Abstract
Whether the 2009 pandemic H1N1 influenza vaccine can induce heterosubtypic cross-protective anti-hemagglutinin (HA) neutralizing antibodies is an important issue. We obtained a panel of fully human monoclonal antibodies from the memory B cells of a 2009 pandemic H1N1 influenza vaccine recipient. Most of the monoclonal antibodies targeted the HA protein but not the HA1 fragment. Among the analyzed antibodies, seven mAbs exhibited neutralizing activity against several influenza A viruses of different subtypes. The conserved linear epitope targeted by the neutralizing mAbs (FIEGGWTGMVDGWYGYHH) is part of the fusion peptide on HA2. Our work suggests that a heterosubtypic neutralizing antibody response primarily targeting the HA stem region exists in recipients of the 2009 pandemic H1N1 influenza vaccine. The HA stem region contains various conserved neutralizing epitopes with the fusion peptide as an important one. This work may aid in the design of a universal influenza A virus vaccine.
Abbreviations: mAb, monoclonal antibody; TCID50, 50% tissue culture infective dose
Keywords: Influenza, H1N1, Fully human monoclonal antibody, Neutralization, Epitope, HA2, Single-cell RT-PCR, Fusion peptide
Introduction
Because of their highly flexible genomes, influenza A viruses cause annual epidemics and sometimes pandemics around the world. For nearly 100 years, influenza A viruses have been a global threat to humans (Palese, 2004). Based on the antigenicity of the hemagglutinin (HA) protein, influenza A viruses are classified into two groups and at least 16 different subtypes (H1-H16). The HA protein is the functional protein that mediates the entry of influenza viruses into susceptible host cells and thus contains various epitopes that are recognized by neutralizing antibodies (Skehel and Wiley, 2000). However, heterosubtypic neutralizing or protective antibody responses are rarely observed in the general population, largely because of the high mutation rate of the HA protein, especially in the globular head (HA1) region, which is the primary target of the humoral immune response. Consequently, when a new reassortant influenza virus emerges that the human immune system has not previously encountered, a pandemic may occur. The 2009 swine-origin H1N1 influenza is an example of such a pandemic.
The 2009 pandemic H1N1 influenza virus contains gene segments that are in both the American and the Eurasia swine genetic linkages (Garten et al., 2009). Nucleotide sequence alignment has shown that the HA sequence of the 2009 pandemic H1N1 influenza virus is divergent from the sequences of the seasonal H1 influenza viruses that have previously been circulating in humans. The antigenicity of the HA in this strain is also highly distinct from that of the previously circulating H1 influenza viruses (Garten et al., 2009, Hancock et al., 2009). People, especially young people, generally lacked protection against this new virus (Hancock et al., 2009), and the 2009 pandemic H1N1 influenza vaccines have been proven effective in inducing neutralizing antibody responses against the pandemic influenza virus (Liang et al., 2010, Zhu et al., 2009).
It is important to determine whether cross-reactive neutralizing antibodies against both seasonal and pandemic influenza viruses are present in individuals who were infected with or vaccinated against 2009 pandemic H1N1 influenza. Recently, Wrammert et al. discovered that plasmablasts from 2009 pandemic H1N1 influenza patients produced cross-subtype neutralizing antibodies that targeted both the HA stalk and the head domain (Wrammert et al., 2011). We examined whether such antibodies existed in individuals vaccinated against pandemic influenza.
In this study, we used the full-length HA protein from the 2009 pandemic H1N1 influenza virus to raise fully human neutralizing mAbs. We obtained 19 monoclonal antibodies from the memory B cells of a 2009 pandemic H1N1 influenza vaccine recipient and confirmed that all 19 of the monoclonal antibodies recognized the lysates of both the pandemic virus and the recently circulating seasonal H1N1 influenza virus. Seven of the human monoclonal antibodies were further found to have apparent neutralizing effects against different subtypes of influenza A viruses, including viruses belonging to both group 1 and group 2 and the pandemic influenza virus. Interestingly, we found that most of the monoclonal antibodies, including the seven neutralizing mAbs, bound to the HA stem region (HA2), which is relatively conserved among different influenza A virus strains. These findings indicate that a broad cross-subtype neutralizing antibody response targeting the HA stem region exists in individuals vaccinated against 2009 pandemic H1N1 influenza and that these broadly reactive memory B cells may be important for protecting humans from infection with different influenza A viruses. A functional analysis revealed that the HA2 region contained several (at least four) conserved neutralizing epitopes that could be recognized by the raised mAbs. Further experiments showed that one of them was a linear epitope (FIEGGWTGMVDGWYGYHH), which was in the region of the fusion peptide on HA2. These results may be helpful in the design of universal influenza vaccines.
Results
Generation of fully human mAbs and their gene usage study
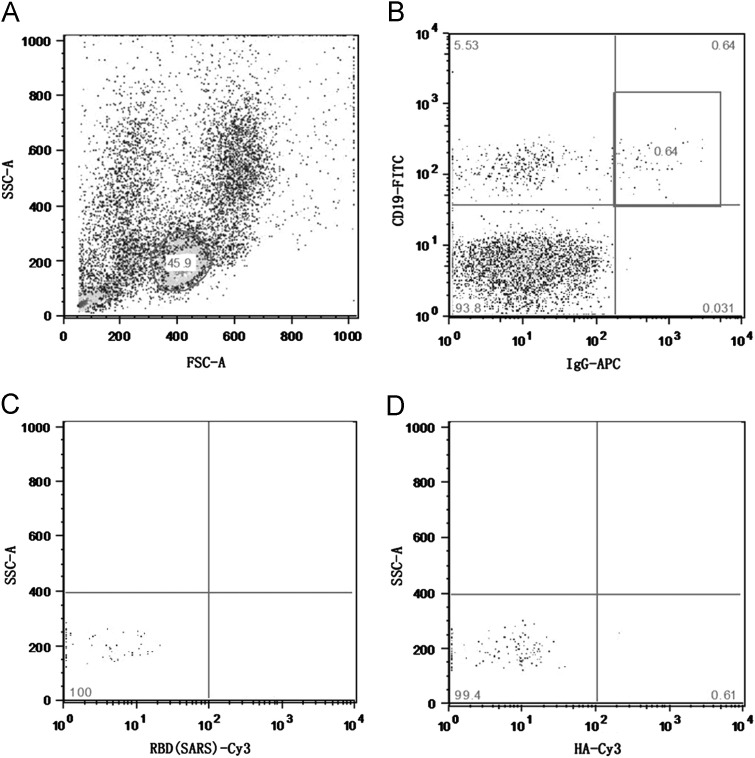
A 27-year-old healthy female adult volunteer who had been vaccinated with a 2009 pandemic H1N1 influenza split-virion vaccine for one month was enrolled in this study. We used flow cytometry to separate pandemic H1N1 HA-specific memory B cells with three surface markers: CD19, IgG, and HA-specific BCR. A baculovirus-expressed HA protein was used for cell sorting. As shown in Fig. 1, the memory B cells accounted for approximately 0.6% of the total peripheral blood small lymphocytes, and less than 1% of the selected memory B cells were HA-specific.
Fig. 1.
The isolation of pandemic H1N1 HA-specific memory B cells. (A) Small lymphocytes were sorted from the human peripheral blood of a vaccinated individual. (B) Memory B cells were sorted using CD19 and IgG as markers. (C) Negative control: cells stained with an unrelated protein (RBD, receptor-binding domain of SARS-CoV). (D) Cells were sorted using the pandemic H1N1 HA protein. HA-specific memory B cells accounted for approximately 0.61% of the total memory B cells.
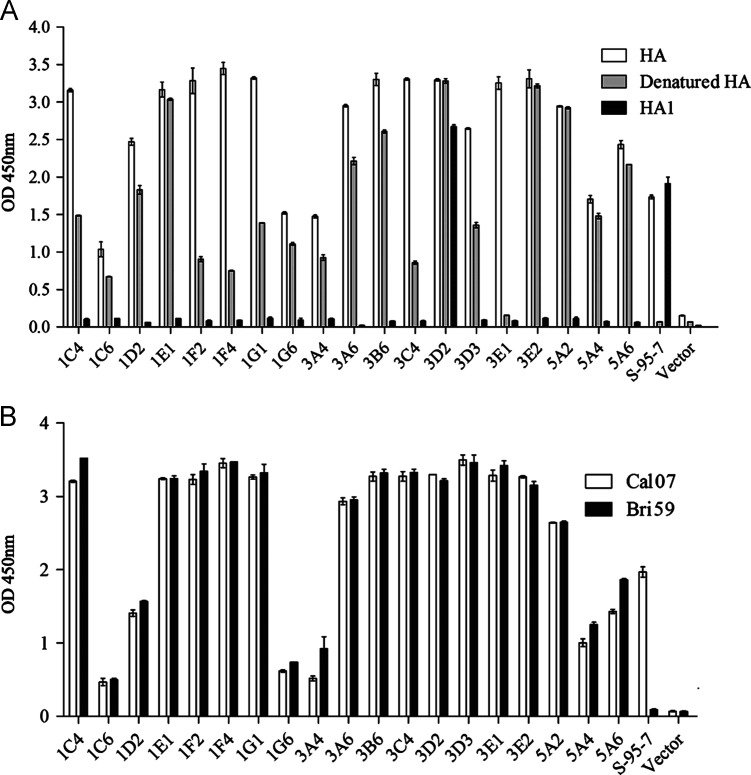
The antibody variable genes of these memory B cells were identified with single-cell RT-PCR and nested PCR (Smith et al., 2009, Wrammert et al., 2008). Nineteen human monoclonal antibodies (constructed using the IgG1 and κ framework) were obtained that bound to the HA protein ( Fig. 2A). These mAbs used different V, D, and J gene segments in their heavy and light chain variable genes; the V, D, and J usage is presented in detail in Table 1. The presence of somatic mutations in the 19 mAbs revealed that most of the B cells producing these mAbs were from the germinal center. Notably, five of the mAbs used the VH1-69 gene; the use of this gene is considered to be a non-exclusive characteristic of heterosubtypic binding to the HA stem region, with a neutralizing effect (Ekiert et al., 2009, Sui et al., 2009). These five mAbs used different DH, VL, and JL genes, indicating that they are from different B cell clones.
Fig. 2.
An ELISA to determine the binding activities of the naturally isolated human monoclonal antibodies. (A) HA, heat-denatured (100 °C, 5 min) HA or the HA1 domain of the 2009 pandemic H1N1 influenza virus was used to coat the wells of the plates for binding activity tests of the 19 human monoclonal antibodies. (B) Influenza virus lysates from both the pandemic and the seasonal H1N1 viruses were used for binding activity tests of the 19 human monoclonal antibodies. Cal07: pandemic H1N1 virus lysate with HA from A/California/7/2009 (H1N1), Bri59: seasonal H1 influenza virus lysate with HA from A/Brisbane/59/2007 (H1N1). S-95-7: a mouse monoclonal antibody recognizing a specific conformational epitope in the HA1 domain of the 2009 pandemic H1N1 influenza virus. Vector: human antibody IgG and Igκ constant regions encoded by the antibody expression plasmid. OD 450 nm: optical density measured at 450 nm.
Table 1.
V gene usage of the cross-strain-reactive human monoclonal antibodies. The germline usage of the heavy and light chain variable genes was defined using the IMGT database.
| mAb | VH | DH | JH | aa mutations | VL | JL | aa mutations |
|---|---|---|---|---|---|---|---|
| 1C4a | 3-30⁎04 | 5-24⁎01 | 6⁎02 | 5/121 | 3-20⁎01 | 2⁎01 | 2/108 |
| 1C6 | 3-11⁎01 | 6-25⁎01 | 4⁎01 | 9/121 | 3-15⁎01 | 2⁎01 | 8/109 |
| 1D2 | 1-69⁎09 | 4-17⁎01 | 4⁎02 | 7/121 | 3-15⁎01 | 2⁎01 | 4/108 |
| 1E1a | 3-23⁎04 | 1-1⁎01 | 4⁎02 | 7/121 | 3-11⁎01 | 5⁎01 | 0/108 |
| 1F2a | 3-23⁎04 | 5-5⁎01 | 5⁎02 | 6/121 | 3-15⁎01 | 5⁎01 | 2/108 |
| 1F4a | 3-23⁎04 | 2-8⁎02 | 3⁎02 | 6/121 | 3-15⁎01 | 4⁎01 | 0/108 |
| 1G1a | 3-23⁎04 | 3-16⁎01 | 4⁎02 | 9/121 | 3-15⁎01 | 4⁎01 | 1/108 |
| 1G6 | 3-23⁎04 | 6-19⁎01 | 5⁎02 | 7/123 | 2D-40⁎01 | 4⁎01 | 3/108 |
| 3A4 | 3-33⁎01 | 5-24⁎01 | 3⁎02 | 6/127 | 3-15⁎01 | 3⁎01 | 3/108 |
| 3A6 | 1-69⁎09 | 3-22⁎01 | 4⁎02 | 5/125 | 3-11⁎01 | 1⁎01 | 2/108 |
| 3B6 | 1-69⁎09 | 5-24⁎01 | 4⁎02 | 5/121 | 3-11⁎01 | 1⁎01 | 6/108 |
| 3C4a | 1-69⁎09 | 1-7⁎01 | 4⁎02 | 4/121 | 3-20⁎01 | 2⁎01 | 7/108 |
| 3D2 | 4-34⁎01 | 3-9⁎01 | 4⁎02 | 0/127 | 3-15⁎01 | 1⁎01 | 0/108 |
| 3D3 | 1-69⁎09 | 3-9⁎01 | 4⁎02 | 4/121 | 3-11⁎01 | 2⁎01 | 1/108 |
| 3E1a | 4-4⁎07 | 3-16⁎02 | 4⁎02 | 4/121 | 1-5⁎03 | 1⁎01 | 0/108 |
| 3E2 | 1-3⁎01 | 3-9⁎01 | 4⁎02 | 1/127 | 3-15⁎01 | 4⁎01 | 3/108 |
| 5A2 | 4-59⁎01 | 1-1⁎01 | 4⁎02 | 3/123 | 4-1⁎01 | 2⁎01 | 4/110 |
| 5A4 | 3-23⁎04 | 3-3⁎01 | 6⁎02 | 5/128 | 1-9⁎01 | 3⁎01 | 1/110 |
| 5A6 | 3-30⁎14 | 2-8⁎02 | 4⁎02 | 6/121 | 4-1⁎01 | 2⁎01 | 3/108 |
mAbs with good neutralizing capacity against the 2009 pandemic H1N1 influenza virus.
Biological features of the mAbs
Further experiments showed that after the HA protein was denatured with heat, four mAbs (1E1, 3D2, 3E2, and 5A2) fully maintained their binding abilities to the antigen, one mAb (3E1) completely lost its binding ability to the antigen, and the rest of the mAbs partially retained their binding abilities to the HA protein (Fig. 2A). All of the mAbs bound to the 2008–09 seasonal H1 influenza (A/Brisbane/59/2007) virus lysate and the 2009 pandemic H1N1 influenza virus lysate, but their binding avidities were different (Fig. 2B). An insect cell-expressed HA1 domain protein from the 2009 pandemic H1N1 influenza virus was also used to analyze the binding activity between HA1 and the mAbs. As shown in Fig. 2A, only one mAb, 3D2, exhibited binding activity to the HA1 domain, whereas all of the other mAbs did not bind to HA1. These results suggest that the other mAbs may bind to the HA2 domain. These results indicate that memory B cells from people vaccinated with the 2009 pandemic H1N1 influenza vaccine can secrete antibodies that primarily target the HA2 region, and these antibodies are cross-reactive with the 2008–09 seasonal H1 influenza virus.
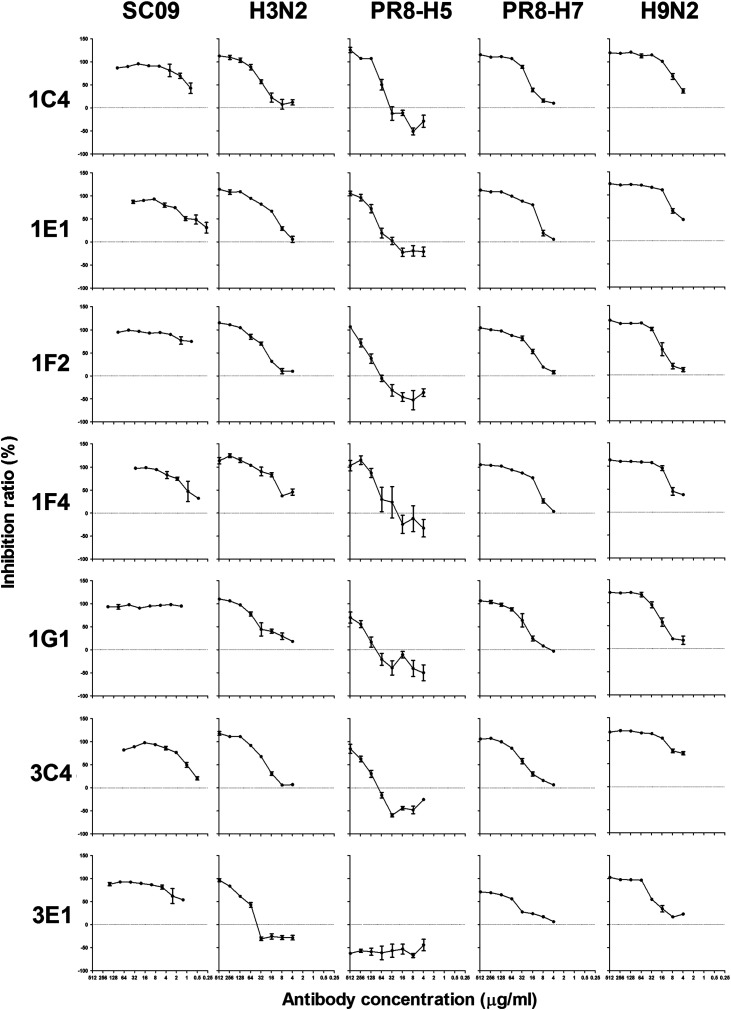
To investigate the neutralizing effect of all of the fully human mAbs, a microneutralization assay was adopted to screen the mAb neutralizing activities. The experiment demonstrated that seven mAbs (1C4, 1E1, 1F2, 1F4, 1G1, 3C4, and 3E1) showed relatively higher neutralizing activities against the 2009 pandemic H1N1 influenza virus. The IC50 of the seven neutralizing mAbs against the pandemic influenza virus SC09 were about 1 μg/ml or less ( Table 2), while the IC50 of the other mAbs against SC09 were more than 200 μg/ml (data not shown). Interestingly, all seven of the mAbs also showed neutralizing activities against several other influenza virus strains, including subtypes of H3 (H3N2), H5 (PR8-H5), H7 (PR8-H7), and H9 (H9N2) ( Fig. 3 and Table 2). Sequence analysis revealed that three mAbs (1F2, 1F4, and 1G1) used the VH3-23 and VL3-15 genes and one mAb, 3C4, used the VH1-69 gene (Table 1).
Table 2.
IC50 of the seven neutralizing mAbs.
| mAb | IC50 (μg/ml)a |
||||
|---|---|---|---|---|---|
| SC09 | H3N2 | PR8-H5 | PR8-H7 | H9N2 | |
| 1C4 | 0.795 | 33.9 | 57.8 | 21.2 | 7.80 |
| 1E1 | 1.26 | 7.51 | 84.2 | 11.7 | 10.1 |
| 1F2 | 0.0935 | 27.9 | 132 | 15.0 | 17.1 |
| 1F4 | 1.20 | 17.5 | 62.7 | 10.2 | 11.9 |
| 1G1 | <0.1 | 49.9 | 130 | 24.7 | 19.1 |
| 3C4 | 0.972 | 27.5 | 111 | 29.8 | 2.94 |
| 3E1 | 0.383 | 56.4 | >1000 | 41.5 | 32.8 |
IC50 was calculated by the GraphPad Prism software.
Fig. 3.
Microneutralization assay of the human monoclonal antibodies. The neutralizing abilities of the seven selected human mAbs against several influenza virus strains of different subtypes, including SC09, H3N2, PR8-H5, PR8-H7, and H9N2, as listed in the “Viruses” section.
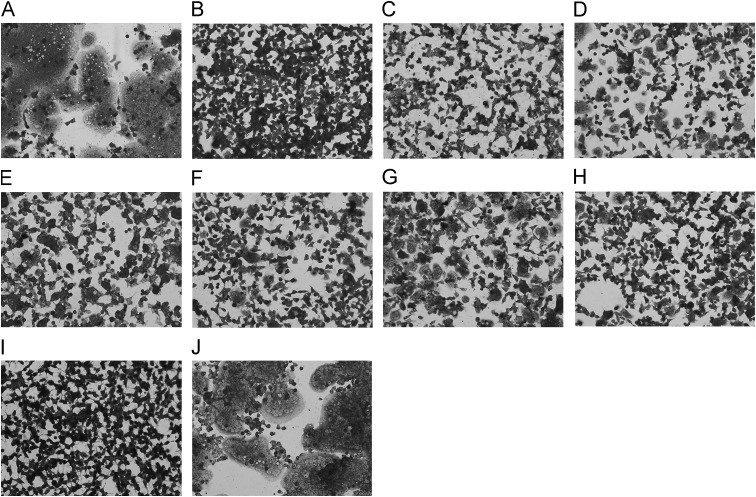
Next, we analyzed the neutralizing mechanism of the broad neutralizing mAbs. A cell–cell fusion experiment was adopted. The results showed that all the seven broad neutralizing mAbs exhibited apparent inhibition activities against syncytia formation of HEK293T cells transfected with the HA expressing plasmids ( Fig. 4), which probably indicated that these mAbs took effect in the virus-cell membrane fusion phase and hindered the function of HA2. The broad neutralizing effect probably resulted from the relatively conservation of HA2.
Fig. 4.
Inhibition of cell–cell fusion by the broadly neutralizing mAbs. HEK293T cells were transfected with plasmids expressing the pandemic H1N1 HA0, then treated with trypsin and pH 5.0. (A) Treatment with no antibody added. (B–H) Treatment in the presence of 100 μg/ml of broadly neutralizing mAbs: 1C4, 3C4, 1E1, 3E1, 1F2, 1F4, and 1G1 represented by B to H in sequence. (I) Cells transfected with plasmids expressing enhanced green fluorescent protein following with the same treatment. (J) Treatment in the presence of 100 μg/ml of a control mAb.
Epitope mapping of the broad neutralizing mAbs
To map the epitopes recognized by the tested mAbs, a competitive ELISA was used. As Fig. 5 shows, 1F2, 1F4, and 1G1, which used the same VH and VL genes, bound to HA at the same site. It is consistent with the sequence analysis results (Table 1). 1C4 and 3C4 were derived from different variable genes but recognized the same epitope. As shown in Fig. 2A, the neutralizing epitope recognized by 1E1 was linear (epit-3), whereas the epitope recognized by 3E1 was completely conformational (epit-4). The other two neutralizing epitopes (recognized by 1F2/1F4/1G1 and 1C4/3C4, respectively) were partially conformational epitopes (epit-1 and epit-2). These results revealed that the HA2 region contained several (at least four) conserved neutralizing epitopes that co-exist in different strains of the influenza virus.
Fig. 5.

A competitive ELISA to detect the binding epitopes of the seven neutralizing mAbs. The percentage of competition between two mAbs in the ELISA binding assay is shown. The percentage of competition was calculated as the reduction in the absorbance when mAb binding competition occurred. Shadowed grids: complete competition (percentage of competition >90%). epit-1 to epit-4: four neutralizing epitopes. The vertical row: non-conjugated mAbs. The horizontal row: biotin-conjugated mAbs.
To further confirm whether the selected neutralizing mAbs are capable of binding to HA2, a series of synthetic peptides (from G345 to P504 of HA) that cover the entire HA2 region with ten-amino-acid overlaps were used to detect the binding sites of the seven mAbs ( Table 3). As shown in Fig. 6, 1F2, 1F4, and 1E1 could specifically recognize peptide no. 2 (FIEGGWTGMVDGWYGYHH) in the ELISA assay, which is in the location of the fusion peptide that is believed to be very conserved among different strains of influenza A viruses (Skehel and Wiley, 2000). Sequence alignment shows that some amino acid residuals in the linear epitope are very conserved among the tested viruses of different subtypes ( Fig. 7). The conserved amino acid residuals may play critical roles in recognition of the epitope by the cross-subtype neutralizing mAbs. Based on this observation, it is concluded that 1F2, 1F4, and 1E1 recognize a linear epitope in the fusion peptide region. The other mAbs (1G1, 1C4, 3C4, and 3E1) showed no significant reaction with the peptides, suggesting that they target conformational epitopes. We will analyze the complex of the HA protein together with the mAbs and use structural biology methods to reveal the characteristics of the epitopes recognized by the mAbs (1G1, 1C4, 3C4, and 3E1).
Table 3.
Overlapping peptides covering the HA2 (G345 to P504).
| 1 | GLFGAIAGFIEGGWTGMVa |
| 2 | FIEGGWTGMVDGWYGYHH |
| 3 | MVDGWYGYHHQNEQGSGY |
| 4 | HHQNEQGSGYAADLKSTQ |
| 5 | GYAADLKSTQNAIDEITN |
| 6 | TQNAIDEITNKVNSVIEK |
| 7 | TNKVNSVIEKMNTQFTAV |
| 8 | EKMNTQFTAVGKEFNHLE |
| 9 | AVGKEFNHLEKRIENLNK |
| 10 | LEKRIENLNKKVDDGFLD |
| 11 | NKKVDDGFLDIWTYNAEL |
| 12 | LDIWTYNAELLVLLENER |
| 13 | ELLVLLENERTLDYHDSN |
| 14 | ERTLDYHDSNVKNLYEKV |
| 15 | SNVKNLYEKVRSQLKNNA |
| 16 | KVRSQLKNNAKEIGNGCF |
| 17 | NAKEIGNGCFEFYHKCDN |
| 18 | CFEFYHKCDNTCMESVKN |
| 19 | DNTCMESVKNGTYDYP |
The peptides are overlapped with 10 amino acid residuals with a total length of 18 amino acids, except peptide no. 19 with a length of 16 amino acids.
Fig. 6.
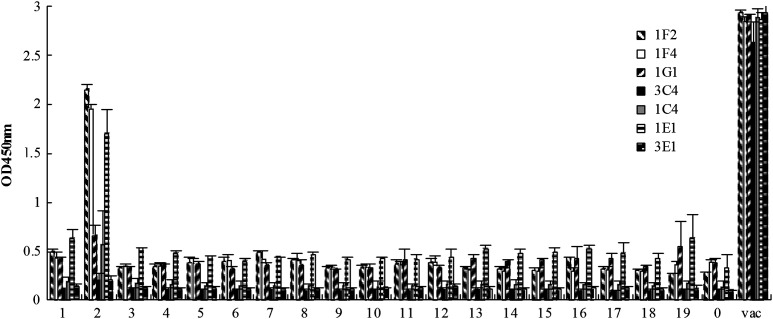
A peptide ELISA to detect the binding epitopes of the seven neutralizing mAbs. A total of 19 overlapping peptides covering the HA2, as listed in Table 3, were tested. No. 1 to 19: the overlapping peptides. 0: biotin without peptide conjugation. vac: the 2009 pandemic H1N1 influenza vaccine. OD 450 nm: optical density measured at 450 nm.
Fig. 7.
Sequence alignment of the linear peptide epitope (FIEGGWTGMVDGWYGYHH). SC09, H3N2, PR8-H5, PR8-H7, and H9N2 referred to the influenza viruses tested in the microneutralization assay as listed in the “Viruses” section. Underlined letters represent the conserved amino acids among the viruses.
Discussion
In this study, we demonstrated that memory B cells that secrete broadly neutralizing heterosubtypic antibodies that target HA2 were present in a recipient of the 2009 pandemic H1N1 influenza vaccine. Corti et al. reported that cross-subtype neutralizing mAbs from immortalized B cells were raised in individuals vaccinated against seasonal influenza, and they confirmed that almost all of the heterosubtypic mAbs recognized epitopes within the HA stem region (Corti et al., 2010). Our results are in line with the observation of Corti et al. (2010) and Wrammert et al. (2011). They show that broadly reactive cross-subtype neutralizing antibody responses are induced in influenza virus infected patients and in individuals who accept vaccination against the pandemic or seasonal influenza viruses.
The influenza HA stem region is crucial for recognition by broadly neutralizing cross-subtype antibodies, and this region can be a promising target for cross-subtype influenza vaccines. It contains a linear B-cell neutralizing epitope and several conformational ones, as shown by this study. Importantly, the fusion peptide is a critical target for the binding of broad neutralizing mAbs. We believe that studies on these broad neutralizing monoclonal antibodies would be beneficial for the development of a broadly effective heterosubtypic influenza vaccine.
We generated seven neutralizing monoclonal antibodies in this study. It is interesting to note that partial V gene differences can influence the neutralizing activity without changing the binding activity; for example, the 3A6, 3B6, and 3C4 mAbs show similar antigen-binding but different neutralizing activities. The reason may be that the neutralizing mAbs target the post-adhesion phase and interfering with HA2 function during the virus-cell membrane fusion process. The binding activity alone may be not sufficient to prevent viral entry into susceptible cells. Additionally, 1C4 and 3C4 used completely different VH genes, but they recognized the same site in the HA2 region.
Our work also indicates that the broadly reactive human neutralizing monoclonal antibodies obtained in this study may be potentially used for the treatment of severe cases of influenza A virus infection (Hung et al., 2011). Several neutralizing monoclonal antibodies targeting different epitopes can be used in a cocktail that covers as many influenza subtypes as possible.
So far, many neutralizing mAbs with the target of the HA stem region have been generated and carefully studied. These antibodies are incompletely enumerated in Table 4. The information obtained is useful for designing broadly protective influenza vaccines. Nevertheless, the antigenic character of the HA stem region is complicated. Including the fusion peptide, there are a number of linear or conformational neutralizing epitopes within this region. Therefore, further research is needed to obtain a deeper and more comprehensive understanding of the interaction between the neutralizing antibodies and the HA stem region. Minimizing the interference of non-neutralizing antibodies will aid in the successful development of a heterosubtypic broadly protective influenza vaccine.
Table 4.
Some neutralizing mAbs targeting the HA stem region.
| Name | Generation methods | Epitope | Neutralizing spectrum | References |
|---|---|---|---|---|
| 1C9 | Hybridoma | Part of the fusion peptide (GLFGAIAGF) | H5N1, cross-clade | Prabhu et al., 2009 |
| PN-SIA28 | EBV transformation | Ile361, Asp362 et al.a | Cross-subtype (both group1 and 2) | Burioni et al., 2010; Clementi et al., 2011 |
| PN-SIA49 | EBV transformation | His25 et al., on HA1 and Met360 et al., on HA2a | Cross-subtype (H1, H2 and H5) | Burioni et al., 2010; De Marco et al., 2012 |
| C179 | Hybridoma | 318 to 322 of HA1 and 47 to 58 of HA2b | Cross-subtype (H1, H2 and H5) | Okuno et al., 1993; Sakabe et al., 2010 |
| CR6261 | Phage display | Helical region in the membrane-proximal stem of HA1 and HA2 | Cross-subtype (H1, H2, H5, H6, H8, and H9) | Ekiert et al., 2009 |
| CR8020 | Cell immortalization | Close to the virus membrane | Most group 2 viruses, including H3N2 and H7N7 | Ekiert et al., 2011 |
| FI6 | Single-cell RT-PCR | F subdomain of the HA trimer | Cross-subtype (both group1 and 2) | Corti et al., 2011 |
Sequence numbering refers to A/Puerto Rico/8/34.
Sequence numbering refers to A/Aichi/2/68.
Single-cell RT-PCR is a rapid method for the production of fully human neutralizing monoclonal antibodies. The CD19, IgG, and HA-specific BCR markers used in this work are suitable for selecting antigen-specific memory B cells, and this screening strategy significantly increased the efficiency of obtaining high-affinity antigen-specific human monoclonal antibodies. The platform established in this study can be applied to the development of fully human mAbs that target other infectious agents, such as HIV, HCV, and HBV.
Materials and methods
Volunteer
This study was approved by the Biomedical Research Ethics Committee of the Shanghai Institutes for Biological Sciences, CAS (ER-SIBS-211101). A 27-year-old healthy female adult who had received the pandemic influenza vaccine was recruited for this study.
Viruses
The influenza virus strains used in this study were as follows: A/Sichuan/1/2009 (H1N1) (referred to as SC09), A/Jiangxi-Donghu/312/2006 (H3N2) (referred to as H3N2), and A/Guangzhou/333/99 (H9N2) (referred to as H9N2) were kindly provided by Prof. Yuelong Shu from the Chinese Center for Disease Control and Prevention, Beijing. PR8-H7 was rescued by reverse genetics from HA and NA genes of low pathogenic A/turkey/Italy/1265/1999 (H7N1) (kindly provided by Prof. H.D. Klenk from Marburg University, Germany) combining with 6 other viral genes of A/Puerto Rico/8/34 in pHW2000 plasmids (kindly provided by Prof. Robert G. Webster from the St. Jude Children's Research Hospital, University of Tennessee). The A/Anhui/1/2005 (H5N1)–PR8-IBCDC-RG5 virus (referred to as PR8-H5) was kindly provided by Prof. Yuelong Shu from the Chinese Center for Disease Control and Prevention, Beijing, for which the four basic amino acid residues at the HA cleavage site were removed and has been confirmed to be non-pathogenic in mammalian (ferrets) and avian (chickens) hosts (Dong et al., 2009).
The H1 subtype virus lysates used in this study were from the 2009 pandemic H1N1 split-virus vaccine (HA from strain A/California/7/2009 (H1N1)) and the 2008–09 trivalent seasonal influenza vaccine (HA from strain A/Brisbane/59/2007 (H1N1)). Both of the vaccines were produced by Hualan Biological Bacterin Company, China.
Protein expression
The HA gene was synthesized by Generay China with the sequence from the influenza strain A/California/4/2009 (H1N1). The full-length HA and the HA1 sub-domain (18–344 aa) were expressed using the bac-to-bac baculovirus expression system (Invitrogen) according to the manufacturer's protocol. HA was purified according to a previously described method (Wang et al., 2006). HA1 (C-terminal fusion with a human IgG Fc tag) was purified using a Protein G Sepharose 4 Fast Flow column (GE) according to the manufacturer's instructions.
Cell sorting
Venous blood was collected from the volunteer in a tube containing 0.4% sodium citrate as an anticoagulant. Lymphocytes and monocytes were prepared using the density separation medium Lympholyte®-H (Cedarlane) according to the manufacturer's instructions. Single-cell sorting was performed using an ARIA II (BD) cell sorter. A FITC-labeled mouse anti-human CD19 antibody and an APC-labeled mouse anti-human IgG antibody were purchased from BD. A streptavidin-Cy3 conjugate was purchased from Sigma-Aldrich. The HA protein was labeled with biotin using the EZ-Link Sulfo-NHS-LC-Biotin biotinylation reagent (Thermo Scientific) according to the manufacturer's instructions. The HA-specific memory B cells were gated and isolated as CD19+/IgG+/HA-specific BCR+ cells. The flow cytometry data were analyzed with FlowJo software.
The generation of human monoclonal antibodies
The generation of human monoclonal antibodies was performed using single-cell RT-PCR as previously described (Smith et al., 2009, Wrammert et al., 2008). Briefly, HA-specific memory B cells were sorted into 96-well PCR plates containing an RNase inhibitor (Promega) at one cell per well. The antibody variable genes (VH and Vκ) were amplified from each cell with RT-PCR and nested PCR using panels of primers as previously described (Smith et al., 2009). Restriction sites were incorporated with PCR with specific primers designed according to the characteristics of the V and J gene families to which the amplified genes belonged. The VH and Vκ genes were cloned into IgG and Igκ expression vectors, respectively. The IgG and Igκ expression plasmids were then co-transfected into HEK293T cells using Lipofectamine transfection reagent (Invitrogen) according to the manufacturer's instructions. The supernatants were harvested, and the human monoclonal antibodies were purified using an rmp Protein A Sepharose Fast Flow column (GE) according to the manufacturer's instructions.
ELISA
Ninety-six microwell plates (Nunc) were coated with viral lysate or recombinant protein at a concentration of 10 μg/ml in 0.1 M sodium carbonate-bicarbonate buffer (pH 9.6) at 4° C overnight. Phosphate-buffered saline (PBS, pH 7.4) containing 10% bovine serum albumin and 0.1% Tween-20 was used as the blocking and dilution buffer. After blocking at 37 °C for two hours, the plates were washed, and human monoclonal antibodies were added. The plates were then incubated at 37 °C for another two hours. Next, a horseradish peroxidase-conjugated goat anti-human IgG (Fc-specific) antibody (Sigma-Aldrich) was added, and the plates were incubated at 37 °C for one hour. Tetramethylbenzidine (Sigma-Aldrich) was used as the substrate, and the optical density was measured at 450 nm (OD 450 nm) with a microwell plate autoreader (Thermo Scientific).
As for the peptide ELISA, 19 overlapped peptides (Table 3) were designed according to the protein sequences of HA2 (G345 to P504) of the pandemic H1N1 influenza virus strain: A/California/4/2009 (H1N1). The peptides (with N-terminal biotinylation and 90% purity) were synthesized by Shanghai Science Peptide, China. Streptavidin was coated onto the 96-microwell plate (Nunc) at 20 μg/ml in 0.1 M sodium carbonate–bicarbonate buffer (pH 9.6) at 4 °C overnight. After washing of unbound streptavidin and blocking at 37 °C for two hours, the overlapping peptides were added at 2 μg/ml and incubated at 37 °C for another one hour. Thereafter, procedures were the same as mentioned above.
Competitive ELISA
The HA protein was used to coat 96-microwell plates (Nunc). Human monoclonal antibodies were labeled with biotin using the EZ-Link Sulfo-NHS-LC-Biotin biotinylation reagent (Thermo Scientific) according to the manufacturer's instructions. After blocking at 37 °C for two hours, excess amounts of human monoclonal antibodies not conjugated to biotin were added to the plates, and the plates were incubated at 37 °C for two hours. The biotin-labeled monoclonal antibodies were then added to the plates, and the plates were incubated at 37 °C for another two hours. Next, the streptavidin–horseradish peroxidase conjugate (BD) was added, and the plates were incubated at 37 °C for one hour. Tetramethylbenzidine (Sigma-Aldrich) was used as the substrate, and the optical density was measured at 450 nm (OD 450 nm) with a microwell plate autoreader (Thermo Scientific). The percentage of competition was calculated as the reduction in the absorbance when mAb binding competition occurred.
Cell–cell fusion inhibition assay
The experiment was performed generally as previously described (Godley et al., 1992, Goto and Kawaoka, 1998, Prabhu et al., 2009). Briefly, HEK293T cells (ATCC) were transfected with pbudCE4.1 (Invitrogen) plasmids expressing the pandemic H1N1 HA0 protein with the sequence from A/California/4/2009. The cells were cultured in 6-well plates and transfected with plasmids using Lipofectamine transfection reagent (Invitrogen). 24 h after transfection, the cells were treated with (tosylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (2.5 μg/ml) at 37 °C for 10 min in order to cleave HA0 into HA1 and HA2. After that, the cells were incubated at 37 °C for 30 min with the mAbs of 100 μg/ml. Afterwards, the cells were washed with PBS, and the pH was decreased to 5.0 at 37 °C for 10 min. Next, the cells were incubated with the mAbs at the concentration of 100 μg/ml in complete medium for 2 h. The cells were then fixed with 4% (wt/vol) paraformaldehyde and stained with 1% (wt/vol) toluidine blue in PBS. Results were observed in optical microscope and photographed.
Microneutralization assay
The microneutralization assay was performed as previously described (Rowe et al., 1999). Briefly, antibodies were serially diluted two-fold in 50 μl and then mixed with influenza virus (100 TCID50 in 50 μl per well). Positive-control wells (virus only) and negative-control wells (without virus) were included on each plate. After a two-hour incubation at 37 °C in a 5% CO2 humidified atmosphere, 100 μl of trypsin–EDTA-suspended MDCK cells (1.5×105/ml) was added to each well. The plates were incubated for 18 h at 37 °C in a 5% CO2 humidified atmosphere. The cell monolayers were washed with PBS and fixed in cold 80% acetone for 10 min. The presence of viral protein was detected with an ELISA as previously described (Rowe et al., 1999, Walls et al., 1986) with a polyclonal antibody against the influenza A NP protein (prepared in our lab). The neutralizing end point was assessed as previously described (Rowe et al., 1999). The inhibition ratio (%) was calculated as [OD (Pos. Control)−OD (Sample)]/[OD (Pos. Control)−OD (Neg. Control)]×100%.
Acknowledgments
We thank Dr. Patrick C. Wilson from the University of Chicago for kindly providing the IgG and Igκ expression vectors. This work was supported by Grants from the National 863 project (2012AA02A404, 2012AA020103), the National Natural Science Foundation of China (31030029, 31100662, 31230024, 81201280), the National Science and Technology Major Project (2011ZX10004-001, 2012ZX10002-007), the National Ministry of Science and Technology (2007DFC31700), the Knowledge Innovation Program of Shanghai Institutes for Biological Sciences, CAS (2010KIP304), the SA-SIBS Discovery Innovation Grant, and the Li Kha Shing Foundation.
Contributor Information
Xiaojun Lin, Email: linxiaojun@hualan.com.
Chao Bian, Email: cbian@sibs.ac.cn.
Bing Sun, Email: bsun@sibs.ac.cn.
References
- Burioni R., Canducci F., Mancini N., Clementi N., Sassi M., De Marco D., Diotti R.A., Saita D., Sampaolo M., Sautto G., Pianezze M., Clementi M. Monoclonal antibodies isolated from human B cells neutralize a broad range of H1 subtype influenza A viruses including swine-origin Influenza virus (S-OIV) Virology. 2010;399:144–152. doi: 10.1016/j.virol.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Corti D., Suguitan A.L., Jr., Pinna D., Silacci C., Fernandez-Rodriguez B.M., Vanzetta F., Santos C., Luke C.J., Torres-Velez F.J., Temperton N.J., Weiss R.A., Sallusto F., Subbarao K., Lanzavecchia A. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi N., De Marco D., Mancini N., Solforosi L., Moreno G.J., Gubareva L.V., Mishin V., Di Pietro A., Vicenzi E., Siccardi A.G., Clementi M., Burioni R. A human monoclonal antibody with neutralizing activity against highly divergent influenza subtypes. PLoS One. 2011;6:e28001. doi: 10.1371/journal.pone.0028001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F., Silacci C., Fernandez-Rodriguez B.M., Agatic G., Bianchi S., Giacchetto-Sasselli I., Calder L., Sallusto F., Collins P., Haire L.F., Temperton N., Langedijk J.P., Skehel J.J., Lanzavecchia A. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- De Marco D., Clementi N., Mancini N., Solforosi L., Moreno G.J., Sun X., Tumpey T.M., Gubareva L.V., Mishin V., Clementi M., Burioni R. A non-VH1-69 heterosubtypic neutralizing human monoclonal antibody protects mice against H1N1 and H5N1 viruses. PLoS One. 2012;7:e34415. doi: 10.1371/journal.pone.0034415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Matsuoka Y., Maines T.R., Swayne D.E., O'Neill E., Davis C.T., Van-Hoven N., Balish A., Yu H.J., Katz J.M., Klimov A., Cox N., Li D.X., Wang Y., Guo Y.J., Yang W.Z., Donis R.O., Shu Y.L. Development of a new candidate H5N1 avian influenza virus for pre-pandemic vaccine production. Influenza Other Respir. Viruses. 2009;3:287–295. doi: 10.1111/j.1750-2659.2009.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert D.C., Bhabha G., Elsliger M.A., Friesen R.H., Jongeneelen M., Throsby M., Goudsmit J., Wilson I.A. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert D.C., Friesen R.H., Bhabha G., Kwaks T., Jongeneelen M., Yu W., Ophorst C., Cox F., Korse H.J., Brandenburg B., Vogels R., Brakenhoff J.P., Kompier R., Koldijk M.H., Cornelissen L.A., Poon L.L., Peiris M., Koudstaal W., Wilson I.A., Goudsmit J. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten R.J., Davis C.T., Russell C.A., Shu B., Lindstrom S., Balish A., Sessions W.M., Xu X., Skepner E., Deyde V., Okomo-Adhiambo M., Gubareva L., Barnes J., Smith C.B., Emery S.L., Hillman M.J., Rivailler P., Smagala J., de Graaf M., Burke D.F., Fouchier R.A., Pappas C., Alpuche-Aranda C.M., López-Gatell H., Olivera H., López I., Myers C.A., Faix D., Blair P.J., Yu C., Keene K.M., Dotson P.D., Jr., Boxrud D., Sambol A.R., Abid S.H., St George K., Bannerman T., Moore A.L., Stringer D.J., Blevins P., Demmler-Harrison G.J., Ginsberg M., Kriner P., Waterman S., Smole S., Guevara H.F., Belongia E.A., Clark P.A., Beatrice S.T., Donis R., Katz J., Finelli L., Bridges C.B., Shaw M., Jernigan D.B., Uyeki T.M., Smith D.J., Klimov A.I., Cox N.J. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godley L., Pfeifer J., Steinhauer D., Ely B., Shaw G., Kaufmann R., Suchanek E., Pabo C., Skehel J.J., Wiley D.C., Wharton S. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68:635–645. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- Goto H., Kawaoka Y. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc. Natl. Acad. Sci. U S A. 1998;95:10224–10228. doi: 10.1073/pnas.95.17.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock K., Veguilla V., Lu X., Zhong W., Butler E.N., Sun H., Liu F., Dong L., DeVos J.R., Gargiullo P.M., Brammer T.L., Cox N.J., Tumpey T.M., Katz J.M. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- Hung I.F., To K.K., Lee C.K., Lee K.L., Chan K., Yan W.W., Liu R., Watt C.L., Chan W.M., Lai K.Y., Koo C.K., Buckley T., Chow F.L., Wong K.K., Chan H.S., Ching C.K., Tang B.S., Lau C.C., Li I.W., Liu S.H., Chan K.H., Lin C.K., Yuen K.Y. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin. Infect. Dis. 2011;52:447–456. doi: 10.1093/cid/ciq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.F., Wang H.Q., Wang J.Z., Fang H.H., Wu J., Zhu F.C., Li R.C., Xia S.L., Zhao Y.L., Li F.J., Yan S.H., Yin W.D., An K., Feng D.J., Cui X.L., Qi F.C., Ju C.J., Zhang Y.H., Guo Z.J., Chen P.Y., Chen Z., Yan K.M., Wang Y. Safety and immunogenicity of 2009 pandemic influenza A H1N1 vaccines in China: a multicentre, double-blind, randomised, placebo-controlled trial. Lancet. 2010;375:56–66. doi: 10.1016/S0140-6736(09)62003-1. [DOI] [PubMed] [Google Scholar]
- Okuno Y., Isegawa Y., Sasao F., Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P. Influenza: old and new threats. Nat. Med. 2004;10:S82–87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- Prabhu N., Prabakaran M., Ho H.T., Velumani S., Qiang J., Goutama M., Kwang J. Monoclonal antibodies against the fusion peptide of hemagglutinin protect mice from lethal influenza A virus H5N1 infection. J. Virol. 2009;83:2553–2562. doi: 10.1128/JVI.02165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T., Abernathy R.A., Hu-Primmer J., Thompson W.W., Lu X., Lim W., Fukuda K., Cox N.J., Katz J.M. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 1999;37:937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Smith K., Garman L., Wrammert J., Zheng N.Y., Capra J.D., Ahmed R., Wilson P.C. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.M., Santelli E., Stec B., Cadwell G., Ali M., Wan H., Murakami A., Yammanuru A., Han T., Cox N.J., Bankston L.A., Donis R.O., Liddington R.C., Marasco W.A. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe S., Iwatsuki-Horimoto K., Horimoto T., Nidom C.A., Le M.Q., Takano R., Kubota-Koketsu R., Okuno Y., Ozawa M., Kawaoka Y. A cross-reactive neutralizing monoclonal antibody protects mice from H5N1 and pandemic (H1N1) 2009 virus infection. Antiviral Res. 2010;88:249–255. doi: 10.1016/j.antiviral.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls H.H., Harmon M.W., Slagle J.J., Stocksdale C., Kendal A.P. Characterization and evaluation of monoclonal antibodies developed for typing influenza A and influenza B viruses. J. Clin. Microbiol. 1986;23:240–245. doi: 10.1128/jcm.23.2.240-245.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Holtz K.M., Anderson K., Chubet R., Mahmoud W., Cox M.M. Expression and purification of an influenza hemagglutinin—one step closer to a recombinant protein-based influenza vaccine. Vaccine. 2006;24:2176–2185. doi: 10.1016/j.vaccine.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J., Smith K., Miller J., Langley W.A., Kokko K., Larsen C., Zheng N.Y., Mays I., Garman L., Helms C., James J., Air G.M., Capra J.D., Ahmed R., Wilson P.C. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J., Koutsonanos D., Li G.M., Edupuganti S., Sui J., Morrissey M., McCausland M., Skountzou I., Hornig M., Lipkin W.I., Mehta A., Razavi B., Del Rio C., Zheng N.Y., Lee J.H., Huang M., Ali Z., Kaur K., Andrews S., Amara R.R., Wang Y., Das S.R., O'Donnell C.D., Yewdell J.W., Subbarao K., Marasco W.A., Mulligan M.J., Compans R., Ahmed R., Wilson P.C. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.C., Wang H., Fang H.H., Yang J.G., Lin X.J., Liang X.F., Zhang X.F., Pan H.X., Meng F.Y., Hu Y.M., Liu W.D., Li C.G., Li W., Zhang X., Hu J.M., Peng W.B., Yang B.P., Xi P., Wang H.Q., Zheng J.S. A novel influenza A (H1N1) vaccine in various age groups. N. Engl. J. Med. 2009;361:2414–2423. doi: 10.1056/NEJMoa0908535. [DOI] [PubMed] [Google Scholar]