).
We sincerely thank the individuals who donate their blood for this study. We also thank Z. Peng for performing flow cytometry assay.
References
- Boots A.M., Benaissa-Trouw B.J., Hesselink W., Rijke E., Schrier C., Hensen E.J. Induction of anti-viral immune responses by immunization with recombinant-DNA encoded avian coronavirus nucleocapsid protein. Vaccine. 1992;10:119–124. doi: 10.1016/0264-410X(92)90028-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Pei D., Jiang L., Song Y., Wang J., Wang H., Zhou D., Zhai J., Du Z., Li B., Qiu M., Han Y., Guo Z., Yang R. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Chem. 2004;50:988–995. doi: 10.1373/clinchem.2004.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson E.W., Pei J., Dzielawa J., Seo S.H. Cytotoxic T lymphocytes are critical in the control of infectious bronchitis virus in poultry. Dev. Comp. Immunol. 2000;24:187–200. doi: 10.1016/s0145-305x(99)00072-5. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Klenk S.H.D., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Falsey A.R., Walsh E.E. Novel coronavirus and severe acute respiratory syndrome. Lancet. 2003;361:1312–1313. doi: 10.1016/S0140-6736(03)13084-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Tamin A., Soloff A., Aiuto L.D, Nwanegbo E., Robbins P.D., Bellini W.J., Barratt-Boyes S., Gambotto A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362:1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E., Lewis M.W., Hansen S.G., Strelow L.I., Nelson J.A., Sexton G.J. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- Harari A., Vallelian F., Meylan P.R., Pantaleo G. Functional heterogeneity of memory CD4 T cell responses in different conditions of antigen exposure and persistence. J. Immunol. 2005;174:1037–1045. doi: 10.4049/jimmunol.174.2.1037. [DOI] [PubMed] [Google Scholar]
- He Y., Zhou Y., Wu H., Kou Z., Liu S., Jiang S. Mapping of antigenic sites on the nucleocapsid protein of the severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:5014–5309. doi: 10.1128/JCM.42.11.5309-5314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A., Wurm T., Wison L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;201:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey J. Updated SARS case definition using laboratory criteria. CMAJ. 2003;168:1566–1567. [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister B., Kiecker F., Tesfa L., Volk H.D., Picker L.J., Kern F. Mapping T cell epitopes by flow cytometry. Methods. 2003;292:270–281. doi: 10.1016/s1046-2023(02)00349-3. [DOI] [PubMed] [Google Scholar]
- Huang J.L., Huang J., Duan Z.H., Wei J., Min J., Luo X.H., Li J.G., Tan W.P., Wu L.Z., Liu R.Y., Li Y., Shao J., Huang B.J., Zeng Y.X., Huang W.L. Th2 predominance and CD8+ memory T cell depletion in patients with severe acute respiratory syndrome. Microbes Infect. 2005;7:427–436. doi: 10.1016/j.micinf.2004.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson J., Cruz J., Ennis F.A. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 1998;72:8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K., Chong X., Chen Z., Chen Z., Kang Y., Ma Y., Zhu K., Xie Q., Tu Y., Yua Y., Wang B. Induction of Th1 type response by DNA vaccinations with N, M, and E genes against SARS-CoV in mice. Biochem. Biophys. Res. Commun. 2005;328:979–986. doi: 10.1016/j.bbrc.2005.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Lee J.H., Hung C.F., Peng S., Roden R., Wang M.C., Viscidi R., Tsai Y.C., He L., Chen P.J., Boyd D.A., Wu T.C. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalvani A., Brookes R.S., Hambleton S., Britton W.J., Hill A.V., McMichael A.J. Rapid effector function in CD8+ memory T cells. J. Exp. Med. 1997;186:859–865. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L.L., Jamieson B.D., Somasundaram T., Ahmed R. Cytotoxic T-cell memory without antigen. Nature. 1994;369:648–652. doi: 10.1038/369648a0. [DOI] [PubMed] [Google Scholar]
- Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- Liang Y., Wan Y., Qiu L., Zhou J., Ni B., Guo B., Zou Q., Zou L., Zhou W., Jia Z., Che X., Wu Y. Comprehensive antibody epitope mapping of the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus: insight into the humoral immunity of SARS. Clin. Chem. 2005;51:1382–1396. doi: 10.1373/clinchem.2005.051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lier R.A.W., Berge I.J.M., Gamadia L.E. Human CD8+ T-cell differentiation in response to viruses. Nat. Rev., Immunol. 2003;3:1–8. doi: 10.1038/nri1254. [DOI] [PubMed] [Google Scholar]
- Lin Y., Shen X., Yang R.F., Li Y.X., Ji Y.Y., He Y.Y., Shi M.D., Lu W., Shi T.L., Wang J., Wang H.X., Jiang H.L., Shen J.H., Xie Y.H., Wang Y., Pei G., Shen B.F., Wu J.R., Sun B. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13:141–145. doi: 10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Mo H.Y., Xu J., Ren X.L., Zeng G.Q., Tan Y.X., Chen R.C., Moira C.Y., Zhong N.S. Evaluation by indirect immunofluorescent assay and enzyme linked immunosorbent assay of the dynamic changes of serum antibody responses against severe acute respiratory syndrome coronavirus. Chin. Med. J. 2005;118:150–446. [PubMed] [Google Scholar]
- Murali-Krishna K., Lau L.L., Sambhara S., Lemonnier F., Altman J., Ahmed R. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science. 1999;286:1377–1381. doi: 10.1126/science.286.5443.1377. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L., Lim W., Nicholls J., Yee W., Yan W., Cheung M., Cheng V., Chan K., Tsang D., Yung R., Ng T., Yuen K. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lenig D., Forster R., Lipp M., Anzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Seder R.A., Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Rev., Immunol. 2003;9:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- Stohlman S.A., Bergmann C., Cua D., Wege H., van der Veen R. Location of antibody epitopes within the mouse hepatitis virus nucleocapsid protein. Virology. 1994;202:146–153. doi: 10.1006/viro.1994.1330. [DOI] [PubMed] [Google Scholar]
- Stohlman S.., Bergmann C.C., Roel D., van der Veen R.C., Hinton D.R. Mouse hepatitis virus nucleocapsid-specific cytotoxic T lymphocytes protect from lethal infection without eliminating virus from the central nervous system. J. Virol. 1995;69:684–694. doi: 10.1128/jvi.69.2.684-694.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Lim S.J., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antivir. Res. 2004;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M., Lam W.K., Seto W.H., Yam L.Y., Cheung T.M., Wong P.C., Lam B., Ip M.S., Chan J., Yuen K.Y., Lai K.N. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- Wesseling J.G., Godeke G.J., Schijns V.E., Prevec L., Graham F.L., Horzinek M.C., Rottier P.J. Mouse hepatitis virus spike and nucleocapsid proteins expressed by adenovirus vectors protect mice against a lethal infection. J. Gen. Virol. 1993;74:2061–2069. doi: 10.1099/0022-1317-74-10-2061. [DOI] [PubMed] [Google Scholar]
- Wherry E., Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78:5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills M.R., Okecha G., Weekes M.P., Gandhi M.K., Sissons P.J.G., Carmichael A.J. Identification of naive or antigen-experienced human CD8+ T cells by expression of costimulation and chemokine receptors: analysis of the human cytomegalovirus-specific CD8+ T cell response. J. Immunol. 2002;168:5455–5464. doi: 10.4049/jimmunol.168.11.5455. [DOI] [PubMed] [Google Scholar]
- Wu Z.D., Wu D., Zhen H.Q, Xu J., Yu X.B. Epidemiological analysis on SARS infection of clinical practice. Zhong guo gong gong wei shen. 2004;20:32–34. [Google Scholar]
- Xu D., Zhang Z., Jin L., Chu F., Mao Y., Wang H., Liu M., Wang M., Zhang L., Gao G.F., Wang F.S. Persistent shedding of viable SARS-CoV in urine and stool of SARS patients during the convalescent phase. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:165–171. doi: 10.1007/s10096-005-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Cao J., Zhao L.J., Qin Z.L., Ke J.S., Pan W., Ren H., Yu J.G., Qi Z.T. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology. 2005;331:128–135. doi: 10.1016/j.virol.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Yang H., Guo Z.F., Sin W.Y.F., Chen W., Xu J., Fu L., Wu J., Mak C.K.G., Cheng C.S.S., Yang Y., Cao S., Wong T.Y., Lai S.T., Xie Y., Guo Z. B-cell responses in patients who have recovered from severe acute respiratory syndrome target a dominant site in the S2 domain of the surface spike glycoprotein. J. Virol. 2005;79:3401–3408. doi: 10.1128/JVI.79.6.3401-3408.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M.S., Pan Y., Chen H.Q., Shen Y., Wang X.C., Sun Y.J., Tao K.H. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol. Lett. 2004;92:237–243. doi: 10.1016/j.imlet.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


