Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV), isolated from humans infected during the peak of epidemic, encodes two accessory proteins termed as 8a and 8b. Interestingly, the SARS-CoV isolated from animals contains an extra 29-nucleotide in this region such that these proteins are fused to become a single protein, 8ab. Here, we compared the cellular properties of the 8a, 8b and 8ab proteins by examining their cellular localizations and their abilities to interact with other SARS-CoV proteins. These results may suggest that the conformations of 8a and 8b are different from 8ab although nearly all the amino acids in 8a and 8b are found in 8ab. In addition, the expression of the structural protein, envelope (E), was down-regulated by 8b but not 8a or 8ab. Consequently, E was not detectable in SARS-CoV-infected cells that were expressing high levels of 8b. These findings suggest that 8b may modulate viral replication and/or pathogenesis.
Keywords: Severe acute respiratory syndrome (SARS), Coronavirus (CoV), Accessory proteins, Envelope (E) protein, 8a, 8b, 8ab
Introduction
A novel coronavirus was identified as the aetiological agent for the recent severe acute respiratory syndrome (SARS) epidemic (Drosten et al., 2003, Poon et al., 2004). In addition to the replicase polyproteins (pp1a and pp1ab) and structural proteins (spike (S), membrane (M), nucleocapsid (N) and envelope (E)), which are common to all members of the genus coronavirus, the SARS-CoV genome also encodes eight putative proteins with no significant sequence homology to viral proteins of other known coronaviruses (i.e., open reading frames (ORFs) 3a, 3b, 6, 7a, 7b, 8a, 8b and 9b) (Marra et al., 2003, Snijder et al., 2003, Tan et al., 2005). Although it was recently demonstrated that most of these so-called accessory proteins are not essential for viral replication in cell culture or in the murine model (Yount et al., 2005), the exact contributions of these proteins to viral replication or pathogenesis in the natural host have not been established.
Interestingly, epidemiological studies have revealed that the part of the viral genome that encodes for two of these accessory proteins, 8a and 8b, shows major variations. In one of these studies, Guan and co-workers (2003) analyzed SARS-CoV isolates obtained from animals in a live-market in Guangdong and found that all the animal isolates contain a 29-nucleotides (nt) sequence, which is absent in most of the human isolates (Fig. 1A). As a result of this, the ORF8a and ORF8b (also termed as ORF10 and ORF11, respectively) in the human isolates become one ORF, termed as ORF8ab. ORF8ab encodes a protein of 122 amino acids (aa), whose N terminus is identical to 8a and C terminus is identical to 8b (Fig. 1B). Another extensive study of 63 SARS-CoV isolates obtained from the SARS epidemic in China also showed that there are major variations in this region of the viral genome (The Chinese SARS Molecular Epidemiology Consortium, 2004). In this study, the course of the epidemic was divided into the early, middle and late phase with the early phase defined as the period of first emergence of SARS in Guangdong Province. The middle phase referred to all events up to the first cluster of SARS cases in the Metropole hotel in Hong Kong and the late phase referred to all cases following this cluster. Interestingly, the clustering of patients with different patterns of variations in ORF8 region was correlated with the different phases of the epidemic. These findings were subsequently verified by researchers who studied the SARS-CoV isolated in different countries (Chiu et al., 2005, Lan et al., 2005, Qin et al., 2003, Wang et al., 2004, Wang et al., 2005).
Fig. 1.
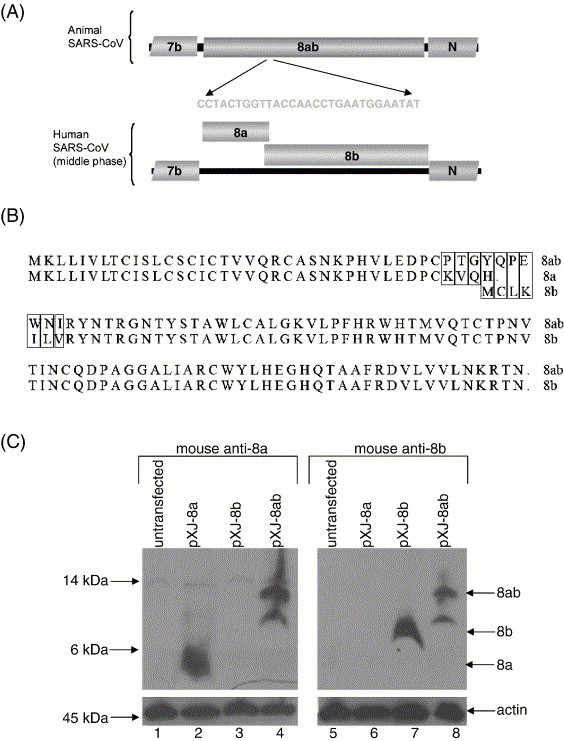
Expressions of SARS-CoV 8a, 8b and 8ab proteins. (A) Schematic diagram showing the genetic differences in the ORF8 region of the SARS-CoV isolated from animals and humans infected during the middle phase of the SARS epidemic in 2003 (modified from Guan et al., 2003). The animal isolates have an extra 29-nucleotides insertion such that the subgenomic RNA encodes for a single protein, termed 8ab, whereas that of the human isolates (from the middle phase) encodes two proteins, 8a and 8b. Human isolates from early and late phases of the epidemic also have the 29-nucleotides insertion found in the animal SARS-CoV. (B) Alignment of the sequences of 8a, 8b and 8ab proteins used in this study. Mismatches between 8a and 8ab or 8b and 8ab are boxed. The 8ab is reconstructed from a human isolate from the middle phase (SIN2774) by insertion of the 29-nucletoides found in a human isolate from the early phase (GZ02). (C) Western blot analysis was performed to detect 8a, 8b and 8ab proteins expressed in Vero E6 cells using cDNA constructs. The experiments were performed with either mouse anti-8a polyclonal antibody (upper panel, lanes 1–4) or mouse anti-8b polyclonal antibody (upper panel, lanes 5–8). Equal amounts of cells were used in each lane as verified by the level of endogenous actin (bottom panel).
Although these mutations in the ORF8 region do not appear to have any adverse effect on the survival of the virus, it is conceivable that the 8a, 8b and 8ab proteins may have different stabilities and/or functions and hence would contribute differently to viral replication and/or pathogenesis in vivo. In order to understand how the changes in the ORF8 region of the viral genome may impact on viral replication or pathogenesis, we compared the cellular properties of the human SARS-CoV 8a and 8b proteins with their counterpart, 8ab, in animal SARS-CoV. Specific antibodies were produced and used to determine the expression of 8a and 8b in SARS-CoV-infected cells. Indirect immunofluorescence and co-immunoprecipitation experiments were performed to compare the cellular localization of the 8a, 8b and 8ab proteins and their abilities to interact with other SARS-CoV proteins. Finally, the specific effects of the 8b protein on the expression of the small structural protein E in cells co-expressing 8b and E, and in SARS-CoV-infected cells were demonstrated.
Results
Polyclonal antibodies to the SARS-CoV unique proteins, 8a and 8b
For SARS-CoV isolated from humans infected in the middle phase of the epidemic, the subgenomic RNA 8 encodes two proteins, 8a and 8b, of 39 and 84 aa, respectively (Figs. 1A and B). Mouse polyclonal antibodies were raised against bacterially expressed GST-fusion 8a and 8b proteins. To determine the specificity of these antibodies, Western blot analysis was performed to detect 8a, 8b and 8ab expressed in transiently transfected Vero E6 cells. As shown in Fig. 1C, mouse anti-8a polyclonal antibody specifically detected 8a and 8ab whereas mouse anti-8b polyclonal antibody detected 8b and 8ab. 8a and 8b migrated close to their predicted molecular weight of 4.5 kDa and 9.6 kDa, respectively, whereas two forms of 8ab, ∼ 14 kDa (major) and ∼ 12 kDa (minor), were detected (Fig. 1C). As the predicted molecular weight of 8ab is 13.8 kDa, the minor form is likely to have arisen from cleavage of the full-length protein.
Expression of 8a and 8b in SARS-CoV-infected Vero E6 cells
SARS-CoV 2003VA2774, an isolate from a SARS patient in Singapore, was used to infect Vero E6 cells as previously described (Tan et al., 2004b) and anti-8a or anti-8b polyclonal antibodies were used in indirect immunofluorescence experiments to determine the expression of 8a and 8b, respectively, in infected cells (Figs. 2A and B). The 8a and 8b proteins were detected in SARS-CoV-infected cells and were found to be localized in the cytoplasm.
Fig. 2.
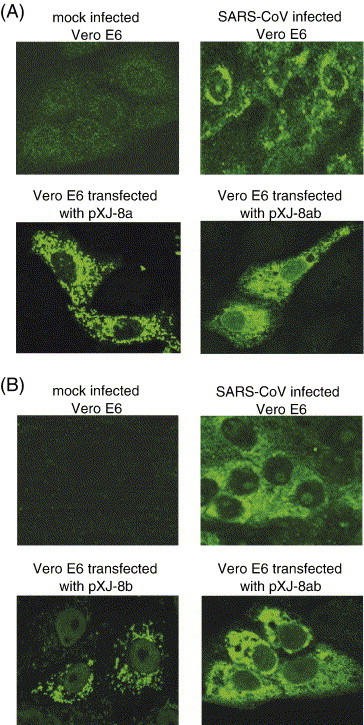
Cellular localizations of 8a and 8b in SARS-CoV-infected cells and Vero E6 cells transfected with DNA constructs for expressing 8a, 8b and 8ab. Specific mouse anti-8a and anti-8b polyclonal antibodies were used in indirect immunofluorescence experiments to determine the expressions of (A) 8a and (B) 8b, respectively. The top two panels showed the specific reactivities of the anti-8a and anti-8b antibodies to proteins expressed in SARS-CoV-infected cells (right panels) as no unspecific staining was observed for the mock-infected cells (left panels). The bottom two panels showed the reactivities of the antibodies to 8a and 8ab (A) or 8b and 8ab (B) expressed in Vero E6 by transfection of cDNA constructs.
The same antibodies were used to examine the detailed cellular localization of 8a, 8b and 8ab, expressed from DNA constructs, in Vero E6 cells (Figs. 2A and B). Whereas 8a and 8b were found in punctuate vesicle-like structures throughout the cytoplasm, 8ab was found to be diffused in the cytoplasm. Hence, there appears to be significant differences in the conformations of 8a and 8ab although 35 out of 39 aa of 8a is present in the 8ab protein (Fig. 1B). Similarly, 77 out of 84 aa of 8b is present in 8ab, but the cellular localization of 8b is distinct from 8ab.
Interaction of 8a, 8b and 8ab with other SARS-CoV proteins
In order to further characterize the cellular properties of 8a, 8b and 8ab, co-immunoprecipitation experiments were performed to determine if these proteins can interact with the SARS-CoV structural proteins, S, M, E and N, as well as two SARS-CoV accessory proteins, 3a and 7a, which were previously shown to be expressed in SARS-CoV-infected cells (Fielding et al., 2004, Tan et al., 2004b). All these proteins were not tagged except for M, where the C terminus was fused with a HA tag because of the lack of a suitable antibody for the detection of M. 8a, 8b and 8ab were fused at their C termini with a myc tag so that it is possible to compare the relative expression of the three proteins in this experiment. N-terminal-tagged myc-GST was used as a negative control. The results showed that 8a-myc interacted strongly with S, 8b-myc interacted strongly with M, E, 3a and 7a and 8ab-myc interacted strongly with S, 3a and 7a (Table 1 and Fig. 3 ). These results showed that the binding profiles of 8a, 8b and 8ab are clearly distinct, suggesting that the conformations of the 8a and 8b proteins may be quite different from the 8ab protein.
Table 1.
The interactions between SARS-CoV 8a, 8b and 8ab with other viral proteins were determined by co-immunoprecipitation experiments
| Bait proteinsa | Interacting partnersb |
|||||
|---|---|---|---|---|---|---|
| S | E | M | N | 3a | 7a | |
| 8a-myc | Strong | Weak | No | No | No | No |
| 8b-myc | No | Strong | Strong | No | Strong | Strong |
| 8ab-myc | Strong | No | Weak | No | Strong | Strong |
These proteins were immunoprecipitated using myc-polyclonal antibody and protein A-agarose.
These proteins were co-expressed with the bait proteins and co-immunoprecipitation experiments were performed to be determined if they could bind the bait proteins. The strengths of the binding were classified as strong, weak or no interaction.
Fig. 3.
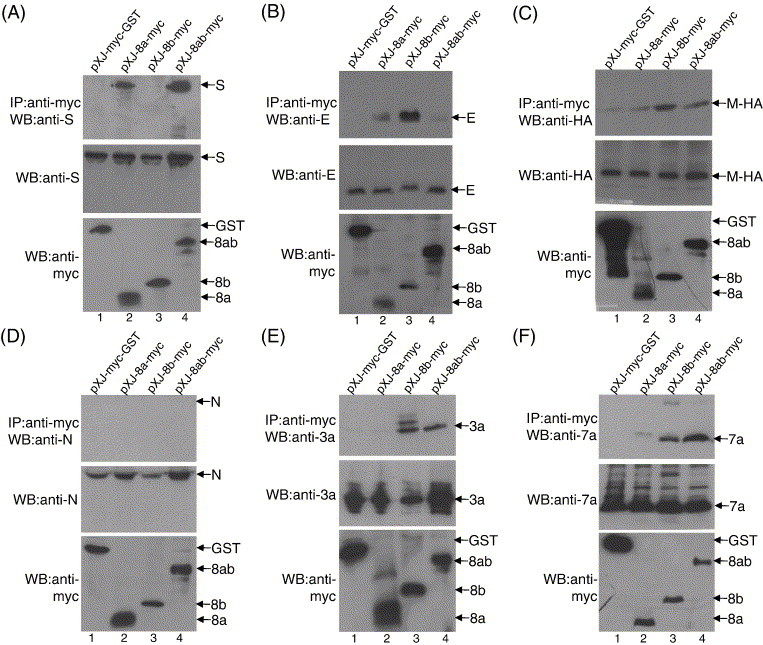
Interactions of 8a, 8b and 8ab with other SARS-CoV proteins. Cell lysates containing myc-GST (lane 1), 8a-myc (lane 2), 8b-myc (lane 3) or 8ab-myc (lane 4) and another SARS-CoV protein (S, E, M-HA, N, 3a or 7a) were immunoprecipitated with an anti-myc polyclonal antibody and protein A-agarose beads. (A) The amounts of S protein co-immunoprecipitated (IP) by the myc-tagged proteins were determined using an anti-S monoclonal antibody (top panel). The amounts of S and myc-tag proteins in the lysates before co-immunoprecipitation were also determined by Western blot (WB) with anti-S and anti-myc monoclonal antibodies, respectively (middle and bottom panels). The same experiments were performed for panels B–F except that different antibodies against the specific viral proteins were used, namely (B) anti-E mouse polyclonal; (C) anti-HA monoclonal (as the M protein fused with a HA tag at the C terminus); (D) anti-N mouse polyclonal; (E) anti-3a mouse polyclonal; (F) anti-7a mouse polyclonal.
Overexpression of 8b down-regulates the expression of E protein
While performing the co-immunoprecipitation experiments, we observed that the expression of E was significantly reduced in the presence of 8b. The co-transfections of pXJ-E and pXJ-8b-myc into 293 T cells were repeated using different amount of the pXJ-8b-myc construct. As shown in Fig. 4A, the down-regulation of E expression was specific and dependent on the expression levels of 8b-myc (lane 3 and lanes 5–10). On the contrary, the expressions of E were similar when pXJ-E was co-transfected with control plasmid (pXJ-myc-GST) (lane 1) or pXJ-8a-myc (lane 2) or pXJ-8ab-myc (lane 4).
Fig. 4.
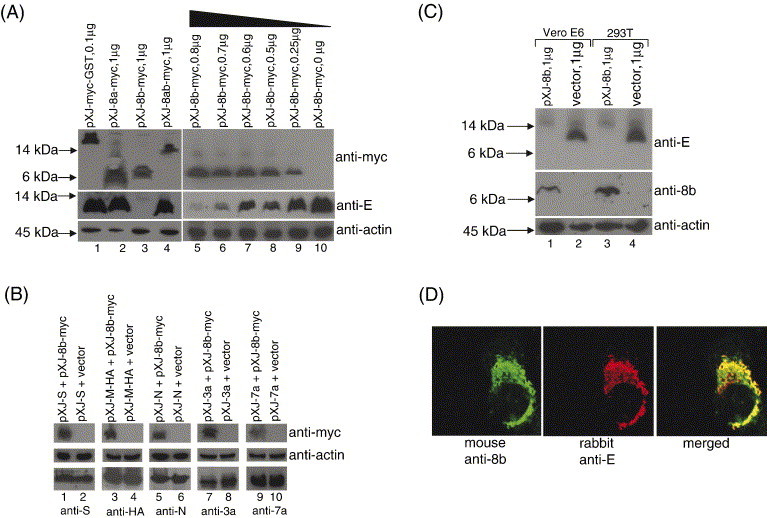
Effects of 8b on the expression of the small structural protein, E. (A) 293T cells were co-transfected with 2 μg of pXJ-E and 0.1 μg of pXJ-myc-GST (lane 1), 1 μg of pXJ-8a-myc (lane 2), pXJ-8b-myc (lane 3), pXJ-8ab-myc (lane 4), or decreasing amount of pXJ-8b-myc (lanes 5–10). Total cell lysates were subjected to Western blot analysis to determine the expression of E (middle panel) and myc-tagged proteins (top panel). Equal amounts of cells were used in each lane as verified by the level of endogenous actin (bottom panel). (B) 293T cells were co-transfected with 1 μg of pXJ-S and 1 μg of either pXJ-8b-myc or empty vector (lanes 1 and 2). Total cell lysates were subjected to Western blot analysis to determine the expression of 8b-myc (top panel) and S (lower panel). Equal amounts of cells were used in each lane as verified by the level of endogenous actin (middle panel). Similar experiments were performed with 1 μg of pXJ-M-HA (lanes 3 and 4), 0.25 μg of pXJ-N (lanes 5 and 6), 0.4 μg of pXJ-3a (lanes 7 and 8) or 0.4 μg of pXJ-7a (lanes 9 and 10). (C) Vero E6 or 293T cells were co-transfected with 2 μg of pXJ-E and 1 μg of pXJ-8b (lanes 1 and 3) or 1 μg of empty vector (lanes 2 and 4). Total cell lysates were subjected to Western blot analysis to determine the expression of E (top panel) and 8b (middle panel). Equal amounts of cells were used in each lane as verified by the level of endogenous actin (bottom panel). (D) Indirect immunofluorescence experiments were performed to determine the cellular localization of 8b and E in Vero E6 cells co-transfected with pXJ-8b and pXJ-E. The expression of 8b is represented by FITC staining (left panel), whereas the expression of E is represented by rhodamine staining (middle panel). The merged images showed that the 8b and E partially colocalized in co-transfected cells (right panel).
In order to determine if 8b has any effect on the expression of other SARS-CoV proteins, 8b was co-expressed with S (Fig. 4B, lane 1) or M-HA (Fig. 4B, lane 3) or N (Fig. 4B, lane 5) or 3a (Fig. 4B, lane 7) or 7a (Fig. 4B, lane 9). The results showed that 8b did not have any significant effect on the expression of these other viral proteins examined here. The down-regulation of E expression by 8b was also observed when untagged forms of 8b and E were co-expressed in Vero E6 and 293 T cells (Fig. 4C, lanes 1 and 3). Indirect immunofluorescence experiments also showed that E and 8b colocalized partially in Vero E6 cells (Fig. 4D).
Expression of 8b did not reduce the transcription of the E gene
In order to determine if the effect of 8b on the expression of E is due to inhibition of the transcription of the E gene, Northern blot analysis was performed to determine the mRNA level of E in the presence or absence of 8b protein. The results showed that the mRNA level of E was not decreased in 293 T cells co-transfected with pXJ-E and pXJ-8b-myc (Fig. 5 , lane 2) when compared to cells transfected with pXJ-E alone (Fig. 5, lane 1), but rather there appeared to be an increase in the mRNA level of E in the presence of 8b. This implies that the down-regulation of E protein expression by 8b is not due to a reduction of the transcription of the E gene and is likely to be post-translational. No signal was detected in untransfected cells (Fig. 5, lane 3), showing that the hybridization probe is highly specific for mRNA of E. The experiment was repeated three times and a representative set of data was presented.
Fig. 5.
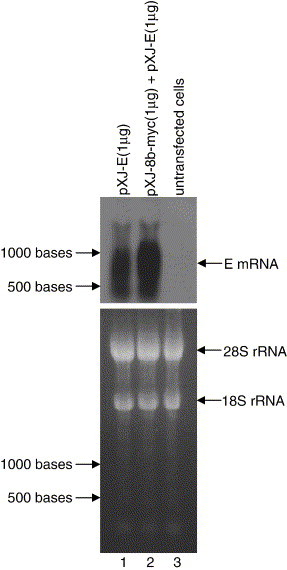
Effects of 8b protein on the transcription of the E gene determined by Northern blot analysis. Equal amount of total RNA (15 μg) isolated from 293T cells transfected with pXJ-E (lane 1), pXJ-8b-myc and pXJ-E (lane 2) cDNA constructs or untransfected 293T (lane 3) was separated on a denaturing agarose gel and transferred to nylon membrane. The amounts of E mRNA present were determined by hybridization with an E gene-specific probe (top panel). In order to verify that equal amounts of total RNA were loaded in each lane of the agarose gel before transfer, the amounts of 18S and 28S ribosomal RNA were visualized under UV light (bottom panel).
The 8b protein can bind E protein in SARS-CoV-infected cells
Co-immunoprecipitation experiment was also performed to determine the interaction between 8b and E in SARS-CoV-infected cells. Lysates from mock-infected or SARS-CoV-infected cells were immunoprecipitated using rabbit anti-8b polyclonal antibody (Fig. 6 , lanes 4 and 6) or an irrelevant antibody (rabbit anti-HA polyclonal antibody, lanes 3 and 5). The results showed that the E protein present in the lysates from SARS-CoV-infected cells could bind specifically to the 8b protein (lane 6). No unspecific binding was observed with the irrelevant antibody (lane 5).
Fig. 6.
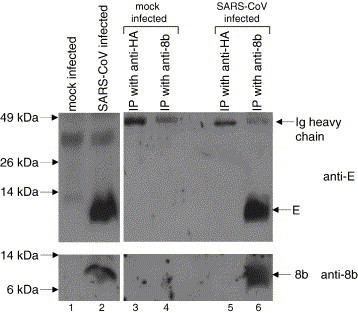
Interaction between E and 8b in SARS-CoV-infected cells determined by co-immunoprecipitation experiment. Lysates from mock-infected or SARS-CoV-infected cells were immunoprecipitated using rabbit anti-8b polyclonal antibody (lanes 4 and 6) or an irrelevant antibody (rabbit anti-HA polyclonal antibody, lanes 3 and 5) and protein A-agarose beads. Western blot analyses were then performed to determine the amount of E (upper panel) or 8b (lower panel) present in the lysates before immunoprecipitation (lanes 1 and 2) and the immunocomplexes on the protein A-agarose beads (lanes 3–6).
Expressions of 8b and E are mutually exclusive in SARS-CoV-infected cells
Indirect immunofluorescence experiments were further performed to determine the localization of E and 8b in SARS-CoV-infected cells. Strikingly, cells that were expressing high levels of 8b did not have detectable levels of E (Fig. 7 ). Two representative sets of data were presented and cells expressing high levels of 8b were marked with white asterisks. Hence, it appears that 8b can down-regulate the expression of E during SARS-CoV infection.
Fig. 7.
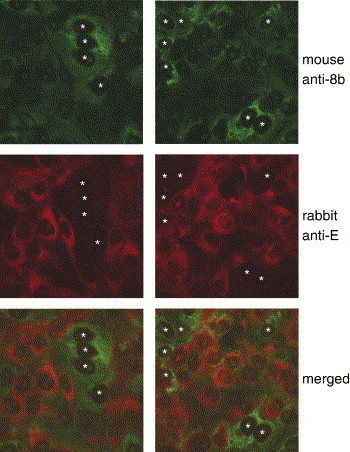
Expressions of E and 8b in SARS-CoV-infected cells determined by indirect immunofluorescence experiments. The expression of 8b was represented by FITC staining (top row), whereas the expression of E was represented by Rhodamine staining (middle row). The merged images showed that the expression of 8b and E were mutually exclusive (bottom row). Two representative sets of data were presented and cells expressing high levels of 8b were marked with white asterisks.
Discussion
When a virus is first introduced into the human population from an animal source, it has to undergo evolution in order to optimize the entry, replication and budding processes as well as to evade immune responses. Thus, genetic and epidemiological studies can yield valuable insights on how viruses cross the species barrier and evolve to cause disease in humans (Webby et al., 2004). Indeed, such studies conducted on the SARS-CoV have revealed that this virus has crossed the animal–human barrier recently (Donnelly et al., 2004, Guan et al., 2003, Lau et al., 2005, Song et al., 2005). Interestingly, the animal strains of SARS-CoV, isolated from a raccoon dog and palm civets in markets/restaurants and from wild bats, contain an extra 29-nt in the ORF8 region (Guan et al., 2003, Lau et al., 2005, Song et al., 2005). This 29-nt sequence is not found in all the human strains that were isolated in the middle phase of the epidemic but is present in most of the human isolates from the earliest outbreaks in Guangdong, China, 2002 (The Chinese SARS Molecular Epidemiology Consortium, 2004). Indeed, human infection in the early phase probably represents the first breach of the animal–human barrier as these isolates share the closest phylogenetic relationship with the animal isolates.
After the WHO's declaration of the end of the SARS epidemic, there were four confirmed SARS patients in Guangzhou, China, in late 2003 to early 2004 (Liang et al., 2004, Song et al., 2005). These patients did not have any contact history with previously documented SARS cases. Sequence analysis of viruses isolated from these patients showed that they were not derived from the preceding epidemic in 2003 but rather suggested that these cases represented new zoonotic transmissions (Song et al., 2005, The Chinese SARS Molecular Epidemiology Consortium, 2004). Like the animal isolates, these viruses also contained the additional 29-nt in the ORF8 region.
These findings clearly indicate that the extra 29-nt sequence in ORF8 is not necessary for the animal–human transmission. Analysis of the variation in the sequences of S protein showed that the SARS-CoV has rapidly evolved during the SARS epidemic and that the virus was undergoing adaptation in the human host (Song et al., 2005, The Chinese SARS Molecular Epidemiology Consortium, 2004). Although it is clear that the S gene was undergoing positive selection (Holmes, 2005), whether the genetic variation in the ORF8 region is a result of viral adaptation or genomic instability remains to be determined. As a consequence of the additional 29-nt in the ORF8 region, the 8a and 8b protein in the human SARS-CoV, circulating during the middle phase of the epidemic, are joined together to form a single protein, 8ab, in the animal SARS-CoV or human SARS-CoV from the early and late phases (Fig. 1B).
In this study, we detected the expression of 8a and 8b in Vero E6 cells infected by a human SARS-CoV isolated from the middle phase (SIN2774; GenBank accession number AY283798) and showed that the cellular localizations of 8a and 8b are distinct from 8ab (Fig. 2). We used co-immunoprecipitation of overexpressed proteins in mammalian cells to determine the abilities of these proteins to interact with different SARS-CoV proteins and showed that the binding profiles of 8a, 8b and 8ab are different (Fig. 3). Although these viral–viral protein interactions need to be verified in infected cells, these observations implied that there are conformational differences between these protein when they are expressed as separate proteins (8a and 8b) and when they are expressed as a single fused protein (8ab).
It has been demonstrated that the palm civets are equally susceptible to the human SARS-CoV isolate BJ01 from the middle phase (with the 29-nt deletion) and the isolate GZ01 from the early phase (Wu et al., 2005). Using reverse genetic methods, Yount and co-workers (2005) also reported similar findings in the mouse model. These results suggested that the 8a, 8b and 8ab proteins are not essential for viral replication or pathogenesis in the mouse and palm civet models. However, we found that the expression of 8b can down-regulate the expression of E in a dose-dependent manner (Fig. 4A) and the expressions of 8b and E in SARS-CoV-infected cells were mutually exclusive (Fig. 7). Interestingly, the expression of E was not affected by either 8a or 8ab (Fig. 4A). In addition, Northern blot analysis showed that the mRNA level of E was not decreased in the presence of the 8b protein, suggesting that the effect of 8b on the expression of the E protein is likely to be post-translational (Fig. 5).
Although the co-expression of SARS-CoV E and M is sufficient for the assembly of viral-like particles in the baculovirus system (Ho et al., 2004, Mortola and Roy, 2004), it was demonstrated by reverse genetic techniques that the E protein is not essential for the replication of SARS-CoV in Vero E6 cells (personal communication from Marta L. DeDiego and Luis Enjuanes, Centro Nacional de Biotecnologia, Madrid, Spain). However, the role of the E protein in SARS-CoV replication in its natural host remains unclear. Interestingly, the E protein is essential for the replication of the porcine transmissible gastroenteritis virus (TGEV) (Ortego et al., 2002) but not for the mouse hepatitis virus (MHV) (Kuo and Masters, 2003). However, for the latter, the deletion of the E gene reduces virus replication significantly. In addition, it was reported that overexpression of SARS-CoV E can induce apoptosis in T cells (Yang et al., 2005); thus, the down-regulation of E may also have an effect on viral pathogenesis.
An interesting question that arises from our observations concerns the regulation of 8b expression during SARS-CoV infection. 8a and 8b are encoded by the bicistronic subgenomic RNA 8 produced in SARS-CoV-infected cells (Marra et al., 2003, Snijder et al., 2003). Because its translation initiation codon is not the first AUG in the subgenomic RNA, the 8b protein is likely to be expressed via an internal ribosomal entry mechanism or by a leaky ribosomal scanning mode of translation, as have been described for viral proteins encoded by other bicistronic or tricistronic coronaviral mRNAs (Liu and Inglis, 1992, Senanayake and Brian, 1997, Thiel and Siddell, 1994). However, in order for 8b to be expressed via such mechanisms, activation of certain host translational machineries may be necessary (Komar and Hatzoglou, 2005, Stoneley and Willis, 2004). Indeed, we observed that 8b was only expressed in a fraction of the SARS-CoV-infected cells (Fig. 7). This means that the effect of 8b on viral replication or pathogenesis is likely to be only modulative as viral replication in those cells that did not express high levels of 8b will be normal. However, this modulating mechanism is not functional in the animal SARS-CoV because 8ab does not appear to have any effect on the expression of E. In future studies, it will be crucial to determine the underlying mechanism regulating the expression of 8b and its temporal expression during the viral replication cycle.
Materials and methods
Materials
All reagents used in this study were purchased from Sigma (St. Louis, MO, USA) unless otherwise stated.
Construction of ORF8-encoded proteins
For the construction of pXJ-8a, pXJ-8a-myc, pXJ-8b and pXJ-8b-myc, the ORF 8a and 8b were amplified by PCR using cDNA prepared from SARS-CoV-infected cells as templates as previously described (Tan et al., 2004a, Tan et al., 2004b). This strain of virus was isolated from a Singapore patient (SIN2774; GenBank accession number AY283798) and contained the 29-nt deletion in the ORF8 region. In order to construct a plasmid for expressing the 8ab protein found in animal SARS-CoVs, we used the cDNA template described above and sequential PCR to insert the 29-nt. Primers were designed based on the early phase human SARS-CoV isolate, GZ02, which has the 29-nt insertion in ORF8 (GenBank accession number AY390556). All sequences were confirmed by sequencing performed by the core facilities at the Institute of Molecular and Cell Biology, Singapore. All primers used in this study were purchased from Research Biolabs, Singapore, and are listed in Table 2 .
Table 2.
Primers used in this study
| Primer | Sequencea | Sense | Application |
|---|---|---|---|
| 8a-F1 | 5′-CGGGATCCGCCACCATGAAACTTCTC-3′ | + | pXJ-8a and pXJ-8ab |
| 8a-F2b | 5′-CGGGATCCACCATGGGAATGAAACTTCTC-3′ | + | pXJ-8a-myc and pXJ-8ab-myc |
| 8a-F3 | 5′-CGGGATCCACCATGGGAATATGCACTGT-3′ | + | pGex4T1-8aΔN15 |
| 8a-R1 | 5′-CCGCTCGAGCTAGTGTTGTACC-3′ | − | pXJ-8a and pGex4T1-8aΔN15 |
| 8a-R2 | 5′-CCGCTCGAGTTGTGTTGTACC-3′ | − | pXJ-8a-myc |
| 8b-F1 | 5′-CGGGATCCGCCACCATGTGCTTGAAG-3′ | + | pXJ-8b |
| 8b-F2b | 5′-CGGGATCCACCATGGGAATGTGCTTGAAG-3′ | + | pXJ-8b-myc |
| 8b-F3 | 5′-CGGGATCCACCATGGGAGTTTTACCTTT-3′ | + | pGex4T1-8bΔN26 |
| 8b-R1 | 5′-CCGCTCGAGTTAATTTGTTCGT-3′ | − | pXJ-8b and pXJ-8ab and pGex4T1-8bΔN26 |
| 8b-R2 | 5′-CCGCTCGAGCCATTTGTTCGTTTATT-3′ | − | pXJ-8b-myc and pXJ-8ab-myc |
| 8ab-1 | 5′-GTTGGTACCCAGTAGGACAAGGATCTTC-3′ | − | Construction of 8ab |
| 8ab-2 | 5′-GGTTACCAACCTGAATGGAATATAAGG-3′ | + | Construction of 8ab |
| 8ab-3 | 5′-TCCATTCAGGTTGGTACCCAG-3′ | − | Construction of 8ab |
| 8ab-4 | 5′-AATGGAATATAAGGTACAACAC-3′ | + | Construction of 8ab |
| 8ab-5 | 5′-CCTTATATTCCATTCAGG-3′ | − | Construction of 8ab |
Restriction sites introduced into primers are shown in bold face.
Six additional base pairs (ATGGGA), which encodes for two additional amino acids (methionine and glycine), were added to the 5′ end of 8a-myc, 8b-myc and 8ab-myc to give a Kozak consensus ribosome binding site for more efficient translation initiation.
The PCR amplicons containing ORF8a, ORF8b and ORF8ab were cloned into the mammalian expression vector pXJ3′HA as previously described (Tan et al., 2004a, Tan et al., 2004b). In order to create a C terminus myc-tag, PCR methods were used to insert a myc-tag (AEEQKLISEEDLLRKH) into the 3′ of the ORFs. These proteins were tagged at their C termini to avoid interference with their post-translational processing as both 8a and 8ab are predicted to contain one signal peptide at their N termini (http://www.cbs.dtu.dk/services/SignalP). The C-terminal HA tag present in the pXJ3′HA vector is not expressed as a stop codon was added before the HA tag coding sequences.
Production of glutathione transferase (GST)-fusion proteins
The cDNA encoding 8a (16–39aa) and 8b (27–84aa) were obtained by PCR methods (see Table 2 for primer sequences) and were cloned into pGEX-4T1 vector (Amersham Biosciences, Uppsala, Sweden) and transformed into E. coli BL21(DE3) cells (Stratagene, La Jolla, CA). Expression and purification of the GST-8aΔN15 were performed as previously described (Tan et al., 2004a), and for long-term storage at − 20 °C, 10% glycerol was added to the purified proteins to prevent aggregation. As for GST-8bΔN26, 2 mM DTT and 1.5% sarkosyl were included in the lysis buffer and after sonication, Triton X-100 was added to a final concentration of 2% before the protein was purified using GSH-sepharose beads. The purified proteins were used to immunize BALB/c mice for the production of antibodies using standard protocols. This was performed by trained personnel at the Biological Resource Centre, Agency for Science, Technology and Research (A*STAR), Singapore. GST-8bΔN26 was also used to raise rabbit polyclonal antibodies as previously described (Keng et al., 2005).
Transient transfections and Western blot analysis
293T and Vero E6 cells were propagated as previously described (Tan et al., 2004b) and transient transfections were performed using Lipofectamine reagent (Invitrogen, Carlsbad, CA), according to manufacturer's protocol. Western blot analysis were performed as previously described (Tan et al., 2004b) and some of the primary antibodies (anti-HA monoclonal (Roche Molecular Biochemicals, Indianapolis, Ind.) and anti-myc monoclonal (Santa Cruz Biotechnology, Santa Cruz, CA)) were purchased. The mouse anti-N, anti-E, anti-3a and anti-7a polyclonal antibodies have been described previously (Fielding et al., 2004, Guan et al., 2004, Tan et al., 2004b), whereas the mouse anti-8a and anti-8b polyclonal antibodies were produced for this study as described above. Anti-S monoclonal antibody (clone 1G10) has been described previously (Lip et al., 2006).
Immunofluorescence and co-immunoprecipitation experiments
Transiently transfected and SARS-CoV-infected Vero E6 cells were subjected to indirect immunofluorescence experiments as previously described (Tan et al., 2004b). For each co-immunoprecipitation experiment, one 6-cm dish of 293T cells was co-transfected with pXJ-myc-GST, pXJ-8a-myc, pXJ-8b-myc or pXJ-8ab-myc and the DNA construct for expressing one of the other viral proteins (S, E, M-HA, N, 3a or 7a). These DNA constructs have been previously described (Fielding et al., 2004, Tan et al., 2004a, Tan et al., 2004b). Untagged forms of S, N, E, 3a and 7a were used whereas a C-terminally HA-tagged M (M-HA) was used because no suitable anti-M antibody was available.
Due to the differences in the binding affinity of the different antibodies used for detection (i.e., anti-S, anti-E, anti-HA, anti-N, anti-3a and anti-7a), the amount of DNA plasmids required for each co-immunoprecipitation experiments was determined experimentally to ensure that good signals were obtained in Western blot analysis. In all cases (except for Fig. 3B, lane 3), the amount of pXJ-myc-GST, pXJ-8a-myc, pXJ-8b-myc and pXJ-8ab-myc used were 0.1 μg, 1 μg, 2 μg and 1 μg, respectively. The amount of pXJ-S (1 μg), pXJ-E (1 μg, except for Fig. 3B, lane 3), pXJ-M-HA (1 μg), pXJ-N (0.25 μg), pXJ-3a (0.4 μg) and pXJ-7a (0.4 μg) used are given in parentheses. Due to the effects of 8b on the expression of E, the amount of DNA plasmids used for Fig. 3B, lane 3, were 0.5 μg of pXJ-8b-myc and 2 μg of pXJ-E.
The cells were harvested at 16 h post-transfection and washed with PBS. Then, the cells were resuspended in 150 μl of IP buffer (50 mM Tris pH 8, 150 mM NaCl, 0.5% NP40, 0.5% deoxycholic acid, 0.005% SDS) supplemented with 0.5% Triton X-114 and subjected to sonication for 45 min using an ultrasonic processor (Sonics, Newtown, CT, USA), followed by freeze–thawing for six times. 100 μl of the lysates were diluted with 100 μl of IP buffer and 5 μl of rabbit anti-myc polyclonal antibody (Santa Cruz Biotechnology) were added and the mixture was subjected to end-over-end mixing at 4 °C for 2 h. Protein A-agarose beads (Roche) were then added and the mixing continued for at least 4 h at 4 °C. Beads were washed three times with cold IP buffer and then 20 μl of Laemmli's SDS buffer were added and the samples boiled at 100 °C for 5 min to release the immunocomplexes. As the M and 3a proteins tend to form large aggregates when boiled, samples containing M-HA or 3a were heated at 50 °C for 30 min, followed by 100 °C for 1 min. Samples were separated on SDS–PAGE and subjected to Western blot analysis. In some cases, tricine gels (BIORAD, Hercules, CA) were used instead for better resolution of low molecular weight proteins.
Co-immunoprecipitation experiments with SARS-CoV-infected cells were performed in a similar manner. Lysates obtained from SARS-CoV-infected Vero E6 cells were subjected to immunoprecipitation with either rabbit anti-8b polyclonal antibody or an unrelated antibody (anti-HA polyclonal, Y11, Santa Cruz Biotechnology) and protein-A agarose beads. Western blot analyses were then performed to detect the amount of E (using mouse anti-E polyclonal antibody) and 8b (using rabbit anti-8b polyclonal antibody) present in the immunocomplexes on the protein-A agarose beads.
Northern blot analysis
Total RNA from 293T cells transfected as described above was extracted using the RNeasy mini kit (Qiagen, Valencia, CA, USA) by following the protocol supplied by the manufacturer. The final RNA pellet was resuspended in diethyl pyrocarbonate-treated H2O and quantified by measuring absorbance at 260 nm. 15 μg of total RNA was separated on 1.2% denaturing agarose-formaldehyde gel (containing ethidium bromide), transferred overnight in 1× saline-sodium citrate (SSC) buffer to nylon membranes Hybond N+ (Amersham Biosciences). The blot was dried at room temperature for 30 min and baked at 80 °C for 30 min. The blot was then pre-hybridized with salmon testes DNA (Sigma) in hybridization buffer containing 6× SSC, 2× Denhardt's Reagent and 0.1% sodium dodecyl sulfate (SDS) for 2 h. The E gene cDNA fragment was cloned into the TA cloning vector pCRII-TOPO (Invitrogen) and linearized with Hind III restriction enzyme. Probes were generated from the linearized plasmid using T7 polymerase from the DIG RNA labeling kit (Roche). Hybridization was performed overnight at 68 °C with the DIG-labeled probe in the hybridization buffer. After hybridization, the blot was washed once for 20 min in 1× SSC buffer with 0.1% SDS followed by 3 washes, each 30 min, in 0.2× SSC with 0.1% SDS. All washes were performed at room temperature. The blot was then probed with alkaline phosphatase-conjugated anti-digoxigenin antibody and developed using chemiluminescence substrate CSPD (DIG luminescent detection kit, Roche). This was performed according to the manufacturer's protocol and the results were obtained by autoradiography. RNA ladders from Fermentas Life Sciences (Ontario, Canada) were used for size determination of mRNA.
Acknowledgments
We thank Puay Yoke Tham, Vithiagaran Gunalan and Kuo-Ming Lip and personnel at the Biological Resource Centre for technical assistance. This work was supported by grants from the Agency for Science, Technology and Research (A*STAR), Singapore.
References
- Chiu R.W., Chim S.S., Tong Y.K., Fung K.S., Chan P.K., Zhao G.P., Lo Y.M. Tracing SARS-coronavirus variant with large genomic deletion. Emerging Infect. Dis. 2005;11:168–170. doi: 10.3201/eid1101.040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C.A., Fisher M.C., Fraser C., Ghani A.C., Riley S., Ferguson N.M., Anderson R.M. Epidemiological and genetic analysis of severe acute respiratory syndrome. Lancet Infect. Dis. 2004;4:672–683. doi: 10.1016/S1473-3099(04)01173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Preiser W., Gunther S., Schmitz H., Doerr H.W. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol. Med. 2003;9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C., Tan Y.J., Shuo S., Tan T.H., Ooi E.E., Lim S.G., Ho W., Goh P.Y. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome (SARS) coronavirus. J. Virol. 2004;78:7311–7318. doi: 10.1128/JVI.78.14.7311-7318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guan M., Chen H.Y., Tan P.H., Shen S., Goh P.-Y., Tan Y.-J., Pang P.H., Lu Y., Fong P.Y., Chin D. Use of viral lysate antigen combined with recombinant protein in Western immunoblot assay as confirmatory test for serodiagnosis of severe acute respiratory syndrome. Clin. Diagn. Lab. Immunol. 2004;11:1148–1153. doi: 10.1128/CDLI.11.6.1148-1153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Lin P.H., Liu C.Y., Lee S.P., Chao Y.C. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 2004;318:833–838. doi: 10.1016/j.bbrc.2004.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. Structural biology. Adaptation of SARS coronavirus to humans. Science. 2005;309:1822–1823. doi: 10.1126/science.1118817. [DOI] [PubMed] [Google Scholar]
- Keng C.-T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H.P., Chou C.-F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.-J. Amino acids 1055 to 1192 in the S2 region of SARS coronavirus S protein induces neutralizing antibodies: implications for the development of vaccine and anti-viral agent. J. Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A.A., Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 2003;77:4597–4608. doi: 10.1128/JVI.77.8.4597-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y.C., Liu H.F., Shih Y.P., Yang J.Y., Chen H.Y., Chen Y.M. Phylogenetic analysis and sequence comparisons of structural and non-structural SARS coronavirus proteins in Taiwan. Infect. Genet. Evol. 2005;5:261–269. doi: 10.1016/j.meegid.2004.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Chen Q., Xu J., Liu Y., Lim W., Peiris J.S., Anderson L.J., Ruan L., Li H., Kan B., Di B., Cheng P., Chan K.H., Erdman D.D., Gu S., Yan X., Liang W., Zhou D., Haynes L., Duan S., Zhang X., Zheng H., Gao Y., Tong S., Li D., Fang L., Qin P., Xu W., SARS Diagnosis Working Group Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerging Infect. Dis. 2004;10:1774–1781. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lip K.M., Shen S., Yang X., Keng C.T., Zhang A., Oh H.L., Li Z.H., Hwang L.A., Chou C.F., Fielding B.C., Tan T.H., Mayrhofer J., Falkner F.G., Fu J., Lim S.G., Hong W., Tan Y.J. Monoclonal antibodies targeting the HR2 domain and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. J. Virol. 2006;80:941–950. doi: 10.1128/JVI.80.2.941-950.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J. Virol. 1992;66:6143–6154. doi: 10.1128/jvi.66.10.6143-6154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Mortola E., Roy P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 2004;576:174–178. doi: 10.1016/j.febslet.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Guan Y., Nicholls J.M., Yuen K.Y., Peiris J.S. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect. Dis. 2004;4:663–671. doi: 10.1016/S1473-3099(04)01172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin E., He X., Tian W., Liu Y., Li W., Wen J., Wang J., Fan B., Wu Q., Chang G., Cao W., Xu Z., Yang R., Wang J., Yu M., Li Y., Xu J., Si B., Hu Y., Peng W., Tang L., Jiang T., Shi J., Ji J., Zhang Y., Ye J., Wang C., Han Y., Zhou J., Deng Y., Li X., Hu J., Wang C., Yan C., Zhang Q., Bao J., Li G., Chen W., Fang L., Li C., Lei M., Li D., Tong W., Tian X., Wang J., Zhang B., Zhang H., Zhang Y., Zhao H., Zhang X., Li S., Cheng X., Zhang X., Liu B., Zeng C., Li S., Tan X., Liu S., Dong W., Wang J., Wong G.K., Yu J., Wang J., Zhu Q., Yang H. A genome sequence of novel SARS-CoV isolates: the genotype, GD-Ins29, leads to a hypothesis of viral transmission in South China. Genomics Proteomics Bioinformatics. 2003;1:101–107. doi: 10.1016/S1672-0229(03)01014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake S.D., Brian D.A. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 1997;48:101–105. doi: 10.1016/S0168-1702(96)01423-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M., Willis A.E. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Tan Y.-J., Goh P.-Y., Fielding B.C., Shen S., Chou C.-F., Fu J.-L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profile of antibody responses against SARS-coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diag. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Teng E., Shen S., Tan T.H.P., Goh P.-Y., Fielding B.C., Ooi E.-E., Tan H.-C., Lim S.G., Hong W. A novel SARS coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Thiel V., Siddell S.G. Internal ribosome entry in the coding region of murine hepatitis virus mRNA 5. J. Gen. Virol. 1994;75:3041–3046. doi: 10.1099/0022-1317-75-11-3041. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Li L.J., Luo Y., Zhang J.Y., Wang M.Y., Cheng S.Y., Zhang Y.J., Wang X.M., Lu Y.Y., Wu N.P., Mei L.L., Wang Z.X. Molecular biological analysis of genotyping and phylogeny of severe acute respiratory syndrome associated coronavirus. Chin. Med. J. (Engl) 2004;117:42–48. [PubMed] [Google Scholar]
- Wang Z.G., Zheng Z.H., Shang L., Li L.J., Cong L.M., Feng M.G., Luo Y., Cheng S.Y., Zhang Y.J., Ru M.G., Wang Z.X., Bao Q.Y. Molecular evolution and multilocus sequence typing of 145 strains of SARS-CoV. FEBS Lett. 2005;579:4928–4936. doi: 10.1016/j.febslet.2005.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby R., Hoffmann E., Webster R. Molecular constraints to interspecies transmission of viral pathogens. Nat. Med., Suppl. 2004;10:77–81. doi: 10.1038/nm1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Tu C., Xin C., Xuan H., Meng Q., Liu Y., Yu Y., Guan Y., Jiang Y., Yin X., Crameri G., Wang M., Li C., Liu S., Liao M., Feng L., Xiang H., Sun J., Chen J., Sun Y., Gu S., Liu N., Fu D., Eaton B.T., Wang L.F., Kong X. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J. Virol. 2005;79:2620–2625. doi: 10.1128/JVI.79.4.2620-2625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xiong Z., Zhang S., Yan Y., Nguyen J., Ng B., Lu H., Brendese J., Yang F., Wang H., Yang X.F. Bcl-xL inhibits T cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 2005;392:135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


