Abstract
Bone marrow stromal antigen 2 (BST2), an interferon-inducible antiviral factor, has been shown to block the release of various enveloped viruses from cells. It has also been identified as an innate immune system component. Most enveloped viruses subject to BST2 restriction bud at the plasma membrane. Here we report our findings that (a) the production of human coronavirus 229E (HCoV-229E) progeny viruses, whose budding occurs at the ER-Golgi intermediate compartment (ERGIC), markedly decreases in the presence of BST2; and (b) BST2 knockdown expression results in enhanced HCoV-229E virion production. Electron microscopy analyses indicate that HCoV-229E virions are tethered to cell surfaces or intracellular membranes by BST2. Our results suggest that BST2 exerts a broad blocking effect against enveloped virus release, regardless of whether budding occurs at the plasma membrane or intracellular compartments.
Keywords: BST2, CD317, Tetherin, Human coronavirus 229E, Virus budding
Highlights
-
•
BST2 knockdown expression results in enhanced HCoV-229E egress.
-
•
HCoV-229E virions are tethered to cell surfaces or intracellular membranes by BST2.
-
•
HCoV-229E infection at high MOI can significantly downregulate HeLa BST2 and rescue HIV-1 egress.
Introduction
Coronaviruses are found in a wide variety of host species, including bats, mice, cats, dogs, pigs, horses, birds, whales and humans (Perlman and Netland, 2009). Infected hosts may develop respiratory, enteric, hepatic or neurological diseases with different degrees of severity. Prior to the 2002–2003 severe acute respiratory syndrome (SARS) outbreak triggered by a previously unknown coronavirus (Drosten et al., 2003, Marra et al., 2003, Rota et al., 2003), human coronaviruses (HCoV) 229E and OC43 were recognized as the two primary coronaviruses causing upper respiratory tract infections associated with severe pulmonary diseases in elderly, newborn, and immunocompromised individuals (Garbino et al., 2006). Results from one study involving a large collection of human nasopharyngeal specimens suggest that HCoV-229E, -NL63, -OC43 and -HKU1 are the only coronaviruses circulating in human populations (Zlateva et al., 2013). However, more recent studies of fatal human infections by a novel SARS-like coronavirus named MERS-CoV have been reported (Zaki et al., 2012). This is a reminder that novel coronaviruses can emerge and cause life-threatening infectious diseases, although most human coronavirus infections result in mild symptoms such as those associated with the common cold.
Most virus infections induce a type 1 interferon (IFN-α⧸β) response that in turn triggers the expression of numerous diverse genes (Sadler and Williams, 2008). IFN-inducible or IFN-stimulated gene (ISGs) expression is considered an important innate defense system against virus infections (Randall and Goodbourn, 2008). Among IFN-inducible expression factors, BST2 (also known as CD317, HM1.24, or tetherin) has been characterized as a host restriction factor capable of impeding the release of multiple viruses, including retroviruses (Gupta et al., 2009, Jia et al., 2009, Le Tortorec and Neil, 2009, Neil et al., 2008, Van Damme et al., 2008, Zhang et al., 2009), filoviruses (Jouvenet et al., 2009, Kaletsky et al., 2009, Sakuma et al., 2009), herpesviruses (Mansouri et al., 2009), arenaviruses (Radoshitzky et al., 2010), and influenza viruses, including virus-like particles (Watanabe et al., 2011, Yondola et al., 2011) and wild-type virions (Mangeat et al., 2012), and the Sendai virus (Bampi et al., 2013).
BST2 is a 30–36 kDa type II transmembrane protein consisting of a cytoplasmic amino-terminal region followed by a transmembrane domain, a coiled-coil extracellular domain, and a carboxy-terminal glycosyl-phosphatidylinositol (GPI) anchor (Kupzig et al., 2003). The protein is known to associate with lipid rafts on cell surfaces and at the internal membrane, likely with the trans-Golgi network (TGN) (Kupzig et al., 2003). One model suggests that BST2 inhibits virion release by tethering nascent virions to cell surfaces via its amino-terminal transmembrane domain and carboxy-terminal GPI anchor (Neil et al., 2008, Perez-Caballero et al., 2009). Treatment with a protease such as subtilisin can support virion release, presumably via proteolytic BST2 cleavages linking budding virions to cell surfaces (Neil et al., 2006).
Most viruses restricted by BST2 in this manner share two common features: they possess envelope proteins and bud from the plasma membrane. Coronavirus assembly and budding occurs at the ER-Golgi intermediate compartment (ERGIC) (Hunter, 2001). Vesicles containing accumulated coronavirus particles are transported to the plasma membrane, where they release virions exocytotically (Masters, 2006). The goal of our research was to determine whether BST2 also affects the release of coronaviruses whose budding processes differ from those of most other enveloped viruses. We selected HCoV-229E, a common human coronavirus, as a representative strain. Our results suggest that BST2 is capable of counteracting HCoV-229E infection via the tethering of nascent progeny virions at the cell surface. Our data also support the idea that BST2 is capable of restricting the virus particle release of different enveloped virus families.
Results
BST2 reduces HCoV-229E virion yields
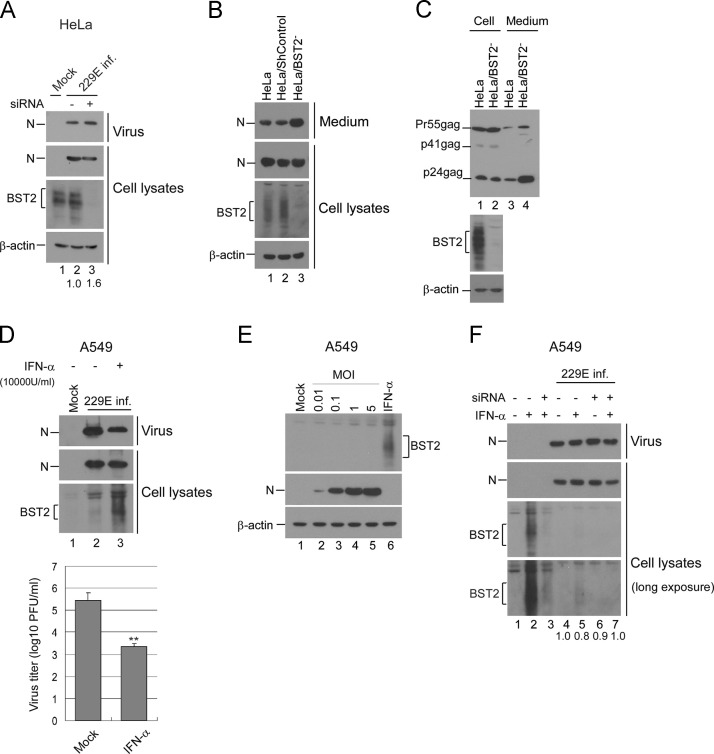
To determine whether BST2 is also capable of inhibiting human coronavirus egress, we examined the supernatants of HCoV-229E-infected HeLa cells following transient BST2 knockdown. As shown in Fig. 1A, BST2-knockdown HeLa cells yielded higher quantities of virions compared to control HeLa cells. We then established a stable BST2 knockdown HeLa cell line (HeLa/BST2-), and observed that it exerted an effect similar to that of the transient BST2 knockdown in terms of promoting HCoV-229E production (Fig. 1B). When tested with a Vpu-deleted HIV-1 clone (known to be restricted by BST2 in the absence of the Vpu viral protein), the BST2 knockdown resulted in the release of a larger quantity of virus particles (Fig. 1C); this is consistent with previous reports (Goffinet et al., 2010, Neil et al., 2008).
Fig. 1.
BST2 inhibits virion production. (A) HeLa cells were transfected with BST2-specific siRNA (5 μg) three times at 54 h, 30 h and 6 h prior to infection with HCoV-229E at 0.1 MOI for 1 h at 37 °C. At 12 h post-infection, cell lysates (bottom) and supernatants (upper) were harvested and subjected to western immunoblotting. Viral proteins were detected with an anti-HCoV229E nucleocapsid (N) antiserum. N proteins from medium or cell samples were quantified by scanning N band density from immunoblots. Ratios of N levels in medium to those in cells were determined and compared with the ratio of mock-infected samples. (B) HeLa cells transduced with shRNA targeting the BST2 gene (HeLa/BST2-) or a non-targeting shRNA (shControl) were infected with HCoV-229E at 0.1 MOI. At 20 h post-infection, cell lysates (bottom) and supernatants (upper) were harvested and subjected to western immunoblotting. (C) HeLa or HeLa/BST2-cells were transfected with the HIV-1 virus-producing vector NL4.3delVpu. At 48 h post-transfection, cells and supernatants were collected, prepared, and analyzed by western immunoblotting. Viral proteins were detected with an anti-HIV-1 p24CA monoclonal antibody. (D) A549 cells were infected with HCoV-229E at 0.1 MOI for 1 h at 37 °C. Cells were treated twice with the indicated concentrations of IFNα at 1 h and 24 h post-infection. At 36 h post-infection, cell lysates (bottom) and supernatants (upper) were harvested and subjected to western immunoblotting. Virus titers were determined by plaque assays of A549 cells. ⁎⁎, p≤0.01. (E) A549 cells were infected with HCoV-229E at the indicated MOIs (lanes 2–5) or treated with IFNα (10,000 U/ml). At 24 h post-infection, cells were collected and analyzed by western immunoblotting. (F) A549 cells were transfected with BST2-specific siRNA twice (at 1 h and 24 h) following infection with HCoV-229E at 5.0 MOI for 1 h at 37 °C. Cells were treated twice with the indicated concentrations of IFNα at 4 h and 28 h post-infection. At 48 h post-infection, cell lysates and supernatants were harvested and subjected to Western immunoblotting. Viral N proteins from medium or cell lysate samples were quantified by scanning N band densities from immunoblots. The ratios of N levels in medium to those in cells were determined and normalized to the group without siRNA transfection and IFN treatment (lane 4). The lower blot was derived from the upper blot by extending the exposure.
Since IFNα is capable of enhancing BST2 expression, we tested the impact of IFNα treatment on HCoV-229E replication. HCoV-229E-infected A549 cells were treated with IFNα. At 24 h post-treatment, culture supernatants were collected and subjected to plaque assays and Western immunoblotting. As expected, the IFNα treatment enhanced BST2 expression (Fig. 1D, lane 3), which is associated with a decrease in infectious progeny virus production (lane 3 vs. lane 2).
To find out if BST2 expression is induced by infections, we inoculated A549 cells with HCoV-229E at different MOIs. Results indicate barely detectable BST2 following HCoV-229E infection (Fig. 1E). To determine if the IFNα-induced decrease in HCoV-229E egress was due to BST2-triggered restrictions, we transiently transfected HCoV-229E-infected A549 cells with BST2-specific siRNA following IFNα treatment. Results indicate that BST2 knockdown is capable of elevating HCoV-229E levels in supernatants when normalized to cell-associated N levels (Fig. 1F, lane 7 vs. lane 5). Similar results from independent experiments were observed. BST2 expression in A549 cells was barely induced by IFNα following HCoV-229E infection (Fig. 1F, lane 5 vs. lane 2), suggesting that HCoV-229E counteracts the capability of IFNα to induce BST2 expression or expresses certain factors against BST2. IFNα does not significantly enhance BST2 expression in HCoV-229E-infected cells to restrict virus egress—in other words, the effect of BST2 knockdown on virus egress enhancement is marginal. According to these data, it is difficult to conclude whether HCoV-229E egress is significantly restricted by IFNα-induced BST2. Our observation of a noticeable reduction in virus egress following IFNα treatment (Fig. 1D) could be due, at least in part, to the time of sample analysis (see “Discussion” section).
BST2 knockdown exerts no significant effects on HCoV-229E entry
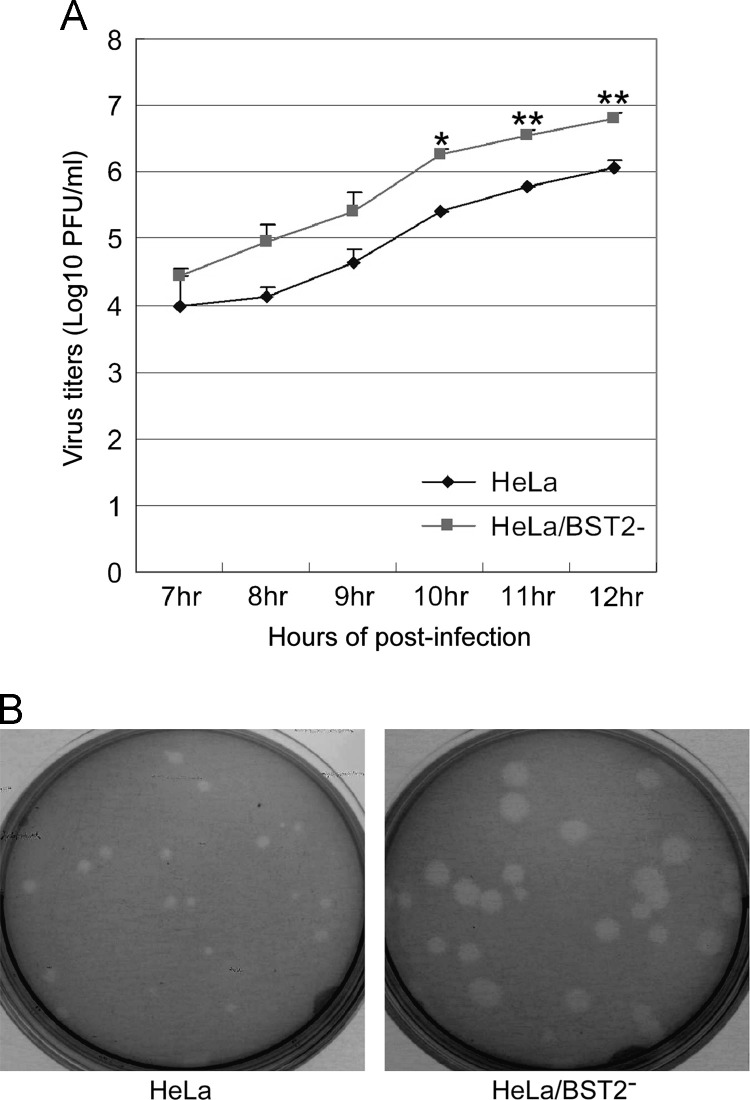
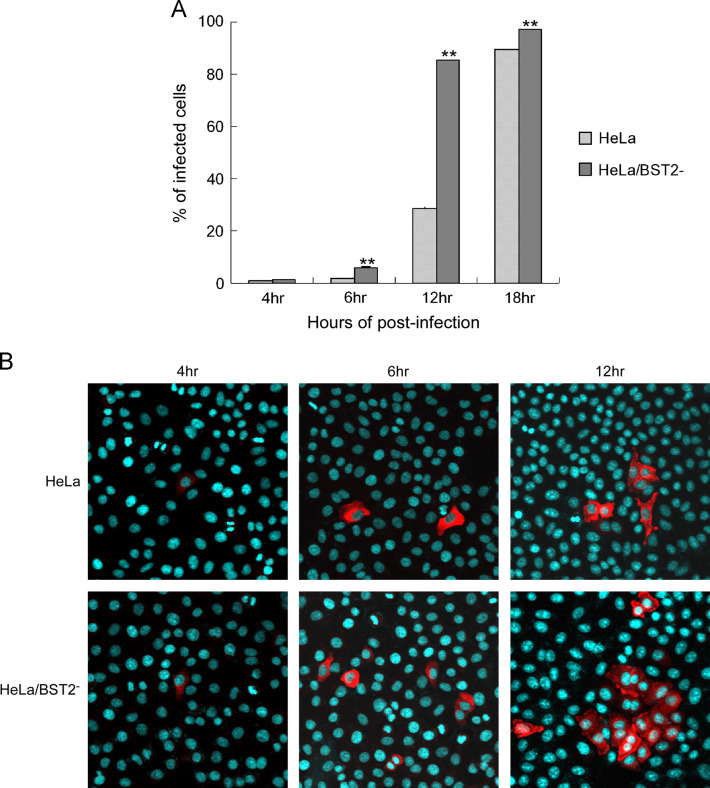
To further determine whether BST2 affects HCoV-229E replication, we assayed HCoV-229E growth kinetics in BST2 knockdown HeLa cells (HeLa/BST2-). HeLa cells with or without BST2 knockdown were inoculated with HCoV-229E at 0.1 MOI. Virus titers at various times post-infection were determined by A549 cell plaque assays. At 10 h post-infection and later, HeLa cells produced viral titers at levels significantly lower than HeLa/BST2- ( Fig. 2A). Between 7 and 9 h post-inoculation, HeLa/BST2- cells produced higher virus titers, but not at a statistically significant level. Plaque morphologies of A549 cells infected with virions released from HeLa/BST2- or HeLa cells were similar (data not shown). However, we repeatedly observed that the sizes of viral plaques on HeLa/BST2‐ cells were larger than those on HeLa cells following HCoV-229E infection, despite the lack of major differences in plaque numbers between the two cell types (Fig. 2B). Larger plaque sizes observed on HeLa/BST2‐ cells were likely the result of the spreading of nascent progeny virions to adjacent cells rather than increased susceptibility to HCoV-229E infection. To verify the lack of major impacts of BST2 knockdown on HCoV-229E susceptibility or entry, we examined viral protein expression in both HeLa/BST2- and HeLa cells at various times post-HCoV-229E infection. As shown in Fig. 3A, HeLa/BST2- and HeLa cells had approximately equal numbers of fluorescent-positive viral nucleocapsid protein cells at 4 h post-infection. However, at 6 h post-infection and later there was a much larger number of fluorescent-positive HeLa/BST2- cells compared to HeLa cells—an increase likely due to the spread of nascent progeny virions. These results were confirmed by immunofluorescence microscopy (Fig. 3B). Combined, the data suggest that BST2 knockdown does not significantly affect HeLa cell susceptibility to HCoV-229E, but markedly affects progeny virus yields.
Fig. 2.
BST2 restricts HCoV-229E replication. (A) HCoV-229E growth kinetics in HeLa and HeLa/BST2− cells. HeLa cells with or without BST2 knockdown were infected with HCoV-229E at 3 MOI. At 7 to 12 h post-infection, serially diluted supernatants were used to infect A549 cells, and viral titers were determined by standard plaque assays (⁎,p≤0.05; ⁎⁎, p≤0.01). (B) Plaque morphologies of HCoV-229E. HeLa, and HeLa/BST2- cells grown in dish plates were inoculated with HCoV-229E at 0.1 MOI for 1 h. Culture dishes were then washed twice with PBS and gently overlaid with 2–4 ml of 0.5% agarose. After five days of incubation, plaques were visualized by staining with 0.03% neutral red.
Fig. 3.
BST2 knockdown has no significant effects on HCoV-229E entry. (A) HeLa and HeLa/BST2− cells were inoculated with HCoV-229E at 0.1 MOI for 1 h at 37 °C. At each indicated time, cells were fixed and expressed viral nucleocapsid (N) proteins were detected with a rabbit anti-N antibody and a secondary FITC-conjugated anti-rabbit antibody. Cells were then analyzed by flow cytometry. Percentages of infected cells were determined by dividing the number of FITC-positive cells by the number of total cells and multiplying by 100. (B) HeLa and HeLa/BST2‐ cells were infected with HCoV-229E at either 0.1 or 2 MOI. At 4, 6, or 12 h post-infection, cells were fixed, probed with a primary anti-229E N antibody and a secondary rhodamine-conjugated anti-rabbit antibody, and observed using a laser confocal microscope. Nuclei were stained with DAPI (blue). Mock-infected cells or cells not exposed to the primary antibody yielded no signal (data not shown).
HCoV-229E virus particles are tethered at cell surfaces
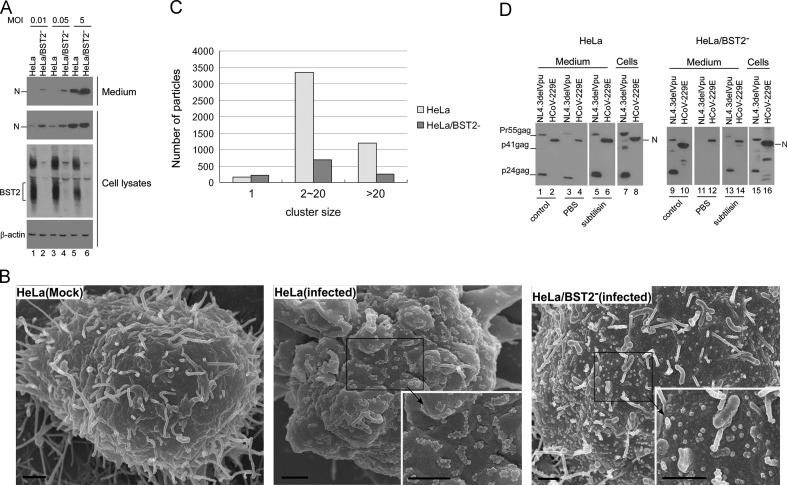
The above findings reflect low-MOI infections; we also ran tests to determine if the same findings occurred under high-MOI conditions. As shown in Fig. 4A, HeLa/BST2‐ cells produced larger quantities of progeny virions compared to HeLa cells regardless of MOI. The finding that HeLa cells produced a relatively low level of N was likely due to a portion of those cells not being infected, in part because of decreased virus spreading in the presence of BST2 (Fig. 3). Another possibility is that viral proteins were not fully expressed at 12 h post-infection. To determine whether BST2 restriction of HCoV-229E occurs in the same manner as in other enveloped viruses (i.e., whether virus particles are tethered at the cell surface), we fixed HeLa and HeLa/BST2‐ cells and observed them with a scanning electron microscope at 12 h post-infection. As shown in Fig. 4B and C, numerous clusters consisting of between 2 and 20 virus particles were found on HeLa cell surfaces. In contrast, most of the virus particles found on HeLa/BST2‐ cell surfaces were singular and scattered. When treated with the protease subtilisin (which triggers the release of cell surface-associated HIV-1 virions by cleaving BST2 (Neil et al., 2007)), much larger quantities of HCoV-229E or HIV-1 (control) virions were recovered in supernatants (Fig. 4D, lanes 5–6 vs. lanes 3–4). No significant increases in virion quantity were found in HeLa/BST2- culture supernatant following subtilisin treatment (Fig. 4D, lane 12 vs. lane 14). We also observed that larger amounts of HCoV-229E virions were recovered from HeLa culture medium than from HeLa/BST2- cell culture medium following subtilisin treatment, suggesting that the HeLa cells had larger quantities of virions trapped at the cell surface (Fig. 4D, lane 6 vs. lane 14). This result is compatible with SEM analysis data showing much smaller quantities of HCoV-229E virions found on the surfaces of HeLa/BST2‐ cells (Fig. 4B); it also provides support for the proposal that HCoV-229E is readily released from cells in the absence of BST2. Combined, these findings support the assumption that BST2 strongly inhibits HCoV-229E progeny virion production by physically tethering budding virions to cell surfaces.
Fig. 4.
BST2 restricts virion release by retaining virus particles on cell surfaces. (A) BST2 restricts HCoV-229E progeny virion release at different MOIs. HeLa and HeLa/BST2− cells were infected with HCoV-229E at 0.01, 0.05 or 5 MOI. At 12 h post-infection, cell lysates (bottom) and supernatants (upper) were harvested and subjected to western immunoblotting. HCoV-229E N was detected with anti-HCoV-229E N antiserum, and BST2 proteins were probed with rabbit anti-BST2 antiserum. (B) Scanning electron microscopy (SEM) analyses of HCoV-229E-infected cells. HeLa or HeLa/BST2− cells were infected with HCoV-229E at 0.1 MOI. At 12 h post-infection, cells were prepared and observed under a scanning electron microscope. The high-power view in the inset shows cell surface-associated virions. Bars, 1 μM. (C) Classification of virus particles tethered to cell surfaces. Cell surface-associated virus particles were counted in randomly selected 50 view-fields of SEM images. Virus particles were classified as 1, 2–20, or >20 virus particles per cluster. (D) Cell surface-associated virions become detached following subtilisin treatment. HeLa and HeLa/BST2− cells were transfected with NL4.3delVpu or infected with HCoV-229E at 0.1 MOI. At 24 h post-infection or post-transfection, supernatants were collected and cells were split equally into two dish plates. At 4 h, culture medium was removed, washed twice with PBS, and incubated with PBS containing subtilisin (1 mg/ml) for 10 min at 37 °C. Supernatants were then harvested and pelleted through 20% sucrose cushions. Pellets and cell lysates were subjected to western immunoblot analyses. HIV-1 was detected with an anti-HIV-1 p24CA antibody, and HCoV-229E was determined with an antibody against the HCoV-229E nucleocapsid (N).
HCoV-229E virions associate with BST2
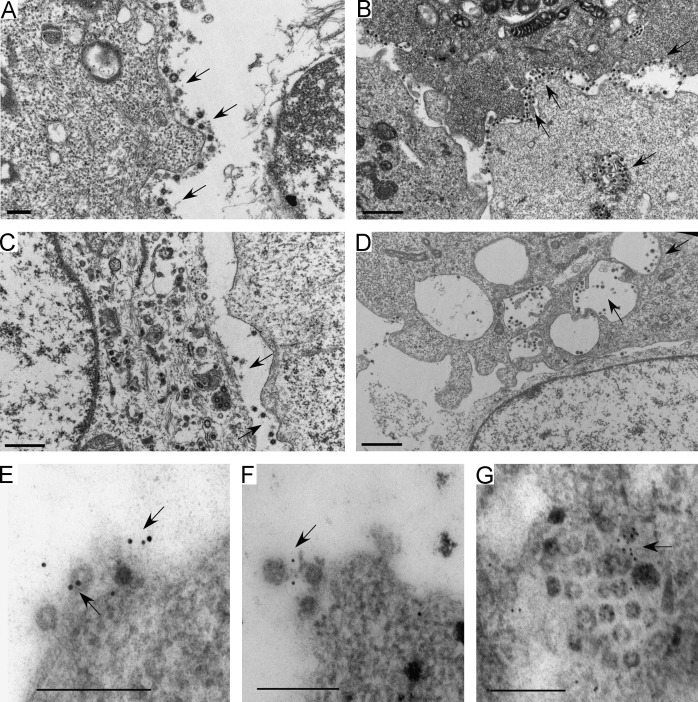
Since coronavirus particle budding occurred at ER/Golgi compartments, it is likely that the budding virions were tethered to vesicle membranes by BST2, and therefore could not be released from cell surfaces post-exocytosis at the plasma membrane. To test this idea, we performed transmission electron microscopy analyses of HCoV-229E-infected cells. Our results indicate much larger quantities of virions on HeLa cell surfaces compared to HeLa/BST2‐ cell surfaces ( Fig. 5A vs. Fig. 5C), which agrees with our SEM data (Fig. 4B). In addition, we consistently observed that virions in HeLa intracellular vesicles tended to cluster at membrane surfaces (Fig. 5B), whereas virions in HeLa/BST2‐ intracellular vesicles were distinctly separate from each other and/or not closely associated with cell membranes (Fig. 5D). Results from the immunogold labeling of BST2 indicate the presence of BST2 molecules between virions and/or between virions and cell membranes (Fig. 5E–G, arrows), giving further support to the proposal that BST2 tethers virions to cell membranes, and suggesting that HCoV-229E virions can be linked via BST2.
Fig. 5.
BST2 associates with HCoV-229E virions. HeLa (panels A, B and E–G) or HeLa/BST2- cells (panels C and D) were infected with HCoV-229E at 0.1 MOI. At 12 h post-infection, cells were fixed, prepared, and viewed with a transmission electron microscope. BST2 was probed with rabbit anti-BST2 antiserum, followed by a secondary 10 nm gold conjugated anti-rabbit antibody (panels E–G). Immunogold particles were barely detectable in HeLa/BST2- cells and in HeLa cells not exposed to the primary antibody (data not shown). Arrows indicate virions found on cell surfaces or in vesicles. Bars, 200 nm.
HCoV-229E infection downregulates BST2 expression level and rescues HIV-1 egress
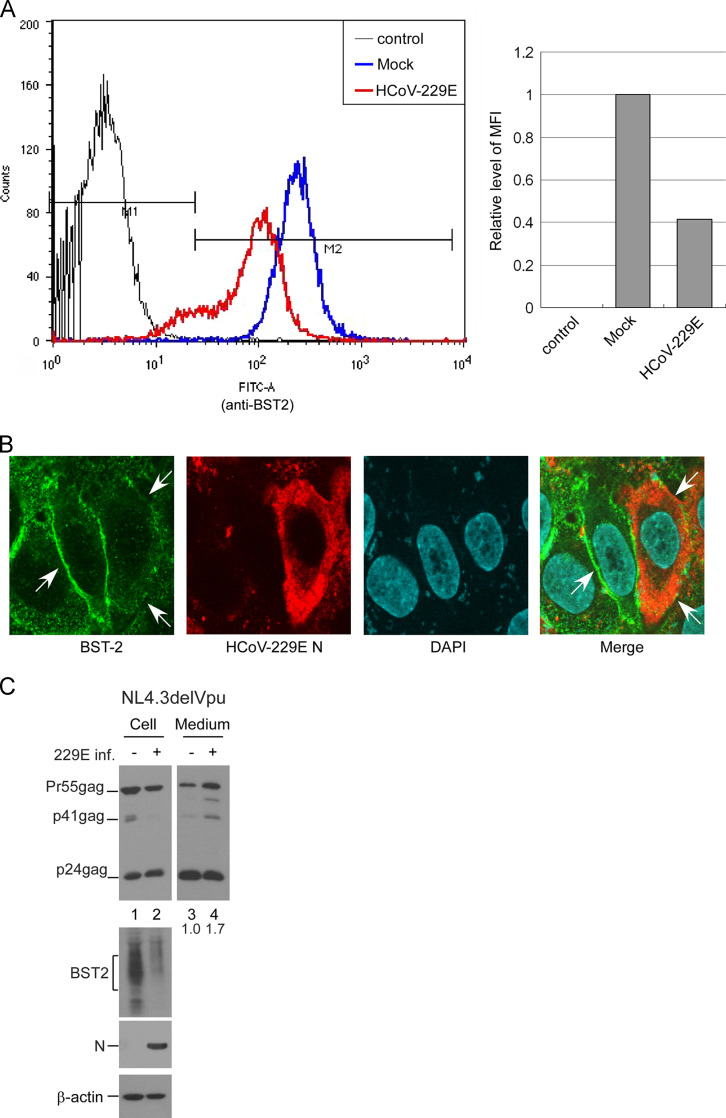
Since viruses such as HIV-1 (Habermann et al., 2010) and influenza (Mangeat et al., 2012) are capable of downregulating BST2 expression levels on cell surfaces, we looked at whether HCoV-229E infection has a similar capability. Flow cytometry results indicate that cell surface BST2 levels were significantly lower following HCoV-229E infection at 5 MOI ( Fig. 6A). Further, confocal microscopy observations show that BST2 signal on the plasma membranes of HCoV-229E-infected cells was noticeably lower compared to uninfected cells (Fig. 6B). Consistent with the results shown in Fig. 4A, flow cytometry and immunofluorescence analyses indicate that HCoV-229E infection at a lower MOI did not result in significant change in overall or surface BST2 expression levels in HeLa cells (data not shown).
Fig. 6.
HCoV-229E infection downregulates BST2 expression levels on cell surfaces and enhances HIV-1delVpu egress. (A) HeLa cells were mock-infected or infected with HCoV-229E at 5 MOI. At 18 h post-infection, cells were fixed and probed with a rabbit anti-BST2 antibody prior to the permeabilization of cell membranes, followed by a secondary FITC-conjugated anti-rabbit antibody. After the permeabilization of cell membranes, HCoV-229E N proteins were probed with a primary anti-HCoV-229E N antibody and a secondary phycoerythrin-conjugated goat anti-rabbit antibody. Cells were then analyzed by flow cytometry. Median fluorescence intensity (MFI) values of HCoV-229E-infected samples were compared to those of mock-infected samples. (B) HeLa cells were infected with HCoV-229E at 0.1 MOI. At 18 h post-infection, cells were fixed and permeabilized. BST2 proteins were detected with a mouse anti-BST2 antibody followed by a secondary FITC-conjugated anti-mouse antibody. HCoV-229E N proteins were probed with a primary anti-HCoV-229E N antibody and a secondary rhodamine-conjugated goat anti-rabbit antibody. BST2 signal (arrows) decreased on the surfaces of HCoV-229E-infected (red) cells. (C) HeLa cells were transfected with NL4.3delVpu. At 4 h post-transfection, cells were replated onto two dish plates and either mock-infected or infected with HCoV-229E at 5 MOI. At 24 h post-infection, cells and culture supernatants were collected and subjected to western immunoblot analysis. Ratios of total Gag protein level in the medium versus cell were calculated by scanning p24gag-associated band densities from immunoblots. The ratio of infected group was normalized to that of mock-infected group.
Given the association between efficient HIV-1 egress and BST2 downregulation, it is possible that HCoV-229E is capable of enhancing HIV-1 NL4.3delVpu release from HeLa cells by counteracting BST2 anti-viral activity. To test this possibility, we transfected HeLa cells with a NL4.3delVpu expression vector prior to HCoV-229E infection. We observed a significant reduction in BST2 expression level tied to a 1.7-fold increase in NL4.3delVpu virus particle production (Fig. 6C). Combined, these results suggest that HCoV-229E infection leads to BST2 downregulation in HeLa cells, thus contributing to greater Vpu-defective HIV-1 egress.
Discussion
Our main finding is that BST expression results in a significant decrease in progeny virus production. This is in agreement with the notion that BST2 is a major component of innate immunity against enveloped virus replicative infections. Both transient and stable expression of BST2 knockdown resulted in the marked enhancement of HCoV-229E progeny virus titer quantities. Electron microscope observations indicate an association with virus particle clusters at cell surfaces in the presence of BST2 (Fig. 4, Fig. 5). Immunogold labeling results indicate that BST2 molecules enhanced the linking of virions to each other or to cell membranes (Fig. 5), suggesting that HCoV-229E virions are restricted to membranes by BST2 during budding in ER/Golgi compartments. The primary site of BST2 antiviral activity is thought to be the cell surface, where many viruses acquire their envelopes by directly budding from the plasma membrane (Le Tortorec et al., 2011, Swiecki et al., 2013). However, some enveloped viruses such as herpesviruses (which complete their final envelopment by obtaining membranes from TGN and/or endosomal compartments and egress via exocytosis), are also subject to BST2 restrictions Mansouri et al., 2009; Mettenleiter, 2002). Results from one recent study suggest that BST2 can moderately restrict the release of hepatitis C virus (HCV), whose assembly takes place in the ER, and whose release from cells occurs via secretory pathways in a fashion similar to that of coronaviruses (Dafa-Berger et al., 2012, Jones and McLauchlan, 2010). It is possible that virions linked to vesicle membranes via BST2 association are retained on cell surfaces following the exocytotic fusion of virion-containing vesicles with plasma membranes. This finding, plus the inhibition of enveloped virus budding at the plasma membrane, suggests that BST2 is also capable of inhibiting virus budding at intracellular membranes.
While Dafa-Berger et al. (2012) report that BST2 restricts hepatis C virus egress, they also found that (a) IFN-induced BST2 expression levels in Huh-7.5 cells remained unchanged when cells were infected with HCV followed by IFN treatment, and (b) HCV infection had little impact on BST2 level in a stable BST2-expressing Huh-7.5 cell line, suggesting that HCV lacks the necessary mechanism or mechanisms to counteract BST2. Our results indicate that HCoV-229E infections can significanly reduce BST2 expression and enhace NL4.3delVpu virion release (Fig. 6), suggesting that HCoV-229E may express anti-BST2 factors. Further, the high BST2 expression level induced by IFNα in mock-infected A549 cells was not observed when cells were infected with HCoV-229E (Fig. 1F, lane 2 vs. lane 5). We have no direct evidence suggesting that HCoV-229E infection can significanly downregulate IFNα-induced BST2; however, our data suggest that HCoV-229E-infected A549 cells can counteract the ability of IFNα to trigger BST2 expression. This scenario is similar to the intial phase of HCoV-229E infection of respiratroy tract cells, which express very low or undetectable levels of basal BST2 (Erikson et al., 2011). Once cells are infected by HCoV-229E, BST2 expression is downregulated, even in the presence of subsequently induced IFN. As a result, HCoV-229E egress is unrestricted.
We consistently observed that HeLa cell infections with HCoV-229E at low MOIs (0.01–1.0) did not trigger significant changes in total or cell surface BST2 expression 24 h post-infection. However, total or surface BST2 levels in HeLa cells were markedly reduced at an infection MOI of 5.0 (Fig. 4A, lane 5 and Fig. 6). This suggests that BST2 downregulation following HCoV-229E infection is MOI-dependent. Bampi et al. have reported the mediation of BST2 degradation by Sendai virus (SeV) glycoproteins; however, significant total BST2 downregulation in HeLa cells at 24 h post-infection requires inoculation at 20 MOI (Bampi et al., 2013). They therefore concluded that BST2 degradation requires a threshold of viral proteins. It is likely that at shorter time periods post-infection and/or at low MOIs, BST2 antagonist expression by HCoV-229E is insufficient to counteract the effects of BST2. Since HCoV-229E egress becomes unrestricted after longer infection periods, the elevation of virus egress by BST2 knockdown becomes unnecessary. This may explain, at least in part, why we did not observe significant improvement in HCoV-229E egress due to the siRNA knockdown of BST2 at 48 h post-infection (Fig. 1F); at this point, IFNα-induced BST2 may have been neutralized by HCoV-229E. In contrast, the impact of IFNα-induced BST2 on virus egress was significant in samples analyzed 24 h post-infection (Fig. 1D).
While BST2 significantly blocks HCoV-229E progeny virus release, it does not completely block HCoV-229E replication. BST2 is one component of innate immune response in the form of restricting enveloped virion release (Sauter et al., 2010). However, many viruses have evolved specific antagonists to counteract BST2 anti-viral activity—examples include HIV-1 Vpu, HIV-2 Env, simian immunodeficiency virus, Nef and Env, Ebola and Sendai virus GP, and Kaposi's sarcoma-associated herpesvirus (KSHV) K5 (Gupta et al., 2009, Jia et al., 2009, Kaletsky et al., 2009, Le Tortorec and Neil, 2009, Mansouri et al., 2009, Neil et al., 2008, Pardieu et al., 2010, Bampi et al., 2013). Our finding that HCoV-229E infection leads to markedly reduced BST2 expression levels similar to those observed in HIV-1 infections suggests that some HCoV-229E factors are capable of triggering BST2 downregulation. In the case of SARS-CoV, IFN expression is suppressed following infection (Narayanan et al., 2008); this may promote virus spreading due to its inability to trigger BST2 expression. However, SARS is considered a systemic disease involving many organs (Holmes, 2003) that constitutively express BST2 (Erikson et al., 2011). It remains to be determined whether SARS-CoV virion release is also restricted by BST2, and whether coronaviruses (including SARS-CoV) have evolved strategies to oppose BST2 anti-viral activity.
In conclusion, our evidence suggests that BST2 is capable of restricting HCoV-229E release from cells by tethering nascent progeny virions to cell surfaces. This also supports the assumption that the release of enveloped viruses whose budding takes place at the plasma membrane or in an intracellular compartment is subject to restriction via BST2.
Materials and methods
Virus, cell culture and transfection
Human coronavirus 229E (HCoV-229E) were propagated in A549 or HeLa cells. 293T, HeLa and A549 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. For transfection, confluent cells were trypsinized, split 1:10, and seeded onto 10 cm dish plates 24 h before transfection. Cells were transfected with 20 μg of plasmid DNA by the calcium phosphate precipitation method, with the addition of 50 μM chloroquine to enhance transfection efficiency. For cotransfection, 10 μg of each plasmid was used. To transfect HeLa cells with BST2-specific siRNA (BST2 siRNA), siRNA was mixed with Lipofectamine 2000 (Invitrogen) at a ratio of 1 μg to 2.5 μl, the transfection procedure was performed according to the manufacture's protocols.
Construction of plasmids and stable BST2 knockdown cell lines
To construct the HIV-1 Vpu deletion mutant (NL4.3delVpu), we employed a PCR-mediated mutagenesis method (Sambrook and Russell, 2001) to introduce two consecutive stop codons in-frame at the N-terminal Vpu region. The primer sequence used to create the delVpu mutation is 5′-ACATGTAATGTACAACCTATAATAG-3′. The delVpu mutation construct was subcloned into NL4.3Env-which has the env coding sequence (BglII-nt.7031 to BglII-nt. 7611) partially deleted. Constructs were confirmed by restriction enzyme digestion and DNA sequencing. Small interfering RNA (siRNA) sequence (5′-CCAggUCUUAAgCgUgAgAAUC-3′) directed against BST-2 was synthesized by MDBio, Inc. To establish BST2 stale knockdown cell lines, pseudotyped lentiviruses containing the BST2 shRNA were generated in 293T cells. Virus-containing supernatants then were used to infect HeLa cells. BST2 knockdown cell lines were selected with puromycin (3 μg/ml) and screened with western immunoblotting. The lentivirus-based shRNA construct targeting the BST2 gene, NM_004335.2-432s1c1 and lentivirus package plasmids including pCMVDR8.91 and pMD.G (VSV-G) were purchased from National RNAi Core Facility Platform (NRC), Academia Sinica, Taiwan.
Western immunoblot
Culture medium from transfected or infected cells was filtered (0.45-μm pore size) and centrifuged through 2 ml of 20% sucrose in TSE (10 mM Tris–HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) plus 0.1 mM phenylmethylsulfonyl fluoride (PMSF) at 4 °C for 40 min at 274,000×g (SW41 rotor at 40,000 rpm). Viral pellets were suspended in IPB (20 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 0.02% sodium azide) plus 0.1 mM PMSF. Cells were collected in 1 ml phosphate-buffered saline (PBS), and pelleted at 3000 rpm for 5 min. Cell pellets were resuspended in 250 μl of IPB plus 0.1 mM PMSF prior to microcentrifugation at 4 °C for 15 min at 13,700×g to remove cell debris. Either supernatant or cell samples were mixed with equal volumes of 2× sample buffer (12.5 mM Tris–HCl [pH 6.8], 2% SDS, 20% glycerol, 0.25% bromophenol blue) and 5% β-mercaptoethanol and boiled for 5 min. Samples were subjected to SDS-PAGE and electroblotted onto nitrocellulose membranes that were subsequently blocked with 5% non fat milk in Tris-buffered saline containing 0.05% Tween 20 (TBST), followed by incubation with the primary antibody in 5% non fat milk-TBST on a rocking platform for 1 h at room temperature. Membranes were then washed three times for 10 min each with TBST and rocked for 30 min with the secondary antibody in 5% non fat milk-TBST. Blots were again washed three times in TBST for 10 min each, followed by enhanced chemiluminescence (ECL) to detect membrane-bound antibody-conjugated enzyme activity.
To detect membrane-bound HCoV-229E N or HIV-1 Gag proteins, we used an anti-HIV-1 p24CA (mouse hybridoma clone 183-H12-5C) monoclonal antibody or a rabbit anti-HCoV229E N serum as the primary antibody. BST2 was detected with a human BST2 mouse antiserum (ab88523, Abcam) or rabbit antiserum (Miyagi et al., 2009). Our secondary antibody was either a rabbit anti-mouse or donkey anti-rabbit (HRP)-conjugated antibody. Manufacturer's protocols were followed for HRP activity detection (Pierce).
Plaque assay
Serial dilutions (10-fold) of the harvested culture medium were used to infect subconfluent monolayers of A549 or HeLa cells grown in 6-well culture plates. Plates were incubated at 37 °C for one hour with gentle rocking every 15 min. Plates then were washed twice with PBS following removal of the culture medium. About 2 to 5 ml of 0.5% agarose were overlaid onto the monolayer cells. After 5 days of incubation, 0.03% neutral red solution was added to plates to visualize the plaque formation.
Laser scanning immunofluresecne microscopy and flow cytometry
HeLa or HeLa/BST2− cells were infected with HCoV-229E at a 0.1 or 2 MOI for 1 h at 37 °C. At 4 to 18 h post-infection, cells were permeabilized at room temperature for 10 min in acetone following fixation with 3.7% formaldehyde at 4 °C for 20 min. Samples were incubated with the primary antibody for 1 h and with the secondary antibody for 30 min. Following each incubation, samples were washes three times with DMEM/calf serum. Primary antibody was an anti-BST2 or anti-HCoV-229E N antibody. A goat anti-rabbit or rabbit anti-mouse FITC-conjugated, phycoerythrin-conjugated, or rhodoamine-conjugated antibody served as the secondary antibody. Cell surface BST2 was probed prior to the permeabilization of cell membranes. Images were observed by a laser scanning confocal microscope (OlympusFluoview FV10i). For flow cytometric analysis, cells were detached from dishes with 0.05% trypsin/EDTA prior to fixation and permeabilization. Human BST2 or HCoV-229E N proteins were probed as described above. Cells were analyzed using a flow cytometer (BD FACSCanto II).
Scanning or transmission electron microscopy
Cells grown on cover glasses were fixed with 2.5% glutaraldehyde (in 0.1 M cacodylate buffer pH 7.4) for 2 h, washed with 0.1 M cacodylate buffer pH 7.4 three times, and fixed again with 1% osmium tetraoxide (OsO4) (in 0.1 M cacodylate buffer pH7.4) for 1 h. Samples then were dehydrated by sequential incubation with 25%, 50%, 70%, 85%, 95% and 100% ethanol for 10 min each, dried in a critical point dryer (Samdri-PVT-3B, Tousimis Research,. Rockville, MD), coated with gold (Jeol JFC-1200), and viewed with a scanning electron microscope (Jeol JSM-7600 F). Images were collected at 10,000× and 20,000×. For transmission electron microscopy (TEM) analysis, cells were harvested 12 h post-infection and fixed in 0.1 M Cacodylate buffer containing 2.5% glutaraldehyde, post-fixed with 1% OsO4, dehydrated in ethanol and embedded in Spurr resin. Ultrathin sections (~70 nm-thick) were cut with an ultramicrotome, stained with 5% uranyl acetate and 0.4% lead citrate. For immunegold labeling of BST2 proteins, samples were embedded in acrylic resin (LR White Resin, London Resin Company) at 4 °C for 48 h following ethanol dehydration. Ultrathin sections were incubated with an anti-BST2 rabbit serum for 2 h and with a secondary 10-nm immunegold-conjugated goat anti-rabbit IgG antibodies for 1 h. Samples were washed with PBS tree times after each incubation, fixed with 2% glutaraldehyde, stained, and viewed with a Joel JEM-2000 EX-II transmission electron microscope. Images were collected at 20,000×, 60,000× or 100,000×.
Acknowledgments
We thank Y.-T. Tseng for reagents and technical assistance. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, HIH: Atni-Bst-2 (cat #11722) from Drs. Klaus Strebel and Amy Andrew. This work was supported by Grants V99C1-013 and V100C-002 from Taipei Veterans General Hospital, by Grants NSC 97-2320-B-010-002-MY3 and 100-2320-B-010-015-MY3 from the National Science Council, Taiwan, and by a grant from the Ministry of Education, Aiming for the Top University Plan.
References
- Bampi C., Rasga L., Roux L. Antagonism to human BST-2/tetherin by Sendai virus glycoproteins. J. Gen. Virol. 2013;94:1211–1219. doi: 10.1099/vir.0.051771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafa-Berger A., Kuzmina A., Fassler M., Yitzhak-Asraf H., Shemer-Avni Y., Taube R. Modulation of hepatitis C virus release by the interferon-induced protein BST-2/tetherin. Virology. 2012;428:98–111. doi: 10.1016/j.virol.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Erikson E., Adam T., Schmidt S., Lehmann-Koch J., Over B., Goffinet C., Harter C., Bekeredjian-Ding I., Sertel S., Lasitschka F., Keppler O.T. In vivo expression profile of the antiviral restriction factor and tumor-targeting antigen CD317/BST-2/HM1.24/tetherin in humans. Proc. Nat. Acad. Sci. U.S.A. 2011;108:13688–13693. doi: 10.1073/pnas.1101684108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbino J., Crespo S., Aubert J.-D., Rochat T., Ninet B., Deffernez C., Wunderli W., Pache J.-C., Soccal P.M., Kaiser L. A prospective hospital-based study of the clinical impact of non-severe acute respiratory syndrome (Non-SARS)–related human coronavirus infection. Clin. Infect. Dis. 2006;43:1009–1015. doi: 10.1086/507898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffinet C., Schmidt S., Kern C., Oberbremer L., Keppler O.T. Endogenous CD317/Tetherin limits replication of HIV-1 and murine leukemia virus in rodent cells and is resistant to antagonists from primate viruses. J. Virol. 2010;84:11374–11384. doi: 10.1128/JVI.01067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.K., Mlcochova P., Pelchen-Matthews A., Petit S.J., Mattiuzzo G., Pillay D., Takeuchi Y., Marsh M., Towers G.J. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc. Nat. Acad. Sci. U.S.A. 2009;106:20889–20894. doi: 10.1073/pnas.0907075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann A., Krijnse-Locker J., Oberwinkler H., Eckhardt M., Homann S., Andrew A., Strebel K., Kräusslich H.-G. CD317/Tetherin is enriched in the HIV-1 envelope and downregulated from the plasma membrane upon virus infection. J. Virol. 2010;84:4646–4658. doi: 10.1128/JVI.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, E., 2001. Virus assembly. In: Knipe, D.M., Howley, P.M. (Eds.), Fundamental Virology, fourth ed.
- Jia B., Serra-Moreno R., Neidermyer W., Jr., Rahmberg A., Mackey J., Fofana I.B., Johnson W.E., Westmoreland S., Evans D.T. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by Tetherin/BST2. PLoS Pathog. 2009;5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.M., McLauchlan J. Hepatitis C virus: assembly and release of virus particles. J. Biol. Chem. 2010;285:22733–22739. doi: 10.1074/jbc.R110.133017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Neil S.J., Zhadina M., Zang T., Kratovac Z., Lee Y., McNatt M., Hatziioannou T., Bieniasz P.D. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 2009;83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R.L., Francica J.R., Agrawal-Gamse C., Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc. Nat. Acad. Sci. U.S.A. 2009;106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupzig S., Korolchuk V., Rollason R., Sugden A., Wilde A., Banting G. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- Le Tortorec A., Neil S.J.D. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 2009;83:11966–11978. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tortorec A., Willey S., Neil S.J.D. Antiviral inhibition of enveloped virus release by Tetherin/BST-2: action and counteraction. Viruses. 2011;3:520–540. doi: 10.3390/v3050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B., Cavagliotti L., Lehmann M., Gers-Huber G., Kaur I., Thomas Y., Kaiser L., Piguet V. Influenza virus partially counteracts restriction imposed by Tetherin/BST-2. J. Biol. Chem. 2012;287:22015–22029. doi: 10.1074/jbc.M111.319996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri M., Viswanathan K., Douglas J.L., Hines J., Gustin J., Moses A.V., Fruh K. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2009;83:9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T.C. Herpesvirus assembly and egress. J. Virol. 2002;76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E., Andrew A.J., Kao S., Strebel K. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Nat. Acad. Sci. U.S.A. 2009;106:2868–2873. doi: 10.1073/pnas.0813223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.-T.K., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S.J., Zang T., Bieniasz P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Neil S.J.D., Eastman S.W., Jouvenet N., Bieniasz P.D. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2:e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S.J.D., Sandrin V., Sundquist W.I., Bieniasz P.D. An interferon-α-induced tethering mechanism inhibits HIV-1 and ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardieu C., Vigan R.l., Wilson S.J., Calvi A., Zang T., Bieniasz P., Kellam P., Towers G.J., Neil S.J.D. The RING-CH Ligase K5 antagonizes restriction of KSHV and HIV-1 particle release by mediating ubiquitin-dependent endosomal degradation of tetherin. PLoS Pathog. 2010;6:e1000843. doi: 10.1371/journal.ppat.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D., Zang T., Ebrahimi A., McNatt M.W., Gregory D.A., Johnson M.C., Bieniasz P.D. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Micro. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshitzky S.R., Dong L., Chi X., Clester J.C., Retterer C., Spurgers K., Kuhn J.H., Sandwick S., Ruthel G., Kota K., Boltz D., Warren T., Kranzusch P.J., Whelan S.P.J., Bavari S. Infectious lassa virus, but not filoviruses, is restricted by BST-2/Tetherin. J. Virol. 2010;84:10569–10580. doi: 10.1128/JVI.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., PeAaranda S., Bankamp B., Maher K., Chen M.-h., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., GA?nther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T., Noda T., Urata S., Kawaoka Y., Yasuda J. Inhibition of lassa and marburg virus production by tetherin. J. Virol. 2009;83:2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. third ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; N.Y: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Sauter D., Specht A., Kirchhoff F. Tetherin: holding on and letting go. Cell. 2010;141:392–398. doi: 10.1016/j.cell.2010.04.022. [DOI] [PubMed] [Google Scholar]
- Swiecki M., Omattage N.S., Brett T.J. BST-2/tetherin: structural biology, viral antagonism, and immunobiology of a potent host antiviral factor. Mol. Immunol. 2013;54:132–139. doi: 10.1016/j.molimm.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Van Damme N., Goff D., Katsura C., Jorgenson R.L., Mitchell R., Johnson M.C., Stephens E.B., Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Leser G.P., Lamb R.A. Influenza virus is not restricted by tetherin whereas influenza VLP production is restricted by tetherin. Virology. 2011;417:50–56. doi: 10.1016/j.virol.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yondola M.A., Fernandes F., Belicha-Villanueva A., Uccelini M., Gao Q., Carter C., Palese P. Budding capability of the influenza virus neuraminidase can be modulated by tetherin. J. Virol. 2011;85:2480–2491. doi: 10.1128/JVI.02188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang F., Wilson S.J., Landford W.C., Virgen B., Gregory D., Johnson M.C., Munch J., Kirchhoff F., Bieniasz P.D., Hatziioannou T. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlateva K., Coenjaerts F.J., Crusio K., Lammens C., Leus F., Viveen M., Ieven M., Spaan W.M., Claas E.J., Gorbalenya A. No novel coronaviruses identified in a large collection of human nasopharyngeal specimens using family-wide CODEHOP-based primers. Arch. Virol. 2013;158:251–255. doi: 10.1007/s00705-012-1487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]