Abstract
Interferons (IFNs) inhibit severe acute respiratory syndrome coronavirus (SARS-CoV) replication and might be valuable for SARS treatment. In this study, we demonstrate that treatment of Vero E6 cells with interleukin-4 (IL-4) decreased the susceptibility of these cells to SARS-CoV infection. In contrast to IFNs, IL-4 did not show antiviral activity when administered immediately after SARS-CoV infection, suggesting that IL-4 acts early during the SARS-CoV replication cycle. Indeed, binding of recombinant SARS-CoV spike protein to Vero E6 cells was diminished on cells treated with IL-4, but also on cells exposed to IFN-γ. Consistent with these observations, IL-4 and IFN-γ downregulated cell surface expression of angiotensin-converting enzyme 2 (ACE2), the SARS-CoV receptor. Besides diminished ACE2 cell surface expression, ACE2 mRNA levels were also decreased after treatment with these cytokines. These findings suggest that IL-4 and IFN-γ inhibit SARS-CoV replication partly through downregulation of ACE2.
Keywords: SARS-CoV, ACE2, Interferon-gamma, Interleukin-4
Introduction
Severe acute respiratory syndrome (SARS) emerged late 2002/early 2003 in Guangdong Province, China, and spread rapidly to several countries in Asia, North America and Europe, causing disease in almost 8000 people, of whom nearly 10% died. Only a few months after the first emergence of SARS, a newly discovered coronavirus (CoV) was identified as its etiological agent (Drosten et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003).
SARS-CoV is a positive stranded RNA virus with a genome of about 30 kb in length that is structurally similar to that of other group 2 coronaviruses (Marra et al., 2003, Rota et al., 2003, Snijder et al., 2003). It enters the host cell by binding of the spike (S) protein S1 domain to angiotensin-converting enzyme 2 (ACE2), the SARS-CoV receptor (Li et al., 2003). Subsequently, the S2 domain fuses with the cellular membrane (Liu et al., 2004). ACE2 is a component of the renin–angiotensin system and mainly involved in the regulation of heart function and blood pressure (Boehm and Nabel, 2002, Crackower et al., 2002). It is expressed in vascular endothelia, heart, kidney and testis, but also in epithelia of the small intestine and in alveolar epithelial cells (Donoghue et al., 2000, Hamming et al., 2004, Harmer et al., 2002). Recently it was shown that ACE2 also acts as a receptor for human coronavirus NL63 (HCoV-NL63) (Hofmann et al., 2005).
SARS-CoV replicates predominantly in the lower respiratory tract and causes diffuse alveolar damage, leading to severe respiratory distress (Kuiken et al., 2003, Nicholls et al., 2003). Treatment of SARS patients was mainly based on the use of ribavirin and corticosteroids, but the efficacy of these drugs has not been proven. Several groups have examined the antiviral effect of interferons (IFNs) against SARS-CoV. In vitro studies have shown that human recombinant IFNs inhibit SARS-CoV replication and that IFN-β and IFN-γ combined have a synergistic antiviral effect against this virus (Cinatl et al., 2003, Morgenstern et al., 2005, Sainz et al., 2004). Furthermore, we have demonstrated that pegylated IFN-α protected type-1 pneumocytes from infection with SARS-CoV in cynomolgous macaques (Haagmans et al., 2004). Usually IFNs achieve their antiviral effect through the induction of various proteins that block viral replication in the infected cell. To further explore the antiviral effect of IFNs and other cytokines specifically against SARS-CoV infection, we treated Vero E6 cells with various amounts of IFN-γ, tumor necrosis factor (TNF)-α or interleukin (IL)-4 and infected these cells with SARS-CoV.
Results
Susceptibility of Vero E6 cells to SARS-CoV or HSV infection after cytokine treatment
Vero E6 cells were treated with 1, 10 or 100 ng/ml of IL-4, IL-10, IFN-γ, TNF-α or a combination of IFN-γ and TNF-α and infected with SARS-CoV or herpes simplex virus (HSV)-1. While treatment with TNF-α (Fig. 1A) or IL-10 (data not shown) had no antiviral effect against SARS-CoV or HSV-1, pretreatment with IFN-γ (Fig. 1B) or a combination of IFN-γ and TNF-α (Fig. 1C) dose dependently reduced the number of infected cells per well after infection with either one of these viruses, as demonstrated previously (Cinatl et al., 2003, Feduchi et al., 1989). Surprisingly, recombinant human IL-4 also exerted SARS-CoV antiviral activity, which was not observed against HSV infection (Fig. 1D), suggesting a virus-specific antiviral activity. We also tested the antiviral activity of IL-4 against HCoV-NL63, another coronavirus that also uses the ACE2 receptor to enter the cell (Hofmann et al., 2005). Compared to untreated control cells, viral excretion from infected Vero E6 cells was reduced by more than 1 log after IL-4 treatment (Fig. 2 ).
Fig. 1.
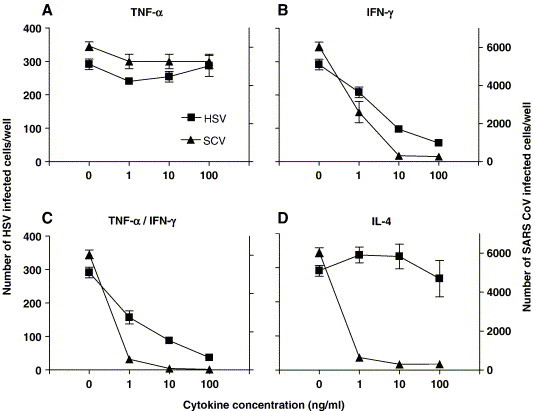
Susceptibility of Vero E6 cells to SARS-CoV or HSV-1 infection after treatment with TNF-α (A), IFN-γ (B), IFN-γ combined with TNF-α (C) or IL-4 (D). Triangle-shaped symbols represent SARS-CoV and rectangular symbols represent HSV-1. The left Y-axis indicates the number of infected cells per well after HSV infection, while the right Y-axis represents the number of infected cells per well after SARS-CoV (SCV) infection. Error bars indicate standard errors.
Fig. 2.
Antiviral activity of IL-4 against HCoV-NL63. Titration of supernatant after infection of Vero E6 cells with 105 TCID50 HCoV-NL63. Error bars indicate standard errors.
Antiviral activity of IL-4 against SARS-CoV
We further evaluated the antiviral effect of IL-4 against SARS-CoV using a SARS-CoV-specific RT-PCR and plaque titration on Vero E6 cells. After pretreatment of the cells with 10 ng/ml IL-4 for 48 h and subsequent infection, excretion of SARS-CoV from the treated cells was diminished by 1 log, just as SARS-CoV RNA levels in these cells (Figs. 3A–B). The antiviral effect of IL-4 was abrogated after preincubation with a neutralizing IL-4 antibody (Figs. 3A, B). These results further demonstrate that IL-4 has a modest antiviral activity against SARS-CoV infection in Vero E6 cells.
Fig. 3.
Antiviral activity of IL-4 against SARS-CoV. Plaque titration of supernatant (A) or amount of SARS-CoV RNA (B) present in mock (control), 10 ng/ml IL-4 or IL-4/α-IL-4-treated cells that were infected with 105 TCID50 SARS-CoV. Susceptibility of IL-4-treated Vero E6 cells to SARS-CoV infection after removal of IL-4 conditioned medium and subsequent washing (C) and susceptibility of fresh Vero E6 cells to SARS-CoV after receiving IL-4 conditioned medium just before infection with 105 TCID50 SARS-CoV (D). Error bars indicate standard errors.
To determine which step of the SARS-CoV life cycle is affected by IL-4, conditioned medium from Vero E6 cells that were treated with IL-4 (10 ng/ml) was removed and the cells were washed before they were infected with SARS-CoV. Removal of IL-4 conditioned medium from the Vero E6 cells after 48 h of treatment still conferred protection against SARS-CoV (Fig. 3C). On the other hand, when this IL-4 conditioned medium was transferred to fresh untreated Vero E6 cells and these cultures were infected 5 min later by adding 105 TCID50 SARS-CoV to the medium, no reduction in the number of SARS-CoV infected cells was observed (Fig. 3D). Therefore, the presence of a soluble SARS-CoV-infection blocking molecule, released by IL-4-treated cells, is unlikely. Because residual IL-4 was still present in this conditioned medium (not shown), IL-4 probably does not inhibit replication of SARS-CoV once it has entered the cell and needs to be added to the cells at an earlier time point. Thus, IL-4 most likely has an antiviral effect on an early step of the SARS-CoV life cycle in Vero E6 cells.
Binding of recombinant SARS-CoV spike protein to cytokine-treated Vero E6 cells
Since IL-4 treatment reduced excretion of virus, viral replication and infection, only when administered before SARS-CoV infection, binding of the SARS-CoV S protein to cytokine-treated Vero E6 cells was analyzed using flow cytometry. As shown in Fig. 4A, treatment with IL-4 reduced the ability of Vero E6 cells to bind recombinant S protein. Because IFN-γ combined with TNF-α also reduced susceptibility of Vero E6 cells to SARS-CoV, we analyzed binding of the S protein to these cells after IFN-γ/TNF-α treatment and demonstrated that 10 ng/ml IFN-γ/TNF-α reduced the binding capacity of the S protein to a background level (Fig. 4B). These data suggest that these cytokines influence expression of the cellular receptor for SARS-CoV, ACE2.
Fig. 4.
Binding of recombinant SARS-CoV S protein to Vero E6 cells after treatment with 10 ng/ml IL-4 (A) or a combination of 10 ng/ml IFN-γ and 10 ng/ml TNF-α (B). Dotted lines represent cytokine-treated cells, while thick lines represent mock-treated control cells. The shaded areas represent background staining.
Effect of cytokine treatment on ACE2 expression
To examine if IL-4 and IFN-γ/TNF-α decrease susceptibility to SARS-CoV infection through an effect on cell surface ACE2 expression, we analyzed ACE2 expression after cytokine treatment. As shown in Fig. 5A, 10 ng/ml IL-4 downregulated ACE2 expression and preincubation with a neutralizing IL-4 antibody abolished the IL-4 effect (Fig. 5B). Similarly, treatment of Vero E6 cells with 100 ng/ml IL-13− also a Th2 cytokine−downregulated ACE2 expression and inhibited SARS-CoV replication (data not shown). Background staining was determined by incubating the cells with normal goat serum followed by the secondary antibody (Fig. 5).
Fig. 5.
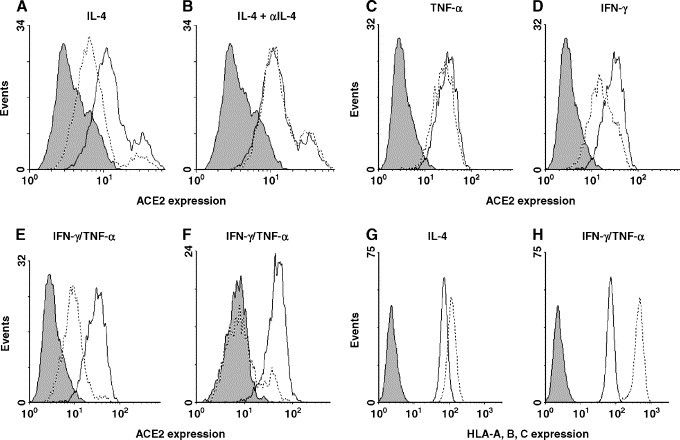
Influence of cytokine treatment on ACE2 expression. ACE2 expression was determined after treatment with 10 ng/ml IL-4 (A), IL-4 preincubated with a neutralizing antibody (B), 10 ng/ml TNF-α (C), 10 ng/ml IFN-γ (D) or 10 ng/ml IFN-γ combined with 10 ng/ml TNF-α (E), all at 48 h. Vero E6 cells were incubated for 96 h in the presence of 10 ng/ml IFN-γ combined with 10 ng/ml TNF-α (F). HLA-A, B, C expression on Vero E6 cells after treatment with 10 ng/ml IL-4 (G) or IFN-γ combined with TNF-α (H). Dotted lines represent cytokine-treated cells, while thick lines represent mock-treated control cells. The shaded areas represent background staining.
A more pronounced decrease in ACE2 expression was observed after treatment with a combination of 10 ng/ml IFN-γ and 10 ng/ml TNF-α compared to treatment with these cytokines separately (Figs. 5C–E). After removal of the cytokines, ACE2 expression was restored only gradually to pretreatment levels (data not shown) whereas addition of fresh IFN-γ and TNF-α at 48 h further decreased ACE2 expression to background level (Fig. 5F). Downregulation of ACE2 was not caused by an overall inhibitory effect on cell surface protein expression; IFN-γ and IL-4 both upregulated MHC class I expression on Vero E6 cells (Figs. 5G and H).
When ACE2 expression on Vero E6 cells was followed in time after treatment with IL-4, a gradual decrease in ACE2 expression was observed, which was maximal at 48 h after treatment (Fig. 6A). The decrease in ACE2 expression over time correlated with decreased susceptibility to SARS-CoV infection (Fig. 6B). These experiments also demonstrate that IL-4 only displayed antiviral activity when administered at least 24 h before infection with SARS-CoV (Fig. 6B). In contrast, treatment with IFN-α 1 h after SARS-CoV infection reduced replication by at least 1 log (not shown). These results further substantiate our hypothesis that IL-4 inhibits SARS-CoV replication through downregulation of ACE2.
Fig. 6.
ACE2 expression and SARS-CoV susceptibility at different time points after IL-4 treatment. ACE2 expression on Vero E6 cells was monitored at different time points after IL-4 treatment relative to control cells (A). Susceptibility of Vero E6 cells to SARS-CoV was monitored at different time points before and after treatment with 10 ng/ml IL-4 (B). Shown is the number of SARS-CoV infected cells per well.
ACE2 mRNA levels after cytokine treatment
The fact that ACE2 downregulation after IL-4 treatment is a gradual process and that ACE2 expression recovers slowly when cytokines are removed suggests that ACE2 is downregulated at the mRNA level. Using an ACE2-specific Taqman, we observed reduced ACE2 mRNA levels in Vero E6 cells after 48 h of IL-4 or IFN-γ/TNF-α treatment (Fig. 7 ). This implicates that downregulation of ACE2 by these cytokines is regulated at transcription level.
Fig. 7.
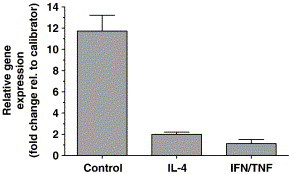
ACE2 mRNA levels in Vero E6 cells after treatment with 10 ng/ml IL-4 or a combination of 10 ng/ml IFN-γ and 10 ng/ml TNF-α compared to ACE2 mRNA levels in untreated control cells. ACE2 mRNA levels in untreated control cell and in IL-4-treated cells are represented as the fold change in gene expression relative to the IFN-γ/TNF-α-treated cells, which were used as the calibrator. GAPDH was used as an endogenous control.
Discussion
It is well established that IFNs inhibit replication of many different viruses, including SARS-CoV (Cinatl et al., 2003, Haagmans et al., 2004, Morgenstern et al., 2005, Sainz et al., 2004). When IFNs bind to their receptors, downstream signaling from these receptors can block one or more steps in the virus life cycle. IL-4 on the other hand does not induce these proteins and may even antagonize the protective effects of IFN (Lohoff et al., 1990). In general, the Th2-derived cytokine IL-4 is not considered to display antiviral activity. In fact, only a few studies have demonstrated IL-4 mediated inhibition of viral replication (Goletti et al., 2002, Lin et al., 2003). Furthermore, expression of IL-4 by recombinant ectromelia virus or vaccinia virus exacerbated infection in vivo and the administration of recombinant IL-4 delayed virus clearance in influenza virus-infected mice (Jackson et al., 2001, Moran et al., 1996, Sharma et al., 1996). In this study, we demonstrate that treatment of Vero E6 cells with IL-4 decreased susceptibility of these cells to SARS-CoV infection by 10-to 100-fold. Interestingly, ACE2 expression on the cell surface was downregulated after treatment with IL-4 or IFN-γ. Therefore, we postulate that IL-4 and IFN-γ may decrease susceptibility to SARS-CoV infection partially through modulation of ACE2 cell surface expression. Cytokine-mediated downregulation of viral receptors has been reported earlier in the case of the coxsackievirus–adenovirus receptor, which can be downregulated synergistically by IFN-γ and TNF-α in vitro (Vincent et al., 2004).
In this study, we show that the antiviral activity of IL-4 was not observed when IL-4 was added after SARS-CoV infection. This suggests that IL-4 inhibits entry of the virus, possibly through an effect on the viral receptor ACE2. In line with these observations, IL-4 does not have an antiviral effect against HSV, but does show antiviral activity against HCoV-NL63, which also utilizes ACE2 as a receptor. Subsequent experiments revealed that ACE2 expression indeed is downregulated by IL-4. Confirmation of ACE2's involvement as an intermediate of the IL-4 antiviral activity is hard to proof because of its essential role in SARS-CoV infection.
ACE2 is a component of the renin–angiotensin system and enzymatically cleaves angiotensin I into angiotensin 1–9 and angiotensin II into angiotensin 1–7 (Donoghue et al., 2000, Vickers et al., 2002). Several studies have reported regulation of ACE2 expression, most likely related to its enzymatic activity. ACE2 protein expression is downregulated in the kidneys of hypertensive or diabetic rats (Crackower et al., 2002, Tikellis et al., 2003), whereas ACE2 upregulation has been demonstrated after blocking of the angiotensin II-receptors in the kidneys of rats during pregnancy, and in the human failing heart (Brosnihan et al., 2003, Crackower et al., 2002, Goulter et al., 2004, Ishiyama et al., 2004, Tikellis et al., 2003). Furthermore, it was recently shown that ACE2 is downregulated in the lungs of mice after acute lung injury, including SARS-CoV infection (Kuba et al., 2005). Most probably, ACE2 can be regulated by several different factors and expression of this protein is dynamic on various cell types. Recently it was reported that ACE2 expression is dependent on the differentiation state of epithelia; ACE2 was poorly expressed on undifferentiated airway epithelial cells, while it was abundantly expressed on well-differentiated cells (Jia et al., 2005). It will be of interest to study the effect of IL-4 and IFN-γ on ACE2 expression on these cells.
The ACE2 downregulation was cytokine-specific; while IFN-γ and IL-4 both downregulated ACE2 expression, IFN-α, a cytokine that inhibits SARS-CoV replication in vivo and in vitro, did not affect ACE2 expression on Vero E6 cells (data not shown). Treatment of Vero E6 cells with IL-13 on the other hand also downregulated ACE2, suggesting the involvement of the IL-4 type II receptor in the IL-4 induced antiviral activity. However, this hypothesis could not be further substantiated because antibodies against the IL-4 type II receptor tested were not reactive on non-human primate Vero E6 cells.
Forced downregulation of ACE2 expression by cytokines may reveal a novel antiviral strategy against SARS-CoV infection. Potentially, addition of cytokines able to downregulate ACE2 expression could further improve the efficacy of IFN-α based anti SARS-CoV therapy. However, downregulation of ACE2 may have a potential negative effects on SARS pathogenesis, since it was recently reported that ACE2 has a protective role in acute lung failure in mice (Imai et al., 2005). Therefore it will be necessary to explore the consequences of ACE2 downregulation on SARS pathogenesis.
The most important finding of this study is the observation that cytokines modulate ACE2 expression. The physiological relevance of the IL-4 induced antiviral activity may be limited because this cytokine is probably not produced in large quantities during natural SARS-CoV infection (Huang et al., 2005, Wong et al., 2004). However, vaccination strategies utilizing inactivated SARS-CoV or subunits in the presence of specific alum based adjuvants may skew immune response to TH2 or TH0, allowing the production of IL-4 upon restimulation (Spruth et al., 2006). The relevance of ACE2 expression modulation by IFN-γ in the pathogenesis of SARS and NL-63 CoV needs further investigation.
Materials and methods
Cells and cytokine treatment
Vero E6 cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS), sodium bicarbonate and 20 mM HEPES buffer. Unless stated otherwise, recombinant human (r-hu) IL-4 (BD Pharmingen), r-hu IFN-γ, r-hu TNF-α, r-hu IL-13, r-hu IL-10 (Peprotech Inc.) and r-hu IFN-α (Roche) were added to cell cultures at a concentration of 10 ng/ml, 48 h before infection. As a specificity control, r-hu IL-4 was preincubated with a neutralizing IL-4 antibody (Ebiosciences) for 30 min at 37 °C before addition to the cells. All treatments were done in quadruplets (96-well experiments) or triplicate (6-well and 24-well experiments).
Infection of Vero E6 cells and immunohistochemistry
Vero E6 cells grown in 96-well plates were infected with 105 50% tissue culture infective doses (TCID50) SARS-CoV (HKU-39849) or HSV-1, by adding the virus directly to Vero E6 cell cultures. After 16 h, cells were fixed with 10% neutral-buffered formalin and treated with 70% ethanol (10 min RT). SARS-CoV infected cells were visualized using purified human IgG from a convalescent SARS patient (CSL), followed by staining with an antibody to human IgG, linked to horseradish peroxidase (Amersham Biosciences). HSV-infected cells were stained using a rabbit anti HSV-1 antiserum, followed by staining with an antibody to rabbit IgG, linked to horseradish peroxidase (DAKO).
In the case of the HCoV-NL63 infection, Vero E6 cells cultured in the presence or absence of IL-4 were infected with 104 TCID50 HCoV-NL63. The inoculum was removed after 1 h and replaced with fresh medium. After 48 h, the supernatant from these cells was titrated on fresh Vero E6 cells.
Antiviral activity of IL-4 against SARS-CoV was determined by plaque titration. Vero E6 cells were grown in 24-well plates in the presence or absence of IL-4 and infected with 105 TCID50 SARS-CoV. After 1 h, the inoculum was removed and fresh medium was added. Supernatant from these cultures was taken at 16 h and plaque titrated by inoculating fresh Vero E6 cells, cultured in 6-well plates, for 1 h. Subsequently, cells were washed and culture medium containing 0.5% low melting point agarose was added. The plates were incubated at 37 °C for 3 days, plaques were counted in wells containing approximately 10–100 plaques and the results were expressed as PFU/ml.
SARS-CoV and ACE2 RT-PCR
RT-PCR with primers and probe specific for the nucleoprotein gene of SARS-CoV was used to quantify SARS-CoV in infected cells, as described earlier (Kuiken et al., 2003). Serial dilutions of the SARS-CoV stock were used as a standard.
ACE2 mRNA levels were determined using the ACE2 Taqman Gene Expression Assay (Applied Biosystems). Differences in ACE2 expression were represented as the fold change in gene expression normalized to a reference, using the 2− ΔΔCt method (Livak and Schmittgen, 2001). The housekeeping gene GAPDH was used as the reference and the IFN-γ/TNF-α-treated cells were used as the calibrator, since those cells had the lowest ACE2 expression.
Spike protein binding studies
Binding of the SARS-CoV S protein was analyzed using flow cytometry. A His-tagged S protein (Protein Sciences) was incubated with cytokine-treated and mock-treated Vero E6 cells for 1 h at 4 °C, followed by αHis-FITC (Invitrogen) staining, to detect binding of the S protein.
ACE2 expression studies
Vero E6 cells, with or without cytokine treatment, were stained with a polyclonal goat αACE2 antibody (R&D Systems) followed by staining with an anti-goat-FITC antibody (Sigma). As a control for background staining of the αACE2 antibody, cells were incubated with normal goat serum (MP Biomedicals). Staining of the ACE2 receptor on Vero E6 cells could be blocked with soluble ACE2 protein (R and D systems), but not with the soluble ACE protein (R&D systems), demonstrating the specificity of the ACE2 antibody (data not shown).
Acknowledgments
We thank R. Lonsdale, B. Martina and R.F. van Lavieren for their assistance with SARS-CoV infections. This work was supported by the Commission of the European Community, FP6 grant 5111060 (SP22-CT-2004).
References
- Boehm M., Nabel E.G. Angiotensin-converting enzyme 2—A new cardiac regulator. N. Engl. J. Med. 2002;347(22):1795–1797. doi: 10.1056/NEJMcibr022472. [DOI] [PubMed] [Google Scholar]
- Brosnihan K.B., Neves L.A., Joyner J., Averill D.B., Chappell M.C., Sarao R., Penninger J., Ferrario C.M. Enhanced renal immunocytochemical expression of ANG-(1–7) and ACE2 during pregnancy. Hypertension. 2003;42(4):749–753. doi: 10.1161/01.HYP.0000085220.53285.11. [DOI] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362(9380):293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E., Oliveira-dos-Santos A.J., da Costa J., Zhang L., Pei Y., Scholey J., Ferrario C.M., Manoukian A.S., Chappell M.C., Backx P.H., Yagil Y., Penninger J.M. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417(6891):822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000;87(5):E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Feduchi E., Alonso M.A., Carrasco L. Human gamma interferon and tumor necrosis factor exert a synergistic blockade on the replication of herpes simplex virus. J. Virol. 1989;63(3):1354–1359. doi: 10.1128/jvi.63.3.1354-1359.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goletti D., Kinter A.L., Coccia E.M., Battistini A., Petrosillo N., Ippolito G., Poli G. Interleukin (IL)-4 inhibits phorbol-ester induced HIV-1 expression in chronically infected U1 cells independently from the autocrine effect of endogenous tumour necrosis factor-alpha, IL-1beta, and IL-1 receptor antagonist. Cytokine. 2002;17(1):28–35. doi: 10.1006/cyto.2001.0989. [DOI] [PubMed] [Google Scholar]
- Goulter A.B., Goddard M.J., Allen J.C., Clark K.L. ACE2 gene expression is up-regulated in the human failing heart. BMC Med. 2004;2(1):19. doi: 10.1186/1741-7015-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Kuiken T., Martina B.E., Fouchier R.A., Rimmelzwaan G.F., van Amerongen G., van Riel D., de Jong T., Itamura S., Chan K.H., Tashiro M., Osterhaus A.D. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 2004;10(3):290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer D., Gilbert M., Borman R., Clark K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532(1–2):107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102(22):7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.J., Su I.J., Theron M., Wu Y.C., Lai S.K., Liu C.C., Lei H.Y. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 2005;75(2):185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama Y., Gallagher P.E., Averill D.B., Tallant E.A., Brosnihan K.B., Ferrario C.M. Upregulation of angiotensin-converting enzyme 2 after myocardial infarction by blockade of angiotensin II receptors. Hypertension. 2004;43(5):970–976. doi: 10.1161/01.HYP.0000124667.34652.1a. [DOI] [PubMed] [Google Scholar]
- Jackson R.J., Ramsay A.J., Christensen C.D., Beaton S., Hall D.F., Ramshaw I.A. Expression of mouse interleukin-4 by a recombinant ectromelia virus suppresses cytolytic lymphocyte responses and overcomes genetic resistance to mousepox. J. Virol. 2001;75(3):1205–1210. doi: 10.1128/JVI.75.3.1205-1210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79(23):14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.J., Shu P.Y., Chang C., Ng A.K., Hu C.P. IL-4 suppresses the expression and the replication of hepatitis B virus in the hepatocellular carcinoma cell line Hep3B. J. Immunol. 2003;171(9):4708–4716. doi: 10.4049/jimmunol.171.9.4708. [DOI] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363(9413):938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lohoff M., Marsig E., Rollinghoff M. Murine IL-4 antagonizes the protective effects of IFN on virus-mediated lysis of murine L929 fibroblast cells. J. Immunol. 1990;144(3):960–963. [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Moran T.M., Isobe H., Fernandez-Sesma A., Schulman J.L. Interleukin-4 causes delayed virus clearance in influenza virus-infected mice. J. Virol. 1996;70(8):5230–5235. doi: 10.1128/jvi.70.8.5230-5235.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B., Michaelis M., Baer P.C., Doerr H.W., Cinatl J., Jr. Ribavirin and interferon-beta synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem. Biophys. Res. Commun. 2005;326(4):905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., Yan K.W., Chan K.H., Tsang N.C., Guan Y., Yuen K.Y., Peiris J.S. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361(9371):1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sainz B., Jr., Mossel E.C., Peters C.J., Garry R.F. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2004;329(1):11–17. doi: 10.1016/j.virol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D.P., Ramsay A.J., Maguire D.J., Rolph M.S., Ramshaw I.A. Interleukin-4 mediates down regulation of antiviral cytokine expression and cytotoxic T-lymphocyte responses and exacerbates vaccinia virus infection in vivo. J. Virol. 1996;70(10):7103–7107. doi: 10.1128/jvi.70.10.7103-7107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruth M., Kistner O., Savidis-Dacho H., Hitter E., Crowe B., Gerencer M., Bruhl P., Grillberger L., Reiter M., Tauer C., Mundt W., Barrett P.N. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine. 2006;24(5):652–661. doi: 10.1016/j.vaccine.2005.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikellis C., Johnston C.I., Forbes J.M., Burns W.C., Burrell L.M., Risvanis J., Cooper M.E. Characterization of renal angiotensin-converting enzyme 2 in diabetic nephropathy. Hypertension. 2003;41(3):392–397. doi: 10.1161/01.HYP.0000060689.38912.CB. [DOI] [PubMed] [Google Scholar]
- Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277(17):14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- Vincent T., Pettersson R.F., Crystal R.G., Leopold P.L. Cytokine-mediated downregulation of coxsackievirus–adenovirus receptor in endothelial cells. J. Virol. 2004;78(15):8047–8058. doi: 10.1128/JVI.78.15.8047-8058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


