Abstract
Autophagy is a conserved eukaryotic mechanism that mediates the removal of long-lived cytoplasmic macromolecules and damaged organelles via a lysosomal degradative pathway. Recently, a multitude of studies have reported that viral infections may have complex interconnections with the autophagic process. These observations strongly imply that autophagy has virus-specific roles relating to viral replication, host innate and adaptive immune responses, virus-induced cell death programs, and viral pathogenesis. Autophagy can supply internal membrane structures necessary for viral replication or may prolong cell survival during viral infections and postpone cell death. It can influence the survival of both infected and bystander cells. This process has also been linked to the recognition of viral signature molecules during innate immunity and has been suggested to help rid the cell of infection. This review discusses interactions between different viruses and the autophagy pathway, and surveys the current state of knowledge and emerging themes within this field.
Keywords: Authophagy, Survival, Autophagosomes, Beclin, LC3, Type II programmed cell death, Innate immunity
Introduction
Macroautophagy (hereafter referred to as autophagy) is a tightly regulated and evolutionarily conserved mechanism for the sequestration, lysosomal degradation, and recycling of discrete intracellular portions of eukaryotic cells, facilitating the removal of materials not typically degraded by the ubiquitin–proteosomal pathway. Regulators of this process include hormones and growth factors that suppress autophagy during cellular growth, as well as intracellular levels of nutrients, oxygen, and energy, allowing the pathway to act as a defense mechanism against inducers of cellular stress (Pattingre et al., 2008, Wullschleger et al., 2006, Yang et al., 2005). Perturbations in autophagy have been correlated with numerous pathological conditions, including oncogenesis and cancer progression, neurodegenerative disorders, liver disease, myopathy, and cardiac disease, highlighting the importance of this pathway in human health and cellular homeostasis (Levine and Kroemer, 2008, Meijer and Codogno, 2006, Mizushima et al., 2008). Autophagy has been shown to play an important role in the pathogenesis of several viral infections and is suggested to act as both an inducer and effector of innate and adaptive immune responses against intracellular pathogens, including viruses. Currently, evidence suggests that viruses have evolved a diverse array of countermeasures to contend with this pathway; some inhibit autophagy and are negatively affected when this interference is abrogated, while others appear to subvert it to their own ends and respond positively when it is induced. However, still other viruses are seemingly unaffected by autophagy, and do not appear to regulate the pathway through any apparent mechanism(s). This review seeks to provide both a synopsis of currently known and suspected viral interactions with the autophagy pathway, and to stimulate a critical discussion concerning the central trends that have been suggested within this field of research.
Overview of the mechanisms and regulation of autophagy
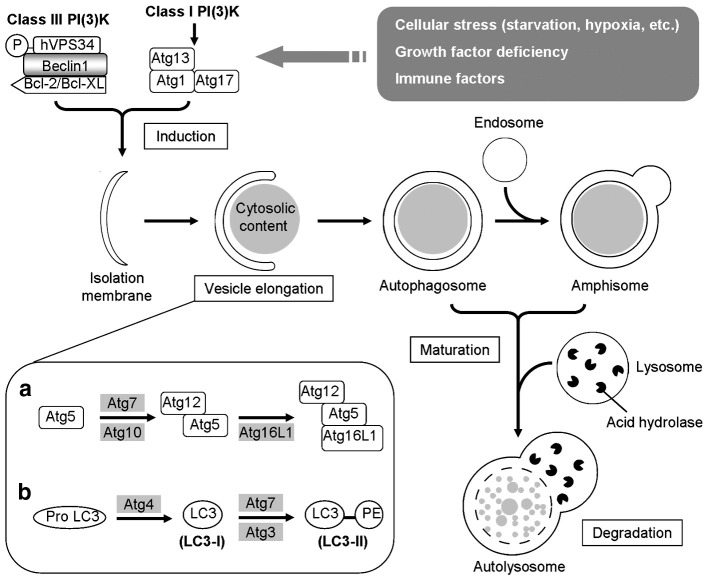
For the sake of brevity, only an overview of the mechanisms and regulation of the autophagy mechanism will be provided. The reader is referred to more detailed reviews concerning specific aspects of autophagy, such as the formation and maturation of the autophagosome (Longatti and Tooze, 2009, Mizushima, 2007, Noda et al., 2009, Reggiori and Klionsky, 2005, Xie and Klionsky, 2007, Yang et al., 2005, Yoshimori and Noda, 2008), regulatory mechanisms of this pathway (Meijer and Codogno, 2004, Pattingre et al., 2008, Wullschleger et al., 2006, Yang et al., 2005), roles in innate antigen recognition and MHC antigen presentation (Delgado et al., 2009, Delgado and Deretic, 2009, Levine and Deretic, 2007, Münz, 2006, Orvedahl and Levine, 2009, Schmid and Münz, 2005, Virgin and Levine, 2009), and the relationship between autophagy and regulated cell death (Codogno and Meijer, 2005, Ferraro and Cecconi, 2007, Kroemer and Levine, 2008, Maiuri et al., 2007, Scarlatti et al., 2009). The core process of autophagy is the de novo synthesis of a double-membrane-bound vesicle capable of fusing with an endosome or lysosome, which ultimately leads to the catabolic degradation of the encapsulated cargo (Fig. 1 ). In mammals, this process begins with the expansion of a small, flat membrane sac of uncertain origin (termed the isolation membrane or phagophore). As autophagy-related (Atg) proteins are recruited to its surface, this membrane sac elongates and curves until the ends merge to form a double-membrane-bound vesicle (autophagosome). Atg proteins are then recovered or disassociate from the autophagosome, and the completed structure fuses with an endosome (amphisome) or lysosome (autolysosome). A brief overview of mammalian genes of particular significance in the regulation and execution of autophagy is provided in Table 1 .
Fig. 1.
Overview of the autophagy process. In response to cellular stimuli such as starvation and immune signals, the class I PI(3)K (phosphoinositide 3-kinases)-induced Atg1 complex and a class III PI(3)K complex involving Beclin-1 activate downstream ATG proteins in a series of steps that guide the induction, elongation, maturation, and degradation of the autophagosome. Two ubiquitin-like conjugation systems involving Atg12 (a) and LC3 (b) direct the vesicle elongation of the isolation membrane, which forms a crescent shape to sequester the cytoplasmic cargo. Upon completion, the autophagosome then undergoes the maturation step through a series of remodeling processes including fusion with endosomes/lysosomes. Fusion with lysosome helps the autophagosome mature into an autolysosome in which the autophagic vacuole along with its content is degraded.
Table 1.
Significant genes in the mammalian autophagy pathway.
| Gene | Important interactions | Protein function/characteristics |
|---|---|---|
| Formation of autophagosomes | ||
| ULK1 (ATG1) | Atg13, FIP200 (Atg17) | Ser/Thr kinase activity important for function; target(s) unknown. Downstream of mTOR signaling. Potentially involved in Atg9 cycling. |
| Beclin-1 (ATG6) | hVps34, Bcl-2/Bcl-xL, UVRAG | Structural regulator of class III PI3 kinase hVps34. Contains BH3-like domain that is down-regulatory when occupied. |
| hVPS34 | Beclin-1, mTOR | Class III PI3 kinase; resulting PtdIns(3)Ps recruit Atg16L multimer/Atg18 to phagophore. Conflictingly activates mTor in response to amino acids. |
| ATG9 | Atg2, Atg18 | Transmembrane protein. Transits between phagophores and trans-Golgi/late endosomes. Possible role(s) in protein recycling and/or membrane transit. |
| ATG12 | Atg5, Atg16L | Covalently bound to Atg5 via mechanism similar to ubquitination. |
| ATG7 | LC3, Atg12 | Functionally similar to E1 ubiquitin activating enzyme (E1-like). Activates C-terminal glycine of both Atg12 and LC3. |
| ATG10 | Atg12, Atg5 | Functionally similar to E2 ubiquitin conjugating enzyme (E2-like). Accepts activated Atg12 and conjugates to internal lysine of Atg5. |
| ATG5 | Atg12, Atg16L | Covalently bound to Atg12; conjugation allows Atg5 to associate with Atg16L. |
| ATG16L | Atg5–Atg12 | Associates with Atg12–Atg5 and dimerizes. Present on outer surface of expanding phagophore; aids membrane curvature and LC3 recruitment (E3-like). Recycled. |
| ATG4 | LC3 | Cysteine protease; exposes C-terminal glycine on LC3 prior to lipidation. Subsequently recycles LC3 from outer membrane of autophagosome. |
| ATG3 | LC3, Atg7 | Functionally similar to E2 ubiquitin conjugating enzyme (E2-like). Conjugates LC3 with phosphatidylethanolamine (PE) phospholipid. |
| MAP1LC3 (ATG8) | Atg4 | Experimental marker of induction. Cytosolic form (LC3-I) conjugated to PE, becoming membrane-associated (LC3-II). Possible role(s) in membrane expansion, autophagosome transit, and lysosomal fusion. Partially recycled by Atg4. |
| Regulation of autophagy | ||
| PI3K (class I) | Produces PtdIns(3)p that activates the Akt/PKB-mTor pathway. | |
| PTEN | Phosphatase that counteracts PI3K by dephosphorylating PtdIns(3)p. | |
| AKT/PKB | PDK1, Tsc 2 | Ser/Thr kinase. Activated by PDK1 in the presence of PtdIns(3)p. Inactivates Tsc 2. |
| REDD1/REDD2 | Transcriptionally up-regulated in response to hypoxia. Inactivates mTor pathway. | |
| AMPK | LKB1, Tsc2 | Activates Tsc2, leading to the induction of autophagy when the AMP/ATP ratio is high. |
| TSC2 | Tsc1, Rheb, Akt/PKB, AMPK | GTPase-activating protein (GAP) with Tsc1; inactivates Rheb. Akt/PKB interferes with function, as does Erk1/2. AMPK enhances activity. |
| Rheb | Tsc1/Tsc2, mTor | Small GTPase. Activates mTor via binding kinase domain in GTP-dependent fashion. Tsc1/Tsc2 GAP activity converts to inactive, GDP-bound form. |
| mTOR | Rheb, raptor, mLST8 | Key regulator of cellular growth. Autophagy induced when mTor inactivated. Ser/Thr kinase. Forms two protein complexes; mTORC1 associated with autophagy. |
| Anti-apoptotic Bcl-2 family | Beclin-1 | Inhibit autophagy via binding with BH3 motif on Beclin-1. JNK1-mediated phosphorylation disrupts interaction and associated inhibition. |
| BH3-only Bcl-2 family | Anti-apoptotic Bcl-2 family | Competitively bind with anti-apoptotic Bcl-2 family members, interfering with their association with Beclin-1. Stimulate autophagy. |
| JNK1 | Anti-apoptotic Bcl-2 family | Phosphorylates anti-apoptotic Bcl-2 family members, inhibiting interaction with Beclin-1. Activity induces autophagy. |
| UVRAG | Bif-1, Beclin-1 | Interacts with Beclin-1's coiled-coil domain, strengthening Beclin-1/hVps34 interactions; promotes autophagy. Possible additional role in lysosome fusion. |
| p53 | Controversial/contradictory role(s) in autophagy. P53-dependent autophagy observed experimentally. However, cytosolic p53 is inhibitory (mechanism unknown). | |
| DRAM | Transmembrane lysosomal protein transcriptionally induced by p53. Stimulates autophagy. Necessary for both p53-dependent autophagy and apoptosis. | |
Central in the regulation of autophagy are two key proteins: mTOR and Beclin-1 (Pattingre et al., 2008, Sinha and Levine, 2008, Wullschleger et al., 2006). mTOR, a conserved serine/threonine kinase, is a component of protein complexes that integrate cellular signals relating to growth factors, nutrient and energy status, and cellular stress (Wullschleger et al., 2006). Important activators of mTOR include the class I PI3K-Akt/PKB signaling pathway and high concentrations of specific amino acids; high AMP/ATP ratios and hypoxia inactivate this pathway (Arsham and Neufeld, 2006, Beugnet et al., 2003, Pattingre et al., 2008, Wullschleger et al., 2006). Activated mTOR suppresses autophagy, enhancing the accumulation of cellular bulk by limiting lysosomal digestion. Downstream of mTOR, Beclin-1 is at the heart of a regulatory complex for the class III PI3K hVps34, whose activity is essential during autophagosome formation. Activators, such as UV radiation resistance associated gene (UVRAG), Bax-interacting factor-1 (Bif-1), and activating molecule in Beclin-1-regulated autophagy-1 (Ambra-1) associate with the Beclin-1 complex and enhance PtdIns(3)P production, while the Bcl-2 family anti-apoptotic proteins such as Bcl-2 and Bcl-xL bind to Beclin-1 and act in an inhibitory fashion (Pattingre et al., 2008, Sinha and Levine, 2008). The Bcl-2-related inhibition of autophagy is abrogated by stress-activated c-Jun N-terminal protein kinase 1 (JNK1) phosphorylation and competition from other BH3 binding domain-containing proteins, providing one of several direct mechanistic links between autophagy and apoptosis (Sinha and Levine, 2008, Wei et al., 2008a, Wei et al., 2008b).
Numerous other cellular factors have been shown or are hypothesized to regulate autophagy, many of which have importance in viral infections. The eukaryotic initiation factor-2 alpha (eIF2α) and the starvation-responsive general control nonderepressible-2 (GCN2) eIF2α kinase are both indispensable for starvation-induced autophagy, suggesting that other eIF2α kinases with important roles in antiviral defense, such as double-stranded RNA (dsRNA)-dependent protein kinase (PKR) and PKR-like ER kinase (PERK), may also up-regulate this pathway in response to cellular stressors (Kouroku et al., 2007, Tallóczy et al., 2002, Tallóczy et al., 2006). Immune signaling molecules can modulate autophagy; type II interferon-γ (IFN-γ) and tumor-necrosis factor-α (TNF-α) are stimulatory, while the TH2-type cytokines interleukin-4 (IL-4) and IL-13 are suppressive (Deretic, 2005, Levine and Deretic, 2007). Certain pathogen-associated molecular patterns (PAMPs) trigger autophagy through their cognate pattern recognition receptors (PRRs), including Toll-like receptor 3 (TLR3), TLR4, and TLR7, although the molecular mechanism(s), physiological function(s), and range of PRRs that induce this pathway are all areas of continuing research (Delgado et al., 2009, Delgado and Deretic, 2009, Orvedahl and Levine, 2009). The p53 protein possesses a dual role in the regulation of autophagy dependent upon its localization; cytoplasmic p53 represses autophagy and must be degraded for autophagy to proceed, whereas nuclear p53 appears to induce it (Maiuri et al., 2009, Tasdemir et al., 2008). Many additional cellular factors, including extracellular signal-regulated kinase (Erk1/2) activation (Pattingre et al., 2003, Shinojima et al., 2007, Wang et al., 2009), intracellular release of calcium (Gordon et al., 1993, Høyer-Hansen and Jäättelä, 2007), increases in reactive oxygen species (ROS) (Djavaheri-Mergny et al., 2007, Djavaheri-Mergny et al., 2006, Scherz-Shouval and Elazar, 2007, Scherz-Shouval et al., 2007), and endoplasmic reticulum (ER) stress (Ding et al., 2007, Kouroku et al., 2007, Ogata et al., 2006, Yorimitsu et al., 2006) have also been shown to trigger the pathway to autophagy.
While numerous experimental approaches including electron microscopy, LC3 lipidation (aggregation and modification), and protein degradation studies, amongst others, can be employed to identify or quantify autophagy in higher eukaryotes, a few warnings should be considered regarding the specific challenges these methods present. First, as autophagy is a process with numerous components, steps, and phases, it is important to clarify whether a given assay measures a step within this pathway (such as LC3 lipidation) or its overall physiological consequence (the aim of protein degradation studies). Since several viruses have been shown to modulate autophagy at multiple stages with varying effects, it is often necessary to combine assays examining both induction and maturation in order to make accurate observations. Second, while autophagy is a responsive cellular process, capable of fluctuation, most assays capture this dynamic process at a single static moment in time. This can pose challenges, as cellular populations are frequently asynchronous and the effects of a viral infection may vary during the course of the cell cycle. Finally, as different cell types have been shown to display different autophagy responses, it can be difficult to make direct comparisons between different virus-cell systems. Particular caution should be used when studying viruses, like HIV-1, which infect more than one cell type. For further information regarding the limitations of current autophagy assays, readers are advised to consult excellent reviews on this subject (Klionsky et al., 2008, Mizushima, 2004).
Viral interactions with the autophagy pathway
Many viruses have been shown to evade, subvert, or exploit autophagy, seemingly to insure their own replication or survival advantage, while others are apparently unaffected by this intrinsic pathway and fail to modulate it by any detectable way. The following discussion concerning viral infection, autophagy, and host immunity has been structured on four emergent themes that have been identified in the research published to date; autophagy as 1) a mechanism for membrane remodeling, 2) a digestive defensive response, 3) a means of cellular surveillance, and 4) a cellular fate-determining process. A comprehensive summary of viruses for which studies have been undertaken is provided in Table 2 .
Table 2.
Brief summary of known interactions between autophagy and viral infections.
| Virus | Known interaction(s) with autophagy | Key references |
|---|---|---|
| Adenovirus | Contradictory results; autophagy may enhance or detract from engineered adenovirus-induced cell death; differences possibly cell-type/virus-specific. | Ito et al., 2006, Baird et al., 2008 |
| Coronavirus | Contradictory evidence. May induce or subvert autophagy membrane remodeling. Autophagy may enhance viral replication. | Prentice et al., 2004a, Zhao et al., 2007 |
| Coxsackievirus | Autophagosome formation enhances CVB3/CVB4 replication in vitro. Autophagy affects virally induced apoptosis. | Wong et al., 2008, Yoon et al., 2008 |
| Cytomegalovirus | Inhibits autophagy through unidentified mechanism. | Chaumorcel et al. (2008) |
| Dengue virus | Induces autophagy. Disruption reduces corresponding viral titers. Membrane co-localization observed with serotype-specific differences. | Lee et al., 2008, Panyasrivanit et al., 2009, Khakpoor et al., 2009 |
| Drosophila C virus | Induces formation of COPI-dependent vesicles. Interactions with autophagy mechanism unknown. | Cherry et al. (2006) |
| Enterovirus 71 | Induces/co-localizes with autophagic membranes. Induction increases viral titer. Mechanism unclear. | Huang et al. (2009) |
| Epstein–Barr virus | Autophagy may aid EBV MHC-II antigen presentation, but contradictory evidence exists. Possibly part of proliferation regulation. | Paludan et al., 2005, Lee and Sugden, 2008 |
| Hepatitis B virus | Encodes transcriptional transactivator HBx, resulting in increased expression of Beclin-1. Sensitizes cells to autophagy signals. | Tang et al. (2009) |
| Hepatitis C virus | Induces autophagy; inhibits maturation. Knockdown of autophagy genes or ER stress response limits replication. Autophagy may aid HCV antigen presentation/TLR detection in some cell types. | Sir et al., 2008, Dreux et al., 2009 |
| Herpes simplex virus type I | Viral encoded ICP34.5 antagonizes pathway through inhibition of PKR/eIF2a induction, as well as Beclin-1 binding. Autophagy effects of minimal importance in vitro, but essential for neurovirulence in vivo. | Tallóczy et al., 2006, Alexander et al., 2007, Orvedahl et al., 2007; English et al. (2009) |
| Human immunodeficiency virus | Autophagy-related mechanism part of envelope protein-induced bystander T cell death and HIV dementia. Pathway dysregulated in some infected cell types, but some components identified as necessary host co-factors. | Espert et al., 2006, Alirezaei et al., 2008, Zhou and Spector, 2008, Espert et al., 2009, Kyei et al., 2009 |
| Influenza virus | Induces autophagosome formation; inhibits maturation. Interference decreases viral yield. May enhance MHC-II antigen presentation. | Gannagé et al., 2009, Zhou et al., 2009 |
| Kaposi's sarcoma-associated herpesvirus | Encodes viral homolog of Bcl-2 that inhibits autophagy through binding interactions with Beclin-1's BH3 domain. Viral FLIP suppresses autophagy through inhibition of Atg3. | Pattingre et al., 2005, Ku et al., 2008, Lee et al., 2009 |
| Parvovirus | Induces formation of autophagosome-like vesicles, possibly to extend host cell survival during viral replication process. | Nakashima et al. (2006) |
| Poliovirus | Induces formation of double-membrane, LC3-positive vesicles. Aids viral replication and release. Viral 2BC protein triggers LC3 lipidation. | Jackson et al., 2005, Taylor and Kirkegaard, 2007 |
| Rhinovirus | Does not induce or modulate autophagy. Replication unaffected by induction or inhibition of pathway. | Jackson et al., 2005, Brabec-Zaruba et al., 2007 |
| Sendai virus | In pDCs, autophagy enhances delivery of viral nucleic acids to endosomes for TLR7 recognition. | Lee et al. (2007) |
| Sindbis virus | In vivo up-regulation of autophagy via Beclin-1 over-expression reduces fatal encephalitis in mice. | Liang et al. (1998) |
| Tobacco mosaic virus | Autophagy necessary to restrict virally induced programmed cell death responses to the site of infection. | Liu et al. (2005) |
| Vaccinia virus | Induces vesicle formation, but mechanism independent of autophagy. | Zhang et al. (2006) |
| Varicella-zoster virus | Induces autophagy. Role in infection unclear. | Takahashi et al. (2009) |
| Vesicular stomatitis virus | Autophagy enhances endocytic detection in pDCs, but may diminish cytosolic detection/antiviral responses in other cell types. | Jounai et al., 2007, Lee et al., 2007; Shelly et al. (2009) |
Autophagy is a mechanism for remodeling internal membranes associated with viral replication
For many viruses, the production of progeny virion is intimately associated with host cell membranes or cytoskeletal elements. As such, many viruses are known to subvert host endosomal and secretory pathways in order to induce host membrane alterations that can then support viral replication and assembly (Miller and Krijnse-Locker, 2008). A number of viruses have been shown to replicate in, or in close association with, multi-membraned vesicles that possess many of the characteristics of autophagosomes. Given the nature and location of these vacuoles, there is strong evidence that autophagosomes may serve as a site of viral replication during some infections and that the autophagy pathway might therefore be exploited by viruses to enhance virion production. Supporting this assertion, the membranes associated with viral replication are often derived from the ER, which has also been suggested as a source of the autophagic double-membrane (Mijaljica et al., 2006). Indeed, the similarities between autophagic vacuoles and some virally induced membrane alterations has lead to increased suspicion, and in some cases proof, of a connection between positive-stranded RNA viral replication and the autophagy mechanism.
A link between poliovirus (PV)-induced double-membrane vesicles and autophagy has long been suggested, and is often cited as the classic example of viral exploitation of the autophagy pathway. Ultrastructural and biochemical analyses have revealed that PV induces massive rearrangements in intracellular membranes, resulting in clusters of double-membrane-bound vesicles capable of supporting viral replication complexes (Bienz et al., 1987, Rust et al., 2001, Schlegel et al., 1996, Suhy et al., 2000). These PV-induced vesicles display the classic autophagosomal marker LC3, which has been hypothesized to be directly recruited by viral proteins (Jackson et al., 2005, Taylor and Kirkegaard, 2007). Subsequent studies have further shown that additional secretory vesicle trafficking molecules are also recruited during the formation of these membranous replicative vesicles (Belov et al., 2007), and that the association of these vesicles with the microtubule network aids in the non-lytic release of PV virion particles (Taylor et al., 2009). Hence, it has been postulated that PV factors initiate elements of both the interrelated secretory trafficking and autophagy pathways to ultimately create a membranous structure capable of supporting both viral replication and virion egress. Besides PV, several other picornaviruses also appear to subvert the autophagy machinery to promote their replication. The group B Coxsackieviruses, CVB3 and CVB4, induce autophagosome formation, and biochemical inhibition of this pathway negatively impacts virion production (Wong et al., 2008, Yoon et al., 2008). Similarly, the enteropathogen EV71 has been shown to trigger autophagy in vitro, and that this induction can significantly increase viral yield (Huang et al., 2009). Apart from the Picornaviridae family, two Dengue virus (DENV) serotypes (DENV-2 and DENV-3) have also been shown to interact with the autophagy machinery to promote their replication (Khakpoor et al., 2009, Lee et al., 2008, Panyasrivanit et al., 2009). Interestingly, the stage of the autophagic process with which DENV is associated varies depending upon the viral serotype involved; DENV-2 translation/replication complex has been shown to specifically locate with pre-lysosomal fusion amphisomes, while DENV-3 requires further vesicle maturation and post-lysosomal fusion vacuoles (autolysosomes) to efficiently produce virions (Khakpoor et al., 2009, Panyasrivanit et al., 2009). The autophagy pathway also appears to be critical in the replication of another Flaviviridae member, namely the hepatitis C virus (HCV), which has also been shown to induce autophagosome formation (Ait-Goughoulte et al., 2008, Dreux et al., 2009, Sir et al., 2008, Tanida et al., 2009). In the case of HCV, autophagy is thought to be responsible for providing membranous support structures during the initial translation and de novo replication of HCV RNA following infection, but subsequently contributes little to virus maturation once an infection is stably established in the host cell (Dreux et al., 2009, Tanida et al., 2009). Finally, basal levels of autophagy have recently been shown to enhance macrophage-tropic human immunodeficiency virus (HIV) viral protein processing and virion production in vitro, suggesting a role for autophagy in HIV biosynthesis in this cell type (Kyei et al., 2009).
While these results are strongly suggestive that viruses can subvert or induce autophagy in order to create advantageous membrane alterations, other research has demonstrated that this effect is not universal. Not all viral infections that induce membrane alterations, including some which are closely related to the examples provided above, modulate or are affected by the autophagy pathway. The replication of human rhinovirus 2, a picornavirus that shares many similarities with PV, is not affected by drugs that either inhibit or induce autophagy, and does not elicit the formation of LC3-positive vesicles (Brabec-Zaruba et al., 2007). Contradictory observations have been made for the Coronaviridae members, murine hepatitis virus (MHV) and severe acute respiratory syndrome-coronavirus (SARS-CoV); some studies have suggested that endogenous LC3 co-localizes with SARS-CoV and MHV vacuole-associated replicase proteins (Prentice et al., 2004a, Prentice et al., 2004b), yet other studies have had difficulty corroborating these results (de Haan and Reggiori, 2008, Snijder et al., 2006, Stertz et al., 2007). In line with these observations, MHV viral replication and release have been shown to be comparable in cells with both normal and defective autophagy mechanisms (Zhao et al., 2007). Likewise, the viral production kinetics of vaccinia virus, which has been shown to utilize double-membrane vesicles for its replication, was similar in wild-type, Atg5−/−, and Beclin-1-deficient cells (Zhang et al., 2006). Thus, subversion of autophagy as a mechanism for inducing membrane alterations may be either cell-type- or virus-dependent. These results suggest that considerable care should be exercised prior to concluding that autophagy is the mechanism responsible for membrane alterations observed under microscopy to ensure that they are biochemically and mechanistically connected to this pathway. While the evidence to date strongly indicates a positive role for autophagy in the replication of some viruses, the observed variation, particularly amongst closely related viruses, suggests that other mechanisms may exist for inducing similar alterations in host cells. One should approach these studies carefully with the correct diagnostic tools for autophagy.
Autophagy can be a mechanism for defense: the digestion or elimination of unwanted viral intruders
As the previous section has indicated, viruses may induce autophagy for their own replicative advantage; yet, the same process may also confine viral replication complexes within vesicles as an innate defense against infection (Wileman, 2006). Furthermore, autophagy has been documented to function as a host cell defense against some intracellular pathogens. This process, which has been termed xenophagy (‘to eat what is foreign’), results in the autophagic-lysosomal destruction of invading pathogens. Several bacterial pathogens including Mycobacterium tuberculosis (Gutierrez et al., 2004), group A Streptococcus (Nakagawa et al., 2004), Shigella flexneri (Ogawa et al., 2005), Legionella pneumophila (Amer and Swanson, 2005), and Yersinia pestis (Pujol et al., 2009) have been shown to either actively inhibit or be degraded through xenophagy. Subsequently, this process has been demonstrated to be particularly important in restricting bacterial escape and survival. Just as xenophagy restricts certain bacterial pathogens, autophagy may capture replicating viruses or newly assembled virions within their host cells, and eliminate them through sequestration and lysosomal degradation (Kirkegaard et al., 2004, Levine, 2005, Wileman, 2007).
The initial hypothesis that autophagy might function as an antiviral defense mechanism was suggested when the autophagy effector and regulatory protein Beclin-1 was shown to be an antiviral effector in mice infected with the neurotropic Sindbis virus (SINV) (Liang et al., 1998). Over-expression of Beclin-1 protected mice from fatal SINV encephalitis, reducing neuronal apoptosis and decreasing SINV viral replication (Liang et al., 1998). The antiviral and anti-apoptotic effects attributed to Beclin-1 in SINV infection in vivo suggested that autophagy may function as a defense against other viral infections. Since these initial observations were first published, further evidence has shown that autophagy may function as a defense against viral infections.
Similar effects to those observed with SINV infection have been demonstrated in HSV-1 encephalitis (Orvedahl et al., 2007). In murine fibroblasts and neurons, mutant HSV-1 deficient in ICP34.5, a viral Beclin-1-interacting protein, but not wild-type virus, induced autophagy upon infection in a PKR-dependent manner (Alexander et al., 2007, Orvedahl et al., 2007, Tallóczy et al., 2002, Tallóczy et al., 2006). This viral induction of autophagy resulted in the observed localization and xenophagic degradation of virions within autophagosomes (Alexander and Leib, 2008, Alexander et al., 2007, Tallóczy et al., 2006). However, the exact significance of increased mutant virions within double-membrane vesicles and xenophagy is questionable, since suppression of autophagy through the use of Atg5−/− mouse embryonic fibroblasts (MEFs) did not significantly alter the replication efficiency of either wild-type or ICP34.5 mutant HSV-1 in vitro (Alexander et al., 2007, Jounai et al., 2007). In the case of HSV-1, it is thought that while ICP34.5 expression can inhibit autophagy in infected cells, the prevention of PKR-mediated translational arrest, rather than regulation of autophagy, may be the pivotal determinant of HSV-1 replicative efficiency in cell culture (Alexander et al., 2007). In contrast to these in vitro observations, the suppression of autophagy in vivo by ICP34.5 appears to be critical in HSV-1 pathogenesis, since Beclin-1-binding-deficient ICP34.5 mutant viruses are neuroattenuated with regards to lethal encephalitis in mice (Orvedahl et al., 2007). Interestingly, this HSV-1 mutant's virulence can be reconstituted if the infection is conducted in mice deficient for the antiviral effector PKR (Orvedahl et al., 2007). The observed discrepancies between in vitro and in vivo results may be due to cell-type dependent differences, and/or the effects of other HSV-1 proteins on the autophagy pathway (Alexander and Leib, 2008).
The observation that a HSV-1 viral protein that abrogates autophagy is necessary in vivo to observe certain pathogenic effects, highlights the potential antiviral function of this pathway. This may be particularly true in the case of neurotropic viruses, since modulation of autophagy has been suggested to influence the development of certain neurological diseases, (Itzhaki et al., 2008, Orvedahl and Levine, 2008). Along these lines, it has been noted that feline, simian, and human immunodeficiency viral infections in vivo can indirectly induce deficits in neuronal autophagy, and that this effect may contribute to the neuro-inflammatory pathology observed in these diseases (Alirezaei et al., 2008, Zhu et al., 2009). Thus, the dysregulation of autophagy by certain neurotropic viruses may not only interfere with their lysosomal clearance, but may also have a significant impact in terms of their pathogenic effect.
One remaining question though is whether xenophagy truly functions as an antiviral effector mechanism, since only HSV-1 viral particles have been observed microscopically within the confines of autophagosomes. Furthermore, this effect may be virus-specific rather than a general defensive mechanism of viral clearance. While it is tempting to speculate that xenophagy, as demonstrated within the field of bacteriology, exists as a general host defense mechanism for the clearance of all intracellular pathogens, including viruses, the limited amount of direct evidence available at this time suggests that caution is warranted. In particular, care should be exercised in differentiating between the incidental inclusion of virion or viral components within autophagosomes due to either their uptake by background or stress-induced activation of this pathway, and enhanced, autophagosome-driven clearance of these materials.
The HSV-1 ICP34.5 protein is known to antagonize PKR by dephosphorylating eIF2α (He et al., 1997), and, as discussed above, also blocks the induction of autophagy in a PKR-dependent fashion (Orvedahl et al., 2007, Tallóczy et al., 2002, Tallóczy et al., 2006). As many viruses employ numerous countermeasures for disrupting the IFN and PKR antiviral systems, these viral disturbances may also have significance in modulating downstream autophagic processes. Indeed, unless a virus can inhibit autophagy it is subject to the effects of immune surveillance which harnesses the autophagy machinery (discussed in Autophagy may function in security surveillance: a watchdog for foreign molecules section).
Autophagy may function in security surveillance: a watchdog for foreign molecules
In addition to xenophagy, autophagy may function as an antiviral pathway in a less direct fashion, sampling and delivering cytoplasmic material to cellular compartments (endosomes and lysosomes). This process may play a significant role in the activation of innate and adaptive immune responses to foreign pathogens. Research has shown that autophagy is involved in the delivery of various antigens (viral, self, and tumor origin) to MHC class II antigen-presenting molecules, which can in turn lead to the activation of CD4+ T lymphocytes (Levine and Kroemer, 2008). As an example, the delivery of Epstein–Barr viral antigens to MHC class II-loading compartment (also known as late endosomes), prior to CD4+ T cell stimulation, has been shown to utilize the autophagy mechanism (Paludan et al., 2005). Constitutive autophagy in immune and epithelial cells has also been observed to participate in the delivery of LC3-tagged influenza matrix proteins to MHC class II-associated endosomal compartments, which resulted in enhanced antigen presentation to CD4+ T cells (Schmid et al., 2007). Furthermore, the importance of autophagy-mediated class II antigen presentation was recently demonstrated in the regulation of HSV-1 pathogenesis by viral ICP34.5 (Leib et al., 2009). As well, it has been shown that the immunization of mice with influenza A (IFA)-infected cells exhibiting enhanced autophagy facilitates more robust MHC class I antigen presentation to the CD8+ T cells (Uhl et al., 2009). This evidence suggests that autophagy may contribute to MHC class I/II-specific viral antigen presentation, aiding the induction of adaptive immune responses. These studies also raise the exciting possibility that it may be feasible to exploit autophagy to deliver viral antigens and enhance MHC class I/II presentation for the development of novel vaccines and adjuvant therapies (Schmid et al., 2007).
The autophagy machinery has also been suggested to play a role in innate immunity by delivering PAMPs to their associated PRRs, including the TLRs. TLRs play critical roles in detecting bacterial and viral signatures and in eliciting appropriate host cell defenses such as IFNs and inflammatory cytokines against bacterial and viral infections (Takeuchi and Akira, 2007). It has been recently shown that the delivery of viral nucleic acids to endosomal TLRs in plasmacytoid dendritic cells (pDCs) can occur through the autophagosomes (Lee et al., 2007). While most TLRs reside on the cell surface, a subset involved in the detection of viral gene products, including TLR3, 7/8, and 9, is located in endosomal compartments and aids in the detection of endocytosed viral material (Barton, 2007). Autophagosomes could theoretically facilitate the sequestration, delivery, and detection of cytoplasmic viral nucleic acids, thereby helping to elicit a classical IFN response (Levine and Deretic, 2007). Indeed, autophagy has been shown to mediate ssRNA virus recognition through TLR7 during vesicular stomatitis virus (VSV) and Sendai virus (SENV) infection. It contributes to the production of interferon-α by pDCs in vitro and in vivo (Lee et al., 2007). Hence, this intrinsic pathway may play an important role in the detection of viral antigens and in the induction of the subsequent IFN response in pDCs.
More recently, evidence supporting the involvement of autophagy in the TLR-related detection of viral PAMPs has increased with the finding that TLRs themselves can induce autophagosome formation. This suggests a potential feedback circuit involving autophagy in the pathogen-triggered immune response. Lipopolysaccharide (LPS)-induced autophagy has been demonstrated in both murine and human macrophages, and requires the TRIF (Toll-interleukin-1 receptor domain-containing adaptor-inducing IFN-β)-dependent TLR4 pathway, whose downstream components include the receptor-interacting protein (RIP1) and the p38 MAPK (Xu et al., 2007). While the physiological importance of this TLR-mediated induction is currently unclear, it suggests that TLR4 can induce autophagy, which may help to limit the spread of pathogenic infections. More importantly, it appears that there is cross-talk between the two pathways and that the autophagic machinery may be regulated by TLRs. TLR3 (dsRNA recognition), TLR7/8 (ssRNA recognition), and TLR9 (recognizes dsDNA with unmethylated CpG motifs) can identify viral signatures (Kawai and Akira, 2006). TLR4 can also recognize mouse mammary tumor virus (MMTV) envelope protein (Rassa et al., 2002) and the respiratory syncytial virus (RSV) fusion protein (Kurt-Jones et al., 2000). In addition, TLR2 recognizes the envelope proteins of HSV-1 (Kurt-Jones et al., 2004) and human cytomegalovirus (Compton et al., 2003), as well as the measles virus hemagglutinin (Bieback et al., 2002). As many of these TLRs share downstream adaptors TRIF and myeloid differentiation primary response gene (88) (MyD88) (Kawai and Akira, 2006), it is possible that apart from TLR4, other TLRs may also trigger autophagy.
On the other hand, autophagy has also been suggested to suppress the innate immune response to VSV infection. The retinoic-acid-inducible gene I (RIG-I)-mediated recognition of VSV in murine fibroblasts is inhibited by over-expression of Atg12–Atg5 (Jounai et al., 2007). Furthermore, MEFs deficient in Atg5 or Atg7 are resistant to VSV infection, more sensitive to polyinosinic:polycytidylic acid (poly:IC) treatment, and produce higher type I IFN responses following either treatment (Jounai et al., 2007, Tal et al., 2009). This evidence suggests that autophagy's effects are likely both cell-type- and PRR-dependent, and further demonstrates the intricate nature of the relationship between autophagy and host cell innate immunity.
Viruses can modulate autophagy to determine cell fate by either postponing or hastening cell death
Autophagy is involved with many biological pathways linked to cellular stress, and the signals that drive the activation of autophagy may ultimately decide the fate of the cell. Indeed, it has been suggested that autophagy possesses a dual role in mediating cell survival and cell death. As a survival mechanism, autophagy sustains cell viability during brief periods of cellular stress, either by providing metabolic substrates in times of shortage, or by removing damaged organelles in order to prevent apoptosis. On the other hand, autophagy has also been shown to induce cellular death under certain circumstances, which has been characterized as a form of programmed cell death (type II PCD) that differs from the more classical apoptotic (type I PCD) and necrotic forms of cell death (Levine and Yuan, 2005). Inhibition of autophagy can trigger apoptosis under starvation conditions (Boya et al., 2005), whereas cells deficient in the pro-apoptotic Bax and Bak proteins undergo autophagic cell death upon treatment with strong pharmacological agents such as etoposide (Shimizu et al., 2004). More importantly, based on autophagy's dual nature with regards to cell fate, it has been suggested that this pathway might have important roles in the development of cancer and tumor suppression (Baehrecke, 2005, Gozuacik and Kimchi, 2007, Kondo et al., 2005, Levine and Yuan, 2005) (Brech et al., 2009, Eisenberg-Lerner et al., 2009, Maiuri et al., 2009, Morselli et al., 2009, Scarlatti et al., 2009).
Recent studies indicate that autophagy may function as a pro-survival mechanism during viral infections. Autophagy has been suggested to extend the survival time of human parvovirus B19-infected erythroid cells during viral expansion (Nakashima et al., 2006). Furthermore, protection against SINV-induced neuronal apoptosis and encephalitis in mice is conferred by concurrent over-expression of Beclin-1, but not anti-apoptotic Bcl-2 (Liang et al., 1998). The hepatitis B virus (HBV)-encoded transcriptional transactivator protein X (HBx) has been shown to up-regulate the expression of Beclin-1 and stimulate autophagy (Tang et al., 2009), an effect that was also observed in HBV-associated hepatocellular carcinomas (HCC) (Song et al., 2004). Interestingly, HBx has long been suggested to be tumorigenic (Kim et al., 1991), while Beclin-1 is commonly regarded as a haplo-insufficient tumor suppressor gene (Qu et al., 2003, Yue et al., 2003). Hence, autophagy could be up-regulated by HBV to prolong cell survival, and this process may help initiate the development of HCC through an as yet uncharacterized mechanism.
As previously indicated, autophagy has been theorized to induce a novel form of programmed cell death, particularly under circumstances when apoptosis is impaired. Hence, while autophagy has been generally characterized as a pro-survival mechanism, certain viruses may counter-intuitively block this process in order to prolong the survival of infected cells. The Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded viral Bcl-2 (vBcl-2) protein inhibits autophagy through its interaction with Beclin-1 (Pattingre et al., 2005) and similar effects have also been observed with the closely related murine γ-herpesvirus-68 (γHV-68)-encoded vBcl-2 molecule M11 (Ku et al., 2008, Sinha et al., 2008). However, in addition to their effects on autophagy, vBcl-2s from γ-herpesviruses also protect infected cells from apoptosis (Choi et al., 2001, Moore and Chang, 2003). While some authors have suggested that vBcl-2 help γ-herpesviruses escape autophagic degradation (Pattingre et al., 2005), given the powerful anti-apoptotic role of vBcl-2 it is difficult to gauge the relative importance of these effects. More recently, the KSHV viral FADD-like interleukin-1-β-converting enzyme (FLICE)/caspase-8-inhibitory protein (vFLIP) was shown to suppress autophagy by interfering with the processing of LC3, and that this effect abrogated autophagy-associated cell death in infected cells (Lee et al., 2009).
Interestingly, viral infections have also been documented to trigger autophagy specifically as mechanism of inducing cell death in uninfected bystander cell populations. During HIV infection, expression of HIV envelope glycoproteins (gp120 and gp41, collectively termed “Env”) induces autophagy in uninfected bystander CD4+ T lymphocytes, which results in a novel cell death program with characteristics of both type I and type II PCD (Espert et al., 2006). Thus, it has been suggested that this autophagy-mediated phenomenon may be significant in contributing to general CD4+ T cell losses during the course of HIV infections (Espert et al., 2006, Espert et al., 2007, Levine and Sodora, 2006). Similarly, HIV gp120 could also enhance autophagy in uninfected neuronal cells, leading to neuronal cell death and potentially contributing to HIV-associated dementia (Spector and Zhou, 2008). Interestingly, in plants, autophagy has been shown to protect bystander non-infected cells from cell death induced by Tobacco mosaic virus (TMV) infection (Liu et al., 2005).
The observed differential regulation of autophagy by viral infection in controlling cellular fate provides some indication to the potential complexity of this intrinsic pathway. It is important to note that it is inherently difficult to distinguish between autophagy as a mechanism of virally induced cell death and a cellular response to infection. Furthermore, there is potential that this may be a pathway whose biology is commonly modified in laboratory cancer cell lines used in virology research (Baird et al., 2008). For example, autophagy has been suggested to regulate the death of brain tumor and prostate cancer cell models infected with engineered adenoviruses (Alonso et al., 2008, Ito et al., 2006, Jiang et al., 2007, Rajecki et al., 2009, Ulasov et al., 2009, Yokoyama et al., 2008), but conflicting results have also suggested that this response may actually be an attempt at survival (Baird et al., 2008). Moreover, as characteristics of both type I and type II PCD are often seen in conjunction, it is often difficult to differentiate whether the observed phenomenon is an executioner mechanism or a futile attempt at cell survival. Nonetheless, from the above discussions it is clear that autophagy can contribute in the regulation of cellular fate, and as such may be specifically regulated by viruses to promote their life cycle.
Discussion
Questions regarding how viral infections interact with, and are impacted by, the autophagy pathway are increasingly gaining prominence within virology. While this relatively new area of research has produced a number of exciting results, limits in our understanding of the mechanism, its regulation, and in the available investigative tools continue to present challenges, as would be expected in any emerging area of research. One complexity for researchers is that the regulation of autophagy is intimately interconnected with cellular growth and survival, as well as numerous cellular stress responses. Hence, considerable caution should be exercised before an effect on viral replication is directly attributed to this pathway. Furthermore, the natural complexity and diversity of viral pathogens and their host cells has resulted in numerous conflicting observations, making it difficult to draw clear and concise conclusions regarding the general role of autophagy in viral infections.
Despite these and other challenges, there is increasing evidence that this ancient cellular process is a significant factor in the fate of numerous different viral infections. During poliovirus infection, autophagy promotes viral replication through its most primitive function: the ability to induce membrane remodeling and vesicle formation. The processes of xenophagy and immune surveillance via cytoplasmic sampling suggest that autophagy may have evolved to act as a defense against intracellular pathogens that viruses must now contend with. Finally, autophagy's interconnections with cell death programs ingrain it in processes that ultimately determine cellular fate, suggesting that viruses may modulate it to influence cell survival and ensure their own reproductive advantage. Thus, autophagy may be implicated in the biological pathogenesis and/or replicative success of many viral infections through a wide variety of possible mechanisms, and can exhibit multiple distinct and cell-type-dependent roles in the course of a single viral infection. This is best illustrated in the case of HIV, where the autophagy machinery is: 1) down-regulated in virally-infected CD4+ T cells (Espert et al., 2009, Zhou and Spector, 2008), which may constitute a strategy for avoiding autophagosome-mediated degradation and/or endosomal detection in order to enhance virion production; 2) engaged in HIV-infected macrophages to promote virion production (Kyei et al., 2009); and 3) activated in bystander neurological and immunological naïve cells to commit cell death by type II PCD, thereby contributing to HIV pathogenesis (CD4+ T cell depletion and neuronal death associated-dementia) (Espert et al., 2006).
The revelation that autophagy can function in immune defense of the host cells, particularly relating to innate immunity, suggests reciprocal regulation (through TLRs and IFN response) aiding in the subsequent development of an adaptive immune response (through enhancement of MHC presentation) to clear pathogens in infected tissues. Further research has the potential to reveal unknown protein interactions and could lead to the development of new pharmacological therapies for treating various virally induced diseases and associated cancers. As new research tools emerge, continuing research will help to clarify the role of autophagy in viral replication, host immune responses, and viral pathogenesis. The intricacies of viral interactions with this ancient and highly conserved pathway strongly suggest that autophagy can function in a myriad of ways, some of which appear contradictory at first glance. Depending upon the circumstances, autophagy can be pro-survival or pro-death, can enhance viral replication or aid in the development of the antiviral response. Autophagy is a double-edged sword that can cut both ways. Clearly a virus would prefer to be on autophagy's good side!!
Acknowledgments
The authors would like to apologize to any investigators whose works were not included during the publication of this work due to space limitation. The authors would also like to thank Dr. Ryan S. Noyce for his critical reading of the manuscript, and Ting-Fen Chin for her technical assistance with Fig. 1. L.-T. Lin is a recipient of the National CIHR Research Training Program in Hepatitis C (NCRTP-HepC) fellowship. P.W.H. Dawson is a recipient of a Frederick Banting and Charles Best master's award from the Canadian Institutes of Health Research (CIHR).
References
- Ait-Goughoulte M., Kanda T., Meyer K., Ryerse J.S., Ray R.B., Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J. Virol. 2008;82(5):2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D.E., Leib D.A. Xenophagy in herpes simplex virus replication and pathogenesis. Autophagy. 2008;4(1):101–103. doi: 10.4161/auto.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D.E., Ward S.L., Mizushima N., Levine B., Leib D.A. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 2007;81(22):12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alirezaei M., Kiosses W., Flynn C., Brady N., Fox H. Disruption of neuronal autophagy by infected microglia results in neurodegeneration. PLoS ONE. 2008;3(8):e2906. doi: 10.1371/journal.pone.0002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso M.M., Jiang H., Yokoyama T., Xu J., Bekele N., Lang F.F., Kondo S., Gomez-Manzano C., Fueyo J. Delta-24-RGD in combination with RAD001 induces enhanced anti-glioma effect via autophagic cell death. Mol. Ther. 2008;16(3):487–493. doi: 10.1038/sj.mt.6300400. [DOI] [PubMed] [Google Scholar]
- Amer A.O., Swanson M.S. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell. Microbiol. 2005;7(6):765–778. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsham A.M., Neufeld T.P. Thinking globally and acting locally with TOR. Curr. Opin. Cell Biol. 2006;18(6):589–597. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Baehrecke E.H. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005;6(6):505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- Baird S., Aerts J., Eddaoudi A., Lockley M., Lemoine N., Mcneish I. Oncolytic adenoviral mutants induce a novel mode of programmed cell death in ovarian cancer. Oncogene. 2008;27(22):3081–3090. doi: 10.1038/sj.onc.1210977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G.M. Viral recognition by Toll-like receptors. Semin. Immunol. 2007;19(1):33–40. doi: 10.1016/j.smim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Belov G.A., Altan-Bonnet N., Kovtunovych G., Jackson C.L., Lippincott-Schwartz J., Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 2007;81(2):558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beugnet A., Tee A., Taylor P., Proud C. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem. J. 2003;372(Pt 2):555–566. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieback K., Lien E., Klagge I.M., Avota E., Schneider-Schaulies J., Duprex W.P., Wagner H., Kirschning C.J., Ter Meulen V., Schneider-Schaulies S. Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J. Virol. 2002;76(17):8729–8736. doi: 10.1128/JVI.76.17.8729-8736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz K., Egger D., Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160(1):220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- Boya P., González-Polo R.A., Casares N., Perfettini J.L., Dessen P., Larochette N., Métivier D., Meley D., Souquere S., Yoshimori T., Pierron G., Codogno P., Kroemer G. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 2005;25(3):1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabec-Zaruba M., Berka U., Blaas D., Fuchs R. Induction of autophagy does not affect human rhinovirus type 2 production. J. Virol. 2007;81(19):10815–10817. doi: 10.1128/JVI.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brech A., Ahlquist T., Lothe R.A., Stenmark H. Autophagy in tumour suppression and promotion. Mol. Oncol. 2009;3(4):366–375. doi: 10.1016/j.molonc.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumorcel M., Souquère S., Pierron G., Codogno P., Esclatine A. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy. 2008;4(1):46–53. doi: 10.4161/auto.5184. [DOI] [PubMed] [Google Scholar]
- Cherry S., Kunte A., Wang H., Coyne C., Rawson R., Perrimon N. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2(10):e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Means R.E., Damania B., Jung J.U. Molecular piracy of Kaposi's sarcoma associated herpesvirus. Cytokine Growth Factor Rev. 2001;12(2–3):245–257. doi: 10.1016/s1359-6101(00)00029-0. [DOI] [PubMed] [Google Scholar]
- Codogno P., Meijer A.J. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- Compton T., Kurt-Jones E.A., Boehme K.W., Belko J., Latz E., Golenbock D.T., Finberg R.W. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J. Virol. 2003;77(8):4588–4596. doi: 10.1128/JVI.77.8.4588-4596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Reggiori F. Are nidoviruses hijacking the autophagy machinery? Autophagy. 2008;4(3):276–279. doi: 10.4161/auto.5241. [DOI] [PubMed] [Google Scholar]
- Delgado M.A., Deretic V. Toll-like receptors in control of immunological autophagy. Cell Death Differ. 2009;16(7):976–983. doi: 10.1038/cdd.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado M., Singh S., De Haro S., Master S., Ponpuak M., Dinkins C., Ornatowski W., Vergne I., Deretic V. Autophagy and pattern recognition receptors in innate immunity. Immunol. Rev. 2009;227(1):189–202. doi: 10.1111/j.1600-065X.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26(10):523–528. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Ding W.-X., Ni H.-M., Gao W., Hou Y.-F., Melan M.A., Chen X., Stolz D.B., Shao Z.-M., Yin X.-M. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. 2007;282(7):4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- Djavaheri-Mergny M., Amelotti M., Mathieu J., Besançon F., Bauvy C., Souquère S., Pierron G., Codogno P. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J. Biol. Chem. 2006;281(41):30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- Djavaheri-Mergny M., Amelotti M., Mathieu J., Besançon F., Bauvy C., Codogno P. Regulation of autophagy by NFkappaB transcription factor and reactives oxygen species. Autophagy. 2007;3(4):390–392. doi: 10.4161/auto.4248. [DOI] [PubMed] [Google Scholar]
- Dreux M., Gastaminza P., Wieland S.F., Chisari F.V. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 2009;106(33):14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A., Bialik S., Simon H., Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16(7):966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- English L., Chemali M., Duron J., Rondeau C., Laplante A., Gingras D., Alexander D., Leib D., Norbury C., Lippé R., Desjardins M. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10(5):480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert L., Denizot M., Grimaldi M., Robert-Hebmann V., Gay B., Varbanov M., Codogno P., Biard-Piechaczyk M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Invest. 2006;116(8):2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert L., Denizot M., Grimaldi M., Robert-Hebmann V., Gay B., Varbanov M., Codogno P., Biard-Piechaczyk M. Autophagy and CD4+ T lymphocyte destruction by HIV-1. Autophagy. 2007;3(1):32–34. doi: 10.4161/auto.3275. [DOI] [PubMed] [Google Scholar]
- Espert L., Varbanov M., Robert-Hebmann V., Sagnier S., Robbins I., Sanchez F., Lafont V., Biard-Piechaczyk M. Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV-1 infection. PLoS ONE. 2009;4(6):e5787. doi: 10.1371/journal.pone.0005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro E., Cecconi F. Autophagic and apoptotic response to stress signals in mammalian cells. Arch. Biochem. Biophys. 2007;462(2):210–219. doi: 10.1016/j.abb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gannagé M., Dormann D., Albrecht R., Dengjel J., Torossi T., Rämer P.C., Lee M., Strowig T., Arrey F., Conenello G., Pypaert M., Andersen J., García-Sastre A., Münz C. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6(4):367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon P.B., Holen I., Fosse M., Røtnes J.S., Seglen P.O. Dependence of hepatocytic autophagy on intracellularly sequestered calcium. J. Biol. Chem. 1993;268(35):26107–26112. [PubMed] [Google Scholar]
- Gozuacik D., Kimchi A. Autophagy and cell death. Curr. Top. Dev. Biol. 2007;78:217–245. doi: 10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119(6):753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- He B., Gross M., Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 1997;94(3):843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyer-Hansen M., Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14(9):1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- Huang S., Chang C., Wang P., Tsai Y., Liu H. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J. Med. Virol. 2009;81(7):1241–1252. doi: 10.1002/jmv.21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Aoki H., Kühnel F., Kondo Y., Kubicka S., Wirth T., Iwado E., Iwamaru A., Fujiwara K., Hess K.R., Lang F.F., Sawaya R., Kondo S. Autophagic cell death of malignant glioma cells induced by a conditionally replicating adenovirus. Cancer Spectr. Knowl. Environ. 2006;98(9):625–636. doi: 10.1093/jnci/djj161. [DOI] [PubMed] [Google Scholar]
- Itzhaki R.F., Cosby S., Wozniak M.A. Herpes simplex virus type 1 and Alzheimer's disease: the autophagy connection. J. Neurovirol. 2008;14(1):1–4. doi: 10.1080/13550280701802543. [DOI] [PubMed] [Google Scholar]
- Jackson W., Giddings T., Taylor M., Mulinyawe S., Rabinovitch M., Kopito R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. Plos Biol. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Gomez-Manzano C., Aoki H., Alonso M.M., Kondo S., McCormick F., Xu J., Kondo Y., Bekele B.N., Colman H., Lang F.F., Fueyo J. Examination of the therapeutic potential of Delta-24-RGD in brain tumor stem cells: role of autophagic cell death. Cancer Spectr. Knowl. Environ. 2007;99(18):1410–1414. doi: 10.1093/jnci/djm102. [DOI] [PubMed] [Google Scholar]
- Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K.Q., Ishii K.J., Kawai T., Akira S., Suzuki K., Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U. S. A. 2007;104(35):14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7(2):131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Khakpoor A., Panyasrivanit M., Wikan N., Smith D.R. A role for autophagolysosomes in dengue virus 3 production in HepG2 cells. J. Gen. Virol. 2009;90(Pt 5):1093–1103. doi: 10.1099/vir.0.007914-0. [DOI] [PubMed] [Google Scholar]
- Kim C.M., Koike K., Saito I., Miyamura T., Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351(6324):317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Taylor M., Jackson W. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2004;2(4):301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., Bamber B.A., Bassham D.C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J.S., Bredesen D.E., Brodsky J.L., Brumell J.H., Brunk U.T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L.-S., Choi A., Chu C.T., Chung J., Clarke P.G.H., Clark R.S.B., Clarke S.G., Clavé C., Cleveland J.L., Codogno P., Colombo M.I., Coto-Montes A., Cregg J.M., Cuervo A.M., Debnath J., Demarchi F., Dennis P.B., Dennis P.A., Deretic V., Devenish R.J., Di Sano F., Dice J.F., Difiglia M., Dinesh-Kumar S., Distelhorst C.W., Djavaheri-Mergny M., Dorsey F.C., Dröge W., Dron M., Dunn W.A., Duszenko M., Eissa N.T., Elazar Z., Esclatine A., Eskelinen E.-L., Fésüs L., Finley K.D., Fuentes J.M., Fueyo J., Fujisaki K., Galliot B., Gao F.-B., Gewirtz D.A., Gibson S.B., Gohla A., Goldberg A.L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R.A., Häussinger D., He Y.-W., Heidenreich K., Hill J.A., Høyer-Hansen M., Hu X., Huang W.-P., Iwasaki A., Jäättelä M., Jackson W.T., Jiang X., Jin S., Johansen T., Jung J.U., Kadowaki M., Kang C., Kelekar A., Kessel D.H., Kiel J.A.K.W., Kim H.P., Kimchi A., Kinsella T.J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A.L., Kroemer G., Kuan C.-Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H.-Y., Lenardo M.J., Levine B., Lieberman A., Lim K.-L., Lin F.-C., Liou W., Liu L.F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K.F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A.J., Meléndez A., Michels P., Miotto G., Mistiaen W.P., Mizushima N., Mograbi B., Monastyrska I., Moore M.N., Moreira P.I., Moriyasu Y., Motyl T., Münz C., Murphy L.O., Naqvi N.I., Neufeld T.P., Nishino I., Nixon R.A., Noda T., Nürnberg B., Ogawa M., Oleinick N.L., Olsen L.J., Ozpolat B., Paglin S., Palmer G.E., Papassideri I., Parkes M., Perlmutter D.H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D.C., Ryan K.M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P.O., Seleverstov O., Settleman J., Shacka J.J., Shapiro I.M., Sibirny A., Silva-Zacarin E.C.M., Simon H.-U., Simone C., Simonsen A., Smith M.A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P.E., Subauste C.S., Sugimoto S., Sulzer D., Suzuki T., Swanson M.S., Tabas I., Takeshita F., Talbot N.J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G.S., Taylor J.P., Terman A., Tettamanti G., Thompson C.B., Thumm M., Tolkovsky A.M., Tooze S.A., Truant R., Tumanovska L.V., Uchiyama Y., Ueno T., Uzcátegui N.L., van der Klei I., Vaquero E.C., Vellai T., Vogel M.W., Wang H.-G., Webster P., Wiley J.W., Xi Z., Xiao G., Yahalom J., Yang J.-M., Yap G., Yin X.-M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R.L. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Kanzawa T., Sawaya R., Kondo S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer. 2005;5(9):726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H., Ogawa S., Kaufman R.J., Kominami E., Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14(2):230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- Kroemer G., Levine B. Autophagic cell death: the story of a misnomer. Nat. Rev. Mol. Cell Biol. 2008;9(12):1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B., Woo J., Liang C., Lee K., Hong H., Xiaofei E., Kim K., Jung J., Oh B. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4(2):e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt-Jones E.A., Popova L., Kwinn L., Haynes L.M., Jones L.P., Tripp R.A., Walsh E.E., Freeman M.W., Golenbock D.T., Anderson L.J., Finberg R.W. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000;1(5):398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- Kurt-Jones E.A., Chan M., Zhou S., Wang J., Reed G., Bronson R., Arnold M.M., Knipe D.M., Finberg R.W. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proc. Natl. Acad. Sci. U. S. A. 2004;101(5):1315–1320. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyei G.B., Dinkins C., Davis A.S., Roberts E., Singh S.B., Dong C., Wu L., Kominami E., Ueno T., Yamamoto A., Federico M., Panganiban A., Vergne I., Deretic V. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009;186(2):255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Lund J.M., Ramanathan B., Mizushima N., Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315(5817):1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Lee D., Sugden B. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene. 2008;27(20):2833–2842. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- Lee Y.R., Lei H.Y., Liu M.T., Wang J.R., Chen S.H., Jiang-Shieh Y.F., Lin Y.S., Yeh T.M., Liu C.C., Liu H. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374(2):240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Li Q., Lee J., Lee S., Jeong J., Lee H., Chang H., Zhou F., Gao S., Liang C., Jung J. FLIP-mediated autophagy regulation in cell death control. Nat. Cell Biol. 2009;11(11):1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib D.A., Alexander D.E., Cox D., Yin J., Ferguson T.A. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J. Virol. 2009;83(23):12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120(2):159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Levine B., Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7(10):767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Sodora D.L. HIV and CXCR4 in a kiss of autophagic death. J. Clin. Invest. 2006;116(8):2078–2080. doi: 10.1172/JCI29447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115(10):2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Kleeman L.K., Jiang H.H., Gordon G., Goldman J.E., Berry G., Herman B., Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998;72(11):8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121(4):567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Longatti A., Tooze S.A. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16(7):956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- Maiuri M., Zalckvar E., Kimchi A., Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8(9):741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Maiuri M., Tasdemir E., Criollo A., Morselli E., Vicencio J., Carnuccio R., Kroemer G. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16(1):87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- Meijer A.J., Codogno P. Regulation and role of autophagy in mammalian cells. Int. J. Biochem. Cell Biol. 2004;36(12):2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Meijer A.J., Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol. Aspects Med. 2006;27(5–6):411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Mijaljica D., Prescott M., Devenish R.J. Endoplasmic reticulum and Golgi complex: contributions to, and turnover by, autophagy. Traffic. 2006;7(12):1590–1595. doi: 10.1111/j.1600-0854.2006.00495.x. [DOI] [PubMed] [Google Scholar]
- Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6(5):363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004;36(12):2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.S., Chang Y. Kaposi's sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu. Rev. Microbiol. 2003;57:609–639. doi: 10.1146/annurev.micro.57.030502.090824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E., Galluzzi L., Kepp O., Vicencio J.M., Criollo A., Maiuri M.C., Kroemer G. Anti- and pro-tumor functions of autophagy. Biochim. Biophys. Acta. 2009;1793(9):1524–1532. doi: 10.1016/j.bbamcr.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Münz C. Autophagy and antigen presentation. Cell. Microbiol. 2006;8(6):891–898. doi: 10.1111/j.1462-5822.2006.00714.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., Hamada S., Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306(5698):1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nakashima A., Tanaka N., Tamai K., Kyuuma M., Ishikawa Y., Sato H., Yoshimori T., Saito S., Sugamura K. Survival of parvovirus B19-infected cells by cellular autophagy. Virology. 2006;349(2):254–263. doi: 10.1016/j.virol.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Noda T., Fujita N., Yoshimori T. The late stages of autophagy: how does the end begin? Cell Death Differ. 2009;16(7):984–990. doi: 10.1038/cdd.2009.54. [DOI] [PubMed] [Google Scholar]
- Ogata M., Hino S.-i., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J.A., Urano F., Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006;26(24):9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307(5710):727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Orvedahl A., Levine B. Autophagy and viral neurovirulence. Cell. Microbiol. 2008;10(9):1747–1756. doi: 10.1111/j.1462-5822.2008.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 2009;16(1):57–69. doi: 10.1038/cdd.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D.A., Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1(1):23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Münz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307(5709):593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- Panyasrivanit M., Khakpoor A., Wikan N., Smith D.R. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J. Gen. Virol. 2009;90(Pt 2):448–456. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- Pattingre S., Bauvy C., Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J. Biol. Chem. 2003;278(19):16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Pattingre S., Espert L., Biard-Piechaczyk M., Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90(2):313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279(11):10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78(18):9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C., Klein K.A., Romanov G.A., Palmer L.E., Cirota C., Zhao Z., Bliska J.B. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect. Immun. 2009;77(6):2251–2261. doi: 10.1128/IAI.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajecki M., Hällström T.A., Hakkarainen T., Nokisalmi P., Hautaniemi S., Nieminen A.I., Tenhunen M., Rantanen V., Desmond R.A., Chen D.T., Guse K., Stenman U.H., Gargini R., Kapanen M., Klefström J., Kanerva A., Pesonen S., Ahtiainen L., Hemminki A. Mre11 inhibition by oncolytic adenovirus associates with autophagy and underlies synergy with ionizing radiation. Int. J. Cancer. 2009;125(10):2441–2449. doi: 10.1002/ijc.24608. [DOI] [PubMed] [Google Scholar]
- Rassa J.C., Meyers J.L., Zhang Y., Kudaravalli R., Ross S.R. Murine retroviruses activate B cells via interaction with toll-like receptor 4. Proc. Natl. Acad. Sci. U. S. A. 2002;99(4):2281–2286. doi: 10.1073/pnas.042355399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D. Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 2005;17(4):415–422. doi: 10.1016/j.ceb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Rust R.C., Landmann L., Gosert R., Tang B.L., Hong W., Hauri H.P., Egger D., Bienz K. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 2001;75(20):9808–9818. doi: 10.1128/JVI.75.20.9808-9818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti F., Granata R., Meijer A.J., Codogno P. Does autophagy have a license to kill mammalian cells? Cell Death Differ. 2009;16(1):12–20. doi: 10.1038/cdd.2008.101. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R., Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17(9):422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A., Giddings T.H., Ladinsky M.S., Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 1996;70(10):6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D., Münz C. Immune surveillance of intracellular pathogens via autophagy. Cell Death Differ. 2005;12(Suppl 2):1519–1527. doi: 10.1038/sj.cdd.4401727. [DOI] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26(1):79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly S., Lukinova N., Bambina S., Berman A., Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30(4):588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004;6(12):1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Shinojima N., Yokoyama T., Kondo Y., Kondo S. Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy. Autophagy. 2007;3(6):635–637. doi: 10.4161/auto.4916. [DOI] [PubMed] [Google Scholar]
- Sinha S., Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S., Colbert C.L., Becker N., Wei Y., Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4(8):989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D., Chen W., Choi J., Wakita T., Yen T.S., Ou J. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48(4):1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Xia S.L., Liao C., Li Y.L., Wang Y.F., Li T.P., Zhao M.J. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. WJG. 2004;10(4):509–513. doi: 10.3748/wjg.v10.i4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector S.A., Zhou D. Autophagy: an overlooked mechanism of HIV-1 pathogenesis and neuroAIDS? Autophagy. 2008;4(5):704–706. doi: 10.4161/auto.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361(2):304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy D.A., Giddings T.H., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 2000;74(19):8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M.N., Jackson W., Laird D.T., Culp T.D., Grose C., Haynes J.I., Benetti L. Varicella-zoster virus infection induces autophagy in both cultured cells and human skin vesicles. Journal of Virology. 2009;83(11):5466–5476. doi: 10.1128/JVI.02670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Signaling pathways activated by microorganisms. Curr. Opin. Cell Biol. 2007;19(2):185–191. doi: 10.1016/j.ceb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Tal M.C., Sasai M., Lee H.K., Yordy B., Shadel G.S., Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U. S. A. 2009;106(8):2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallóczy Z., Jiang W., Virgin H., Leib D.A., Scheuner D., Kaufman R.J., Eskelinen E.L., Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 2002;99(1):190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallóczy Z., Virgin H., Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2(1):24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- Tang H., Da L., Mao Y., Li Y., Li D., Xu Z., Li F., Wang Y., Tiollais P., Li T., Zhao M. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49(1):60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- Tanida I., Fukasawa M., Ueno T., Kominami E., Wakita T., Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5(7) doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- Tasdemir E., Maiuri M., Galluzzi L., Vitale I., Djavaheri-Mergny M., D'amelio M., Criollo A., Morselli E., Zhu C., Harper F., Nannmark U., Samara C., Pinton P., Vicencio J.M., Carnuccio R., Moll U., Madeo F., Paterlini-Brechot P., Rizzuto R., Szabadkai G., Pierron G., Blomgren K., Tavernarakis N., Codogno P., Cecconi F., Kroemer G. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008;10(6):676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M., Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 2007;81(22):12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M., Burgon T.B., Kirkegaard K., Jackson W. Role of microtubules in extracellular release of poliovirus. J. Virol. 2009;83(13):6599–6609. doi: 10.1128/JVI.01819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl M., Kepp O., Jusforgues-Saklani H., Vicencio J., Kroemer G., Albert M.L. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 2009;16(7):991–1005. doi: 10.1038/cdd.2009.8. [DOI] [PubMed] [Google Scholar]
- Ulasov I., Sonabend A., Nandi S., Khramtsov A., Han Y., Lesniak M. Combination of adenoviral virotherapy and temozolomide chemotherapy eradicates malignant glioma through autophagic and apoptotic cell death in vivo. Br. J. Cancer. 2009;100(7):1154–1164. doi: 10.1038/sj.bjc.6604969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin H., Levine B. Autophagy genes in immunity. Nat. Immunol. 2009;10(5):461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Whiteman M.W., Lian H., Wang G., Singh A., Huang D., Denmark T. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J. Biol. Chem. 2009;284(32):21412–21424. doi: 10.1074/jbc.M109.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30(6):678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Sinha S., Levine B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy. 2008;4(7):949–951. doi: 10.4161/auto.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006;312(5775):875–878. doi: 10.1126/science.1126766. [DOI] [PubMed] [Google Scholar]
- Wileman T. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 2007;61:149–167. doi: 10.1146/annurev.micro.57.030502.090836. [DOI] [PubMed] [Google Scholar]
- Wong J., Zhang J., Si X., Gao G., Mao I., McManus B.M., Luo H. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 2008;82(18):9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xie Z., Klionsky D. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Xu Y., Jagannath C., Liu X.D., Sharafkhaneh A., Kolodziejska K.E., Eissa N.T. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27(1):135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Liang Z., Gu Z., Qin Z. Molecular mechanism and regulation of autophagy. Acta Pharmacol. Sin. 2005;26(12):1421–1434. doi: 10.1111/j.1745-7254.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- Yokoyama T., Iwado E., Kondo Y., Aoki H., Hayashi Y., Georgescu M., Sawaya R., Hess K., Mills G., Kawamura H., Hashimoto Y., Urata Y., Fujiwara T., Kondo S. Autophagy-inducing agents augment the antitumor effect of telerase-selve oncolytic adenovirus OBP-405 on glioblastoma cells. Gene Ther. 2008;15(17):1233–1239. doi: 10.1038/gt.2008.98. [DOI] [PubMed] [Google Scholar]
- Yoon S.Y., Ha Y.E., Choi J.E., Ahn J., Lee H., Kweon H.S., Lee J.Y., Kim D.H. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J. Virol. 2008;82(23):11976–11978. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Nair U., Yang Z., Klionsky D.J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006;281(40):30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T., Noda T. Toward unraveling membrane biogenesis in mammalian autophagy. Curr. Opin. Cell Biol. 2008;20(4):401–407. doi: 10.1016/j.ceb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A.J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Monken C.E., Zhang Y., Lenard J., Mizushima N., Lattime E.C., Jin S. Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy. 2006;2(2):91–95. doi: 10.4161/auto.2.2.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E., Mizushima N.N., Denison M.R., Virgin H. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3(6):581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- Zhou D., Spector S.A. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22(6):695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Jiang X., Liu D., Fan Z., Hu X., Yan J., Wang M., Gao G.F. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5(3):321–328. doi: 10.4161/auto.5.3.7406. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Vergote D., Pardo C., Noorbakhsh F., McArthur J.C., Hollenberg M.D., Overall C.M., Power C. CXCR3 activation by lentivirus infection suppresses neuronal autophagy: neuroprotective effects of antiretroviral therapy. FASEB Journal. 2009;23(9):2928–2941. doi: 10.1096/fj.08-128819. [DOI] [PMC free article] [PubMed] [Google Scholar]