Abstract
Among 489 bats of 11 species in China, three novel paramyxoviruses [Tuhokovirus 1, 2 and 3 (ThkPV-1, ThkPV-2 and ThkPV-3)] were discovered in 15 Leschenault's rousettes. Phylogenetically, the three viruses are most closely related to Menangle and Tioman virus. Genome analysis showed that their 3'-leader sequences are unique by possessing GA instead of AG at the 5th and 6th positions. Unlike Menangle and Tioman virus, key amino acids for neuraminidase activity characteristic of rubulavirus attachment proteins are present. The genome of ThkPV-1 represents the largest rubulavirus genome. Unique features between the three viruses include perfect complementary 5'-trailer and 3'-leader sequence and a unique cysteine pair in attachment protein of ThkPV-1, G at + 1 position in all predicted mRNA sequences of ThkPV-2, and amino acid substitutions in the conserved N-terminal motif of nucleocapsid of ThkPV-3. Analysis of phosphoprotein gene mRNA products confirmed mRNA editing. Antibodies to the viruses were detected in 48–60% of Leschenault's rousettes.
Keywords: Paramyxovirus, Tuhokovirus, Bat, Genome, Novel, Phylogeny
Introduction
Since three quarters of all emerging infectious disease agents in humans are zoonotic in origin, the study of animal diseases and their emerging potential has been considered increasingly important (Taylor et al., 2001, Woo et al., 2006a). This has been best exemplified in the emergence of the recent swine-origin influenza, avian influenza and severe acute respiratory syndrome (SARS) epidemics (Guan et al., 2003, Lau et al., 2005a, Lau et al., 2009a, Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team et al., 2009, Peiris et al., 2003, Woo et al., 2004, Yuen et al., 1998). For SARS coronavirus (SARS-CoV), the etiological agent of SARS, horseshoe bats are the natural reservoir of SARS-CoV-like viruses while palm civets were only amplification hosts for animal to human transmission (Guan et al., 2003, Lau et al., 2005a, Li et al., 2005). In retrospect, such findings are not surprising, as bats are known to be reservoir of many emerging infections in humans (Wong et al., 2007).
Paramyxoviruses are enveloped, negative-stranded RNA viruses that are divided into two subfamilies, Paramyxovirinae and Pneumovirinae. Viruses in the subfamily Paramyxovirinae have been associated with a number of emerging diseases in humans and various animals in the past two decades (Barrett, 1999, Chua et al., 2000, Halpin et al., 2000, Moreno-López et al., 1986, Osterhaus et al., 1995, Philbey et al., 1998, Tidona et al., 1999, Young et al., 1996). There are currently five genera within the subfamily Paramyxovirinae, namely Respirovirus, Rubulavirus, Morbillivirus, Henipavirus and Avulavirus, although some members of the subfamily remain unclassified. Among members of Paramyxovirinae, measles virus, mumps virus, and human parainfluenza viruses 1–4 are most well known human paramyxoviruses that cause outbreaks of respiratory to systemic infections (Lau et al., 2005b, Lau et al., 2009b, Virtue et al., 2009).
Little was known about the importance of paramyxoviruses in bats until the recent emergence of zoonotic infections caused by paramyxoviruses of bat origin. Before the emergence of Nipah virus (NipPV) and Hendra virus (HenPV) which belong to the genus Henipavirus (Chua et al., 2000), only two bat paramyxoviruses, both belonging to the genus Rubulavirus, were known to exist (Henderson et al., 1995, Hollinger and Pavri, 1971, Karabatsos, 1985, Pavri et al., 1971). The first was a bat parainfluenza virus isolated from a Rousettus leschenaulti bat in India in 1966 (Hollinger and Pavri, 1971), whereas the other was Mapuera virus (MapPV) isolated from the salivary glands of another fruit bat, Sturnira lilium, captured in the tropical rainforest of Brazil in 1979 (Karabatsos, 1985). In 1994, outbreaks of fatal respiratory disease and meningoencephlaitis occurred in horses and humans in Australia. A novel paramyxovirus, HenPV, was found to be the etiological agent which was originated from fruit bats of the genus Pteropus (Halpin et al., 2000, Young et al., 1996). Menangle virus (MenPV), first isolated from stillborn piglets in a commercial piggery in Australia in 1997, has also been detected from fruit bats of the genus Pteropus (Chant et al., 1998, Philbey et al., 1998). In 1999, another novel paramyxovirus, NipPV, was identified as the causative agent of outbreaks of fatal encephalitis in pig-farmers in Malaysia (Chua et al., 1999, Chua et al., 2000). The virus has also been isolated from fruit bats of the genus Pteropus which are likely the natural reservoir (Enserink, 2000). During the search for the animal reservoir of NipPV, a new paramyxovirus, Tioman virus (TioPV), was isolated from pooled urine samples of fruit bats, Pteropus hypomelanus, in Tioman Island, Malaysia in 2001 (Chua et al., 2001). MenPV and TioPV are antigenically related and belong to the genus Rubulavirus (Chua et al., 2001).
In view of the previous findings of several rubulaviruses in bats, we attempted to study the presence of previously undescribed rubulaviruses among bats in southern China, which may have the potential for zoonotic emergence. Three different paramyxoviruses belonging to the genus Rubulavirus were identified. The complete genome sequences of each of the three paramyxoviruses were determined and compared to those of other paramyoviruses with genome sequence available. Based on the results of genome sequence comparison, we propose three novel bat paramyxoviruses among the genus Rubulavirus.
Results
Identification of three novel paramyxoviruses from bats
A total of 484 respiratory and 477 alimentary specimens from 489 bats of 11 different species were obtained (Supplementary Table 1). RT-PCR for a 408-bp fragment in the L gene of paramyxoviruses was positive in samples from 15 (4.5%) bats of the species R. leschenaulti (Leschenault's rousette), including 13 alimentary and 4 respiratory specimens (Table 1 , Fig. 1 and Supplementary Table 1). Sequencing results suggested the presence of three different paramyxoviruses in these samples, most closely related to members of the genus Rubulavirus (Table 1 and Fig. 1). The sequences of these three paramyxoviruses possessed less than 76% nucleotide identities to those of known rubulaviruses, suggesting three novel paramyxovirsues in the genus Rubulavirus. Samples from all other tested bat species were negative for rubulaviruses by RT-PCR.
Table 1.
RT-PCR, quantitative RT-PCR and Western blot assays of the 23 Leschenault's rousette bats with available serum samples.
| Bat no. | Sampling dates | ThkPV-1 |
ThkPV-2 |
ThkPV-3 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RT-PCR (copies/mg by quantitative RT-PCR) |
Western blot | RT-PCR (copies/mg by quantitative RT-PCR) |
Western blot | RT-PCR (copies/mg by quantitative RT-PCR) |
Western blot | |||||
| Alimentary | Respiratory | Alimentary | Respiratory | Alimentary | Respiratory | |||||
| 006 | Oct 6 2005 | − | − | − | − | − | + | − | + (40) | ++ |
| 022 | Oct 6 2005 | − | − | − | − | − | − | − | − | − |
| 024 | Oct 6 2005 | − | − | + | − | − | + | − | − | − |
| 071 | Oct 6 2005 | − | − | + | − | − | − | − | − | − |
| 141 | Oct 6 2005 | − | − | − | − | − | − | − | − | + |
| 291 | Jan 22 2006 | + (5.2 × 102) | − | ++ | − | − | ++ | − | − | + |
| 330 | Feb 18 2006 | − | − | − | + (5.9 × 102) | − | − | − | − | − |
| 336 | Feb 18 2006 | + (2.2 × 104) | + (68) | ++ | − | − | ++ | − | − | ++ |
| 337 | Feb 18 2006 | − | − | − | − | − | + | + (24) | − | ++ |
| 339 | Feb 18 2006 | − | − | + | − | − | ++ | − | − | + |
| 341 | Feb 18 2006 | − | − | − | − | − | + | + (37) | − | ++ |
| 343 | Feb 18 2006 | − | − | − | − | − | − | − | − | − |
| 357 | Feb 18 2006 | + (1.6 × 102) | − | ++ | − | − | ++ | − | − | − |
| 396 | Mar 18 2006 | − | + (1.2 × 102) | − | − | − | − | − | − | + |
| 397 | Mar 18 2006 | − | − | − | − | − | + | − | − | + |
| 405 | Mar 18 2006 | − | − | − | + (2.9 × 103) | − | − | − | − | − |
| 407 | Mar 18 2006 | − | − | − | − | − | + | + (2.0 × 104) | − | ++ |
| 412 | Mar 18 2006 | − | − | + | + (5.7 × 102) | − | + | − | − | ++ |
| 415 | Mar 18 2006 | − | − | + | − | − | + | + (48) | − | + |
| 416 | Mar 18 2006 | − | − | + | − | − | +++ | − | − | +++ |
| 485 | Apr 10 2006 | − | − | +++ | − | − | +++ | + (2.5 × 102) | − | +++ |
| 488 | Apr 10 2006 | + (7.2 × 102) | − | + | − | − | − | − | − | − |
| 500 | Apr 10 2006 | + (71) | + (24) | +++ | − | − | +++ | − | − | +++ |
Fig. 1.
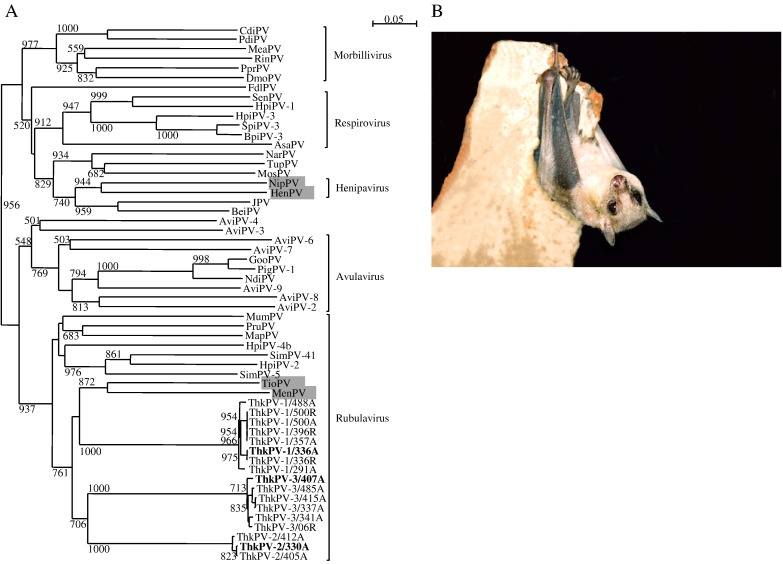
(A) Phylogenetic analysis of amino acid sequences of the 502-bp fragment of L gene of paramyxoviruses identified from bats in the present study. The tree was constructed by neighbor-joining method using Jukes-Cantor correction and bootstrap values calculated from 1000 trees. The scale bar indicates the estimated number of substitutions per 20 amino acids. The different strains of ThkPV-1, ThkPV-2 and ThkPV-3 from different bat samples are named as ThkPV/batA or ThkPV/batR representing alimentary or respiratory sample from the corresponding bat number, respectively. The three strains each from ThkPV-1, ThkPV-2 and ThkPV-3 with genome sequences determined are shown in bold. Paramyxoviruses from bats are shaded in gray. CdiPV, Canine distemper virus (NC_001921); PdiPV, Phocine distemper virus (Y09630); MeaPV, Measles virus (NC_001498); RinPV, Rinderpest virus (NC_006296); PprPV, Peste-des-petits ruminants virus (NC_006383); DmoPV, Dolphin morbillivirus (NC_005283); FdlPV, Fer-de-lance virus (NC_005084); SenPV, Sendai virus (NC_001552); HpiPV-1, Human parainfluenza virus 1 (NC_003461); HpiPV-3, Human parainfluenza virus 3 (NC_001796); SpiPV-3 (EU439429), Swine parainfluenza 3; BpiPV-3, Bovine parainfluenza virus 3 (NC_002161); AsaPV, Atlantic salmon paramyxovirus (EU156171); NarPV, Nariva virus (FJ362497); TupPV, Tupaia paramyxovirus (NC_002199); MosPV, Mossman virus (NC_005339); NipPV, Nipah virus (NC_002728); HenPV, Hendra virus (NC_001906); JPV, J-virus (NC_007454); BeiPV, Beilong virus (NC_007803); AviPV-4, Avian paramyxovirus 4 (FJ177514); AviPV-3, Avian paramyxovirus 3 (EU403085); AviPV-6, Avian paramyxovirus 6 (NC_003043); AviPV-7, Avian paramyxovirus 7 (FJ215864); GooPV, Goose paramyxovirus SF02 (NC_005036); PigPV-1, Pigeon paramyxovirus 1 (AJ880277); NdiPV, Newcastle disease virus (NC_002617); AviPV-9, Avian paramyxovirus 9 (EU910942); AviPV-8, Avian paramyxovirus 8 (FJ215864); AviPV-2, Avian parayxovirus 2 (EU338414); MumPV, Mumps virus (NC_002200); PruPV, Porcine rubulavirus (NC_009640); MapPV, Mapuera virus (NC_009489); HpiPV-4b, Human parainfluenza virus 4b (EU627591); SimPV-41, Simian virus 41 (NC_006428); HpiPV-2, Human parainfluenza virus 2 (NC_003443); SimPV-5, Simian virus 5 (NC_006430); TioPV, Tioman virus (NC_004074); MenPV, Menangle virus (NC_007620). (B) A Leschenault's rousette bat with grayish brown, soft, fine and silky fur, long and narrow muzzle, large and conspicuous eyes, and simple ears without antitragus or tragus.
Quantitative RT-PCR
Quantitative RT-PCR showed that the amount of ThkPV-1, ThkPV-2 and ThkPV-3 RNA ranged from 24 to 2.2 × 104 copies per mg of tissue (Table 1).
Genome organization and coding potential of ThkPV-1, ThkPV-2 and ThkPV-3
Since analysis of the 408-bp fragment of the L gene suggested that the presence of three novel bat paramyxoviruses, complete genome sequence data of each strain of the three novel paramyxoviruses were obtained by assembly of the sequences of the RT-PCR products from the corresponding individual alimentary specimens from three bats (ThkPV-1/336A, ThkPV-2/330A and ThkPV-3/407A).
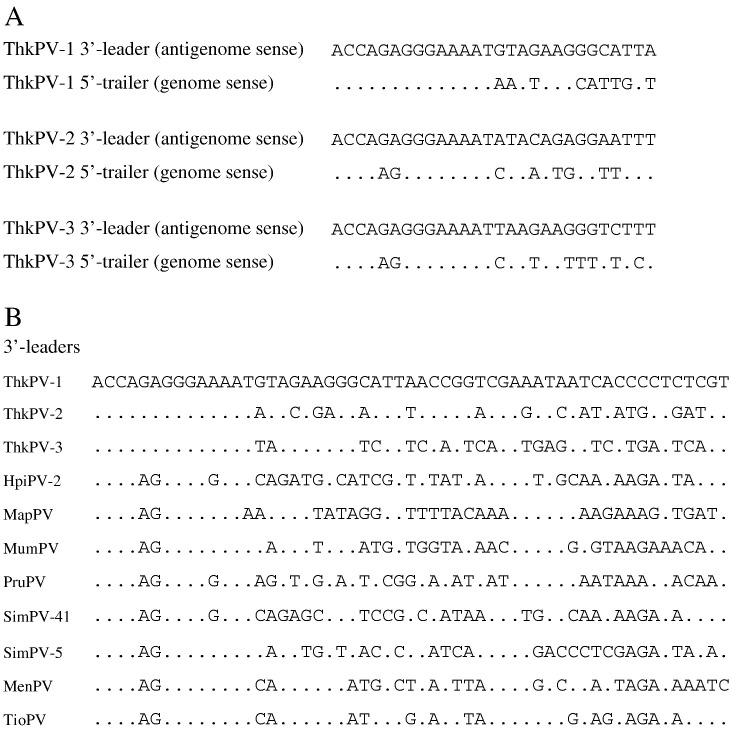
The genome size of ThkPV-1, ThkPV-2 and ThkPV-3 are 15,888, 15,432 and 15,234 bases, and their G + C contents are 40.6%, 40.6% and 41.8% respectively. The sequences of ThkPV-1, ThkPV-2 and ThkPV-3 conform to the rule of six as in other paramyxovirus genomes. Their genome organizations are similar to other rubulaviruses, containing six open reading frames (ORFs) coding for putative nucleocapsid (N), phosphoprotein (P/V), matrix (M), fusion (F), attachment (HN), and large (L) proteins (Supplementary Table 2). Pairwise alignment of the predicted gene products among ThkPV-1, ThkPV-2, ThkPV-3 and other paramyxoviruses showed the highest amino acid identities with the genus Rubulavirus (Table 2 ). The lengths, positions and characteristics of the major structural genes and intergenic regions (IGRs) are summarized in Table 3 and Supplementary Table 2. Genomes of Paramyxovirinae typically contain a 55-nt 3' leader sequence and a 5' trailer sequence of variable length. The terminal 11–14 nt of the 3' leader and 5' trailer sequences are complementary and highly conserved within each genus, which constitute critical elements of the promoter sequences for initiation of transcription and replication (Lamb and Parks, 2007). The genomes of ThkPV-1 also contains a 14-nt complementary 3' leader and 5' trailer sequence, while there is a GA → AG substitution in the genome sense at the 5th and 6th position of the 5' trailer sequence in ThkPV-2 and ThkPV-3, similar to that observed in porcine rubulavirus (NC_009640.1) (Fig. 2 ). Notably, within the first 14-nt 3'-leader sequences conserved among rubulaviruses, a unique AG → GA substitution was observed in the three novel viruses (Fig. 2).
Table 3.
Nucleotide sequences of intergenic regions (IGR) and transcriptional start and stop signals of ThkPV-1, ThkPV-2 and ThkPV-3 compared to those in TioV and MenPV.
| Virus | Start | Gene | Stop | IGR |
|
|---|---|---|---|---|---|
| Sequence | Size (nt) | ||||
| ThkPV-1 | AGGCCCGAAAGT | TTTAATAAAAAA | TAGGAATTATT...AAAGTGCAAGAT | 35 | |
| ThkPV-2 | GGGCCCGAAAGT | TTTTTAAGAAAAAA | CAAGGGT | 7 | |
| ThkPV-3 | GGGCCCGGAAGT | N | TTTAAGAAAAAA | CTATAGT | 7 |
| TioPV | GAGCCCAGAAGT | TTTTAAGAAAAAA | CAGAAATTAA…CAAAAGCCGGT | 41 | |
| MenPV | GAGCCCAGAAGT | TTTTAAAGAAAA | CTTAGAAAAAAGACAGAT | 18 | |
| ThkPV-1 | AGGCCCGAAC | TTTAAGAAAAAA | CAGAACAAT…ATAGAAAAAAC | 54 | |
| ThkPV-2 | GGGCCCGAAG | TTTAAGAAAAAA | TCAAAAT | 7 | |
| ThkPV-3 | AGGCCCGAAG | P/V | TTTAAGAAAAAA | CTAGGCT | 7 |
| TioPV | GAGCCCGAAT | TTTAAGAAAAAA | CTTAAAAT | 8 | |
| MenPV | GAGCCCGAAC | TTTAAGAAAAAA | TAAGT | 5 | |
| ThkPV-1 | AGGCCCGAAT | TTTAATAAAAAA | TTCCCTTACAT…CCAACAAC | 119 | |
| ThkPV-2 | GGGTCCGAAC | TTTAAGAAAAAA | CTAATATTAT…CAGGAACATT | 59 | |
| ThkPV-3 | GGGGTCGAAC | M | TTTAAGAAAAAA | CTGCAAA...TGAAAGAC | 36 |
| TioPV | GGGTCCGAAC | TTTTAATAAAAAA | CTAAGGGGTA...CCATGGCTA | 35 | |
| MenPV | GAGCCCGAAC | TTTAATAAAAAA | TTAGAGAGGT...ACCGGAAAG | 38 | |
| ThkPV-1 | AGGCCCGAAC | TTTATTCAAAA | CACACTATATTG…GAGCATGTT | 47 | |
| ThkPV-2 | GGGCCCGAAC | TTTTAATAAAAAA | CCTAGTGGACTGGAAGCCT | 19 | |
| ThkPV-3 | GGGGGCGAAC | F | TTTTTAATAAAAAA | CTTGACAT…AGCCCTACCC | 22 |
| TioPV | GAGCCCGAAC | TTTAAGGAAAA | CTAGGCATAT...AACAGCGCTTG | 55 | |
| MenPV | GAGCACGAAC | TTCAAGAAAAAA | CAAATATGAGC…GCAGTGAAGT | 85 | |
| ThkPV-1 | AGGCCCGAAT | TTTTAATAAAAAAA | TGATAATTG…AATGCCATCAT | 124 | |
| ThkPV-2 | GGGCCCGACC | TTTAAGAAAAAA | CTGAATT | 7 | |
| ThkPV-3 | GGGCCCGACC | HN | TTTAAGAAAAA | CAAT | 4 |
| TioPV | GAGCCCGACT | TTTAAGAAAAAA | CTAAAGGGGA...CAAGTCCAAGT | 70 | |
| MenPV | GAGCCCGAAT | TTTAAGAAAAAA | GAAAATCAAG...GAGAAAAAGGT | 68 | |
| ThkPV-1 | GGGCCAGAAT | TTAAGAAAAAA | CAATGCTTATTTA… 5' trailer sequence | ||
| ThkPV-2 | GGGCCAGAAT | TTTTAAGAAAAAA | CCCATTTAGATTTT… 5' trailer sequence | ||
| ThkPV-3 | GGGCCAGAAT | L | TTTAAGAAAAAA | TTATTGATTTTCCCC…5' trailer sequence | |
| TioPV | GGGCCAGAAT | TTTAAGAAAAAAA | CCTATATTGA…5' trailer sequence | ||
| MenPV | GGGCCAGAAT | TTTTTTAAGAAAAA | CATTAAAATTTT…5' trailer sequence | ||
Fig. 2.
Alignment of genome terminal sequences of ThkPV-1, ThkPV-2 and ThkPV-3. Dots indicate identical residues. (A) Alignment of the 3' leader and 5' trailer sequences of ThkPV-1, ThkPV-2 and ThkPV-3. (B) Alignment of 3' leader sequences of ThkPV-1, ThkPV-2, ThkPV-3 and other rubulaviruses.
Rubulaviruses and avulaviruses are unique among the subfamily Paramyxovirinae in having IGRs of variable lengths and sequences. While the longest IGR is 70 nt between the HN and L genes for TioPV and 85 nt between the F and HN genes for MenV, the longest IGR among all paramyxoviruses is observed in ThkPV-1 with 124 nt between HN and L genes (Table 3). TioPV and MenPV were previously found to be the only rubulaviruses which use G as the + 1 nucleotide in all mRNAs rather than A which is more commonly observed (Chua et al., 2002), one of our strains, ThkPV-2, also possessed the G residue at + 1 position in all its predicted mRNA sequences (Table 3).
The nucleocapsid (N) gene
The N proteins of ThkPV-1, ThkPV-2 and ThkPV-3 were most closely related to those of other rubulaviruses, with the highest amino acid identities to those of MenPV and TioPV (Table 2). Notably, the conserved N-terminal motif MSSV in rubulaviruses was absent in ThkPV-3, which contained the sequence MASL as a result of S → A (U → G at first codon position) and V → L (G → C at first codon position) substitution at the second and fourth amino acids, respectively (Supplementary Fig. 1). Similar to N proteins of other paramyxoviruses, the N proteins of ThkPV-1, ThkPV-2 and ThkPV-3 were divided into three domains based on sequence variability or two structural domains, with the central domain (amino acid residues 160–400) being the most conserved (Buchholz et al., 1993, Chua et al., 2001, Lamb and Parks, 2007, Yu et al., 1998). Within the central domain, three highly conserved motifs previously identified in all members of Paramyxovirinae (Morgan, 1991) are also found in ThkPV-1, ThkPV-2 and ThkPV-3 (Supplementary Fig. 1).
Table 2.
Pairwise amino acid identities of predicted gene products of ThkPV-1, ThkPV-2 and ThkPV-3 compared to other paramyxoviruses.
| Virus | Percentage of amino acid sequence identity |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ThkPV-1 |
ThkPV-2 |
ThkPV-3 |
||||||||||||||||||||
| N | P | V | M | F | Aa | L | N | P | V | M | F | A | L | N | P | V | M | F | A | L | ||
| Rubulavirus | ThkPV-1 | – | – | – | – | – | – | – | 57 | 45 | 43 | 53 | 43 | 26 | 60 | 57 | 42 | 40 | 51 | 37 | 26 | 56 |
| ThkPV-2 | 57 | 45 | 43 | 53 | 43 | 26 | 60 | - | - | - | - | - | - | - | 64 | 46 | 44 | 55 | 46 | 37 | 62 | |
| ThkPV-3 | 57 | 42 | 40 | 51 | 37 | 26 | 56 | 64 | 46 | 44 | 55 | 46 | 37 | 62 | - | - | - | - | - | - | - | |
| MenPV | 62 | 38 | 37 | 55 | 42 | 25 | 56 | 64 | 41 | 38 | 58 | 47 | 28 | 57 | 61 | 40 | 34 | 54 | 43 | 25 | 55 | |
| TioPV | 62 | 40 | 37 | 54 | 45 | 25 | 55 | 61 | 39 | 40 | 60 | 46 | 28 | 57 | 59 | 43 | 36 | 54 | 43 | 28 | 56 | |
| HpiPV-2 | 44 | 27 | 26 | 37 | 34 | 22 | 51 | 43 | 29 | 28 | 37 | 33 | 22 | 51 | 42 | 30 | 29 | 33 | 36 | 21 | 50 | |
| MapPV | 49 | 26 | 25 | 45 | 36 | 23 | 51 | 45 | 26 | 25 | 46 | 36 | 24 | 52 | 44 | 27 | 26 | 42 | 37 | 21 | 51 | |
| MumPV | 47 | 28 | 22 | 44 | 37 | 21 | 52 | 48 | 28 | 29 | 40 | 35 | 24 | 53 | 45 | 30 | 31 | 40 | 36 | 24 | 52 | |
| PruPV | 48 | 24 | 21 | 45 | 36 | 21 | 50 | 46 | 23 | 26 | 44 | 40 | 22 | 51 | 47 | 26 | 25 | 40 | 37 | 23 | 50 | |
| SimPV-41 | 45 | 27 | 29 | 38 | 33 | 21 | 51 | 44 | 28 | 26 | 39 | 35 | 21 | 51 | 42 | 28 | 28 | 34 | 34 | 22 | 51 | |
| SimPV-5 | 49 | 30 | 27 | 38 | 39 | 19 | 50 | 48 | 30 | 30 | 38 | 37 | 22 | 50 | 43 | 28 | 27 | 33 | 35 | 19 | 50 | |
| HpiPV-4a | 44 | 21 | 19 | 44 | 35 | 21 | NAb | 43 | 23 | 20 | 44 | 35 | 22 | NA | 42 | 23 | 22 | 41 | 35 | 21 | NA | |
| HpiPV-4b | 44 | 20 | 20 | 44 | 36 | 20 | NA | 43 | 21 | 20 | 45 | 36 | 23 | NA | 41 | 22 | 23 | 41 | 35 | 21 | NA | |
| Non-rubulaviruses | ||||||||||||||||||||||
| Avulavirus | AviPV-6 | 33 | 21 | NA | 26 | 30 | 20 | 36 | 32 | 19 | NA | 24 | 27 | 22 | 37 | 32 | 22 | NA | 27 | 30 | 20 | 36 |
| NdiPV | 34 | 19 | NA | 24 | 29 | 20 | 35 | 32 | 18 | NA | 24 | 29 | 21 | 35 | 34 | 20 | NA | 24 | 27 | 21 | 34 | |
| Henipavirus | HenPV | 27 | 12 | 14 | 19 | 25 | 15 | 26 | 28 | 12 | 13 | 18 | 24 | 18 | 27 | 29 | 11 | 14 | 18 | 24 | 20 | 26 |
| NipPV | 28 | 12 | 14 | 18 | 26 | 14 | 27 | 27 | 12 | 10 | 18 | 25 | 16 | 28 | 28 | 10 | 13 | 19 | 24 | 19 | 27 | |
| Morbillivirus | CdiPV | 23 | 16 | NA | 18 | 19 | 11 | 28 | 25 | 13 | NA | 19 | 18 | 13 | 29 | 23 | 16 | NA | 18 | 19 | 13 | 28 |
| MeaPV | 21 | 14 | NA | 19 | 24 | 12 | 27 | 26 | 13 | NA | 19 | 24 | 11 | 29 | 25 | 13 | NA | 18 | 25 | 13 | 28 | |
| Respirovirus | BpiPV-3 | 21 | 12 | NA | 18 | 24 | 18 | 28 | 20 | 13 | NA | 21 | 25 | 20 | 29 | 19 | 11 | NA | 18 | 23 | 19 | 27 |
| HpiPV-1 | 16 | 12 | NA | 16 | 21 | 17 | 27 | 19 | 11 | NA | 19 | 23 | 17 | 28 | 21 | 11 | NA | 17 | 22 | 21 | 27 | |
| HpiPV-3 | 20 | 13 | NA | 18 | 25 | 18 | 28 | 19 | 11 | NA | 20 | 25 | 19 | 29 | 19 | 11 | NA | 17 | 24 | 22 | 28 | |
| SenPV | 18 | 13 | NA | 15 | 23 | 18 | 28 | 18 | 12 | NA | 19 | 21 | 20 | 29 | 20 | 11 | NA | 17 | 21 | 22 | 28 | |
| Unclassified Paramyxovirinae | BeiPV | 24 | 16 | 18 | 17 | 26 | 15 | 29 | 24 | 17 | 19 | 19 | 27 | 14 | 28 | 25 | 13 | 19 | 19 | 24 | 15 | 29 |
| FdlPV | 22 | 11 | 16 | 18 | 24 | 18 | 29 | 24 | 13 | 20 | 19 | 27 | 20 | 30 | 22 | 13 | 23 | 20 | 25 | 20 | 27 | |
| JPV | 25 | 15 | 18 | 17 | 26 | 15 | 28 | 22 | 14 | 19 | 18 | 27 | 17 | 29 | 25 | 13 | 19 | 20 | 22 | 17 | 28 | |
| MosPV | 25 | 14 | 19 | 17 | 23 | 15 | 28 | 26 | 13 | 18 | 16 | 22 | 15 | 28 | 25 | 13 | 16 | 21 | 24 | 14 | 27 | |
| TupPV | 24 | 15 | 20 | 16 | 26 | 15 | 27 | 24 | 13 | 18 | 17 | 25 | 15 | 27 | 25 | 15 | 21 | 18 | 26 | 15 | 27 | |
Attachment protein: HN for rubulaviruses, avulaviruses, respiroviruses and the unclassified FdlPV; H for morbilliviruses and the unclassified TupPV; G for henipaviruses and other unclassified paramyxoviruses.
NA, sequence not available for comparison.
The phosphoprotein (P/V) gene
The P/V genes of most paramyxoviruses encode different proteins as a result of mRNA editing and hence alternative reading frames. The P protein is a structural protein component of the active polymerase complex, while the V protein may be involved in blocking host interferon defense mechanisms (Kurath et al., 2004, Poole et al., 2002). Other proteins such as W, D and I proteins have been found in some paramyxoviruses. The function of W protein, present in MenPV and TioPV (Bowden et al., 2001, Chua et al., 2001), is not yet clearly established, although an inhibitory role in viral RNA synthesis has been suggested in Sendai virus (Horikami et al., 1996). The predicted P proteins of ThkPV-1, ThkPV-2 and ThkPV-3 were most closely related to those of MenPV and TioPV (Table 2). The predicted V proteins of ThkPV-1, ThkPV-2 and ThkPV-3 are also most closely related to those of MenPV and TioPV, with a highly conserved cysteine-rich C-terminal domain among rubulaviruses.
Most members of the Paramyxovirinae contain a conserved UC-rich editing site that allows for the addition of non-templated G residues to mRNA products during P/V gene transcription, resulting in the production of different proteins with a common N-terminal region (Thomas et al., 1988). This common N-terminal region consists of 173 aa in ThkPV-1, and 163 aa in ThkPV-2 and ThkPV-3. Analysis of each of the P/V gene sequences of the three genomes revealed the presence of a stretch of poly-G residues that could potentially be used as RNA editing sites (Lamb and Parks, 2007). To determine the exact location of the P gene editing site and the number and frequency of G residue insertions, a pair of primers were designed to amplify a small cDNA fragment and the resulting PCR products were cloned and the number of G insertions determined by sequence determination of different clones. Since the amount of mRNA from sample ThkPV-2/330 was inadequate, another strain of ThkPV-2/405 was used for mRNA isolation and cloning for ThkPV-2. A total of 40, 28 and 32 independent clones were obtained for ThkPV-1, ThkPV-2 and ThkPV-3, respectively. From all three samples, mRNAs encoding for two different proteins, V protein (mRNA sequence the same as genomic RNA sequence without G insertion) and P protein (2-G insertion), were identified (Table 4 ). For ThkPV-1, among the 40 sequenced clones, 28 (70%) contained the sequence TTTAAGAGGGG (without G insertion) (encoding V protein), 10 (25%) contained the sequence TTTAAGAGGGGGG (2-G insertion) (encoding P protein), while one of the remaining clones contained 3-G insertion (encoding V protein) and the other remaining clone contained 5-G insertion (encoding P protein). For ThkPV-2, among the 28 sequenced clones, the majority of clones (23 clones, 82%) contained the sequence TTTTAAGAGGGGGGG (2-G insertion) (encoding P protein) while 5 (18%) contained the sequence TTTTAAGAGGGGG (without G insertion) (encoding V protein). For ThkPV-3, among the 32 sequenced clones, 13 (41%) contained the sequence TTTAAGAGGGGG (without G insertion) (encoding V protein), 17 (53%) contained the sequence TTTAAGAGGGGGGG (2-G insertion) (encoding P protein), while the remaining two clones contained 3-G insertion (encoding V protein). The sequence TTTAAGA preceding the poly-G residues is identical to that observed in other rubulaviruses as the putative transcriptional termination signal (Chua et al., 2001). None of the clones were found to contain 1-G insertion that encodes the W protein which is also rarely transcribed in MenPV and TioPV (Table 4) (Bowden et al., 2001, Chua et al., 2001).
Table 4.
Transcription products of P/V genes as a result of mRNA editing in ThkPV-1, ThkPV-2, ThkPV-3 as compared to those reported in MenPV and TioPV.
| Transcription products | No. (%) of clones |
||||
|---|---|---|---|---|---|
| ThkPV-1 | ThkPV-2 | ThkPV-3 | MenPV (3) | TioPV (8) | |
| Total | 40 | 28 | 32 | 35 | 60 |
| V proteina | 29/40 (72.5%) | 5/28 (18%) | 15/32 (46.9%) | 27/35 (77%) | 27/60 (45%) |
| P proteinb | 11/40 (27.5%) | 23/28 (82%) | 17/32 (53.1%) | 7/35 (20%) | 30/60 (50%) |
| W proteinc | 0/40 (0%) | 0/28 (0%) | 0/32 (0%) | 1/35 (3%) | 3/60 (5%) |
mRNAs of ThkPV-1, ThkPV-2 and ThkPV-3 possessed no G insertion at the editing site, except one clone from ThkPV-1 and two clones from ThkPV-3 with 3 G insertions.
mRNAs of ThkPV-1, ThkPV-2 and ThkPV-3 possessed 2 G insertions at the editing site, except one clone from ThkPV-1 with 5 G insertions.
mRNAs of ThkPV-1, ThkPV-2 and ThkPV-3 expected to possess single G insertion at the editing site.
The matrix (M) gene
The predicted M proteins of ThkPV-1, ThkPV-2 and ThkPV-3 were most closely related to those of other rubulaviruses, with the highest amino acid identities to those of MenPV and TioPV (Table 2).
The fusion (F) gene
The predicted F proteins of ThkPV-1, ThkPV-2 and ThkPV-3 were most closely related to those of other rubulaviruses, with the highest amino acid identities to those of MenPV and TioPV (Table 2). Similar to F proteins of other paramyxoviruses, the F proteins of ThkPV-1, ThkPV-2 and ThkPV-3 are predicted to be a type I membrane protein, with a transmembrane domain located at the C-terminus and a short cytoplasmic tail of 18 aa in ThkPV-1, 29 aa in ThkPV-2 and 30 aa in ThkPV-3. Similar to other rubulaviruses except HpiPV-4, the F proteins of ThkPV-1, ThkPV-2 and ThkPV-3 have multibasic protein cleavage site with furin recognition site (R-X-K/R-R) followed by a highly conserved fusion peptide (Supplementary Fig. 2). The furin recognition site allows cleavage in the trans-Golgi membranes and delivery of F proteins to the plasma membrane as active F proteins (Morrison, 2003). F proteins with single-basic residues at the cleavage site, as observed in HpiPV-4, must be cleaved by an extracellular host enzyme usually found exclusively in the respiratory tract, rendering infections by these paramyxoviruses limited to the respiratory tract (Morrison, 2003). Two heptad repeat sequences similar to those in F proteins of other paramyxoviruses were also identified in the F1 of ThkPV-1, ThkPV-2 and ThkPV-3. The F proteins of ThkPV-1, ThkPV-2 and ThkPV-3 also contain the 10 Cys residues that are highly conserved in other rubulaviruses and six to nine potential N-glycosylation sites, most of which located in the F1 peptide.
The attachment (HN) gene
The predicted HN proteins of ThkPV-1, ThkPV-2 and ThkPV-3 possessed low homologies among themselves and to those of other paramyxoviruses (Table 2). The HN proteins of ThkPV-1, ThkPV-2 and ThkPV-3 were predicted to be type II integral membrane proteins, with a putative N-terminal transmembrane domain (at amino acid residues 29–51 for ThkPV-1, 38–60 for ThkPV-2 and 34–53 for ThkPV-3 by TMHMM analysis). There were six potential N-linked glycosylation sites in the HN gene of ThkPV-1 and ThkPV-2 and seven sites in ThkPV-3, as compared to six sites in TioPV and five sites in MenPV (Chua et al., 2002).
Comparison of the sequences corresponding to the extracellular globular head domain of paramyxovirus attachment proteins revealed that six of the seven key residues important for neuraminidase activity (Langedijk et al., 1997) were conserved in ThkPV-1, ThkPV-2, and ThkPV-3. This is in contrast to that observed in TioPV and MenPV where only two residues were conserved (Bowden et al., 2001, Chua et al., 2002). Similar to TioPV and MenPV, the globular head domains of ThkPV-1, ThkPV-2 and ThkPV-3 contained eight pairs of cysteine residues, which are potential sites of disulfide bond (Crennell et al., 2000). Of note is that one pair of cysteine residues is only found in ThkPV-1 and the absence of one pair common to ThkPV-2, ThkPV-3, MenPV and TioPV. Similar to TioPV and MenPV (Bowden et al., 2001, Chua et al., 2002), the NRKSCS motif that is highly conserved in other rubulaviruses is not found in ThkPV-1, ThkPV-2 and ThkPV-3.
The large protein (L) gene
The predicted L proteins of ThkPV-1, ThkPV-2 and ThkPV-3 showed the highest homologies to those of rubulaviruses (Table 2). Similar to other rubulaviruses, this ORF is preceded by a relatively short (8 nt) 5' UTR in ThkPV-1, ThkPV-2 and ThkPV-3. A six-domain structure has been recognized in the RNA polymerases of viruses in the order Mononegavirales including paramyxoviruses. Among these six domains, conserved motifs have been recognized in domain II, III and VI, which are believed to play important roles in RNA polymerization, RNA template recognition and binding (Bowden and Boyle, 2005, Chua et al., 2002, Müller et al., 1994, Poch et al., 1990). Multiple alignment of the corresponding domains in the L proteins of ThkPV-1, ThkPV-2 and ThkPV-3 with those of other rubulaviruses showed that these motifs are also conserved in the three novel paramyxoviruses (Supplementary Fig. 3). However, two unique amino acid residues (one in domain III and one in domain VI) were identified among ThkPV-1, ThkPV-2 and ThkPV-3 at positions that are conserved in other rubulaviruses (Supplementary Fig. 3).
Phylogenetic analyses
Phylogenetic trees were constructed using the predicted coding sequences of the N, P, M, F, HN and L genes of ThkPV-1, ThkPV-2, ThkPV-3 and other members of Paramyxoviridae. In all six trees, the three viruses were clustered with MenPV and TioPV, with bootstrap values of 88–100% in all cases, forming a distinct subgroup. Based on the present results, we propose to name these three new viruses as Tuhoko (The University of Hong Kong) virus 1 (ThkPV-1), ThkPV-2 and ThkPV-3 after its place of discovery, within the family Paramyxoviridae.
Antigenicity of recombinant N proteins
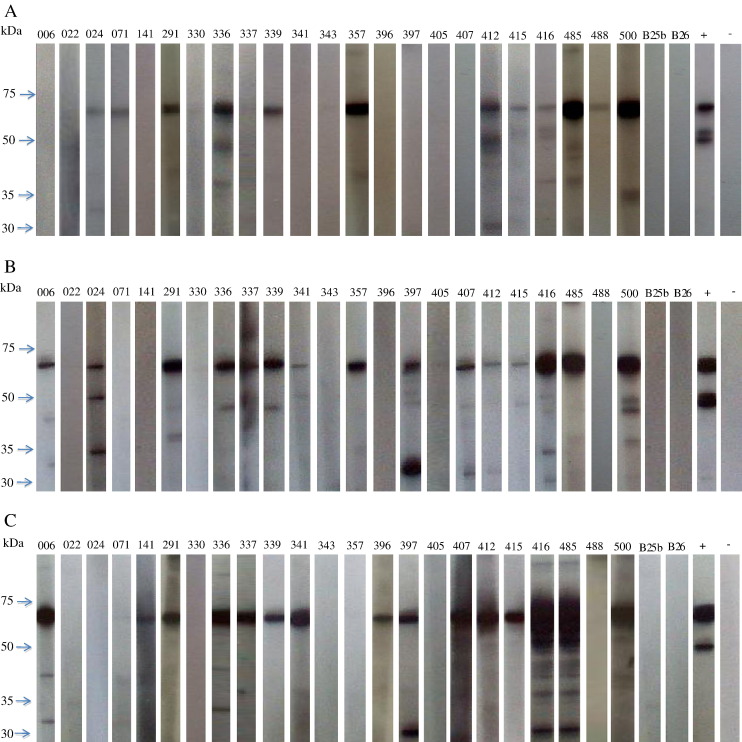
Antibody detection on serum samples of 23 Leschenault's rousette bats with sufficient quantities was performed by recombinant N protein-based Western blot analysis. Among tested sera from the 23 Leschenault's rousette bats, 12 (52%) were positive for ThkPV-1 antibody, and 15 (65%) were positive for ThkPV-2 and ThkPV-3 antibody (Fig. 3 and Table 1). Of the six bats positive for ThkPV-1 by RT-PCR, five were positive for ThkPV-1 antibody. Of the three bats positive for ThkPV-2 by RT-PCR, only one was positive for ThkPV-2 antibody. Of the six bats positive for ThkPV-3 by RT-PCR, all were positive for ThkPV-3 antibody. The presence of multiple bands in some lanes was likely due to degradation of the N proteins. Antibodies were not detected in serum samples from other bat species.
Fig. 3.
Western blot analysis against purified (His)6-tagged recombinant N protein from ThkPV-1 (A), ThkPV-2 (B), and ThkPV-3 (C). Western blot analysis was performed using serum samples from 23 Leschenault's rousette bats and two non-Leschenault's rousette bats (B25b and B26). Prominent immunoreactive protein bands of about 65 kDa, consistent with the expected size of 66.2, 66.7 and 65.5 kDa of the recombinant N proteins of ThkPV-1, ThkPV-2 and ThkPV-3, respectively, were detected with serum samples from Lechenault's rousette bats but not with the other two non-Leschenault's rousette bats. Lane numbers correspond to the bat numbers in Table 1.
Viral culture
No CPE effect was observed in all passages for any of the cell lines inoculated with bat specimens positive for ThkPV-1, ThkPV-2 or ThkPV-3 with or without trypsin. Since RT-PCR assay on the culture supernatants and cell lysates to monitor the presence of viral replication also showed negative results, immunostaining against nucleoprotein was not performed. No cell line toxicity was observed throughout the course of incubation.
Discussion
In the present study, three novel paramyxoviruses, ThkPV-1, ThkPV-2 and ThkPV-3, from Leschenault's rousette bats are described. The three viruses formed three distinct branches within the genus Rubulavirus and are most closely related to two other bat paramyxoviruses, MenPV and TioPV, upon phylogenetic analysis of a 408-bp fragment of the L gene sequence. The amino acid sequences in the various predicted proteins of the three novel viruses also possessed higher amino acid identities to those of other rubulaviruses than to those of non-rubulaviruses, supporting their classification under the genus Rubulavirus. Complete genome sequencing of each strain from each of the three novel viruses also showed distinct genome features between the three viruses and different from other rubulaviruses. The 3'-leader sequence of the three viruses are unique among rubulaviruses by possessing the GA residues instead of AG residues at the 5th and 6th positions. ThkPV-1 is different from ThkPV-2 and ThkPV-3 by having a perfect complementary 5' trailer and 3' leader sequence, instead of GA → AG substitution found only in ThkPV-2, ThkPV-3 and porcine rubulavirus among all rubulaviruses (Fig. 2). ThkV1 possesses the largest genome size among all rubulaviruses with the longest reported IGR sequence (124 nt) between HN and L genes. Moreover, the HN protein sequences of ThkV1 contains a unique pair of cysteine at a position different from the one conserved among ThkPV-2, ThkPV-3, TioPV and MenPV. As for ThkPV-2, it is different from ThkPV-1 and ThkPV-3 by the presence of the G residue instead of A residue at + 1 position in all its predicted mRNA sequences. ThkPV-3 is unique among rubulaviruses by the absence of the conserved N-terminal motif of the N protein, MSSV, which is substituted by MASL. These results support that ThkPV-1, ThkPV-2 and ThkPV-3 represent three separate novel rubulavirus species. Nevertheless, phylogenetic trees constructed using N, P, M, F, A and L gene sequences all showed that ThkPV-1, ThkPV-2 and ThkPV-3 are more closely related to each other than to other rubulaviruses. Together with their same host bat species, it is likely that the three novel bat rubulaviruses have originated from the same common ancestor, and have subsequently diverged into three separate species.
The pathogenic potential of the present three novel bat paramyxoviruses remains to be determined. Since the emergence of HenPV, five other novel paramyxoviruses have been identified in the past few years, including MenPV, NipPV, Tupaia paramyxovirus (TupPV), Salem virus (SalPV) and TioPV. Besides HenPV and NipPV, only MenPV among these novel paramyxoviruses was known to have potential of causing disease in human. Serological evidence of human MenPV infection has been demonstrated in at least two cases in close contact with infected pigs and developed influenza-like illness (Bowden et al., 2001, Chant et al., 1998). In contrast, the pathogenicity of the other three new viruses in both animals and humans was less well understood. TupPV was first isolated from the kidneys of an apparently healthy tree shrew from Bangkok in 1999 (Tidona et al., 1999). SalPV was incidentally isolated from horse mononuclear cells during search for an unrelated equine illness, whereas TioPV was isolated from urine samples of fruit bats with no reported disease association (Chua et al., 2001, Renshaw et al., 2000). In the present study, all the Leschenault's rousette bats positive for the three novel paramyxoviruses appeared well without evidence of disease, as in the case of many other bat viruses (Lau et al., 2005a, Lau et al., 2007, Wong et al., 2007, Woo et al., 2006c, Woo et al., 2007). Nevertheless, it is likely that they may have a wide tissue tropism, as ThkPV-1 and ThkPV-3 were detected in both alimentary and respiratory specimens. Western blot analysis using the recombinant N proteins showed that antibodies to ThkPV-1, ThkPV-2 and ThkPV-3 were present in 52–65% of serum samples from Leschenault's rousette bats, supporting that the bat species is the natural reservoir. This is in line with the finding of high prevalence (up to 46%) of neutralizing antibodies to MenPV in flying foxes (Pteropus spp.) in Australia (Philbey et al., 2008). Antibodies against TioPV has been detected in serum samples from fruit bats in Madagascar, including two from the genus Pteropus and one from the species R. madagascariensis (Iehlé et al., 2007). Possible serological evidence of human infection by TioPV has also been demonstrated in 3 (1.8%) of 169 human serum samples from inhabitants of Tioman Island, Malaysia (Yaiw et al., 2007). It has been shown that MenPV and TioPV are antigenically related, with recombinant N and V proteins of TioPV cross-reacting with porcine anti-MenPV sera in Western blot assay (Chua et al., 2001). Since the N proteins of ThkPV-1, ThkPV-2 and ThkPV-3 share 57–64% amino acid identities, cross-reactivity among the present assays cannot be excluded. Further studies on human serum samples, different bat tissue samples such as urine and development of cell culture isolation may provide better insight into the pathogenicity of the three novel paramyxoviruses.
The megachiropteran frugivorous bat, R. leschenaulti, is an important bat species associated with a number of emerging viruses, including flaviviruses, rabies virus, coronaviruses and paramyxoviruses. R. leschenaulti is a large fruit bat species with a wingspan of over 400 mm and is widely distributed in Southern China and South and Southeast Asia (http://www.afcd.gov.hk/english/conservation/hkbiodiversity/database/popup_record.asp?id=3306&lang=en). As early as in the 1960s, antibodies against Kyasanur Forest disease virus of the tick-borne encephalitis virus group under Flaviviridae family were detected in Leschenault's rousette bats near Poona, India (Pavri and Singh, 1965). West Nile virus was also subsequently isolated from the same bat species in India (Paul et al., 1970). In a recent seroprevalence study, Leschenault's rousette bats from China were also found to possess antibody against rabies virus (Jiang et al., 2009). The presence of coronavirus in Leschenault's rousette bats is unknown until our recent description of two novel coronaviruses, bat-CoV HKU9, under a novel subgroup 2d of group 2 coronaviruses; and bat-CoV HKU10, a group 1 coronavirus, in this bat species in China (Woo et al., 2007). Bat-CoV HKU9 and bat-CoV HKU10 was detected in the alimentary specimens of 42 (12%) and 1 (0.3%) of 350 tested Leschenault's rousette bats (Woo et al., 2007). Interestingly, five of the 15 samples positive for ThkPV-1, ThkPV-2 or ThkPV-3 from Leschenault's rousette bats were found to carry bat-CoV HKU9 (unpublished data). As for paramyxoviruses, a bat parainfluenza virus, antigenically related to simian virus 41, was isolated from a Leschenault's rousette bat collected at Poona, India in 1966 (Hollinger and Pavri, 1971, Pavri et al., 1971). The virus demonstrated pathogenicity in suckling mice, check embryos and white leghorn chicks (Hollinger and Pavri, 1971). However, further studies on paramyxoviruses in Leschenault's rousette bat were lacking until the present finding of three novel rubulaviruses most closely related to MenPV and TioPV. Continuous surveillance of this bat species in the region is important in understanding their role in emerging infectious diseases.
We speculate that there may still be unidentified paramyxoviruses in various bat populations. Bats are the only flying mammals which account for 20% of the > 4800 mammalian species in the world. In addition to SARS-CoV, they are important reservoir of other emerging zoonotic viruses, such as Hendra and Nipah viruses, Ebola virus, rabies virus and other lyssaviruses, and fungi such as Histoplasma capsulatum (Leroy et al., 2005, Lyon et al., 2004, Mackenzie and Field, 2004). For SARS-CoV, Hendra, Nipah, and Ebola virus, bats are the natural reservoir and human infection only occurred after cross-species transmission through an intermediate amplifying host such as palm civet in the case of SARS-CoV. However, bats may also transmit viruses directly to humans, as in the case of rabies virus and other lyssaviruses. Among the 60 viral species reported to be associated with bats, 59 are RNA viruses, which are associated with a high mutation and/or recombination rates that may be important in the adaptation to new hosts and emergence in humans (Lau et al., 2005a, Wong et al., 2007, Woo et al., 2006b). For example, the existence of coronaviruses in bats is unknown until a search for the animal reservoir of SARS-CoV during which a previously unknown diversity of bat coronaviruses including bat-SARS-CoV were identified (Lau et al., 2005a, Lau et al., 2007, Li et al., 2005, Poon et al., 2005, Tang et al., 2006, Woo et al., 2006c, Woo et al., 2007, Woo et al., 2009a, Woo et al., 2009b). The finding of Leschenault's rousette bats as reservoir for ThkV1, ThkV2 and ThkV3 and other fruit bats as reservoir for MenPV, TioPV, HenPV and NipPV suggested that there may be other unknown paramyxovirues in bats especially fruit bats. Although novel paramyxoviruses were only identified in Leschenault's rousette bats in the present study, the total number of bats from other species that were screened was just half the number of Leschenault's rousette bats. Further studies in bats of other species and in other countries may help better understanding of the molecular evolution of paramyxoviruses among these animals and their emerging potential.
Materials and methods
Sample collection
Four hundred and eighty-nine bats, of 11 species, were captured from various locations in the Guangdong province of Southern China over a 7-month period (October 2005–April 2006). Their respiratory, alimentary and blood samples were collected using procedures described previously (Lau et al., 2005a, Yob et al., 2001).
RNA extraction
Viral RNA was extracted from the respiratory and alimentary specimens using QIAamp Viral RNA Mini Kit (QIAgen, Hilden, Germany). The RNA was eluted in 50 μl of AVE buffer and was used as the template for RT-PCR.
RT-PCR of L gene of rubulaviruses using conserved primers and DNA sequencing
Paramyxovirus detection was performed by amplifying a 408-bp fragment of the L gene of rubulaviruses using conserved primers (LPW9648 5'-GCCAATCATGCWGGNAARTT-3' and LPW9649 5'-GTTGAATGGATCACCNACATA-3') designed by multiple alignments of the nucleotide sequences of available L gene of rubulaviruses including MenPV, TioPV, MumPV and HpiPV-4b. Reverse transcription was performed using the SuperScript III kit (Invitrogen, San Diego, CA, USA). The PCR mixture (25 μl) contained cDNA, PCR buffer (10 mM Tris–HCl pH 8.3, 50 mM KCl, 2 mM MgCl2 and 0.01% gelatin), 200 μM of each dNTPs and 1.0 U Taq polymerase (Applied Biosystem, Foster City, CA, USA). The mixtures were amplified in 40 cycles of 94 °C for 1 min, 50 °C for 1.5 min and 72 °C for 2 min and a final extension at 72 °C for 10 min in an automated thermal cycler (Applied Biosystem, Foster City, CA, USA). Standard precautions were taken to avoid PCR contamination and no false-positive was observed in negative controls.
The PCR products were gel-purified using the QIAquick gel extraction kit (QIAgen, Hilden, Germany). Both strands of the PCR products were sequenced twice with an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), using the two PCR primers. The sequences of the PCR products were compared with known sequences of the L genes of paramyxoviruses in the GenBank database.
Quantitative RT-PCR
Quantitative RT-PCR to detect the L genes of ThkPV-1, ThkPV-2 and ThkPV-3 was performed on the 17 positive samples using TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and primers and probes listed in Table 5 with StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The reaction mix was thermal-cycled at 50 °C for 2 min and 95 °C for 10 min followed by 50 cycles of 95 °C for 15 s and 60 °C for 1 min. Three plasmids containing the target sequences were used for generating the standard curves.
Table 5.
Primers and probes used for quantitative RT-PCR of ThkPV-1, ThkPV-2 and ThkPV-3.
| Strains | Primers/probes | Sequences | Product length |
|---|---|---|---|
| ThkPV-1 | Forward | 5'-TGTTAAGCCTAGCACACATGCA-3' | 69 bp |
| Reverse | 5'-GTAAGAAAGCAGGCAGCAATTTC-3' | ||
| Probe | 5'-[FAM] TACCAAAGCCAGATGAGA [MGB]-3' | ||
| ThkPV-2 | Forward | 5'-GGCGTGCTGCAAGAGAAAA-3' | 67 bp |
| Reverse | 5'-TGTGTCGGATTATTGTTTCTTGATTT-3' | ||
| Probe | 5'-[FAM]ATCACTGTAGTCTCACATCT[MGB]-3' | ||
| ThkPV-3 | Forward | 5'-GCAATGTTTGCCAATTCTATGAAC-3' | 68 bp |
| Reverse | 5'-CGTGGGTGTATCCACTCAAAAA-3' | ||
| Probe | 5'-[FAM] AACTGTATGGATATCCC [MGB]-3' |
Complete genome sequencing
Three complete genomes, ThkPV-1, ThkPV-2 and ThkPV-3 discovered in the present study, were amplified and sequenced using the RNA extracted directly from the alimentary specimens as templates. The RNA was converted to cDNA by a combined random-priming and oligo(dT) priming strategy. As the initial results revealed that these paramxyoviruses were rubulaviruses, the cDNA was amplified by degenerate primers designed by multiple alignment of the genomes of Menangle virus (GenBank accession no. NC_007620), Tioman virus (GenBank accession no. NC_004074) and mumps virus (GenBank accession no. NC_002200) and additional primers designed from the results of the first and subsequent rounds of sequencing. These primer sequences are available on request. The 5' ends of the viral genomes were confirmed by rapid amplification of cDNA ends using the 5'/3' RACE kit (Roche, Germany), SMART RACE cDNA amplification kit (Clontech, USA), SMARTer RACE cDNA amplificaion kit (Clontech, USA). Sequences were assembled and manually edited to produce final sequences of the viral genomes.
Genome analysis
The nucleotide sequences of the three genomes and the deduced amino acid sequences of the open reading frames (ORFs) were compared to those of other paramyxoviruses. Homology searches were conducted using the BLAST server at the National Center for Biotechnology Information (NCBI). The ORF start codons were predicted in consideration of favorable Kozak context for initiation of translation (Kozak, 1991). Amino acid sequence identities were determined by pairwise alignment using ClustalW2 (Larkin et al., 2007). Phylogenetic tree construction was performed using neighbor-joining method with Jukes-Cantor correction in Mega 4 (Tamura et al., 2007). The attachment protein genes were aligned by ClustalX2 (Larkin et al., 2007) in Profile Alignment mode, while all other multiple alignments for the construction of phylogenetic trees were performed by ClustalW embedded in Mega 4 (Tamura et al., 2007). Prediction of transmembrane domains was performed using TMHMM 2.0 server (Krogh et al., 2001). The membrane protein types of F and HN proteins were identified from the protein topology predicted from SOSUI system (Hirokawa et al., 1998) and TMHMM 2.0 server (Krogh et al., 2001). Heptad repeats were identified by Multicoil program (Wolf et al., 1997) and COILS (Lupas et al., 1991). The isoelectric point (pI) and molecular weight (Mw) of proteins were calculated by ExPasy Compute pI/Mw tool (Gasteiger et al., 2005).
Analysis of P mRNA editing
To examine the number of G insertions at the P mRNA editing site, mRNA from original specimens was extracted using the Oligotex mRNA Mini kit (QIAgen, Hilden, Germany). First strand cDNA synthesis was performed using SuperScript III kit (Invitrogen, San Diego, CA, USA) with the random hexamer primers supplied. Primers (5'-AGGCCCGAACTGTGGTCCTG-3' and 5'-TCCCACTCCAGACAAAGTTGAA-3' for ThkPV-1, 5'-GACTAAAGATCCATAGAGCCAA-3' and 5'-TCAAACTCTCGTCTATGCCTT-3' for ThkPV-2, 5'-CCCACTCATCAAGCTGAGAA-3' and 5'-GACTATTCTGAGCCAGTACTT-3' for ThkPV-3) were used to amplify 702-bp, 577-bp, and 416-bp products respectively for ThkPV-1, ThkPV-2 and ThkPV-3 respectively covering the putative editing site. PCR was then performed, with PCR mixture (25 μl) containing cDNA, PCR buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl, 2 mM MgCl2 and 0.01% gelatin), 200 μM of each dNTPs and 1.0 U Taq polymerase (Applied Biosystems, Foster City, CA, USA). The mixtures were amplified in 40 cycles of 94 °C for 1 min, 50 °C for 1 min and 72 °C for 1 min and a final extension at 72 °C for 10 min in an automated thermal cycler (Applied Biosystem, Foster City, CA, USA). The products were then purified and cloned using the TOPO TA Cloning Kit (Invitrogen, San Diego, CA, USA). Colonies were picked randomly for sequencing analysis.
Cloning and purification of (his)6-tagged recombinant nucleocapsid protein from Escherichia coli
To produce a fusion plasmid for protein purification, primers (5'-CTAGCTAGCATGTCTTCAGTCTTTAAA-3' and 5'-CTAGCTAGCTTAAGAGTCAAGATCTC-3' for ThkPV-1, 5'-CTAGCTAGCATGTCGTCAGTTTT-3' and 5'-CTAGCTAGCTCATTCAACCAAATCA-3' for ThkPV-2, 5'-CTAGCTAGCATGGCATCACTCTT-3' and 5'-CTAGCTAGCCTACTCAATTAGGTT-3' for ThkPV-3) were used to amplify the gene encoding the nucleocapsid protein of the three novel paramyxoviruses by RT-PCR. The sequence coding for a total of 520, 524 and 514 amino acid residues of the nucleocapsid protein was amplified respectively for ThkPV-1, ThkPV-2 and ThkPV-3 and cloned into the NheI sites of expression vector pET-28b(+) (Novagen, Madison, WI, USA) in frame and downstream of the series of six histidine residues. The (His)6-tagged recombinant nucleocapsid protein was expressed and purified using the Ni2+-loaded HiTrap Chelating System (GE Healthcare, Buckinghamshire, UK) according to the manufacturer's instructions. Approximately 3 mg of purified protein was routinely obtained from one liter of E. coli carrying the fusion plasmid.
Western blot analysis
Western blot analysis was performed according to our published protocols, with slight modifications. Briefly, 3700 ng of purified (His)6-tagged recombinant nucleocapsid protein was loaded into each well of a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel and subsequently electroblotted onto a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). The blot was cut into strips and the strips were incubated separately with 1:1000 dilution of bat serum samples. Horse radish peroxidase conjugated goat anti-bat IgG antibody (1:4000 dilution) (Bethyl laboratories Inc., TX, USA) was used as the secondary antibody. Antigen-antibody interaction was detected with an ECL fluorescence system (GE Healthcare, Buckinghamshire, UK). Anti-His antibody and skim milk were used as positive and negative controls respectively. To evaluate the specificity of the recombinant N protein-based western blot assay, two bat serum samples from two other species negative for rubulaviruses were also subject to Western blot assay.
Viral culture
Two hundred microlitres of samples in viral transport medium stored at −80 °C were retrieved for each of the samples positive for ThkPV-1, ThkPV-2 or ThkPV-3. After thawing, they were immediately clarified by centrifugation. The clarified inoculum was added to culture tubes by adsorption inoculation which involved decanting of the culture medium and direct application of the inoculum to the cell monolayer. After 1 h of adsorption at 37 °C in a horizontal position, excess inoculum is discarded and replaced by 1.5 ml of MEM with 1% fetal calf serum or serum free MEM supplemented by 2 μg/ml of l-1-tosylamide-2-phenylethyl chloromethyl ketone-treated trypsin (TPCK-trypsin; Sigma Immunochemical Co., St. Louis, Mo.). The cultures were incubated at 37 °C with 5% carbon dioxide in stationary slanted racks. They were inspected daily by inverted microscopy for cytopathic effects (CPE) such as rounding or syncytia formation. After 10 days of incubation, subculturing to fresh cell line was performed. Subculturing was performed twice even if there were no CPE. The cell lines included Hep2C (human laryngeal carcinoma), BHK (baby hamster kidney), LLC-Mk2 (rhesus monkey kidney), MRC-5 (human lung fibroblast), FRhK-4 (rhesus monkey kidney), Huh-7.5 (human hepatoma), Vero E6 (African green monkey kidney), and HRT-18 (colorectal adenocarcinoma) cells. When no CPE was observed at the end of incubation, the cells were frozen and thawed once and the culture lysate were collected for RNA extraction. Tuhokovirus RNA was detected by RT-PCR as described. Hemadsorption and immunostaining were not performed if the RT-PCR were negative.
Nucleotide sequence accession numbers
The genome sequences of ThkPV-1, ThkPV-2 and ThkPV-3 have been lodged within the GenBank sequence database under accession no. GU128080–GU128082.
Acknowledgments
We thank Chung-Tong Shek and the HKSAR Department of Agriculture, Fisheries, and Conservation Mammal Working Group for their help in taking the photograph of a Leschenault's rousette bat. We are grateful to the generous support of Mrs. Carol Yu, Professor Richard Yu, Mr. Hui Hoy and Mr. Hui Ming in the genomic sequencing platform. This work is partly supported by the Research Grant Council Grant and University Development Fund, The University of Hong Kong; The Tung Wah Group of Hospitals Fund for Research in Infectious Diseases; the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau; the Providence Foundation Limited in memory of the late Dr. Lui Hac Minh; and Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2010.03.049.
Appendix A. Supplementary data
Multiple alignments of the first 400 residues of the N proteins (Ncore) of ThkPV-1, ThkPV-2 and ThkPV-3 compared to those of other members of the Rubulavirus genus, showing the conserved MSSV motif, central domain and the three conserved motifs. The conserved motifs were marked in open box and reported consensus sequences indicated above the alignment (where x represents any amino acid residue, Ø represents an aromatic amino acid and either of the residues in parentheses can be present at that position). Amino acid residue numbers for each protein are shown to the right of each sequence. Dots indicate identical residues and dashes indicate gaps. Amino acid substitutions different from previously reported consensus sequences in the conserved motifs are highlighted.
Alignment of the F protein cleavage and fusion peptides of ThkPV-1, ThkPV-2 and ThkPV-3 compared to those of other rubulaviruses. The arrow indicates the cleavage site. To the left of the arrow are single or multibasic cleavage sequences in which basic residues are shaded and the furin recognition sites are boxed. To the right of the arrow are the N-terminals of hydrophobic fusion peptides of F1 generated by cleavage of F0.
Multiple alignment of the L protein domains II, III and VI of ThkPV-1, ThkPV-2 and ThkPV-3 and other rubulaviruses. The conserved motifs (Pre-A and A to D) are marked in open boxes. Amino acid residues that are conserved among other rubulaviruses but unique in one or more of ThkPV-1, ThkPV-2 and ThkPV-3 are indicated by asterisks placed below the sequences. Amino acid residues that are conserved among other rubulaviruses but unique and conserved among ThkPV-1, ThkPV-2 and ThkPV-3 are indicated by triangles placed below the sequences. The unique residues at these positions are shaded.
References
- Barrett T. Morbillivirus infections, with special emphasis on morbilliviruses of carnivores. Vet. Microbiol. 1999;69:3–13. doi: 10.1016/s0378-1135(99)00080-2. [DOI] [PubMed] [Google Scholar]
- Bowden T.R., Boyle D.B. Completion of the full-length genome sequence of Menangle virus: characterisation of the polymerase gene and genomic 5' trailer region. Arch. Virol. 2005;150:2125–2137. doi: 10.1007/s00705-005-0552-7. [DOI] [PubMed] [Google Scholar]
- Bowden T.R., Westenberg M., Wang L.F., Eaton B.T., Boyle D.B. Molecular characterization of Menangle virus, a novel paramyxovirus which infects pigs, fruit bats, and humans. Virology. 2001;283:358–373. doi: 10.1006/viro.2001.0893. [DOI] [PubMed] [Google Scholar]
- Buchholz C.J., Spehner D., Drillien R., Neubert W.J., Homann H.E. The conserved N-terminal region of Sendai virus nucleocapsid protein NP is required for nucleocapsid assembly. J. Virol. 1993;67:5803–5812. doi: 10.1128/jvi.67.10.5803-5812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant K., Chan R., Smith M., Dwyer D.E., Kirkland P. Probable human infection with a newly described virus in the family Paramyxoviridae. The NSW Expert Group. Emerg. Infect. Dis. 1998;4:273–275. doi: 10.3201/eid0402.980215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.B., Goh K.J., Wong K.T., Kamarulzaman A., Tan P.S., Ksiazek T.G., Zaki S.R., Paul G., Lam S.K., Tan C.T. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet. 1999;354:1257–1259. doi: 10.1016/S0140-6736(99)04299-3. [DOI] [PubMed] [Google Scholar]
- Chua K.B., Bellini W.J., Rota P.A., Harcourt B.H., Tamin A., Lam S.K., Ksiazek T.G., Rollin P.E., Zaki S.R., Shieh W., Goldsmith C.S., Gubler D.J., Roehrig J.T., Eaton B., Gould A.R., Olson J., Field H., Daniels P., Ling A.E., Peters C.J., Anderson L.J., Mahy B.W. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Chua K.B., Wang L.F., Lam S.K., Crameri G., Yu M., Wise T., Boyle D., Hyatt A.D., Eaton B.T. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology. 2001;283:215–229. doi: 10.1006/viro.2000.0882. [DOI] [PubMed] [Google Scholar]
- Chua K.B., Wang L.F., Lam S.K., Eaton B.T. Full length genome sequence of Tioman virus, a novel paramyxovirus in the genus Rubulavirus isolated from fruit bats in Malaysia. Arch. Virol. 2002;147:1323–1348. doi: 10.1007/s00705-002-0815-5. [DOI] [PubMed] [Google Scholar]
- Crennell S., Takimoto T., Portner A., Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- Enserink M. Emerging diseases. Malaysian researchers trace Nipah virus outbreak to bats. Science. 2000;289:518–519. doi: 10.1126/science.289.5479.518. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. In: Protein Identification and Analysis Tools on the ExPASy Server, The Proteomics Protocols Handbook. Walker John M., editor. Humana Press; 2005. [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Halpin K., Young P.L., Field H.E., Mackenzie J.S. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- Henderson G.W., Laird C., Dermott E., Rima B.K. Characterization of Mapuera virus: structure, proteins and nucleotide sequence of the gene encoding the nucleocapsid protein. J. Gen. Virol. 1995;76:2509–2518. doi: 10.1099/0022-1317-76-10-2509. [DOI] [PubMed] [Google Scholar]
- Hirokawa T., Boon-Chieng S., Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Hollinger F.B., Pavri K.M. Bat parainfluenza virus. Immunological, chemical, and physical properties. Am. J. Trop. Med. Hyg. 1971;20:131–138. [PubMed] [Google Scholar]
- Horikami S.M., Smallwood S., Moyer S.A. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology. 1996;222:383–390. doi: 10.1006/viro.1996.0435. [DOI] [PubMed] [Google Scholar]
- Iehlé C., Razafitrimo G., Razainirina J., Andriaholinirina N., Goodman S.M., Faure C., Georges-Courbot M.C., Rousset D., Reynes J.M. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg. Infect. Dis. 2007;13:159–161. doi: 10.3201/eid1301.060791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Wang L., Lu Z., Xuan H., Han X., Xia X., Zhao F., Tu C. Seroprevalence of rabies virus antibodies in bats from Southern China. Vector Borne Zoonotic Dis. 2010;10:177–181. doi: 10.1089/vbz.2008.0212. [DOI] [PubMed] [Google Scholar]
- Karabatsos N. 3rd ed. American Society of Tropical Mediciene and Hygiene; San Antonio, Texas: 1985. International Catalogue of Arboviruses. [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kurath G., Batts W.N., Ahne W., Winton J.R. Complete genome sequence of Fer-de-Lance virus reveals a novel gene in reptilian paramyxoviruses. J. Virol. 2004;78:2045–2056. doi: 10.1128/JVI.78.4.2045-2056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.A., Parks G.D. Paramyxoviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields virology. 5th ed. Lippincott Williams and Wilkins; Philadelphia, Pa: 2007. pp. 1449–1496. [Google Scholar]
- Langedijk J.P., Daus F.J., van Oirschot J.T. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J. Virol. 1997;71:6155–6167. doi: 10.1128/jvi.71.8.6155-6167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., To W.K., Tse P.W., Chan A.K., Woo P.C., Tsoi H.W., Leung A.F., Li K.S., Chan P.K., Lim W.W., Yung R.W., Chan K.H., Yuen K.Y. Human parainfluenza virus 4 outbreak and the role of diagnostic tests. J. Clin. Microbiol. 2005;43:4515–4521. doi: 10.1128/JCM.43.9.4515-4521.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Wang M., Lam C.S., Xu H., Guo R., Chan K.H., Zheng B.J., Yuen K.Y. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Chan K.H., Yip C.C., Ng T.K., Tsang O.T., Woo P.C., Yuen K.Y. Confirmation of the first Hong Kong case of human infection by novel swine origin influenza A (H1N1) virus diagnosed using ultrarapid, real-time reverse transcriptase PCR. J. Clin. Microbiol. 2009;47:2344–2346. doi: 10.1128/JCM.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Li K.S., Chau K.Y., So L.Y., Lee R.A., Lau Y.L., Chan K.H., Lim W.W., Woo P.C., Yuen K.Y. Clinical and molecular epidemiology of human parainfluenza virus 4 infections in Hong Kong: subtype 4B as common as subtype 4A. J. Clin. Microbiol. 2009;47:1549–1552. doi: 10.1128/JCM.00047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Délicat A., Paweska J.T., Gonzalez J.P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lyon G.M., Bravo A.V., Espino A., Lindsley M.D., Gutierrez R.E., Rodriguez I., Corella A., Carrillo F., McNeil M.M., Warnock D.W., Hajjeh R.A. Histoplasmosis associated with exploring a bat-inhabited cave in costa rica, 1998–1999. Am. J. Trop. Med. Hyg. 2004;70:438–442. [PubMed] [Google Scholar]
- Mackenzie J.S., Field H.E. Emerging encephalitogenic viruses: lyssaviruses and henipaviruses transmitted by frugivorous bats. Arch. Virol. Suppl. 2004;18:97–111. doi: 10.1007/978-3-7091-0572-6_8. [DOI] [PubMed] [Google Scholar]
- Moreno-López J., Correa-Girón P., Martinez A., Ericsson A. Characterization of a paramyxovirus isolated from the brain of a piglet in Mexico. Arch. Virol. 1986;91:221–231. doi: 10.1007/BF01314282. [DOI] [PubMed] [Google Scholar]
- Morgan E.M. Evolutionary relationships of Paramyxovirus nucleocapsid-associated proteins. In: Kingsbury D.W., editor. The Paramyxoviruses. Plenum Press; New York: 1991. pp. 163–179. [Google Scholar]
- Morrison T.G. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta. 2003;1614:73–84. doi: 10.1016/s0005-2736(03)00164-0. [DOI] [PubMed] [Google Scholar]
- Müller R., Poch O., Delarue M., Bishop D.H., Bouloy M. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J. Gen. Virol. 1994;175:1345–1352. doi: 10.1099/0022-1317-75-6-1345. [DOI] [PubMed] [Google Scholar]
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Dawood F.S., Jain S., Finelli L., Shaw M.W., Lindstrom S., Garten R.J., Gubareva L.V., Xu X., Bridges C.B., Uyeki T.M. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- Osterhaus A.D., de Swart R.L., Vos H.W., Ross P.S., Kenter M.J., Barrett T. Morbillivirus infections of aquatic mammals: newly identified members of the genus. Vet. Microbiol. 1995;44:219–227. doi: 10.1016/0378-1135(95)00015-3. [DOI] [PubMed] [Google Scholar]
- Paul S.D., Rajagopalan P.K., Sreenivasan M.A. Isolation of the West Nile virus from the frugivorous bat, Rousettus leschenaulti. Indian J. Med. Res. 1970;58:1169–1171. [PubMed] [Google Scholar]
- Pavri K.M., Singh K.R. Demonstration of antibodies against the virus of Kyasanur Forest disease (KFD) in the frugivorous bat Rousettus leschenaulti, near Poona, India. Indian J. Med. Res. 1965;53:956–961. [PubMed] [Google Scholar]
- Pavri K.M., Singh K.R., Hollinger F.B. Isolation of a new parainfluenza virus from a frugivorous bat, Rousettus leschenaulti, collected at Poona, India. Am. J. Trop. Med. Hyg. 1971;20:125–130. doi: 10.4269/ajtmh.1971.20.125. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philbey A.W., Kirkland P.D., Ross A.D., Davis R.J., Gleeson A.B., Love R.J., Daniels P.W., Gould A.R., Hyatt A.D. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg. Infect. Dis. 1998;4:269–271. doi: 10.3201/eid0402.980214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philbey A.W., Kirkland P.D., Ross A.D., Field H.E., Srivastava M., Davis R.J., Love R.J. Infection with Menangle virus in flying foxes (Pteropus spp.) in Australia. Aust. Vet. J. 2008;86:449–454. doi: 10.1111/j.1751-0813.2008.00361.x. [DOI] [PubMed] [Google Scholar]
- Poch O., Blumberg B.M., Bougueleret L., Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 1990;71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- Poole E., He B., Lamb R.A., Randall R.E., Goodbourn S. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-ß. Virology. 2002;303:33–46. doi: 10.1006/viro.2002.1737. [DOI] [PubMed] [Google Scholar]
- Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw R.W., Glaser A.L., Van Campen H., Weiland F., Dubovi E.J. Identification and phylogenetic comparison of Salem virus, a novel paramyxovirus of horses. Virology. 2000;270:417–429. doi: 10.1006/viro.2000.0305. [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tang X.C., Zhang J.X., Zhang S.Y., Wang P., Fan X.H., Li L.F., Li G., Dong B.Q., Liu W., Cheung C.L., Xu K.M., Song W.J., Vijaykrishna D., Poon L.L., Peiris J.S., Smith G.J., Chen H., Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.H., Latham S.M., Woolhouse M.E. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.M., Lamb R.A., Paterson R.G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidona C.A., Kurz H.W., Gelderblom H.R., Darai G. Isolation and molecular characterization of a novel cytopathogenic paramyxovirus from tree shrews. Virology. 1999;258:425–434. doi: 10.1006/viro.1999.9693. [DOI] [PubMed] [Google Scholar]
- Virtue E.R., Marsh G.A., Wang L.F. Paramyxoviruses infecting humans: the old, the new and the unknown. Future Microbiol. 2009;4:537–554. doi: 10.2217/fmb.09.26. [DOI] [PubMed] [Google Scholar]
- Wolf E., Kim P.S., Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S., Lau S., Woo P., Yuen K.Y. Bats as a continuing source of emerging infections in humans. Rev. Med. Virol. 2007;17:67–91. doi: 10.1002/rmv.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Chan K.H., Wong B.H., Che X.Y., Tam V.K., Tam S.C., Cheng V.C., Hung I.F., Wong S.S., Zheng B.J., Guan Y., Yuen K.Y. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363:841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Yuen K.Y. Infectious diseases emerging from Chinese wet-markets: zoonotic origins of severe respiratory viral infections. Curr. Opin. Infect. Dis. 2006;19:401–407. doi: 10.1097/01.qco.0000244043.08264.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Yip C.C., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Li K.S., Poon R.W., Wong B.H., Tsoi H.W., Yip B.C., Huang Y., Chan K.H., Yuen K.Y. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Wang M., Lau S.K., Xu H., Poon R.W., Guo R., Wong B.H., Gao K., Tsoi H.W., Huang Y., Li K.S., Lam C.S., Chan K.H., Zheng B.J., Yuen K.Y. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lai K.K., Huang Y., Lee P., Luk G.S., Dyrting K.C., Chan K.H., Yuen K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Yaiw K.C., Crameri G., Wang L., Chong H.T., Chua K.B., Tan C.T., Goh K.J., Shamala D., Wong K.T. Serological evidence of possible human infection with Tioman virus, a newly described paramyxovirus of bat origin. J. Infect. Dis. 2007;196:884–886. doi: 10.1086/520817. [DOI] [PubMed] [Google Scholar]
- Yob J.M., Field H., Rashdi A.M., Morrissy C., van der Heide B., Rota P., bin Adzhar A., White J., Daniels P., Jamaluddin A., Ksiazek T. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001;7:439–441. doi: 10.3201/eid0703.010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P.L., Halpin K., Selleck P.W., Field H., Gravel J.L., Kelly M.A., Mackenzie J.S. Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg. Infect. Dis. 1996;2:239–240. doi: 10.3201/eid0203.960315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Hansson E., Shiell B., Michalski W., Eaton B.T., Wang L.F. Sequence analysis of the Hendra virus nucleoprotein gene: comparison with other members of the subfamily Paramyxovirinae. J. Gen. Virol. 1998;79:1775–1780. doi: 10.1099/0022-1317-79-7-1775. [DOI] [PubMed] [Google Scholar]
- Yuen K.Y., Chan P.K., Peiris M., Tsang D.N., Que T.L., Shortridge K.F., Cheung P.T., To W.K., Ho E.T., Sung R., Cheng A.F. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignments of the first 400 residues of the N proteins (Ncore) of ThkPV-1, ThkPV-2 and ThkPV-3 compared to those of other members of the Rubulavirus genus, showing the conserved MSSV motif, central domain and the three conserved motifs. The conserved motifs were marked in open box and reported consensus sequences indicated above the alignment (where x represents any amino acid residue, Ø represents an aromatic amino acid and either of the residues in parentheses can be present at that position). Amino acid residue numbers for each protein are shown to the right of each sequence. Dots indicate identical residues and dashes indicate gaps. Amino acid substitutions different from previously reported consensus sequences in the conserved motifs are highlighted.
Alignment of the F protein cleavage and fusion peptides of ThkPV-1, ThkPV-2 and ThkPV-3 compared to those of other rubulaviruses. The arrow indicates the cleavage site. To the left of the arrow are single or multibasic cleavage sequences in which basic residues are shaded and the furin recognition sites are boxed. To the right of the arrow are the N-terminals of hydrophobic fusion peptides of F1 generated by cleavage of F0.
Multiple alignment of the L protein domains II, III and VI of ThkPV-1, ThkPV-2 and ThkPV-3 and other rubulaviruses. The conserved motifs (Pre-A and A to D) are marked in open boxes. Amino acid residues that are conserved among other rubulaviruses but unique in one or more of ThkPV-1, ThkPV-2 and ThkPV-3 are indicated by asterisks placed below the sequences. Amino acid residues that are conserved among other rubulaviruses but unique and conserved among ThkPV-1, ThkPV-2 and ThkPV-3 are indicated by triangles placed below the sequences. The unique residues at these positions are shaded.