Abstract
The spike (S) protein of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) is not only responsible for receptor binding, but also a major antigenic determinant capable of inducing protective immunity. In this study, we demonstrated that the receptor-binding domain (RBD) of S protein is an important immunogenic site in patients with SARS and rabbits immunized with inactivated SARS-CoV. Serum samples from convalescent SARS patients and immunized rabbits had potent neutralizing activities against infection by pseudovirus expressing SARS-CoV S protein. Depletion of RBD-specific antibodies from patient or rabbit immune sera by immunoadsorption significantly reduced serum-mediated neutralizing activity, while affinity-purified anti-RBD antibodies had relatively higher potency neutralizing infectivity of SARS pseudovirus, indicating that the RBD of S protein is a critical neutralization determinant of SARS-CoV during viral infection and immunization. Two monoclonal antibodies (1A5 and 2C5) targeting at the RBD of S protein were isolated from mice immunized with inactivated SARS-CoV. Both 1A5 and 2C5 possessed potent neutralizing activities, although they directed against distinct conformation-dependant epitopes as shown by ELISA and binding competition assay. We further demonstrated that 2C5, but not 1A5, was able to block binding of the RBD to angiotensin-converting enzyme 2 (ACE2), the functional receptor on targeted cells. These data provide important information for understanding the antigenicity and immunogenicity of SARS-CoV and for designing SARS vaccines.
Keywords: SARS-CoV, Spike protein, Receptor-binding domain, Neutralizing antibodies, Monoclonal antibodies, Vaccines
Introduction
The global emergency of severe acute respiratory syndrome (SARS) was caused by a new coronavirus (SARS-CoV) (Drosten et al., 2003, Ksiazek et al., 2003, Marra et al., 2003, Peiris et al., 2003, Rota et al., 2003). Similar to other coronaviruses, SARS-CoV features a large positive-stranded RNA genome that harbors open-reading frames (ORFs) encoding a large polyprotein required for virus replication, four structural proteins (spike, S; envelop, E; membrane, M; and nucleocapsid, N), and eight additional polypeptides of unknown function (Marra et al., 2003, Rota et al., 2003). SARS-CoV infection can trigger humoral and cellular immune responses against viral proteins in humans. The high rate of recovery from acute illness of SARS in the absence of effective medical therapy and the low rate of re-infection by SARS-CoV suggest that protective immunity is achievable. Recent studies have demonstrated that sera from convalescent-phase SARS patients contain high titers of neutralizing antibodies against SARS-CoV (Nie et al., 2004, Simmons et al., 2004). The S protein of SARS-CoV, like those of other coronaviruses, has been shown to be a major antigenic determinant that induces neutralizing antibodies and protective immunity (Bisht et al., 2004, Buchholz et al., 2004, Bukreyev et al., 2004, Yang et al., 2004). A DNA vaccine encoding the S protein induced SARS-CoV neutralization and protective immunity in mice (Yang et al., 2004). Depletion of T cells and antibody transfer experiments revealed that protection was mediated by neutralizing antibodies (Subbarao et al., 2004, Yang et al., 2004), and protection can be achieved by sole administration of neutralizing monoclonal antibodies specific for S protein (Traggiai et al., 2004). Furthermore, vaccination of animals with recombinant viruses, such as attenuated vaccinia virus (MVA) and parainfluenza virus (BHPIV3), that express S protein can elicit neutralizing antibodies to protect animals against SARS-CoV challenge (Bisht et al., 2004, Buchholz et al., 2004, Bukreyev et al., 2004). Infection by pseudovirus expressing the SARS-CoV S protein can be effectively neutralized by convalescent sera from SARS patients (Nie et al., 2004, Simmons et al., 2004). These data suggest that the S protein of SARS-CoV is a promising candidate for developing SARS vaccines.
The S protein of SARS-CoV, a large class I transmembrane glycoprotein consisting of S1 domain (residues 15–680) and S2 domain (residues 681–1255), mediates attachment of virions to susceptible cells and fusion of the viral and cellular membranes (Hofmann and Pohlmann, 2004, Holmes, 2003). A 193-amino acid small fragment (residues 318–510) in the S1 region was characterized as the minimal receptor-binding domain (RBD) of SARS-CoV (Babcock et al., 2004, Wong et al., 2004, Xiao et al., 2003). RBD mediates S protein binding to angiotensin-converting enzyme 2 (ACE2), the functional receptor for SARS-CoV on susceptible cells (Dimitrov, 2003, Li et al., 2003, Prabakaran et al., 2004, Wang et al., 2004). Most recently, we demonstrated that the recombinant RBD of SARS-CoV S protein could induce potent neutralizing antibodies (He et al., 2004a, He et al., 2004b). In this report, we showed that the RBD of S protein is a critical neutralization determinant of SARS-CoV in SARS patients and SARS-CoV-immunized rabbits. We further identified two neutralization epitopes in the RBD by monoclonal antibodies (MAbs) isolated from mice immunized with inactivated SARS-CoV.
Results
The RBD of S protein is an immunogenic site of SARS-CoV in infected patients
It has been demonstrated that the RBD of S protein is highly immunogenic in mice and rabbits immunized with inactivated SARS-CoV (He et al., 2004b). In this study, 40 serum samples were collected from SARS patients at convalescent phase to investigate the immunogenicity of RBD in humans. All the sera were positive for SARS-CoV as detected by ELISA with commercially available diagnostic kits, in which the mixture of proteins purified from viral lysates of SARS-CoV was used as coating antigen (data not shown). To determine antibodies specific for the RBD of S protein in the SARS convalescent sera, we used recombinant RBD-C9 as a coating antigen in ELISA. As shown in Fig. 1 , all of the 40 convalescent sera from SARS patients reacted significantly with RBD-C9, whereas none of the sera from healthy blood donors was reactive to this antigen, suggesting that the RBD of SARS-CoV S protein is highly immunogenic during virus infection.
Fig. 1.
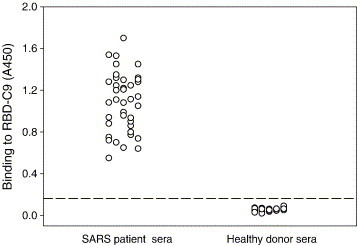
Measurement of the RBD-specific antibodies in sera from convalescent-phase SARS patients by ELISA. Recombinant RBD-C9 was used for coating plates and the sera from 40 SARS patients and 30 healthy blood donors were tested at 1:50 dilution. Sera were considered positive when the optical density values were above the cutoff value (the mean absorbance at 450 nm of sera from healthy blood donors plus three standard deviations).
Isolation of RBD-specific antibodies from convalescent sera of SARS patients
The RBD-Fc fusion protein was used to isolate RBD-specific antibodies by immunoaffinity chromatography from the convalescent sera of SARS patients. Serum samples from four SARS patients (SARS A–D) prescreened for high levels of reactivity against RBD-C9 and a control serum from health blood donor were adsorbed on resins immobilized with RBD-Fc fusion protein, and bound antibodies (anti-RBD) were eluted with pH 2.5 buffer. The efficiencies of depletion and recovery of the RBD-specific antibodies were monitored by measuring the reactivity of the starting human sera, the corresponding flowthroughs, and eluted antibody fractions by ELISA against the RBD-C9 and S1-C9. As shown in Fig. 2A, the reactivity of anti-RBD in the sera of SARS patients was efficiently removed by RBD-Fc affinity column. The eluted anti-RBD antibodies could significantly react with RBD-C9. All samples corresponding to the starting sera, flowthroughs (anti-RBD depleted), and eluates (anti-RBD antibodies) were reactive to the S1-C9 in ELISA (Fig. 2B). These suggest that the convalescent sera of SARS patients contain abundant antibodies specific for S protein and the RBD-specific antibodies can be specifically isolated by immunoaffinity chromatography.
Fig. 2.
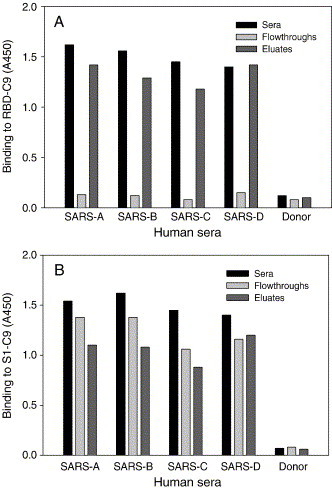
Isolation of RBD-specific antibodies from sera of convalescent-phase SARS patients by immunoaffinity chromatography. Serum samples were adsorbed to RBD-Fc columns and bound antibodies were eluted at pH 2.5. The patient sera, flowthroughs, and eluates were tested, respectively, against RBD-C9 (A) and S1-C9 (B) at 1:50 dilution by ELISA. One serum sample from a healthy blood donor was used as control.
The RBD of S protein is a potent inducer of neutralizing antibodies in infected patients
Entry of SARS-CoV into target cells is initiated by binding of its S protein via RBD in the S1 region to ACE2, a cellular receptor. It has been shown that a fusion protein (RBD-Fc) containing the RBD of S protein is a potent inducer of neutralizing antibodies against SARS-CoV in immunized animals (He et al., 2004a). It is interesting to know whether the RBD of S protein can mediate antibody responses responsible for virus neutralization in infected SARS patients. Therefore, the neutralizing activity of the samples of the starting sera, flowthroughs, and eluates was determined using SARS pseudovirus. Strikingly, the neutralizing activity of patient sera was significantly reduced after depletion of anti-RBD antibodies. The anti-RBD antibodies in the eluates possessed higher potency than the antibodies in the flowthroughs to neutralize SARS pseudovirus (Fig. 3 and Table 1 ), suggesting that more than 50% neutralizing activity in the convalescent sera of SARS patients was contributed by the RBD-specific antibodies. In contrast, the serum samples including the starting sera, flowthroughs, and eluates from the healthy blood donor had no neutralizing activities to same pseudovirus (data not shown). These data indicate that the RBD of S protein is an important inducer of neutralizing antibodies during viral infection.
Fig. 3.
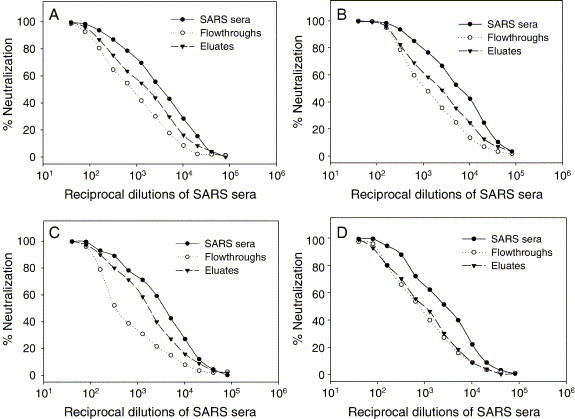
Neutralizing activity of the RBD-specific antibodies isolated from the sera of SARS patients. (A) SARS-A; (B) SARS-B; (C) SARS-C; and (D) SARS-D. Inhibition of SARS pseudovirus infectivity in 293T/ACE2 cells by patient sera, flowthroughs, and eluates, respectively, at a series of 2-fold dilutions was determined and the percentage of neutralization was calculated for each sample.
Table 1.
Neutralizing activity of serum samples against SARS pseudovirus
| Sera sample | Neutralization titer (ND50)a |
||
|---|---|---|---|
| Starting sera | Flowthroughs | Eluates | |
| Human sera | |||
| SARS-A | 2812 | 945 | 1694 |
| SARS-B | 4999 | 1718 | 2670 |
| SARS-C | 3031 | 796 | 1822 |
| SARS-D | 2614 | 907 | 1052 |
| Rabbit sera | |||
| A | 2202 | 750 | 1250 |
| B | 1966 | 845 | 1125 |
| C | 2142 | 772 | 1050 |
| D | 1868 | 698 | 980 |
ND50: Titer of a serum sample to yield 50% neutralization.
The RBD of S protein is a critical antigenic site to induce neutralizing antibodies in rabbits immunized with inactivated SARS-CoV
We previously demonstrated that the inactivated SARS-CoV was able to induce neutralizing antibodies in the immunized rabbits (He et al., 2004b). To further elucidate neutralization determinants of the SARS-CoV, the RBD-specific antibodies were isolated by immunoaffinity chromatography from four rabbit antisera (A–D). Similarly, the reactivity of anti-RBD antibodies in the rabbit antisera could be efficiently depleted by RBD-Fc affinity column, since the flowthroughs did not bind to RBD-Fc while the eluted anti-RBD antibodies significantly reacted with the RBD-Fc (Fig. 4A). Samples corresponding to the starting sera, flowthroughs (anti-RBD depleted sera), and eluates containing anti-RBD antibodies had similar reactivity with the S1-C9 (Fig. 4B). The neutralizing activity of anti-RBD antibody-depleted rabbit antisera was much lower than the starting sera, while anti-RBD antibodies possessed more potent activity to neutralize SARS pseudovirus (Fig. 4C and Table 1). These data suggest that the RBD of S protein is an effective inducer of neutralizing antibodies in virus-immunized rabbits.
Fig. 4.
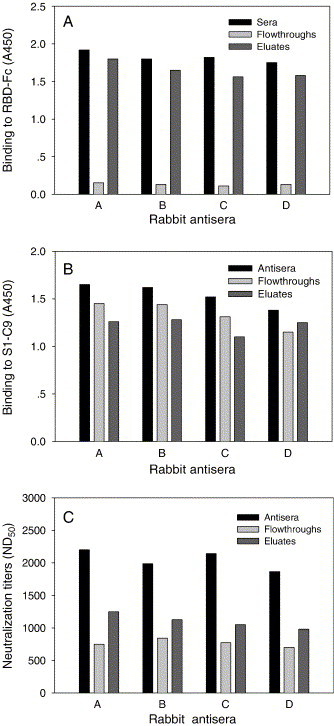
Isolation and characterization of RBD-specific antibodies from rabbit antisera by immunoaffinity chromatography. Reactivities of rabbit antibodies with RBD-Fc (A) and S1-C9 (B) were tested at 1:50 dilution by ELISA. (C) Neutralizing activity of the RBD-specific antibodies isolated from rabbit antisera. Inhibition of SARS pseudovirus infection in 293T/ACE2 cells by rabbit antisera, flowthroughs, and eluates, respectively, at a series of 2-fold dilutions was determined and the percent neutralization was calculated for each sample.
Isolation and characterization of MAbs against the RBD of S protein
It has been shown that inactivated SARS-CoV can induce high titers of antibodies specific for the RBD of S protein in immunized mice (He et al., 2004b). To identify neutralization epitopes of SARS-CoV, two MAbs (1A5 and 2C5) targeting at the RBD of S protein were isolated from mice immunized with inactivated SARS-CoV. Epitope specificities of 1A5 (IgG1/κ) and 2C5 (IgG1/κ) were initially determined by ELISAs using RBD-Fc, DTT-reduced RBD-Fc, S1-C9, and DTT-reduced S1-C9 as coating antigens (Figs. 5A and B). Both MAbs were reactive with native RBD-Fc and S1-C9, but not DTT-reduced RBD-Fc and S1-C9, indicating that they were directed against disulfide bond-dependent conformational epitopes expressed on the RBD of S protein. These two MAbs did not compete each other in an ELISA-based binding competition assay (Fig. 5C), suggesting their epitopes locate at different sites in the RBD.
Fig. 5.
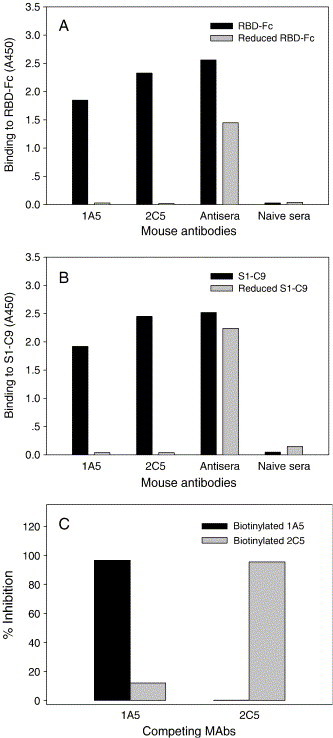
Characterization of the RBD-specific MAbs. The reactivities of MAbs against RBD-Fc (A) and S1-C9 (B) were tested by ELISA. MAbs were tested at 10 μg/ml, and control sera were tested at 1:100 dilution. (C) Inhibition of biotinylated MAbs binding to RBD-Fc by RBD-specific MAbs measured by competitive ELISA. Competing MAbs were tested at 100 μg/ml.
RBD-Fc could efficiently bind to ACE2 expressed on 293T/ACE2 cells as measured by flow cytometry. We tested whether the RBD-specific MAbs inhibit binding of RBD-Fc to cell-associated ACE2. As shown in Fig. 6 , 2C5, but not 1A5, was able to block RBD-Fc binding to 293T/ACE2 cells. This result suggests that 2C5 recognizes an epitope that may overlap with the receptor-binding sites in the S protein, while 1A5 directs against a distinct conformation-dependant epitope that is not involve in receptor binding. With a single-cycle infection assay, we tested the neutralizing activities of these two MAbs against SARS pseudovirus. Strikingly, 1A5 and 2C5 potently neutralized SARS pseudovirus with 50% neutralization dose (ND50) at 0.048 μg/ml and 0.097 μg/ml, respectively (Fig. 7 ), suggesting that both conformation-dependant epitopes in the RBD of S protein can elicit potent neutralizing antibodies.
Fig. 6.
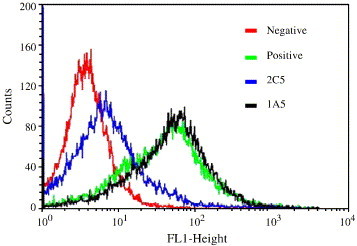
Inhibition of RBD-Fc binding to ACE2 by MAbs. Inhibitory activity of MAbs on RBD-Fc binding to cell-associated ACE2 expressed on 293T/ACE2 cells was measured by flow cytometry. RBD-Fc and MAbs were used at 1 and 50 μg/ml, respectively.
Fig. 7.
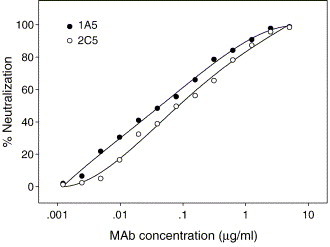
Neutralization of SARS pseudovirus infection in 293T/ACE2 cells by MAbs. Each of the MAbs was tested at a series of 2-fold dilutions and percent neutralization was calculated.
Discussion
The S protein of SARS-CoV is able to induce protective immunity in immunized animals (Bisht et al., 2004, Buchholz et al., 2004, Bukreyev et al., 2004, Yang et al., 2004). Using a set of overlapping peptides that cover the entire sequence of S protein, we have identified several immunodominant domains within S protein. However, none of these domains modeled by linear peptides were able to elicit neutralizing antibodies (He et al., 2004c). In contrast, the RBD (residues 318–510) in the S1 region of S protein can induce highly potent neutralizing antibodies against the SARS-CoV (He et al., 2004a, He et al., 2004b). Here, we have further demonstrated that the RBD of SARS-CoV S protein is an immunogenic site to induce antibody responses in SARS-CoV-infected patients and immunized rabbits. With an immunoabsorption assay, we have demonstrated that anti-RBD antibodies function as a main population of neutralizing antibodies, suggesting that the RBD of S protein is an important neutralization determinant of SARS-CoV.
The first essential step of coronavirus infection is the interaction of S protein via the RBD with specific cellular receptor. The RBDs on the S proteins of other coronaviruses, such as mouse hepatitis virus (MHV), transmissible gastroenteritis virus (TGEV), and human coronavirus (HCoV-229), also contain major antigenic determinants capable of inducing neutralizing antibodies (Bonavia et al., 2003, Godet et al., 1994, Kubo et al., 1994). We previously demonstrated that inactivated SARS-CoV vaccine was capable of inducing high titers of RBD-specific antibodies that block receptor binding and virus entry (He et al., 2004b). In the present study, two neutralization epitopes in the RBD of S protein have been identified with MAbs isolated from mice immunized with inactivated SARS-CoV. Therefore, the RBD of S protein may serve as an important target site for developing SARS vaccines and immunotherapeutics.
Inactivated SARS-CoV has been used as one of the major vaccine candidates and is currently being tested in clinical trial in China (Marshall and Enserink, 2004). However, caution should be taken in using the inactivated SARS-CoV as vaccine since it contains a number of viral components that may induce harmful immune and/or inflammatory responses (Enserink, 2004, He et al., 2004c, Oba, 2003, Wang and Lu, 2004). It was recently reported that an inactivated SARS-CoV vaccine triggered an autoimmune response against the carbohydrate moieties in an abundant human serum glycoprotein asialo-orosomucoid (Wang et al., 2004). Recombinant viruses or DNA vaccines that express full-length S protein have also been considered as SARS vaccine candidates (Bisht et al., 2004, Buchholz et al., 2004, Bukreyev et al., 2004, Yang et al., 2004). However, the full-length S protein also contains several linear immunodominant domains that induce a high titer of non-neutralizing antibodies (He et al., 2004c). It is unclear whether or not these non-neutralizing antibodies may enhance SARS-CoV infection or mediate harmful immune responses. It was reported that vaccination of ferrets with vaccinia virus-based SARS vaccine expressing full-length S protein aggravated liver damage caused by SARS-CoV infection (Enserink, 2004, Weingartl et al., 2004). It has been demonstrated that the S protein of feline coronavirus (FIPV) expressed by recombinant vaccinia can cause antibody-dependant enhancement of disease if the vaccinated animals are subsequently infected with wild-type virus (Corapi et al., 1992, Olsen et al., 1993, Vennema et al., 1990).
Therefore, we propose to use the RBD of SARS-CoV S protein for developing an effective and safe subunit vaccine, since the RBD of S protein is not only a functional domain that mediates virus-receptor binding, but also a critical neutralization determinant of SARS-CoV. If an effective RBD-based SARS vaccine can be developed, it may provide a promising example for designing vaccines against other viruses.
Materials and methods
Expression of recombinant S proteins
Recombinant S proteins of SARS-CoV (Tor2 strain) were expressed as previously described (He et al., 2004a, He et al., 2004b). Plasmids encoding a 193-amino acid fragment of RBD (residues 318–510) linked to the Fc domain of human IgG1 (designated RBD-Fc) or tagged with C9 (designated RBD-C9) and plasmid encoding residues 12–672 corresponding to the S1 domain of S protein tagged with C9 at the C-terminus (designated S1-C9) were kindly provided by Dr. M. Farzan at the Harvard Medical School (Li et al., 2003, Wong et al., 2004). RBD-Fc, RBD-C9, and S1-C9 proteins were expressed in 293T cells transfected with the plasmids using Fugene 6 reagents (Boehringer Mannheim, Indianapolis, IN) according to the manufacturer's protocol. Supernatants were harvested 72 h post-transfection. RBD-C9 and S1-C9 were purified by affinity chromatography with anti-C9 MAb 1D4 (National Cell Culture Center, Minneapolis, MN). RBD-Fc was purified by Protein A Sepharose 4 Fast Flow (Amersham Biosciences, Piscataway, NJ).
Serum samples from SARS patients
Serum samples were collected from 40 convalescent SARS patients 30–60 days after the onset of symptoms based on clinical diagnosis during the SARS epidemics in Beijing in 2003. The diagnostic criteria for SARS-CoV infection followed the clinical description of SARS released by WHO. All of the sera were heat-inactivated at 56 °C prior to performing the experiments and verified to be positive for SARS-CoV as detected by immunofluorescence assay (IFA) and enzyme-linked immunoabsorbed assay (ELISA) using commercially available diagnostic kits (Beijing Genomics Institute, Beijing). Sera collected from 30 healthy blood donors were used as controls.
Immunization of rabbits with inactivated SARS-CoV
Inactivated SARS-CoV was prepared as described previously (He et al., 2004b, He et al., 2004c). Briefly, SARS-CoV strain BJ01 was propagated in Vero-E6 cells and inactivated by β-propiolactone. The inactivated virus was purified by desalting with Sephadex G-50, concentrated with PEG-8000, and filtrated with Sepharose-CL 2B sequentially. The purified viral particles were at >95% purity by HPLC analysis. Four NZW rabbits were primarily immunized intradermally with 30 μg of purified inactivated viruses as immunogen in the presence of complete Freund's adjuvant (FCA) and boosted twice with same amount freshly prepared emulsion of the immunogen and Freund's incomplete adjuvant (FIA) at 2-week intervals. Pre-immune sera were collected before the primary immunization and antisera were collected 5 days after the second boost.
Immunoaffinity chromatography
Immunoaffinity resins were prepared by coupling the RBD-Fc fusion protein to cyanogen bromide-activated Sepharose beads (Pharmacia, Piscataway, NJ) according to the manufacturer's instructions. For immunoadsorption, human or rabbit serum samples were diluted 10-fold with PBS and incubated with the RBD-Fc resin overnight at 4 °C with constant rotation. Resins were then packed into columns and the flowthroughs (anti-RBD depleted) were collected. After washing with 10 column volumes of PBS, the bound antibodies (anti-RBD) were eluted in 0.2 M glycine–HCl buffer, pH 2.5. The low-pH eluates were neutralized immediately with pH 9.0 Tris buffer, and bovine serum albumin (100 μg/ml) was added for stabilization. Buffer was exchanged with PBS by several cycles of dilution and concentration on Amicon Ultra-15 centrifugal filter device (Millipore Corporation, Bedford, MA). All samples were adjusted to the original volume of serum and sterilized with 0.2-μm pore size microspin filters (Millipore).
Isolation of monoclonal antibodies (MAbs)
BALB/C mice were immunized with inactivated SARS-CoV as described previously (He et al., 2004b, He et al., 2004c). Hybridomas for producing MAbs were generated using standard protocol. Briefly, the splenocytes from immunized mice were harvested and fused with SP2/0 myeloma cells. Cell culture supernatants from wells containing hybridoma colonies were screened by IFA. Cells from positive wells were expanded and retested. Cultures that remained positive were subcloned to generate stable hybridoma cell lines. Mouse ascites was generated by injecting BALB/c mice with hybridomas, and the MAbs were purified from mouse ascites by Protein A Sepharose 4 Fast Flow (Amersham Biosciences). The isotypes of mAbs were determined with Mouse MonoAb-ID Kit (Zymed, South San Francisco, CA).
ELISA and binding competition
Human or rabbit serum samples and mouse MAbs were tested against the recombinant S proteins (RBD-C9, RBD-Fc, or S1-C9) by ELISA, respectively. Briefly, 1 μg/ml of each recombinant protein was coated to 96-well microtiter plates (Corning Costar, Acton, MA) in 0.1 M carbonate buffer (pH 9.6) at 4 °C overnight. After blocking with 2% non-fat milk, diluted samples were added and incubated at 37 °C for 1 h, followed by three washes with PBS containing 0.1% Tween 20. Bound antibodies were detected with HRP-conjugated goat anti-human IgG or goat anti-rabbit IgG or goat anti-mouse IgG (Zymed, South San Francisco, CA), respectively, at 37 °C for 1 h, followed by washes. The reaction was visualized by addition of the substrate 3,3′,5,5′-tetramethylbenzidine (TMB) and absorbance at 450 nm (A450) was measured by an ELISA plate reader (Tecan US, Research Triangle Park, NC).
To determine the effect of disulfide bond reduction on the binding of RBD-specific MAbs, ELISA plate was coated with recombinant RBD-Fc or S1-C9 at a concentration of 1 μg/ml and then treated for 1 h at 37 °C with dithiothreitol (DTT) at a concentration of 10 mM, followed by washes. Then the wells were treated with 50 mM iodoacetamide for 1 h at 37 °C. After washes, a standard ELISA was performed as described above.
A competitive ELISA was performed to determine the inhibitory activity of the RBD-specific MAbs on binding of the biotinylated MAbs to RBD-Fc. Briefly, the wells of ELISA plates were coated with RBD-Fc at 1 μg/ml as described above. A mixture of 50 μg/ml of an unlabeled MAb and 1 μg/ml of a biotinylated MAb was added, followed by incubation at 37 °C for 1 h. Binding of the biotinylated MAbs was detected after addition of HRP-conjugated streptavidin (Zymed) and TMB sequentially. Biotinylation of MAbs was performed using the EZ-link NHS-PEO Solid Phase Biotinylation Kit (Pierce, Rockford, IL) according to the manufacturer's protocol.
Inhibition of receptor-binding
Inhibition of MAbs on RBD-Fc binding to ACE2-expressing cells was measured by flow cytometry. Briefly, 106 293T/ACE2 cells were detached, collected, and washed with Hank's balanced salt solution (HBSS) (Sigma, St. Louis, MO). RBD-Fc was added to the cells to a final concentration of 1 μg/ml in the presence or absence of 50 μg/ml MAbs, followed by incubation at room temperature for 30 min. Cells were washed with HBSS and incubated with anti-human IgG-FITC conjugate (Zymed) at 1:50 dilution at room temperature for an additional 30 min. After washing, cells were fixed with 1% formaldehyde in PBS and analyzed in a Becton FACSCalibur flow cytometer (Mountain View, CA) using CellQuest software.
Neutralization of SARS pseudovirus infection
SARS pseudovirus was prepared as previously described (He et al., 2004a, He et al., 2004b). In brief, 293T cells were co-transfected with a plasmid encoding codon-optimized S protein of SARS-CoV (Tor2 strain) and a plasmid encoding Env-defective, luciferase-expressing HIV-1 genome (pNL4-3.luc.RE) using Fugene 6 reagents (Boehringer Mannheim). Supernatants containing pseudovirus bearing SARS-CoV S protein were harvested 48 h post-transfection and used for single-cycle infection of ACE2-transfected 293T cells. Briefly, ACE2-expressed 293T cells were plated (104 cells/well) in 96-well tissue-culture plates and grown overnight. The pseudovirus was preincubated with 2-fold serially diluted serum samples or MAbs at 37 °C for 1 h before addition to cells. The culture was re-fed with fresh medium 24 h later and incubated for an additional 48 h. Cells were washed with PBS and lysed using lysis reagent included in the luciferase kit (Promega, Madison, WI). Aliquots of cell lysates were transferred to 96-well Costar flat-bottom luminometer plates (Corning Costar, Corning, NY), followed by addition of luciferase substrate (Promega). Relative light units were determined immediately on the Ultra 384 luminometer (Tecan US).
Acknowledgment
We thank Dr. Michael Farzan at the Department of Medicine, Harvard Medical School, for providing plasmids encoding RBD-Fc, RBD-C9, and S1-C9.
References
- Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78(9):4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia A., Zelus B.D., Wentworth D.E., Talbot P.J., Holmes K.V. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 2003;77(4):2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. U.S.A. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M., Murphy B.R., Subbarao K., Collins P.L. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363(9427):2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corapi W.V., Olsen C.W., Scott F.W. Monoclonal antibody analysis of neutralization and antibody-dependent enhancement of feline infectious peritonitis virus. J. Virol. 1992;66(11):6695–6705. doi: 10.1128/jvi.66.11.6695-6705.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115(6):652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der W.S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Enserink M. Infectious diseases. One year after outbreak, SARS virus yields some secrets. Science. 2004;304(5674):1097. doi: 10.1126/science.304.5674.1097. [DOI] [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68(12):8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Siddiqui P., Jiang S. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 2004;325(2):445–452. doi: 10.1016/j.bbrc.2004.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Wu H., Luo B., Chen J., Li W., Jiang S. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173(6):4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Pohlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12(10):466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Invest. 2003;111(11):1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68(9):5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Marshall E., Enserink M. Medicine. Caution urged on SARS vaccines. Science. 2004;303(5660):944–946. doi: 10.1126/science.303.5660.944. [DOI] [PubMed] [Google Scholar]
- Nie Y., Wang G., Shi X., Zhang H., Qiu Y., He Z., Wang W., Lian G., Yin X., Du L., Ren L., Wang J., He X., Li T., Deng H., Ding M. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J. Infect. Dis. 2004;190(6):1119–1126. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba Y. The use of corticosteroids in SARS. N. Engl. J. Med. 2003;348(20):2034–2035. doi: 10.1056/NEJM200305153482017. [DOI] [PubMed] [Google Scholar]
- Olsen C.W., Corapi W.V., Jacobson R.H., Simkins R.A., Saif L.J., Scott F.W. Identification of antigenic sites mediating antibody-dependent enhancement of feline infectious peritonitis virus infectivity. J. Gen. Virol. 1993;74(Pt. 4):745–749. doi: 10.1099/0022-1317-74-4-745. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran P., Xiao X., Dimitrov D.S. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem. Biophys. Res. Commun. 2004;314(1):235–241. doi: 10.1016/j.bbrc.2003.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 2004;101(12):4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78(7):3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10(8):871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., de Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C., Spaan W.J. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J. Virol. 1990;64(3):1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Lu J. Glycan arrays lead to the discovery of autoimmunogenic activity of SARS-CoV. Physiol. Genomics. 2004;18(2):245–248. doi: 10.1152/physiolgenomics.00102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen J., Zheng A., Nie Y., Shi X., Wang W., Wang G., Luo M., Liu H., Tan L., Song X., Wang Z., Yin X., Qu X., Wang X., Qing T., Ding M., Deng H. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 2004;315(2):439–444. doi: 10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J., Smith G., Jones S., Proulx R., Deschambault Y., Grudeski E., Andonov A., He R., Li Y., Copps J., Grolla A., Dick D., Berry J., Ganske S., Manning L., Cao J. Immunization with modified vaccinia virus ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 2004;78(22):12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312(4):1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]


