Abstract
Analyses of turkey coronavirus (TCoV), an enteric disease virus that is highly similar to infectious bronchitis virus (IBV) an upper-respiratory tract disease virus in chickens, were conducted to determine the adaptive potential, and genetic changes associated with emergence of this group 3 coronavirus. Strains of TCoV that were pathogenic in poults and nonpathogenic in chickens did not adapt to cause disease in chickens. Comparative genomics revealed two recombination sites that replaced the spike gene in IBV with an unidentified sequence likely from another coronavirus, resulting in cross-species transmission and a pathogenicity shift. Following emergence in turkeys, TCoV diverged to different serotypes through the accumulation of mutations within spike. This is the first evidence that recombination can directly lead to the emergence of new coronaviruses and new coronaviral diseases, emphasizing the importance of limiting exposure to reservoirs of coronaviruses that can serve as a source of genetic material for emerging viruses.
Keywords: Turkey coronavirus; Pathogenicity; Serology; Virus neutralization; Recombination, Molecular evolution; Real-time reverse transcriptase polymerase chain reaction; Comparative genomics; Sequencing
Introduction
Coronaviruses are worldwide in distribution, highly infectious, and extremely difficult to control. They can cause respiratory, enteric, and in some cases hepatic and neurological diseases in a wide variety of animals and in humans. Three groups have historically been recognized based on antigenic and genetic characteristics. However, after the emergence of severe acute respiratory syndrome coronavirus (SARS-CoV), the group 2 coronaviruses were split into subgroup 2a containing previously recognized group 2 viruses and subgroup 2b containing SARS-CoV (Gorbalenya et al., 2004). Studies to identify the animal reservoir for SARS-CoV led to the discovery of a diverse population of coronaviruses related to groups 1 and 2 in bats (Chiroptera) (Vijaykrishna et al., 2007, Woo et al., 2006).
Coronaviruses similar to the group 3 coronaviruses, which include the avian coronaviruses infectious bronchitis virus (IBV) and turkey coronavirus (TCoV), have been found in a variety of avian species including thrush (Turdus spp.), munia (Lonchura spp.), bulbul (Hypsipetes spp.), teal (Anas crecca), duck (Anseriformes), pigeon (Columbiformes), peafowl (Galliformes), pheasant (Galliformes), red knot (Calidris canutus), goose (Anserinae), and whooper swan (Cygnus cygnus) as well as in a beluga whale (Delphinapterus leucas) (Cavanagh et al., 2002, Hughes et al., 2009, Jonassen et al., 2005, Liu et al., 2005, Mihindukulasuriya et al., 2008, Muradrasoli et al., 2009, Woo et al., 2009b). These novel group 3 coronaviruses led to the proposal of subgroups 3a, 3b, and 3c (Woo et al., 2009b). Group 3-related viruses have also been found in an Asian leopard cat (Prionailurus bengalensis) and Chinese ferret badgers (Melogale moschata), but not in bats (Dong et al., 2007, Woo et al., 2006). It was suggested that wild bird species are the reservoir for the group 3 coronaviruses, which are primarily avian in origin, whereas bats are the reservoir for the group 1 and 2 mammalian coronaviruses (Woo et al., 2009a). This observation fits well with the view that, in addition to close contact, RNA viruses more readily undergo cross-species transmission when the two host species are closely related (Holmes and Rambaut, 2004). These studies indicate that an enormous, previously unrecognized reservoir for coronaviruses exists among animals, which is not unlike the reservoir that exists for influenza viruses in animals.
Coronavirus diversity is due to adaptive evolution driven by high mutation rates and genetic recombination (Holmes, 2009). Given the vast capacity for coronaviruses to change, it is not surprising that host switching leading to new diseases has occurred among coronaviruses (Li et al., 2005, Rest and Mindell, 2003, Stavrinides and Guttman, 2004, Woo et al., 2009a, Zhang et al., 2005), most notably the abrupt emergence of SARS-CoV in people in southern China in 2002, which quickly spread to 27 countries (including the United States) and resulted in 8096 cases and 744 deaths (http://www.who.int/csr/sars/country/table2004_04_21/en/index.html). However, recombination events in coronaviruses are not thought to play a key role in cross-species transmission since it appears that recombination events in the SARS-CoV occurred prior to the jump into humans (Hon et al., 2008). Rather, the emergence of new coronaviruses and coronaviral diseases are believed to be due to adaptive evolution (Holmes and Rambaut, 2004).
Previously unrecognized coronaviruses have emerged in animals. In 1995, a coronavirus was isolated from the enteric tract of turkeys with diarrhea in North Carolina and quickly spread throughout commercial turkeys in the USA (Guy, 2008). That virus was found to be similar to IBV based on cross-reactive antibodies (Guy, 2000). IBV causes a highly contagious upper-respiratory tract disease in chickens and is not known to infect or cause disease in turkeys (Cavanagh and Gelb, 2008). Several studies conducted on various TCoV isolates showed that the order of the genes at the 3′ end of the genome was similar to IBV (Breslin et al., 1999, Cavanagh et al., 2001, Guy, 2000, Li et al., 2005, Stephensen et al., 1999, Velayudhan et al., 2003). One of those genes, the coronavirus spike glycoprotein gene, codes for a membrane-bound protein found on the surface of the virus that is involved in attachment and entry into the host cell and therefore plays a key role in host specificity (Holmes et al., 1993). In the group 3 coronaviruses, the spike glycoprotein is post-translationally cleaved into an S1 subunit that contains the receptor binding domain and an S2 subunit that is non-covalently bound to S1 and contains a transmembrane domain that anchors the spike to the viral membrane. Since IBV does not infect and cause disease in turkeys, we sequenced the TCoV spike glycoprotein gene and found that it was not related to IBV (GenBank accession nos. AY342356 and AY342357). More recently, TCoV was reported to be closely related to IBV across the entire genome with the exception of the spike gene, suggesting that TCoV emerged from IBV (Cao et al., 2008, Gomaa et al., 2008) and Lin et al. (2004) was the first to suggest that recombination may account for the unique spike gene in TCoV. However, the origin of the spike gene sequence in TCoV is unknown.
In this study, we examine the pathogenicity and serologic relatedness of different TCoV strains to better understand the disease potential of this novel coronavirus. We identified genetic changes associated with antigenic differences and with propagation in embryonating eggs, and we analyzed the full-length genomic sequences for several TCoV strains to reconstruct the genetic changes that led to a host shift from chickens to turkeys and a pathogenicity shift from the upper-respiratory tract to the enteric tract.
Results
In vivo experiments show that TCoV is pathogenic in poults, nonpathogenic in chickens, and does not adapt to chickens
The TCoV strains used in this study (Table 1 ) were pathogenic for 1-day-old poults with diarrhea developing by 48 h post-inoculation (PI) and lesions in the gastrointestinal (GI) tract observed in most of the birds by 72 and 96 h PI (Table 2 ). Lesions in the GI tracts consisted of pale, flaccid intestines with thin walls, and gaseous watery contents. The ceca were bloated and filled with air and watery contents. Real-time RT-PCR conducted on pooled GI tracts from poults infected with the TCoV/TX-1038/98 strain had cycle threshold (Ct) values of 21.7, 16.7, 20.1, and 37.6 at 24, 48, 72, and 96 h PI, respectively (Table 2), indicating that the most virus (lowest real-time RT-PCR Ct value) was detected in poults at 48 h PI. Based on this data, the pooled GI tracts from poults given the other TCoV isolates were examined at 48 h PI and the data are presented in Table 2.
Table 1.
TCoV viruses examined in this study.
| Isolate | Location | Year of isolation | Accession numbers | Virus titera |
|---|---|---|---|---|
| TCoV/VA-74/03 | Virginia | 2003 | GQ427173 | 1 × 101.5 |
| TCoV/TX-GL/01 | Texas | 2001 | GQ427174 | NDb |
| TCoV/TX-1038/98 | Texas | 1998 | GQ427176 | 1 × 103 |
| TCoV/IN-517/94 | Indiana | 1994 | GQ427175 | 1 × 104 |
Titers represent 50% poult infectious doses as determined in 1-day-old turkey poults.
ND = not done.
Table 2.
Pathogenicity of TCoV in 1-day-old turkeys.
| Virus | No. with lesions/total examined |
|||
|---|---|---|---|---|
| 24 h PIa | 48 h PI | 72 h PI | 96 h PI | |
| TCoV/VA-74/03 | 0/4 | 3/4 (13.7) | 4/4 | 4/4 |
| TCoV/TX-GL/01 | 0/4 | 2/4 (15.4) | 3/4 | 4/4 |
| TCoV/IN-517/94 | 0/4 | 0/4 (13.9) | 4/4 | 4/4 |
| TCoV/TX-1038/98 | 0/4 (21.7)b | 1/4 (16.7) | 3/4 (20.1) | 4/4 (37.6) |
PI = post-inoculation.
Real-time RT-PCR cycle threshold value.
No clinical signs of infection were observed in the upper-respiratory tract or in the GI tract when the TCoV/TX-1038/98 or the TCoV/TX-GL/01 strains were given to chickens. To determine if TCoV could revert back to pathogenicity in chickens we passaged the viruses in 1-day-old chicks by infecting the birds, re-isolating the virus from the GI tract and giving it to additional 1-day-old chicks. No clinical signs or lesions were observed when either virus was passaged in 1-day-old chickens. The TCoV/TX-1038/98 virus was detected in pooled GI tracts on day 3 for passages 1 and 2, and on day 5 for passage 3. The TCoV/TX-1038/98 virus could not be detected in the pooled GI tracts from the fourth passage so the experiment was terminated. The TCoV/TX-GL/01 virus was detected on day 3 after the first passage in 1-day-old SPF chickens but could not be detected in the subsequent passage. No clinical signs, lesions, or virus were detected in any of the contact exposed chickens for any of the passages. Finally, the last virus-positive passage of TCoV/TX-1038/98 (third passage) and TCoV/TX-GL/01 (first passage) in chickens were examined for pathogenicity in 1-day-old poults and both were found to induce clinical signs and lesions similar to the original virus before passage in chickens (data not shown). The above data show that TCoV does not adapt or revert to cause disease in chickens.
Sequence analysis following virus passage in embryonating eggs reveals changes in the spike gene
We wanted to determine the capacity of this virus to change at the molecular level. The spike glycoprotein gene was examined for nucleotide and amino acid changes associated with passage of TCoV in 19-day-old embryonating turkey eggs. No sequence changes were observed between the initial sequence determined for passage 1 and passages 8, 10, 13, 14, 17, 18, 20, and 30 (Table 3 ). Between passages 30 and 40, we found 7 nucleotide changes in the spike glycoprotein gene with 6 changes in S1 and 1 change in S2 (G2269A resulting in amino acid change Val757Ile). Only two nucleotide changes, both in S1, resulted in non-synonymous changes at residues Ser65Pro and Asn84Lys. Between passages 40 and 49, we observed that 5 of the 6 changes detected in passage 30 were conserved, one reverted to the original sequence, and there were 4 new changes. Between passages 49 and 50, we observed 7 conserved changes, 2 reversions, and 4 new changes. Between passages 50 and 52, we observed 7 conserved changes, 6 reversions to nucleotides observed in previous passages, and 2 new changes.
Table 3.
Nucleotide and amino acid changes in spike following passage of TCoV in embryonating turkey eggs.
| Strain and passage no. | Nucleotidea, Amino Acidb changes | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TCoV/TX-1038/98 Pass 40c | C183T | A189C | T193C, Ser65Pro | C198T | C225T | C252A, Asn84Lys | G2269A, Val757Ile | |||||||||
| TCoV/TX-1038/98 Pass 49 | T237C | A252C, Lys84Asn | A271T, Thr91Ser | G380A, Gly127Asp | A403C, Asn135 His | |||||||||||
| TCoV/TX-1038/98 Pass 50 | C237T | C246T | C252T | C403A, His135Asn | C411T | T413C, Ile138 Thr | ||||||||||
| TCoV/TX-1038/98 Pass 52 | T246C | T252C | T271A, Ser91Thr | G379A | A380G, Asp127Ser | A403C Asn135His | T411C | C413T, Thr138Ile | A716G, Gln239Arg | |||||||
| TCoV/TX-R/98 Pass 24d | G416A, Gly139Asp | G540T, Gln180His | ||||||||||||||
Positions are relative to the ATG start site in the spike gene with the A being number 1, and the change is synonymous if no amino acid change is indicated.
Positions are relative to the beginning of the spike glycoprotein with methionine being number 1.
Using passage 1 as the base sequence, no changes were observed for passages 8, 10, 13, 14, 17, 18, 20, and 30.
Using passage 1 as the base sequence, no changes were observed for passage 20 and no additional changes from passage 24 were observed for passage 27 and passage 30.
Predicted residue changes in S1 between passages 30 and 40 included 2 changes (Ser65Pro and Asn84Lys); between passages 40 and 49, there were 1 conserved (Pro65), 1 reversion (Lys84Asn), and 3 new changes (Thr91Ser, Gly127Asp, Asn135His); between passages 49 and 50, there were 4 conserved (Pro65, Asn84, Ser91, and Asp127), 1 reversion (His135Asn), and 1 new change (Ile138Thr); and between passages 50 and 52, there were 2 conserved (Pro65 and Asn84), 3 reversions (Ser91Thr, Asn135His, and Thr138Ile), and 2 new changes (Asp127Ser and Gln239Arg). We sequenced all structural genes for passages 8 and 40 of TCoV/TX-1038/98 and found only 1 nucleotide change in a structural protein gene other than spike, which was a synonymous change that occurred in the nucleocapsid gene (data not shown).
We sequenced the spike glycoprotein gene from a second virus (TCoV/TX-R/98) passaged in embryonating turkey eggs and found no changes between passages 1 and 20 (Table 3). Between passages 20 and 24, two changes both non-synonymous occurred resulting in a Gly139Asp and Gln180His change. The changes were conserved and were the only changes observed through 30 passages. Taken together, these data indicate that TCoV has the capacity to change through replication, but the tolerated presumably advantageous mutations are only observed following repeated passages in the host.
Real-time RT-PCR-based serum neutralization tests revealed the presence of different serotypes of TCoV
To determine if TCoV has diverged to different serotypes, we conducted virus neutralization (VN) testing. Because TCoV does not produce lesions reliably when inoculated into embryonating eggs and thereby cannot be used to evaluate the endpoint of a VN assay, we used a real-time RT-PCR quantitative test to measure the neutralizing activities of sera against TCoV in turkey poults (Bostic et al., 1999, Saito et al., 2003, van Santen et al., 2004). Antiserum prepared against 3 different TCoV strains induced a statistically significant reduction (Student's t-test, P < 0.01) in virus replication in turkey poults as determined by real-time RT-PCR when the homologous virus was used in the assay. The antiserum titers representing the highest dilution of serum that protected 50% of the poults from infection were determined to be 64 for TCoV/VA-74/03, 256 for TCoV/TX-1038/98, and 256 for TCoV/IN-517/94. When the Archetti and Horsfall (1950) antigenic relatedness values were calculated using the homologous and heterologous virus neutralization titers (Table 4 ), the relatedness values were 2.2 for TCoV/VA-74/03 and TCoV/TX-1038/98, 1.6 for TCoV/VA-74/03 and TCoV/IN-517/94, and 3.1 for TCoV/TX-1038/98 and TCoV/IN-517/94, indicating that the viruses were not serologically related.
Table 4.
Virus neutralization titers based on clinical signs in turkey poults and detection of virus by the real-time RT-PCR test.
| Antisera | Virus |
||
|---|---|---|---|
| TCoV/VA-74/03 | TCoV/TX-1038/98 | TCoV/IN-517/94 | |
| TCoV/VA-74/03 | 64a | 2 | 2 |
| TCoV/TX-1038/98 | 4 | 256 | 2 |
| TCoV/IN-517/94 | 2 | 32 | 256 |
Highest dilution of antiserum that protected 50% of poults from infection as determined by the real-time RT-PCR test and calculated using the Reed and Muench test (Thayer and Beard, 2008).
Whole virus genome sequencing and comparative genomics show that recombination led to the emergence of TCoV and different serotypes result from molecular divergence within the spike gene
Strand displacement amplification was successfully used to amplify the viral genomic RNA from 4 different strains of TCoV, and the cloned fragments were used to generate two different libraries for each virus. Sequences generated from both libraries resulted in 5 to 10× coverage of the genomes. The organization of the genome and location and size of the open reading frames for TCoV/VA-74/03, TCoV/TX-GL/01, TCoV/TX-1038/98, and TCoV/IN-517/94 are presented in Table 5 . The genome sizes of TCoV/VA-74/03, TCoV/TX-GL/01, TCoV/TX-1038/98, and TCoV/IN-517/94 are 27,771, 27,619, 27,782, and 27,665 respectively. The order of the genes was the same for all of the viruses examined; 5′ UTR-1a (1ab)-spike-3a-3b-envelope-membrane-ORF X-5a-5b-Nucleocapsid-3′ UTR. The size of the 5′ UTR was 531 nucleotides (nt) for each of the strains except for the TCoV/IN-517/94 strain, which had a 528-nt 5′ UTR. The 3′ UTR was 502 nt for TCoV/VA-74/03 and TCoV/TX-1038/98 whereas TCoV TX-GL/01 and TCoV/IN-517/94 had a 330-nt and 373-nt 3′ UTR, respectively. The ranges and sizes of the other genes were similar among the 4 viruses (Table 5).
Table 5.
Genes and coding regions for 4 strains of turkey coronavirus examined in this study.
| ORFa | TCoV/VA-74/03 |
TCoV/TX-GL/01 |
TCoV/TX-1038/98 |
TCoV/IN-517/94 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size |
Size |
Size |
Size |
|||||||||
| Location | ntb | aac | Location | nt | aa | Location | nt | aa | Location | nt | aa | |
| 5′ UTR | 1–531 | 531 | – | 1–531 | 531 | – | 1–531 | 531 | – | 1–528 | 528 | – |
| 1a | 532–12,357 | 11,844 | 3,947 | 532–12,381 | 11,850 | 3,949 | 532–12,384 | 11,853 | 3,950 | 529–12,387 | 11,859 | 3,952 |
| 1ab | 532–20,321 | 19,790 | 6,596 | 532–20,339 | 19,808 | 6,602 | 532–20,339 | 19,808 | 6,602 | 529–20,345 | 19,817 | 6,605 |
| Spike | 20,345–24,025 | 3681 | 1,226 | 20,363–24,040 | 3678 | 1,225 | 20,363–24,037 | 3,675 | 1,224 | 20,369–24,049 | 3,681 | 1,226 |
| 3a | 23,973–24,146 | 174 | 57 | 23,988–24,161 | 174 | 57 | 23,985–24,158 | 174 | 57 | 23,997–24,170 | 174 | 57 |
| 3b | 24,146–24,340 | 195 | 64 | 24,161–24,355 | 195 | 64 | 24,158–24,352 | 195 | 64 | 24,170–24,364 | 195 | 64 |
| Envelope | 24,321–24,620 | 300 | 99 | 24,336–24,635 | 300 | 109 | 24,333–24,632 | 300 | 99 | 24,345–24,644 | 300 | 99 |
| Membrane | 24,622–25,293 | 672 | 223 | 24,637–25,314 | 678 | 225 | 24,634–25,305 | 672 | 223 | 24,646–25,317 | 672 | 223 |
| X | 25,294–25,578 | 285 | 94 | 25,315–25,599 | 285 | 94 | 25,306–25,590 | 285 | 94 | 25,318–25,602 | 285 | 94 |
| 5a | 25,654–25,851 | 198 | 65 | 25,675–25,872 | 198 | 65 | 25,666–25,863 | 198 | 65 | 25,678–25,875 | 198 | 65 |
| 5b | 25,848–26,096 | 249 | 82 | 25,869–26,117 | 249 | 82 | 25,830–26,108 | 279 | 92 | 25,872–26,120 | 249 | 82 |
| Nucleocapsid | 26,039–27,268 | 1230 | 409 | 26,060–27,289 | 1230 | 409 | 26,051–27,280 | 1230 | 409 | 26,063–27,292 | 1230 | 409 |
| 3′ UTR | 27,269–27,771 | 502 | – | 27.290–27,619 | 330 | – | 27,281–27,782 | 502 | – | 27,293–27,665 | 373 | – |
ORF = open reading frame.
nt = nucleotide.
aa = amino acid.
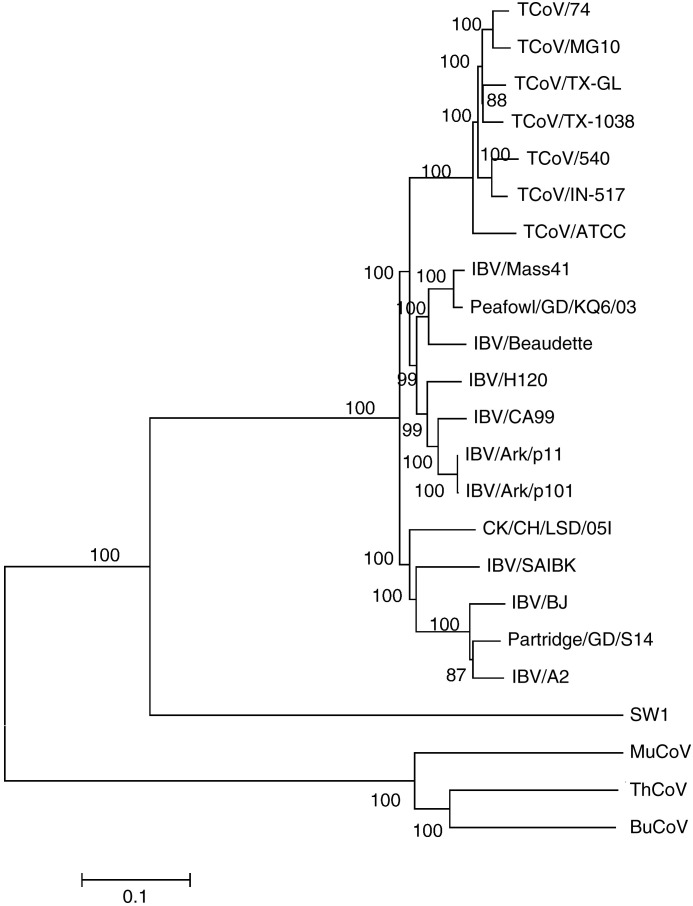
Whole genome alignments were generated and phylogenetic trees were constructed with the Neighbor-Joining method, Minimum Evolution method, Maximum Parsimony method, and UPGMA. All of the trees had similar topography with significant bootstrap support. A tree containing the 4 TCoV isolates sequenced herein as well as other available full-length genomes for the group 3 coronaviruses is shown in Fig. 1 . Three major clades are observed. The TCoV isolates form a monophyletic group that nests within a lineage of IBV isolates. This is supported by the percent similarity between the TCoV and IBV isolates for the full-length genomes (Table 6 , upper triangle), which are greater than 86%. Three viruses, MuCoV HKU13, ThCoV HKU12, and BuCoV HKU11, make up a second distinct lineage and a single virus SW1 from a beluga whale separates into a third.
Fig. 1.
Neighbor-Joining method used to infer evolutionary history using full genomic sequence data of the available group 3 coronaviruses. The bootstrap consensus tree was constructed from 1000 replicates (percentage of replicate trees in which associated strains clustered together are presented at nodes). The p-distance scale is presented at the bottom of the figure.
Table 6.
Sequence percent similarity for the full-length genome (upper triangle) and the spike glycoprotein (lower triangle) alignmentsa.
| TCoV/VA-74 | TCoV/TX-GL | TCoV/IN-517 | TCoV/TX-1038 | TCoV/540 | TCoV/ATCC | TCoV/MG10 | IBV/Mass 41 | IBV/Ark | BuCoV | ThCoV | MuCoV | SW1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TCoV/VA-74 | – | 95.5 | 95.1 | 95.7 | 93.7 | 92.8 | 97.0 | 86.6 | 89.3 | 48.3 | 49.3 | 47.9 | 51.2 |
| TCoV/TX-GL | 93.5 | – | 94.4 | 96.0 | 93.3 | 92.4 | 95.6 | 87.3 | 89.5 | 48.4 | 49.1 | 47.9 | 51.3 |
| TCoV/IN-517 | 97.2 | 93.9 | – | 94.6 | 96.2 | 93.4 | 95.8 | 87.0 | 88.8 | 48.3 | 49.2 | 47.9 | 51.3 |
| TCoV/TX-1038 | 98.0 | 93.9 | 95.8 | – | 93.5 | 93.0 | 95.3 | 87.1 | 90.4 | 48.3 | 49.1 | 47.9 | 51.2 |
| TCoV/540 | 94.4 | 92.8 | 95.0 | 94.5 | – | 92.8 | 94.1 | 86.5 | 87.8 | 48.2 | 49.0 | 47.8 | 51.1 |
| TCoV/ATCC | 92.4 | 91.0 | 92.6 | 92.3 | 92.4 | – | 92.9 | 86.9 | 88.1 | 48.1 | 49.0 | 47.7 | 51.1 |
| TCoV/MG10 | 99.9 | 93.5 | 97.2 | 98.0 | 94.4 | 92.4 | – | 86.8 | 88.3 | 48.4 | 49.2 | 47.9 | 51.4 |
| IBV/Mass41 | 34.9 | 34.9 | 35.1 | 34.9 | 35.5 | 35.6 | 34.9 | – | 91.3 | 48.0 | 48.8 | 47.6 | 50.8 |
| IBV/Ark | 35.2 | 35.2 | 35.6 | 35.1 | 35.7 | 35.7 | 35.2 | 86.8 | – | 47.9 | 48.7 | 47.4 | 50.8 |
| BuCoV | 28.5 | 28.9 | 28.4 | 28.7 | 28.2 | 28.8 | 28.5 | 27.7 | 28.0 | – | 75.3 | 71.2 | 45.5 |
| ThCoV | 26.6 | 26.9 | 26.7 | 26.4 | 26.4 | 26.5 | 26.6 | 26.8 | 26.6 | 47.2 | – | 70.4 | 46.3 |
| MuCoV | 28.8 | 28.9 | 28.9 | 28.6 | 28.8 | 29.0 | 28.8 | 27.7 | 27.9 | 48.4 | 48.4 | – | 45.0 |
| SW1 | 30.3 | 30.5 | 30.6 | 30.3 | 30.5 | 30.8 | 30.3 | 28.6 | 28.7 | 28.1 | 26.8 | 29.1 | – |
Alignments were assembled using ClustalW in the MegAlign program (DNASTAR, Inc.).
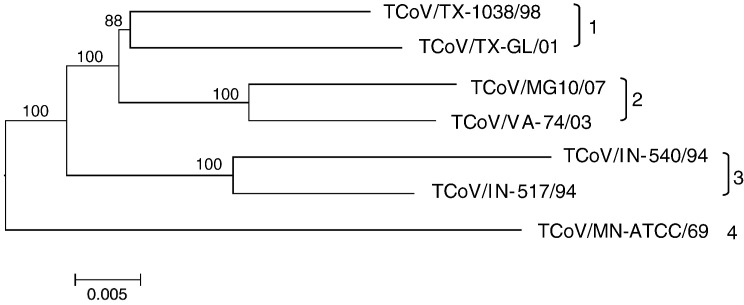
The sub-branch of the full genomic sequence tree containing the TCoV isolates is shown in Fig. 2 . Four genetic groups were observed with TCoV/TX-GL/01 and TCoV/TX-1038/98 grouping together, TCoV/MG10 and TCoV/VA-74/03 forming a second group, TCoV/IN-517/94 and TCoV/540 making up a third group, and TCoV/ATCC alone in a fourth group. All viral genomes are at least 92.4% similar at the nucleotide level. Similarity between TCoV groups at the nucleotide level varies from 92.4% to 95.7% and within group similarity ranges from 96.0% to 97.0% at the nucleotide level, which was the basis for recognizing 4 groups.
Fig. 2.
Neighbor-Joining method used to infer evolutionary history using full genomic sequence data for available TCoV isolates. The bootstrap consensus sub-tree was constructed from 1000 replicates (percentage of replicate trees in which associated strains clustered together are presented at nodes). The p-distance scale is presented at the bottom of the figure. Different genetic groups are indicated at the right of the figure.
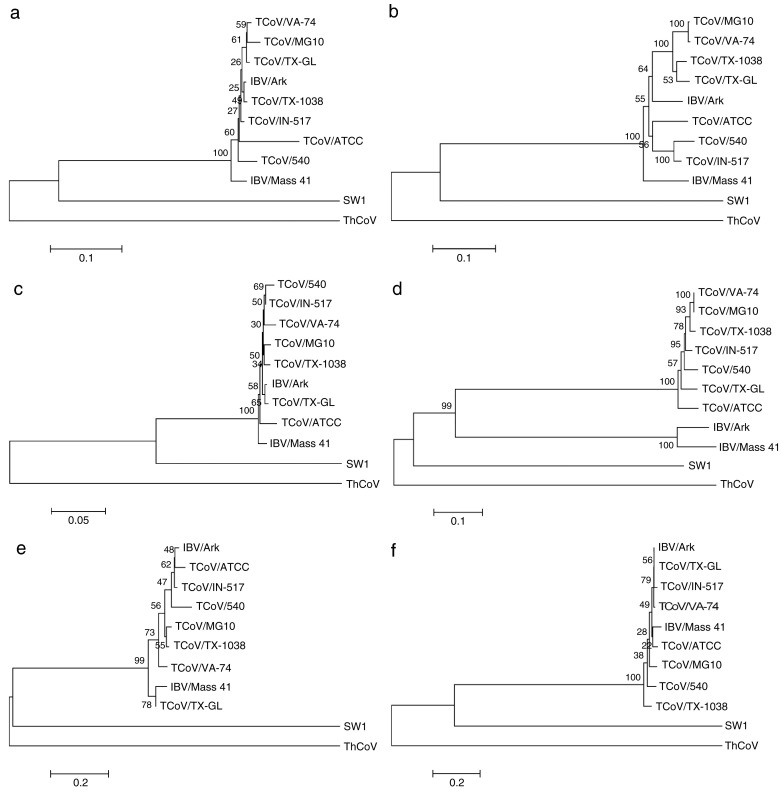
Phylogenetic analysis of the protein sequences for spike, membrane, RNA-dependent RNA polymerase (RdRp), nucleocapsid, 3CLpro, and helicase computed using Neithbor-Joining and the Nei-Gojobori method are presented in Fig. 3 for TCoV/VA-74/03, TCoV/TX-GL/01, TCoV/IN-517/94, TCoV/TX-1038/98, TCoV/MG10, TCoV/540, TCoV/ATCC, and other representative group 3 coronaviruses including IBV/Arkp11, beluga whale/CoV/SW1, ThrushCoV/HKU12, and IBV/Mass41. Bayesian analysis and maximum likelihood methods for reconstructing evolutionary trees yield similar topologies (data not shown). For all of the proteins, the beluga whale and thrush CoVs do not cluster with the other viruses or with each other. In addition, the TCoV's cluster with IBV/Mass41 and IBV/Arkp11 for all of the proteins except spike. For the spike protein (Fig. 3d), the IBV isolates clearly fall outside the cluster of TCoV viruses. This is supported by the high percent similarities for the spike glycoproteins among TCoV isolates, which are greater than 90%, whereas between TCoV and IBV they are less than 36% (Table 6, lower triangle).
Fig. 3.
Phylogenetic relationships of virus proteins. Phylogenetic trees showing amino acid sequence relatedness computed using Neighbor-Joining and the Nei-Gojobori method. (a) Main protease (3CLpro) coding region (residues 2778-3084 in the 1ab protein). (b) RNA-dependent RNA polymerase (RdRp) coding region (residues 3927-4866 in the 1ab protein). (c) Helicase coding region (residues 4867–5465 in the 1ab protein). (d) Spike glycoprotein. (e) Membrane glycoprotein. (f) Nucleocapsid protein. Viruses included in the analysis are TCoV/74 (GQ427173), TCoV/TX-GL (GQ427174), TCoV/TX-1038 (GQ427176), TCoV/IN-517 (GQ427175), TCoV/MG10 (EU095850), TCoV/540 (EU022525), TCoV/ATCC (EU022526), IBV/Ark (EU418976), IBV/Mass41 (AY851295), SW1 (NC010646), and ThCoV (FJ376621). The amino acid sequences were aligned with ClustalW (MEGA 4.0.2, Tamura et al., 2007), and the evolutionary distances are shown for each of the trees. Residue positions listed above are relative to the TCoV/TX-GL/01 strain.
A codon-based Z-test for positive selection, MEGA4 (http://www.megasoftware.net/index.html) was used to analyze the numbers of non-synonymous and synonymous substitutions per site (dN/dS ratio) in the spike gene and the test showed that positive selection was occurring between the TCoV/TX-1038/98 and the TCoV/VA-74/03 and TCoV/MG10/07 isolates (dN/dS = 2.784 and 2.674, respectively, and P < 0.003 and P < 0.004 respectively, Table 7 ).
Table 7.
Codon-based Z-test for positive selectiona in the spike gene.
| TCoV/TX-1038/98 | TCoV/IN-517/94 | TCoV/TX-GL/01 | TCoV/VA-74/03 | TCoV/MG10/07 | TCoV/MN-ATCC/69 | TCoV/IN-540/94 | |
|---|---|---|---|---|---|---|---|
| TCoV/TX-1038/98 | – | − 0624 | − 3.299 | 2.784 | 2.674 | − 3.690 | − 2.036 |
| TCoV/IN-517/94 | 1.000 | – | − 2.862 | − 2.176 | − 2.258 | − 4.532 | − 2.842 |
| TCoV/TX-GL/01 | 1.000 | 1.000 | – | − 2.791 | − 2.850 | − 4.635 | − 2.847 |
| TCoV/VA-74/03 | 0.003 | 1.000 | 1.000 | – | 1.032 | − 3.786 | − 2.891 |
| TCoV/MG10/07 | 0.004 | 1.000 | 1.000 | 0.152 | – | − 3.743 | − 2.953 |
| TCoV/MN-ATCC/69 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | – | − 4.744 |
| TCoV/IN-540/94 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | – |
The probabilities (P) of rejecting the null hypothesis of strict neutrality (dN = dS) in favor of the alternative hypothesis (dN > dS) is shown below the diagonal. Values of P < 0.05 are considered significant. The test statistic values shown above the diagonal are the numbers of synonymous dS and non-synonymous dN substitutions per site, respectively. The variance of the difference was computed using the bootstrap method (1000 replicates).
To determine how TCoV isolates are related to IBV (IBV/Mass41 strain), an analysis using the SimPlot software (http://sray.med.som.jhmi.edu/SCRoftware/simplot/) was conducted and the results are shown in Fig. 4 . Two major recombination events were observed, one in the 3′ end of gene 1ab (at nucleotide 20,173, average for all TCoVs examined) and one in the 3′ end of spike (at nucleotide 23,849, average for all TCoVs examined). That result was supported by the amino acid alignments (data not shown) used to construct the phylogenetic trees shown in Fig. 3. Subsequently, BLASTN and BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) analyses using all and parts of the recombined region were conducted. No matches to the GenBank or Swissprot databases were found. In addition, we conducted a conserved domain search using RPS-BLAST (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), which identified conserved domains between TCoV spike and four group 1 coronaviruses (canine coronavirus P36300, feline infectious peritonitis virus Q66951, porcine enteric disease virus Q84712, and human coronavirus-229E P15423) as well as IBV. Taken together, these data show that recombination replaced the spike gene in IBV leading to the emergence of TCoV, and that the spike glycoprotein gene subsequently underwent selection following cross-species transmission.
Fig. 4.
Simplot analysis of full-length genomic sequence for IBV/Mass 41, TCoV/VA-74/03, TCoV/TX-GL/01, TCoV/IN-517/94, and TCoV/TX-1038/98 showing recombination between nucleotides 20,173 and 23,849. The query sequence is TCoV/VA-74/03. Bars at the top represent relative position of the coding regions for 1a, 1ab, spike, membrane (M), and nucleocapsid (N).
Discussion
A previously unrecognized group 3 coronavirus isolated from turkeys with enteric disease was found to be genetically similar to IBV, a coronavirus that causes an upper-respiratory tract disease in chickens (Cao et al., 2008, Lin et al., 2004). In this study, we determined the full-length genome sequence of 4 different TCoV strains and showed that the genomes were closely related to IBV except in the spike glycoprotein gene. All 4 strains of TCoV were found to be pathogenic for 1-day-old turkeys but were not pathogenic for 1-day-old chickens, indicating that the spike glycoprotein of this coronavirus was responsible for the host shift from chickens to turkeys and the pathogenicity shift from upper-respiratory disease to enteric disease.
To determine if TCoV could adapt to cause disease in chickens, we passaged two different strains of TCoV in 1-day-old chicks. Virus could not be detected in the chicks following the 3rd passage of TCoV/TX-1038/98 or after only 1 passage of TCoV/TX-GL/01. Furthermore, TCoV was not transmissible among chickens since it was not detected in any of the contact control birds. These data indicate that exposure/passage of TCoV in chickens did not result in selection for changes sufficient to maintain infection and replication in chickens nor did it lead to transmission of the virus among chickens. Based on this finding and the observation that the TCoV spike glycoprotein only bears about 35% similarity to the IBV spike glycoprotein, it appears likely that a recombination event that replaced the spike glycoprotein gene rather than selection of a virus subpopulation or genetic changes was responsible for the emergence of TCoV. However, until a source for the TCoV spike has been identified, alternate explanations for the observed differences between TCoV and IBV spike should be considered.
It is well known that many different serotypes of IBV exist, none of which cross-protect (Cavanagh and Gelb, 2008, Fabricant, 1998). Herein, three TCoV strains with genetically distinct spike glycoprotein genes were examined by the real-time RT-PCR based serum neutralization test and were found to be serologically unrelated. Thus, it appears that the spike glycoprotein gene of TCoV has diverged to encode at least 3 different serotypes. These serologically distinct strains of TCoV could be the result of adaptive evolution driven by the host immune response. Adaptive evolution is the process by which genetic changes in the viral genome leading to a more fit virus population become fixed over time and has been reported for many coronaviruses (Hasoksuz et al., 2007, Lee and Jackwood, 2001, Shi et al., 2006, Tang et al., 2009, Zhang et al., 2006). Although all coronaviruses undergo adaptive evolution, it is interesting that only the group 3 coronaviruses, IBV and now based on our data TCoV, have thus far been reported to diverge within the spike glycoprotein gene to generate multiple different serotypes. Studies on divergent SARS-CoVs showed that pseudoviruses expressing different human derived SARS-CoV spike glycoproteins were all equally neutralized with the exception of one virus (GD03), indicating that possibly two serotypes may be present (Enjuanes et al., 2008). In addition, feline enteric coronavirus (FECoV) has been reported to have two serologically distinct types, but nearly all of the naturally occurring FECoV viruses are type I (Tekes et al., 2008). It is not clear why divergence to multiple different serotypes appears to be a unique characteristic for at least two of the group 3 coronaviruses, but elucidating the advantage of this phenomenon could lead to better vaccination control strategies.
Typically, IBV is passaged in embryonating eggs to attenuate the virus for use as a modified live vaccine in chickens. To determine structural protein sequence changes associated with multiple passages in eggs, we passaged two different TCoV strains in 18-day-old embryonating turkey eggs and found that few or no amino acid changes were observed for the first 20 to 30 passages (Table 3). Between 30 and 52 passages in embryonating eggs, we found 4 amino acid changes that became fixed, 3 that reverted, and 1 that was variable in the S1 subunit of the spike glycoprotein. No non-synonymous changes and only 1 synonymous change (nucleocapsid gene) were observed in the other structural proteins of the virus. It is not clear why no changes were observed in the virus for the earlier passages in embryonating eggs. Coronaviruses have a high mutation rate that has been estimated to be 1 × 10− 3 synonymous substitutions/site/year (Holmes, 2009). Studies on the genetic changes associated with adaption of the SARS-CoV to palm civets and subsequently to humans showed that mutations were being fixed at a high rate following cross-species transmission then slowed in the later stages of the epidemic (Kan et al., 2005, Song et al., 2005). Assuming the evolutionary dynamics of coronaviruses result in an increase in observed sequence variation when introduced into a new host, one would expect to see a high number of changes in the first few passages in embryonating eggs. It is possible that changes occurred in the 1a and 1ab polyprotein genes since we did not examine that area of the genome, but it is well known that the highest rate of change is observed in the spike glycoprotein because it is involved in attachment and entry into the host cell, so some level of adaptive change would be expected in spike (Cavanagh et al., 2005). We did not examine the sequence of the TCoV strains prior to the first passage in embryonating eggs because there was not enough of the virus present. Therefore, it is possible that selection of a viral subpopulation following only one passage in embryonating eggs could have occurred leading to a reasonably fit population of viruses that then underwent adaptive evolution. In a previous study, we found that a more fit subpopulation of an embryonating egg-adapted vaccine virus for IBV was selected after only one passage in chickens (McKinley et al., 2008). Since deleterious as well as advantageous changes can occur with extremely high mutation rates, such as those rates found in coronaviruses, it is possible that minor more fit coronavirus subpopulations would then take several passages before becoming the predominant species (Holmes, 2009). It is interesting to note that attenuation of pathogenic IBV strains by passage in embryonating eggs takes more than 50 passages and can take up to 75 or 100 passages (Jackwood et al., 2003).
Strand displacement amplification was used to randomly amplify the viral genomic RNA of 4 TCoV strains and two cDNA libraries for each virus were produced. The genome organization, location and size of the open reading frames for each of the viruses were similar to previously published data for TCoVs (Cao et al., 2008, Gomaa et al., 2008, Lin et al., 2004). Two different alignment methods and four different methods of constructing phylogenetic trees, resulted in trees with nearly identical topology. When other available group 3 coronavirus full-length genomes were examined (Fig. 1), three major clades were observed supporting recently published data indicating that the group 3 coronaviruses can be divided into three subgroups (Woo et al., 2009b); subgroup 3a containing TCoV and IBV isolates, subgroup 3b containing viruses isolated from a bulbul, a thrush, and a munia, and subgroup 3c containing a virus isolated from a beluga whale (SW1). We further divide the TCoV isolates into 4 genetic groups (Fig. 2), and confirmed that viruses from 3 of the genetic groups represent different serotypes (Table 4).
The topology of the spike glycoprotein trees compared to other protein coding sequences (Fig. 3) showed the TCoV spike glycoprotein to be clearly different from that of IBV. Recombination detection analysis on TCoV isolates (Fig. 4) showed that recombination occurred in the 3′ end of gene 1ab (nt. position 20,173) and at about 177 nucleotides from the 3′ end of spike (nt. position 23,849), essentially inserting a different spike gene into TCoV. That result was supported by the amino acid alignments used to construct the phylogenetic relationships of virus proteins shown in Fig. 3. Similarities between spike glycoprotein sequences and the locations of the crossover sites among TCoVs isolated over time indicate that this was a one-time event, which led to the emergence of TCoV from its closest IBV ancestor. This is the first evidence that recombination in coronaviruses directly led to cross-species transmission and the emergence of a new disease. Although SARS-CoV was shown to be a recombinant virus, this recombination event was reported to have occurred before the cross-species spread to intermediate hosts (raccoon dog, Nyctereutes procyonoides and Asian palm civet, Paradoxurus hermaphroditus) and ultimately to humans (Hon et al., 2008). Therefore, recombination is likely not the direct cause of emergence of SARS.
Examining the dN/dS ratios for the spike gene in TCoV showed that positive selection was occurring between viruses isolated in 1998 and 2003/2007, after the emergence of the disease in turkeys. Similar to what was observed for the emergence and evolution of the SARS-CoV, a high rate of molecular evolution is observed when viruses switch hosts (Song et al., 2005). This result appears to contradict our observation above that little or no change in the structural proteins of TCoV occurs during the first 30 to 50 passages in eggs. Perhaps a low rate of molecular evolution occurred in the eggs because the cells lining the GI tract of an 18-day-old turkey embryo are not substantially different from the cells in the GI tract of a poult or because there are other yet to be identified constraints on these molecules.
We conducted BLAST analyzes on the nucleic acid and amino acid sequences within the recombination sites as well as on different overlapping segments of the TCoV spike sequence but could not find similar sequences in any of the databases examined. Although the source of the genetic material making up the TCoV spike is not known at this time, it is likely from another coronavirus because the makeup of the TCoV spike resembles that of other coronavirus surface glycoproteins, including a receptor binding domain, cleavage site, fusion peptide, heptad repeats, and a transmembrane domain (Bosch et al., 2003). Our data clearly show that the mechanism driving cross-species transmission in this group 3 coronavirus was a double crossover event that replaced the spike gene, followed by adaptive evolution to create different serotypes. Thus, it appears that emergence of new coronaviruses and coronaviral diseases can occur through recombination as well as selection upon mutations (Holmes, 2009, Holmes and Rambaut, 2004). Preventing the emergence of new coronaviruses therefore requires that we identify and limit exposure to reservoirs of coronaviruses that can serve as a source of genetic material for emerging viruses. Elucidating the molecular events associated with cross-species transmission and pathogenicity shifts provides valuable insight into the mechanisms behind coronaviral evolutionary biology. With that knowledge, we will be able to develop better control measures that prevent vaccine failures, antiviral drug resistance, and the emergence of new coronaviral diseases in animals and humans.
Materials and methods
Viruses and specific serotyping antisera
Four isolates of TCoV (Table 1.) designated TCoV/VA-74/03, TCoV/TX-GL/01, TCoV/TX-1038/98, and TCoV/IN-517/94 (Lin et al., 2002) were obtained from Mr. Tom Hooper (Purdue University, West Lafayette, IN, USA). The viruses were propagated in 1-day-old specific pathogen-free (SPF) turkeys and in 18- to 22-day-old embryonating turkey eggs (a gift from Dr. M. J. Pantin-Jackwood, Southeast Poultry Research Laboratory, USDA, ARS, Athens, GA and from Dr. Y. M. Saif, FAHRP, OARDC, The Ohio State University, Wooster, OH). When viruses were propagated in 1-day-old turkeys, the birds were inoculated with 100 μl of virus per os. The birds were necropsied 48 h post-inoculation and the gastrointestinal (GI) tracts were harvested and frozen at – 80 C. When virus was propagated in embryonating turkey eggs, the chorioallantoic sac was inoculated with 100 μl of virus. The eggs were incubated for 48 h then the GI tracts of the embryos were extracted and frozen at – 80 C. The frozen GI tracts from embryos and poults were thawed, homogenized, clarified by low speed centrifugation (200×g for 10 min), and filtered through a 0.8-μm filter then a 0.22-μm filter (Millipore, Bedford, MA).
Serotyping antiserum was prepared against each of the TCoV isolates in SPF turkeys using previously published methods with a few modifications (Gelb and Jackwood, 2008). Briefly, 2-week-old SPF turkeys (Southeast Poultry Research Laboratory, Athens, GA) held in positive pressure Horsfal isolation units were given at least 1 × 103 50% poult infectious doses of virus per os. Two weeks post-exposure, the birds were injected intravenously with 1 × 103 50% poult infectious doses and serum was collected 2 weeks post intravenous injection. The serum was heat inactivated at 56 C for 30 min and stored at – 20 C.
Pathogenicity testing in turkeys and chickens
One-day-old turkeys were given 100 μl of TCoV per os then examined for clinical signs and lesions in the GI tract at 24, 48, 72, and 96 h after exposure. Each of the TCoV isolates TCoV/VA-74/03, TCoV/TX-GL/01, TCoV/IN-517/94, and TCoV/TX-1038/98 were given to 4 poults per sample time for each virus. In addition, the GI tracts for TCoV/TX-1038/98 were harvested at 24, 48, 72, and 96 h PI, pooled per sample time, and frozen at – 80 C. The GI tracts of all the other viruses were harvested at 48 h PI, pooled and frozen at − 80 C. The pooled GI tracts were processed for RNA extraction and real-time RT-PCR as described below.
Ten, 1-day-old SPF chickens (Charles River Laboratories, Inc. SPAFAS, North Franklin, CT) were each given 100 μl of either the TCoV/TX-1038/98 virus or the TCoV/TX-GL/01 virus per os and examined daily for clinical signs of disease. In addition, 3 non-inoculated contact exposed chickens were included to determine transmissibility. At 3 and 5 days PI, 3 exposed birds and 1 contact control bird and at 10 days PI, 4 exposed birds and 1 contact control bird for each virus were killed and examined for lesions in the GI tract. The GI tracts were harvested, pooled per virus/treatment/necropsy day, and frozen at – 80 C. The pooled GI tracts were thawed, homogenized, clarified by low speed centrifugation (200×g for 10 min), and filtered through a 0.8-μm filter then a 0.22-μm filter (Millipore) to prepare inoculums for further passage in chicks. Real-time RT-PCR was used to verify that virus was present in the inoculums before passing the virus in 10 additional infected and 3 contact exposed 1-day-old chickens. The virus was passed until it could no longer be recovered from the chickens. To verify the viruses were still pathogenic for poults, the last passage of the viruses reisolated from the chickens was given to ten 1-day-old turkeys. The birds were examined for clinical signs daily then necropsied at 5 days PI and examined for lesions.
Real-time RT-PCR-based serum neutralization test
The serum neutralization test was conducted using the constant-virus diluted-serum (beta) method as previously described (Gelb and Jackwood, 2008). Ten-fold serial dilutions of the viruses were titrated in 1-day-old turkeys by inoculating 5 poults per dilution with 100 μl of virus per os. The birds were necropsied 48 h post-inoculation and the gastrointestinal (GI) tracts were examined for lesions. One-hundred, 50% poult infectious doses of virus were mixed with an equal volume of serotyping antisera diluted 2-fold in PBS (pH 7.4). The mixtures were incubated at room temperature for 30 min then 200 μl was inoculated per os into 5 poults per dilution. Poults were held in isolation units for 48 h then necropsied and the GI tracts were examined for signs of TCoV infection, harvested, and processed as described above for the real-time RT-PCR serum neutralization test. Positive and negative control groups of 5 poults each were also maintained.
A real-time RT-PCR quantitative analysis on the processed GI tracts was conducted as previously described (Callison et al., 2006) and used to evaluate the neutralizing immune reactions (Bostic et al., 1999, Saito et al., 2003, van Santen et al., 2004). Briefly, RNA was extracted using the High Pure RNA Isolation Kit (Roche, Indianapolis, IN) and the real-time RT-PCR was conducted in a SmartCycler (Cepheid, Sunnyvale, CA). Forward primer (IBV5′GU391 5′-GCTTTTGAGCCTAGCGTT-3′), reverse primer (IBV5′GL533 5′GCCATGTTGTCACTGTCTATTG-3′), and Taqman® dual-labeled probe (IBV5′G probe 5′-FAM-CACCACCAGAACCTGTCACCTC-BHQ1-3′) were used to amplify and detect a 143-bp fragment at the 5′ end of the TCoV genome. A standard curve based on 10-fold serial dilutions of each TCoV isolate titrated in poults was developed and used to calculate the amount of TCoV genomes present in each GI tract from poults in the neutralization test. The neutralization assay data was tested using the null hypothesis and Student's t-test statistic as previously described (van Santen et al., 2004). Antibody titers were recorded as the highest serum dilution that neutralized the virus (negative real-time RT-PCR test) in at least 50% of the samples collected within a group as calculated by the Reed and Muench Test (Thayer and Beard, 2008). Antigenic relatedness values were calculated using the Archetti and Horsfall (1950) test.
Sequence analysis following passage in embryonating eggs
The genes encoding structural proteins of TCoV/TX-1038/98 virus passages 8 and 40 and TCoV/TX-GL/01 virus passages 11 and 30 in embryonating turkey eggs were sequenced to identify sequence changes. Amplicons from RT-PCR reactions using specific overlapping primer pairs listed in Table 8 were generated and purified as previously described (Lee, Hilt, and Jackwood, 2000) and sequenced directly at the Molecular Genetics Instrumentation Facility (University of Georgia, Athens, GA) with the Prism( DyeDeoxy terminator cycle sequencing kit according to the manufacturer's recommendations (Perkin-Elmer, Foster City, CA). The sequences were assembled using SeqMan Pro, and genome annotation was conducted with SeqBuilder (DNASTAR, Inc., v.8.0.2, Madison, WI). Nucleotide and deduced amino acid alignments were assembled using Clustal W in the MegAlign program (DNASTAR, Inc.).
Table 8.
Primer pairs used in RT-PCR reactions to sequence the 3′ end of TCoV isolates passaged in embryonating eggs.
| Primer pairs | Locationsa | Sequence |
|---|---|---|
| TCoV S1 5′ | 20,338–20,358 | 5′-AAGAGTGGCAAGTTGTTAGTT-3′ |
| TCoV S1 3′ | 22,017–22,037 | 5′-CCATAGCTGCAAATAGACCTA-3′ |
| TCoV 1511 5′ | 21,882–21,901 | 5′-CTGCATCTTGTCGGTTAGCC-3′ |
| TCoV 3809 3′ | 24,179–24,198 | 5′-TGCTCACCAGTTTCAATAAT-3′ |
| TCoV 3565 5′ | 23,936–23,956 | 5′-ACTTTTGATAATGATGTGGTG-3′ |
| TCoV 5538 3′ | 25,963–25,983 | 5′-GCTCTGCTTGTCCTGCTTTGT-3′ |
| TCoV 5046 5′ | 25,378–25,397 | 5′-AAGAGTATTTATTTTGAGGA-3′ |
| Oligo-dT 3′ | 27,621–27,638 | 5′-TTTTTTTTTTTTTTTTTT-3′ |
Primer pair nucleotide locations are based on the sequence for TCoV/TX-GL/01 (accession number GQ427174).
Whole genome nucleotide and deduced amino acid sequence analysis
Viral RNA was purified with the High Pure RNA extraction kit (Roche Diagnostics Corporation, Indianapolis, IN) and used directly in the amplification reaction.
The TaKaRa RNA LA PCR kit (Takara Bio Inc., Otsu, Shiga, Japan), which utilizes avian myeloblastosis virus (AMV) reverse transcriptase (RT), was used in a strand displacement amplification reaction to randomly amplify the viral genomic RNA. A random amplification primer (5′-AGCGGGGGTTGTCGAATGTTTGANNNNN-3′) and an amplification primer is (5′AGCGGGGGTTGTCGAATGTTTGA-3′) were used in the reaction.
A GeneAmp System 9600 thermocycler (Perkin-Elmer Applied Biosystems, Foster City, CA) was used for the RT-PCR reactions, which included an RNA denaturing step at 65 C for 1 min followed by 30 C for 10 min then 42 C for 60 min. The RT reaction was heat denatured at 99 C for 5 min then cooled to 5 C. The PCR amplification reaction was 30 cycles of 94 C for 30 s, 60 C for 30 s, and 72 C for 3 min.
The expected result is a smear of amplified DNAs that range in size from 500 to 1500 bp in length. The amplified products were gel purified, cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced.
Plasmid DNA from the libraries of cloned cDNA fragments for each virus was isolated using an alkaline lysis method modified for the 96-well format and incorporating both Hydra and Tomtek robots (http://www.intl-pag.org/11/abstracts/P2c_P116_XI.html). Cycle sequencing reactions were performed using the BigDye™ Terminator® Cycle Sequencing Kit Version 3.1 (Applied Biosystems, Foster City, CA) and MJ Research (Watertown, MA) thermocyclers. Finished reactions were filtered through Sephadex filter plates into Perkin-Elmer MicroAmp Optical 96-well plates. A 1/12-strength sequencing reaction on an ABI 3730 was used to sequence each clone from both the 5′ and 3′ ends. Each viral genome was shotgun sequenced to 10× coverage.
Chromatogram files and trace data were read and assembled using SeqMan Pro, and genome annotation was conducted with SeqBuilder (DNASTAR, Inc.). Whole genome alignments were generated and phylogenetic trees were constructed with the Neighbor-Joining method, Minimum Evolution method, Maximum Parsimony method, and UPGMA with 1000 bootstrap replicates (MEGA4, http://www.megasoftware.net/index.html) (Tamura et al., 2007). To reconstruct evolutionary trees, maximum likelihood (Tree-Puzzle, http://www.tree-puzzle.de/) and Bayesian analysis (BEAST 1.4, http://beast.bio.ed.ac.uk/) were conducted. A codon-based test of positive selection (Z-test, MEGA4) was used to analyze the numbers of non-synonymous and synonymous substitutions per site (dN/dS ratio). To determine how TCoV isolates are related to IBV (IBV/Mass41 strain), an analysis using the SimPlot software (http://sray.med.som.jhmi.edu/SCRoftware/simplot/) was conducted with TCoV/VA-74/03 as the query sequence and using a 400-bp window and a 100-bp step. Finally, BLASTN and BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) analyses using all and overlapping (100 nt and 33 amino acid) fragments of the recombined region were conducted using the GenBank or Swissprot databases.
GenBank accession numbers
Sequences generated in this study for TCoV were submitted to GenBank and assigned the following accession numbers: TCoV/VA-74/03 (GQ427173); TCoV/TX-GL/01 (GQ427174); TCoV/TX-1038/98 (GQ427176); TCoV/IN-517/94 (GQ427175); TCoV/TX-1038/98 Pass 40 (GQ469644); TCoV/TX-1038/98 Pass 49 (GU213199); TCoV/TX-1038/98 Pass 50 (GU213200); TCoV/TX-1038/98 Pass 52 (GU213201); TCoV/TX-R/98 Pass 24 (GU213202). Accession numbers for reference sequences used in this study are TCoV/MG10 (EU095850); TCoV/IN-540/94 (EU022525); TCoV/MN-ATCC (EU022526); TCoV/Canada-MG10 (EU095850); IBV/CK/Mass 41 (AY851295); IBV/CK/Arkp11 (EU418976); IBV/CK/Arkp101 (EU418975); IBV/CK/Beaudette (NC001451); IBV/CK/H120 (FJ888351); IBV/CK/CA99 (AY514485); IBV/CK/CH/LSD/051 (EU637854); IBV/CK/SAIBK (DQ288927); IBV/CK/BJ (AY319651); Partridge/GD/S14/03 (AY646283); IBV/CK/A2 (EU526388); beluga whale/CoV/SW1 (NC010646); ThrushCoV/HKU12 (FJ376621); MuniaCoV/HKU13 (FJ376622); Bulbul/CoV/HKU11 (FJ376619).
Acknowledgments
This research was supported by USDA, CSREES award number 2007-35600-17786.
References
- Archetti I., Horsfall F.L. Persistent antigentic variation of influenza A viruses after incomplete neutralization in vivo with heterologous immune serum. J. Exp. Med. 1950;92:441–462. doi: 10.1084/jem.92.5.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostic J.R., Brown K.E., Young N.S., Koenig S. Quantitative analysis of neutralizing immune responses to human parvovirus B19 using a novel reverse transcriptase-polymerase chain reaction-based assay. J. Infect. Dis. 1999;179:619–626. doi: 10.1086/314648. [DOI] [PubMed] [Google Scholar]
- Breslin J.J., Smith L.G., Fuller F.J., Guy J.S. Sequence analysis of the turkey coronavirus nucleocapsid protein gene and 3′ untranslated region identifies the virus as a close relative of infectious bronchitis virus. Virus Res. 1999;65:187–193. doi: 10.1016/S0168-1702(99)00117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callison S.A., Hilt D.A., Boynton T.O., Sample B.F., Robison R., Swayne D.E., Jackwood M.W. Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J. Virol. Methods. 2006;138:60–65. doi: 10.1016/j.jviromet.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wu C.C., Lin T.L. Complete nucleotide sequence of polyprotein gene 1 and genome organization of turkey coronavirus. Virus Res. 2008;136:43–49. doi: 10.1016/j.virusres.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Gelb J. Infectious Bronchitis. In: Saif Y.M., Fadly A.M., Glisson J.R., McDougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th ed. Blackwell Publishing; Ames, Iowa: 2008. pp. 117–135. [Google Scholar]
- Cavanagh D., Mauditt K., Sharma M., Drury S.E., Ainsworth H.L., Britton P., Gough R.E. Detection of coronavirus from turkey poults in Europe genetically related to infectious bronchitis virus of chickens. Avian Pathol. 2001;30:355–368. doi: 10.1080/03079450120066368. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Mawditt K., Welchman Dde B., Britton P., Gough R.E. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys. Avian Pathol. 2002;31:81–93. doi: 10.1080/03079450120106651. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Picault J.P., Gough R., Hess M., Mawditt K., Britton P. Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs. Avian Pathol. 2005;34:20–25. doi: 10.1080/03079450400025414. [DOI] [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L., Zhang S.Y., Peiris J.S., Smith G.J., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007;81:6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Dediego M.L., Alvarez E., Deming D., Sheahan T., Baric R. Vaccines to prevent severe acute respiratory syndrome coronavirus-induced disease. Virus Res. 2008;133:45–62. doi: 10.1016/j.virusres.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabricant J. The early history of infectious bronchitis. Avian Dis. 1998;42:648–650. [PubMed] [Google Scholar]
- Gelb J., Jackwood M.W. Infectious Bronchitis. In: Dufour-Zavala L., Swayne D.E., Glisson J.R., Pearson J.E., Reed W.M., Jackwood M.W., Woolcock P., editors. A Laboratory Manual for the Isolation, Identification, and Characterization of Avian Pathogens. 5th ed. American Association of Avian Pathologists; Kennett Square, PA: 2008. pp. 146–149. [Google Scholar]
- Gomaa M.H., Barta J.R., Ojkic D., Yoo D. Complete genomic sequence of turkey coronavirus. Virus Res. 2008;135:237–246. doi: 10.1016/j.virusres.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Snijder E.J., Spaan W.J. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78:7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J.S. Turkey coronavirus is more closely related to avian infectious bronchitis virus than to mammalian coronaviruses: a review. Avian Pathol. 2000;29:207–212. doi: 10.1080/03079450050045459. [DOI] [PubMed] [Google Scholar]
- Guy J.S. Turkey coronavirus enteritis. In: Saif Y.M., Fadly A.M., Glisson J.R., McDougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th ed. Blackwell Publishing; Ames, Iowa: 2008. pp. 330–338. [Google Scholar]
- Hasoksuz M., Alekseev K., Vlasova A., Zhang X., Spiro D., Halpin R., Wang S., Ghedin E., Saif L.J. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J. Virol. 2007;81:4981–4990. doi: 10.1128/JVI.02361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C. The evolution and emergence of RNA viruses. In: Harvey P.H., May R.M., editors. Oxford Series in Ecology and Evolution. Oxford University Press Inc.; New York: 2009. [Google Scholar]
- Holmes E.C., Rambaut A. Viral evolution and the emergence of SARS coronavirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:1059–1065. doi: 10.1098/rstb.2004.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V., Dveksler G., Gagneten S., Yeager C., Lin S.H., Beauchemin N., Look A.T., Ashmun R., Dieffenbach C. Coronavirus receptor specificity. Adv. Exp. Med. Biol. 1993;342:261–266. doi: 10.1007/978-1-4615-2996-5_40. [DOI] [PubMed] [Google Scholar]
- Hon C.C., Lam T.Y., Shi Z.L., Drummond A.J., Yip C.W., Zeng F., Lam P.Y., Leung F.C. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L.A., Savage C., Naylor C., Bennett M., Chantrey J., Jones R. Genetically diverse coronaviruses in wild bird populations of northern England. Emerg. Infect. Dis. 2009;15:1091–1094. doi: 10.3201/eid1507.090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hilt D.A., Brown T.P. Attenuation, safety, and efficacy of an infectious bronchitis virus GA98 serotype vaccine. Avian Dis. 2003;47:627–632. doi: 10.1637/6094. [DOI] [PubMed] [Google Scholar]
- Jonassen C.M., Kofstad T., Larsen I.L., Lovland A., Handeland K., Follestad A., Lillehaug A. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos) J. Gen. Virol. 2005;86:1597–1607. doi: 10.1099/vir.0.80927-0. [DOI] [PubMed] [Google Scholar]
- Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Cui B., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Qi X., Chen K., Du L., Gao K., Zhao Y.T., Zou X.Z., Feng Y.J., Gao Y.F., Hai R., Yu D., Guan Y., Xu J. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Hilt D.A., Jackwood M.W. Redesign of primer and application of the reverse transcriptase-polymerase chain reaction and restriction fragment length polymorphism test to the DE072 strain of infectious bronchitis virus. Avian Dis. 2000;44:650–654. [PubMed] [Google Scholar]
- Lee C.W., Jackwood M.W. Origin and evolution of Georgia 98 (GA98), a new serotype of avian infectious bronchitis virus. Virus Res. 2001;80:33–39. doi: 10.1016/s0168-1702(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lin T.L., Loa C.C., Wu C.C. Complete sequences of 3′ end coding region for structural protein genes of turkey coronavirus. Virus Res. 2004;106:61–70. doi: 10.1016/j.virusres.2004.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.L., Loa C.C., Wu C.C., Bryan T., Hooper T., Schrader D. Antigenic relationship of turkey coronavirus isolates from different geographic locations in the United States. Avian Dis. 2002;46:466–472. doi: 10.1637/0005-2086(2002)046[0466:AROTCI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Liu S., Chen J., Chen J., Kong X., Shao Y., Han Z., Feng L., Cai X., Gu S., Liu M. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas) J. Gen. Virol. 2005;86:719–725. doi: 10.1099/vir.0.80546-0. [DOI] [PubMed] [Google Scholar]
- McKinley E.T., Hilt D.A., Jackwood M.W. Avian coronavirus infectious bronchitis attenuated live vaccines undergo selection of subpopulations and mutations following vaccination. Vaccine. 2008;26:1274–1284. doi: 10.1016/j.vaccine.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihindukulasuriya K.A., Wu G., St Leger J., Nordhausen R.W., Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 2008;82:5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muradrasoli S., Mohamed N., Hornyak A., Fohlman J., Olsen B., Belak S., Blomberg J. Broadly targeted multiprobe QPCR for detection of coronaviruses: coronavirus is common among mallard ducks (Anas platyrhynchos) J. Virol. Methods. 2009;159:277–287. doi: 10.1016/j.jviromet.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rest J.S., Mindell D.P. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 2003;3:219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Munakata Y., Fu Y., Fujii H., Kodera T., Miyagawa E., Ishii K., Sasaki T. Evaluation of anti-parvovirus B19 activity in sera by assay using quantitative polymerase chain reaction. J. Virol. Methods. 2003;107:81–87. doi: 10.1016/s0166-0934(02)00195-7. [DOI] [PubMed] [Google Scholar]
- Shi P., Yu L., Fu Y.X., Huang J.F., Zhang K.Q., Zhang Y.P. Evolutionary implications of avian infectious bronchitis virus (AIBV) analysis. Cell Res. 2006;16:323–327. doi: 10.1038/sj.cr.7310041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephensen C.B., Casebolt D.B., Gangopadhyay N.N. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 1999;60:181–189. doi: 10.1016/S0168-1702(99)00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tang X., Li G., Vasilakis N., Zhang Y., Shi Z., Zhong Y., Wang L.F., Zhang S. Differential stepwise evolution of SARS coronavirus functional proteins in different host species. BMC Evol. Biol. 2009;9:52. doi: 10.1186/1471-2148-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Hofmann-Lehmann R., Stallkamp I., Thiel V., Thiel H.J. Genome organization and reverse genetic analysis of a type I feline coronavirus. J. Virol. 2008;82:1851–1859. doi: 10.1128/JVI.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer S.G., Beard C.W. Serologic procedures. In: Dufour-Zavala L., Swayne D.E., Glisson J.R., Pearson J.E., Reed W.M., Jackwood M.W., Woolcock P., editors. A Laboratory Manual for the Isolation, Identification, and Characterization of Avian Pathogens. 5th ed. American Association of Avian Pathologists; Kennett Square, PA: 2008. pp. 222–229. [Google Scholar]
- van Santen V.L., Kaltenboeck B., Joiner K.S., Macklin K.S., Norton R.A. Real-time quantitative PCR-based serum neutralization test for detection and titration of neutralizing antibodies to chicken anemia virus. J. Virol. Methods. 2004;115:123–135. doi: 10.1016/j.jviromet.2003.09.022. [DOI] [PubMed] [Google Scholar]
- Velayudhan B.T., Shin H.J., Lopes V.C., Hooper T., Halvorson D.A., Nagaraja K.V. A reverse transcriptase-polymerase chain reaction assay for the diagnosis of turkey coronavirus infection. J. Vet. Diagn. Invest. 2003;15:592–596. doi: 10.1177/104063870301500616. [DOI] [PubMed] [Google Scholar]
- Vijaykrishna D., Smith G.J., Zhang J.X., Peiris J.S., Chen H., Guan Y. Evolutionary insights into the ecology of coronaviruses. J. Virol. 2007;81:4012–4020. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lai K.K., Huang Y., Lee P., Luk G.S., Dyrting K.C., Chan K.H., Yuen K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Li K.S., Poon R.W., Wong B.H., Tsoi H.W., Yip B.C., Huang Y., Chan K.H., Yuen K.Y. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.Y., Wei J.F., He S.H. Adaptive evolution of the spike gene of SARS coronavirus: changes in positively selected sites in different epidemic groups. BMC Microbiol. 2006;6:88. doi: 10.1186/1471-2180-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.W., Yap Y.L., Danchin A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch. Virol. 2005;150:1–20. doi: 10.1007/s00705-004-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]