Abstract
Programmed − 1 ribosomal frameshifting (− 1 PRF) is one viral translation strategy to express overlapping genes in positive-strand RNA viruses. Red clover necrotic mosaic virus (RCNMV) uses this strategy to express its replicase component protein p88. In this study, we used a cell-free translation system to map cis-acting RNA elements required for − 1 PRF. Our results show that a small stem-loop structure adjacent to the cap-independent translation element in the 3′ untranslated region (UTR) of RCNMV RNA1 is required for − 1 PRF. Site-directed mutagenesis experiments suggested that this stem-loop regulates − 1 PRF via base-pairing with complementary sequences in a bulged stem-loop adjacent to the shifty site. The existence of RNA elements responsible for − 1 PRF and the cap-independent translation of replicase proteins in the 3′ UTR of RNA1 might be important for switching translation to replication and for regulating the ratio of p88 to p27.
Keywords: Red clover necrotic mosaic virus, Dianthovirus, Positive-strand RNA virus, Frameshifting, Cap-independent translation, RNA-dependent RNA polymerase, Long distance RNA–RNA interaction, Bulged stem-loop, RNA structure
Introduction
The genomic RNAs of positive-strand RNA viruses are often polycistronic. Therefore, these viruses must have some strategies to translate the downstream open reading frame (ORF). The production of subgenomic RNAs (sgRNAs) is one of these strategies and is used by many viruses (Miller and Koev, 2000). Another strategy is a translational read-through mechanism, in which a stop codon is skipped in-frame by a suppressor tRNA that can recognize the stop codon, and translation continues to produce a C-terminally extended protein (Lobanov et al., 2010).
Programmed − 1 ribosomal frameshifting (− 1 PRF) is a strategy to express a downstream ORF. In response to certain signals encoded in mRNA, a small percentage of ribosomes are induced to move back by one nucleotide and to continue translation in the new (− 1) frame (Brierley, 1995, Giedroc and Cornish, 2009). As a result, a stop codon in the first ORF is skipped, and a C-terminally extended protein is produced at a certain ratio. This mechanism is used by many viruses, including those in Retroviridae, Nidovirales, astroviruses, Totiviridae, and Luteoviridae, to produce viral replicase protein (Brierley, 1995). Typically, − 1 PRF requires two cis-acting RNA elements. The first element is a heptanucleotide sequence where the reading frame shifts. This sequence usually fits the consensus X XxY YYZ (the zero frame is indicated by a space; X is any identical base, Y is A or U, Z is not G, and lowercase indicates a consensus with some exceptions) (Dreher and Miller, 2006, Jacks et al., 1988). A second element is an RNA secondary structure immediately downstream from the shifty site (Brierley et al., 1989, Brierley and Pennell, 2001). This RNA structure is regarded as a physical barrier to stop translating ribosomes and to shift the reading frame (Namy et al., 2006). A pseudoknot or a very stable RNA structure is usually proposed for the second element (Giedroc and Cornish, 2009). In addition to these RNA elements, Barley yellow dwarf virus (BYDV) requires a third RNA element that is located in the 3′ untranslated region (UTR) (Paul et al., 2001). This far-downstream element is thought to base-pair with a bulge-loop in a stem-loop adjacent to the shifty site, where it facilitates frameshifting (Barry and Miller, 2002).
Red clover necrotic mosaic virus (RCNMV) is a member of the genus Dianthovirus in the family Tombusviridae. Its genome comprises two positive-sense single-stranded RNA molecules, RNA1 and RNA2. Both genomic RNAs lack a cap structure at the 5′ end (Mizumoto et al., 2003), and a poly(A) tail at the 3′ end (Lommel et al., 1988, Mizumoto et al., 2002, Xiong and Lommel, 1989). Instead, RNA1 contains an essential RNA element (3′TE-DR1) that is required for cap-independent translation (Mizumoto et al., 2003). RNA1 encodes replicase component protein p27 and its N-terminally coincident but C-terminally distinct protein p88 (Xiong and Lommel, 1989). Both p27 and p88 are required for RNA replication and are contained in RCNMV RNA replicase complexes (Bates et al., 1995, Mine et al., 2010a, Mine et al., 2010b). RNA1 also encodes a coat protein that is expressed from sgRNA (Xiong et al., 1993a, Zavriev et al., 1996). RNA2 encodes a cell-to-cell movement protein (Xiong et al., 1993a).
p27 is an auxiliary replicase protein that plays essential roles in multiple steps in RCNMV RNA replication: specific recognition of viral RNA and recruitment of viral RNAs to the endoplasmic reticulum membrane, the site of RNA replication (Hyodo et al., 2011, Mine et al., 2010b). p88 has an RNA-dependent RNA polymerase motif (Koonin, 1991) and is required in cis for the replication of RNA1 in a translation-coupled manner (Iwakawa et al., 2011, Okamoto et al., 2008). p88 is translated via a − 1 PRF event, which occurs in less than 10% of translations from RNA1 in plant and rabbit reticulocyte lysate (Kim and Lommel, 1994, Kim and Lommel, 1998, Xiong et al., 1993b). Several previous works identified two cis-acting RNA elements that are necessary and sufficient for − 1 PRF in RCNMV RNA1. One element is the heptanucleotide sequence G GAU UUU, where frameshifting takes place (Kim and Lommel, 1994, Xiong et al., 1993b). The other element is a bulged stem-loop structure predicted to be adjacent to the shifty site (Kim and Lommel, 1998). In addition to these elements, our previous study suggested that the third cis-acting RNA element required for − 1 PRF could exist in the 3′ UTR of RCNMV RNA1 (Iwakawa et al., 2007).
In this study, using a cell-free extract in vitro assay system prepared from evacuolated tobacco BY-2 protoplast lysate (BYL; Komoda et al., 2004), we mapped the third cis-acting RNA element required for efficient − 1 PRF. Our results show that a small stable stem-loop structure adjacent to 3′TE-DR1 in the 3′ UTR of RNA1 is required for efficient − 1 PRF and that this stem-loop structure promotes − 1 PRF via base-pairing with a bulge of the stem-loop structure adjacent to the shifty site.
Results
Mapping of the regions required for − 1 PRF in the 3′ UTR of RCNMV RNA1
Our previous study showed that several capped RNA1 mutants with deletions in the 3′ UTR supported the accumulation of negative-strand RNA2 less efficiently than did the wild-type (wt) RNA1 in BYL, although these RNA1 mutants produced p27 to similar or even higher levels than did wt RNA1 (Iwakawa et al., 2007). This result suggests that the deleted regions are involved in the production of p88, which is translated via − 1 PRF. To investigate whether the 3′ UTR of RNA1 has the cis-acting RNA element(s) required for − 1 PRF and the production of p88, we tested RNA1 mutants with a series of deletions in their 3′ UTR (Figs. 1A and B, Iwakawa et al., 2007) for their ability to produce p88 in BYL. We used capped viral RNA transcripts because the 3′ UTR of RNA1 contains RNA elements essential for cap-independent translation of both p88 and p27 (Iwakawa et al., 2007, Mizumoto et al., 2003). Capped RNA1 mutants were incubated in BYL at 17 °C for 4 h, and the accumulated levels of p27 and p88 were analyzed by western blotting using an anti-p27 antiserum. For this assay, we used the membrane fraction of BYL obtained after centrifugation at 20,000 × g for 10 min because the level of p88 accumulated in BYL was below the detectable threshold (data not shown). An RNA1 mutant (R1-SM) that has mutations in the heptanucleotide shifty site was used as a negative control. d3′SLB, d3′SLC, and d3′SLDE-5′ accumulated lower levels of p88 than those accumulated in wt RNA1, whereas all the RNA1s tested accumulated p27 to similar levels (Fig. 1C, lanes 3–5). d3′SLF that lacks the 3′ terminal SL required for negative-strand synthesis (Iwakawa et al., 2007) accumulated p88 as efficiently as wt RNA1 (Fig. 1C, lane 7), suggesting that RNA replication, including negative-strand RNA synthesis and accumulations of dsRNAs, has no or little effects on the production of p88 via − 1 PRF in BYL. These results suggest that the regions, including SLB, SLC, and SLDE, contain cis-acting RNA elements required for − 1 PRF. To delimit the regions required for − 1 PRF, we constructed RNA1 mutants with a series of deletions in the regions (Figs. 1A and D) and tested them as described above. Western blot analysis showed that the accumulated levels of p88 were much lower in RNA1 mutants with deletions in the stem of SLC than in wt RNA1 and in other RNA1 mutants despite their ability to accumulate p27 to a level similar to that in wt RNA1 (Fig. 1E, lanes 4 and 6). Deletions in SLB and SLDE affected the accumulation of p88 mildly (Fig. 1E, lanes 2, 3, 7, and 8). Interestingly, deletion of 3′TE-DR1c did not affect the accumulation of p88 (Fig. 1E, lane 5). This result suggests that the region of SLC, except for 3′TE-DR1c, is involved in efficient − 1 PRF.
Fig. 1.
Identification of the regions required for − 1 PRF in RCNMV RNA1. (A) Schematic diagram of RCNMV RNA1 and the secondary structure predicted in the 3′ UTR (Iwakawa et al., 2007). Numbers indicate nucleotide positions in RCNMV RNA1. (B and D) Schematic representation of deleted regions in the 3′ UTR of RCNMV RNA1. Boldface lines indicate the virus-derived sequences with the nucleotide numbers at the 5′ and 3′ ends. Bent lines indicate the deleted regions. (C and E) The effects of deletions in the 3′ UTR of RCNMV RNA1 on − 1 PRF. RNA1 and its mutants were incubated in BYL for 4 h, and the accumulated levels of p27 and p88 were analyzed by western blotting using an anti-p27 antiserum. Coomassie brilliant blue (CBB)-stained cellular proteins are shown below the western blottings as a loading control. The accumulated levels of both p27 and p88 were quantified with the Image Gauge program (Fuji Photo Film, Tokyo), respectively. The ratios of p88 to p27 were calculated, and the mean values with standard errors relative to the wild type from at least three independent experiments are shown below the CBB-stained proteins.
To delineate the regions required for − 1 PRF more precisely, we tested RNA1 mutants with smaller deletions in SLC (Figs. 2A and B). Deletions of two-thirds of the 5′ stem of SLC and half of the 3′ stem of SLC severely decreased the accumulated levels of p88 without affecting the accumulation of p27 (Fig. 2C, lanes 2, 3, and 7). These results suggest that the nucleotide sequences and/or RNA structures in the basal region of SLC are important for − 1 PRF.
Fig. 2.
The basal stem region of SLC is required for − 1 PRF in RCNMV RNA1. (A) Predicted secondary structure of SLC in the 3′ UTR of RCNMV RNA1. Numbers indicate nucleotide positions in RCNMV RNA1. (B) Schematic representation of deleted regions in SLC. (C) The effects of deletions in SLC on the translation of p88 in BYL. For others, see the legend to Fig. 1.
A small stem-loop in SLC is required for − 1 PRF
To identify the cis-acting RNA elements required for − 1 PRF in SLC, we analyzed the RNA secondary structures predicted in this limited region using the Mfold program (version 3.4; Zuker, 2003). A basal stem of SLC and a small stem-loop structure (named here SLCsSL) are predicted here (Fig. 3A). In particular, SLCsSL is expected to be a stable structure (ΔG = − 15.52 kcal/mol at 17 °C), and we focused on SLCsSL.
Fig. 3.
Both the stem structure and the loop sequence of the small stem-loop in SLC are essential for − 1 PRF in RCNMV RNA1. (A) Predicted secondary structure of the small stem-loop and basal stem in SLC. Boldface italics indicate disrupted and restored stem structures and altered loop sequences. Numbers show nucleotide positions in RCNMV RNA1. (B) Effects of mutations introduced in SLC on − 1 PRF in BYL. For other conditions, see the legend to Fig. 1. (C) Predicted secondary structures of the small stem-loop in the region within SLC required for − 1 PRF in dianthoviruses using the Mfold program (Zuker, 2003). Conserved nucleotides are denoted by boldface letters with asterisks. SCNMV and RCNMV-Can mean Sweet clover necrotic mosaic virus and RCNMV Canadian strain, respectively. For others, see the text. (D) Effects of mutations introduced in SLC on viral replication in BY-2 protoplasts. BY-2 protoplasts were inoculated with viral RNA transcripts and incubated for 16 h. Total RNA and protein were extracted and used for northern and western blot analysis, respectively. Western blotting was performed using an anti-p27 antiserum, and northern blotting was performed using appropriate digoxigenin-labeled RNA probes. Coomassie brilliant blue (CBB)-stained cellular proteins are shown below the western blotting as a loading control. EtBr-stained rRNAs are shown below the northern blotting as a loading control.
To investigate the role of SLCsSL in − 1 PRF, we tested RNA1 mutants with nucleotides substituted in the loop or the stem of SLCsSL using the method described above. Disruption of the stem of SLCsSL reduced the accumulation of p88 markedly, and restoration of the stem structure by compensatory mutations restored the ability to accumulate p88 (Fig. 3B, lanes 2–4). 3′SLCsSL-mLoop, which has nucleotide substitutions in the loop of SLCsSL, accumulated p88 at very low levels (Fig. 3B, lane 5). Nucleotide changes in the basal short stem of SLC that disrupt and restore the stem structure did not affect the accumulated levels of p88 (Fig. 3B, lanes 6–8). All the RNA1 mutants used here accumulated p27 to levels similar to that of wt RNA1 (Fig. 3B). These results strongly suggest that both the stem structure and the loop sequence of SLCsSL are required for − 1 PRF in BYL.
To investigate the roles of SLCsSL in RCNMV replication in vivo, we performed protoplast experiments. BY-2 protoplasts were inoculated with uncapped transcripts of RNA1 mutants with mutations in SLCsSL used in BYL experiments (Fig. 3A) together with RNA2, and RCNMV replication was assessed by analyzing the accumulation of viral RNAs and p27 by northern and western blot analyses using appropriate RNA probes and an anti-p27 antiserum. RNA1 mutants with the stem structure of SLCsSL disrupted, or the mutant with nucleotides substituted in its loop did not replicate efficiently and failed to support the efficient replication of RNA2, whereas the mutant with the stem structure of SLCsSL restored replicated and supported the replication of RNA2 as efficiently as wt RNA1 (Fig. 3D, lanes 2–5). All these uncapped transcripts accumulated p27 to similar levels in BYL (data not shown). Together with the results in BYL, these results suggest that both the stem structure and the loop sequence of SLCsSL are required for − 1 PRF in RCNMV RNA1.
We next compared the nucleotide sequence and the secondary structure of the SLCsSL among dianthoviruses. The nucleotide sequences in the region were highly conserved in RCNMV Canadian strain and Sweet clover necrotic mosaic virus, but were less conserved in Carnation ringspot virus (CRSV) (Fig. 3C). However, the RNA secondary structures predicted there by the Mfold program were highly conserved among all dianthoviruses (Fig. 3C). These findings support the idea that the RNA secondary structure of SLCsSL, rather than its nucleotide sequences, is important for − 1 PRF.
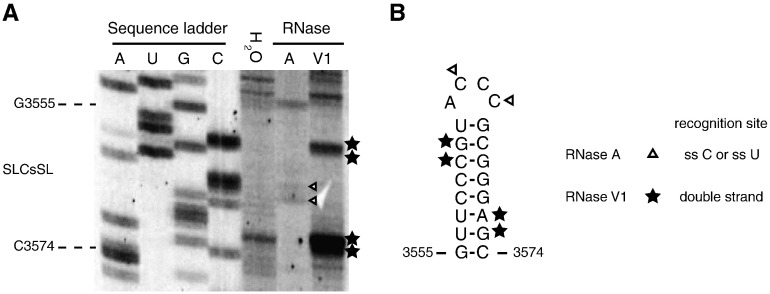
In addition, to verify the computer-predicted structure of SLCsSL, we performed enzymatic probing in solution. In this assay, in vitro transcribed full-length wt RNA1 was used as a template for the reactions. The signals of RNase V1, which preferentially digests double-stranded regions, were found in the predicted stem of SLCsSL, and those of RNase A, which preferentially cleaves phosphodiester bonds after the 3′ phosphates of unpaired cytosine and uracil, were found in the predicted loop of SLCsSL (Figs. 4A and B). These results support the predicted structures of SLCsSL.
Fig. 4.
Enzymatic probing analysis supports the predicted RNA secondary structures of SLCsSL. (A) Enzymatic structure probing of SLCsSL in vitro. RNA transcripts were subjected to enzymatic modifications with RNase A and RNase V1, and the products generated were analyzed by primer extension using a primer 3′SLCsSL-2forSP. The products were separated in a 5% polyacrylamide gel in the presence of 8 M urea along with a sequence ladder generated with the same primer used for reverse transcription. Specific bands of the RNase A treated lane are indicated by white arrowheads and those of the RNase V1 treated lane are indicated by black stars. (B) The results of the structure probing are mapped onto the predicted secondary structure model of the SLCsSL. Different reactivities with residues are indicated by various symbols that are defined in the model.
A long-distance RNA–RNA interaction between SLCsSL and a bulged stem-loop adjacent to the slippage site is required for − 1 PRF in RCNMV RNA1
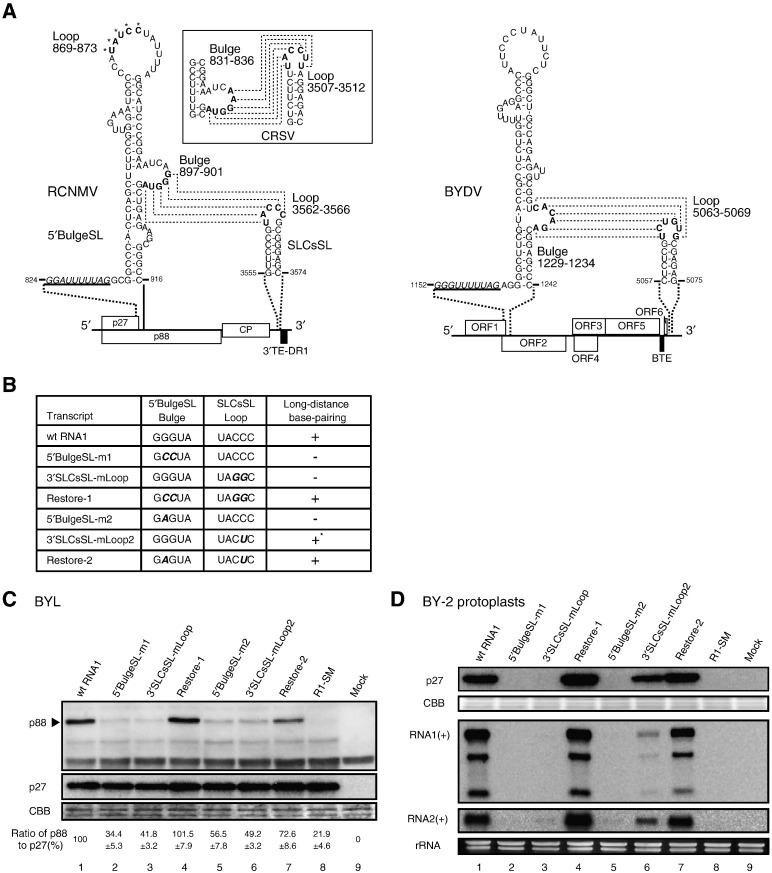
We next investigated how SLCsSL, which is located more than 2.5 kb from the shifty site, participates in − 1 PRF. We hypothesized that SLCsSL stimulates − 1 PRF via an interaction with the upstream signal. To address this issue, we examined the nucleotide sequences of the upstream cis-acting RNA element that is complementary to the loop sequence of SLCsSL. Five bases 897-GGGUA-901 in the bulge of the stem-loop adjacent to the shifty site (named here 5′BulgeSL) can potentially base-pair to five bases 3562-UACCC-3566 in the loop sequence of SLCsSL (Fig. 5A). These long-distance base-pairings are possible in all dianthoviruses, although the sequences in the 5′BulgeSL (831-AAGGUA-836) and the SLCsSL (3507-UACCUU-3512) of CRSV slightly differ from those of other dianthoviruses (Fig. 5A, data not shown). Interestingly, as shown in Fig. 5A, the RNA secondary structures of both 5′BulgeSL and SLCsSL are quite similar to those of a bulged stem-loop adjacent to the shifty site and a stem-loop predicted in the 3′ UTR of BYDV genomic RNA (Barry and Miller, 2002). Moreover, in BYDV, the long-distance base-pairing between them is required for efficient − 1 PRF (Barry and Miller, 2002).
Fig. 5.
A long-distance RNA–RNA interaction between 5′BulgeSL and SLCsSL is essential for − 1 PRF in RCNMV RNA1. (A) Predicted secondary structures of RCNMV RNA1 and BYDV genomic RNA showing the base-pairing interaction. Potential base-pairings in CRSV RNA1 is shown in the box. Dotted lines indicate potential base-pairings between 5′ bulged stem-loop and the stem-loop in the 3′ UTR. Underlined italics indicate a shifty heptanucleotide sequence followed by the stop codon of p27 or ORF1, respectively. Numbers show nucleotide positions in RCNMV RNA1, CRSV RNA1, and BYDV genomic RNA, respectively. In RCNMV, letters labeled with asterisks in the apical loop of 5′BulgeSL indicate the nucleotide sequences complementary to those in the bulge of 5′BulgeSL (Kim and Lommel, 1998). Black boxes indicate the cap-independent translation enhancer element of RCNMV and BYDV. (B) Mutations introduced into the bulge of 5′BulgeSL, the loop of SLCsSL, or both. Boldface italics indicate altered nucleotides. +: complete base-pairing, −: incomplete base-pairing by one or two mismatches, +⁎ : complete base-pairing including one non-Watson–Crick base-pairing. Effects of the mutations introduced into 5′BulgeSL, SLCsSL or both on − 1 PRF in BYL (C) and in BY-2 protoplasts (D). For others, see the legends to Fig. 1, Fig. 3.
To test if the possible long-distance RNA–RNA interaction is required for − 1 PRF, we constructed RNA1 mutants with mutations that disrupt and restore the potential base-pairings. These RNA1 mutants were analyzed in both BYL and BY-2 protoplasts as described above. 5′BulgeSL-m1 and 5′BulgeSL-m2 have mutations in the bulge of 5′BulgeSL, 3′SLCsSL-mLoop and 3′SLCsSL-mLoop2 have mutations in the loop sequence of SLCsSL, and Restore-1 and Restore-2 have mutations in both the 5′ bulge and the 3′ loop, which restore the potential base-pairings between them (Fig. 5B). 5′BulgeSL-m1 has the mutation in the central two bases that alters the amino acid sequence of p88, arginine to serine and valine to leucine at positions 259 and 260, respectively, but the mutation in 5′BulgeSL-m2 does not alter it. All the RNA1 mutants with mutations that disrupt the 5′–3′ long-distance RNA–RNA interaction accumulated much less p88 in BYL (Fig. 5C, lanes 2, 3, 5, and 6) and failed to support the efficient accumulation of viral genomic RNAs in BY-2 protoplasts (Fig. 5D, lanes 2, 3, 5, and 6). By contrast, RNA1 mutants with mutations that restored the 5′–3′ long-distance RNA–RNA interaction accumulated p88 as efficiently as did wt RNA1 in BYL (Fig. 5C, lanes 4 and 7) and supported the accumulation of viral genomic RNAs in BY-2 protoplasts (Fig. 5D, lanes 4 and 7). It should be noted that, although the C to U mutation in 3′SLCsSL-mLoop2 still allows non-Watoson–Crick base-pairing (G–U), this mutation greatly reduced the accumulated level of p88 (Fig. 5C, lane 6). These results suggest that the stable long-distance RNA–RNA base-pairing between 5′BulgeSL and SLCsSL is required for efficient − 1 PRF in RCNMV RNA1.
Discussion
In this paper, we show that a cis-acting RNA element in the 3′ UTR of RCNMV RNA1 facilitates − 1 PRF through base-pairing with a bulge sequence in the stem-loop structure (5′BulgeSL) predicted to be just downstream of the shifty site. Two cis-acting RNA elements, a heptanucleotide sequence and 5′BulgeSL, are required for − 1 PRF in RCNMV RNA1 (Kim and Lommel, 1994, Kim and Lommel, 1998, Xiong et al., 1993b). In the predicted structure of 5′BulgeSL (Fig. 5A; Kim and Lommel, 1998), the five-nucleotide sequence (897-GGGUA-901) involved in the long-distance interaction can potentially base-pair to five bases 869-UAUCC-873 in the large loop sequence of the 5′BulgeSL and could form an apical loop-internal loop interaction (Mazauric et al., 2008). The putative pseudoknot played no role in − 1 PRF in an assay using rabbit reticulocyte lysate and in infectivity assays using Nicotiana benthamiana (Kim and Lommel, 1998), supporting the role of the five-nucleotide sequence in the bulge in a long-distance interaction. However, nucleotide substitutions in the bulge sequences in 5′BulgeSL caused no deleterious effects on the accumulation of p88 in rabbit reticulocyte lysate or on the infectivity in N. benthamiana (Kim and Lommel, 1998). These results contrast with our data showing that nucleotide substitutions in the same bulge sequences almost abolished the ability of RNA1 to accumulate p88 in BYL and to replicate and support RNA2 replication in BY-2 protoplasts (Figs. 5C and D). Such differences might arise from different assay systems and host plants. The requirement of RNA elements in the 3′TE-DR1-mediated cap-independent translation of RCNMV RNA1 differs between host plants (Sarawaneeyaruk et al., 2009). Alternatively, the six nucleotides changed in the previous research might produce other long-distance interactions to promote − 1 PRF.
Roles of a long-distance RNA–RNA interaction in − 1 PRF in the production of p88
Long-distance RNA–RNA interactions regulate various steps in the viral life cycle. For example, BYDV and Tomato bushy stunt virus (TBSV) require RNA–RNA base-pairings between the 5′ UTR and the 3′ UTR to initiate translation (Fabian and White, 2004, Guo et al., 2001, Nicholson et al., 2010, Treder et al., 2008). Bacteriophage Qβ and TBSV require a long-distance RNA–RNA interaction for RNA replication (Klovins et al., 1998, Panavas and Nagy, 2005). Viruses such as Potato virus X, Flock house virus, and Coronavirus require long-distance base-pairings for sgRNA transcription (Kim and Hemenway, 1999, Lindenbach et al., 2002, Moreno et al., 2008). In the case of RCNMV, an intermolecular RNA–RNA interaction plays an essential role in sgRNA transcription (Sit et al., 1998). In addition to these roles, a long-distance RNA–RNA interaction is required for − 1 PRF in BYDV (Barry and Miller, 2002, Paul et al., 2001) and in RCNMV (Fig. 5). In many viruses, RNA pseudoknot structures predicted to be adjacent to the heptanucleotide shifty site alone seem sufficient for − 1 PRF (Giedroc and Cornish, 2009). Such RNA pseudoknot structures are regarded as a physical barrier to stop translating ribosomes and facilitate − 1 PRF (Namy et al., 2006). It is unclear why the downstream complex stem-loop structure (5′BulgeSL) alone is not enough to facilitate − 1 PRF in RCNMV RNA1 or in BYDV. Because the potential base-pairing between the bulge sequence and the large loop sequence within the 5′BulgeSL, which could form an atypical pseudoknot, has no role in − 1 PRF in RCNMV RNA1 (Kim and Lommel, 1998), 5′BulgeSL might not be strong enough to stall the translating ribosomes efficiently. Kissing RNA–RNA interactions between two stem-loops are more stable than a simple RNA helix of the same sequence (Weixlbaumer et al., 2004). Taking this into account, the long-distance base-pairings between the stem-loop in the 3′ UTR and the 5′BulgeSL seem to be required to stabilize the 5′BulgeSL to allow for the stalling of the elongating ribosomes over the shifty site to promote − 1 PRF in RCNMV RNA1.
Switching from translation to replication
The existence of RNA elements responsible for − 1 PRF and the cap-independent translation of replicase proteins in the 3′ UTR of RNA1 (Mizumoto et al., 2003; this paper) might be important for switching translation to replication. A model for the switch from translation to replication has been reported in BYDV, in that the passage of RNA replicase on the 3′ UTR of the viral genome disrupts the structures of the 3′ RNA elements, which are needed for two sets of the long-distance base-pairing required for cap-independent translation and − 1 PRF (Barry and Miller, 2002). However, a long-distance RNA–RNA base-pairing between the 5′ and 3′ UTRs seems not to be essential for 3′TE-DR1-mediated cap-independent translation in RCNMV RNA1 (Sarawaneeyaruk et al., 2009).
RCNMV RNA1 requires p88 in cis for its replication (Iwakawa et al., 2011, Okamoto et al., 2008). This suggests that only RNA1 molecule, in which − 1 PRF occurs, can be a template for RNA replication. p88 binds to RNA1 and its 3′ UTR fragment (named SR1f) (Iwakawa et al., 2008) through a puromycin-insensitive translation-coupled mechanism (Iwakawa et al., 2011). The p88 bound in the 3′ UTR might become a core for assembling the 480-kDa RNA replicase complexes with p27 and host proteins to initiate negative-strand RNA synthesis (Mine et al., 2010a, Mine et al., 2010b). The p88 binding or the formation of the 480-kDa RNA replicase complexes in the 3′ UTR of RNA1 might disrupt the structures of the 3′ RNA elements required for 3′TE-DR1-mediated cap-independent translation and − 1 PRF. Structural rearrangement of RNA elements is reported in Turnip crinkle virus, in that the binding of RNA-dependent RNA-polymerase to the 3′ UTR of the viral genomic RNA induces a conformational shift of the RNA element required for efficient ribosome binding, causing a transient switch from translation to replication (Yuan et al., 2009).
In contrast to p88, p27 binds to RCNMV RNA1 through a puromycin-sensitive translation-coupled mechanism, and it does not bind to the 3′ UTR fragment SR1f (Iwakawa et al., 2011). These previous results suggest that p27 is associated mainly with translating RNA1 with polyribosomes because puromycin, a peptidyl acceptor antibiotic, causes polypeptide chain termination and induces the dissociation of polyribosomes from mRNA (Blobel and Sabatini, 1971, Lehninger et al., 1993), and suggest that p27 does not bind to the 3′ UTR of RNA1. The lack of the association between p27 and the 3′ UTR of RNA1 might allow translating RNA1 to continue to be a template for further translation for p27 until − 1 PRF occurs by an unknown mechanism(s) that regulates the frequency of − 1 PRF. Thus, the existence of RNA elements responsible for − 1 PRF in the translation of p88 in the 3′ UTR of RNA1 and the distinct binding properties between p27 and p88 might be important for regulating the ratio of p88 and p27.
In summary, we present a model for regulation of translation and replication in RCNMV RNA1 (Fig. 6 ). In this model, p27 interacts with its template RNA1 except for the 3′ UTR. The production of sufficient amounts of p27 allows 5′BulgeSL to access SLCsSL by an unknown mechanism. The formation of base-pairings between 5′BulgeSL and SLCsSL facilitates the translation of p88 via − 1 PRF. p88 interacts with the 3′ UTR of its template RNA1. The interaction of p88 or the formation of the 480-kDa replicase complex disrupts the structures of the 3′ RNA elements required for both cap-independent translation and − 1 PRF, causing a switch from translation to replication of RCNMV RNA1.
Fig. 6.
A model for regulation of translation and replication. (A) At the early replication step, an auxiliary replicase protein p27 is produced via 3′TE-DR1-mediated cap-independent translation. p27 interacts with translating RNA1 except for its 3′ UTR (Iwakawa et al., 2011). The lack of the association of p27 with the 3′ UTR containing 3′TE-DR1 allows translating RNA1 to continue to be a template for p27. (B) Sufficient amounts of p27 on RNA1 allow 5′BulgeSL to access SLCsSL by an unknown mechanism. (C) The interaction between 5'BulgeSL and SLCsSL facilitates the production of p88 via − 1 PRF. (D) p88 binds to the 3′ UTR of RNA1 through a translation-coupled manner by an unknown mechanism (Iwakawa et al., 2011). The p88 binding or the formation the 480-kDa replicase complex in the 3′ UTR of RNA1 disrupts RNA structures required for both cap-independent translation and − 1 PRF. The conformational change of RNA structure causes a switch from translation to replication of RNA1.
Materials and methods
Plasmids construction
pUCR1 and pRC2|G are full-length cDNA clones of RNA1 and RNA2 of RCNMV Australian strain, respectively (Takeda et al., 2005, Xiong and Lommel, 1991). Constructs described previously that were used in this study included: pUCR1-d3′SLA, pUCR1-d3′SLB, pUCR1-d3′SLC, pUCR1-d3′SLD (we renamed as pUCR1-d3′SLDE-5′ in this paper), pUCR1-d3′SLE (we renamed as d3′SLDE-3′ in this paper), pUCR1-d3′SLF, and pUCR1-d3′TE (Iwakawa et al., 2007). All constructs were verified by sequencing. The primers used in this study are listed in Table 1 .
Table 1.
List of primer and their sequences used for PCR to generate constructs.
| Primers | Sequence |
|---|---|
| R1_EcoRI+ | CCTCAGTAAATGAATTCTTCG |
| R1_XhoI- | CCACCTTCTCGAGTACATCG |
| Slippery+ | CAAATCCCTTGAGGACTTCTAGGCGGCCCACTCAGCTTTC |
| Slippery- | GGCCGCCTAGAAGTCCTCAAGGGATTTGAACCCAGC |
| A1+3380 | TGCAGTTTTCAGGTTCC |
| M4 | GTTTTCCCAGTCACGAC |
| dSLB-5′+ | AAGAGGGGAACAACAGTAAAATTGCAAAAAATAGAG |
| dSLB-5′- | GCAATTTTACTGTTGTTCCCCTCTTGCAACTCG |
| dSLB-3′+ | CCCTGTTGGCAATAGGAGTAGTTCCCGTACCC |
| dSLB-3′- | ACTACTCCTATTGCCAACAGGGTCGGCGAG |
| dSLC-5′+ | AATAGAGTGCGACCCTGGGAAACAGGTACC |
| dSLC-5′- | CCCAGGGTCGCACTCTATTTTTTGCAATTTTACTG |
| dSLC-3′+ | GTGCGCACGTTTTTCTTTTAGGTAGGAGCAC |
| dSLC-3′- | CCTAAAAGAAAAACGTGCGCACAACCACACAGAGG |
| dSLD-5′+ | GTTATTTCCTTACCTCTGGTAAAACAAAATTGGC |
| dSLD-5′- | ACCAGAGGTAAGGAAATAACTACAACAGTGAG |
| dSLD-3′+ | AGAGGGCGCAAACTCAGGTTAATAAAACAG |
| dSLD-3′- | AACCTGAGTTTGCGCCCTCTGGAGCAAGTGC |
| dSLC-5′a+ | AATAGAGTGCGCGGGAGCAAGACCCTACTAC |
| dSLC-5′a- | CTTGCTCCCGCGCACTCTATTTTTTGCA |
| dSLC-5′b+ | TCCCGTACCCCAGTAGACGAACCGGCATCG |
| dSLC-5′b- | GTTCGTCTACTGGGGTACGGGAACTACTCCTAG |
| dSLC-5′c+ | GACCCTACTAGACCCTGGGAAACAGGTACC |
| dSLC-5′c- | GTTTCCCAGGGTCTAGTAGGGTCTTGCTCCCGC |
| dSLC-3′a+ | GTGCGCACGTCTGTTGTAGTTATTTCCTTTTTC |
| dSLC-3′a- | ACTACAACAGACGTGCGCACAACCACACAG |
| dSLC-3′b+ | GAAGACTCTCATTTTCTTTTAGGTAGGAGCAC |
| dSLC-3′b- | CTAAAAGAAAATGAGAGTCTTCCGACAACGAC |
| SLC sSL5′b+ | GAGTGCTAGGAGTAGTTGGGGTACCCGCGGGAGCAAGACCC |
| SLC sSL5′b- | GTCTTGCTCCCGCGGGTACCCCAACTACTCCTAGCACTCTAT |
| SLC sSL3′b+ | GGAGTAGTTCCCGTACCCGCCCCAGCAAGACCCTACTACAGTAG |
| SLC sSL3′b- | CTGTAGTAGGGTCTTGCTGGGGCGGGTACGGGAACTACTCC |
| SLC sSLmLoop+ | GCTAGGAGTAGTTCCCGTAGGCGCGGGAGCAAGACCCTACTAC |
| SLC sSLmLoop- | GTAGGGTCTTGCTCCCGCGCCTACGGGAACTACTCCTAGC |
| SLC sSLmR+ | GAGTGCTAGGAGTAGTTGGGGTACCCGCCCCAGCAAGACCCTACTACAGTAG |
| SLC sSLmR- | CTGTAGTAGGGTCTTGCTGGGGCGGGTACCCCAACTACTCCTAGCACTCTAT |
| SLC largeSL5′b+ | GCAAAAAATAGAGTGCTACCAGTAGTTCCCGTACCCGCGG |
| SLC largeSL5′b- | GCGGGTACGGGAACTACTGGTAGCACTCTATTTTTTGCAA |
| SLC largeSL3′b+ | CTCACTGTTGTAGTTATTTGGTTTTTCTTTTAGGTAGGAGC |
| SLC largeSL3′b- | GCTCCTACCTAAAAGAAAAACCAAATAACTACAACAGTGAG |
| slipSLm1+ | GGCATCCCGGAAATCAGCCTAGCTGAGAAGCGGGCCAGTAG |
| slipSLm1- | GGCCCGCTTCTCAGCTAGGCTGATTTCCGGGATGCCTAAAATAG |
| slipSLm2+ | GGCATCCCGGAAATCAGAGTAGCTGAGAAGCGGGCCAGTAG |
| slipSLm2- | GGCCCGCTTCTCAGCTACTCTGATTTCCGGGATGCCTAAAATAG |
| SLC sSLmLoop2+ | GCTAGGAGTAGTTCCCGTACTCGCGGGAGCAAGACCCTACTAC |
| SLC sSLmLoop2- | GTAGGGTCTTGCTCCCGCGAGTACGGGAACTACTCCTAGC |
| 3′SLCsSL-2forSP | TCCGACAACGACGTGCGCAC |
DNA fragments were amplified by PCR from pUCR1 using primer A1 + 3380 plus one each of following: dSLB-5′-, dSLB-3′-, dSLC-5′-, dSLC-3′-, dSLD-5′-, dSLD-3′-, dSLC-5′a-, dSLC-5′b-, dSLC-5′c-, dSLC-3′a-, dSLC-3′b-, SLC sSL5′b-, SLC sSL3′b-, SLC sSLmR-, SLC sSLmLoop-, SLC largeSL5′b-, SLC largeSL3′b-, and SLC sSLmLoop2-, respectively. Another primer M4 was used together with one each following: dSLB-5′+, dSLB-3′+, dSLC-5′+, dSLC-3′+, dSLD-5′+, dSLD-3′+, dSLC-5′a+, dSLC-5′b+, dSLC-5′c+, dSLC-3′a+, dSLC-3′b+, SLC sSL5′b+, SLC sSL3′b+, SLC sSLmR+, SLC sSLmLoop+, SLC largeSL5′b+, SLC largeSL3′b+, and SLC sSLmloop2+, respectively. Then a PCR fragment was amplified from a mixture of these two fragments using the primers A1 + 3380 and M4, digested with MluI and SphI and used to replace the corresponding region of pUCR1.
pUCR1-3′SLCbSL-R
DNA fragments were amplified by PCR from pUCR1 using three sets of primers, A1 + 3380 plus SLC largeSL5′b-, SLC largeSL5′b+ plus SLC largeSL3′b-, and SLC largeSL3′b+ plus M4. The amplified DNA fragments were mixed and further amplified by PCR using the primer pair A1 + 3380 and M4. The amplified DNA fragments were digested with MluI and SphI, and used to replace the corresponding region of pUCR1.
pUCR1-SM, pUCR1-5′BulgeSL -m1, and pUCR1-5′BulgeSL -m2
DNA fragments were amplified by PCR from pUCR1 using primer R1_EcoRI+ plus one each following: Slippery-, slipSLm1-, and slipSLm2-, respectively. Another primer R1_XhoI- was used together with one each of following: Slippery+, slipSLm1+, and slipSLm2+, respectively. Recombinant PCR products were amplified with the primer pair R1_EcoRI+ and R1_XhoI+, digested with EcoRI and XhoI and used to replace the corresponding region of pUCR1.
pUCR1-Restore-1 and pUCR1-Restore-2
DNA fragments were amplified by PCR from pUCR1 using primer R1_EcoRI+ plus one each following: slipSLm1- and slipSLm2-, respectively. Another primer R1_XhoI- was used together with one each of following: slipSLm1+ and slipSLm2+, respectively. Recombinant PCR products were amplified with the primer pair R1_EcoRI+ and R1_XhoI+, digested with EcoRI and XhoI and used to replace the corresponding region of pUCR1-3′SLCsSL-mLoop and pUCR1-3′SLCsSL-mLoop2, respectively.
Protoplast experiments
BY-2 protoplast experiments were performed as described previously (Iwakawa et al., 2007). Briefly, RNA1 (1.1 pmol) or its derivatives with RNA2 (1.1 pmol) was suspended in 0.2 ml cold MES buffer and mixed with 0.6 ml of BY-2 protoplast solution (1.67 × 106 cells/ml) before electroporation using a Pulse Controller Plus (Bio-Rad). Protoplasts were incubated at 17 °C for 16 h in the dark. Total RNAs were subjected to northern blot analysis as described previously (Iwakawa et al., 2007). The probe-specific RNA signals were detected using a luminescent image analyzer (LAS 1000 Plus; Fuji Photo Film). Total proteins were subjected to western blot analysis using an anti-p27 antiserum, and the signals were detected using a luminescent image analyzer (LAS 1000 Plus).
BYL experiments
Preparation of cell-free extracts of evacuolated tobacco BY-2 protoplasts and in vitro translation/replication reaction were as described previously (Komoda et al., 2004). Western blot analysis was performed essentially as described previously (Iwakawa et al., 2007). Briefly, capped RNA1 (1.5 μg) or its mutants were incubated in 25 μl BYL at 17 °C for 4 h together with uncapped RNA2 (100 ng). After incubation, BYL was centrifuged at 20,000 × g for 10 min at 4 °C to obtain membrane fractions. The proteins from membrane fractions were analyzed by western blotting using an anti-p27 antiserum. The signals were detected with a luminescent image analyzer (LAS 1000 Plus), and the signal intensities were quantified with the Image Gauge program (Fuji Photo Film).
Enzymatic probing of the RNA secondary structures
Enzymatic probing was performed essentially as described previously (Fabian and White, 2008, Wu et al., 2001). Briefly, in vitro transcribed wt RNA1 (approximately 14 pmol) was treated by heating at 65 °C for 2 min, 37 °C for 10 min, and 25 °C for 10 min in reaction buffer (10 mM Tris [pH 7.0], 100 mM KCl, 10 mM MgCl2) with 3 μg of yeast RNA in 20 μl volume. One microliter of 0.1 U/μl RNase V1 (Ambion) or 0.1 ng/μl RNase A (Qiagen) was added to each of the pretreated RNAs and incubated at 25 °C for 1 min. As a control, 1 μl of nuclease-free water was added to the pretreated RNA and incubated at 25 °C for 1 min. The reaction was terminated by phenol-chloroform extraction, and ethanol precipitated, washed with 70% ethanol, and dried by vacuum. The cleavage products were resuspended in 8 μl nuclease-free water. One microliter of each enzymatically treated RNA transcripts was mixed with 0.5 μl of 10 pmol/μl primer, incubated at 90 °C for 1 min and then transferred to room temperature for 5 min. The extension reaction was carried out in a final volume of 9.5 μl that included reverse transcriptase (Superscript III, Invitrogen), a 1× concentration of the buffer provided by the manufacturer, 0.05 mM each of dCTP, dGTP and dTTP, 5 mM of DTT, 0.5 μl of 40 U/μl RNase inhibitor (TOYOBO), and 10 μCi [α-35S] dATP. The reaction was incubated at 55 °C for 5 min, and then 0.5 μl of each of the four dNTPs (10 mM) was added. The mixture was incubated at 55 °C for an additional 20 min, and then the reaction was stopped by the addition of formamide loading dye. The extension products were separated in a 5% (w/v) polyacrylamide gel in the presence of 8 M urea. Dideoxy sequencing reactions were prepared using Sequenase™ Version 2.0 DNA sequencing Kit (USB), and the products were separated along with the primer extension products.
Acknowledgments
The authors thank S. A. Lommel for the original RNA1 and RNA2 cDNA clones of RCNMV Australian strain. This work was supported in part by a Grant-in-Aid for Scientific Research (A) (18208004) and a Grant-in-Aid for Scientific Research (A) (22248002) from the Japan Society for the Promotion of Science.
References
- Barry J.K., Miller W.A. A − 1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl Acad. Sci. U. S. A. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates H.J., Farjah M., Osman T.A., Buck K.W. Isolation and characterization of an RNA-dependent RNA polymerase from Nicotiana clevelandii plants infected with red clover necrotic mosaic dianthovirus. J. Gen. Virol. 1995;76:1483–1491. doi: 10.1099/0022-1317-76-6-1483. [DOI] [PubMed] [Google Scholar]
- Blobel G., Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl Acad. Sci. U. S. A. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Brierley I., Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic − 1 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T.W., Miller W.A. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M.R., White K.A. 5'-3' RNA–RNA interaction facilitates cap- and poly(A) tail-independent translation of Tomato bushy stunt virus mRNA: a potential common mechanism for Tombusviridae. J. Biol. Chem. 2004;279:28862–28872. doi: 10.1074/jbc.M401272200. [DOI] [PubMed] [Google Scholar]
- Fabian M.R., White K.A. Solution structure probing of RNA structures. Methods Mol. Biol. 2008;451:243–250. doi: 10.1007/978-1-59745-102-4_17. [DOI] [PubMed] [Google Scholar]
- Giedroc D.P., Cornish P.V. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Allen E.M., Miller W.A. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- Hyodo K., Mine A., Iwakawa H.O., Kaido M., Mise K., Okuno T. Identification of amino acids in auxiliary replicase protein p27 critical for its RNA-binding activity and the assembly of the replicase complex in Red clover necrotic mosaic virus. Virology. 2011;413:300–309. doi: 10.1016/j.virol.2011.02.017. [DOI] [PubMed] [Google Scholar]
- Iwakawa H.O., Kaido M., Mise K., Okuno T. cis-Acting core RNA elements required for negative-strand RNA synthesis and cap-independent translation are separated in the 3′-untranslated region of Red clover necrotic mosaic virus RNA1. Virology. 2007;369:168–181. doi: 10.1016/j.virol.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Iwakawa H.O., Mizumoto H., Nagano H., Imoto Y., Takigawa K., Sarawaneeyaruk S., Kaido M., Mise K., Okuno T. A viral noncoding RNA generated by cis-element-mediated protection against 5′ → 3′ RNA decay represses both cap-independent and cap-dependent translation. J. Virol. 2008;82:10162–10174. doi: 10.1128/JVI.01027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H.O., Mine A., Hyodo K., An M., Kaido M., Mise K., Okuno T. Template recognition mechanisms by replicase proteins differ between bipartite positive-strand genomic RNAs of a plant virus. J. Virol. 2011;85:497–509. doi: 10.1128/JVI.01754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Hemenway C.L. Long-distance RNA–RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA. 1999;5:636–645. doi: 10.1017/s1355838299982006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Lommel S.A. Identification and analysis of the site of − 1 ribosomal frameshifting in red clover necrotic mosaic virus. Virology. 1994;200:574–582. doi: 10.1006/viro.1994.1220. [DOI] [PubMed] [Google Scholar]
- Kim K.H., Lommel S.A. Sequence element required for efficient − 1 ribosomal frameshifting in red clover necrotic mosaic dianthovirus. Virology. 1998;250:50–59. doi: 10.1006/viro.1998.9358. [DOI] [PubMed] [Google Scholar]
- Klovins J., Berzins V., van Duin J. A long-range interaction in Qbeta RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA. 1998;4:948–957. doi: 10.1017/s1355838298980177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoda K., Naito S., Ishikawa M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl Acad. Sci. U. S. A. 2004;101:1863–1867. doi: 10.1073/pnas.0307131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- Lehninger A.L., Nelson D.L., Cox M.M. World Publishers; New York, NY: 1993. Principles of Biochemistry; pp. 927–928. [Google Scholar]
- Lindenbach B.D., Sgro J.Y., Ahlquist P. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 2002;76:3905–3919. doi: 10.1128/JVI.76.8.3905-3919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov A.V., Turanov A.A., Hatfield D.L., Gladyshev V.N. Dual functions of codons in the genetic code. Crit. Rev. Biochem. Mol. Biol. 2010;45:257–265. doi: 10.3109/10409231003786094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel S.A., Weston-Fina M., Xiong Z., Lomonossoff G.P. The nucleotide sequence and gene organization of red clover necrotic mosaic virus RNA-2. Nucleic Acids Res. 1988;16:8587–8602. doi: 10.1093/nar/16.17.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazauric M.H., Licznar P., Prere M.F., Canal I., Fayet O. Apical loop-internal loop RNA pseudoknots: a new type of stimulator of − 1 translational frameshifting in bacteria. J. Biol. Chem. 2008;283:20421–20432. doi: 10.1074/jbc.M802829200. [DOI] [PubMed] [Google Scholar]
- Miller W.A., Koev G. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- Mine A., Takeda A., Taniguchi T., Taniguchi H., Kaido M., Mise K., Okuno T. Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of red clover necrotic mosaic virus. J. Virol. 2010;84:6070–6081. doi: 10.1128/JVI.00054-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine A., Hyodo K., Takeda A., Kaido M., Mise K., Okuno T. Interactions between p27 and p88 replicase proteins of Red clover necrotic mosaic virus play an essential role in viral RNA replication and suppression of RNA silencing via the 480-kDa viral replicase complex assembly. Virology. 2010;407:213–224. doi: 10.1016/j.virol.2010.07.038. [DOI] [PubMed] [Google Scholar]
- Mizumoto H., Hikichi Y., Okuno T. The 3′-untranslated region of RNA1 as a primary determinant of temperature sensitivity of Red clover necrotic mosaic virus Canadian strain. Virology. 2002;293:320–327. doi: 10.1006/viro.2001.1275. [DOI] [PubMed] [Google Scholar]
- Mizumoto H., Tatsuta M., Kaido M., Mise K., Okuno T. Cap-independent translational enhancement by the 3′ untranslated region of red clover necrotic mosaic virus RNA1. J. Virol. 2003;77:12113–12121. doi: 10.1128/JVI.77.22.12113-12121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J.L., Zuniga S., Enjuanes L., Sola I. Identification of a coronavirus transcription enhancer. J. Virol. 2008;82:3882–3893. doi: 10.1128/JVI.02622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Moran S.J., Stuart D.I., Gilbert R.J., Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B.L., Wu B., Chevtchenko I., White K.A. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA. 2010;16:1402–1419. doi: 10.1261/rna.2135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Nagano H., Iwakawa H., Mizumoto H., Takeda A., Kaido M., Mise K., Okuno T. cis-Preferential requirement of a − 1 frameshift product p88 for the replication of Red clover necrotic mosaic virus RNA1. Virology. 2008;375:205–212. doi: 10.1016/j.virol.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T., Nagy P.D. Mechanism of stimulation of plus-strand synthesis by an RNA replication enhancer in a tombusvirus. J. Virol. 2005;79:9777–9785. doi: 10.1128/JVI.79.15.9777-9785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul C.P., Barry J.K., Dinesh-Kumar S.P., Brault V., Miller W.A. A sequence required for − 1 ribosomal frameshifting located four kilobases downstream of the frameshift site. J. Mol. Biol. 2001;310:987–999. doi: 10.1006/jmbi.2001.4801. [DOI] [PubMed] [Google Scholar]
- Sarawaneeyaruk S., Iwakawa H.O., Mizumoto H., Murakami H., Kaido M., Mise K., Okuno T. Host-dependent roles of the viral 5′ untranslated region (UTR) in RNA stabilization and cap-independent translational enhancement mediated by the 3′ UTR of Red clover necrotic mosaic virus RNA1. Virology. 2009;391:107–118. doi: 10.1016/j.virol.2009.05.037. [DOI] [PubMed] [Google Scholar]
- Sit T.L., Vaewhongs A.A., Lommel S.A. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998;281:828–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- Takeda A., Tsukuda M., Mizumoto H., Okamoto K., Kaido M., Mise K., Okuno T. A plant RNA virus supresses RNA silencing through viral RNA replication. EMBO J. 2005;24:3147–3157. doi: 10.1038/sj.emboj.7600776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treder K., Kneller E.L., Allen E.M., Wang Z., Browning K.S., Miller W.A. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14:134–147. doi: 10.1261/rna.777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A., Werner A., Flamm C., Westhof E., Schroeder R. Determination of thermodynamic parameters for HIV DIS type loop–loop kissing complexes. Nucleic Acids Res. 2004;32:5126–5133. doi: 10.1093/nar/gkh841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Vanti W.B., White K.A. An RNA domain within the 5′ untranslated region of tomato bushy stunt virus genome modulates viral RNA replication. J. Mol. Biol. 2001;305:741–756. doi: 10.1006/jmbi.2000.4298. [DOI] [PubMed] [Google Scholar]
- Xiong Z., Lommel S.A. The complete nucleotide sequence and genome organization of red clover necrotic mosaic virus RNA-1. Virology. 1989;171:543–554. doi: 10.1016/0042-6822(89)90624-7. [DOI] [PubMed] [Google Scholar]
- Xiong Z., Lommel S.A. Red clover necrotic mosaic virus infectious transcripts synthesized in vitro. Virology. 1991;193:213–221. doi: 10.1016/0042-6822(91)90687-7. [DOI] [PubMed] [Google Scholar]
- Xiong Z., Kim K.H., Giesman-Cookmeyer D., Lommel S.A. The roles of the red clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology. 1993;192:27–32. doi: 10.1006/viro.1993.1004. [DOI] [PubMed] [Google Scholar]
- Xiong Z., Kim K.H., Kendall T.L., Lommel S.A. Synthesis of the putative red clover necrotic mosaic virus RNA polymerase by ribosomal frameshifting in vitro. Virology. 1993;193:213–221. doi: 10.1006/viro.1993.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Shi K., Meskauskas A., Simon A.E. The 3′ end of Turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA. 2009;15:1849–1864. doi: 10.1261/rna.1708709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavriev S.K., Hickey C.M., Lommel S.A. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology. 1996;216:407–410. doi: 10.1006/viro.1996.0076. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]