Abstract
Global predictions of the secondary structure of coronavirus (CoV) 5′ untranslated regions and adjacent coding sequences revealed the presence of conserved structural elements. Stem loops (SL) 1, 2, 4, and 5 were predicted in all CoVs, while the core leader transcription-regulating sequence (L-TRS) forms SL3 in only some CoVs. SL5 in group I and II CoVs, with the exception of group IIa CoVs, is characterized by the presence of a large sequence insertion capable of forming hairpins with the conserved 5′-UUYCGU-3′ loop sequence. Structure probing confirmed the existence of these hairpins in the group I Human coronavirus-229E and the group II Severe acute respiratory syndrome coronavirus (SARS-CoV). In general, the pattern of the 5′ cis-acting elements is highly related to the lineage of CoVs, including features of the conserved hairpins in SL5. The function of these conserved hairpins as a putative packaging signal is discussed.
Keywords: Coronavirus, SARS-CoV, RNA secondary structure, Packaging signal, 5′ Untranslated region
Introduction
The emergence of the Severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 has boosted related research and led to the discovery of many novel coronaviruses (CoVs) from different hosts such as equines, whales, birds, and bats; the latter species are considered as the potential reservoir of SARS-CoV (Guan et al., 2003, Ksiazek et al., 2003, Li et al., 2005, Marra et al., 2003, Mihindukulasuriya et al., 2008, Woo et al., 2007, Woo et al., 2009, Zhang et al., 2007). In the past few years, also two novel human CoVs, NL63 and HKU1, have been identified causing rather severe symptoms in infants and the elderly (van der Hoek et al., 2004, Woo et al., 2005). The discovery of so many novel CoVs calls for a better understanding of the phylogeny of CoVs.
Based on serological patterns and genome organization, the genus Coronavirus has been classified into three major groups: group I, II and III (Lai and Cavanagh, 1997, Brian and Baric, 2005). More recently, these groups have been further subdivided into, in total, 9 subgroups, based upon amino acid similarity of structural and non-structural proteins (nsp) (Snijder et al., 2003, Woo et al., 2006, Woo et al., 2007, Woo et al., 2006, Woo et al., 2007). However, other studies propose at least 5 distinct lineages (Tang et al., 2006, Dong et al., 2007, Vijaykrishna et al., 2007), and even for SARS-CoV there is discussion whether it represents a separate lineage (Rota et al., 2003) or is an early split-off of group II CoVs (Snijder et al., 2003, Gibbs et al., 2004). Thus, in addition to the conventional pair-wise comparison of viral protein sequences, other genetic or structural features may be helpful in the classification of CoVs.
In the genome of CoVs, like that of most RNA viruses, the 5′ and 3′ untranslated regions (UTRs) usually harbor important structural elements which are involved in replication and/or translation (Chang et al., 1994, Raman et al., 2003; Raman and Brian, 2005, Goebel et al., 2007, Züst et al., 2008, Liu et al., 2009). In Mouse hepatitis virus (MHV), a group II CoV, a bulged stem–loop and a pseudoknot structure were identified in the 3′ UTR (Goebel et al., 2004a). Similar pseudoknot structures were found in other group I and II CoVs, showing structural conservations of the CoV 3′ UTR (Goebel et al., 2004a). However, the 3′ UTR of MHV could be functionally replaced by the 3′ UTR of group II SARS-CoV but not by that of the group I Transmissible gastroenteritis virus (TGEV) or the group III Avian infectious bronchitis virus (IBV), indicating certain group-specific functions for the 3′ UTR (Goebel et al., 2004b).
In this study the secondary structures of the 5′ UTRs and the 5′-proximal sequences of the ORF1ab gene in all known CoVs were predicted. The structural features of this region turned out to reflect the known grouping of CoVs, which is based on amino acid similarity. The unique and conserved features were further investigated in detail.
Results and discussion
The clustering of the 5′-proximal sequence of CoV RNAs shows group specificity
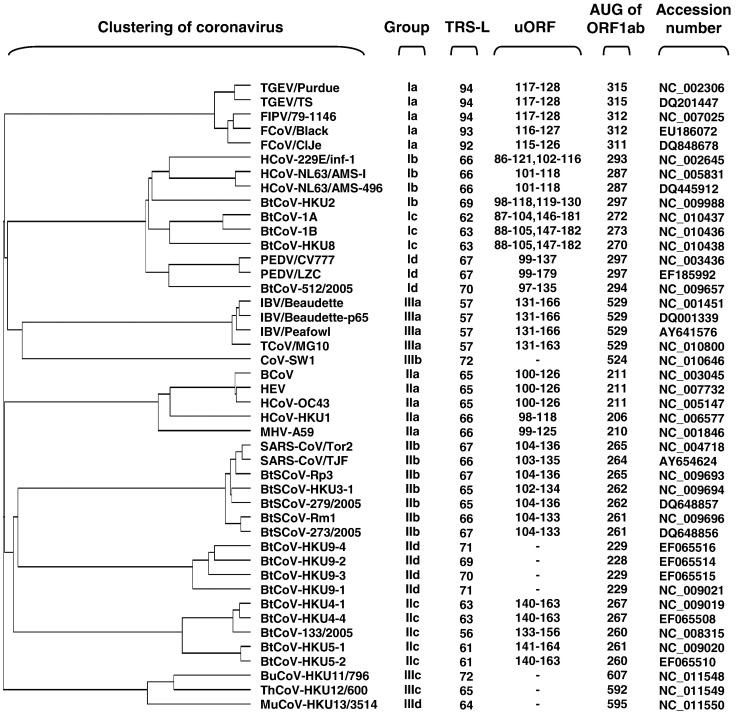
The clustering of the CoV 5′-proximal 420 nucleotides (nts) obtained from the Kalign webserver (see Materials and methods) basically resembled the current grouping system for CoVs (Fig. 1 ), though group I CoVs may be further subdivided into 4 subgroups, groups Ia to Id, according to their relatively large phylogenetic distances (Fig. 1). Sequence comparison further showed conserved and unique features for each CoV group, including: (i) the relative location of the core sequence of the leader transcription-regulating sequence (L-TRS) is quite conserved in all CoVs, except for the one in group Ia CoVs which has a rather long leader sequence upstream of the core TRS; (ii) the potentially translatable short ORF upstream of the genomic ORF1ab, the uORF, is present in most CoVs except for group IId, IIIb, IIIc, and IIId CoVs; (iii) the 5′ UTR in group III CoVs is substantially longer than that in group I and II CoVs, while group IIa CoVs have an exclusively short 5′ UTR (Fig. 1). It has to be noted that in order to obtain a higher threshold of the phylogenetic distance, strains with the highest sequence variation were used for analysis (selected from the genomic sequences of all CoVs available in GenBank). This made it more promising if homology was found within a cluster. To further examine if particular features found in the RNA sequence in each group are relevant to specific organization of the 5′ cis-acting elements, we globally predicted the secondary structures of the CoV 5′ UTRs, predominantly using computational calculations at the mfold webserver (Zuker, 2003). We have identified several conserved stem–loop (SL) structures in this region, some of which are organized in a group-specific manner (see Fig. 2, Fig. 3, Fig. 4 ).
Fig. 1.
Clustering and general features of the 5′ 420 nucleotides of CoVs. The tree is based on a multiple sequence alignment using ClustalW2 at the European Bioinformatics Institute webserver. The phylogenetic group, the start of core TRS-L, the region of upstream ORF (uORF), the start of ORF1ab, and GenBank accession number of each CoV are listed.
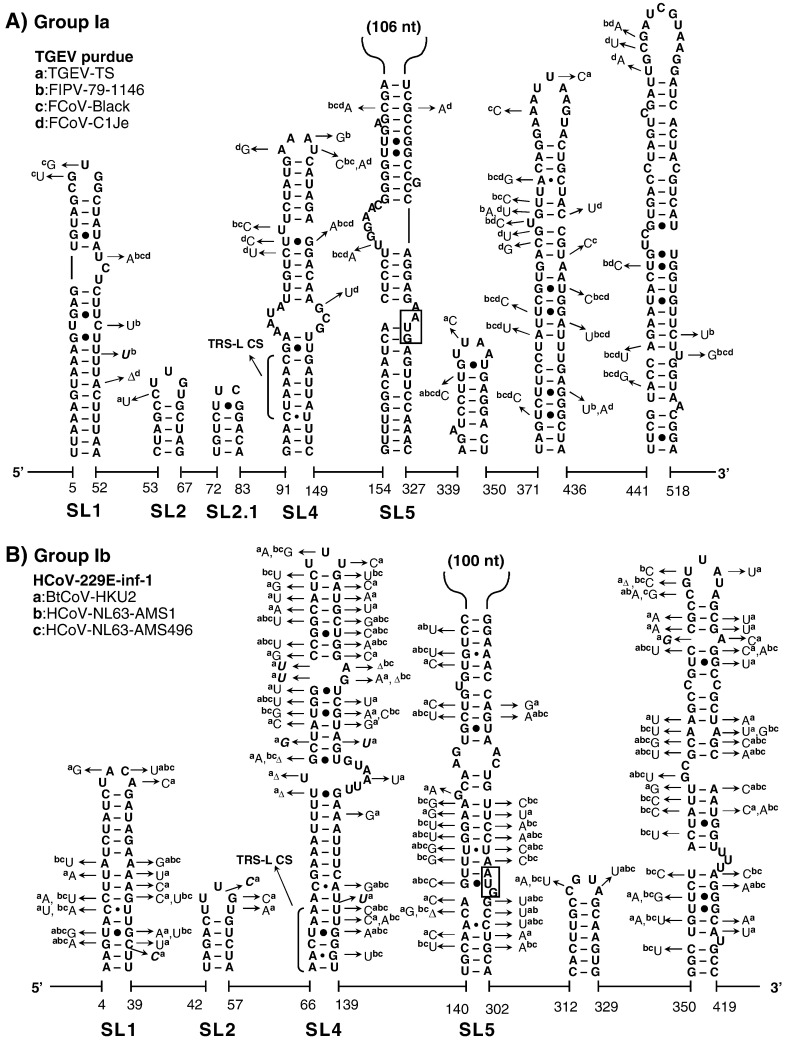
Fig. 2.
The structural–phylogenetic analysis of the 5′-proximal sequences in group I CoVs. The predicted secondary structures of the 5′-proximal sequence of (A) group Ia TGEV-purdue, (B) group Ib HCoV-229E-inf-1, (C) group Ic PEDV-CV777, and (D) group Id BtCoV-1A coronaviruses are shown. Nucleotide variations located in the conserved elements in the other representative CoVs of each subgroup are indicated. The start codon of the ORF1ab is boxed, the core sequence of the transcription-regulating leader (TRS-L CS) is bracketed, and the length of the sequence insertion in SL5 is indicated.
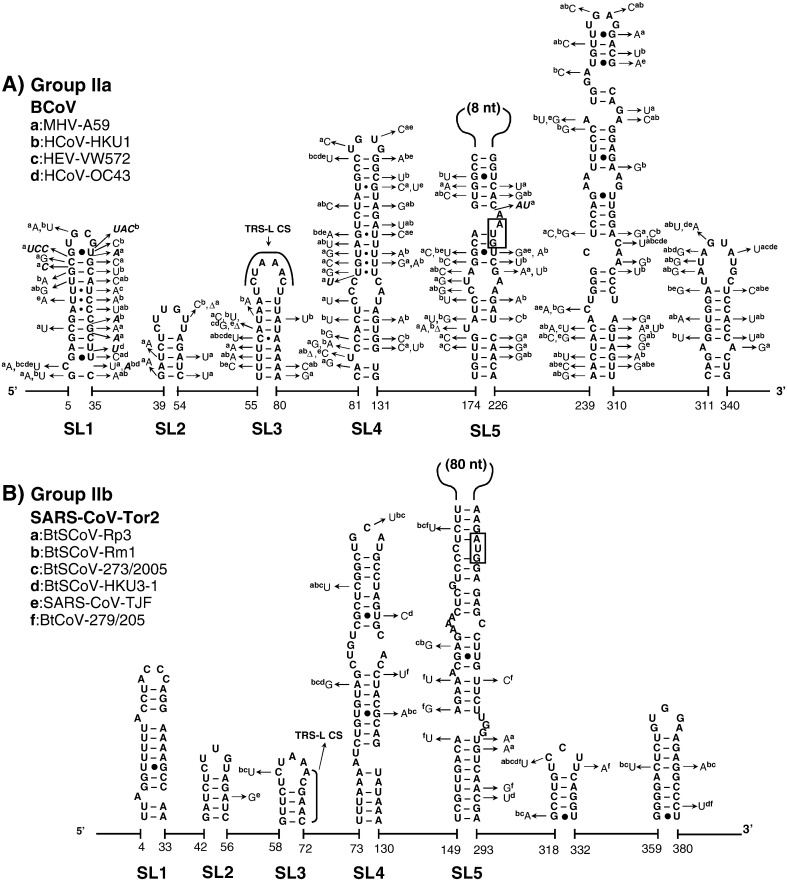
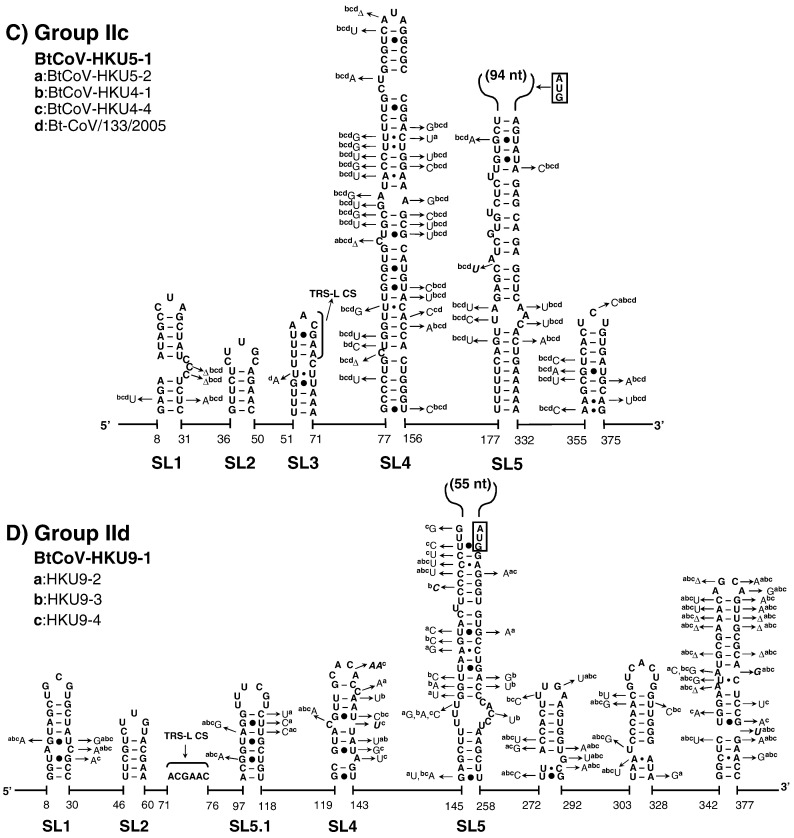
Fig. 3.
The structural–phylogenetic analysis of the 5′-proximal sequences in group II CoVs. The predicted secondary structures of the 5′-proximal sequence of (A) group IIa BCoV, (B) group IIb SARS-CoV-Tor2, (C) group IIc BtCoV-HKU5-1, and (D) group IId BtCoV-HKU9-1 are shown. For details see Fig. 2.
Fig. 4.
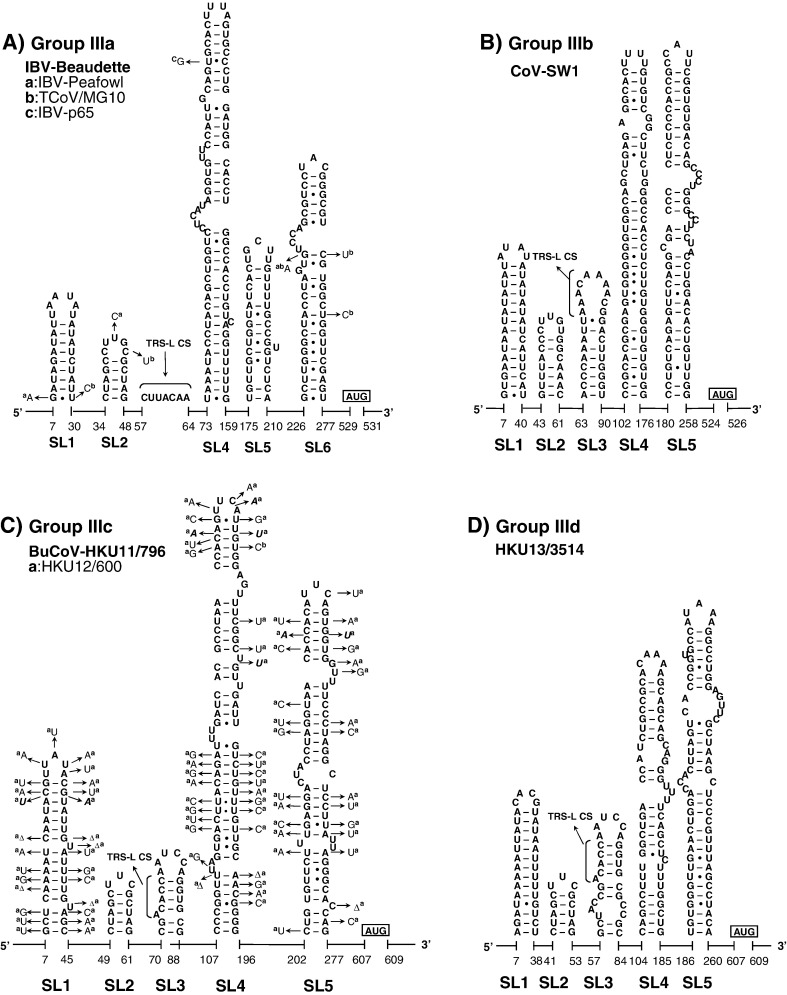
The structural–phylogenetic analysis of the 5′-proximal sequences in group III CoVs. The predicted secondary structures of the 5′-proximal sequence of (A) group IIIa IBV-Beaudette, (B) group IIIb CoV-SW1, (C) group IIIc BuCoV-HKU11/796, and (D) group IIId HKU13/3514 are shown. For further details see Fig. 2.
The universal presence of SL1 and SL2 in CoV 5′ UTR
The very 5′ nts of CoV RNAs fold into a hairpin of low thermodynamic stability, SL1, which is supported by many co-variations (Fig. 2, Fig. 3, Fig. 4), particularly in group IIa and IIIc CoVs. The loop sequences are not strongly conserved although a YRYR tetra-loop seems to be preferred in most SL1s. A general feature of SL1 is the presence of mismatches, bulges (e.g. in group I and II CoV RNAs) and a high number of A–U and U–A base pairs (bps) (e.g. in group IIIa, b, and d CoV RNAs). Recent data by Li et al. (2008) suggest that the low thermodynamic stability of SL1 is important for the replication of MHV.
Another conserved hairpin is SL2 which consists of a 5-bp stem and a highly conserved loop sequence, 5′-CUUGY-3′, which has an important role in MHV replication (Liu et al., 2007), though the motif is less conserved in SL2 of group I and III CoVs (Fig. 2, Fig. 4). Downstream of SL2, an additional hairpin, SL2.1, with the stable UUCG tetra-loop, was predicted in group Ia CoVs. Interestingly, the CUUGY loop was recently shown to adopt the YNMG-type of tetra-loop-folds (Liu et al., 2009).
The diversity of SL3 and SL4 in CoVs
Previously, the core L-TRS in CoVs has either been proposed to be non-structured (Stirrups et al., 2000, Wang and Zhang, 2000) or to form a hairpin structure (Shieh et al., 1987, Chang et al., 1996). We found that the core L-TRS and the adjacent sequence may fold into SL3 in some CoVs, e.g. the group II Bovine coronavirus (BCoV), SARS-CoV and Bat coronavirus HKU4 (BatCoV-HKU4), and the group III coronavirus SW1 (CoV-SW1), Bulbul coronavirus HKU11 (BuCoV-HKU11), and Munia coronavirus HKU13 (MuCoV-HKU13) (Fig. 3, Fig. 4). However, the sequence variations found in group IIa CoVs are partially in conflict with the lower part of SL3, while in other CoVs there are no co-variations to support the formation of SL3. Thus, the CoV SL3 may not structurally resemble the L-TRS Hairpin (LTH) found in the related arterivirus, the Equine arteritis virus (EAV), which directs discontinuous transcription (van den Born et al., 2004, van den Born et al., 2005). In some other CoVs, e.g. TGEV and the Human coronavirus-229E (HCoV-229E), the core L-TRS was predicted to participate in the stem of SL4 (Figs. 2A and B), although sequence variations found in group Ib CoVs do not strongly support the involvement of the core L-TRS in the SL4 stem (Fig. 2B). All in all, based on the structural–phylogenetic survey, it can be concluded that the core L-TRS and the flanking sequences are poorly structured in CoVs.
Downstream of the L-TRS, a long hairpin, SL4, was predicted for all CoVs (Fig. 2, Fig. 3, Fig. 4). The presence of a large number of co-variations seems to support the existence of SL4 strongly, particularly the upper half of this structure. Raman et al. (2003) have shown that the structural integrity, in positive or negative strands or both, of the upper part of SL4 (the SL-III in their study) is important for replication of BCoV DI RNA. We also found that the uORF predominantly terminates within the SL4 (data not shown), even for those uORFs that are in-frame with the downstream ORF1ab (Fig. 1).
There has no direct evidence for the translation of uORF in CoV infected cells, although Raman et al. (2003) have suggested a positive correlation between maintenance of the uORF and maximal BCoV DI RNA accumulation. They have also shown that a DI RNA in which this uORF was replaced by a totally unrelated uORF could be replicated. Our phylogenetic analysis showed that the sequence variations located in SL4, which were found to maintain the integrity of the RNA secondary structure, are not always silent at the amino acid level (data not shown). Although features of uORFs seem to be conserved and group-specific (Fig. 1), the necessity of translation of this ORF needs to be determined in the future to understand why certain groups of CoVs do need uORF for their propagation and others do not.
We noticed that the sequence of SL4 is included in the hotspot of the 5′-proximal genomic acceptor (Wu et al., 2006), suggesting that SL4 may play a role in directing the subgenomic RNA synthesis and thereby compensates for the absence of a structured L-TRS hairpin (see above).
Features of the inserted sequence in SL5 reflect the lineage of CoVs
A fifth structural element, SL5, was predicted downstream of SL4 in all CoVs (Fig. 2, Fig. 3, Fig. 4). SL5 is a homologue of SL-IV of BCoV reported by Brian and coworkers (Raman and Brian, 2005, Brown et al., 2007) and is supported by co-variations in almost all CoV groups with the exception of group Ia, and IIIa, b, and d CoVs, where sequence variation is low. Compared to group IIa and III CoVs, the other CoVs have sequence insertions in the top of SL5, which are about 110-nt long in group I CoVs and between 55 and 94 nt in group IIb, c, and d CoVs (Fig. 2, Fig. 3, Fig. 4). Secondary structure predictions of these inserts revealed hairpins displaying the conserved 5′-UUYCGU-3′ loop motif (Fig. 5 ). We note that some of these hairpins resemble the predicted structures for four group I CoVs and SARS-CoV reported by Raman and Brian (2005), which were proposed to be homologues of BCoV SL5 (SL-IV in their report). Nevertheless, our comprehensive structural–phylogenetic analysis indicates that these conserved structural motifs are not SL5 homologues as such but are substructural hairpins within SL5 (Fig. 2, Fig. 3, Fig. 5).
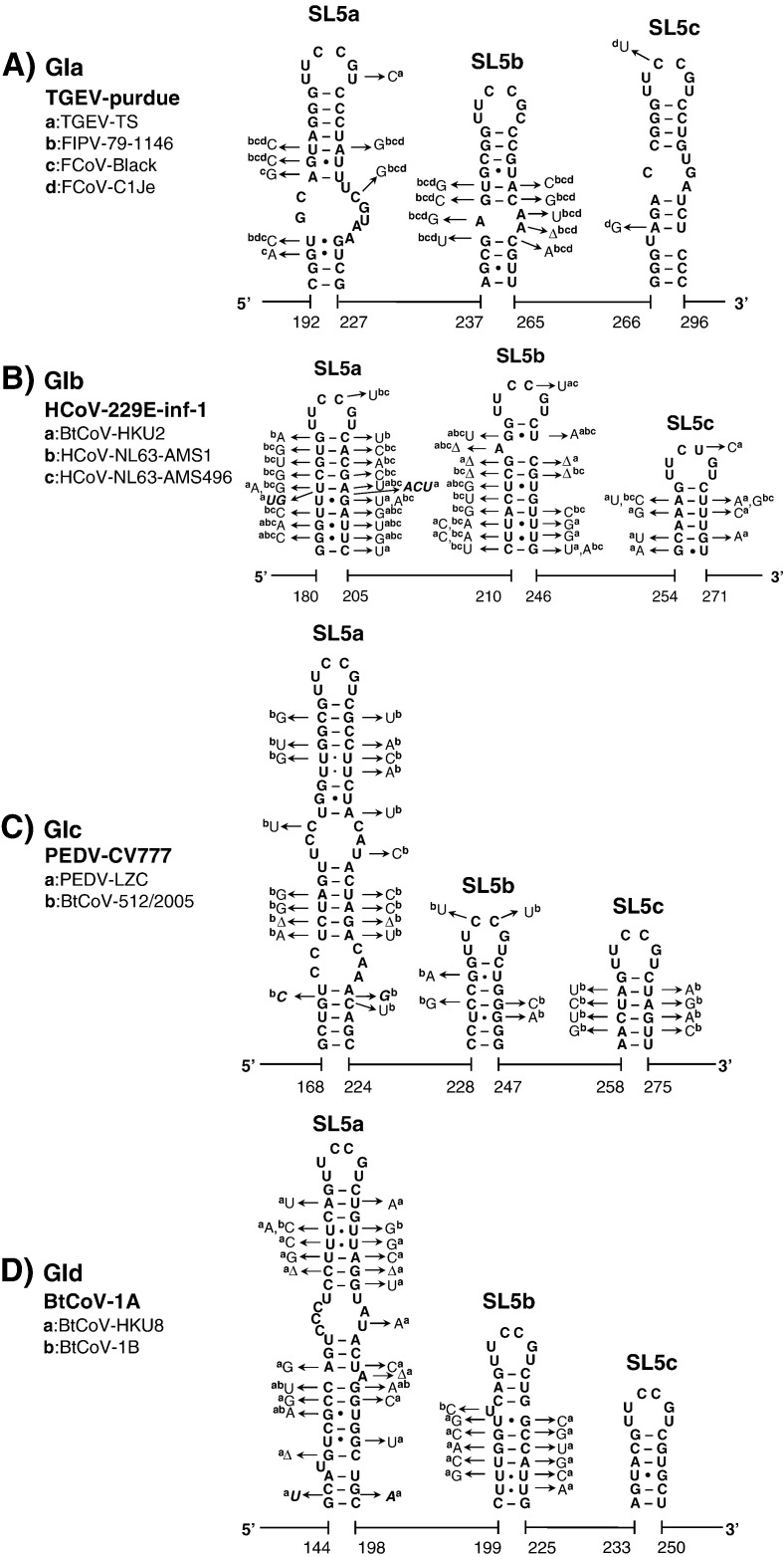
Fig. 5.
The substructural hairpins of SL5 in group I and II CoVs. The secondary structure of the SL5 substructural hairpins, SL5a–c, in (A) group Ia TGEV-purdue, (B) group Ib HCoV-229E-inf-1, (C) group Ic PEDV-CV777, (D) group Id BtCoV-1A, (E) group IIa BCoV, (F) group IIb SARS-CoV-Tor2, (G) group IIc BtCoV-HKU5-1, and (H) group IId BtCoV-HKU9-1 are shown. The start codon of the BtCoV-HKU5-1 ORF1ab is located in SL5b as indicated. SL5.1 which is located upstream of SL5 in BtCoV-HKU9-1 also contains the conserved UUUCGU motif.
In group I CoVs, a large number of co-variations, particularly in group Ib CoVs, was observed, supporting the existence of these substructural hairpins at the top of SL5 (Fig. 5). We noticed that 4 different patterns of the SL5 substructural hairpins were found in group I CoVs. This finding supports the idea that group I CoVs may be clustered into 4 subgroups, groups Ia to d. Nonetheless, the structural homology of SL5 within the lineage of the group I CoVs is still higher than that of the group II CoVs; three hairpins, SL5a, b, and c, with mainly the conserved 5′-UUCCGU-3′ loop sequence, were found in all group I CoVs. This is in agreement with the shorter phylogenetic distances found between each subgroup (group Ia–d) in group I CoVs compared to group II CoVs, which feature more diverse sequence insertions, in terms of length, the presence of 5′-UUYCGU-3′ motifs, and secondary structure. The greater structural variation in SL5 of group II CoVs is as follows: (i) the substructural hairpins are replaced by an 8-nt sequence in group IIa CoVs (Fig. 5E); (ii) one of the three substructural hairpins in SL5, SL5c, contains a GNRA tetra-loop sequence (group IIb) or a non-conserved hepta-loop sequence (group IIc) but not the UUYCGU motif (Figs. 5F and G); (iii) only two substructural hairpins are folded on top of SL5 in group IId CoVs, yet an additional conserved UUCCGU motif is present in SL5.1 located further upstream, in between the L-TRS and SL4 (Fig. 3, Fig. 5). Thus, the pattern of the SL5 substructures is strongly related to the lineage and the phylogenetic distance of the group I and II CoVs.
Similar hairpins with a conserved loop motif could not be identified in group III CoVs (Fig. 4). Here, SL5 has a rod-like shape as in group IIa. Also in the remainder of the 5′ UTR of group III CoVs no hairpins could be identified that featured a UUYCGU sequence or another motif.
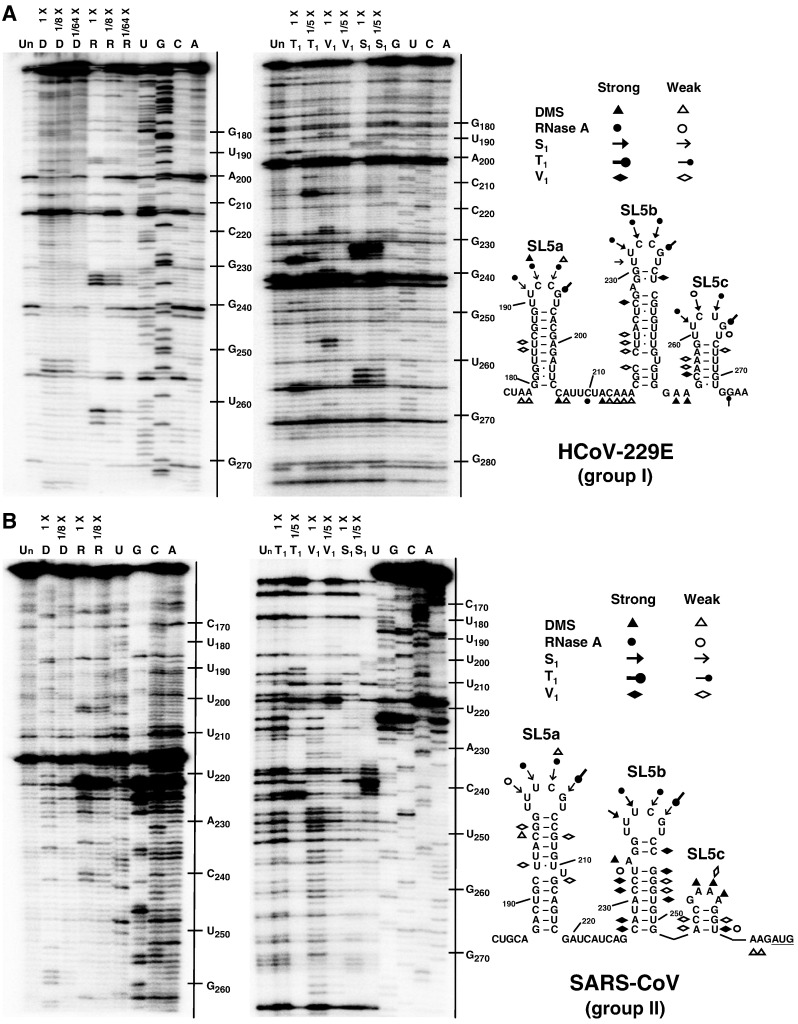
Structure probing of the SL5 substructure in HCoV-229E and SARS-CoV
To verify the secondary structures of the proposed substructural hairpins in group I and II CoVs, the corresponding RNA transcripts of HCoV-229E and SARS-CoV were subjected to enzymatic and chemical structure probing (see Materials and methods). Clearly, the single-stranded 5′-UUUCGU-3′-hexa-loop sequences in HCoV-229E SL5a, SL5b, and SL5c can be recognized by the single-strand specific probes, DMS, RNase A, S1 nuclease and/or RNase T1 (Fig. 6A), suggesting that these nucleotides are unpaired. The presence of RNase V1 cuts, an enzyme that cuts double-stranded RNA, in the predicted stem regions is also in agreement with the model. Probing results of SARS-CoV were also in agreement with the existence of SL5a, b, and c (Fig. 6B).
Fig. 6.
Structure probing of the inserted sequences in SL5 of group Ib HCoV-229E and group IIb SARS-CoV-Tor2. The secondary structures of the SL5 substructural hairpins of (A) the HCoV-229E and (B) the SARS-CoV are analyzed by enzymatic and chemical structure probing. Annotation of the denaturing electrophoresis: Un, untreated; D, DMS treated; R, RNase A treated; T1, RNase T1 treated; V1, RNase V1 treated; S1, S1 nuclease treated; G, U, C and A, the RNA sequencing ladder.
Notably, the U:U mismatches located in the stems of these substructures seem to form non-canonical base pairs since RNAse V1 recognized U222 and U221 in HCoV-229E SL5b, as well as U193 in SARS-CoV SL5a. In fact, several (tandem) U:U mismatches were identified in the SL5 substructural hairpins, e.g. the SL5a in the group Ic Porcine epidemic diarrhea virus (PEDV) (Fig. 5C) and the group Id Bat coronavirus 1A (BtCoV-1A) (Fig. 5D), as well as in other 5′ cis-acting elements, e.g. the MHV SL1 (Li et al., 2008). Interestingly, co-variations were frequently found at the positions of these tandem U:U mismatches, e.g. SL4 (Fig. 2, Fig. 3, Fig. 4) and SL5a–c (Figs. 5B, C, D, and H). This suggests the formation of (tandem) U:U base pairs similar to what has been reported for the 5′-CU-3′/5′-UU-3′ non-canonical base pairs found in the Y stem of polio-like enterovirus 3′ UTRs (Lescrinier et al., 2003).
Are the SL5 substructural hairpins the counterparts of the group IIa packaging signal?
It has been generally found that a strong packaging signal (PS) or encapsidation signal, which directs specific packaging or encapsidation of genomic RNA, usually encompasses repetitions of conserved (structural) motifs (Hellendoorn et al., 1996, Chen et al., 2007). This leads us to propose that the SL5 substructures bearing the highly conserved UUYCGU repeats function as genomic PS for group I and II CoVs, including SARS-CoV.
Studies of the genomic PSs in CoVs have been mainly focused on group IIa CoVs in the past, e.g. MHV and BCoV (Fosmire et al., 1992, Makino et al., 1990, van der Most et al., 1991, Woo et al., 1997, Chen et al., 2007, Cologna and Hogue, 2000). For other groups of CoVs, e.g. SARS-CoV, the identification of a putative PS has been reported by Hsieh et al. (2005). This PS was thought to be a homologue of the MHV PS located in the corresponding region near the 3′ end of ORF1ab. However, it has to be noted that the specificity of the proposed SARS-CoV PS to direct RNA packaging was not determined in their study, and the predicted secondary structure of their “homologue of MHV PS” lacks the conserved features of the MHV PS structure reported by Chen et al. (2007). Also we doubt the possibility of identifying a MHV-like PS in the “corresponding region” of SARS-CoV genomic RNA because an alignment of nsp15 sequences clearly shows that the sequence corresponding to the MHV PS is absent in SARS and other non-group IIa CoVs (Fig. 7 ).
Fig. 7.
Multiple alignment of the CoV nsp15 sequence corresponding to the group IIa packaging signal. The amino acid sequences of the group IIa CoV nsp15 are aligned with the sequences of other CoV groups, showing the underlined sequence insertion of the packaging signal corresponding region in group IIa CoVs.
Interestingly, the presence or absence of the region corresponding to the group IIa PS may not interfere with the function of nsp15 as the functional domains remain intact in both MHV and SARS-CoV nsp15 (Joseph et al., 2007). There seems to be however a strong correlation between the lack of a MHV PS-corresponding region and the presence of SL5 substructures and vice versa (Fig. 5). This correlation strongly suggests that the SL5 substructural hairpins located in the 5′ UTR are the counterparts of the genomic PS present in group IIa CoVs, and presumably the UUCCGU structural repeats (Fig. 5A) are responsible for the packaging activity reported by Escors et al. (2003) for the first 649 nts of TGEV genomic RNA.
Conclusions
The diversity of the genomic RNA sequence provides a wealth of structural and phylogenetic information on the lineage of CoVs and improves our understanding of the evolution of the 5′ cis-acting elements. We have shown that the pattern of these cis-acting elements in the 5′ UTR is highly related to the phylogenetic distance based on the viral protein sequences, suggesting that the viral proteins and the RNA sequence evolved simultaneously, possibly to maintain functional RNA–protein interactions.
The unique and conserved features of the 5′ UTR and SL5 highlight the role of RNA structure in the evolution of CoVs and may serve as a roadmap for further studies. Future experiments should also verify whether the conserved UUYCGU motifs in SL5 function as PS in group I and II CoVs by interacting with nucleocapsid and/or membrane proteins (Molenkamp and Spaan, 1997, Narayanan and Makino, 2001, Narayanan et al., 2003). The absence of these or other conserved motifs in the 5′ UTR of group III CoVs suggests that their PSs are located elsewhere in the genome. This possibility is currently being explored.
Materials and methods
Structural–phylogenetic analysis
Multiple alignment of all CoV 5′-proximal sequences available in GenBank was used to select coronaviruses with the highest sequence diversity. Sequences of the 5′ 420 nts of these variants were clustered by ClustalW2 on EBI webserver (Larkin et al., 2007). Secondary structures of this region were predicted by the Mfold webserver (Zuker, 2003). The alignment of CoV nsp15 was done by Kalign webserver (Lassmann and Sonnhammer, 2006) (Fig. 7).
Structure probing and primer extension
The RNA transcripts encompassing the entire HCoV-229E and SARS-CoV SL5 region (about 180 nt) were synthesized in vitro using RibomaxTM RNA production system (Promega). The corresponding cDNA templates with an upstream T7 promoter were amplified by PCR using oligo-nucleotides 5′-TAATACGACTCACTATAGGGCATGCCTAGTGCACCTACGCAG-3′ (the T7 promoter sequence is underlined) and 5′-CAAACTGAGTTGGACGTGTG-3′ for SARS-CoV SL5 and oligo-nucleotides 5′-TAATACGACTCACTATAGGGTAATTGAAATTTCATTTGGG-3′ (the T7 promoter sequence is underlined) and 5′-GTGTGACACTTGCCGTAGC-3′ for HCoV-229E SL5. Purified RNA transcripts were subjected to chemical and enzymatic probing as described in Chen et al. (2007). In general, 0.001% dimethylsulfate (DMS), 1 pg Rnase A, 0.001 U RNase T1, 0.1 U RNase V1, and 0.8 U S1 nuclease were used for the probing reactions (1×), followed by serial dilutions with a factor 1/5 (1/5× and 1/25×) or 1/8 (1/8× and 1/64×). The primer extension was carried out with 0.01 µg of treated transcripts, 0.5 µl of a 0.1 mM concentration of the MHV1 primer, 1 µl of 5 mM dGAT, 1 µl of 25 µM dCTP, 0.1 µl of α-32P-labeled dCTP (10 mCi/ml), 1 µl of 5× reverse transcriptase buffer, and 20 U of Moloney murine leukemia virus reverse transcriptase (Promega).
Acknowledgments
We acknowledge Eric Snijder for providing us with partial cDNA corresponding to the 5′-proximal region of HCoV-229E and SARS-CoV. This research was supported by a VIDI grant from the Netherlands Organisation for Scientific Research (NWO) awarded to R.C.L.O.
References
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.G., Nixon K.S., Senanayake SDChang R.Y., Hofmann M.A., Sethna P.B., Brian D.A. An RNA stem–loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J. Virol. 2007;81:7716–7724. doi: 10.1128/JVI.00549-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R.Y., Hofmann M.A., Sethna P.B., Brian D.A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 1994;68(12):8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R.Y., Krishnan R., Brian D.A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 1996;70(5):2720–9272. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., van den Born E., van den Worm S.H., Pleij C.W., Snijder E.J., Olsthoorn R.C. New structure model for the packaging signal in the genome of group IIa coronaviruses. J. Virol. 2007;81(12):6771–6774. doi: 10.1128/JVI.02231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cologna R., Hogue B.G. Identification of a bovine coronavirus packaging signal. J. Virol. 2000;74(1):580–583. doi: 10.1128/jvi.74.1.580-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L., Zhang S.Y., Peiris J.S., Smith G.J., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007;81(13):6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Izeta A., Capiscol C., Enjuanes L. Transmissible gastroenteritis coronavirus packaging signal is located at the 5′ end of the virus genome. J. Virol. 2003;77(14):7890–7902. doi: 10.1128/JVI.77.14.7890-7902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosmire J.A., Hwang K., Makino S. Identification and characterization of a coronavirus packaging signal. J. Virol. 1992;66(6):3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A.J., Gibbs M.J., Armstrong J.S. The phylogeny of SARS coronavirus. Arch. Virol. 2004;149(3):21–624. doi: 10.1007/s00705-003-0244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Hsue B., Dombrowski T.F., Masters P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004;78(2):669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Taylor J., Masters P.S. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J. Virol. 2004;78(14):7846–7851. doi: 10.1128/JVI.78.14.7846-7851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Miller T.B., Bennett C.J., Bernard K.A., Masters P.S. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J. Virol. 2007;81(3):1274–1287. doi: 10.1128/JVI.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Hellendoorn K., Michiels P.J., Buitenhuis R., Pleij C.W.A. Protonatable hairpins are conserved in the 5′-untranslated region of tymovirus RNAs. Nucleic Acids Res. 1996;24(24):4910–4917. doi: 10.1093/nar/24.24.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., Huang T.H., Chang M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79(22):13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 2007;81(12):6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. ClustalW and ClustalX version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lassmann T., Sonnhammer E.L.L. Kalign, Kalignvu and Mumsa: web servers for multiple sequence alignment. Nucleic Acids Res. 2006;34:W596–W599. doi: 10.1093/nar/gkl191. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescrinier E.M., Tessari M., van Kuppeveld F.J., Melchers W.J., Hilbers C.W., Heus H.A. Structure of the pyrimidine-rich internal loop in the poliovirus 3′-UTR: the importance of maintaining pseudo-2-fold symmetry in RNA helices containing two adjacent non-canonical base-pairs. J. Mol. Biol. 2003;331(4):759–769. doi: 10.1016/s0022-2836(03)00787-3. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li L., Kang H., Liu P., Makkinje N., Williamson S.T., Leibowitz J.L., Giedroc D.P. Structural lability in stem–loop 1 drives a 5′ UTR–3′ UTR interaction in coronavirus replication. J. Mol. Biol. 2008;377(3):790–803. doi: 10.1016/j.jmb.2008.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li L., Millership J.J., Kang H., Leibowitz J.L., Giedroc D.P. A U-turn motif-containing stem–loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA. 2007;13(5):763–780. doi: 10.1261/rna.261807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li L., Keane S.C., Yang D., Leibowitz J.L., Giedroc D.P. Mouse hepatitis virus stem–loop 2 adopts a uYNMG(U)a-like tetraloop structure that is highly functionally tolerant of base substitutions. J. Virol. 2009;83(23):12084–12093. doi: 10.1128/JVI.00915-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Yokomori K., Lai M.M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J. Virol. 1990;64(12):6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Mihindukulasuriya K.A., Wu G., St Leger J., Nordhausen R.W., Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 2008;82(10):5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenkamp R., Spaan W.J. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239(1):78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Makino S. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 2001;75(19):9059–9067. doi: 10.1128/JVI.75.19.9059-9067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77(5):2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Brian D.A. Stem–loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2005;79(19):12434–12446. doi: 10.1128/JVI.79.19.12434-12446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Bouma P., Williams G.D., Brian D.A. Stem–loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2003;77(12):6720–6730. doi: 10.1128/JVI.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Peñaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Günther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Shieh C.K., Soe L.H., Makino S., Chang M.F., Stohlman S.A., Lai M.M. The 5′-end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology. 1987;156(2):321–330. doi: 10.1016/0042-6822(87)90412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirrups K., Shaw K., Evans S., Dalton K., Cavanagh D., Britton P. Leader switching occurs during the rescue of defective RNAs by heterologous strains of the coronavirus infectious bronchitis virus. J. Gen. Virol. 2000;81(Pt 3):791–801. doi: 10.1099/0022-1317-81-3-791. [DOI] [PubMed] [Google Scholar]
- Tang X.C., Zhang J.X., Zhang S.Y., Wang P., Fan X.H., Li L.F., Li G., Dong B.Q., Liu W., Cheung C.L., Xu K.M., Song W.J., Vijaykrishna D., Poon L.L.M., Peiris J.S.M., Smith G.J.D., Chen H., Guan Y. Prevalance and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80(15):7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E., Gultyaev A.P., Snijder E.J. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA. 2004;10(3):424–437. doi: 10.1261/rna.5174804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E., Posthuma C.C., Gultyaev A.P., Snijder E.J. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 2005;79(10):6312–6324. doi: 10.1128/JVI.79.10.6312-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R.G., Bredenbeek P.J., Spaan W.J. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J. Virol. 1991;65(6):3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D., Smith G.J., Zhang J.X., Peiris J.S., Chen H., Guan Y. Evolutionary insights into the ecology of coronaviruses. J. Virol. 2007;81(8):4012–4020. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang X. The leader RNA of coronavirus mouse hepatitis virus contains an enhancer-like element for subgenomic mRNA transcription. J. Virol. 2000;74(22):10571–10580. doi: 10.1128/jvi.74.22.10571-10580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K., Joo M., Narayanan K., Kim K.H., Makino S. Murine coronavirus packaging signal confers packaging to nonviral RNA. J. Virol. 1997;71(1):824–827. doi: 10.1128/jvi.71.1.824-827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.Y., Chu C.M. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Li K.S., Poon R.W., Wong B.H., Tsoi H.W., Yip B.C., Huang Y., Chan K.H., Yuen K.Y. Molecular diversity of coronaviruses in bats. Virology. 2006;351(1):180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Wang M., Lau S.K.P., Xu H., Poon R.W., Guo R., Wong B.H., Gao K., Tsoi H.W., Huang Y., Li K.S., Lam C.S., Chan K.H., Zheng B.J., Yuen K.Y. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007;81(4):1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lai K.K.Y., Huang Y., Lee P., Luk G.S.M., Dyrting K.C., Chan K.H., Yuen K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009;83(2):908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Ozdarendeli A., Brian D.A. Bovine coronavirus 5′-proximal genomic acceptor hotspot for discontinuous transcription is 65 nucleotides wide. J. Virol. 2006;80(5):2183–2193. doi: 10.1128/JVI.80.5.2183-2193.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Guy J.S., Snijder E.J., Denniston D.A., Timoney P.J., Balasuriya U.B. Genomic characterization of equine coronavirus. Virology. 2007;369(1):92–104. doi: 10.1016/j.virol.2007.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Miller T.B., Goebel S.J., Thiel V., Masters P.S. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J. Virol. 2008;82(3):1214–1228. doi: 10.1128/JVI.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]