Abstract
The human coronavirus HCoV-OC43 causes a significant fraction of upper respiratory tract infections. Most coronaviruses show a strong species specificity, although the SARS-Coronavirus crossed species from palm civet cats to infect humans. Similarly, HCoV-OC43, likely a member of the same coronavirus group as SARS-CoV, readily crossed the species barrier as evidenced by its rapid adaptation to the murine brain [McIntosh, K., Becker, W.B., Chanock, R.M., 1967. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci U.S.A. 58, 2268–73]. Herein, we investigated two consequences of this plasticity in species tropism. First, we showed that HCoV-OC43 was able to infect cells from a large number of mammalian species. Second, we showed that virus that was passed exclusively in suckling mouse brains was highly virulent and caused a uniformly fatal encephalitis in adult mice. The surface glycoprotein is a major virulence factor in most coronavirus infections. We identified three changes in the HCoV-OC43 surface glycoprotein that correlated with enhanced neurovirulence in mice; these were located in the domain of the protein responsible for binding to host cells. These data suggest that some coronaviruses, including HCoV-OC43 and SARS-CoV, readily adapt to growth in cells from heterologous species. This adaptability has facilitated the isolation of HCoV-OC43 viral variants with markedly differing abilities to infect animals and tissue culture cells.
Keywords: Coronavirus, Rodent model, Encephalitis
Introduction
Until recently, human coronaviruses were mostly associated with mild upper respiratory tract infections (“the common cold”), and occasionally with outbreaks of gastroenteritis (Vabret et al., 2003). However, with the recognition that the Severe Acute Respiratory Syndrome (SARS) was caused by a coronavirus, it became apparent that coronaviruses could also cause more significant disease in the human population (Drosten et al., 2003, Fouchier et al., 2003). Since then, two additional coronaviruses, HCoV-NL63 and HCoV-HKU1, have been identified; these agents cause upper and lower respiratory tract diseases that are much less severe than SARS (van der Hoek et al., 2004, Woo et al., 2005).
HCoV-OC43 and HCoV-229E are the etiological agents for many coronavirus-induced upper respiratory tract infections. HCoV-OC43, harvested from a patient with an upper respiratory tract infection, was originally isolated after passage in human embryonic tracheal organ cultures; this virus caused neurological disease after only one passage in suckling mice and encephalitis within 2–4 passages (McIntosh et al., 1967) (termed HCoV-OC43NV). HCoV-OC43NV was then propagated in tissue culture cells generating a tissue culture-adapted variant (termed HCoV-OC43TC).
HCoV-OC43 showed increasing neurovirulence with passage through the murine brain (McIntosh et al., 1967); however, most recent studies have used CNS-adapted viruses that were further propagated, at least for a few passages, in tissue culture cells. For example, Talbot and co-workers showed, using the mouse-adapted virus after passage in tissue culture cells (termed HCoV-OC43QUE herein;) that mice infected intranasally with 104–105 TCID50 developed encephalitis if inoculated 8 days but not 21 days postnatally (Jacomy and Talbot, 2003). HCoV-OC43QUE was passaged 5–6 times in tissue culture prior to use in mice (personal communication, Dr. Pierre Talbot, INRS-Institut Armand-Frappier, Laval, Quebec) and consequently may be less virulent than virus isolated directly from infected suckling mouse brains. Consistent with this possibility, we observed, in preliminary experiments, that virus directly harvested from suckling mouse brains caused a lethal infection after intranasal inoculation of 5- to 8-week-old mice. Mice died 9–11 days after inoculation, a time when the adaptive immune response to another coronavirus, mouse hepatitis virus (MHV), is maximal (Bergmann et al., 1999). To begin to understand these differences in virulence, we initiated a more complete study of the disease caused by the neurovirulent strain of HCoV-OC43.
This ability of HCoV-OC43 to cross species barriers to infect mice and to gain virulence in the new host contrasts with the strict species specificity exhibited by most coronaviruses. For example, the group I coronavirus HCoV-229E does not readily infect mice, even transgenic mice expressing human aminopeptidase N, the virus receptor for HCoV-229E (Wentworth et al., 2005). However, the ability of the group II coronavirus HCoV-OC43 to adapt easily to replication within the murine brain suggests that it may be more lax in its species specificity than other coronaviruses. In that sense, it resembles another group II coronavirus, SARS-CoV, which likely crossed the species barrier from animals such as palm civet cats to infect humans (Chinese SARS Molecular Epidemiology Consortium, 2004, Guan et al., 2003, Song et al., 2005). Unlike other coronaviruses, HCoV-OC43 and the closely related bovine coronavirus (BCoV) appear to bind to cells via N-acetyl-neuraminic acid (Schultze and Herrler, 1992, Vlasak et al., 1988), although there are data to suggest that HCoV-OC43 can also employ MHC class I antigen as a host cell receptor (Collins, 1993). This use of a sugar moiety for entry would also be consistent with the ability to infect a broader range of species than most coronaviruses. This possibility was investigated by infecting tissue culture cells from several different animal species with HCoV-OC43, using both the mouse-adapted and the tissue culture-adapted strains.
The ability to rapidly gain virulence after passage in the murine brain is likely to occur via selection of mutations in the S protein that optimize binding and entry to target cells. Passage of neurovirulent variants of MHV, in tissue culture, selects for viruses that are attenuated in vivo but enhanced for replication in vitro. These changes map to the surface (S) glycoprotein (Gallagher and Buchmeier, 2001, Tsai et al., 2003). In addition, infection of rats with uncloned stocks of MHV resulted in the selection of virulent strains of virus; again, virulence correlated with changes in the S protein (Taguchi et al., 1985). Also, replacement of the S gene in the moderately virulent A59 strain of MHV with the gene encoding the S protein of the virulent JHM strain resulted in a gain in virulence in mice (Phillips et al., 1999). In other studies, several groups showed that the adaptation of the SARS-CoV to humans during the 2003 epidemic included several mutations in the S protein. These mutations were shown to enhance binding to human angiotensin converting enzyme 2 (ACE 2), the host cell receptor for SARS-CoV (Kan et al., 2005, Li et al., 2005). Therefore, to investigate the role of the S protein in HCoV-OC43 pathogenesis, we sequenced the S genes of HCoV-OC43TC and HCoV-OC43NV and compared the results to published sequences of the S genes of several other HCoV-OC43 isolates.
Results
Intranasal inoculation of HCoV-OC43NV, but not HCoV-OC43TC, is uniformly fatal to 5- to 8-week-old C57BL/6 (B6) mice
In confirmation of our preliminary results, intranasal inoculation of HCoV-OC43NV resulted in 100% mortality in mice ranging from 5 to 8 weeks old (Fig. 1A). Mice developed signs of acute encephalitis, including hunched posture, lethargy and wasting by days 6–7. Mortality was associated with a 30–35% loss of body mass (Fig. 1B). Severe clinical encephalitis was associated with widespread mononuclear cell infiltration including perivascular cuffing and with loss of CNS architecture (data not shown). In contrast, intranasal inoculation of 5-week-old C57BL/6 mice with HCoV-OC43TC did not cause any clinical disease, including any weight loss (Fig. 1). Consistent with the uniformly lethal outcome observed in mice infected with HCoV-OC43NV, we detected high titers of virus in the CNS of infected mice. HCoV-OC43NV grew poorly in tissue culture cells and we could only reliably titer infectious virus using suckling mice, as described in Materials and methods. Virus titers increased from days 5 to 7 (Fig. 2 ). To confirm these results, we also measured viral loads using a real-time RT-PCR assay. As shown in Figs. 2A and B, there was a strong positive correlation between recovery of infectious virus and the detection of OC43 nucleocapsid RNA in the brains of mice. Also, and in agreement with the results of Jacomy et al. (Jacomy and Talbot, 2003), HCoV-OC43 infected other organs to a small extent, such as the lungs and intestines, when viral RNA was assayed by real-time RT-PCR (Fig. 2C). Notably, the results also suggested that virus was in the process of clearance at the time of death since virus titers/RNA levels declined between days 7 and 9 p.i. (Fig. 2C).
Fig. 1.
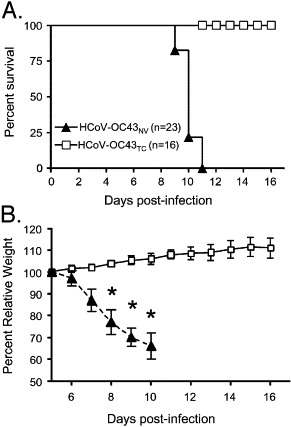
HCoV-OC43NV is uniformly lethal to wild type C57BL/6 mice. 5-week-old mice were inoculated intranasally with 30 LD50 HCoV-OC43NV or 106 TCID50 HCoV-OC43TC as described in Materials and methods. Mice were monitored daily for survival (A) and weight loss (B). (* Indicates P < 0.0002).
Fig. 2.
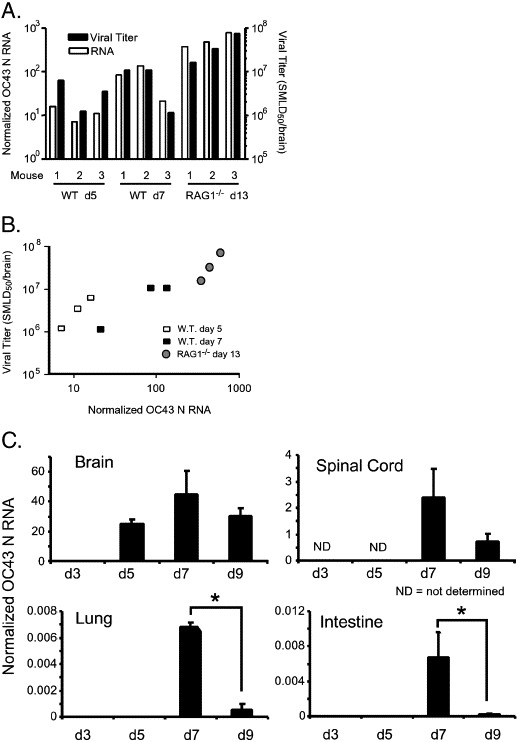
Viral titers and viral RNA burdens in the brains of HCoV-OC43NV-infected mice. Wild type and RAG1−/− C57BL/6 mice were infected intranasally with 30 LD50 HCoV-OC43NV. (A and B) Whole brains were isolated at the indicated time points and titers of infectious virus and virus RNA burden were determined as described in Materials and methods. Titers from 3 representative mice are shown for each time point demonstrating correlation between recovery of infectious virus and viral RNA burden. (C) HCoV-OC43 RNA titers increase until day 7 p.i. then decline by day 9. Whole RNA was isolated from the indicated tissue and assayed for virus burden with real-time RT-PCR. Data in panel C are expressed as mean ± SEM for 4–6 mice per time point. Note the different scales in “Brain” and “Spinal Cord” versus “Lung” and “Intestine.” (*Indicates P < 0.05).
In mice infected intranasally with neurovirulent strains of MHV, virus enters the CNS via the olfactory nerves with subsequent transneuronal retrograde dissemination to distant connections of the olfactory bulb (Perlman et al., 1989, Perlman et al., 1990). Using in situ hybridization and immunohistochemistry to track HCoV-OC43 RNA and antigen, respectively, we detected overlap between the pathways of entry and spread used by MHV and those used by HCoV-OC43 after intranasal inoculation. HCoV-OC43 was detected in the olfactory bulb and the olfactory nucleus at 3 days p.i. but was cleared from these structures by day 5 p.i. At later time points (days 7 and 9 p.i.), HCoV-OC43 RNA and antigen were detected primarily in the brainstem and the spinal cord (Fig. 3A, data not shown). Unlike MHV, which is known to infect oligodendrocytes and is present throughout the white matter in the infected CNS, HCoV-OC43 was not detected in the white matter at any time point examined (shown at day 9 p.i. in Figs. 3A, B).
Fig. 3.
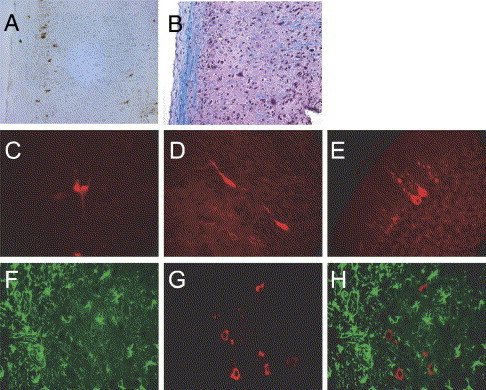
Neurons are the primary target of HCoV-OC43NV in vivo. Mice were inoculated intranasally with 30 LD50 of HCoV-OC43NV and brains and spinal cords were harvested 7 or 9 days p.i. Tissue samples were prepared for in situ hybridization, antigen staining or Luxol Fast Blue (LFB) staining and slides were examined with standard light, fluorescence or confocal microscopy as described in Materials and methods. (A–B) HCoV-OC43 antigen is localized in the gray matter of spinal cords. Panel A depicts immunohistochemical detection of HCoV-OC43 using the anti-OC43 S hybridoma O.4.3. Panel B depicts a serial section stained with LFB to demarcate the white matter. (C–E) Morphology of HCoV-OC43NV-infected cells is consistent with that of neurons. Images are representative coronal sections of brain stems from mice harvested 7–9 days p.i. Sections were stained with O.4.3 followed by Cy3-labeled goat anti-mouse. (F–H) Combination in situ hybridization for HCoV-OC43 nucleocapsid RNA (Cy3-labeled antisense probe, red) and immunohistochemistry for the astrocyte marker GFAP (FITC-labeled anti-GFAP, green). Panel H is a merged image of panels F and G. Original images are 20× magnification for panels A and B, or 40× magnification for panels C–H.
HCoV-OC43NV infection of mouse CNS is restricted to neurons
The ability of HCoV-OC43NV to cause lethal encephalitis in adult animals contrasted with that reported for HCoV-OC43QUE. Jacomy et al. reported that HCoV-OC43QUE infection was primarily restricted to neurons in vivo (Jacomy and Talbot, 2003). Thus, one possible explanation for the enhanced neurovirulence of HCoV-OC43NV, relative to HCoV-OC43QUE, is an expanded cell tropism. To more directly assess this possibility, we used combined in situ hybridization and immunohistochemistry to identify cell types harboring viral RNA and antigen. We found that viral product was restricted to cells with morphologies consistent with neurons (Figs. 3C, D). Moreover, in some animals evaluated at late times postinfection (day 9), we could clearly identify Purkinje cells that harbored virus antigen (Fig. 3E). When we evaluated whether astrocytes were infected by HCoV-OC43NV in vivo, we found no evidence of colocalization of HCoV-OC43 RNA and the astrocyte-specific marker, GFAP antigen (Figs. 3F–H). We also performed a series of colocalization experiments for viral RNA and F4/80, a macrophage/microglia-specific cell marker, but could not identify F4/80+ cells that were positive for viral product. To confirm these results, we sorted F4/80+CD45hi (macrophages) and F4/80+CD45int (microglia) cells from the CNS of infected mice at 7 days postinfection, collected them onto glass slides and stained them for virus antigen. Neither macrophages nor microglia stained positive for viral antigen, while infected tissue culture cells processed in parallel stained positively (data not shown). These data suggest that unlike MHV, HCoV-OC43 does not infect astrocytes or macrophages/microglia. Based on the morphology of the infected cells as well as the lack of virus staining in the white matter or in GFAP+ or F4/80+cells, we conclude that the predominant, if not sole, targets for HCoV-OC43NV are neurons.
Host adaptive immune response contributes to HCoV-OC43-induced morbidity and mortality
Viral RNA burden in the CNS was diminishing at the time of death (Fig. 2C), consistent with a role for the host immune response in both virus clearance and disease. When we immunophenotyped mononuclear cell infiltrates from HCoV-OC43NV-infected mice 7 days postinfection, we found that a large fraction consisted of CD4 and CD8 T cells (Fig. 4A). One HCoV-OC43-specific CD4 T cell epitope recognized in C57BL/6 mice is known, spanning residues 133–147 of the transmembrane (M) protein (epitope M133). Identification of this epitope was based on sequence homology with MHV. We found that 1–3% of the infiltrating CD4 T cells at day 9 recognized this epitope (Fig. 4B). By contrast, 20–25% of CD4 T cells in the MHV-infected CNS responded to epitope M133 at 7 days p.i. (Haring et al., 2001). No HCoV-OC43 CD8 T cell epitopes have been identified yet so the magnitude of the virus-specific CD8 T cell response could not be determined.
Fig. 4.
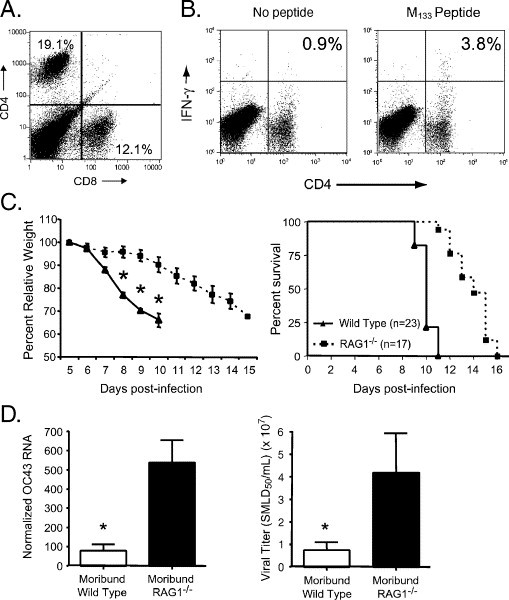
T cell infiltration into the HCoV-OC43NV-infected CNS contributes to morbidity and mortality. (A) Both CD4 and CD8 T cells infiltrate the brains of HCoV-OC43NV-infected mice. Mononuclear cells were harvested 7 days p.i., surface stained for CD4 and CD8 and subjected to FACS analysis as described in Materials and methods. Data from one representative mouse are shown. (B) Two color FACS analysis of in vitro peptide-stimulated mononuclear cells demonstrating infiltration of OC43-specific CD4 T cells. Whole brains were isolated from HCoV-OC43NV-infected mice 7 days p.i. Mononuclear cells were surface stained for CD4 and intracellularly stained for IFN-γ ex vivo after no stimulation (left panel) or stimulation with M133 peptide (right panel). Data from one representative mouse are shown. (C) HCoV-OC43NV-infected RAG1−/− C57BL/6 mice lose weight (left panel) and succumb to infection (right panel) with delayed kinetics relative to infected wild type mice (*Indicates P < 0.05). (D) Relative burden of HCoV-OC43 RNA (left panel) and infectious virus (right panel) in brains of moribund wild type and RAG1−/− mice. Data in panel C are expressed as mean ± SEM for 4–6 mice per group. (*Indicates P < 0.05).
To probe the role of the host adaptive immune response in pathogenesis, we infected immunodeficient mice lacking normal T and B cell responses (mice with genetic disruption of the recombination activating gene 1 (RAG1−/−)) and monitored these mice for weight loss and survival. HCoV-OC43NV-infected RAG1−/− mice developed signs of encephalitis (lethargy, hunching and weight loss) similar to those observed in infected wild type mice, but with delayed kinetics (Fig. 4C, left panel). Infected RAG1−/− mice also survived longer than did their B6 counterparts (Fig. 4C, right panel). At the time of death, virus loads were 5–7-fold higher than detected in moribund B6 mice, when measured by real-time RT-PCR (Fig. 4D, left panel) or infectious virus titers (Fig. 4D, right panel). At this time, viral antigen or RNA, detected by immunohistochemistry or in situ hybridization, was detected primarily in the brainstem although more cells were infected than in B6 mice. As in wild type C57BL/6 mice, neurons were the primary target for infection, suggesting that the antiviral immune response was not responsible for the lack of infection of glial cells (data not shown).
To confirm the pathological role of T cells, we adoptively transferred HCoV-OC43-immune cells to RAG1−/− mice that had been infected with 30 LD50 HCoV-OC43NV intranasally 4 days earlier. HCoV-OC43NV-infected RAG1−/− animals that received no cells died by days 16–18, in line with experiments described above. In contrast, the adoptive transfer of HCoV-OC43-immune splenocytes to RAG1−/− mice resulted in death of recipient animals by days 11–13 (data not shown).
Divergent spike glycoprotein gene sequences between mouse CNS-adapted, tissue culture-adapted and primary clinical isolates of HCoV-OC43
While HCoV-OC43NV causes lethal encephalitis that is enhanced by the antiviral T cell response, the results described above show that HCoV-OC43TC is severely attenuated in mice (Fig. 1). HCoV-OC43QUE was reported to cause less severe disease than we observed in mice infected with HCoV-OC43NV. These differences in neurovirulence prompted us to determine the sequence of the S glycoproteins of HCoV-OC43NV and HCoV-OC43TC and compare them to the published HCoV-OC43QUE sequence, since the coronavirus S protein is often associated with virulence (Thorp and Gallagher, 2004). Included in this comparison are all available sequences submitted for a number of HCoV-OC43 isolates. These additional isolates represent strains that were reported as having been minimally or extensively passaged in tissue culture cells, as well as several primary clinical isolates that were never passaged in tissue culture cells (Supplementary Fig. 1). The sequence alignment revealed substantial divergence in the primary amino acid sequence of these HCoV-OC43 isolates. Of note, there were three amino acid substitutions wholly unique to HCoV-OC43NV (Table 1 ). Multiple other differences between the HCoV-OC43TC and HCoV-OC43QUE or HCoV-OC43NV strains are also present in both the S1 and S2 domains (Supplementary Fig. 1). The large number of differences will make it difficult to determine which substitutions are critical for tissue culture cell adaptation. Of note, sequencing of the 3′ terminal end of HCoV-OC43NV, including genes encoding non-structural 12.9 kDa, the small envelope (E), the nucleocapsid (N) and the transmembrane (M) proteins and the 3′ untranslated region, revealed no nucleotide differences between HCoV-OC43NV and HCoV-OC43QUE.
Table 1.
Comparison of S protein sequences from HCoV-OC43NV, HCoV-OC43QUE and HCoV-OC43TC
| S protein residuea | HCoV-OC43NV | HCoV-OC43QUEb | HCoV-OC43TC |
|---|---|---|---|
| 33 (31,33) | Serine | Threonine | Threonine |
| 491 (489,495) | Histidine | Asparagine | Asparagine |
| 759 (757,765) | Histidine | Arginine | Arginine |
| Virus passage history | Only in suckling mouse brain | 5–6 times in HRT-18 cells | Extensively (>10) in HRT-18 cells |
| Neurovirulence | Uniformly lethal to adult mice | Lethal only to young (<21 days) micec | Avirulent in miced |
S protein residue numbering is based on HCoV-OC43NV strain, with the corresponding residues for HCoV-OC43QUE and HCoV-OC43TC in parentheses. S1 and S2 encompass amino acids 1–759 and 760–1353 to 1365, respectively.
The sequence for HCoV-OC43QUE corresponds to GenPept accession number AAT84362, submitted by St-Jean et al. (St-Jean et al., 2004).
Broad species and cell-type tropism of HCoV-OC43TC and HCoV-OC43NV
As described above, HCoV-OC43 differs from most strains of coronaviruses in that it readily crossed species barriers to infect mice (McIntosh et al., 1967). This observation raised that possibility that HCoV-OC43, unlike most other coronaviruses, is able to infect a wide variety of species without substantial adaptation. To gain insight into this possibility, we infected a variety of tissue culture cells from a wide range of species (hamster, pig, human, mouse, rat, monkey and cat) with HCoV-OC43NV or HCoV-OC43TC. As shown in Fig. 5 , immunocytochemical staining for viral antigen revealed that the tissue culture-adapted virus infected hamster, pig, human, mouse, monkey and cat cells, but not FRT rat epithelium cells. The neurovirulent strain also exhibited a wide host range specificity, infecting hamster, pig, human, monkey, cat and mouse cells. For some cells lines, such as murine 17CL-1, and human KB and HT1080 cells, we detected infection only by the tissue culture-adapted variant. Thus, both HCoV-OC43NV and HCoV-OC43TC showed broad species specificity, and not surprisingly, HCoV-OC43TC showed enhanced ability to infect all types of tissue culture cells. Whether animals from these various species are also infectable by HCoV-OC43 remains to be determined, although data suggesting a close relationship between HCoV-OC43 and BCoV (Vijgen et al., 2005b) make it likely that the virus would be able to infect other species after minimal adaptation.
Fig. 5.
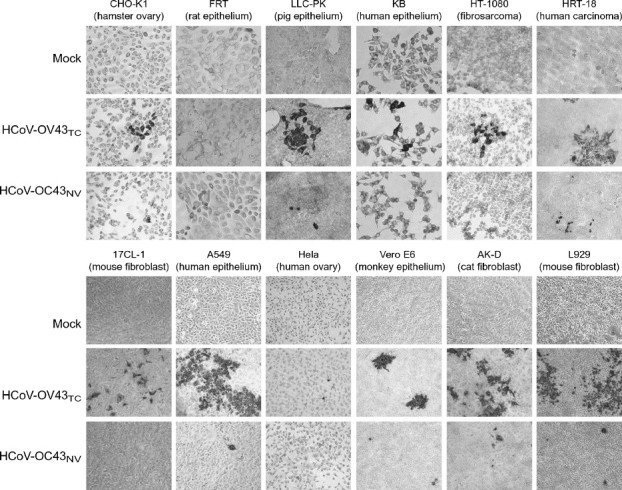
Broad tissue and cell type tropism of HCoV-OC43. A wide variety of tissue culture cells from hamster, rat, pig, human, mouse, monkey and cat were infected with 10 TCID50 (105 SMLD50) of HCoV-OC43NV, 30 TCID50 of HCoV-OC43TC or mock-infected as described in Materials and methods. After 3 days, virus-infected cells were detected with immunocytochemistry as described in Materials and methods. Nearly all of the cell lines were infected with both viruses, demonstrating that HCoV-OC43 exhibits a broad species specificity in vitro. Original images are 20× magnification.
Discussion
HCoV-OC43, as demonstrated herein, infects many cell types and readily adapts to cause a virulent infection in mice (Jacomy and Talbot, 2001, Jacomy and Talbot, 2003) (Fig. 1). BCoV, which is closely related to HCoV-OC43, also infects mice (Akashi et al., 1981, Barthold et al., 1990); phylogenetic analyses suggest that the two viruses diverged as recently as 1891 (Vijgen et al., 2005b). These results suggest that HCoV-OC43 or BCoV crossed species from either humans or bovine to infect the other species, although the direction of spread is not known. This ability to cross species is also shared by SARS-CoV. SARS-CoV spread from exotic animals, most likely palm civet cats, to humans during contacts in “wet markets” in China (Peiris et al., 2004). Also, as part of efforts to study the pathogenesis of this virus and to develop vaccines, several animal species, including mice, cats, ferrets and monkeys were shown to be susceptible to infection with the virus (Peiris et al., 2004). SARS-CoV did not cause reproducible disease in any of these species, suggesting that further adaptation was required for optimal virus replication in these heterologous species. HCoV-OC43 and SARS-CoV differ from many coronaviruses, exemplified by HCoV-229E, in their ability to cross species. HCoV-229E, a human respiratory virus, does not infect mice or even mice transgenic for human aminopeptidase N (hAPN), the HCoV-229E host cell receptor (Wentworth et al., 2005). However, infection occurs, albeit with mild clinical disease if virus is adapted for growth in murine cells expressing hAPN and if mice are deficient in type I interferon responses (Lassnig et al., 2005).
SARS-CoV and HCoV-OC43, while both able to cross species, differ in regard to host cell receptor usage. The primary receptor for SARS-CoV is angiotensin converting enzyme 2 (ACE2) and other molecules such as DC-SIGN serve to enhance binding to target cells (Jeffers et al., 2004, Li et al., 2003, Marzi et al., 2004). Adaptation to growth in foreign species involves mutations that enhance binding to heterologous ACE2 (Li et al., 2005). No proteinaceous receptor for HCoV-OC43 has been conclusively identified; rather, O-acetylated sialic acid or a closely related compound appears to serve as the host cell receptor (Schultze and Herrler, 1992, Vlasak et al., 1988). While little is known about the changes required for efficient replication of HCoV-OC43 in the murine CNS, it is likely that this process includes mutations that affect binding to host cell sugar moieties. Infection with the prototypic alphavirus, Sindbis virus (SINV), or with a picornavirus, Theiler's murine encephalomyelitis virus (TMEV), is initiated by binding to a polyanionic polysaccharide, heparin sulfate (Byrnes and Griffin, 1998, Klimstra et al., 1998, Reddi and Lipton, 2002). SINV with enhanced binding to heparin sulfate is selected after passage in tissue culture cells. HCoV-OC43 may undergo a similar set of changes during the course of adaptation to growth in tissue culture cells. The S protein of HCoV-OC43TC differs from that of HCoV-OC43NV at 20 positions (Supplementary Fig. 1), making it difficult to determine which ones might contribute to enhanced ability to replicate in tissue culture cells.
By contrast, HCoV-OC43NV includes a limited number of amino acids that are not present in any S proteins described in the literature (Table 1 and Supplementary Fig. 1). In all laboratory isolates of HCoV-OC43, viruses encoding these proteins were passaged at least a few times in tissue culture cells (Kunkel and Herrler, 1993b, Kunkel and Herrler, 1996, Mounir and Talbot, 1993, St-Jean et al., 2004, Vijgen et al., 2005b). While no two sequences are identical, three substitutions, T33S, N491H and R759H, are unique to the HCoV-OC43NV strain (Table 1 and Supplementary Fig. 1). The receptor binding domain of HCoV-OC43 has not been defined, but both T33S and N491H are in regions important for receptor binding by other coronaviruses (e.g., Saeki et al., 1997, Xiao et al., 2003). Thus, these two changes could potentially alter the association or stability between HCoV-OC43NV and host cell receptors in the murine CNS.
In other coronaviruses, cleavage of S into S1 and S2 domains is required for optimal virus replication. For example, disruption of the S1–S2 cleavage site in MHV resulted in a viable virus that was attenuated for growth in tissue culture cells and mice (Stauber et al., 1993, Taguchi, 1993). On the other hand, studies in BCoV revealed no correlation between the presence of the furin cleavage motif and virulence (Zhang et al., 1991). Of the several laboratory isolates of HCoV-OC43 that have been sequenced, most contain the sequence RRSRG at the putative furin cleavage site. Kunkel and Herrler showed that S proteins containing this motif are not cleaved in infected cells, whereas two strains (designated OC43-CU and OC43-VA, Supplementary Fig. 1) in which the S gene encoded a prototypic cleavage site (RRSRR) were cleaved (Kunkel and Herrler, 1993a). The sequences of seven recently described primary clinical isolates also share the RRSRR motif (Supplemental Fig. 1) (Vijgen et al., 2005a). Whether or not these primary isolates are cleaved and whether this motif contributes to clinical disease in humans are not known. As reported herein, the S protein of HCoV-OC43NV contains the sequence RRSHG and presumably is not cleaved to a significant extent because it contains a glycine at position 5; it is even less likely that HCoV-OC43TC, in which the sequence at this site is IRSRG, is cleaved. Although the R759H substitution is unlikely to affect cleavability of HCoV-OC43NV S, as this is a conserved (charged-basic to charged-basic) change, it is formally possible that this unique substitution could alter cleavage and contribute to the enhanced virulence of HCoV-OC43NV.
The position of these 3 mutations (T33S, N491H, and R759H) is consistent with a role in virulence, but to prove definitively their role in disease enhancement, it will be necessary to introduce these changes into the HCoV-OC43 genome. This will require a reverse genetics system. An infectious clone for HCoV-OC43 has not yet been reported, but it is likely that one or more infectious cDNA clones will be available in the near future.
While a comparison of our results with those of Jacomy et al. suggest that HCoV-OC43NV is more virulent than HCoV-OC43QUE, both strains show a tropism for neurons (Jacomy and Talbot, 2003). Direct virus destruction of infected neurons is clearly important for much of the disease observed in mice infected with HCoV-OC43NV or HCoV-OC43QUE; even though death is delayed in infected RAG1−/− mice compared to wild type mice, all mice still die by 16 days p.i. (Fig. 4). However, the delayed death of infected RAG1−/− mice suggests that HCoV-OC43-induced encephalitis is in part mediated by the antiviral T cell response. Of note, Jacomy et al. previously concluded that the adaptive immune response did not contribute to pathology associated with HCoV-OC43 infection, based on the observation that treatment of mice with cyclosporin (CsA) prior to infection resulted in more rapid onset of disease and an increase in the percentage of mice that succumbed to the virus (Jacomy and Talbot, 2003). These contrasting results may result from the different mechanisms of immunosuppression observed in RAG1−/− and CsA-treated mice.
Neurons do not normally express MHC class I or II antigen and express only low levels of the machinery required for loading peptide onto MHC class I antigen (Joly and Oldstone, 1992). However, electrically silent or damaged neurons do express MHC class I antigen (Medana et al., 2000, Neumann et al., 1995, Neumann et al., 1997) and it is possible that infection with HCoV-OC43 makes neurons into suitable targets for CD8 T cells. Of note, even damaged neurons have never been reported to express MHC class II antigen, making a direct effect of CD4 T cells on infected neurons unlikely. However, CD4 T cells are required for optimal function of CD8 T cells in the MHV-infected CNS (Stohlman et al., 1998). Sindbis virus (SINV), a prototypic alphavirus, also primarily affects neurons. While virus clearance is largely mediated by antiviral antibodies, T cells, by secreting IFN-γ, are critical for clearance from spinal cord neurons (Binder and Griffin, 2001). Similar mechanisms may be involved in virus clearance from HCoV-OC43 neurons, with subsequent immunopathology. Future studies, directed at determining whether antiviral CD4 and CD8 T cell responses contribute to both virus clearance and to severe disease in HCoV-OC43-infected mice, may also be relevant to understanding disease outcome in patients with SARS, since neurons are infected in some patients (Gu et al., 2005).
Materials and methods
Viruses and cell lines
HCoV-OC43NV and HCoV-OC43TC (VR-759 and VR-1558, respectively), as well as the cell lines 17Cl-1, L929, HeLa and AK-D, were obtained from the ATCC (Manassas, VA). FRT, CHO-K1 and LLCK-PK cells were a generous gift from Dr. Michael Welsh, University of Iowa. HT-1080 and KB cells were a generous gift from Dr. Paul McCray, University of Iowa. HCoV-OC43 VR-759 was propagated in suckling mouse brain. HCoV-OC43 VR-1558 was propagated in HRT-18 cells.
Mice
Pathogen-free male C57BL/6 mice were purchased from the National Cancer Institute (Bethesda, MD). RAG1−/− and suckling Swiss mice were obtained from breeding colonies maintained by our laboratory. For intranasal infection, 5- to 8-week-old mice were lightly anesthetized with halothane and droplets containing 30 LD50 (107 SMLD50) of HCoV-OC43NV or 106 TCID50 of HCoV-OC43TC were administered to the nares. All procedures used in this study were approved by the University of Iowa Institutional Animal Care and Use Committee.
Antibodies and reagents
Monoclonal antibodies recognizing murine glial fibrillary acidic protein (GFAP) or murine F4/80 were purchased from Dako (Carpinteria, CA) and Caltag (Burlingame, CA), respectively. Monoclonal antibody (mAb) directed against the surface glycoprotein of OC43 was prepared from the hybridoma O.4.3, a generous gift from Dr. John O. Fleming, University of Wisconsin, Madison, WI.
Cell culture infection
Cells were maintained in the appropriate growth medium at 37 °C in 5% CO2. For OC43 infection, cells were washed twice with PBS, and virus was adsorbed for 2 h at 33 °C in Dulbucco's Modified Eagle's Medium (serum-free DMEM) + 15 mM HEPES, pH 7.5. Cells were infected with 30 TCID50 of HCoV-OC43TC (based on infection of HRT-18 cells) or 105 SMLD50 (approximately 10 TCID50 based on infection of HRT-18 cells). Virus suspensions were aspirated and medium containing 2% fetal bovine serum was added to cells. Cells were then cultured for an additional 2–5 days at 33 °C.
Determination of viral titers
Whole brains were aseptically removed from mice at various times postinfection, homogenized in sterile PBS and clarified by centrifugation. 20 μl of serial log10 dilutions of brain homogenates was inoculated intracranially into 2- or 3-day-old Swiss pups. Survival was monitored and viral titers were calculated using the formula of Karber: Negative log of the lowest dilution − [(sum of percentage positive / 100) − 0.5] × log interval. Virus titers are expressed as the reciprocal of the highest dilution where the virus suspension killed 50% of inoculated suckling mice (SMLD50).
Virus antigen detection
For immunohistochemistry, brain and spinal cord sections were processed as previously described (Pewe et al., 2002). Primary antibody was mouse anti-S mAb (O.4.3) and secondary antibody was biotinylated goat anti-mouse. Sections were developed by sequential incubation with streptavidin–horseradish peroxidase (HRP) conjugate and DAB (Sigma, St Louis, MO) or streptavidin–Cy3 reagent (Jackson ImmunoResearch, West Grove, PA). For immunocytohemistry of OC43-infected adherent cells, samples were fixed for 30 min in 10% formalin, rinsed with 100 mM glycine and permeabilized in 0.5% Triton X-100. Wells were blocked with 10% NGS and incubated with O.4.3 overnight at 4 °C. Wells were developed as described above.
In situ hybridization
8 μm brain and spinal cord sections were prepared as described above and in situ hybridization for viral RNA was performed using a protocol adapted from Gene Detect (Auckland, New Zealand). Briefly, formalin fixed, paraffin embedded tissue sections were permeabilized for 30 min at 37 °C in 250 μg/ml pepsin in 200 mM HCl and then blocked with prehybridization buffer for 2 h at 37 °C. 3′-biotinylated, HCoV-OC43-specific oligonucleotide probes (antisense 5′–3′, GTATTGACATCAGCCTGGTTGCTAG; sense 5′–3′, CTAGCAACCAGGCTGATGTCAATAC) were added to a final concentration of 200 ng/ml and sections were incubated for 18 h at 37 °C. After several washes of increasing stringency, streptavidin–HRP conjugate was added for 1 h at 25 °C followed by tyramide signal amplification (TSA) cyanine 3 reagent (Perkin Elmer, Boston, MA). For some experiments, slides were also stained with anti-GFAP followed by FITC-conjugated goat anti-rabbit secondary antibody prior to developing with TSA-Cy3 reagent. Sections were examined with a Bio-Rad (Hercules, CA) LaserSharp2000 confocal microscope.
Real-time RT-PCR
Total RNA was isolated using Tri Reagent (Molecular Research Center, Cincinnati, OH) following the manufacturer's instructions. 2 μg of total RNA was reverse transcribed to cDNA using RETROscript RT-PCR Kit (Ambion, Austin, TX) according to the manufacturer's instructions. The resulting cDNA was subjected to PCR as follows. 2 μl of cDNA was added to a 23 μl PCR cocktail containing 2× SYBR Green Master Mix (Applied Biosystems, Foster City, CA) and 0.2 μM of each sense and antisense primers (Intergrated DNA Technologies, Coralville, IA). Amplification was then performed in an Applied Biosystems Prism 7700 thermocycler. Specificity of the amplification was confirmed using melting curve analysis. Data were collected and recorded by the Prism 7700 software and expressed as a function of Threshold Cycle (C t). Specific primer sets used for HCoV-OC43 and murine housekeeping gene are as follows (5′ to 3′); HCoV-OC43 nucleocapsid forward, GGTCTCAACCCCCAGCTAGT; OC43 nucleocapsid reverse, TGATGCTCTTTAGGCTTTCCA; HPRT forward, CCTCATGGACTGATTATGGAC; HPRT reverse, CAGATTCAACTTGCGCTCATC. HCoV-OC43 nucleocapsid RNA abundance was calculated using methods described previously (Pewe et al., 2005).
Sequencing
Total RNA from infected murine brain or A549 cells was reverse transcribed to cDNA. Primers were designed (based on GenBank accession number AY585228) to generate overlapping amplimers, ensuring complete coverage of the S genes of HCoV-OC43NV and HCoV-OC43TC, as well as 3′ terminal end of the HCoV-OC43NV genome. PCR products were sequenced directly by the University of Iowa DNA Core.
Flow cytometry
Single cell suspensions of mononuclear cells from whole brain homogenates were prepared as previously described (Pewe et al., 2005). Fc receptors were blocked with normal rat serum and anti-CD16/CD32 (clone 2.4G2, BD Biosciences, San Jose, CA). Antibodies used to phenotype cells were fluorescein isothiocyanate-labeled anti-mouse CD4 and phycoerythrin-labeled anti-mouse CD8 (clones GK1.5 and 53–6.7, BD Biosciences, Mountain View, CA). CD4 T cells recognizing an HCoV-OC43-specific CD4 T cell epitope M133 (spanning residues 133–147 of the M protein) were identified using intracellular cytokine staining as previously described (Wu et al., 2000). Samples were analyzed on a FACScan flow cytometer (BD Biosciences, Mountain View, CA).
Statistics
Statistical analysis was done with unpaired (two-tailed) t tests. Values in figures are expressed as mean ± SEM. Values of P < 0.05 were considered significant and are indicated by an asterisk (*) in figures.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2005.11.044.
Appendix A. Supplementary data
Supplementary Fig. 1.
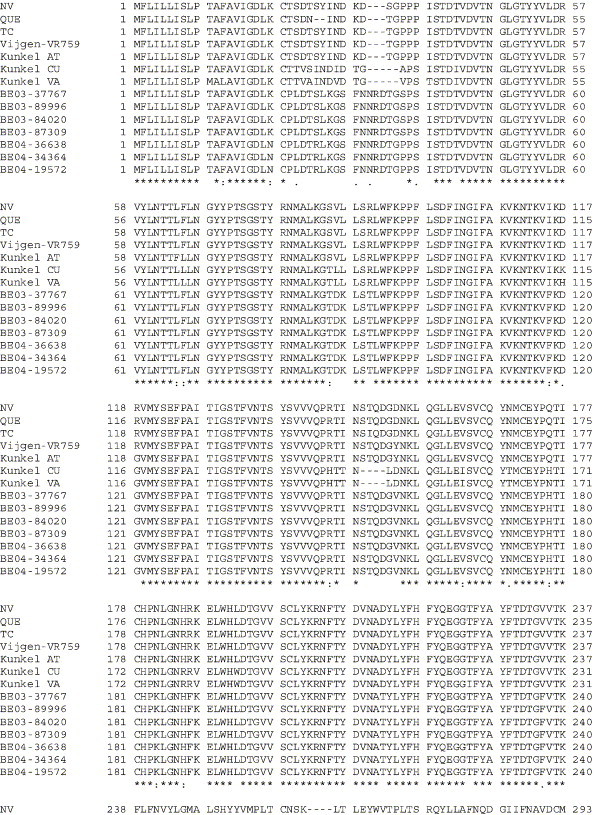
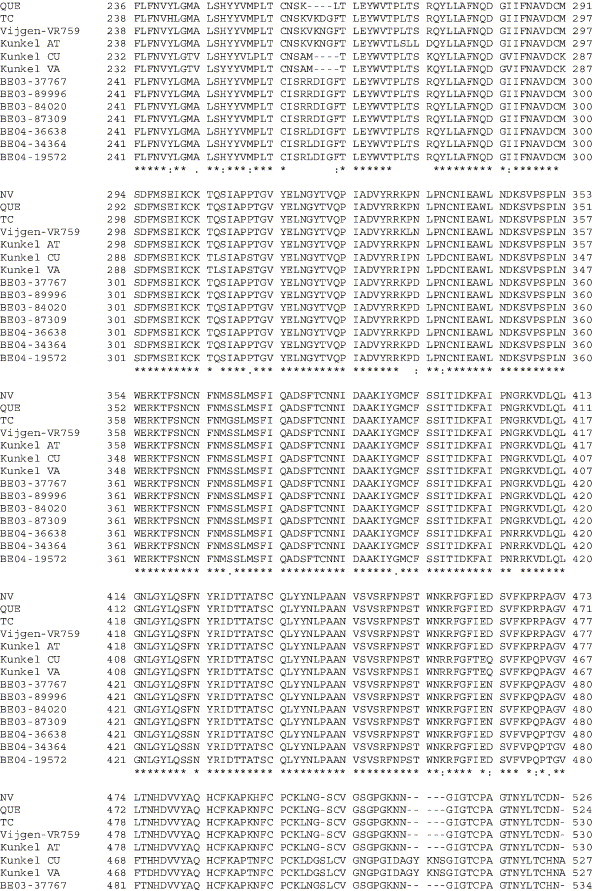
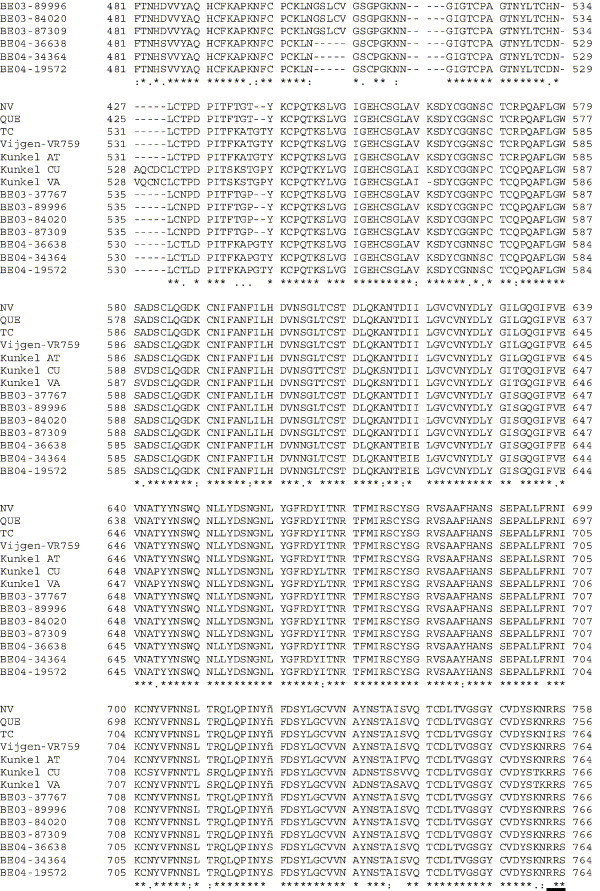
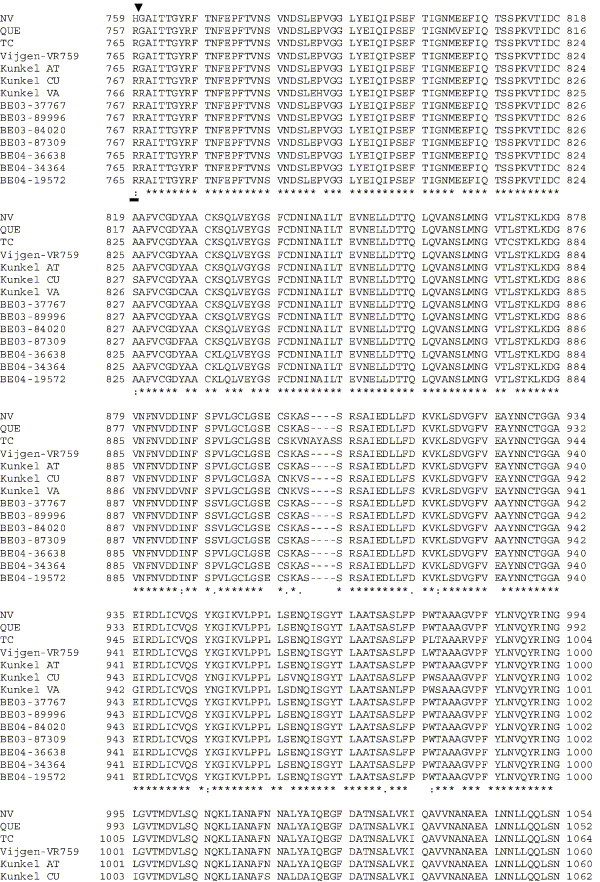
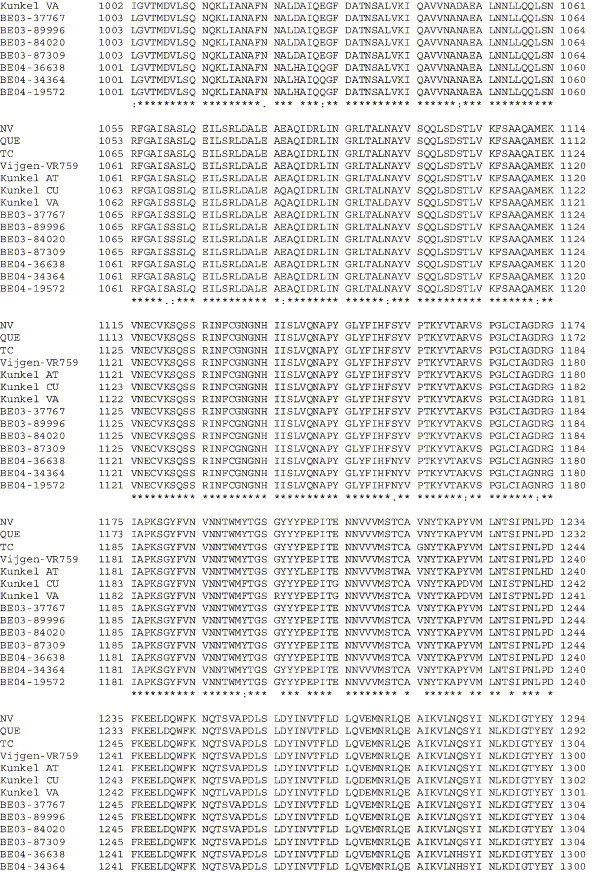
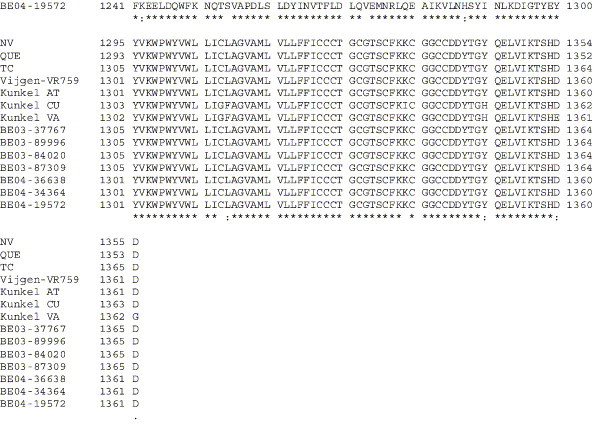
Amino acid alignment of S protein sequences for HCoV-OC43NV, HCoV-OC43QUE, HCoV-OC43TC and the published sequence of several HCoV-OC43 isolates. Asterisks indicate identical residues, colons represent conserved changes and blank spaces denote non-conserved substitutions. The putative furin recognition site is underlined and the cleavage site is marked with an arrowhead. For HCoV-OC43NV and HCoV-OC43TC, total RNA was reverse transcribed to cDNA and the S gene was amplified by PCR. PCR products were directly sequenced as described in Materials and methods. The sequence for HCoV-OC43QUE corresponds to GenPept accession number AAT84362, passed 5–6 times in HRT-18 cells (St-Jean et al., 2004); the sequence for Vijgen VR-759 was translated from GenBank accession number AY391777, passed an unknown number of times in HRT-18 cells (Vijgen et al., 2005b); the sequence for Kunkel AT corresponds to GenPept accession number AAB27260, passed 3 times in HRT-18 cells (Kunkel and Herrler, 1993b); the sequences for Kunkel CU and Kunkel VA correspond to GenPept accession numbers CAA83660 and CAA83661, passed 3 times in MDCK cells and 12 times in Vero E6 cells, respectively (Kunkel and Herrler, 1996); and sequences for HCoV-OC43 clinical isolates BE03-37767, BE03-89996, BE03-84020, BE03-87309, BE04-36638, BE04-34364 and BE04-19572 correspond to GenPept accession numbers AAX84794, AAX84791, AAX84793, AAX85674, AAX84795, AAX84792 and AAX85678, respectively, submitted by Vijgen et al. (2005a).
References
- Akashi H., Inaba Y., Miura Y., Sato K., Tokuhisa S., Asagi M., Hayashi Y. Propagation of the Kakegawa strain of bovine coronavirus in suckling mice, rats and hamsters. Arch. Virol. 1981;67(4):367–370. doi: 10.1007/BF01314841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold S.W., de Souza M.S., Smith A.L. Susceptibility of laboratory mice to intranasal and contact infection with coronaviruses of other species. Lab. Anim. Sci. 1990;40(5):481–485. [PubMed] [Google Scholar]
- Bergmann C.C., Altman J.D., Hinton D., Stohlman S.A. Inverted immunodominance and impaired cytolytic function of CD8+T cells during viral persistence in the central nervous system. J. Immunol. 1999;163:3379–3387. [PubMed] [Google Scholar]
- Binder G.K., Griffin D.E. Interferon-gamma-mediated site-specific clearance of alphavirus from CNS neurons. Science. 2001;293(5528):303–306. doi: 10.1126/science.1059742. [DOI] [PubMed] [Google Scholar]
- Byrnes A.P., Griffin D.E. Binding of Sindbis virus to cell surface heparan sulfate. J. Virol. 1998;72(9):7349–7356. doi: 10.1128/jvi.72.9.7349-7356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303(5664):1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Collins A.R. HLA class I antigen serves as a receptor for human coronavirus OC43. Immunol. Invest. 1993;22:95–105. doi: 10.3109/08820139309063393. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z., Zhuang H., Wu B., Zhong H., Shao H., Fang W., Gao D., Pei F., Li X., He Z., Xu D., Shi X., Anderson V.M., Leong A.S. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haring J.S., Pewe L.L., Perlman S. High-magnitude, virus-specific CD4 T-cell response in the central nervous system of coronavirus-infected mice. J. Virol. 2001;75(6):3043–3047. doi: 10.1128/JVI.75.6.3043-3047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy H., Talbot P.J. Susceptibility of murine CNS to OC43 infection. Adv. Exp. Med. Biol. 2001;494:101–107. doi: 10.1007/978-1-4615-1325-4_16. [DOI] [PubMed] [Google Scholar]
- Jacomy H., Talbot P.J. Vacuolating encephalitis in mice infected by human coronavirus OC43. Virology. 2003;315(1):20–33. doi: 10.1016/S0042-6822(03)00323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., Demartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101(44):15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly E., Oldstone M.B. Neuronal cells are deficient in loading peptides onto MHC class I molecules. Neuron. 1992;8(6):1185–1190. doi: 10.1016/0896-6273(92)90138-4. [DOI] [PubMed] [Google Scholar]
- Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Cui B., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Qi X., Chen K., Du L., Gao K., Zhao Y.T., Zou X.Z., Feng Y.J., Gao Y.F., Hai R., Yu D., Guan Y., Xu J. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79(18):11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra W.B., Ryman K.D., Johnston R.E. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 1998;72(9):7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel F., Herrler G. Bovine coronavirus hemagglutinin protein. Virus Res. 1993;2:53–61. doi: 10.1016/0168-1702(85)90059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel F., Herrler G. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology. 1993;195(1):195–202. doi: 10.1006/viro.1993.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel F., Herrler G. Structural and functional analysis of the S proteins of two human coronavirus OC43 strains adapted to growth in different cells. Arch. Virol. 1996;141(6):1123–1131. doi: 10.1007/BF01718615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassnig C., Sanchez C.M., Egerbacher M., Walter I., Majer S., Kolbe T., Pallares P., Enjuanes L., Muller M. Development of a transgenic mouse model susceptible to human coronavirus 229E. Proc. Natl. Acad. Sci. U.S.A. 2005 doi: 10.1073/pnas.0408589102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., Steinkasserer A., Becker S., Bates P., Hofmann H., Pohlmann S. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78(21):12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Becker W.B., Chanock R.M. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. U.S.A. 1967;58:2268–2273. doi: 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medana I.M., Gallimore A., Oxenius A., Martinic M.M., Wekerle H., Neumann H. MHC class I-restricted killing of neurons by virus-specific CD8+T lymphocytes is effected through the Fas/FasL, but not the perforin pathway. Eur. J. Immunol. 2000;30(12):3623–3633. doi: 10.1002/1521-4141(200012)30:12<3623::AID-IMMU3623>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Mounir S., Talbot P.J. Molecular characterization of the S protein gene of human coronavirus OC43. J. Gen. Virol. 1993;74:1981–1987. doi: 10.1099/0022-1317-74-9-1981. [DOI] [PubMed] [Google Scholar]
- Neumann H., Cavalie A., Jenne D., Wekerle H. Induction of MHC class I genes in neurons. Science. 1995;269:549–552. doi: 10.1126/science.7624779. [DOI] [PubMed] [Google Scholar]
- Neumann H., Schmidt H., Cavalie A., Jenne D., Wekerle H. Major histocompatibility complex (MHC) class I gene expression in single neurons of the central nervous system: differential regulation by interferon (IFN)-gamma and tumor necrosis factor (TNF)-alpha. J. Exp. Med. 1997;185:305–316. doi: 10.1084/jem.185.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10(12 Suppl.):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Jacobsen G., Afifi A. Spread of a neurotropic murine coronavirus into the CNS via the trigeminal and olfactory nerves. Virology. 1989;170:556–560. doi: 10.1016/0042-6822(89)90446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Evans G., Afifi A. Effect of olfactory bulb ablation on spread of a neurotropic coronavirus into the mouse brain. J. Exp. Med. 1990;172:1127–1132. doi: 10.1084/jem.172.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewe L., Haring J., Perlman S. CD4 T-cell-mediated demyelination is increased in the absence of gamma interferon in mice infected with mouse hepatitis virus. J. Virol. 2002;76:7329–7333. doi: 10.1128/JVI.76.14.7329-7333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewe L., Zhou H., Netland J., Tangadu C., Olivares H., Shi L., Look D., Gallagher T.M., Perlman S. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 2005;79:11335–11342. doi: 10.1128/JVI.79.17.11335-11342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.J., Chua M.M., Lavi E., Weiss S.R. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 1999;73:7752–7760. doi: 10.1128/jvi.73.9.7752-7760.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi H.V., Lipton H.L. Heparan sulfate mediates infection of high-neurovirulence Theiler's viruses. J. Virol. 2002;76(16):8400–8407. doi: 10.1128/JVI.76.16.8400-8407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki K., Ohtsuka N., Taguchi F. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J. Virol. 1997;71(12):9024–9031. doi: 10.1128/jvi.71.12.9024-9031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B., Herrler G. Bovine coronavirus uses N-acetyl-9-O-acetylneuraminic acid as a receptor determinant to initiate the infection of cultured cells. J. Gen. Virol. 1992;73:901–906. doi: 10.1099/0022-1317-73-4-901. [DOI] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A. 2005;102(7):2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber R., Pfleidera M., Siddell S. Proteolytic cleavage of the murine coronavirus surface glycoprotein is not required for fusion activity. J. Gen. Virol. 1993;74:183–191. doi: 10.1099/0022-1317-74-2-183. [DOI] [PubMed] [Google Scholar]
- St-Jean J.R., Jacomy H., Desforges M., Vabret A., Freymuth F., Talbot P.J. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 2004;78(16):8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohlman S.A., Bergmann C.C., Lin M.T., Cua D.J., Hinton D.R. CTL effector function within the central nervous system requires CD4+T Cells. J. Immunol. 1998;160:2896–2904. [PubMed] [Google Scholar]
- Taguchi F. Fusion formation by the uncleaved spike protein of murine coronavirus JHMV variant cl-2. J. Virol. 1993;67:1195–1202. doi: 10.1128/jvi.67.3.1195-1202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Siddell S., Wege H., ter Meulen V. Characterization of a variant virus selected in rat brains after infection by coronavirus MHV JHM. J. Virol. 1985;54:429–435. doi: 10.1128/jvi.54.2.429-435.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp E.B., Gallagher T.M. Diversity of coronavirus spikes: relationship to pathogen entry and dissemination. In: Fischer W., editor. Viral membrane Proteins: Structure, function and Drug Design. Kluwer Academic/Plenum Publishers; New York: 2004. [Google Scholar]
- Tsai J.C., de Groot L., Pinon J.D., Iacono K.T., Phillips J.J., Seo S.H., Lavi E., Weiss S.R. Amino acid substitutions within the heptad repeat domain 1 of murine coronavirus spike protein restrict viral antigen spread in the central nervous system. Virology. 2003;312(2):369–380. doi: 10.1016/S0042-6822(03)00248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Mourez T., Gouarin S., Petitjean J., Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis. 2003;36:985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Moes E., Li S., Vandamme A.M., Van Ranst M. Circulation of genetically distinct contemporary human coronavirus OC43 strains. Virology. 2005;337(1):85–92. doi: 10.1016/j.virol.2005.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79(3):1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak R., Luytjes W., Spaan W., Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth D.E., Tresnan D.B., Turner B.C., Lerman I.R., Bullis B., Hemmila E.M., Levis R., Shapiro L.H., Holmes K.V. Cells of human aminopeptidase N (CD13) transgenic mice are infected by human coronavirus-229E in vitro, but not in vivo. Virology. 2005;335(2):185–197. doi: 10.1016/j.virol.2005.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.F., Dandekar A.A., Pewe L., Perlman S. CD4 and CD8 T cells have redundant but not identical roles in virus-induced demyelination. J. Immunol. 2000;165:2278–2286. doi: 10.4049/jimmunol.165.4.2278. [DOI] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312(4):1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.M., Kousoulas K.G., Storz J. Comparison of the nucleotide and deduced amino acid sequences of the S genes specified by virulent and avirulent strains of bovine coronaviruses. Virology. 1991;183(1):397–404. doi: 10.1016/0042-6822(91)90154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


