Abstract
High throughput screening has rendered new inhibitors of eukaryotic protein synthesis. One such molecule, 4EGI-1 has been reported to selectively block the initiation factor eIF4E. We have investigated the action of this inhibitor on translation directed by several viral mRNAs which, in principle, do not utilize eIF4E. We found that 4EGI-1 inhibits translation directed by poliovirus IRES, in rabbit reticulocyte lysates, to a similar extent as capped mRNA. Moreover, 4EGI-1 inhibits translation driven by poliovirus IRES, both in vitro and in cultured cells, despite cleavage of eIF4G by picornavirus proteases. Finally, translation of vesicular stomatitis virus mRNAs and Sindbis virus subgenomic mRNA is blocked by 4EGI-1 in infected cells to a similar extent as cellular mRNAs. These findings cast doubt on the selective action of this inhibitor, and suggest that this molecule may affect other steps in protein synthesis unrelated to cap recognition by eIF4E.
Keywords: IRES translation, 4EGI-1, IRES inhibitors, Picornavirus translation, eIF4E
Highlights
-
•
4EGI-1 inhibits translation driven by different IRES as well as other viral mRNAs.
-
•
This inhibition is observed even after cleavage of eIF4G by picornavirus proteases.
-
•
These IRES-containing mRNAs do not utilize eIF4E when eIF4G has been cleaved.
-
•
Mechanism of action 4EGI-1 could be different from described previously.
-
•
This molecule may partially affect the elongation phase of protein synthesis.
Introduction
The initiation of protein synthesis is a complex process that is a major target for the regulation of gene expression at the translational level. Two major mechanisms for the initiation of mRNA translation are known in eukaryotic cells: cap-dependent and internal ribosome entry site (IRES)-driven translation. The first one of these mechanisms involves the recognition of the methylated cap structure m7GpppN located at the 5′ end of eukaryotic mRNAs by the initiation factor eIF4E, that together with eIF4G and eIF4A form the eIF4F multiprotein complex (Jackson et al., 2010, Sonenberg and Hinnebusch, 2009). After binding of this complex to the cap structure, the interaction of the small ribosomal subunit 40S containing several initiation factors (eIFs), such as eIF1, eIF1A, eIF3 and eIF2 is promoted. The initiator Met-tRNAi Met interacts with eIF2 and together with GTP form the ternary complex that is located close to the P site of the 40S ribosomal subunit. Binding of eIF3 to the middle domain of eIF4G establishes the interaction of the preinitiation complex 43 S at the 5′ end of mRNAs bearing the cap structure (Lorsch and Dever, 2010). Afterwards, the 40S subunit scans the 5′-untranslated region (5′ UTR) of the mRNA until an AUG initiation codon is found within a good initiation context (Hinnebusch, 2011, Valasek, 2012). The secondary structure of the 5′ UTR is melted during the scanning process in part by the helicase activity of eIF4A (Parsyan et al., 2011). The eIF4F complex represents a key target for the regulation of translation and most particularly the interaction between eIF4G and eIF4E, that takes place by a motif sequence Y(X)4LF, where X is variable and F is hydrophobic (Mader et al., 1995, Morino et al., 2000). The rate-limiting factor of the eIF4F complex is eIF4E, whose involvement in translation is regulated by the PI3K/Akt/mTOR pathway (Gingras et al., 1999, Mamane et al., 2006). The activity of eIF4E is controlled by several proteins able to bind to this factor, i.e. the eIF4E binding proteins (4E-BPs). This interaction is regulated by the mTOR pathway that induces 4E-BP phosphorylation, blocking in this manner the interaction between eIF4E and 4E-BP and leaving free eIF4E to bind eIF4G. Thus, the interaction sites of eIF4G and 4E-BPs on eIF4E are overlapping (Marcotrigiano et al., 1999, Richter and Sonenberg, 2005).
In contrast to cap-dependent translation, the process of IRES-driven protein synthesis does not require the recognition of a cap structure at the 5′ end, and initiation takes place by the direct interaction of the 40S ribosomal subunit, or even the 80S ribosome, to an internal region of the 5′ UTR located upstream to the AUG initiation codon (Belsham, 2009, Fitzgerald and Semler, 2009). Picornaviruses, such as encephalomyocarditis virus (EMCV) or poliovirus (PV), mRNA do not contain a cap structure and therefore does not require eIF4E to initiate translation. Instead, the C-terminal two-thirds of eIF4G bound to eIF4A are necessary for the initiation of translation of picornavirus mRNA (Gingras et al., 1999). Other animal viruses that contain capped mRNAs may not require eIF4E for their translation in infected cells. This is the case for vesicular stomatitis virus (VSV) mRNAs (Connor and Lyles, 2002, Welnowska et al., 2009) or Sindbis virus (SV) subgenomic 26S mRNA (sgmRNA) (Castello et al., 2006, Sanz et al., 2009).
Selective inhibitors of protein synthesis have proven to be useful tools to help unravel the exact mechanism of mRNA translation (Vazquez, 1979). Classically, these inhibitors were derived from natural compounds, while in recent years more elegant and sophisticated screening methods are being used to identify inhibitors of the initiation step of translation (Cencic et al., 2011b, Moerke et al., 2007, Novac et al., 2004). High throughput screening of small molecule libraries has provided novel molecules that selectively block the interaction between eIF4G and eIF4E. One of these molecules, 4EGI-1, identified as an inhibitor of cap-dependent translation, not only interferes with the binding of eIF4E to eIF4G but also paradoxically increases the interaction of 4E-BP1 with eIF4E (Moerke et al., 2007). This commercially available compound is being employed in several laboratories to analyze biological processes related to protein synthesis. For example, 4EGI-1 impairs long-term associative memory consolidation (Hoeffer et al., 2011, Hoeffer et al., 2013), and may provide beneficial effects for the autistic-like behavior observed in eIF4E-transgenic mice (Santini et al., 2013). Furthermore, eIFs are overexpressed or activated in several human cancers, and inhibition of translation by 4EGI-1 induces apoptosis-mediated cell death in a number of myeloma cells lines (Descamps et al., 2012). In addition, 4EGI-1 has been shown to inhibit growth of human breast and melanoma cancer xenografts without apparent toxicity (Chen et al., 2012). However, 4EGI-1 abrogated the growth and induced apoptosis of human lung cancer cells by a mechanism independent from the interference with cap-dependent translation (Fan et al., 2010). Protein synthesis in cultured cells is strongly reduced by 4EGI-1 leading to accumulation of 80S ribosomes, a phenomenon not observed after depletion of eIF4E (Mokas et al., 2009). In a thorough study on the action of 4EGI-1 on protein synthesis, it was demonstrated that inhibition of translation by this compound was unrelated to its interference with the interaction between eIF4E and eIF4G (McMahon et al., 2011). Thus, the mTOR inhibitor Torin1 only partially blocked overall translation in primary human cells after disruption of eIF4E binding to eIF4G, whereas 4EGI-1 potently blocked cellular protein synthesis without interfering with the eIF4F complex. Notably, this inhibition was readily reversible even after a prolonged incubation of several days, despite the fact that protein synthesis was strongly diminished. In addition, 4EGI-1 potently blocks poxvirus replication as well as both the reactivation and lytic phases of herpesvirus infection (McMahon et al., 2011). In the present work we have tested the efficacy of 4EGI-1 to inhibit viral mRNA translation. Curiously, VSV and SV protein synthesis, that do not employ eIF4E, are inhibited to a similar extent as cellular mRNA translation. In addition, PV(IRES)-driven translation is also blocked by 4EGI-1, even after cleavage of eIF4G by PV 2Apro. These findings demonstrate that 4EGI-1 can inhibit translation even when this process is independent of eIF4E.
Results
Inhibition of mRNA translation by 4EGI-1 in cell free systems
As 4EGI-1 is thought to be a selective inhibitor of the translation initiation factor eIF4E, we tested its efficacy against different mRNA templates. Initially, the action of this molecule was assayed in the translation of cap-luc, PV(IRES)-luc, EMCV(IRES)-luc and CrPV IGR-luc mRNAs. In principle, only cap-luc requires eIF4E for translation, while the other mRNAs do not. RRL programmed with mRNA was thus incubated in the presence of increasing concentrations of 4EGI-1 and assayed for synthesis of luciferase protein. As shown in Fig. 1, synthesis of luciferase directed by cap-luc mRNA was sensitive to 4EGI-1, with a 40% reduction in translation at 60 μM, and greater than 90% reduction at 120 μM. In contrast, translation directed by mRNAs bearing EMCV IRES or CrPV IGR was more resistant to 4EGI-1, however, there was a significant reduction in translation, of approximately 50–60%, at 120 μM. Interestingly, CrPV IGR translation does not require any of the canonical eIFs (Deniz et al., 2009, Wilson et al., 2000) and so this partial inhibition of translation by 4EGI-1 was unexpected. Most probably, the rather high concentrations of 4EGI-1 used to interfere with other translation processes, independent from initiation. Surprisingly, luciferase synthesis directed by PV(IRES)-luc mRNA was more sensitive to 4EGI-1 inhibition than cap-luc mRNA (Fig. 1) with an inhibition of 85% at 90 μM.
Fig. 1.
4EGI-1 inhibits IRES-driven translation in a cell-free system. 50 ng of mRNA was incubated at 30 °C for 90 min with increasing concentrations of 4EGI-1 as indicated. Samples were then processed to measure luciferase activity. A, B, C and D represent values obtained from PV(IRES)-luc, EMCV(IRES)-luc, CrPV IGR(IRES)-luc and cap-luc mRNAs, respectively. The RLUs obtained in the absence of inhibitor were 7.9×105, 7.2×106, 5.8×105 and 3.2×106, respectively. These values were considered as 100% of control in the graphs. Error bars indicate standard deviations (SD) obtained from at least three independent experiments.
It has previously been suggested that the partial inhibition of EMCV IRES-driven translation by 4EGI-1 may be due to the fact that this mRNA uses the eIF4F complex for its translation, and the inhibition of eIF4E binding to eIF4G may alter the functionality of this complex devoid of eIF4E (Moerke et al., 2007). To test this possibility, we analyzed the effect of 4EGI-1 after cleavage of eIF4G by picornavirus proteases. It is well established that EMCV and PV IRESs are efficiently translated when eIF4G has been cleaved by picornavirus proteases 2Apro or Lpro (Castello et al., 2011, Gingras et al., 1999). Under these conditions, the N-terminal moiety of eIF4G that interacts with eIF4E does not participate in translation. Therefore, protein synthesis directed by those mRNAs bearing the picornavirus IRESs, does not employ eIF4E after eIF4G cleavage. As expected, pretreatment of RRL with FMDV Lpro or with HRV 2Apro led to cleavage of eIF4G ( Fig. 2A and B, lower panels). Under these conditions, translation directed by PV(IRES)-luc mRNA was inhibited by 4EGI-1 to a similar extent as when eIF4G remained intact (Fig. 2A and B). Therefore, the blockade of PV(IRES)-luc mRNA translation by this compound is not due to the lack of interaction of eIF4E with eIF4G.
Fig. 2.
4EGI-1 inhibits PV IRES-driven translation despite cleavage of eIF4G by HRV 2Apro or FMDV Lpro in RRL. RRL was incubated with purified HRV 2Apro (A) or FMDV Lpro (B) for 20 min at 30 °C to ensure eIF4G cleavage. Then, 50 ng PV(IRES)-luc mRNA was added, together with the different concentrations of 4EGI-1. Samples were then incubated at 30 °C for 90 min. Finally, samples of RRL were removed to measure luciferase activity. The values of luciferase activity obtained are represented in the graphs A and B. Error bars indicate SD obtained from at least three independent experiments. Western blot analysis of eIF4GI cleavage, using specific antibodies against eIF4GI (lower panels).
As a control we examined the integrity and stability of PV(IRES)-luc mRNA in the presence of 4EGI-1. To this end, RNA was extracted after incubation of PV(IRES)-luc mRNA with different concentrations of the compound in RRL. The integrity of extracted PV(IRES)-luc mRNA, as measured by quantitative RT-PCR, together with its capacity to direct luciferase synthesis in RRL in the absence of inhibitor was unaffected by incubation with 4EGI-1 (data not shown). Thus, 4EGI-1 does not affect the stability or functionality of PV(IRES)-luc in cell free systems.
Action of 4EGI-1 on picornavirus IRES-driven translation in BHK-21 cells
To analyze the activity of 4EGI-1 in cells, the different mRNAs were next individually transfected into BHK-21 fibroblasts. Transfection was carried out for 2 h, followed by a medium exchange with addition of 4EGI-1, and a further 90 min of incubation. Under these experimental conditions, translation of cap-luc, EMC(IRES)-luc and PV(IRES)-luc mRNAs were inhibited by approximately 50% with 150 μM 4EGI-1, while CrPV IGR-luc mRNA was inhibited by approximately 20% ( Fig. 3A). Notably, we have found that when 4EGI-1 is present during transfection, the inhibition of luc synthesis is much stronger than if the molecule is added after the transfected cells are washed in fresh medium. Under these conditions, translation of cap-luc mRNA and PV(IRES)-luc is inhibited by 50% with 15 μM 4EGI-1. Similarly, a 50% inhibition of CrPV IGR-luc mRNA translation occurs with 15 μM 4EGI-1, and EMCV(IRES)-luc mRNA translation is blocked by 25% with the same concentration of inhibitor. Increasing the inhibitor concentration to 30 μM abolished translation of all mRNAs except in the case of CrPV IGR-luc mRNA, which was inhibited by 80%. At 60 μM 4EGI-1, translation is totally abrogated with all mRNAs tested when the inhibitor is added to the transfection mixture (Fig. 3B). A possible explanation for this result could be that 4EGI-1 enters more efficiently into cells undergoing transfection.
Fig. 3.
4EGI-1 inhibits IRES-driven translation in BHK-21 cells. (A) BHK-21 cells were transfected for 2 h with PV(IRES)-luc, EMCV(IRES)-luc, CrPV IGR(IRES)-luc or cap-luc mRNA. Cells were then washed and incubated in DMEM 5% FCS with the absence or presence of different concentrations of the 4EGI-1 for 90 min. Then, cells were recovered and luciferase activity was measured. The RLUs obtained in the absence of inhibitor were 3.8×105, 3.6×106, 1.5×105 and 8.4×105, respectively. These values were considered as 100% of control in the graphs. Error bars indicate SD obtained from at least three independent experiments. (B) BHK-21 cells were transfected for 1 h with PV(IRES)-luc, EMCV(IRES)-luc, CrPV IGR(IRES)-luc or cap-luc mRNA and different concentrations of 4EGI-1 were added to the transfection mixture. Then, cells were washed and were incubated with fresh medium in the absence or presence of inhibitor 4EGI-1, for 90 min. Cells were then recovered and luciferase activity was measured. The RLUs obtained in the absence of inhibitor were 5.8×105, 1.4×106, 4.5×105 and 6.5×105, respectively. These values were considered as 100% of control in the graphs. Error bars indicate SD obtained from at least three independent experiments. (C) BHK-21 cells were transfected with EMC (IRES)-2Apro mRNA in the presence or absence of 90 μM 4EGI-1 for 1 h to induce eIF4GI cleavage. Then, cells were transfected with PV(IRES)-luc for 1 h under the same conditions. Then, in both cases cells were washed and incubated with 90 μM 4EGI-1 for 90 min. Finally, cells were harvested to measure luciferase activity. The RLUs obtained in absence of inhibitor were 4.6×105 in the absence of PV 2Apro and 3.2×106 in the presence of the viral protease. These values were considered as 100% of control in the graphs (black bars). Gray bars represent incubation with inhibitor after transfection and white bars represent transfection in the presence of the inhibitor. Error bars indicate that SD is obtained from at least three independent experiments. (D) Western blot analysis using specific antibodies against eIF4GI was carried out to assess eIFGI cleavage.
To ensure that the eIF4F complex, devoid of eIF4E, was inhibitory for translation, the eIF4G protein present in BHK-21 cells was cleaved by PV 2Apro. To this end, BHK-21 cells were transfected with mRNA encoding PV 2Apro for 2 h, after which virtually all eIF4G is cleaved (Fig. 3D). Transfection of PV(IRES)-luc mRNA, followed by addition of 4EGI-1 to 90 μM, indicates that mRNA translation is strongly inhibited (approximately 70%) in the absence of PV 2Apro. However, when eIF4G has been cleaved, the inhibition is reduced to approximately 40%. When 4EGI-1 is added during transfection, there is a profound arrest of PV(IRES)-luc mRNA translation, irrespective of the presence or absence of PV 2Apro (Fig. 3C). Therefore, 4EGI-1 could block translation of PV(IRES)-luc mRNA in cells despite eIF4G cleavage, indicating that this inhibition takes place by a mechanism different from the abrogation of eIF4E activity.
Action of 4EGI-1 on translation of different animal viruses in BHK-21 cells
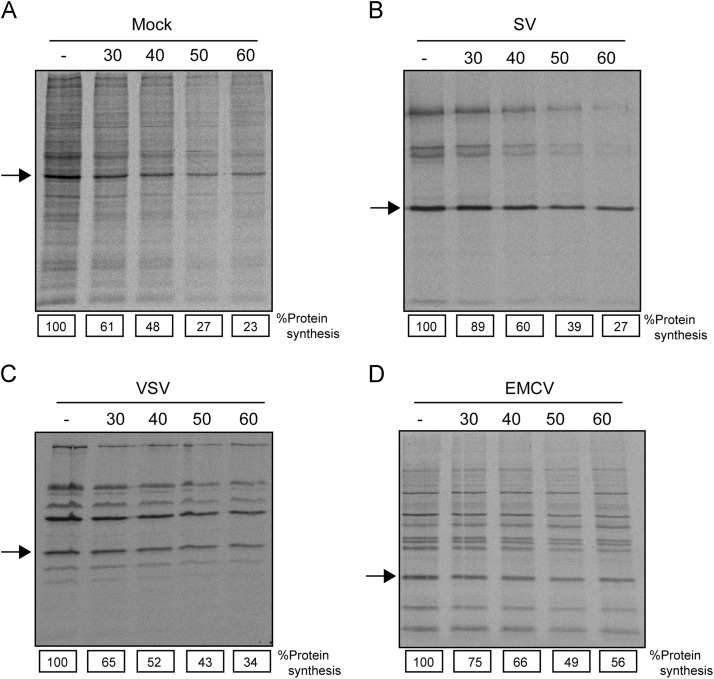
The requirements for eIFs may be different in cells infected with viruses as compared to the translation of their mRNAs in transfected cells or in cell free systems (Sanz et al., 2009, Welnowska et al., 2011). Therefore, the effect of 4EGI-1 was next analyzed in cells infected with animal viruses that, in principle, do not employ eIF4E. For this purpose, BHK-21 cells were infected with SV, VSV and EMCV. In this case, we employed concentrations of 4EGI-1 between 30 and 60 μM, which is the range in which cellular mRNA translation is sensitive to inhibition. It should be noted that these cells have not been treated with lipofectamine. As indicated previously, the inhibitor presumably enters into cells much better in the presence of the transfectant, whereas cells are more refractory to 4EGI-1 entry after lipofectamine treatment (Fig. 3). Fig. 4A shows that 30 μM 4EGI-1 diminished protein synthesis in control BHK-21 cells by 40% and this inhibition increased to more than 70% at 60 μM 4EGI-1. A similar inhibition of translation was found with SV and VSV (Fig. 4B and C, respectively) while EMCV protein synthesis was more resistant (Fig. 4D). However, a 50% inhibition of EMCV translation was observed at 50–60 μM 4EGI-1. Since none of these three animal viruses requires eIF4E during infection, we can conclude that the inhibition of viral mRNA translation by 4EGI-1 is not due to the lack of interaction between eIF4E and eIF4G.
Fig. 4.
Activity of eIF4GI on BHK cells infected with different animal viruses. BHK-21 cells were mock infected or infected with SV, VSV or EMCV at a multiplicity of infection of 10 pfu/cell, in DMEM without serum, for 1 h at 37 °C. Afterwards, the medium was removed and cell monolayers were washed with PBS and infection continued in DMEM with 5% FCS at the same temperature. At 6 hpi (in the case of mock, SV and VSV) or at 4 hpi for EMCV, cells were treated with different concentrations of 4EGI-1 as indicated in Fig. 1 and labeled with [35S]Met/Cys for 90 min in DMEM without methionine. Samples were analyzed by SDS-PAGE (15%) followed by fluorography and autoradiography. A, B, C and D represent infections from mock, SV, VSV and EMCV, respectively. Numbers below each lane represent the percentage of inhibition obtained by densitometry of the indicated protein (arrows).
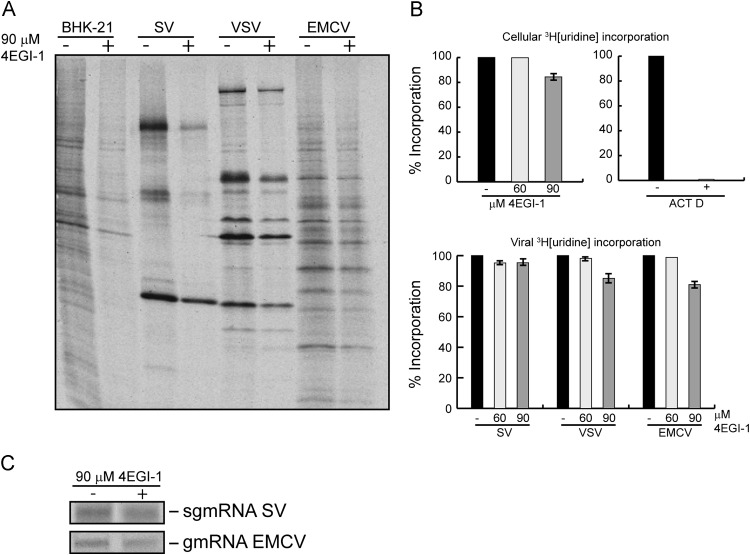
To test the possibility that 4EGI-1 was affecting the synthesis or stability of viral mRNAs, we examined the incorporation of [3H]uridine in SV, VSV and EMCV-infected cells in the presence of actinomycin D. Viral protein synthesis was strongly inhibited in SV-infected cells (approximately 90%) and to a lesser extent both in VSV and EMCV-infected cells at 90 μM 4EGI-1 ( Fig. 5A). At this concentration, the compound had little effect on cellular RNA synthesis in control cells in the absence of actinomycin D, or in viral RNA synthesis in the presence of this inhibitor (Fig. 5B upper and lower panels, respectively). Moreover, the synthesis and stability of SV sgmRNA that is translated late in infection or EMCV RNA is similar in BHK-21 cells treated or not with 90 μM 4EGI-1. In conclusion, the inhibition of SV or EMCV protein synthesis by this compound is not due to the blockade of the synthesis of viral mRNAs or to their degradation.
Fig. 5.
Viral mRNA stability in BHK-21 infected cells. (A) BHK-21 cells were infected for 1 h with 20 pfu/cell for SV and VSV, or 30 pfu/cell for EMCV. At 7 hpi (SV, VSV) or 5 hpi (EMCV) cells were labeled with [35S]Met/Cys for 1 h in the presence or absence of 90 µM 4EGI-1. Samples were analyzed by SDS-PAGE (17.5%) followed by fluorography and autoradiography. (B) BHK-21 cells were labeled with 40 μCi/ml [3H]uridine in the presence of 60 or 90 μM 4EGI-1 or with 5 μg/ml actinomycin D for 90 min. Also, BHK-21 cells were infected as in (A). At 5 hpi (SV, VSV) or 3 hpi (EMCV) cells were treated with actinomycin D for 2 h. Then, RNA was labeled with 40 μCi/ml [3H]uridine in the absence or presence of 60 or 90 μM 4EGI-1 for 90 min. Finally, total RNA from mock or infected cells was extracted. Incorporation of [3H]uridine in the RNA was measured in a liquid scintillation counter. Values obtained from mock (upper panels) or infected cells (bottom panel) are represented in the graphs as percentage, being 100% the value obtained in the absence of the inhibitors. Error bars indicate that SD is obtained from three independent experiments. (C) BHK-21 cells were infected with SV and EMCV as detailed before. At 5 hpi (SV) or 3 hpi (EMCV) cells were treated with 5 μg/ml actinomycin D for 2 h. Then, cells were labeled with 40 μCi/ml [3H]uridine in the presence or absence of 90 µM 4EGI-1 for 2 h. Finally, RNA was extracted and labeled RNA was analyzed in 0.8% agarose gels subjected to fluorography and autoradiography.
Analysis of the components of the eIF4F complex in BHK-21 cells treated with 4EGI-1
Previous work demonstrated that cellular protein synthesis was inhibited by 4EGI-1 at concentrations that did not change the interaction of several factors with eIF4E after binding to 7-methyl-GTP-Sepharose (McMahon et al., 2011). We found that treatment of BHK-21 cells with different concentrations of 4EGI-1 did not alter the amount or stability of the three components of eIF4F, namely eIF4G, eIF4A and eIF4E ( Fig. 6A). In addition, immunoprecipitation of eIF4G using specific anti-eIF4G rabbit polyclonal antibodies co-immunoprecipitated eIF4E from BHK cell extracts. No co-precipitation of eIF4E was observed when anti-eIF4G antibodies were omitted (Fig. 6B). Furthermore, a partial inhibition of eIF4E co-immunoprecipitation was found at 90 μM 4EGI-1, whereas potent blockade of eIF4G-eIF4E interaction resulted at a concentration of 120 μM (Fig. 6B and C). Lower concentrations of this compound had no effect on the interaction of eIF4E with eIF4G in BHK-21 cells, despite the fact that cellular translation was strongly blocked at concentrations of 60 μM 4EGI-1 (Fig. 4A). Finally, the cellular distribution of eIF4G and eIF4E was examined in cells treated with two different concentrations of the inhibitor: 60 and 120 μM. As observed in Fig. 6D, there was a co-localization of eIF4G and eIF4E in BHK cells, even after treatment with 120 μM 4EGI-1, indicating that the prevention of eIF4E to interact with eIF4G did not alter their intracellular distribution.
Fig. 6.
Effect of 4EGI-1 on the stability of eIF4F components and on the interaction between eIF4G and eIF4E in BHK-21 cells. (A) BHK-21 cells were treated with 4EGI-1 at the indicated concentrations for 90 min and then cell monolayers were dissolved in sample buffer and analyzed by SDS-PAGE. eIF4GI, eIF4A and eIF4E proteins were analyzed by Western blot using specific antibodies as described in Materials and methods. (B) In parallel, cells were treated with 4EGI-1 as before for 90 min and then cell lysates were subjected to immunoprecipitation with an anti-eIF4GI antibody. The specific association of eIF4E with eIF4G was analyzed by Western blot using anti-eIF4E antibody (upper panel). (C) The bar chart represents quantification of immunoprecipitated proteins in the Western blots of eIF4E (upper panel) and eIF4GI (not shown). (D) BHK cells were seeded on glass coverslips and treated or not for 90 min with 120 µM 4EGI-1. Cells were then fixed, permeabilized and processed for immunofluorescence using anti-eIF4GI (green) and anti-eIF4E (red) antibodies. Nuclei were stained with To-Pro-3 (blue). Images were acquired on a confocal microscope and subsequently processed with Huygens 4.3 software.
4EGI-1 has a partial effect on the elongation phase of translation in BHK-21 cells
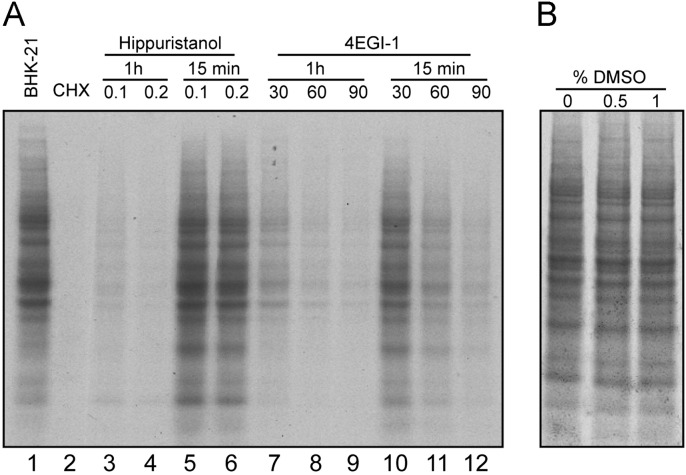
Previous work, as well as our current findings, indicated that 4EGI-1 interfered with translation directed by IRES-containing mRNAs (Moerke et al., 2007). These results, together with the observations that cellular protein synthesis was blocked by 4EGI-1 at concentrations below those affecting the interaction of eIF4E with eIF4G (McMahon et al., 2011), suggest that this inhibitor functions at a still unidentified step in initiation or elongation. To test if 4EGI-1 inhibited the elongation phase of translation in BHK-21 cells, we measured the synthesis of cellular proteins during a short time period (15 min). Under these conditions, protein labeling with radioactive methionine is due mainly to elongation. Thus, cycloheximide potently blocked protein synthesis under these conditions ( Fig. 7, lane 2), while hippuristanol at concentrations of 0.1 or 0.2 μM did not affect translation (Fig. 7, lanes 5 and 6). Preincubation of BHK cells with hippuristanol for 1 h before radioactive labeling strongly blocked cellular protein synthesis (Fig. 7, lanes 3 and 4). When 4EGI-1 was present during only 15 min, there was a significant inhibition of translation, and this inhibition was greater when the compound was preincubated for 1 h prior to protein labeling (Fig. 7, lanes 10–12 and lanes 7–9, respectively). Thus, 30 μM 4EGI-1 diminished protein synthesis by 46%, and this inhibition was above 90% when the compound was preincubated for 1 h. These findings show that 4EGI-1 has a partial inhibitory effect on the elongation of protein synthesis, aside from its blockade on the initiation of mRNA translation.
Fig. 7.
Effect of 4EGI-1 on the elongation phase of translation. (A) BHK-21 cells were pre-treated for 1 h with 0.1 or 0.2 µM hippuristanol (lanes 3 and 4) or increasing concentrations of 4EGI-1 (lanes 7–9). After pre-treatment, proteins were labeled for 15 min. Alternatively, the compounds were added together with [35S]Met-Cys and incubated for 15 min (lanes 5–6 and 10–12), as indicated in the figure. As a control, cells were treated with 100 µM cycloheximide (lane 2) and labeled during 15 min. Samples were analyzed by SDS-PAGE (17.5%) followed by fluorography and autoradiography. (B) BHK-21 cells were pre-treated for 1 h with different concentrations of dimethylsulfoxide (DMSO), as indicated in the figure. Then, cells were labeled with [35S]Met-Cys for 15 min in the presence of DMSO. Finally, samples were analyzed as in panel A.
Discussion
The search for new translation inhibitors has produced in recent years several promising compounds that selectively block this process at the initiation level. This is the case for hippuristanol which specifically abrogates eIF4A activity (Bordeleau et al., 2006, Garcia-Moreno et al., 2013, Lindqvist et al., 2008). Other recently described inhibitors of initiation include 4E1RCat and 4E2RCat, which interfere with the binding of eIF4G–eIF4E, and also abrogate the interaction between eIF4E and 4E-BP1 (Cencic et al., 2011a). 4E2RCat also decreases coronavirus replication (Cencic et al., 2011b). In the present work, we have analyzed the efficacy of the protein synthesis inhibitor 4EGI-1 on mRNA translation of different animal viruses. Curiously, it has been described that this inhibitor blocks eIF4G-eIF4E interaction, but stimulates binding of 4E-BP1 to eIF4E (Moerke et al., 2007). These results suggest that the binding region of 4E1RCat and 4EGI-1 to eIF4E may overlap, but are distinct (Cencic et al., 2011a). Therefore, the exact mechanism of action of each of these inhibitors with regards to the eIF4G-eIF4E interaction as well as their consequent biological effects, may differ. It is important to precisely establish the mechanism of action of these compounds during protein synthesis, since they are increasingly used to analyze several cellular and viral functions. Indeed, 4EGI-1 exhibits interesting antiviral and antitumor activities (Chen et al., 2012, McMahon et al., 2011). Moreover, this compound has also promising activity in a mice model for autism (Santini et al., 2013). Previous work to interpret the mode of action of 4EGI-1 revealed that this compound blocks translation at concentrations that do not disrupt the eIF4F complex (McMahon et al., 2011). In good agreement with those results, we also describe in this work that cellular translation is blocked at concentrations lower than those required to block eIF4G-eIF4E interaction. In this regard, our present findings clearly demonstrate that 4EGI-1 restricts protein synthesis despite the fact that eIF4E supposedly does not participate in translation of some viral mRNAs. As we have shown, this inhibitor strongly blocks in vitro translation directed by PV(IRES)-luc mRNA, a process in which eIF4E would not be necessary. Furthermore, this inhibition is similar when eIF4G remains intact or after its cleavage by picornavirus proteases.
Several domains have been recognized in eIF4G through molecular analysis (Gingras et al., 1999, Marcotrigiano et al., 1999). The N-terminal one-third of eIF4G is responsible for its interaction with eIF4E, while the other two-thirds can participate in IRES-driven translation by several mRNAs (De Gregorio et al., 1999, Pestova et al., 2001). Some picornavirus proteases, such as HRV or PV 2Apro, proteolytically cleave eIF4G releasing the N-terminal one third of this factor (Belsham, 2009, Castello et al., 2011). Translation of mRNAs bearing EMCV or PV IRES takes place efficiently in the presence of the distal two-thirds-containing C-terminus of eIF4G (Castello et al., 2011, Hundsdoerfer et al., 2005, Pestova et al., 2001). Under these conditions, eIF4E is not required for this translation. Therefore, picornavirus proteases are particularly useful to analyze selective inhibitors of the eIF4E–eIF4G interaction. Our present observations demonstrating that 4EGI-1 impairs PV IRES-driven translation in the presence of picornavirus 2Apro clearly indicate that this molecule affects other steps in the translation process different to its activity against eIF4E. In addition, the finding that 4EGI-1 blocks VSV and SV sgmRNA translation, adds further support to this assertion. It has been well established that initiation of mRNA translation in VSV-infected cells is independent of eIF4E and an intact eIF4F complex (Connor and Lyles, 2002, Welnowska et al., 2009). This has also been observed for translation of SV sgmRNA (Castello et al., 2006, Sanz et al., 2009). It has been proposed that the inhibitory activity of 4EGI-1 could be mediated by the accumulation of phosphorylated eIF2α in initiation complexes (McMahon et al., 2011). The presence of inactive eIF2 in initiation complexes, together with eIF4F complex may reflect the impairment in the recycling of eIFs. If so, inhibition of the recycling of eIFs may account for the inhibitory effect of PV IRES-driven translation, as described in this work. Translation of SV sgmRNA takes place even when phosphorylation of eIF2α is induced by several compounds (Sanz et al., 2009). Moreover, picornavirus translation can occur even when eIF2α becomes phosphorylated, particularly when eIF4G has been cleaved by picornavirus proteases (Redondo et al., 2012, Redondo et al., 2011, Welnowska et al., 2011). Even though translation of these mRNAs is independent of eIF2, 4EGI-1 potently blocks SV and picornavirus mRNA translation. In part, this inhibition could be due to the interference of this inhibitor with the elongation phase of protein synthesis. Also, the interference with the recycling of initiation factors due to the accumulation of initiation complexes bearing phosphorylated eIF2 could account for the inhibitory effect of 4EGI-1 on the initiation phase. On the other hand, the activity of 4EGI-1 on elongation can account for the decrease observed in translation directed by IRESs from CrPV or EMCV (Moerke et al., 2007).
The knowledge that low concentrations of 4EGI-1 block the initiation of translation would suggest that two distinct processes are taking place: one process would be the blockade of eIF4E-eIF4G interaction at high concentrations of 4EGI-1, while the other step involves an inhibition by a mechanism which remains to be determined. Our future studies will be directed to uncover the exact mode of action of 4EGI-1, in addition to assessing the activity of recently described selective translation inhibitors on viral protein synthesis.
Materials and methods
Cell line and viruses
Baby hamster kidney-21 (BHK-21) cells were obtained from ATCC. The viruses employed for infection were Sindbis virus (SV), vesicular stomatitis virus (VSV) and encephalomyocarditis virus (EMCV). Infections were carried out at a multiplicity of infection of 10 pfu/cell. Cells were grown at 37 °C, 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal calf serum (FCS). Viral infection of BHK-21 cells was carried out in DMEM without serum for 1 h at 37 °C. The medium was then removed, and cells were washed once with PBS Infection was continued in DMEM with 5% FCS at 37°C for 5 h and 30 min in the case of mock, SV and VSV infections, or 3 h and 30 min for EMCV infection.
Plasmids and transfections
The plasmid encoding EMCV and PV(IRES)-luc has been described previously (Redondo et al., 2011). Plasmid pTM1 bears the EMCV IRES element before the corresponding gene. Plasmid T7 Rluc ΔEMC IGR-Fluc (pIGR CrPV-luc) was kindly provided by P. Sarnow (Stanford University, USA). BHK-21 cells were transfected using Lipofectamine 2000 (Invitrogen). Cells were transfected or co-transfected as indicated with in vitro transcribed mRNA. RNAs were added together with 2 μl Lipofectamine per well in Opti-MEM medium (Invitrogen) and incubated for 2 h at 37 °C. Lipofectamine-containing medium was then removed and the cells were supplemented with fresh medium (5% FCS).
Analysis of protein synthesis
Protein synthesis was analyzed at the indicated times by replacing the growth media with DMEM without methionine/cysteine and supplemented with EasyTag™ EXPRESS 35S Protein Labeling mix, [35S]Met/Cys (Perkin Elmer). The cells were then collected in sample buffer, boiled for 5 min and analyzed by autoradiography of SDS-polyacrylamide gels (15%). Protein synthesis was quantified by densitometry using a GS-800 Calibrated Densitometer (Bio-Rad).
In vitro transcription and translation
pPV-luc, pTM1-luc, pKS-luc and pIGR CrPV-luc were linearized prior to in vitro transcription with T7 RNA polymerase (BioLabs), according to manufacturer’s instructions. Plasmid pKS-luc was incubated with cap analog to obtain cap-luc mRNA. In vitro translation was carried out in rabbit reticulocyte lysate (RRL, Promega). To ensure the cleavage of eIF4G, the lysates were pre-incubated for 20 min at 30 °C with 20 ng/ml of purified human rhinovirus (HRV) 2Apro or foot-and-mouth disease virus (FMDV) Lpro proteins as indicated. Purified proteases were kindly provided by T. Skern (Max F. Perutz laboratories, Austria). Extracts were then treated with different concentrations of 4EGI-1 (Calbiochem) plus the addition of 50 ng of mRNA, followed by incubation for 90 min at 30 °C. Protein synthesis was estimated by measuring luciferase activity. The cleavage of eIF4G was determined by Western blot.
Analysis of viral RNA synthesis by radioactive labeling
[3H]uridine incorporation in cells infected with SV, VSV or EMCV was determined as described (Sanz et al., 2010). Briefly, cells were infected and treated with actinomycin D (5 μg/ml) at 5 hpi in the case of SV and VSV or at 3 hpi for EMCV. Then, [3H]uridine was added for 2 h in the presence or absence of 4EGI-1. Finally, total RNA was extracted using the RNAeasy kit (Qiagen). RNA was processed to detect viral RNA synthesis by agarose gel electrophoresis or by measuring radioactivity in a scintillation counter.
Immunoprecipitation assay
Following 4EGI-1 treatment at the indicated concentrations for 90 min, cells were washed in PBS and lysed in RIPA buffer (0.01 M Tris HCl, pH 7.4, 0.15 M NaCl, 0.1% SDS, 1% Triton X-100, 0.4 mM PMSF, 1% sodium desoxycholate). Cell lysates were then immunoprecipitated with an anti-eIF4GI antibody (Feduchi et al., 1995) at 1:100 dilution using Dynabeads coupled to Protein A (Invitrogen), according to manufacturer’s directions.
Immunofluorescence microscopy
Fixation, permeabilization and confocal microscopy were performed as described (Madan et al., 2008), employing a confocal LSM510 lens coupled to an Axio Imager Z1 microscope (Zeiss) with a 63×/1.4 oil Plan-Apochromat objective. Primary antibodies used were a rabbit polyclonal anti-eIF4GI (Feduchi et al., 1995) and a mouse monoclonal anti-eIF4E (sc-9976, Santa Cruz Biotechnology, Inc.), at 1:100 dilution. Specific antibodies conjugated to Alexa 488 or Alexa 555 (A-21202 and A-21432, respectively; Invitrogen) were used as secondary antibodies at 1:500 dilution. To-Pro-3 (Invitrogen) was used at 1:1000 dilution. Images were processed with Huygens 4.3 software (Scientific Volume Imaging B.V.).
Western blotting
Transfected cells were collected in sample buffer, boiled and analyzed by SDS-PAGE. After electrophoresis, proteins were transferred to a nitrocellulose membrane as described previously (Barco and Carrasco, 1995). To detect eIF4GI, rabbit antibodies against the N-terminal and C-terminal moieties of this protein (Aldabe et al., 1995) were used at 1:1000 dilution. To detect eIF4A and eIF4E, mouse monoclonal antibodies (Santa Cruz Biotechnology, Inc) were used at 1:500 dilution. Incubation with primary antibodies was performed for 2 h at 4 °C. The membrane was then washed three times with PBS containing 0.2% Tween-20 and incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit IgG antibodies (Amersham) at a 1:5000 dilution. After washing three times, protein bands were visualized with the ECL detection system (Amersham).
Measurement of luciferase activity
Cells were recovered in a buffer containing 25 mM glycylglycine (pH 7.8), 0.5% Triton X-100 and 1 mM dithiothreitol. Luciferase activity was determined using the luciferase assay system (Promega) with a Monolight 2010 Luminometer (Analytical Luminescence Laboratory), as described previously (Alvarez et al., 2003, Ventoso et al., 2001).
Acknowledgments
This study was supported by a DGICYT (Direccion General de Investigación Cientifica y Técnica) project BFU2012-31861. MGM is the holder of a FPU Fellowship. An Institutional Grant awarded to the Centro de Biología Molecular “Severo Ochoa” (CSIC-UAM) by the Fundación Ramón Areces is acknowledged.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2013.06.008.
Appendix A. Supplementary materials
Supplementary Material
References
- Aldabe R., Feduchi E., Novoa I., Carrasco L. Efficient cleavage of p220 by poliovirus 2Apro expression in mammalian cells: effects on vaccinia virus. Biochem. Biophys. Res. Commun. 1995;215(3):928–936. doi: 10.1006/bbrc.1995.2553. [DOI] [PubMed] [Google Scholar]
- Alvarez E., Menendez-Arias L., Carrasco L. The eukaryotic translation initiation factor 4GI is cleaved by different retroviral proteases. J. Virol. 2003;77(23):12392–12400. doi: 10.1128/JVI.77.23.12392-12400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A., Carrasco L. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO. J. 1995;14(14):3349–3364. doi: 10.1002/j.1460-2075.1995.tb07341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham G.J. Divergent picornavirus IRES elements. Virus. Res. 2009;139(2):183–192. doi: 10.1016/j.virusres.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2(4):213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- Castello A., Alvarez E., Carrasco L. The multifaceted poliovirus 2A protease: regulation of gene expression by picornavirus proteases. J. Biomed. Biotechnol. 2011;2011:369648. doi: 10.1155/2011/369648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A., Sanz M.A., Molina S., Carrasco L. Translation of Sindbis virus 26S mRNA does not require intact eukariotic initiation factor 4G. J. Mol. Biol. 2006;355(5):942–956. doi: 10.1016/j.jmb.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Cencic R., Desforges M., Hall D.R., Kozakov D., Du Y., Min J., Dingledine R., Fu H., Vajda S., Talbot P.J., Pelletier J. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J. Virol. 2011;85(13):6381–6389. doi: 10.1128/JVI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R., Hall D.R., Robert F., Du Y., Min J., Li L., Qui M., Lewis I., Kurtkaya S., Dingledine R., Fu H., Kozakov D., Vajda S., Pelletier J. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc. Natl. Acad. Sci. U. S. A. 2011;108(3):1046–1051. doi: 10.1073/pnas.1011477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor J.H., Lyles D.S. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 2002;76(20):10177–10187. doi: 10.1128/JVI.76.20.10177-10187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Aktas B.H., Wang Y., He X., Sahoo R., Zhang N., Denoyelle S., Kabha E., Yang H., Freedman R.Y., Supko J.G., Chorev M., Wagner G., Halperin J.A. Tumor suppression by small molecule inhibitors of translation initiation. Oncotarget. 2012;3(8):869–881. doi: 10.18632/oncotarget.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Preiss T., Hentze M.W. Translation driven by an eIF4G core domain in vivo. Embo. J. 1999;18(17):4865–4874. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz N., Lenarcic E.M., Landry D.M., Thompson S.R. Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA. 2009;15(5):932–946. doi: 10.1261/rna.1315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descamps G., Gomez-Bougie P., Tamburini J., Green A., Bouscary D., Maiga S., Moreau P., Le Gouill S., Pellat-Deceunynck C., Amiot M. The cap-translation inhibitor 4EGI-1 induces apoptosis in multiple myeloma through Noxa induction. Br. J. Cancer. 2012;106(10):1660–1667. doi: 10.1038/bjc.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Li Y., Yue P., Khuri F.R., Sun S.Y. The eIF4E/eIF4G interaction inhibitor 4EGI-1 augments TRAIL-mediated apoptosis through c-FLIP Down-regulation and DR5 induction independent of inhibition of cap-dependent protein translation. Neoplasia. 2010;12(4):346–356. doi: 10.1593/neo.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feduchi E., Aldabe R., Novoa I., Carrasco L. Effects of poliovirus 2A(pro) on vaccinia virus gene expression. Eur. J. Biochem. 1995;234(3):849–854. doi: 10.1111/j.1432-1033.1995.849_a.x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K.D., Semler B.L. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta. 2009;1789(9–10):518–528. doi: 10.1016/j.bbagrm.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno M., Sanz M.A., Pelletier J., Carrasco L. Requirements for eIF4A and eIF2 during translation of Sindbis virus subgenomic mRNA in vertebrate and invertebrate host cells. Cell. Microbiol. 2013;15(5):823–840. doi: 10.1111/cmi.12079. [DOI] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiology and Molecular Biology Reviews. 2011;75(3):434–467. doi: 10.1128/MMBR.00008-11. first page of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer C.A., Cowansage K.K., Arnold E.C., Banko J.L., Moerke N.J., Rodriguez R., Schmidt E.K., Klosi E., Chorev M., Lloyd R.E., Pierre P., Wagner G., LeDoux J.E., Klann E. Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proc. Natl. Acad. Sci. U. S. A. 2011;108(8):3383–3388. doi: 10.1073/pnas.1013063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer C.A., Santini E., Ma T., Arnold E.C., Whelan A.M., Wong H., Pierre P., Pelletier J., Klann E. Multiple components of eIF4F are required for protein synthesis-dependent hippocampal long-term potentiation. J. Neurophysiol. 2013;109(1):68–76. doi: 10.1152/jn.00342.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundsdoerfer P., Thoma C., Hentze M.W. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. U. S. A. 2005;102(38):13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist L., Oberer M., Reibarkh M., Cencic R., Bordeleau M.E., Vogt E., Marintchev A., Tanaka J., Fagotto F., Altmann M., Wagner G., Pelletier J. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One. 2008;3(2):e1583. doi: 10.1371/journal.pone.0001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch J.R., Dever T.E. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem. 2010;285(28):21203–21207. doi: 10.1074/jbc.R110.119743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., Castello A., Carrasco L. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell. Microbiol. 2008;10(2):437–451. doi: 10.1111/j.1462-5822.2007.01057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader S., Lee H., Pause A., Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995;15(9):4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y., Petroulakis E., LeBacquer O., Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25(48):6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell. 1999;3(6):707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- McMahon R., Zaborowska I., Walsh D. Noncytotoxic inhibition of viral infection through eIF4F-independent suppression of translation by 4EGi-1. J. Virol. 2011;85(2):853–864. doi: 10.1128/JVI.01873-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke N.J., Aktas H., Chen H., Cantel S., Reibarkh M.Y., Fahmy A., Gross J.D., Degterev A., Yuan J., Chorev M., Halperin J.A., Wagner G. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128(2):257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Mokas S., Mills J.R., Garreau C., Fournier M.J., Robert F., Arya P., Kaufman R.J., Pelletier J., Mazroui R. Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell. 2009;20(11):2673–2683. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino S., Imataka H., Svitkin Y.V., Pestova T.V., Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 2000;20(2):468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novac O., Guenier A.S., Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32(3):902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W.C., Sonenberg N. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell. Biol. 2011;12(4):235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G., Lomakin I.B., Pilipenko E.V., Shatsky I.N., Agol V.I., Hellen C.U. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 2001;98(13):7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo N., Sanz M.A., Steinberger J., Skern T., Kusov Y., Carrasco L. Translation directed by hepatitis A virus IRES in the absence of active eIF4F complex and eIF2. PLoS One. 2012;7(12):e52065. doi: 10.1371/journal.pone.0052065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo N., Sanz M.A., Welnowska E., Carrasco L. Translation without eIF2 promoted by poliovirus 2A protease. PLoS One. 2011;6(10):e25699. doi: 10.1371/journal.pone.0025699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J.D., Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433(7025):477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Santini E., Huynh T.N., MacAskill A.F., Carter A.G., Pierre P., Ruggero D., Kaphzan H., Klann E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493(7432):411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz M.A., Castello A., Ventoso I., Berlanga J.J., Carrasco L. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS One. 2009;4(3):e4772. doi: 10.1371/journal.pone.0004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz M.A., Welnowska E., Redondo N., Carrasco L. Translation driven by picornavirus IRES is hampered from Sindbis virus replicons: rescue by poliovirus 2A protease. J. Mol. Biol. 2010;402(1):101–117. doi: 10.1016/j.jmb.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L.S. 'Ribozoomin'--translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs) Curr. Protein Pept. Sci. 2012;13(4):305–330. doi: 10.2174/138920312801619385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez D. Inhibitors of protein biosynthesis. Mol. Biol. Biochem. Biophys. 1979;30(i-x):1–312. doi: 10.1007/978-3-642-81309-2. [DOI] [PubMed] [Google Scholar]
- Ventoso I., Blanco R., Perales C., Carrasco L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl. Acad. Sci. U. S. A. 2001;98(23):12966–12971. doi: 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welnowska E., Castello A., Moral P., Carrasco L. Translation of mRNAs from vesicular stomatitis virus and vaccinia virus is differentially blocked in cells with depletion of eIF4GI and/or eIF4GII. J. Mol. Biol. 2009;394(3):506–521. doi: 10.1016/j.jmb.2009.09.036. [DOI] [PubMed] [Google Scholar]
- Welnowska E., Sanz M.A., Redondo N., Carrasco L. Translation of viral mRNA without active eIF2: the case of picornaviruses. PLoS One. 2011;6(7):e22230. doi: 10.1371/journal.pone.0022230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.E., Pestova T.V., Hellen C.U., Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102(4):511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material