Abstract
Viral factories are complex structures in the infected cell where viruses compartmentalize their life cycle. Rubella virus (RUBV) assembles factories by recruitment of rough endoplasmic reticulum (RER), mitochondria and Golgi around modified lysosomes known as cytopathic vacuoles or CPVs. These organelles contain active replication complexes that transfer replicated RNA to assembly sites in Golgi membranes. We have studied the structure of RUBV factory in three dimensions by electron tomography and freeze-fracture. CPVs contain stacked membranes, rigid sheets, small vesicles and large vacuoles. These membranes are interconnected and in communication with the endocytic pathway since they incorporate endocytosed BSA-gold. RER and CPVs are coupled through protein bridges and closely apposed membranes. Golgi vesicles attach to the CPVs but no tight contacts with mitochondria were detected. Immunogold labelling confirmed that the mitochondrial protein p32 is an abundant component around and inside CPVs where it could play important roles in factory activities.
Keywords: Rubella virus, Virus factory, Replication complex, RNA virus, Cytopathic vacuole, Togavirus, Electron tomography, 3D EM, High-pressure freezing, Freeze-fracture
Introduction
The replication of many viruses is associated with specific intracellular compartments called virus factories. These are thought to provide a physical scaffold to concentrate viral components for genome replication and morphogenesis (Netherton et al., 2007, Novoa et al., 2005). The formation of virus factories often results in rearrangement of cellular membranes, reorganization of the cytoskeleton, and recruitment of mitochondria. One of the early events in factory formation is the assembly of replication complexes (RCs). RNA viruses modify specific membranes of the factory to concentrate viral replicases and necessary co-factors. Their RCs include the viral-RNA-dependent RNA polymerase (RdRp), further accessory non-structural proteins, viral RNA and host cell factors (Miller and Krijnse-Locker, 2008). Studies of individual non-structural proteins have revealed that the replication complexes are targeted to the respective organelle by the non-structural proteins rather than RNA (Salonen et al., 2005). These complexes could have a sophisticated organization for a more efficient replication of the viral genome (Dye et al., 2005, Fontana et al., 2007, Lyle et al., 2002, Mackenzie, 2005, Spagnolo et al., 2010, Wang et al., 2002). Why different viruses choose different subcellular membranes for their replication, how the viral proteins are targeted to those membranes and the host factors involved are currently under intensive investigations (Barajas et al., 2009, Goff, 2008, Nagy, 2008, Ortin & Parra, 2006, Sessions et al., 2009).
Rubella virus (RUBV) is an important human teratogenic virus and the only member of the genus Rubivirus in the family Togaviridae (Frey, 1994). RUBV anchors its RNA synthesis in membranes of a cell organelle known as the “cytopathic vacuole” or CPV that derives from modified endosomes and lysosomes (Fontana et al., 2007, Lee et al., 1994, Magliano et al., 1998). This is a very unique feature of RUBV shared with alphaviruses, the other genus of the family (Froshauer et al., 1988, Kujala et al., 2001). RER cisternae, mitochondria and Golgi stacks are recruited around CPVs to build RUBV factories. Such structures are likely to help the virus to evade host cell defense responses as well as connecting viral replication with assembly and maturation of new viral particles in recruited Golgi membranes (Risco et al., 2003).
The RUBV genome is a single-stranded, “plus-sense” RNA of ~ 10 kb in length. The genome RNA contains two long open reading frames (ORFs): a 5' proximal or non-structural (NS)-ORF encodes the two non-structural proteins, P150 and P90, involved in viral RNA replication and a 3' proximal ORF or structural protein (SP)-ORF encodes the structural proteins (capsid or C, and two envelope glycoproteins, E1 and E2). The nonstructural proteins (NSPs) are translated from the genome RNA while the SP-ORF is translated from a subgenomic (SG) RNA that is co-terminal with the 3' end of the genomic RNA. From a RUBV infectious cDNA clone, a replicon derivative was generated in which the 3' proximal ORF was replaced with reporter genes and drug resistance markers. In transfected cells, replicons create CPVs and factories ultra-structurally identical to those formed during viral infection and thus the P150 and P90 replicase proteins are sufficient to generate these structures (Fontana et al., 2007). Cell signaling pathways involved in RUBV factory construction, as well as the nature of contacts between organelles within the structure, are basically unknown.
RUBV has genomic coding and replication strategies similar to alphaviruses, but the two genera are only distantly related, sharing no sequence homology at either the nucleotide or amino acid level. However, CPVs assembled by RUBV and alphaviruses are very similar: they have been described as structures of 600–2000 nm in size with a surface consisting of small vesicular invaginations or spherules (the sites of viral RNA replication) that have a diameter of approximately 50 nm and line the vacuole membrane at regular intervals (Froshauer et al., 1988, Griemley et al., 1972, Lee et al., 1994, Magliano et al., 1998). The recent use of improved specimen preparation techniques for electron microscopy of cells, such as freeze-substitution before embedding, showed that CPVs are indeed filled with electron-dense material, small and large vesicles and elongated “straight elements” which had not been observed previously (Fontana et al., 2007). In cells transfected with RUBV replicons, these structures were shown to contain P150, P90 and dsRNA, and small amounts of the mitochondrial p32 protein, as shown by immunoelectron microscopy. In the present work we have gone one step further in preservation by applying high-pressure freezing on live cells and subsequently avoiding chemical treatments before fast freezing and freeze-substitution. High-pressure freezing is considered the best method of cryo-immobilization for obtaining the greatest depth of optimal ultra-structural cell preservation for electron microscopy (Studer et al., 2008). With highly preserved cells processed by high-pressure freezing and freeze-substitution we have then applied methods for imaging in three dimensions (3D). Random thin-sectioning and electron microscopy (EM) has missed many of the fine features in RUBV factories such as connections between recruited organelles and the relationships between different membranous compartments inside CPVs and with the surrounding cytosol. The main technical options for 3D characterization of cells include reconstruction from serial sections, which is particularly useful for very large structures such as whole eukaryotic cells (Fiala, 2005, Fontana et al., 2008, Romao et al., 2008), metal replication after freeze-fracture or freeze-etching that provides planar views of the internal organization of membranes (Cabezas & Risco, 2006, Risco & Pinto da Silva, 1998, Severs, 2007, Severs & Robenek, 2008), and electron tomography (ET), a process involving rotation of the specimen in the electron beam, capturing images at incremental rotations, and using the images for 3D reconstruction. Cellular electron tomography (ET) is a powerful technique that can provide molecular resolution of cell ultra-structure if combined with “close-to-native” preservation of biological samples (Lucić et al., 2005). Nowadays, ET is becoming a mainstream research tool in cell biology and structural virology (Cyrklaff et al., 2005, McIntosh et al., 2005, Miller & Krijnse-Locker, 2008, Steven & Aebi, 2003).
Here we present the structural characterization of RUBV factories by electron tomography and metal replication after freeze-fracture followed by freeze-etching. We have found that recruited organelles (RER, mitochondria and Golgi) exhibit a variety of contacts with the CPVs, whose internal membranes are interconnected and in communication with the endocytic pathway. Immunogold labelling showed the presence of significant amounts of the mitochondrial protein p32 inside and around the CPVs, which suggests a role for this protein in the assembly and activities of the viral factory.
Results
2D organization of highly preserved RUBV factories
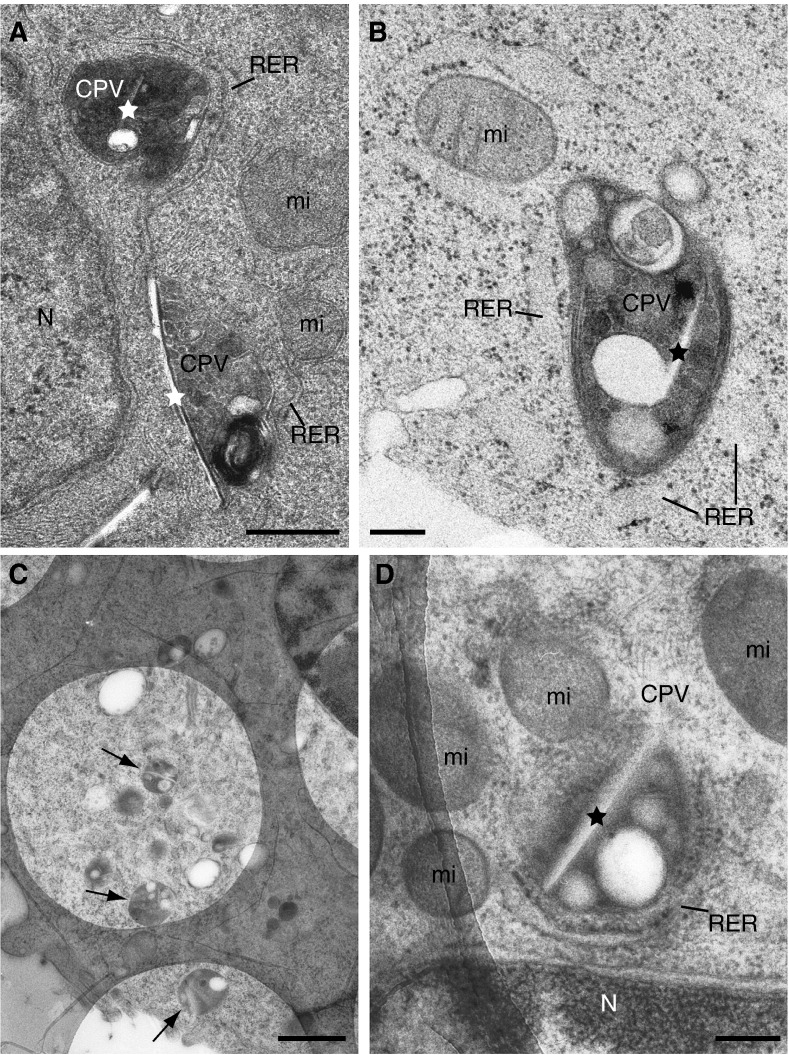
Thin-section electron microscopy shows the basic features of Rubella virus factories built around modified lysosomes or CPVs (Fig. 1 ). Both lysosomes and CPVs are round-shaped organelles filled with lamellar membranes and vesicles but as described in previous work from our lab CPVs have a more complex content with different types of vesicles, one or several larger vacuoles and rigid sheets (Fontana et al., 2007). RER cisternae, mitochondria and sometimes one Golgi stack surround the CPV as assembled in RUBV-infected cells and in cells transfected with RUBV replicons. These general features are appreciated in thin-sections of cells transfected with the RUBrep/GFP/neo RUBV replicon and conventionally dehydrated and embedded in an epoxy-resin before TEM (Fontana et al., 2007) (Fig. 1A). For three-dimensional studies at higher resolution a superior preservation of the structure is needed and for this purpose we processed our samples by high-pressure freezing. Structures associated with RUBV factories are better preserved after high-pressure freezing of live cells followed by freeze-substitution and thin-sectioning (Fig. 1B). For example, RER cisternae are thicker and closer to CPVs, mitochondrial membranes are smooth, and the interior of CPVs exhibits a variety of spherical vesicles and vacuoles. Semi-thick sections (~ 250–300 nm) were then obtained to encompass a larger proportion of the viral factories. High contrast was obtained by testing the most adequate substitution medium and resin (see Materials and methods). Specimen stability in the electron beam was optimized by collecting the sections on holey carbon grids (Fig. 1C and D). CPVs were easily identified on the holes at low magnification (Fig. 1C, arrows) which helped in selection of representative factories (CPVs with associated organelles) for electron tomography (Fig. 1D).
Fig. 1.
Two-dimensional ultrastructure of RUBV factories: high-pressure freezing, freeze-substitution and sectioning. (A) Conventional EM shows two CPVs (frontal view for the one on the top and side view for the CPV on the bottom) in the perinuclear area of a replicon-transfected cell. Both CPVs exhibit internal vesicles and straight elements (stars) and locally recruited RER and mitochondria. (B to D) Suspensions of BHK-21 cells stably transfected with a RUBV replicon were cryofixed by HPF without cryoprotectants. Cell in (B) was freeze-substituted in 1% OsO4, 0.25% glutaraldehyde in acetone and embedded in EML-812 while cells in (C) and (D) were freeze-substituted in 0.5% UA in acetone and embedded in Lowicryl HM23. (B) Detail of a CPV surrounded by RER cisternae and one mitochondrion. The CPV exhibits a very heterogeneous content with numerous vesicles of variable size and density, vacuoles and a rigid membrane (star). (C) Low magnification view showing large areas of the cell and several CPVs (arrows) over the holes of the carbon film. (D) Image at 0º of a CPV surrounded by RER and mitochondria ready for ET. Ultra-thin (~ 50 nm) post-stained sections are shown in (A) and (B), while (C) and (D) show semi-thick (~ 200 nm) sections collected on Quantifoil® holey carbon grids and visualized without post-staining. RER, rough endoplasmic reticulum; mi, mitochondria; N, nucleus. Bars, 200 nm in A, B, and D; 1 μm in C.
Three-dimensional analysis of RUBV factories: General organization and contacts with recruited organelles
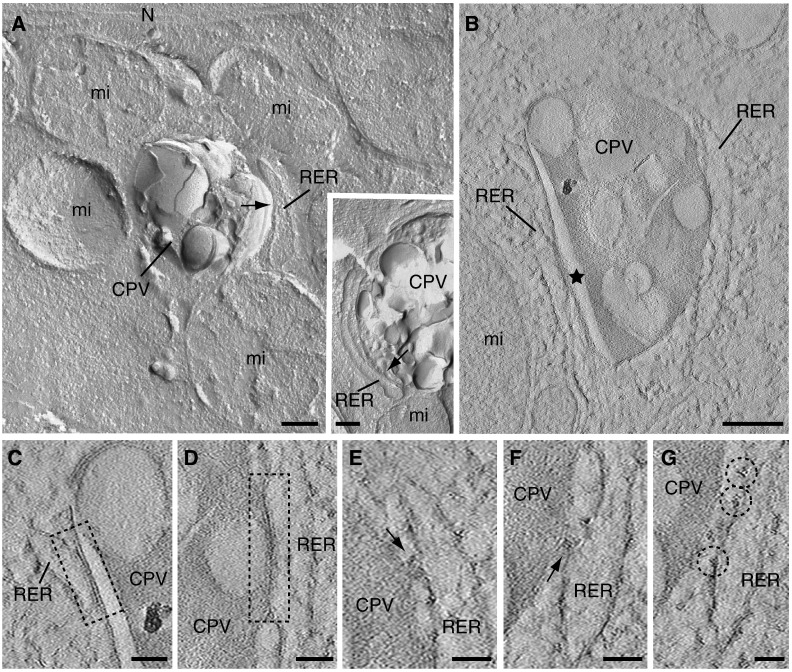
Freeze-fractured RUBV factories were submitted to short etching and replicated with Ta/W for higher resolution (see Materials and methods). TEM of these metal replicas shows particular details of the factories, such as crowding of organelles around CPVs that are completely surrounded by mitochondria (Fig. 2A). Fine details of the CPV are also seen, such as the variety of sizes for the internal vesicles and the heterogeneity in the protein content of these membranes, visualized as small particles corresponding to integral membrane proteins. These images also reveal the need for electron tomography to study the structure with higher resolution: it is not possible to appreciate if organelles contact or how they attach to the CPV's surface in images from metal replicas (arrows in Fig. 2A, mainfield and inset). Electron tomography has solved this dilemma by showing where real contacts between organelles are established. Fig. 2B–G show low magnification (B) and high magnification (C to G) views of several areas from a selected computational tomographic slice corresponding to one of the eleven reconstructions obtained by ET and image processing.
Fig. 2.
Three-dimensional views of RUBV factories as visualized by freeze-fracture and electron tomography. (A) Metal replica of BHK-21 cells stably transfected with the RUBrep/GFP/neo replicon and processed by freeze-fracture followed by etching. Mitochondria and RER cisternae surround the CPVs. Although these organelles are very close to the CPVs no clear contacts are detected (arrows in mainfield and inset). (B to G) A computational tomographic slice (~ 1.2 nm) corresponding to the tomogram of a semi-thick (~ 250 nm) section obtained from suspensions of trypsinized cells processed by HPF, FS and embedding in Lowicryl HM23 and visualized without post-staining. (B) CPV with a characteristic rigid membrane (star) surrounded by RER. (C to G) Expanded areas from the CPV shown in (B). (C and D) RER–CPV contacts seen as closely apposed membranes (dashed rectangles). (E and F) Protein bridges between RER and CPV are also observed (arrows). (G) Close to these complexes several ribosomes facing the CPV are found (dashed circles). RER, rough endoplasmic reticulum; mi, mitochondria. Bars, 200 nm in A and B; 50 nm in C to G.
This analysis showed two types of contacts between RER and CPVs: closely apposed membranes without evident fusion (Fig. 2C and D) together with protein bridges (Fig. 2E and F). The macromolecular complexes in the bridges fill a gap of ~ 10–15 nm. Ribosomes bound to recruited RER usually face the CPV but they do not seem to attach to its surface (Fig. 2G).
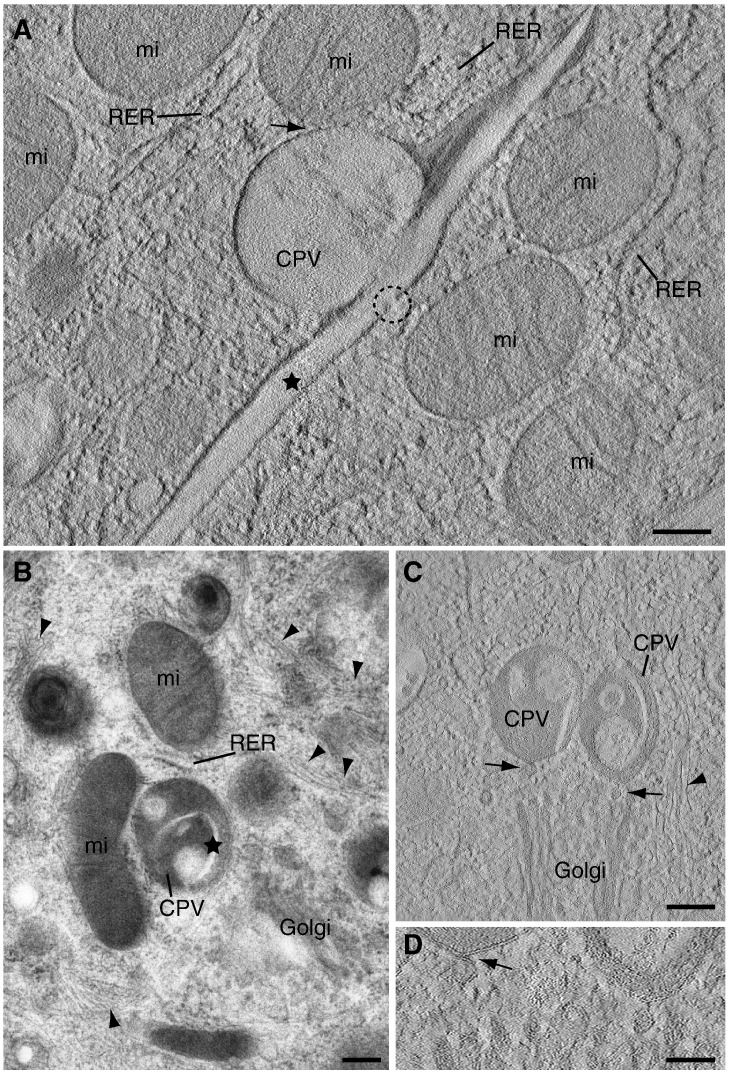
Electron tomography showed that mitochondria get very close to the CPV's surface but tight contacts such as those detected for RER cisternae were not seen. Instead, a fuzzy material occupies the small gap between both organelles (Fig. 3A, arrow). Most mitochondria remain at a distance from the CPVs, sometimes facing pores open in their peripheral membranes (Fig. 3A, dashed circle). Just one Golgi stack near each CPV was observed (Fig. 3B). The stack interacts laterally with the CPV through small peripheral vesicles that contact the CPV's surface as detected in the tomograms (Fig. 3C and D, arrows). Arrays of filaments similar to intermediate filaments visualized in ultra-thin sections of cells are frequently detected around CPVs although they do not seem to establish physical contacts with them (Fig. 3B and C, arrowheads).
Fig. 3.
CPV contacts with recruited mitochondria and Golgi stacks. (A, C and D) Computational tomographic slices (~ 1.2 nm) and (B) semi-thick (~ 200 nm) section visualized by TEM. (A) Side view of a CPV completely surrounded by mitochondria. Holes or channels in the CPV rigid membrane (dashed circle) apparently open the CPV to the cytoplasm. (B) Characteristic RUBV factory with a CPV containing rigid membranes (the star labels one of them) surrounded by RER, mitochondria and Golgi. The Golgi stack approaches laterally to the CPV but no clear contacts are detected. (C and D) Computational tomographic slice of a similar factory showing contacts between Golgi vesicles and the CPVs (arrows). Arrowheads in B and C point to filaments around the factories. RER, rough endoplasmic reticulum; mi, mitochondria. Bars, 200 nm in A to C; 100 nm in D.
Internal structure of the CPVs and communication with the endo-lysosomal pathway
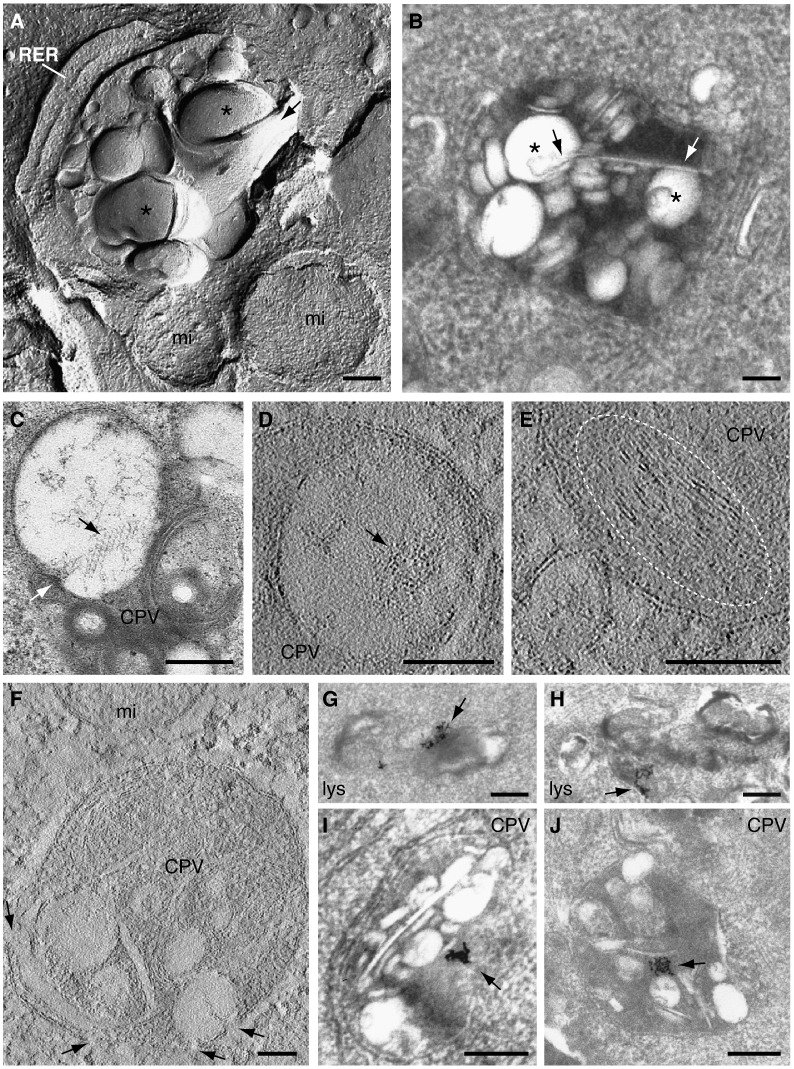
Inside CPVs a variety of vesicles co-exist with large vacuoles and membrane sheets as appreciated by freeze-fracture and freeze-etching (Fig. 4A). The smallest vesicles are usually attached to the periphery of the CPV and large sheets are frequently seen wrapping the vacuoles (Fig. 4A, arrow). These membrane sheets, seen as rigid membranes in thin sections are in contact with the large vacuoles in many CPVs (Fig. 4B, arrows). This contact, which is highlighted by freeze-fracture, is a generally observable feature (Fig. 1, Fig. 2, Fig. 3, Fig. 4). Serial sections revealed that rigid membranes are frequently attached to one or two vacuoles (not shown). When the section goes along the straight sheet, a potential connection with the periphery of the vacuole is detected (Fig. 4C, white arrow) as well as some internal order that is compatible with a close-packing of particles (Fig. 4C, black arrow). Looking for similar views in the tomograms we have distinguished groups of particles associated with vacuoles (Fig. 4D, arrow) but a clear internal order was not obvious. Closely packed membranes are sometimes seen in tomograms of CPVs (Fig. 4E) that always reveal numerous openings to the cytosol (Fig. 4F). To investigate if CPVs communicate not only with the surrounding organelles and the cytosol, but also with the endo-lysosomal pathway from where they derive, we treated both control and transfected cells with BSA-gold particles. When added to cell culture medium, these colloidal gold conjugates are rapidly endocytosed and are detectable in lysosomes within 1 h (Kleijmeer et al., 1997). Electron microscopy of thin-sections of non-transfected BHK-21 cells confirms that BSA-gold accumulates in lysosomes 3 h after adding the conjugate to the cell cultures (Fig. 4G and H). In RUBV-replicon stably-transfected BHK-21 cells, BSA-gold particles reach the CPVs and accumulate in the dense internal content but never inside vesicles or large vacuoles (Fig. 4I and J). A quantitative analysis revealed that after 3 h exposure to BSA-gold 21 out of 115 lysosome-like organelles in control, non-transfected cells were positive for BSA-gold (18 %). In transfected cells 90 out of 257 CPVs contained BSA-gold particles (35 %). Thus, although CPVs were surrounded by recruited organelles they retained communication with the endo-lysosomal pathway.
Fig. 4.
Complex internal organization of CPVs and connection with the endo-lysosomal pathway. (A) Metal replica of a RUBV factory processed by freeze-etching showing the interior of a CPV with a rigid membrane (black arrow) wrapping two large vacuoles (asterisks). (B and C) Ultra-thin sections of CPVs as visualized by TEM after staining. (B) Rigid membrane (arrows) contacting two vacuoles (asterisks). (C) A section along the rigid membrane showing features compatible with a close packing of particles (black arrow). The white arrow points to the spot where the rigid membrane interacts with the membrane of the large vacuole. (D to F) Computational slices (~ 0.6 nm for D and E, ~ 1.2 nm for F) of two tomograms of CPVs filtered with three rounds of a median filter. (D) Groups of particles associated with a large vacuole (arrow) and (E) stacked membranes (white dashed ellipse) are shown. (F) Openings to the cytosol (arrows) in the peripheral membrane of a CPV (frontal view). Thin-sections of lysosomes from control BHK-21 cells (G and H) and CPVs from stably transfected cells (I and J) after treatment with BSA-gold in culture and posterior fixation and embedding. Both lysosomes (lys) from control cells and CPVs contain BSA-gold particles (arrows). RER, rough endoplasmic reticulum; mi, mitochondria; Bars, 200 nm.
Analysis of complex ET reconstructions
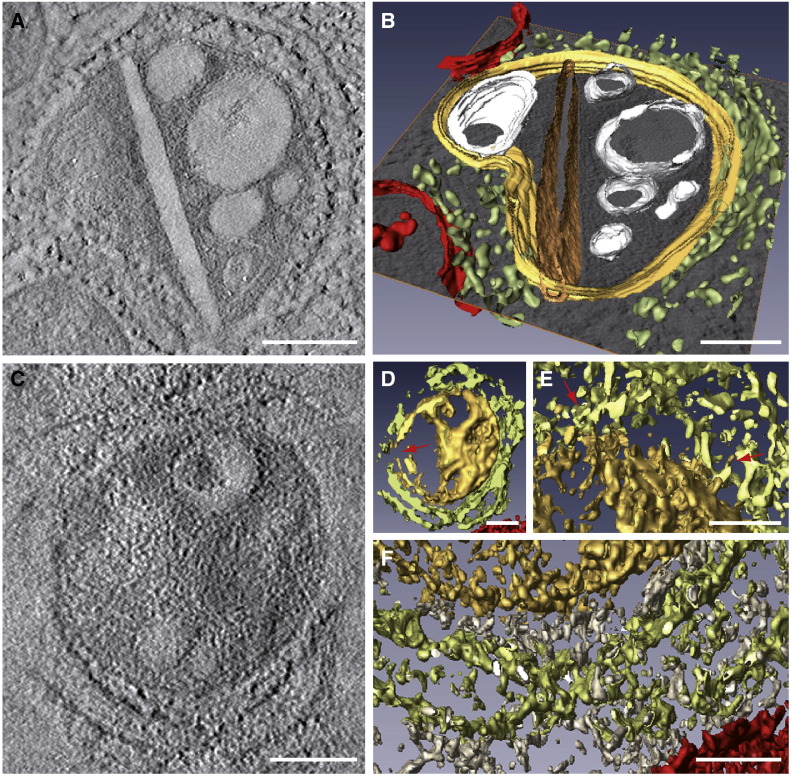
The detailed analysis of thin computational sections covering the whole volume for each of the tomograms included in this study was used to make the main conclusions summarized in Fig. 2, Fig. 3, Fig. 4. In electron tomography all useful 3D information is actually extracted from this analysis of planes. In addition the volumetric representation of reconstructed factories using programs such as Amira helps us to summarize our findings and to highlight some important features of RUBV factories (Fig. 5 ). The extraordinary complexity of volumes generated by ET of highly-preserved cells is apparent in these representations. In order to facilitate their interpretation, noise reduction and segmentation have been applied to display the most important features (see Methods). In Fig. 5A and B, a computational tomographic slice and a 3D model, respectively, are shown. In the 3D model RER cisternae (segmented in green) surround the CPV (yellow); vesicles and vacuoles inside the CPV are represented in white, a rigid straight sheet is segmented in brown and nearby mitochondria are shown in red. Vacuoles, vesicles and the rigid sheet are connected with the periphery of the CPV. Fig. 5C shows a computational tomographic slice from a different CPV and two 3D models (Fig. 5D and E–F). The CPV is filled with abundant material, open to the cytosol and almost completely surrounded by RER (Fig. 5D). The model in Fig. 5E and F shows the complex material surrounding the CPV. Physical contacts between the RER and the CPV's surface are seen (Fig. 5E) but the gap between CPV and RER is frequently filled with additional molecules of unknown nature (Fig. 5F, white).
Fig. 5.
Analysis of ET volumes. Computational tomographic slices (A and C) and 3D models after segmentation and visualization with AMIRA (B and D to F). Color code is as follows: CPV, yellow; straight sheet, brown; RER, light green; mitochondria, red; vesicles and vacuoles, white; cytoplasm, grey. (A) A computational tomographic slice and the corresponding 3D model (B) of a CPV surrounded by the RER and containing a number of vacuoles, vesicles and a rigid straight sheet that is connected with the periphery of the CPV. The 3D model was built using a combination of masking, isosurface, and manual tracing using Amira segmentation tools. (C) A computational tomographic slice and two 3D models (D, E-F) of a CPV almost completely surrounded by the RER and filled with abundant material. The 3D model in (D) was built with a combination of masking and isosurface after strong filtering of the volume with nonlinear diffusion. The internal material of the CPV was solid rendered, keeping the vacuoles hollow, thus revealing the dense content of the vacuole at the top. This representation also shows that the CPV is opened to the cytosol (red arrow). The other 3D model in (E–F) shows contacts RER-CPV (red arrows in E, this area corresponds to the upper part of the model in (D)) and the “cytosolic macromolecules” (grey) filling the gap between the RER and the CPV (F, corresponding to the lower part of the model in (D)). This model was prepared as that in (D), but without nonlinear diffusion, to show the complex material surrounding the CPV. Bars, 200 nm in A–D; 100 nm in E and F.
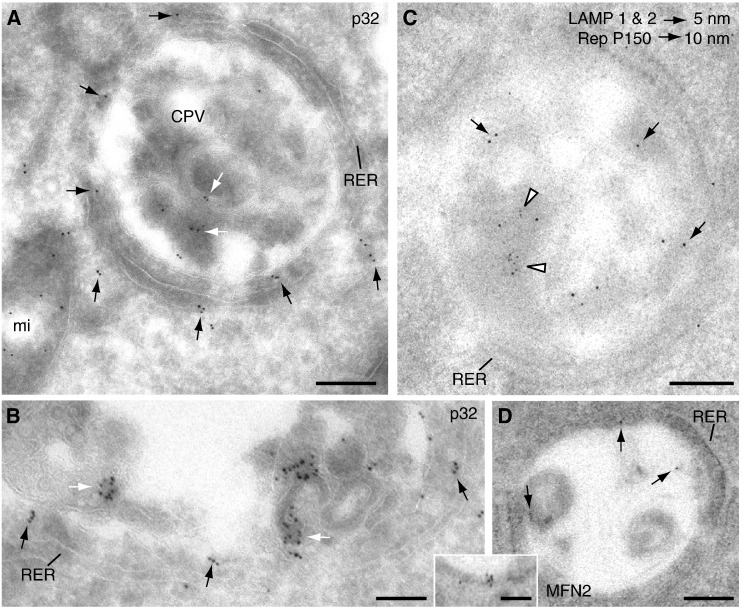
Immunogold labelling of CPVs with antibodies specific for the RUBV replicase, dsRNA, lysosomal markers LAMP 1 and LAMP 2 and the mitochondrial protein p32 was performed on cryosections of stably transfected cells. The procedure was complicated by the fact that the content of the CPVs was particularly delicate and difficult to preserve in cryosections even with glutaraldehyde fixation, something already observed for CPVs assembled by alphaviruses (Kujala et al., 2001). Probably because of that we have obtained weak to moderate signals associated to replicase, lysosomal markers and dsRNA in internal membranes of the CPVs, similar to those obtained with resin sections in a previous study by our group (Fontana et al., 2007). However, labelling on cryosections recovered more intense signals for the mitochondrial protein p32 (Fig. 6A), a protein that is incorporated into CPVs and hypothesized to participate in RUBV RNA replication. In addition to mitochondria, this antibody labelled the interior of the CPVs (Fig. 6B, white arrows) as well as their periphery (Fig. 6B, black arrows). The fuzzy material between mitochondria and CPV observed in the computational sections from tomograms (Fig. 3A) could be p32 as suggested by immunogold labelling results. Although this is difficult to quantify in thin sections, we have observed that mitochondria close to CPVs seem to have reduced signals of matrix protein p32 than mitochondria far away from viral factories. A similar observation was reported recently in RUBV- infected cells where the levels of immunogold labelled mitochondrial matrix protein p32 were reduced when compared with mock-infected cells (Ilkow et al., 2010). Also our own previous observations showed that immunolabelled p32 was more abundant in the cytosol of cells transfected with RUBV replicons when compared with non-transfected cells (Fontana et al., 2007 and our unpublished results). Thus signalling from RUBV or RUBV replicons seems to cause the release of p32 protein from the mitochondria into the cytosol. A significant amount of this protein incorporates into CPVs, where labelling signal is clearly associated to their internal membranes (Fig. 6B). An interesting feature is the significant presence of gold markers in the gap between RER and CPVs (Fig. 6A and B, black arrows) where particles are regularly spaced in the gap rather than accumulated in particular zones. Double immunogold labelling showed weak signals associated to RUBV P150 replicase component and lysosomal LAMP 1 and LAMP 2 proteins (Fig. 6C) and on the CPV's peripheral membrane when using an antibody specific for the mitochondrial protein mitofusin 2 (MFN2) (Fig 6D, main field and inset). In addition, arrays of filaments surrounding the factory were strongly labelled with this antibody (not shown).
Fig. 6.
Immunogold labeling of CPVs. (A and B) correspond to Tokuyasu cryosections while (C and D) are Lowicryl resin sections. (A) Labeling with an antibody specific for the mitochondrial protein p32 showing signals in the CPVs' internal membranes (white arrows) and in the gap between the CPV and the RER (black arrows). (B) Higher magnification view of a CPV labeled with anti-p32 antibodies showing intense signals in CPV's internal membranes (white arrows) and around the CPV (black arrows). (C) Double labeling with a mixture of two mouse monoclonal antibodies specific for lysosomal markers (LAMP1 and LAMP2, white arrowheads) and a rabbit polyclonal antibody specific for P150 protein, one of the two components of the RUBV replicase complex (black arrows). Labeling concentrates in the interior of CPVs. (D) Immunogold labelling with an antibody specific for Mitofusin 2 (MFN2) (mainfield and inset). A weak signal is observed on the internal periphery of CPVs (arrows). Bars, 200 nm in A, C and D; 100 nm in B and inset in H.
Discussion
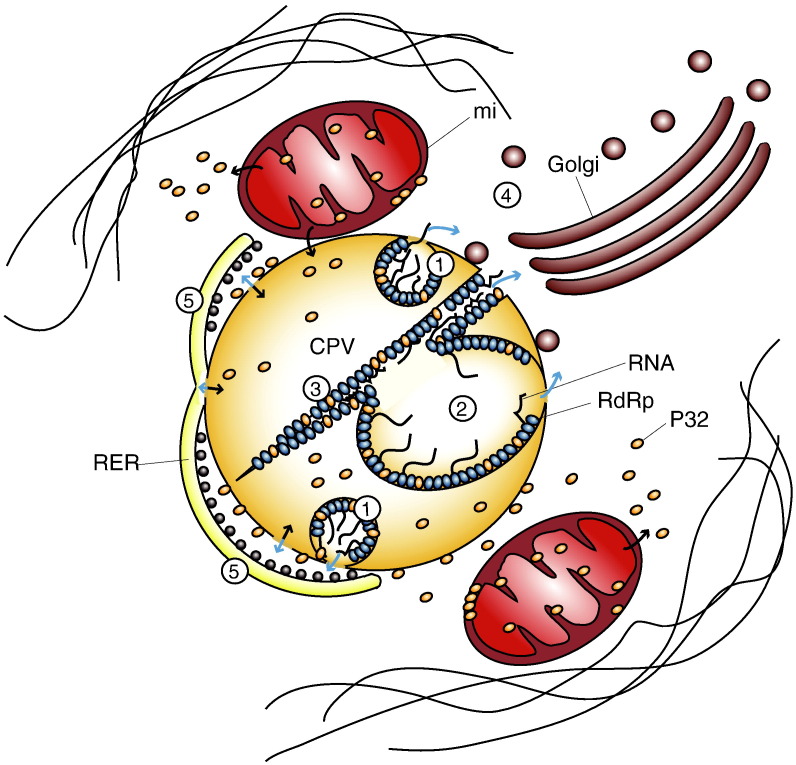
Our model for RUBV factory is shown in Fig. 7 . Similar to previous models, we propose that RNA synthesis occurs on vesicular membranes within the CPV which are in communication with the cytosol (1). The viral replicase molecules are associated with vesicles that grow with time into large vacuoles (2) and straight elements (3). This is supported by immunogold detection of replicase and dsRNA (Fontana et al., 2007, Lee et al., 1994, Magliano et al., 1998). Recruitment of RER, Golgi, and mitochondria forms the whole factory (Fontana et al., 2007), allowing for immediate contact of the newly synthesized genome RNA and SG RNA with the Golgi (4) and RER (5), respectively, for encapsidation and protein synthesis, as previously reported (Froshauer et al., 1988, Risco et al., 2003). We speculate that large vacuoles could store the replicated viral RNA and that capsid molecules would remove RNA molecules to transfer them to the assembly sites in Golgi membranes. As shown here, organelles recruited around the CPV interact through well-defined protein bridges (potential mitofusin-like proteins), closely apposed membranes (RER and Golgi) or fuzzy material (mitochondria). Immunogold labeling shows that the mitochondrial protein p32 is present in the material that fills the gap between RER and CPV and inside the CPV. It is tempting to speculate that p32 could participate in the activities of the factory by playing structural or regulatory functions. The incorporation of endocytosed BSA-gold particles demonstrates that in spite of the multiple interactions with recruited organelles, these drastically modified lysosomes maintain their communication with the plasma membrane through the endo-lysosomal pathway. Filaments around the structure resemble those seen in vimentin cages, described for factories of DNA viruses that build large factories in the cytosol.
Fig. 7.
A model for RUVB factory organization. CPVs and recruited organelles build functional factories that would work as follows: Viral RNA would replicate in vesicles (1), vacuoles (2) and rigid membranes (3). Newly synthesized RNA molecules would be transported to the Golgi as RNPs (ribonucleoproteins) with the help of capsid protein molecules for the assembly of new viruses (4) and to the RER for the synthesis of new viral proteins (5). Recruited mitochondria will provide energy and co-factors such as p32 while cytoskeletal elements would build a cage-like structure around the viral factory. Blue arrows: movements of RNA/RNPs. Black arrows: movements of proteins. CPV, cytopathic vacuole; RdRp, RNA dependent- RNA polymerase; RER, rough endoplasmic reticulum; mi, mitochondria. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Adding the third dimension to our structural studies is contributing substantially to revisions of current models for viral factory construction and virus morphogenesis. Electron tomography is having a great impact in cell biology and this 3D imaging technology is being also applied successfully to the study of virus-cell interactions. Interesting examples include the first study of the replication complex of an RNA virus by electron tomography that showed the organization of RC-containing spherules in mitochondria and their open connections to the cytoplasm (Kopek et al., 2007), the study of Dengue virus replication and assembly sites whose architecture could explain the coordination of distinct steps of the flavivirus replication cycle (Welsch et al., 2009) or the extensive virus-induced reorganization of host cell membranes into a network that is used to compartmentalize viral replication of SARS-coronavirus (Knoops et al., 2008). ET is also providing new insights into the structure and morphogenesis of viruses, such as herpes simplex virus, the first virus studied by ET (Grünewald et al., 2003), vaccinia (Chlanda et al., 2009, Cyrklaff et al., 2005), HIV (Carlson et al., 2008), or the giant mimivirus (Zauberman et al., 2008).
Our present study on the 3D architecture of RUBV factories has shown a variety of interactions between the CPV, a structure that harbors the viral replication complexes, and the extensively recruited organelles. Freeze-fracture showed interesting details of the variety of membranes inside CPVs while electron tomography revealed the existence of physical contacts between organelles and between membranes inside the CPVs as well as the presence of pores on the CPV's surface. With an estimated resolution of 3–4 nm, our tomograms can distinguish one or two membranes where organelles contact the CPV and we have not been able to detect the existence of membrane fusion in those areas where organelles interact. The necessary exchange of molecules between CPV and RER, that is subgenomic RNA towards the RER and newly synthesized proteins towards the CPV, must be facilitated by the apposed membranes and protein bridges that we have also described. Particularly interesting are the protein bridges whose width (~ 10–15 nm) is compatible with the size described for macromolecular complexes involving mitofusins (Csordás et al., 2006, Merkwirth & Langer, 2008). These dynamin-related proteins are thought to stabilize interactions between mitochondria and to be involved in tethering mitochondria with RER (de Brito & Scorrano, 2008, Ishihara et al., 2004) and it has been proposed that mitofusins could participate in co-localising other organelles as well (Parekh, 2009). Interestingly there is a great similarity between the grape-like clusters of mitochondria near the nucleus of cells over-expressing mitofusins (Santel et al., 2003) and the perinuclear clustering of mitochondria that is one of the hallmarks in RUBV-infected cells (Beatch et al., 2005) and viral factories in general (Novoa et al., 2005).
While mitofusins could have been brought to the factory passively as components of recruited organelles, mitochondrial p32 clearly represents a different case: its massive recruitment into CPVs strongly suggests a function. Considering that we are labelling the surface of cryosections, that are single planes of a big structure, the signals associated to p32 suggest that a significant amount of the molecules inside and around CPVs and visualized by ET correspond to this mitochondrial protein. Interactions between p32 and other virus proteins are known to be important for virus replication (reviewed in Ghebrehiwet et al., 2001). Binding of p32 to RUBV capsid appears to be important for transcription of the RUBV subgenomic RNA while overexpression of p32 enhances replication of RUBV by an unknown mechanism (Beatch et al., 2005, Mohan et al., 2002). P32 was originally identified as a protein that co-purified with alternative splicing factors but subsequent studies showed that it interacts with a wide variety of cellular and viral proteins that are not involved in splicing (Ghebrehiwet et al., 2001). Among its various cellular functions, p32 is known to participate in a number of apoptotic pathways. Although the bulk of this protein resides in the matrix of mitochondria p32 cycles between mitochondria and the nucleus and in most cases, interactions between virus-encoded proteins and p32 occur at non-mitochondrial sites. Immunogold and EM studies of RUBV-infected cells and of cells transfected with replicons showed that p32 seems to exit mitochondria to accumulate in the cytosol (Fontana et al., 2007, Ilkow et al., 2010) and as shown here, to be targeted to CPVs. Our results suggest that p32 could be a structural component of the RUBV factory since it is translocated into the CPVs in the absence of capsid. Further studies will be necessary to confirm this hypothesis.
While Golgi stacks touch RUBV CPVs at very discrete zones, most likely to receive new RNPs for virus assembly, mitochondria do not seem to touch the surface of CPVs. We have not been able to detect tight contacts between mitochondria and CPVs by ET although we cannot discard that they could establish interactions that are short lasting. Functional connections between mitochondria and lysosomes have been documented (Kiselyov and Muallem, 2008) but to our knowledge no specific tethering proteins have been described to physically connect these organelles in cells.
The presence of filaments around RUBV factories could be a consequence of their pre-existing interaction with lysosomes (Styers et al., 2004). Alternatively, they could have been dragged to the factory because of their binding to recruited mitochondria or Golgi (Tang et al., 2008). However, they could be also related to vimentin cages, known to participate in the construction of viral factories built by some DNA viruses that use the aggresome pathway (Netherton et al., 2007, Stefanovic et al., 2005). Although the formation of vimentin cages has not been reported for RNA viruses, this possibility cannot be excluded.
In spite of the many organelles surrounding and interacting with the CPV its communication with the endocytic pathway is maintained since these structures incorporate endocytosed BSA-gold particles. This result is consistent with a flow of RCs between the cell surface and CPVs as proposed for alphaviruses: short BSA-gold pulses in cells infected with Semliki Forest Virus (SFV) revealed that CPVs were in continuous contact with the medium even late in infection (Kujala et al., 2001). These results suggested that the RC-containing spherules arise by primary assembly of the RNA replication complexes at the plasma membrane. Endosomal recycling and fusion with CPV can circulate spherules between the plasma membrane and the endosomal–lysosomal compartment (Kujala et al., 2001). This has been confirmed in a recent work with the first detailed description of the sequential stages of alphavirus RC transport (Spuul et al., 2010). In the case of RUBV immunolocalization of dsRNA by light microscopy has revealed signals in CPVs but also in additional locations such as the cell periphery (Fontana et al., 2007, Matthews et al., 2009) and we have observed characteristic spherules on the surface of RUBV-infected cells studied by thin-section electron microscopy (unpublished results). These data are consistent with movements of RUBV RCs in cells like those described for alphaviruses. CPVs could then maintain communication with the endocytic pathway in order to incorporate new RCs from their primary assembly site, the plasma membrane.
The analysis of hundreds of RUBV factories in thin-sections, by freeze-fracture and electron tomography revealed one of the most interesting features of these structures: the complexity of the CPV's interior. Internal membranes do not seem to be just an accumulation of vesicles but they are well organized: small vesicles are preferentially located on the periphery, while internal rigid sheets usually have one or two vacuoles attached. Membranes seem to be interconnected and exhibit pores open to the cytosol. From our previous studies (Fontana et al., 2007) we know that the viral replicase is present in all these membranes and we have been looking for some internal order in them, feature sporadically suggested by thin-sectioning (Fig. 4C). The formation of 2D arrays of viral polymerases has been previously proposed for polioviruses (Lyle et al., 2002, Spagnolo et al., 2010). We have been looking for higher resolution views of potential polymerase molecules arrays in membranes inside CPVs, in particular in rigid sheets. However we have not been able to visualize a clear order in these membranes by freeze-etching and electron tomography. Nowadays it is becoming evident that new methods for macromolecular mapping of 3D volumes are needed. Together with many other groups we have tested immunogold to study viruses, cells and macromolecular complexes and we know about their usefulness and limitations (Hayat, 2002, Peters & Pierson, 2008, Risco & Pinto da Silva, 1991, Risco & Pinto da Silva, 1995, Risco et al., 1995, Risco et al., 2002, Severs & Robenek, 2008). When using antibodies a variable percentage of molecules is detected depending on antigen preservation and protein interactions in vivo. Further studies will use a clonable metallothionein tag for EM (Diestra et al., 2009a, Diestra et al., 2009b) to locate the polymerase molecules in membranes unequivocally.
Materials and methods
Cells and replicons
BHK-21 cells supplied by the American Type Culture Collection (ATCC) and stably-transfected with the RUBrep/GFP/neo replicon were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% FCS (Reactiva S.A., Barcelona, Spain) and 1.2 mg/ml of geneticin (Gibco). RUBrep/GFP/neo replicon as well as in vitro transcription and transfection protocols have been previously described (Fontana et al., 2007).
Treatment with BSA-gold and conventional embedding
Control and transfected cells were incubated 3 h with medium containing 15 μg/ml of BSA-gold (Biocell International, Cardiff, UK) and chemically fixed with a mixture of 2% glutaraldehyde and 2% tannic acid in 0.4 M Hepes buffer (pH 7.4), post-fixed 1 h at 4 °C with a mixture of 1% osmium tetroxide and 0.8% potassium ferricyanide in distilled water and incubated 1 h more with 2% uranyl acetate in distilled water also at 4 °C. Dehydration was done with increasing concentrations of acetone (50%, 70%, 90%, and 100%, 10 min each) at 4 °C. Infiltration in the epoxy-resin EML-812 (Taab Laboratories, UK) was done at room temperature for 1 day. Samples were then polymerized 3 days at 60 °C. Ultra-thin (~ 50 nm) sections were obtained in a Leica EM UC6 ultramicrotome. After staining 30 min with saturated uranyl acetate and 2 min with lead citrate, samples were studied in a JEOL JEM-1011 (Jeol, Japan) electron microscope operating at 100 kV.
High pressure freezing and freeze-substitution
We have tested several protocols for an optimal procedure to process cultured cell monolayers prior to ET. While there are other reported protocols for this approach, our final procedure focuses on: (1) avoiding the use of cryoprotectants during HPF; (2) minimizing the use of chemical fixatives during FS and the deleterious influence of the solvent on ultra-structural preservation by employing 0.5 % UA in acetone as the freeze-substitution (FS) medium; (3) attaining maximum contrast of the sections during tilt series acquisition by embedding samples in the transparent resin Lowicryl HM23; (4) seeking maximum stability of the sections while avoiding plastic coated grids by collecting sections on holey carbon grids; and (5) avoiding section staining. Thus, cellular monolayers were trypsinized 5 min at 37 °C following standard procedures with 0.25% trypsin supplemented with 0.02% EDTA. Cell suspensions were collected using Dulbecco's modified Eagle's medium supplemented with 10% FCS and centrifuged in a table-top centrifuge (1,400 r.p.m., 5 min at 4 °C) to obtain a pellet. This pellet was loaded into flat specimen carriers using only the medium as carrier filler and frozen in the Leica EMPact HPF unit. Once cryofixed, cells were maintained in liquid nitrogen.
For FS, frozen cells were transferred to the AFS (automatic freeze substitution) unit (Leica Microsystems, Vienna) precooled at − 160 °C, and warmed to − 90 °C at 20 °C/h. FS was carried out for 24 h at this temperature employing: (1) 1% OsO4, 0.25% glutaraldehyde in acetone to analyze the quality of vitrification and sample preservation or (2) 0.5% uranyl acetate (UA) in acetone for electron tomography. UA was dissolved in acetone as described by Hawes et al. (2007) Samples substituted in medium 1 were warmed to − 30 °C at 1 °C/h and then transferred to ice. After three washes in acetone, infiltration was carried out in crescent concentrations of epoxy resin EML-812 diluted in acetone (acetone:EML-812 3:1, 1:1 and 1:3) and finally in pure EML-812. Washing in acetone was carried out at 4 °C and resin infiltration at 22 °C. Polymerization was done at 60 °C for 72 h. Samples substituted in medium 2 were warmed to − 40 °C at 20 °C/h and washed twice with pre-cooled ethanol. Infiltration was carried out at − 40 °C in increasing concentrations of the acrylic resin HM23 diluted in ethanol (ethanol:HM23 3:1, 1:1 and 1:3), and finally in pure HM23. After infiltration, polymerization was done with UV light for 48 h at − 40 °C and for another 48 h at room temperature (temperature was raised at 10 °C/h). After polymerization carriers were removed by immersion in liquid nitrogen and samples were sectioned in a Leica Ultracut EM-UC6 cryoultramicrotome (Leica Microsystems, Vienna) operating at room temperature. Ultra-thin (~ 50 nm) or semi-thick (~ 200–300 nm) sections were collected on uncoated 300 mesh copper grids (Taab Laboratories, UK) or on holey carbon Quantifoil® grids (R 3.5/1 Cu/Rh, Quantifoil Micro Tools, Jena, Germany) and studied by electron microscopy. Optimal freezing in the whole cell volume was assessed by studying all cell planes in oriented ultra-thin serial sections collected on Formvar-coated, parallel-bar copper grids (Taab Laboratories, UK) and stained with saturated uranyl acetate for 30 min and 2 min more with lead citrate. For electron tomography, semi-thick (~ 250–300 nm) sections were collected on Quantifoil® grids and used without staining.
Freeze fracture, etching and metal replication
Freeze-etching of BHK-21 cells transfected with RUBrep/GFP/neo replicon was done as described in Cabezas and Risco (2006) Tantalum/Wolfram (Ta/W) was used instead of Platinum/carbon (Pt/C) to make the metal replicas of exposed surfaces because it is known that Ta forms thinner layers of fine grain showing a detectable increase in resolution of a variety of biological structures (Cabezas and Risco, 2006). Briefly, cells where fixed with 1% glutaraldehyde in PBS, cryo-protected with glycerol and fast-frozen in liquid ethane. Samples where then transferred to a Bal Tec BAF 060 unit (Leica, Vienna) precooled at − 150 °C with a vacuum of 10− 7 mbar and fractured under these conditions. Etching (15 min at − 90 °C) was done to sublimate the layer of ice from the surface exposed by the fracture and gain more detail on membrane surfaces. Shadowing at 45º with a 2-nm layer of tantalum (evaporated at 1.85 kV and 110 mA) and at 90º with a 20 nm layer of carbon (evaporated at 1.85 kV and 80 mA) then followed. Replicas were floated in commercial bleach and maintained overnight before intensive washing in distilled water. They were finally picked up in uncoated copper EM grids of 400 mesh (Taab Laboratories, UK) and dried at room temperature before electron microscopy. Ta/W wires and carbon rods were supplied by Bal Tec.
Immunogold labelling
Cryosections obtained by the standard Tokuyasu method (Salanueva et al., 2003, Fontana et al., 2008) and ultra-thin Lowicryl HM23 resin sections obtained after HPF and FS as described in this paper were both used for immunogold labeling and EM. For cryosectioning cells were fixed with 4% paraformaldehyde with or without 0.1% glutaraldehyde in PBS, washed with 0.15 M glycine in PBS and embedded in 10% gelatine in PBS prior to their infiltration overnight in 2.3 M sucrose. Cells were then vitrified in liquid nitrogen and sectioned in a cryoultramicrotome Leica EM UC6/FC6 (Leica, Vienna) operating at − 120 °C. The sections were retrieved in formvar coated gold EM grids of 300 mesh (Taab Laboratories, UK) employing a mixture of 2.3 M sucrose and 2% methyl cellulose (1:1). For immunogold labeling on cryosections, grids were incubated 20 min at 37 °C in 2% gelatine, then in 50 mM glycine in PBS (4 × 3 min), in 5 % BSA in PBS, and finally in saturation buffer (1% BSA in PBS) (2 × 5 min). Antibodies and gold conjugates were diluted in saturation buffer as follows: the rabbit polyclonal anti-p32 antibody, a kind gift of Dr W.C. Russell (University of St Andrews, Scotland), was diluted 1:100; the rabbit polyclonal anti-Mitofusin 2 antiserum (Abcam, Cambridge, UK) was diluted 1:200; The GU3 rabbit polyclonal antiserum specific for the RUBV P150 protein (Fontana et al., 2007) was diluted 1:100; The mouse monoclonal antibodies specific for the lysosomal proteins LAMP 1 (UH1) and LAMP 2 (UH3) were supplied by the DSHB (Developmental Studies Hybridoma Bank at the University of Iowa) and diluted 1:100; the 10 nm protein A-gold conjugate (BioCell International, Cardiff, UK) was diluted 1:90, and secondary antibodies (goat anti-mouse and goat anti-rabbit) conjugated with 5 or 10 nm colloidal gold particles were diluted 1:40; Samples were incubated 1 h with primary antibodies, washed with 0.1% BSA in PBS (7 × 2 min) and then incubated 1 h with protein A conjugated with colloidal gold. For double immunogold labelling samples were incubated with a mixture of primary antibodies followed by a second incubation with a mixture of secondary antibodies. After washing with 0.1 % BSA in PBS (7 × 2 min) grids where finally post-fixed 5 min with 1% glutaraldehyde in PBS, washed in distilled water (7 × 2 min) and stained with 2% methyl cellulose-saturated uranyl acetate (9:1) for 10 min. Immunogold labeling on resin sections was done following standard procedures as described by Fontana et al. (2008).
Electron tomography and image processing
Semi-thick (~ 200–300 nm) sections collected on 300 mesh Quantifoil® holey carbon coated grids were studied without post-staining in a FEI TecnaiG2 FEG200 (FEI company, Eindhoven, The Netherlands) electron microscope operating at 200 kv using a Gatan side-entry cryo-holder. Single axis tilt series were obtained following a Saxton scheme with angle ranges between − 70° and + 70º and an angular interval of 1.35º. The nominal defocus employed was − 3 μm. The images were recorded on an Eagle 4 k × 4 k slow-scan charge-coupled device (FEI Company) using FEI software. Recorded images were binned to 1024 × 1024 or to 2048 × 2048 pixels (1.2 and 0.6 nm/pixel, respectively). The images were aligned and the tomograms reconstructed using the IMOD software package (Kremer et al., 1996). The resolution of the tomograms was around 3 nm, as estimated with the Bsoft software (Heymann et al., 2008) using the FSCe/o criterion (Cardone et al., 2005). In order to facilitate their interpretation, some tomograms were subjected to noise filtering with the iterative median filter (van der Heide et al., 2007). The 3D models of the viral factories were prepared by a combination of masking and isosurface, following a strategy similar to the one described by Cyrklaff et al. (2005) and manual tracing. In some cases, strong filtering with nonlinear diffusion (Fernández, 2009) was applied to the volume in order to remove noise and fine details, and clearly show the global shape of the CPV and its connections to the cytosol and organelles. On the other hand, manual tracing was particularly useful to highlight the boundaries of the CPV and reveal its internal content (vacuoles, sheets). Segmentation and 3D visualization were done with AMIRA (TGS Europe, Merignac, France). Only the elements with unequivocal identity were included in the final 3D model.
Acknowledgments
We thank Javier Chichón and Elisenda Coll for assistance with electron tomography acquisition, Pilar Cabezas and Gloria Calderita for technical assistance with freeze-fracture and BSA-gold studies and Sonia Ruíz and Gema Martínez for technical support with freeze-substitution and cryosectioning. The polyclonal anti-p32 antibody was kindly provided by Dr W.C. Russell (University of St Andrews, Scotland). This work has been supported by grants BFU2006-04584/BMC and BIO2009-07255 from the Ministry of Science and Innovation of Spain (to C.R.) and by grants MCI-TIN2008-01117 and CSIC-PIE200920I075 (to J.J.F.) J. F. has been recipient of a contract from the Comunidad de Madrid.
Contributor Information
José J. Fernández, Email: jj.fernandez@cnb.csic.es.
Cristina Risco, Email: crisco@cnb.csic.es.
References
- Barajas D., Jiang J., Nagy P.D. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 2009;5(12):e1000705. doi: 10.1371/journal.ppat.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatch M.D., Everitt J.C., Law L.J., Hobman T.C. Interactions between Rubella virus capsid and host protein p32 are important for virus replication. J. Virol. 2005;79:10807–10820. doi: 10.1128/JVI.79.16.10807-10820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas P., Risco C. Studying cellular architecture in three dimensions with improved resolution: Ta replicas revisited. Cell Biol. Int. 2006;30:747–754. doi: 10.1016/j.cellbi.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Cardone G., Grünewald K., Steven A.C. A resolution criterion for electron tomography based on cross-validation. J. Struct. Biol. 2005;151:117–129. doi: 10.1016/j.jsb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Carlson L.-A., Briggs J.A.G., Glass B., Riches J.D., Simon M.N., Johnson M.C., Müller B., Grünewald K., Kräusslich H.-G. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlanda P., Carbajal M.A., Cyrklaff M., Griffiths G., Krijnse-Locker J. Membrane rupture generates single open membrane sheets during vaccinia virus assembly. Cell Host Microbe. 2009;6:81–90. doi: 10.1016/j.chom.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K.F., Balla T., Mannella C.A., Hajnóczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyrklaff M., Risco C., Fernández J.J., Jimenez M.V., Esteban M., Baumeister W., Carrascosa J.L. Cryo-electron tomography of vaccinia virus. Proc. Nat. Acad. Sci. U.S.A. 2005;102:2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito O.M., Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–611. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Diestra E., Fontana J., Guichard P., Marco S., Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J. Struct. Biol. 2009;165:157–168. doi: 10.1016/j.jsb.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Diestra E., Cayrol B., Arluison V., Risco C. Cellular electron microscopy imaging reveals the localization of the Hfq protein close to the bacterial membrane. PLoS ONE. 2009;4:e8301. doi: 10.1371/journal.pone.0008301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B.T., Miller D.J., Ahlquist P. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J. Virol. 2005;79:8909–8919. doi: 10.1128/JVI.79.14.8909-8919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández J.J. Tomobflow: feature-preserving noise filtering for electron tomography. BMC Bioinform. 2009;10:178. doi: 10.1186/1471-2105-10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala J.C. Reconstruct: a free editor for serial section microscopy. J. Microsc. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- Fontana J., Tzeng W.P., Calderita G., Fraile-Ramos A., Frey T.K., Risco C. Novel replication complex architecture in rubella replicon-transfected cells. Cell. Microbiol. 2007;9:875–890. doi: 10.1111/j.1462-5822.2006.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J., Lopez-Montero N., Elliott R.M., Fernandez J.J., Risco C. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell. Microbiol. 2008;10:2012–2028. doi: 10.1111/j.1462-5822.2008.01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey T.K. Molecular biology of rubella virus. Adv. Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froshauer S., Kartenbeck J., Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebrehiwet B., Lim B.L., Kumar R., Feng X., Peerschke E.I. gC1q-R/p33, a member of a new class of multifunctional and multicompartmental cellular proteins, is involved in inflammation and infection. Immunol. Rev. 2001;180:65–77. doi: 10.1034/j.1600-065x.2001.1800106.x. [DOI] [PubMed] [Google Scholar]
- Goff S.P. Knockdown screens to knockout HIV-1. Cell. 2008;135:417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griemley P.M., Levin J.G., Berezesky I.K., Friedman R.M. Specific membranous structures associated with the replication of group A arboviruses. J. Virol. 1972;10:492–503. doi: 10.1128/jvi.10.3.492-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald K., Desai P., Winkler D.C., Heymann J.B., Belnap M., Baumeister W., Steven A.C. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- Hawes P., Netherton C.L., Mueller M., Wileman T., Monaghan P. Rapid freeze-substitution preserves membranes in high-pressure frozen tissue culture cells. J. Microsc. 2007;226:182–189. doi: 10.1111/j.1365-2818.2007.01767.x. [DOI] [PubMed] [Google Scholar]
- Hayat M.A. Microscopy, Immunohistochemistry, and Antigen Retrieval Methods for Light and Electron Microscopy. first ed. Kluwer Academic/Plenum Publishers; New York: 2002. [Google Scholar]
- Heymann J.B., Cardone G., Winkler D.C., Steven A.C. Computational resources for cryo-electron tomography in Bsoft. J. Struct. Biol. 2008;161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilkow C.S., Weckbecker D., Cho W.J., Meier S., Beatch M.D., Goping I.S., Herrmann J.M., Hobman T.C. The Rubella virus capsid protein inhibits mitochondrial import. J. Virol. 2010;84:119–130. doi: 10.1128/JVI.01348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Eura Y., Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- Kiselyov K., Muallem S. Mitochondrial Ca2+ homeostasis in lysosomal storage diseases. Cell Calcium. 2008;44:103–111. doi: 10.1016/j.ceca.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijmeer M.J., Morkowski S., Griffith J.M., Rudensky A.Y., Geuze H.J. Major histocompatibility complex class II compartments in human and mouse B lymphoblasts represent conventional endocytic compartments. J. Cell Biol. 1997;139:639–649. doi: 10.1083/jcb.139.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., van den Worm S.H.E., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek B.G., Perkins G., Millar D.J., Ellisman M.H., Ahlquist P. Three-dimensional analysis of a viral replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5(9):e220. doi: 10.1371/journal.pbio.0050220. doi:10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kujala P., Ikäheimonen A., Ehsani N., Vihinen H., Auvinen P., Kääriäinen L. Biogenesis of the Semliki Forest virus RNA replication complexes. J. Virol. 2001;75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., Marshall J.A., Bowden D.S. Characterization of Rubella virus replication complexes using antibodies to double-stranded RNA. Virology. 1994;200:307–312. doi: 10.1006/viro.1994.1192. [DOI] [PubMed] [Google Scholar]
- Lucić V., Förster F., Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Lyle J.M., Bullitt E., Bienz K., Kirkegaard K. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science. 2002;296:2218–2222. doi: 10.1126/science.1070585. [DOI] [PubMed] [Google Scholar]
- Mackenzie J. Wrapping things up about virus RNA replication. Traffic. 2005;6:967–977. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliano D., Marshall J.A., Bowden D.S., Vardaxis S., Meanger J., Lee J.-Y. Rubella virus replication complexes are virus modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- Matthews J.D., Tzeng W.-P., Frey T.K. Determinants of subcellular localization of the rubella virus non-structural replicase proteins. Virology. 2009;390:315–323. doi: 10.1016/j.virol.2009.05.019. [DOI] [PubMed] [Google Scholar]
- McIntosh R., Nicastro D., Mastronarde D. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 2005;15:43–51. doi: 10.1016/j.tcb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Merkwirth C., Langer T. Mitofusin 2 builds a bridge between ER and mitochondria. Cell. 2008;135:1165–1167. doi: 10.1016/j.cell.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan K.V., Ghebrehiwet B., Atreya C.D. The N-terminal conserved domain of rubella virus capsid interacts with the C-terminal region of cellular p32 and overexpression of p32 enhances the viral infectivity. Virus Res. 2002;85:151–161. doi: 10.1016/s0168-1702(02)00030-8. [DOI] [PubMed] [Google Scholar]
- Nagy P.D. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 2008;46:217–242. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed] [Google Scholar]
- Netherton C., Moffat K., Brooks E., Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa R.R., Calderita G., Arranz R., Fontana J., Granzow H., Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortin J., Parra F. Structure and function of RNA replication. Annu. Rev. Microbiol. 2006;60:305–326. doi: 10.1146/annurev.micro.60.080805.142248. [DOI] [PubMed] [Google Scholar]
- Parekh A. Calcium signalling: mitofusins promote interorganellar crosstalk. Curr. Biol. 2009;19:R200–R203. doi: 10.1016/j.cub.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Peters P.J., Pierson J. Immunogold labelling of thawed cryosections. Methods Cell Biol. 2008;88:131–149. doi: 10.1016/S0091-679X(08)00408-1. [DOI] [PubMed] [Google Scholar]
- Risco C., Pinto da Silva P. Binding of bacterial endotoxins to the macrophage surface: visualization by fracture-flip and immunocytochemistry. J. Histochem. Cytochem. 1991;41:601–608. doi: 10.1177/41.4.8450199. [DOI] [PubMed] [Google Scholar]
- Risco C., Pinto da Silva P. Cellular functions during activation and damage by pathogens: immunogold studies of the interaction of bacterial endotoxins with target cells. Mic. Res. Tech. 1995;31:141–150. doi: 10.1002/jemt.1070310206. [DOI] [PubMed] [Google Scholar]
- Risco C., Pinto da Silva P. The fracture-flip technique reveals new structural features of the Escherichia coli cell wall. J. Microsc. 1998;189:213–218. doi: 10.1046/j.1365-2818.1998.00316.x. [DOI] [PubMed] [Google Scholar]
- Risco C., Menendez-Arias L., Copeland T.D., Pinto da Silva P., Oroszlan S. Intracellular transport of the murine leukemia virus during acute infection of NIH 3 T3 cells: nuclear import of nucleocapsid protein and integrase. J. Cell Sci. 1995;108:3039–3050. doi: 10.1242/jcs.108.9.3039. [DOI] [PubMed] [Google Scholar]
- Risco C., Rodríguez J.R., López-Iglesias C., Carrascosa J.L., Esteban M., Rodríguez D. Endoplasmic reticulum-Golgi intermediate compartment membranas and vimentin filaments particiate in vaccinia virus assembly. J. Virol. 2002;76:1839–1855. doi: 10.1128/JVI.76.4.1839-1855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C., Carrascosa J.L., Frey T.K. Structural maturation of rubella virus in the Golgi complex. Virology. 2003;312:261–269. doi: 10.1016/S0042-6822(03)00384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao M., Tanaka K., Sibarita J.-B., Ly-Hartig N.T.B., Tanaka T.U., Antony C. Three-dimensional electron microscopy analysis of ndc10-1 mutant reveals an aberrant organization of the mitotic spindle and spindle pole body defects in Saccharomyces cerevisiae. J. Struct. Biol. 2008;163:18–28. doi: 10.1016/j.jsb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Salanueva I.J., Novoa R.R., Cabezas P., López-Iglesias C., Carrascosa J.L., Elliott R.M., Risco C. Polymorphism and structural maturation of bunyamwera virus in Golgi and post-Golgi compartments. J. Virol. 2003;77:1368–1381. doi: 10.1128/JVI.77.2.1368-1381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A., Ahola T., Kääriäinen L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A., Frank S., Gaume B., Herrler M., Youle R.J., Fuller M.T. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J. Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- Sessions O.M., Barrows N.J., Souza-Neto J.A., Robinson T.J., Hershey C.L., Rodgers M.A., Ramirez J.L., Dimopoulos G., Yang P.L., Pearson J.L., Garcia-Blanco M.A. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severs N.J. Freeze-fracture electron microscopy. Nat. Protoc. 2007;2:547–576. doi: 10.1038/nprot.2007.55. [DOI] [PubMed] [Google Scholar]
- Severs N.J., Robenek H. Freeze-fracture cytochemistry in cell biology. Methods Cell Biol. 2008;88:181–204. doi: 10.1016/S0091-679X(08)00411-1. [DOI] [PubMed] [Google Scholar]
- Spagnolo J.F., Rossignol E., Bullitt E., Kirkegaard K. Enzymatic and nonenzymatic functions of viral RNA-dependent RNA polymerases within oligomeric arrays. RNA. 2010;16:382–393. doi: 10.1261/rna.1955410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuul, P., Balistreri, G., Kääriäinen, L., Ahola, T., 2010. PI3K-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. (in press). [DOI] [PMC free article] [PubMed]
- Stefanovic S., Windsor M., Nagata K.-i., Inagaki M., Wileman T. Vimentin rearrangement during African swine fever virus infection involves retrograde transport along microtubules and phosphorylation of vimentin by calcium calmodulin kinase II. J. Virol. 2005;79:11766–11775. doi: 10.1128/JVI.79.18.11766-11775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven A.C., Aebi U. The next ice age: cryo-electron tomography of intact cells. Trends Cell Biol. 2003;13:107–110. doi: 10.1016/s0962-8924(03)00023-0. [DOI] [PubMed] [Google Scholar]
- Studer D., Humbel B.M., Chiquet M. Electron microscopy of high-pressure frozen samples: bridging the gap between cellular ultrastructure and atomic resolution. Histochem. Cell Biol. 2008;130:877–889. doi: 10.1007/s00418-008-0500-1. [DOI] [PubMed] [Google Scholar]
- Styers M.L., Salazar G., Love R., Peden A.A., Kowalczyk A.P., Faundez V. The endo-lysosomal sorting machinery interacts with the intermediate filament cytoskeleton. Mol. Biol. Cell. 2004;15:5369–5382. doi: 10.1091/mbc.E04-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.L., Lung H.L., Wu K.C., Phan Le A.-H., Tang H.M., Fung M.C. Vimentin supports mitochondrial morphology and organization. Biochem. J. 2008;410:141–146. doi: 10.1042/BJ20071072. [DOI] [PubMed] [Google Scholar]
- van der Heide P., Xu X.P., Marsh B.J., Volkmann N. Efficient automatic noise reduction of electron tomographic reconstructions based on iterative median filtering. J. Struct. Biol. 2007;158:196–204. doi: 10.1016/j.jsb.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Hockman M.A., Staschke K., Johnson R.B., Case C.A., Lu J., Parsons S., Zhang F., Rathnachalam R., Kirkegaard K., Colacino J.M. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2002;76:3865–3872. doi: 10.1128/JVI.76.8.3865-3872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Millar S., Romero-Brey I., Merz A., Bleck C.K.E., Walter P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the Dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauberman N., Mutsafi Y., Halevy D.B., Simón E., Klein E., Xiao C., Sun S., Minsky A. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6(5):e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]