Abstract
In addition to the major ectodomain, the gp41 transmembrane glycoprotein of HIV-1 is now known to have a minor ectodomain that is part of the long C-terminal tail. Both ectodomains are highly antigenic, carry neutralizing and non-neutralizing epitopes, and are involved in virus-mediated fusion activity. However, data have so far been biologically based, and derived solely from T cell line-adapted (TCLA), B clade viruses. Here we have carried out sequence and theoretically based structural analyses of 357 gp41 C-terminal sequences of mainly primary isolates of HIV-1 clades A, B, C, and D. Data show that all these viruses have the potential to form a tail loop structure (the minor ectodomain) supported by three, β-sheet, membrane-spanning domains (MSDs). This means that the first (N-terminal) tyrosine-based sorting signal of the gp41 tail is situated outside the cell membrane and is non-functional, and that gp41 that reaches the cell surface may be recycled back into the cytoplasm through the activity of the second tyrosine-sorting signal. However, we suggest that only a minority of cell-associated gp41 molecules – those destined for incorporation into virions – has 3 MSDs and the minor ectodomain. Most intracellular gp41 has the conventional single MSD, no minor ectodomain, a functional first tyrosine-based sorting signal, and in line with current thinking is degraded intracellularly. The gp41 structural diversity suggested here can be viewed as an evolutionary strategy to minimize HIV-1 envelope glycoprotein expression on the cell surface, and hence possible cytotoxicity and immune attack on the infected cell.
Keywords: HIV-1, Envelope protein, gp41 envelope protein, gp41 C-terminal tail, Structure, Conformation, Membrane spanning domain, Tyrosine-sorting signal
Introduction
Viruses of the Lentivirus genus of the Retroviridae have a single virus-encoded, envelope glycoprotein (Env). This is a type I membrane protein with the N-terminus on the outside of the cell. Env mRNA is synthesized in the nucleus, and exported with the help of the viral Rev protein to the cytoplasm where it is translated as a gp160 precursor. Gp160 is then translocated through the rough endoplasmic reticulum (ER) (Dettenhofer and Yu, 2001), and folded with the assistance of the chaperone proteins, calreticulin, and calnexin (Otteken et al., 1996). Glycans are added in the Golgi, and subsequently trimmed (Dash et al., 1994, Fenouillet and Jones, 1995). Gp160 oligomerizes in the ER, forming a non-covalently linked trimer (Earl et al., 1991, Willey et al., 1988). Most gp160 is degraded, but about 5–15% is cleaved into gp120 (distal) and gp41 (membrane anchor) components. These remain associated non-covalently and are targeted to the cell surface (see below). Gp120-gp41 trimers are incorporated into progeny virions. Gp120 can be shed from the surface of cells and virions (Willey et al., 1988, Willey et al., 1991).
Enzymatic cleavage of gp160 to gp120-gp41 is essential for Env function. Uncleaved gp160 cannot induce syncytium formation, and virions with uncleaved gp160 are not infectious. Gp160 is rarely incorporated into virions, probably because so little reaches the cell surface (Duensing et al., 1995, Pal et al., 1991, Pfeiffer et al., 1997, Willey et al., 1988). HIV-1 gp160 is cleaved between residues 518R and 519A by the subtilisin/kexin-like Ca2+-dependent convertases such as furin, PACE4, PC5/6-B, and PC1 (Morikawa et al., 1993, Moulard et al., 1994, Vollenweider et al., 1996). Gp160 is probably cleaved by more than one cellular protease.
Gp160 cleavage occurs in the trans-Golgi network (TGN) or after it exits the TGN (Bultmann et al., 2000, Pal et al., 1991, Pfeiffer et al., 1997, Stein and Engelman, 1990). Brefeldin A, A1Fn, monensin, and tunicamycin inhibit gp160 cleavage even when gp160 is allowed to accumulate in the TGN, suggesting that cleavage occurs in the late compartment of the Golgi or after gp160 has exited from the Golgi (Dewar et al., 1989, Kantanen et al., 1995, Pal et al., 1991, Pal et al., 1988). Cleavage may occur in an acidic compartment, as it was inhibited by NH4Cl and partially inhibited by chloroquine (Courageot et al., 1999, Willey et al., 1988). However, these inhibitors may prevent gp160 from reaching the correct intracellular site for cleavage. Methionine methyl ester failed to inhibit gp160 cleavage, indicating that cleavage may occur in a non-lysosomal compartment (Willey et al., 1988). Only a small proportion of gp160 (5–40%) is cleaved into gp120-gp41, and this depends on the host cell (Bird et al., 1990, Hallenberger et al., 1993, Jose et al., 1997, Kimura et al., 1996, Kozarsky et al., 1989, Moulard et al., 1999, Pfeiffer et al., 1997, Willey et al., 1988, Willey et al., 1991). For example, 10–20% of gp160 is cleaved in peripheral blood lymphocytes (Willey et al., 1988).
Uncleaved, trimeric gp160 is degraded in the ER and lysosomes. However, the proportion degraded in each location is not known. According to Willey et al., most gp160 leaves the ER and reaches the Golgi, and the minority that remains in the ER is degraded there (Willey et al., 1988). However, others find that most gp160 remains in the ER and is degraded (Bultmann et al., 2000, Courageot et al., 1999, Hallenberger et al., 1993, Jabbar and Nayak, 1990, Pfeiffer et al., 1997), or is rapidly sent to lysosomes and degraded (Jabbar and Nayak, 1990, Pfeiffer et al., 1997). Whichever the location, degradation is relatively rapid, and only 10–20% of gp160 remains after 8 h (Willey et al., 1988). When prevented from leaving the ER, gp160 accumulates there and is degraded (Courageot et al., 1999, Pal et al., 1991, Willey et al., 1991). Gp160 degradation has also been observed in proteosomes (Bultmann et al., 2000). The consensus view is that most gp160 is degraded in the ER, and only a minority reaches the Golgi. Of the latter, most (85–95%) is degraded in lysosomes, and the remainder is targeted as gp120-gp41 to the cell surface (Willey et al., 1988). Gp120 is relatively stable, and 30–50% can be detected as secreted or intracellular protein 24 h later (Bird et al., 1990, Willey et al., 1988, Willey et al., 1991). The amount of gp160 secreted from the cell is less than 10% of the gp120 secreted (Willey et al., 1988), although the amount varies with the type of host cell (Moulard et al., 1999). Tyrosine-dependent sorting signals (Yxxφ, where x represents any amino acid residue and φ represents a bulky hydrophobic residue), and possibly di-leucine-sorting signals are found in the C-terminal tail of gp41, and have been implicated in transport of gp160 and gp120-gp41 (see Results and discussion section and Berlioz-Torrent et al., 1999, Boge et al., 1998, Bu et al., 2004, Deschambeault et al., 1999, Egan et al., 1996, Lodge et al., 1997, Ohno et al., 1997, Owens et al., 1991, Rowell et al., 1995, West et al., 2002, Wyss et al., 2001).
Gp120 is the distal part of the envelope glycoprotein trimer, and recognizes the primary CD4 receptor, and coreceptors on the target cell. Gp41 anchors the envelope glycoprotein in the cell or virion membrane, and mediates the virion fusion entry process that is activated by receptor binding. The N-terminal peptide of gp41 is inserted into the membrane of the target cell and leads to fusion of the virion and cell membranes, entry of the virus genome and associated proteins into the cell, and infection. Gp41 comprises an ectodomain, a single membrane-spanning domain (MSD), and a long C-terminal tail that in the past has been viewed as being entirely contained inside the cell or virion (Gallaher, 1987, Gallaher et al., 1989, Gonzalez-Scarano et al., 1987, Levy, 1998, White, 1990). However, there is abundant evidence that HIV-1 virions can be neutralized by antibodies directed to an epitope in the C-terminal tail (Buratti et al., 1998, Chanh et al., 1986, Cleveland et al., 2000a, Cleveland et al., 2000b, Cleveland et al., 2003, Dalgleish et al., 1988, Durrani et al., 1998, Ho et al., 1987, McInerney et al., 1999, McLain et al., 1995, McLain et al., 1996, McLain et al., 2001, Newton et al., 1995, Reading et al., 2003). Since antibodies do not cross lipid bilayers, this means that part of the tail is exposed on the virion surface. We have called this exposed region of the gp41 tail the minor ectodomain, to distinguish it from the better known, and larger, major ectodomain. Non-neutralizing antibodies specific for the minor ectodomain have also been shown to bind virions (Cleveland et al., 2003, McLain et al., 2001), providing further evidence of the externalization of the minor ectodomain. Binding of antibody is abrogated by pre-treatment of virions with protease, again supporting the external location of this part of the tail (Cleveland et al., 2003). More recently, we demonstrated for the first time that neutralizing and non-neutralizing antibodies to the minor ectodomain bind to infected cells and that the neutralizing antibodies inhibit fusion of infected and non-infected cells (Cheung et al., 2005, Heap et al., 2005). Thus, part of the gp41 C-terminal tail is exposed on the surface of both infected cells and virions.
Until now, evidence for the exposure of part of the gp41 C-terminal tail has been based on the study of HIV-1 B clade, T cell line-adapted (TCLA) viruses, and the generality of the existence of the minor ectodomain, and its structural basis, have not been explored. To remedy this, we have here analyzed 357 gp41 C-terminal tail database sequences from clades A, B, C, and D. This analysis shows that all these could potentially have a minor ectodomain of approximately 40 residues, supported by three MSDs, and an internal tail of approximately 100 residues. We shall further suggest that this 3-MSD form of gp41 coexists with the accepted 1-MSD form of gp41, and that the 3-MSD form represents a minor population of intracellular gp41 that is destined for incorporation into virions. In contrast, 1-MSD gp41 is the majority form of intracellular gp41, most of which is degraded.
Results and discussion
Sequence comparisons
Regions of conservation in the gp41 sequence 690–793 of HIV-1 clades A to D are summarized in Fig. 1 , with residues numbered according to Ratner et al. (1985). The only very highly conserved region throughout all clades is 713NRVRQGYSPLSFQ725, which contains the most N-terminal (first) tyrosine-dependent sorting signal (719YSPL722). As expected, the region comprising the accepted MSD, 690KIFIMIVGGLIGLRIVFAVLSIV712, is highly conserved and forms part of a larger highly conserved sequence 690KIFIMIVGGLIGLRIVFAVLSIVNRVRQGYSPLSFQ725, which includes the region surrounding the first tyrosine-dependent sorting signal. However, there are conservative substitutions in individual clade consensus sequences of 700I-V (clade B), 705V-I (clade C), 711I-L (clade D), and 712V-I (clade A). The region 767RSLCLFSYHRLR779 containing the second potential tyrosine-dependent sorting signal (775YHRL778) is highly conserved. The highly conserved 790ELLG793 contains a potential di-leucine signal, but the only di-leucine signal known to be functional in gp41 is 862LL863 (Wyss et al., 2001).
Fig. 1.
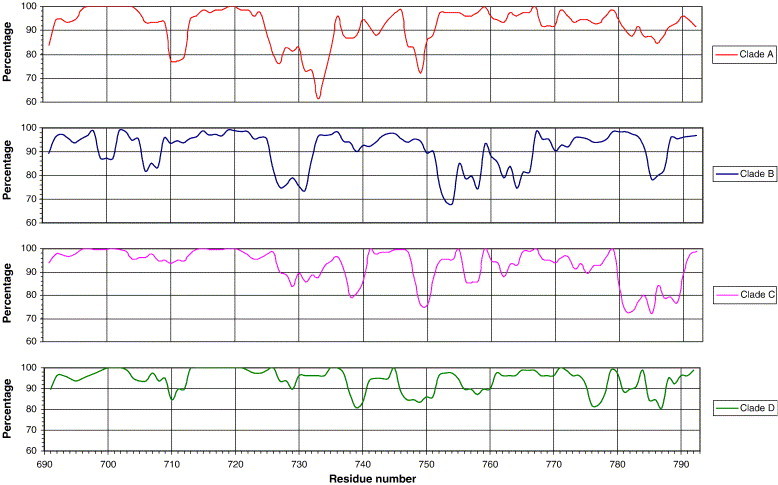
Summary of the conservation of gp41 C-terminal amino acid residues 690–793 of HIV-1 clades A to D, using a 7-residue moving window.
The Kennedy sequence (731PRGPDRPGRIEEEGGEQDRDRS752) in clade B viruses contains the neutralizing epitope core sequence 746ERDRD750, and two non-neutralizing epitopes (734PDRPEG739 and 740IEEE743) (Chanh et al., 1986, Dalgleish et al., 1988, Evans et al., 1989, Ho et al., 1987, Kennedy et al., 1986, Niedrig et al., 1992, Vella et al., 1993). Overall the Kennedy sequence is only poorly to moderately conserved. Specifically 731PRGPDRPGRI740 and 747QDRDRS752 are poorly to moderately conserved, but 741EEEGGE746 is moderately to highly conserved. 741EEEGGE746 is notable in having 67% acidic residues. 746ERDRD750 is present only in clade B and the recombinant CRF03_AB clade. Clades A and C have 746EQDRD750 and clade D has 746EQGRG750. The exchange of 747R (basic, ionizable, hydrophilic, with a long side chain) for Q (polar, hydrophilic, amidic, with a shorter side chain) might create a different epitope, and 746ERDRD750-specific neutralizing antibody may not recognize 746EQDRD750.
The gp41 tail reading frame overlaps the second exons of tat (+1 relative to env) and rev (+2). Both of the latter start at the same position, equivalent to the codon for residue 725 of Env. Tat and Rev stop at the equivalent of the codons for residues 740 and 816 of Env, respectively. This is the only part of the env sequence where all three ORFs are used, yet counter-intuitively, conservation of 725–740 of Env ranges from poor to high. It may be that the tat and/or rev sequences are conserved at the expense of env. All clades have a very highly conserved 745G (100%), 758G (99.9%), and 764W (99.4%). The conservation of 745G and 758G may be a consequence of the reading frame shared with Rev. The last two bases of a rev codon are the first two bases of the env codon. Thus, if rev requires a tryptophan in its second exon, the overlapping codon of env has to be glycine, since tryptophan is only encoded by UGG, while the codon for glycine is GGx. The reason why 764W is very highly conserved is not clear, but any change in its codon results in a different amino acid residue or a stop codon (UGA or UAG). Regions of conservation for clades A to D consensus sequences of gp41 residues 690–793, and an overall consensus sequence are summarized in Fig. 2 .
Fig. 2.

Consensus sequences for gp41 C-terminal amino acid residues 690–793 of HIV-1 clades A to D combined (top line), and for clades A to D individually. The potential tyrosine-dependent sorting signals are in red. A potential di-leucine signal is pink. Residues common to the majority of sequences are in black, while residues that vary are in green. Where two residues are equally represented between the consensus sequences of the four clades, the overall consensus sequence is based on the larger number of sequences analyzed. The proposed first, second, and third MSDs are overlined. Underlined is the antigenically active Kennedy sequence.
Structure predications
The consensus sequences of HIV-1 clades A to D derived above were analyzed individually (see Materials and methods), but as the predicted structures were virtually identical, only clade A data are presented. According to Kyte and Doolittle (1982), four regions (690–699, 702–711, 758–763, and 783–787) have hydropathy values of >1.6, indicating that they are potential MSDs (Fig. 3a). There are two single point peaks of 1.6 at residues 771 and 781, but the values of surrounding residues are too low for these to form an MSD. The Kennedy region (731–752) is highly hydrophilic, with values of mainly −1, suggesting that it is exposed to solvent. Essentially the same conclusions were reached (data not shown) using other hydropathy prediction algorithms (Eisenberg et al., 1984, Hopp and Woods, 1981, Sweet and Eisenberg, 1983).
Fig. 3.
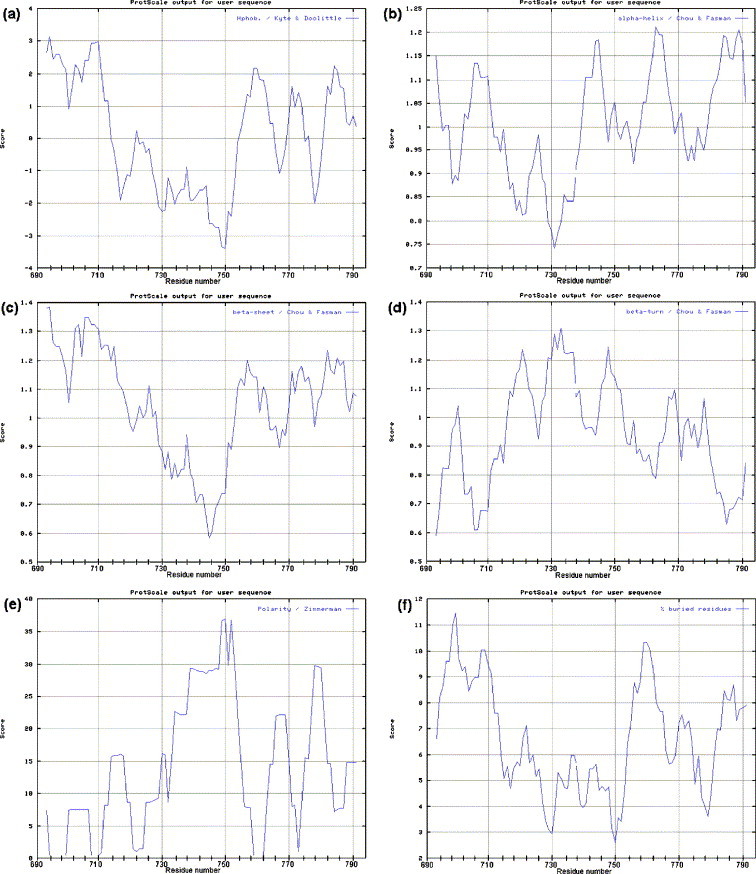
Analysis of the structure of the gp41 C-terminal residues 690–793 of HIV-1 clade A using various algorithms: (a) hydropathy prediction, where high values represent the more hydrophobic regions; (b) α-helix prediction, where high values indicate a greater probability of α-helical structure; (c) β-sheet prediction, where high values indicate a greater probability of β-sheet structure; (d) β-turn prediction, where high values indicate a greater probability of a β-turn; (e) polarity prediction, where high values suggest high polarity; and (f) buried residue prediction, where high values suggest buried residues.
Five regions (690–694, 704–711, 740–747, 758–766, and 779–793) have α-helix potentials of >1.03 (Fig. 3b), and four regions (690–719, 753–761, 770–777, and 779–788) have β-sheet values of >1.05 (Fig. 3c). The most likely conformation is that with the highest predicted value (Chou and Fasman, 1978). Thus, 690–719 (1.34) is likely to be β-sheet. In this region there is a dip to 1.05 at residue 702, suggesting that 690–719 may comprise two discrete regions (690–701 and 703–719). This is consistent with the two hydropathic regions predicted above. There are two regions with different conformations adjacent to each other at 753–766, with 753–761 showing higher β-sheet potential and 761–766 showing greater α-helix potential. 779–788 could be either α-helix or β-sheet. The Kennedy region (731–752) shows no potential for β-sheet formation with values <0.95. However, 740–747 of the Kennedy region that in clade B TCLA viruses contains the highly immunogenic and antigenic epitope 740IEEE743 (Cleveland et al., 2000a) may form an α-helix (>1.03).
There are potential β-turns at 702, 721, 731, 733, 748, 768, and 778 (Fig. 3d). The β-turn at 702 is consistent with the hydropathy andβ-sheet predictions that this links two discrete MSDs. Residue 721P is present in most of the first tyrosine-dependent sorting signals (719YSPL722) and is discussed later. The Kennedy region contains potential β-turns at 731, 733, and 748, and with the prediction of only one short region of α-helix and no β-sheet, the region is probably unstructured. The potential β-turn at 749R might be important for maintaining the complex conformational neutralizing epitope 746ERDRD750 found in clade B virions (Buratti et al., 1998, Cheung et al., 2005, Cleveland et al., 2000a, Cleveland et al., 2000b, Cleveland et al., 2003, Heap et al., 2005, McLain et al., 2001, Reading et al., 2003, Vella et al., 1993). β-turns at 768 and 778 would occur after the putative third MSD (see below).
There are four highly non-polar regions (694–699, 708–711, 720–725, and 759–762) (Fig. 3e). Regions 694–699 and 708–711 coincide with the MSD 1 and 2 predicted above. The unequal polarity of these two domains adds to the suggestion that they are separate entities. Residues 720–725 contain the first tyrosine-dependent sorting signal, but the significance of its lack of polarity is not clear. Residues 759–762 coincide with the possible location of the third MSD predicted above (754–763). Region 780–793 is moderately polar and unlikely to form an MSD. The Kennedy region is the most polar region in the analyzed sequence, consistent with it being exposed to aqueous solvent and with its antigenic properties.
Regions 694–712 (containing the predicted MSD 1 and 2 at 691–700 and 703–712) and 755–763 are likely to be inaccessible to aqueous solvent (Fig. 3f). The dip at the center of 694–712 (residue 703) supports the prediction that this region comprises two MSDs. Region 755–763 coincides with the predicted third MSD (754–763). The Kennedy region is accessible to solvent.
Taken together, these data suggest that the gp41 of HIV-1 clades A, B, C, and D all have the potential to have three MSDs in a β-sheet conformation. MSD 1 and 2 are connected by a short β-turn (701GL702), and MSD 2 and 3 support the highly antigenic Kennedy sequence on the outside of the cell or virion. Further the MSDs all have significant parallel and anti-parallel β-strand potential (Lifson and Sander, 1979; data not shown). The 1- and 3-MSD forms of gp41 are shown schematically in Figs. 4a and b. The significance of the new positioning of the potential tyrosine-sorting signals is discussed below. While MSDs are typically α-helical and approximately 20 residues in length (Sabatini et al., 1982, Singer, 1990), β-sheets as short as 7 residues form MSDs as part of transmembrane β-barrel proteins of bacteria, choroplasts, and mitochondria (Schultz, 2003). Short β-turns connect sequential MSDs and do not normally occur within MSDs (Jahnig, 1990), but the β-turn may intrude into the membrane, as suggested for herpes simplex virus glycoprotein B (Pellett et al., 1985). An arginine residue is unlikely to occur in the middle of an MSD (Singer, 1990), and R703 of the 3-MSD form of gp41 is predicted to be on the membrane surface, where the polar head groups of membrane lipids can neutralize its positive charge. However, we shall suggest below that the 3-MSD gp41 is a minority form that is selectively incorporated into the plasma membrane and virions, and that most intracellular gp41 exists in the 1-MSD conformation. Finally preliminary sequence and structural analysis suggests that other primate lentiviruses (HIV-2, SIV) have the potential to form a 3-MSD structure (unpublished data).
Fig. 4.
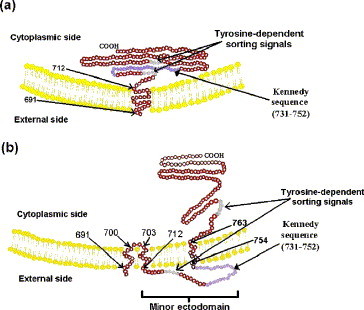
Schematic showing the suggested membrane insertion of a monomer of (a) the 1-MSD and (b) the 3-MSD forms of gp41 of HIV-1 clades A to D. The positions of the two potential tyrosine-sorting signals and the minor ectodomain containing the Kennedy sequence are shown. The major ectodomain at the N-terminus of gp41 and gp120 have been omitted. The distance shown in (b) between MSD 2 and 3 (reading from left to right) is conjectural.
Conclusions
Gp41 molecules of HIV-1 clades A to D all have the potential to form three short MSDs, which most likely have a β-sheet conformation. The position of the third MSD places the first potential tyrosine-dependent sorting signal outside the membrane, where it is non-functional. All viruses have a hydrophilic, unstructured region of 41 residues supported by MSDs 2 and 3 that probably equates to the antigenically and biologically active minor ectodomain of HIV-1 clade B, TCLA viruses. The definition and values of the peaks obtained in the above analysis make the data highly significant. The position and number of residues in the MSDs and other regions of interest are summarized in Table 1 and Fig. 5 . The latter also shows possible interactions of elements of the proposed new 3-MSD conformation.
Table 1.
Predicted location of residues in the 3-MSD conformation of the gp41 of HIV-1 clades A to D in the virion or cell membrane
| Position | Number of residues | |
|---|---|---|
| Major ectodomain | 512–690 | 179 |
| MSD 1 | 691–700 | 10 |
| Turn | 701–702 | 2 |
| MSD 2 | 703–712 | 10 |
| Minor ectodomain (external loop) | 713–753 | 41 |
| MSD 3 | 754–763 | 10 |
| Internala | 764–863 | 100 |
Comprises 30 residues analyzed here, and 70 not analyzed.
Fig. 5.
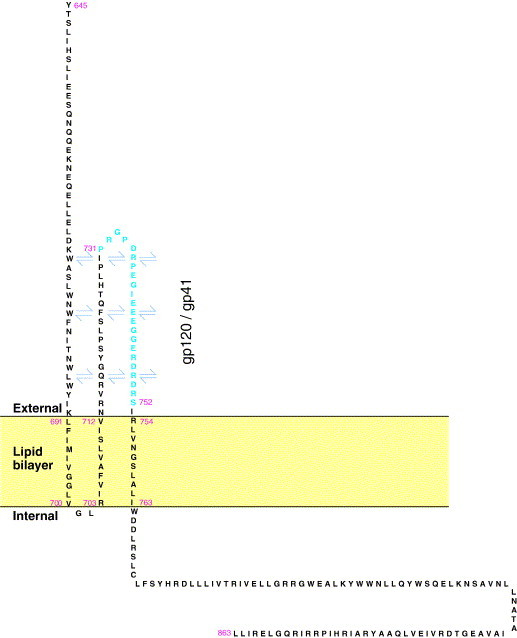
Representation of some possible interactions of the minor ectodomain (residues 713–753) of the 3-MSD form of gp41 with other elements of the Env trimer. Shown are residues 645–863, comprising part of the main ectodomain, the MSDs, the minor ectodomain, and the entire C-terminal tail of the gp41 of HIV-1 NL-4.3 (clade B). Only a monomer is represented. There may be interactions between MSD 1, 2, and 3, between the minor ectodomain and the gp41 major ectodomain, between the minor ectodomain and elements of gp120, or with the other gp41 monomers that form the trimer (arrows). The nine MSDs of the trimer could also interact with each other. The antigenically active Kennedy sequence (731–752) containing neutralizing and non-neutralizing epitopes is shown as the outer face of the minor ectodomain.
Possible implications of the existence of multiple MSDs on the functioning of tyrosine-dependent sorting signals in the C-terminal tail of gp41
The HIV-1 gp41 tail has an (N-terminal or first) tyrosine signal (719YSPL722) that functions in endocytosis (Berlioz-Torrent et al., 1999, Boge et al., 1998, Deschambeault et al., 1999, Egan et al., 1996, Ohno et al., 1997, Rowell et al., 1995, West et al., 2002) and basolateral sorting (Deschambeault et al., 1999, Lodge et al., 1997, Owens et al., 1991). The signal sequence and upstream residues are highly to very highly conserved in clades A to D: 716R = 100%, 717Q = 96.4%, 718G = 99.95%, 719Y = 99.7%, 720S = 99.5%, 721 P = 99.8%, 722L = 96.9%. The glycine immediately before a tyrosine signal signifies that it can function in the TGN to target the protein to lysosomes. Also for signal functionality there is a strict requirement that the tyrosine residue is the 7th–11th residue from the membrane (Rohrer et al., 1996). Gp41 is also involved in basolateral localization of envelope protein in the plasma membrane of polarized cells (Owens et al., 1991), a property that is lost when the tyrosine of the first signal is substituted (Deschambeault et al., 1999, Lodge et al., 1997). In the 1-MSD model of gp41, Gallaher et al. (1989) have proposed that the MSD ends at residue 712V, and thus 719Y will be the 7th residue from the membrane, and within the required distance for optimum function. Much depends on knowing precisely which residue ends the MSD. Often a charged residue that acts as a stopper defines the end, and it is possible that the gp41 MSD could end at 714R. If so, 719Y would be 5 residues from the membrane, and this might compromise the endocytosis, basolateral sorting, and lysosomal targeting functions of the signal. The minimum distance of the tyrosine-signal from the membrane is required for both direct lysosomal targeting and endocytosis (Collawn et al., 1990, Pytowski et al., 1995, Trowbridge and Collawn, 1992, Trowbridge et al., 1993). However, only lysosomal targeting signals have a strict maximum distance from the membrane (Collawn, 1990; Rohrer et al., 1996, Trowbridge and Collawn, 1992, Trowbridge et al., 1993). The fact that the first tyrosine signal functions in endocytosis (as referenced above) argues that it is indeed at the required distance from the membrane, but at the time of writing there are no experimental data to show if the signal is active in directing gp41 to lysosomes. The 3-MSD model, as stated above, puts the first tyrosine signal outside the membrane where it is non-functional for any transport function (Fig. 5).
Adaptor protein (AP) complexes interact with tyrosine-sorting signals. AP-1 and AP-3 complexes are mainly found in the TGN and function in lysosomal targeting, while AP-2 is predominantly localized to the plasma membrane and functions in endocytosis. They have distinct preferences for specific residues or combinations of residues of the tyrosine signals, although there is overlap, particularly with AP-1 and AP-3 complexes (Table 2 ) (Boll et al., 1996, Ohno et al., 1996, Ohno et al., 1998). Table 2 shows that the consensus sequence of the first tyrosine-sorting signal of the gp41 tail of clades A to D most closely matches the preferences of AP-1 and AP-3 complexes. The arginine residue at position Y − 3 of the signal is 100% conserved, and there is almost complete conservation of the glycine at Y − 1 (99.95%), the tyrosine itself (99.7%), and the proline at Y + 2 (99.8%). The leucine residue at Y + 3 is 96.9% conserved, but if leucine, isoleucine, and valine at Y + 3 (all with similar properties and tolerated at this position of the tyrosine signal) are summed, conservation reaches 98.4%. Thus, the first tyrosine-sorting signal (719YSPL722) could interact with the AP-1 and AP-3 complexes in the TGN and target gp41 to the lysosomes. However, as stated above, this signal functions at the plasma membrane (Berlioz-Torrent et al., 1999, Boge et al., 1998, Deschambeault et al., 1999, Egan et al., 1996, Ohno et al., 1997, Rowell et al., 1995, West et al., 2002), and is not known to be active in the TGN. It may be that the amount of gp41 synthesized saturates the lysosomal targeting system in the TGN, allowing gp41 to reach the cell surface and the first tyrosine-sorting signal to function as an endocytosis signal. The possibility that the signal targets gp41 to the lysosomes is consistent with the observation that the majority of gp160 that reaches the Golgi is degraded in the lysosomes (Willey et al., 1988). AP-2 complexes have the broadest specificity range and associate with the same signals as AP-1 and AP-3 complexes (Ohno et al., 1998). The endocytosis function of the first tyrosine-sorting signal may enable plasma membrane gp41 to be re-directed to the lysosomes if it escapes that route initially (Ohno et al., 1998).
Table 2.
Comparison of the preferences of AP complexes for cellular tyrosine-dependent sorting signalsa with the consensus sequence for the first and second potential tyrosine-dependent sorting signals of the gp41 of HIV-1 clades A to D
| Position (relative to Y) | AP-1 preferences | AP-2 preferences | AP-3 preferences | HIV-1 tyrosine-dependent sorting signal sequences |
|
|---|---|---|---|---|---|
| First | Second | ||||
| − 3 | R | G | R | R | L |
| − 2 | S | F | Np | Q | F |
| − 1 | L/D | P | D/E/G | G | S |
| 0 (Y) | Y | Y | Y | Y | Y |
| 1 (x) | Np | Np | E | S | H |
| 2 (x) | P | P/R | P | P | R |
| 3 (φ) | L | L | I | L | L |
Np, no preference known at this position; x, any amino acid residue; φ, a bulky hydrophobic residue.
The second tyrosine-dependent sorting signal (775YHRL778) has no preceding glycine, and is non-functional in the context of the 1-MSD model of gp41 (Boge et al., 1998, Rowell et al., 1995). It is situated 63 and 12 residues from the membrane in the 1-MSD and 3-MSD models, respectively, the latter being close to the optimal 7-11 residue distance. The residues of the second signal are variably conserved: 772L = 94.5%, 773F = 96.8%, 774S = 86.0%, 775Y = 99.7%, 776H = 89.3%, 777R = 86.5%, and 778L = 99.8%. The F at position Y − 2, R at Y + 2, and L at Y + 3 suggest that the signal interacts with the AP-2 complex (Table 2). The lack of an R at Y − 3, glycine at Y − 1, and proline at Y + 2, and the fact that it is not within the favored distance from the membrane (Rohrer et al., 1996), indicate that this signal is not optimal for interacting with AP-1 or AP-3 complexes. AP-3 complexes disfavor serine at position Y − 1 and makes this interaction less likely (Ohno et al., 1996, Ohno et al., 1998). As yet there is no evidence that this signal is functional in gp41 (Boge et al., 1998, Rowell et al., 1995). A peptide containing the signal interacted strongly with the medium subunit of AP-2 (Boge et al., 1998, Ohno et al., 1997), but the presence of a major upstream sequence in the 1-MSD model could alter its environment and its possible interaction with the AP-2 complex.
Conclusions
All HIV-1 gp41 tail sequences have a potential N-terminal GYxxϕ-sorting signal of the type that would be expected to interact strongly with AP-1 and AP-3 complexes. Thus, with the critical 7-residue spacing from the membrane, the signal is likely to be functionally important in targeting TGN gp41 to lysosomes. If the signal was required only for endocytosis, it is unlikely that the G at position Y − 1, R at Y − 3, and the 7-residue spacing from the membrane would be so highly conserved. However, this would not preclude it from functioning as an endocytosis signal at the cell surface. The second Yxxϕ sequence is not so well conserved, is 63 residues from the membrane in the 1-MSD model, and is unlikely to interact with AP-1 and AP-3 complexes, or to be involved in lysosomal targeting from the TGN. However, the Y is almost completely conserved and the bulky hydrophobic residue is highly conserved. In the 3-MSD structure of gp41, only the second signal is on the cytoplasmic side of the membrane, where it could be functional.
In the infected cell there may be populations of gp41 with one MSD and three MSDs
Until recently all primate lentivirus envelope protein anchor components were viewed as having a single MSD (691–712 in HIV-1; Fig. 4a) (Gallaher, 1987, Gallaher et al., 1989, Gonzalez-Scarano et al., 1987, Levy, 1998, White, 1990). However, published work and the discussion above suggest that the gp41 tail crosses the membranes of HIV-1 virions and infected cells three times (Cheung et al., 2005, Cleveland et al., 2003, McLain et al., 2001). To resolve this apparent conflict, we shall argue here that the 1- and 3-MSD forms of gp41 coexist in the infected cell, with the 1-MSD version being the major form. However, we shall propose that only the minor 3-MSD form is incorporated into virions. These proposals are consistent with, and rationalize the observed degradation of the majority (85–95%) of the1-MSD form of cellular gp160 (Bultmann et al., 2000, Courageot et al., 1999, Jabbar and Nayak, 1990, Pfeiffer et al., 1997, Willey et al., 1988), the apparently contradictory evidence of a functioning first tyrosine-sorting signal (Berlioz-Torrent et al., 1999, Boge et al., 1998, Deschambeault et al., 1999, Egan et al., 1996, Ohno et al., 1997, Rowell et al., 1995, West et al., 2002) and basolateral-sorting signal (Deschambeault et al., 1999, Lodge et al., 1997, Owens et al., 1991), with the immunogenic and antigenic properties of the Kennedy region (Chanh et al., 1986, Dalgleish et al., 1988, Evans et al., 1989, Ho et al., 1987, Kennedy et al., 1986, Niedrig et al., 1992).
The C-terminal tail of the 1-MSD form of HIV-1 gp41 starts at residue 713 (Fig. 4a). As already stated, the majority of this type of gp41 (85–95%) is degraded intracellularly. The Y residue of the first tyrosine-dependent sorting signal (718GYSPL722) is situated precisely 7 residues from the membrane and conforms closely to the requirements for cellular GYxxϕ lysosomal targeting signals (see above). In the 1-MSD form of gp41, the second tyrosine-dependent sorting signal is apparently not needed, and we suggest may not be functional.
The 3-MSD form of HIV-1 gp41 has MSDs at 691–700, 703–712, and 754–763, and its 718GYSPL722 sequence is outside the virion. This model is supported by antigenic and other data showing that residues of the C-terminal tail are exposed on the surface of the virion (Buratti et al., 1998, Chanh et al., 1986, Cleveland et al., 2000a, Cleveland et al., 2000b, Cleveland et al., 2003, Dalgleish et al., 1988, Durrani et al., 1998, Ho et al., 1987, McInerney et al., 1999, McLain et al., 1995, McLain et al., 1996, McLain et al., 2001, Newton et al., 1995, Reading et al., 2003) and the infected cell (Cheung et al., 2005, Heap et al., 2005). Furthermore, we propose that this form of gp41 represents the 5–15% of the TGN gp160 that is directed to the cell surface. Its GYxxϕ signal is outside the cell membrane and cannot function in lysosomal targeting or endocytosis, but the second potential tyrosine-dependent sorting signal (775YHRL778), which has none of the requirements for lysosomal targeting (see above), is 12 residues from the membrane, and well situated to function in endocytosis and recycling of cell membrane-inserted gp41. Because of its location this gp41, in association with gp120, is the major gp41 form in the cell membrane, and the major gp41form incorporated into virions. Co-existence of two forms of gp41 raises questions, which will require further work to answer. The mechanism by which two forms of gp41 arise is not known, although translocational pausing may be involved in formation of the multiple MSDs (Dettenhofer and Yu, 2001). The 1- and 3-MSD forms are proposed to be α-helix and β-sheet, respectively, and it is noted above that structure predications allow for either conformation. Conformation may be determined by the length of unbroken MSD sequence, as a long sequence of lower value α-helix, can take precedence over a higher value, shorter β-sheet region (Chou and Fasman, 1978). The conventional 1-MSD model still has the problem of 703R being centrally located in the membrane with no counter charge. However, a recent suggestion that the 703R equivalent in SIV is situated in a position where it can react with the polar lipid head groups may provide a solution (West et al., 2001).
Degradation of the 1-MSD gp41 and recycling of the 3-MSD form would both act to reduce the surface expression of gp120-gp41. This may be important, as high intracellular concentrations of gp41 can be cytotoxic (Arroyo et al., 1995, Chernomordik et al., 1994, Comardelle et al., 1997, Gawrisch et al., 1993, Miller et al., 1993, Zhang et al., 1996), and provoke immune responses against the infected cell. It may be that such post-translational control measures, utilizing tyrosine-dependent sorting signals and the host cell's degradation pathways, have evolved as envelope expression is not easily controlled at a genetic level due to the overlap of the env, tat, and rev ORFs.
The suggestion above that a membrane-inserted viral protein can have different numbers of MSDs is not unique. For example, the GL envelope protein of equine arteritis virus is proposed to have 1 or 3 MSDs (Snijder and Meulenberg, 1998), the M protein of transmissible gastroenteritis coronavirus and equine arteritis virus, and the S antigen of hepatitis B virus are proposed to have three or four MSDs (Prange and Streeck, 1995, Risco et al., 1995, Snijder and Meulenberg, 1998), and the herpes simplex virus glycoprotein B (Pellett et al., 1985), and the Epstein–Barr virus 58 kDa latent protein (Fennewald et al., 1984, Hennessy et al., 1984) both have multiple MSDs.
Summary
Based on an analysis of their sequence and structure, we propose that the gp41 transmembrane region and C-terminal tail of all HIV-1 clades A to D can exist in two conformations, with either 1 MSD (the conventional structure) or with 3 MSDs. We suggest that these are, respectively, the majority and minority forms of intracellular Env. In the 3-MSD form, MSD 1 and MSD 2 are separated by a highly conserved beta turn, while the MSD 2 and MSD 3 support an unstructured hydrophilic loop/minor ectodomain of 41 residues that in clade B strains is highly antibody-reactive and involved in fusion. All viruses have two potential tyrosine-dependent sorting signals within the region analyzed. In the 1-MSD model it is likely that only the N-terminal signal is functional, and that this interacts with AP-1 and AP-3 to direct Env from the TGN to degradation in the lysosomes. In the 3-MSD version, the N-terminal signal is situated outside the membrane and non-functional, thus allowing this form of gp41 to reach the cell membrane. Thus, it seems that the 3-MSD form is the majority species on the cell surface and hence in virions. We propose that the second signal is functional in the 3-MSD gp41, and controls recycling of cell surface Env. The 1- and 3-MSD strategy can be seen as an evolutionary adaptation that allows HIV-1-infected cells to evade the immune system or to avoid gp41-induced cytotoxicity.
Materials and methods
Sequence analysis
HIV-1 gp41 sequences from infectious viruses, molecular clones, and PCR products from blood samples from infected individuals were obtained from http://hiv-web.lanl.gov/. We analyzed residues 690–793 of HIV-1 (numbering according to Ratner et al., 1985) of the following number of sequences: clade A, n = 25; clade B, n = 245; clade C, n = 61; clade D, n = 26. The sequence analyzed comprises the MSD and approximately two thirds of the C-terminal tail. Conserved regions of sequence were aligned, with spaces as necessary to maintain alignment. The residue occupying each position was recorded as a percentage, and consensus sequences constructed for each clade. Sequence conservation is defined here as poor (<80%), moderate (80–89.9%), high (90–96.4%), and very high (96.5–100%).
Structure predications
For structure predications we used the consensus sequences derived in this report for the gp41 MSD and C-terminal region of HIV-1 clades A to D. Hydropathy values assigned to the amino acids are based on water vapor transfer free energies and the interior–exterior distribution of amino acid side chains (Kyte and Doolittle, 1982). This system predicts that a region with a value >1.6 is likely to be an MSD, and that a region with a value >1.09 is likely to be sequestered inside the protein. α-Helices and β-sheets were predicted according to Chou and Fasman (1978), where P α is the helix conformational parameter and P β is the β-sheet conformational parameter. Any segment of ≥6 residues with (P α) ≥1.03 and (P α) > (P β) is predicted to be α−helical, and any segment of three residues or longer in a native protein with (P β) ≥1.05 and (P β) > (P α) is predicted to be β-sheet. A segment containing overlapping α- and β- residues is resolved through conformational boundary analysis so that (P α) > (P β) is α-helical, and (P β) > (P α) is β-sheet. A β-turn occurs where a polypeptide folds back on itself by nearly 180° and typically requires four consecutive residues (Chou and Fasman, 1978). However, a β-turn of two residues occurs between two MSDs of herpes simplex virus glycoprotein B (Pellett et al., 1985). The lower cut-off value for β-turns of 0.75 was used here (Chou and Fasman, 1978). Polarity was determined according to Zimmerman et al. (1968). Accessibility of a region of a protein to solvent was predicated using the percentage buried residues index (Janin, 1979). Low values indicate a region that is likely to be accessible to solvent and hence surface exposed, and high values indicate regions not accessible to the solvent. Predications used a moving window of 7 residues.
Acknowledgments
We thank Richard Compans for reading the manuscript and his advice and are pleased to acknowledge the financial support of AVERT, UK.
References
- Arroyo J., Boceta M., González M.E., Michel M., Carrasco L. Membrane permeabilization by different regions of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J. Virol. 1995;69:4095–4102. doi: 10.1128/jvi.69.7.4095-4102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlioz-Torrent C., Shacklett B.L., Erdtmann L., Delamarre L., Bouchaert I., Sonigo P., Dokhelar M.C., Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adapter complexes modulate intracellular and cell surface expression of envelope glycoprotein. J. Virol. 1999;73:1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird C., Burke J., Gleeson P.A., McCluskey J. Expression of human immunodeficiency virus type 1 (HIV-1) envelope gene products transcribed from a heterologous promoter. J. Biol. Chem. 1990;265:19151–19157. [PubMed] [Google Scholar]
- Boge M., Wyss S., Bonifacino J.S., Thali M. A membrane-proximal signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- Boll W., Ohno H., Songyang Z., Rapoport U., Cantley L.C., Bonifacino J.S. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 1996;15:5789–5795. [PMC free article] [PubMed] [Google Scholar]
- Bu Z., Ye L., Vzorov A., Taylor D., Compans R.W., Yang C. Enhancement of immunogenicity of an HIV Env DNA vaccine by mutation of the Tyr-based endocytosis motif in the cytoplasmic tail. Virology. 2004;328:62–73. doi: 10.1016/j.virol.2004.06.041. [DOI] [PubMed] [Google Scholar]
- Bultmann A., Eberle J., Haas J. Ubiquitination of the human immunodeficiency virus type 1 Env protein. J. Virol. 2000;74:5373–5376. doi: 10.1128/jvi.74.11.5373-5376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E., McLain L., Tisminetzky S.G., Cleveland S.M., Dimmock N.J., Baralle F.E. The neutralizing antibody response against a conserved region of HIV-1 gp41 (amino acid residues 731–752) is uniquely directed against a conformational epitope. J. Gen. Virol. 1998;79:2709–2716. doi: 10.1099/0022-1317-79-11-2709. [DOI] [PubMed] [Google Scholar]
- Chanh T.C., Dreesman G., Kanda P., Linette G.P., Sparrow J.T., Ho D.D., Kennedy R.C. Induction of anti-HIV neutralizing antibodies by synthetic peptides. EMBO J. 1986;5:3065–3071. doi: 10.1002/j.1460-2075.1986.tb04607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L., Chanturiya A.N., Suss-Toby E., Nora E., Zimmerberg J. An amphipathic peptide from the C-terminal region of the human immunodeficiency virus envelope glycoprotein causes pore formation in membranes. J. Virol. 1994;68:7115–7123. doi: 10.1128/jvi.68.11.7115-7123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung L., McLain L., Hollier M.J., Reading S.A., Dimmock N.J. Part of the C-terminal tail of the envelope gp41 transmembrane protein of human immunodeficiency virus type 1 (HIV-1) is exposed on the cell surface and is involved in virus-mediated cell-cell fusion. J. Gen. Virol. 2005;86:131–138. doi: 10.1099/vir.0.80439-0. [DOI] [PubMed] [Google Scholar]
- Chou P.Y., Fasman G.D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. 1978;47:45–147. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cleveland S.M., Buratti E., Jones T.D., North P., Baralle F.E., McLain L., McInerney T.L., Durrani Z., Dimmock N.J. Immunogenic and antigenic dominance of a non-neutralizing epitope over a highly conserved neutralizing epitope in the gp41 transmembrane envelope glycoprotein of HIV-1: its deletion leads to a strong neutralizing antibody response. Virology. 2000;266:66–78. doi: 10.1006/viro.1999.0041. [DOI] [PubMed] [Google Scholar]
- Cleveland S.M., Jones T.D., Dimmock N.J. Properties of a neutralizing antibody that recognises a conformational form of epitope ERDRD in the C-terminal tail of human immunodeficiency virus type 1. J. Gen. Virol. 2000;81:1251–1260. doi: 10.1099/0022-1317-81-5-1251. [DOI] [PubMed] [Google Scholar]
- Cleveland S.M., McLain L., Cheung L., Jones T., Hollier M., Dimmock N.J. A region of the C-terminal tail of the gp41 envelope glycoprotein of human immunodeficiency virus type 1 contains a neutralizing epitope: evidence for its exposure on the surface of the virion. J. Gen. Virol. 2003;84:591–602. doi: 10.1099/vir.0.18630-0. [DOI] [PubMed] [Google Scholar]
- Collawn J.F., Stangel M., Kuhn L.A., Esekogwu V., Jing S., Trowbridge I.S., Tainer J.A. Transferrin receptor internalization sequence YXRF implicates a tight turn as the structural recognition motif for endocytosis. Cell. 1990;63:1061–1072. doi: 10.1016/0092-8674(90)90509-d. [DOI] [PubMed] [Google Scholar]
- Comardelle A.M., Norris C.H., Plymale D.R., Gatti P.J., Choi B., Fermin C.D., Haislip A.M., Tencza S.B., Mietzner T.A., Montelaro R.C., Garry R.F. A synthetic peptide corresponding to the carboxyterminus of human immunodeficiency virus type 1 transmembrane protein induces alterations in the ionic permeability of Xenopus laevis oocytes. AIDS Res. Hum. Retroviruses. 1997;13:1525–1532. doi: 10.1089/aid.1997.13.1525. [DOI] [PubMed] [Google Scholar]
- Courageot J., Fenouillet E., Bastiani L., Miquelis R. Intracellular degradation of the HIV-1 envelope glycoprotein. Evidence for, and some characteristics of, an endoplasmic reticulum pathway. Eu. J. Biochem. 1999;260:482–489. doi: 10.1046/j.1432-1327.1999.00193.x. [DOI] [PubMed] [Google Scholar]
- Dalgleish A.G., Chanh T.C., Kennedy R.C., Kanda P., Clapham P.R., Weiss R.A. Neutralization of diverse strains of HIV-1 by monoclonal antibodies raised against a gp41 synthetic peptide. Virology. 1988;165:209–215. doi: 10.1016/0042-6822(88)90674-5. [DOI] [PubMed] [Google Scholar]
- Dash B., McIntosh A., Barrett W., Daniels R. Deletion of a single N-linked site from the transmembrane envelope protein of human immunodeficiency virus type 1 stops cleavage and transport of gp160 preventing Env-mediated fusion. J. Gen. Virol. 1994;75:1389–1397. doi: 10.1099/0022-1317-75-6-1389. [DOI] [PubMed] [Google Scholar]
- Deschambeault J., Lalonde J.-P., Cervantes-Acpsta G., Lodge R., Cohen E.A., Lemay G. Polarized human immunodeficiency virus budded in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J. Virol. 1999;73:5010–5017. doi: 10.1128/jvi.73.6.5010-5017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettenhofer M., Yu X.-F. Characterization of the biosynthesis of human immunodeficiency virus type 1 Env from infected T-cells and the effects of glucose trimming of Env on virion infectivity. J. Biol. Chem. 2001;276:5985–5991. doi: 10.1074/jbc.M008933200. [DOI] [PubMed] [Google Scholar]
- Dewar R.L., Vasudevachari M.B., Natarajan V., Salzman N.P. Biosynthesis and processing of human immunodeficiency virus type 1 envelope glycoproteins: effects of monensin on glycosylation and transport. J. Virol. 1989;63:2452–2456. doi: 10.1128/jvi.63.6.2452-2456.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing T.-D., Fang H., Dorward D.W., Pincus S.H. Processing of the envelope protein gp160 in immunotoxin-resistant cell lines chronically infected with human immunodeficiency virus type 1. J. Virol. 1995;69:7122–7131. doi: 10.1128/jvi.69.11.7122-7131.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrani Z., McInerney T.L., McLain L., Jones T., Bellaby T., Brennan F.R., Dimmock N.J. Intranasal immunization with a plant virus expressing a peptide from HIV-1 gp41 stimulates better mucosal and systemic HIV-1-specific IgA and IgG than oral immunization. J. Immunol. Methods. 1998;220:93–103. doi: 10.1016/s0022-1759(98)00145-8. [DOI] [PubMed] [Google Scholar]
- Earl P.L., Moss B.A., Doms R.W. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M.A., Carruth L.M., Rowell J.F., Yu X., Siliciano R.F. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor. J. Virol. 1996;70:6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komarony M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Evans D.J., McKeating J.A., Meredith J.M., Burke K.L., Katrak K., Ferguson M., Minor P.D., Weiss R.A., Almond J.W. An engineered poliovirus chimaera elicits broadly reactive HIV-1 neutralizing antibodies. Nature (London) 1989;339:385–388. doi: 10.1038/339385a0. [DOI] [PubMed] [Google Scholar]
- Fennewald S., van Santen V., Kieff E. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J. Virol. 1984;51:411–419. doi: 10.1128/jvi.51.2.411-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenouillet E., Jones I.M. The glycosylation of human immunodeficiency virus type 1 transmembrane protein (gp41) is important for the efficient transport of the envelope precursor gp160. J. Gen. Virol. 1995;76:1509–1514. doi: 10.1099/0022-1317-76-6-1509. [DOI] [PubMed] [Google Scholar]
- Gallaher W.R. Detection of fusion peptide sequence in the transmembrane peptide of human immunodeficiency virus. Cell. 1987;50:327–328. doi: 10.1016/0092-8674(87)90485-5. [DOI] [PubMed] [Google Scholar]
- Gallaher W.R., Ball J.M., Garry R.F., Griffin M.C., Montelaro R.C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res. Hum. Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- Gwrisch K., Han K.-H., Yang J., Bergelson L.D., Ferretti J.A. Interaction of peptide fragment 828–848 of the envelope glycoprotein of human immunodeficiency virus type 1 with lipid bilayers. Biochemistry. 1993;32:3112–3118. doi: 10.1021/bi00063a024. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Scarano F., Waxham M.N., Ross A.M., Hoxie J.A. Sequence similarities between human immunodeficiency virus gp41 and paramyxovirus fusion proteins. AIDS Res. Hum. Retroviruses. 1987;3:245–252. doi: 10.1089/aid.1987.3.245. [DOI] [PubMed] [Google Scholar]
- Hallenberger S., Tucker S.P., Owens R.J., Berstein H.B., Compans R.W. Secretion of a truncated form of the human immunodeficiency virus type 1 envelope glycoprotein. Virology. 1993;193:510–514. doi: 10.1006/viro.1993.1156. [DOI] [PubMed] [Google Scholar]
- Heap C.J., Reading S.A., Dimmock N.J. An antibody specific for the C-terminal tail of gp41 of HIV-1 mediates post-attachment neutralization probably through inhibiting virus-cell fusion. J. Gen. Virol. 2005;86:1501–1509. doi: 10.1099/vir.0.80414-0. [DOI] [PubMed] [Google Scholar]
- Heilker R., Spiess M., Crottet P. Recognition of sorting signals by clathrin adapters. BioEssays. 1999;7:558–567. doi: 10.1002/(SICI)1521-1878(199907)21:7<558::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Hennessy K., Fennewald S., Hummel M., Cole T., Kieff E. A membrane protein encoded by Epstein–Barr virus in latent growth-transforming infection. Proc. Natl. Acad. Sci. U.S.A. 1984;81:7207–7211. doi: 10.1073/pnas.81.22.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D.D., Sarngadharan M.G., Hirsch M.S., Schooley R.T., Rota T.R., Kennedy R.C., Chanh T.C., Sato V.L. Human immunodeficiency virus virus-neutralizing antibodies recognise several conserved domains on the envelope glycoprotein. J. Virol. 1987;61:2024–2028. doi: 10.1128/jvi.61.6.2024-2028.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höning S., Griffith J., Geuze H.J., Hunziker W. The tyrosine-based lysosomal targeting signal in lamp-1 mediates sorting into Golgi-derived clathrin-coated vesicles. EMBO J. 1996;15:5230–5239. [PMC free article] [PubMed] [Google Scholar]
- Hopp T.P., Woods K.R. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. U.S.A. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbar M.A., Nayak D.P. Intracellular interaction of human immunodeficiency virus type (ARV-2) envelope glycoprotein gp160 with CD4 blocks the movement and maturation of CD4 to the plasma membrane. J. Virol. 1990;64:6297–6304. doi: 10.1128/jvi.64.12.6297-6304.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnig F. Structure predictions of membrane proteins are not that bad. Trends Biochem. Sci. 1990;15:93–95. doi: 10.1016/0968-0004(90)90188-h. [DOI] [PubMed] [Google Scholar]
- Janin J. Surface and inside volumes in globular proteins. Nature (London) 1979;277:491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- Jose E.S., Munoz-Fernandez M.A., Alarcón B. Megalomycin inhibits HIV-1 replication and interferes with gp160 processing. Virology. 1997;239:303–314. doi: 10.1006/viro.1997.8872. [DOI] [PubMed] [Google Scholar]
- Kantanen M.L., Leinikki P., Kuismanen E. Endoproteolytic cleavage of HIV-1 gp160 envelope precursor occurs after exit from the trans-Golgi network (TGN) Arch. Virol. 1995;140:1441–1449. doi: 10.1007/BF01322670. [DOI] [PubMed] [Google Scholar]
- Kennedy R.C., Henkel R.D., Pauletti D., Allan J.S., Lee T.H., Essex M., Dreesman G.R. Antiserum to a synthetic peptide recognizes the HTLV-III envelope glycoprotein. Science. 1986;231:1556–1559. doi: 10.1126/science.3006246. [DOI] [PubMed] [Google Scholar]
- Kimura T., Nishikawa M., Fukisawa J.-I. Uncleaved env gp160 of human immunodeficiency virus type 1 is degraded within the Golgi apparatus but not lysosomes in COS-1 cells. FEBS Lett. 1996;390:15–20. doi: 10.1016/0014-5793(96)00614-x. [DOI] [PubMed] [Google Scholar]
- Kozarsky K., Penman M., Basiripour L., Haseltine W., Sodroski J., Krieger M. Glycosylation and processing of the human immunodeficiency virus type 1 envelope glycoprotein. J. AIDS. 1989;2:163–169. [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Levy J.A. 2nd ed. ASM Press; Herndon,VA: 1998. HIV and the Pathogenesis of AIDS. [Google Scholar]
- Lifson S., Sander M. Antiparallel and parallel beta-strands differ in amino acid residue preferences. Nature (London) 1979;282:109–111. doi: 10.1038/282109a0. [DOI] [PubMed] [Google Scholar]
- Lodge R., Lalonde J.-P., Lemay G., Cohen E.A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney T.L., Brennan F.R., Jones T.D., Dimmock N.J. Analysis of the ability of five adjuvants to enhance immune responses to a chimeric plant virus displaying a HIV-1 peptide. Vaccine. 1999;17:1359–1368. doi: 10.1016/s0264-410x(98)00388-0. [DOI] [PubMed] [Google Scholar]
- McLain L., Porta C., Lomonossoff G.P., Durrani Z., Dimmock N.J. Human immunodeficiency virus type 1 neutralizing antibodies raised to a gp41 peptide expressed on the surface of a plant virus. AIDS Res. Hum. Retroviruses. 1995;11:327–334. doi: 10.1089/aid.1995.11.327. [DOI] [PubMed] [Google Scholar]
- McLain L., Durrani Z., Wisniewski L.A., Porta C., Lomonossoff G.P., Dimmock N.J. Stimulation of neutralizing antibodies to human immunodeficiency virus type 1 in three strains of mice immunized with a 22-mer amino acid peptide expressed on the surface of a plant virus. Vaccine. 1996;14:799–810. doi: 10.1016/0264-410x(95)00229-t. [DOI] [PubMed] [Google Scholar]
- McLain L., Brown J.L., Cheung L., Reading S.A., Parry C., Jones T.D., Cleveland S.M., Dimmock N.J. Different effects of a single amino acid substitution on three epitopes in the gp41 C-terminal loop of a neutralizing antibody escape mutant of human immunodeficiency virus type 1. Arch. Virol. 2001;146:157–166. doi: 10.1007/s007050170199. [DOI] [PubMed] [Google Scholar]
- Miller M.A., Cloyd M.W., Liebmann J., Rinaldo C.R., Islam K.R., Wang S.Z.S., Mietzner T.A., Montelaro R.C. Alterations in cell membrane permeability by the lentiviral peptide (LLP-1) of HIV-1 transmembrane protein. Virology. 1993;196:89–100. doi: 10.1006/viro.1993.1457. [DOI] [PubMed] [Google Scholar]
- Morikawa Y., Barsov E., Jones I. Legitimate and illegitimate cleavage of human immunodeficiency virus glycoproteins by furin. J. Virol. 1993;67:3601–3604. doi: 10.1128/jvi.67.6.3601-3604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulard M., Achstetter T., Kieny M.-P., Montagnier L., Bahraoui E. Kex2p: a model for cellular endopeptidase processing human immunodeficiency virus type 1 envelope glycoprotein precursor. Eur. J. Biochem. 1994;225:565–572. doi: 10.1111/j.1432-1033.1994.00565.x. [DOI] [PubMed] [Google Scholar]
- Moulard M., Hallenberger S., Garten W., Klenk H.-D. Processing and routage of HIV glycoproteins by furin to the cell surface. Virus Res. 1999;60:55–65. doi: 10.1016/s0168-1702(99)00002-7. [DOI] [PubMed] [Google Scholar]
- Newton S.M.C., Joys T.M., Anderson S.A., Kennedy R.C., Hovi M.E., Stocker B.A.D. Expression and immunogenicity of an 18-residue epitope of HIV-1 gp41 inserted in the flagellar protein of a Salmonella live vaccine. Res. Microbiol. 1995;146:203–216. doi: 10.1016/0923-2508(96)80276-2. [DOI] [PubMed] [Google Scholar]
- Niedrig M., Bröker M., Walter G., Stüber W., Harthus H.-P., Mehdi S., Gelderblom H.R., Pauli G. Murine monoclonal antibodies directed against the transmembrane protein gp41 of human immunodeficiency virus type 1 enhance its infectivity. J. Gen. Virol. 1992;73:951–954. doi: 10.1099/0022-1317-73-4-951. [DOI] [PubMed] [Google Scholar]
- Ohno H., Stewart J., Fournier M.-C., Bosshart H., Rhee I., Miyatake S., Saito T., Galluser A., Kirchhausen T., Bonifacino J.S. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Ohno H., Fournier M.-C., Poy G., Bonifacino J.S. Structural determinants of interaction of tyrosine-based sorting signals with the adaptor medium chains. J. Biol. Chem. 1996;271:29009–29015. doi: 10.1074/jbc.271.46.29009. [DOI] [PubMed] [Google Scholar]
- Ohno H., Aguilar R.C., Fournier M.-C., Hennecke S., Cosson P., Bonifacino J.S. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- Ohno H., Aguilar R.C., Yeh D., Taura D., Daito T., Bonifacino J.S. The medium subunits of adaptor complexes recognise distinct but overlapping sets of tyrosine-based sorting signals. J. Biol. Chem. 1998;273:25915–25921. doi: 10.1074/jbc.273.40.25915. [DOI] [PubMed] [Google Scholar]
- Ooi C.E., Moreira J.E., Dell'Angelica E.C., Poy G., Wassarman D.A., Bonifacino J.S. Altered expression of a novel adaptin leads to a defective pigment granule biogenesis in the Drosophila eye color mutant garnet. EMBO J. 1997;16:4508–4518. doi: 10.1093/emboj/16.15.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otteken A., Earl P.L., Moss B. Folding, assembly, and intracellular trafficking of the human immunodeficiency virus type 1 envelope glycoprotein analyzed with monoclonal antibodies recognizing maturational intermediates. J. Virol. 1996;70:3407–3415. doi: 10.1128/jvi.70.6.3407-3415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.J., Evans P.R. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1317–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R.J., Dubay J.W., Hunter E., Compans R.W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R., Gallo R.C., Sarngadharan M.G. Processing and secretion of envelope glycoprotein of human immunodeficiency virus type 1 in the presence of trimming glucosidase inhibitor deoxynojirimycin. Intervirology. 1988;30:27–35. doi: 10.1159/000150073. [DOI] [PubMed] [Google Scholar]
- Pal R., Mumbauer S., Hoke G.M., Takatsuki A., Sarngadharan M.G. Brefeldin A inhibits the processing and secretion of envelope glycoprotein of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses. 1991;7:707–712. doi: 10.1089/aid.1991.7.707. [DOI] [PubMed] [Google Scholar]
- Pellett P.E., Kousoulas K.G., Pereira L., Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 1985;53:243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T., Zentgraf H., Freyaldenhoven B., Bosch V. Transfer of endoplasmic reticulum and Golgi retention signals to human immunodeficiency virus type 1 gp160 inhibits intracellular transport and proteolytic processing of viral glycoprotein but does not influence the cellular site of virus particle budding. J. Gen. Virol. 1997;78:1745–1753. doi: 10.1099/0022-1317-78-7-1745. [DOI] [PubMed] [Google Scholar]
- Prange R., Streeck R.E. Novel transmembrane topology of the hepatitis B envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pytowski B., Judge T.W., McGraw T.E. An internalization motif is created in the first cytoplasmic domain of the transferrin receptor by substitution of a tyrosine at the first position of a predicted turn. J. Biol. Chem. 1995;270:9067–9073. doi: 10.1074/jbc.270.16.9067. [DOI] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak R., Starcich B., Josephs S.F., Doran E.R., Rafalski J.A., Whitehorn E.A., Baumeister K., Ivanoff L., Petteway S.R., Pearson M.L., Lautenberger J.A., Papas T.S., Ghrayeb J., Chang N.T., Gallo R.C., Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature (London) 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Reading S.A., Heap C.J., Dimmock N.J. A novel monoclonal antibody specific for the C-terminal tail of the gp41 envelope transmembrane protein of human immunodeficiency virus type 1 that preferentially neutralizes virus after it has attached to the target cell and inhibits the production of infectious progeny. Virology. 2003;315:362–372. doi: 10.1016/s0042-6822(03)00533-6. [DOI] [PubMed] [Google Scholar]
- Risco C., Antón I.M., Suñé̦ A., Pedregosa A.M., Martin-Alonso F., Parra F., Carrasco L., Enjuanes L. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J. Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer J., Schweizer J.-R., Esteban M. The targeting signal of lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J. Cell Biol. 1996;132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell J.F., Stanhope P.E., Siliciano R.F. Endocytosis of endogenously synthesized HIV-1 envelope protein: mechanism and role in processing for association with class II MHC. J. Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- Sabatini D.D., Kreibich G., Morimoto T., Adesnik M. Mechanisms for the incorporation of proteins in membranes and organelles. J. Cell Biol. 1982;92:1–22. doi: 10.1083/jcb.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz G. Transmembrane β-barrel proteins. Adv. Protein Chem. 2003;63:47–70. doi: 10.1016/s0065-3233(03)63003-2. [DOI] [PubMed] [Google Scholar]
- Simpson F., Peden A.A., Christopolou L., Robinson M.S. Characterization of the adaptor-related protein complex, AP-3. J. Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S.J. The structure and insertion of integral proteins in membrane. Annu. Rev. Cell Biol. 1990;6:247–296. doi: 10.1146/annurev.cb.06.110190.001335. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Meulenberg J.J.M. The molecular biology of arteriviruses. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Stein B.S., Engelman E.G. Intracellular processing of the gp160 HIV-1 envelope precursor. Endoproteolytic cleavage occurs in a cis or medial compartment of the Golgi complex. J. Biol. Chem. 1990;265:2640–2649. [PubMed] [Google Scholar]
- Stephens D.J., Banting G. Specificity of interaction between adaptor-complex medium chains and the tyrosine-based sorting signals of TGN38 and Igp120. Biochem. J. 1998;335:567–572. doi: 10.1042/bj3350567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp J.D., Huang K., Lemmon S.K. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet R.M., Eisenberg D. Correlation of sequence hydrophobicities measures similarity in three dimensional protein structure. J. Mol. Biol. 1983;171:479–488. doi: 10.1016/0022-2836(83)90041-4. [DOI] [PubMed] [Google Scholar]
- Trowbridge I.S., Collawn J.F. Structural requirements for high efficiency endocytosis of the human transferrin receptor. J. Inorg. Biochem. 1992;47:209–217. doi: 10.1016/0162-0134(92)84066-v. [DOI] [PubMed] [Google Scholar]
- Trowbridge I.S., Collawn J.F., Hopkins C.R. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 1993;9:139–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- Vella C., Ferguson M., Dunn G., Meloen R., Langedijk H., Evans D., Minor P.D. Characterization and primary structure of a human immunodeficiency virus type 1 (HIV-1) neutralization domain as presented by a poliovirus type 1/HIV-1 chimera. J. Gen. Virol. 1993;74:2603–2607. doi: 10.1099/0022-1317-74-12-2603. [DOI] [PubMed] [Google Scholar]
- Vollenweider F., Benjannet S., Decroly E., Savaria D., Lazure C., Thomas G., Chretien M., Seidah N.G. Comparative cellular processing of the human immunodeficiency virus (HIV-1) envelope glycoprotein gp160 by the mammalian subtilisin/kexin-like convertases. Biochem. J. 1996;314:521–532. doi: 10.1042/bj3140521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.T., Johnston P.B., Dubay S.R., Hunter E. Mutations within the putative membrane-spanning domain of the simian immunodeficiency virus transmembrane glycoprotein define the minimal requirements for fusion, incorporation and infectivity. J. Virol. 2001;75:9601–9612. doi: 10.1128/JVI.75.20.9601-9612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.T., Weldon S.K., Wyss S., Lin X., Yu Q., Thali M., Hunter E. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J. Virol. 2002;76:3338–3349. doi: 10.1128/JVI.76.7.3338-3349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M. Viral and cellular membrane fusion reactions. Annu. Rev. Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- Willey R.L., Bonifacino J.S., Potts B.J., Martin M.A., Klausner R.D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus type 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey R.L., Klimkait T., Frucht D.M., Bonifacino J.S., Martin M.A. Mutations with the human immunodeficiency virus type 1 gp160 glycoprotein alter its intracellular transport and processing. Virology. 1991;184:319–329. doi: 10.1016/0042-6822(91)90848-6. [DOI] [PubMed] [Google Scholar]
- Wyss S., Berlioz-Torrent C., Boge M., Blot G., Höning S., Berarous R., Thali M. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor. J. Virol. 2001;75:2982–2992. doi: 10.1128/JVI.75.6.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Dornadula G., Alur P., Laughlin M.A., Pomerantz R.J. Amphipathic domains in the C terminus of the transmembrane protein (gp41) permeabilize HIV-1 virions: a molecular mechanism underlying natural endogenous reverse transcription. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12519–12524. doi: 10.1073/pnas.93.22.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman J.M., Eliezer N., Simha R. The characterization of amino acids in proteins by statistical methods. J. Theor. Biol. 1968;21:170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]


