Abstract
Ebola (EBOV) and Marburg virus (MARV) cause severe hemorrhagic fever. The host cell proteases cathepsin B and L activate the Zaire ebolavirus glycoprotein (GP) for cellular entry and constitute potential targets for antiviral intervention. However, it is unclear if different EBOV species and MARV equally depend on cathepsin B/L activity for infection of cell lines and macrophages, important viral target cells. Here, we show that cathepsin B/L inhibitors markedly reduce 293T cell infection driven by the GPs of all EBOV species, independent of the type II transmembrane serine protease TMPRSS2, which cleaved but failed to activate EBOV-GPs. Similarly, a cathepsin B/L inhibitor blocked macrophage infection mediated by different EBOV-GPs. In contrast, MARV-GP-driven entry exhibited little dependence on cathepsin B/L activity. Still, MARV-GP-mediated entry was efficiently blocked by leupeptin. These results suggest that cathepsins B/L promote entry of EBOV while MARV might employ so far unidentified proteases for GP activation.
Keywords: Entry, Glycoprotein, Proteolytic activation, Cathepsin, Ebola
Introduction
Ebola virus (EBOV) and Marburg virus (MARV), negative strand RNA viruses of the filoviridae family, can cause severe hemorrhagic fever in humans. A single species of MARV, Lake Victoria marburgvirus, has been identified (Hartman et al., 2010), while EBOVs are separated into four species, Zaire ebolavirus (ZEBOV), Sudan ebolavirus (SEBOV), Cote d'Ivoire ebolavirus (CIEBOV) and Reston ebolavirus (REBOV) (Feldmann and Geisbert, 2011, Hartman et al., 2010). A fifth species, Bundibugyo ebolavirus (BEBOV), has been proposed (Towner et al., 2008). Outbreaks of ZEBOV, SEBOV, CIEBOV, BEBOV and MARV occur sporadically in Africa and are associated with case-fatality rates of up to 90%. In contrast, REBOV has been detected in pigs in the Philippines and seems to be apathogenic in humans (Feldmann and Geisbert, 2011, Hartman et al., 2010).
EBOV and MARV bear a single glycoprotein (GP) in their envelope which mediates infectious viral entry into host cells and constitutes an important target for prevention and therapy. Many glycoproteins related to the EBOV-GP, like the HIV envelope protein (Env), are synthesized as inactive precursor proteins in the constitutive secretory pathway of infected cells and require activation by host cell proteases to transit into an active form (Bertram et al., 2010b). Both EBOV-GP and HIV Env are cleaved by furin in the Golgi apparatus of infected cells (Hallenberger et al., 1997, Volchkov et al., 1998) and cleavage at the furin consensus site is essential for HIV infectivity (McCune et al., 1988). In contrast, GP cleavage by furin is dispensable for EBOV spread in cell culture and in animals (Neumann et al., 2002, Neumann et al., 2007, Wool-Lewis and Bates, 1999), indicating that furin does not activate GP. Work by Chandran et al. (2005) and subsequent studies (Kaletsky et al., 2007, Schornberg et al., 2006) suggest that ZEBOV-GP is activated by the pH-dependent cysteine proteases cathepsin B and L (termed cathepsins B/L in the remainder of the manuscript) upon viral entry into host cell endosomes. In addition, a link between α5β1 integrin expression, cathepsin B/L activity and GP-mediated entry has been revealed (Schornberg et al., 2009), in line with a previous report indicating that β1 integrins promote EBOV-GP-dependent host cell entry (Takada et al., 2000). However, it is at present unclear if all EBOV species and MARV are comparably dependent on cathepsin B/L activity for viral entry and it is unknown if cathepsin B/L activity is required for GP-driven entry into macrophages, central target cells of filoviruses.
The cell entry of the severe acute respiratory syndrome coronavirus (SARS-CoV) also requires activation of the viral envelope protein spike (S) by cathepsins B and L (Simmons et al., 2005). However, expression of the type II transmembrane serine protease (TTSP) TMPRSS2, a cell-associated protease known to activate influenza virus (Böttcher et al., 2006, Chaipan et al., 2009) and human metapneumovirus (Shirogane et al., 2008), allows cathepsin B/L-independent host cell entry, indicating that SARS-CoV can use different proteases to ensure activation of its S-protein (Glowacka et al., 2011, Matsuyama et al., 2010, Shulla et al., 2011). These findings and the observation that TMPRSS2 is expressed by type II pneumocytes, major viral target cells (Glowacka et al., 2011, Matsuyama et al., 2010), suggest that inhibition of cathepsin B/L activity might not appreciably reduce viral spread in infected patients. Whether TTSPs can also cleave and activate EBOV-GP for cathepsin B/L-independent host cell entry is unknown.
Here, we show that dependence on cathepsin B/L-activity for entry into 293T cells is conserved between the GPs of the four EBOV species and is not modulated by expression of TMPRSS2. Macrophage infection driven by different EBOV-GPs was also dependent on cathepsin B/L activity, in line with the findings made with 293T cells. In contrast, MARV-GP-mediated infection exhibited little dependence on cathepsin B/L activity, indicating that MARV-GP can use other proteases to ensure its activation.
Results
Inhibition of cathepsin B and L activity by protease inhibitors
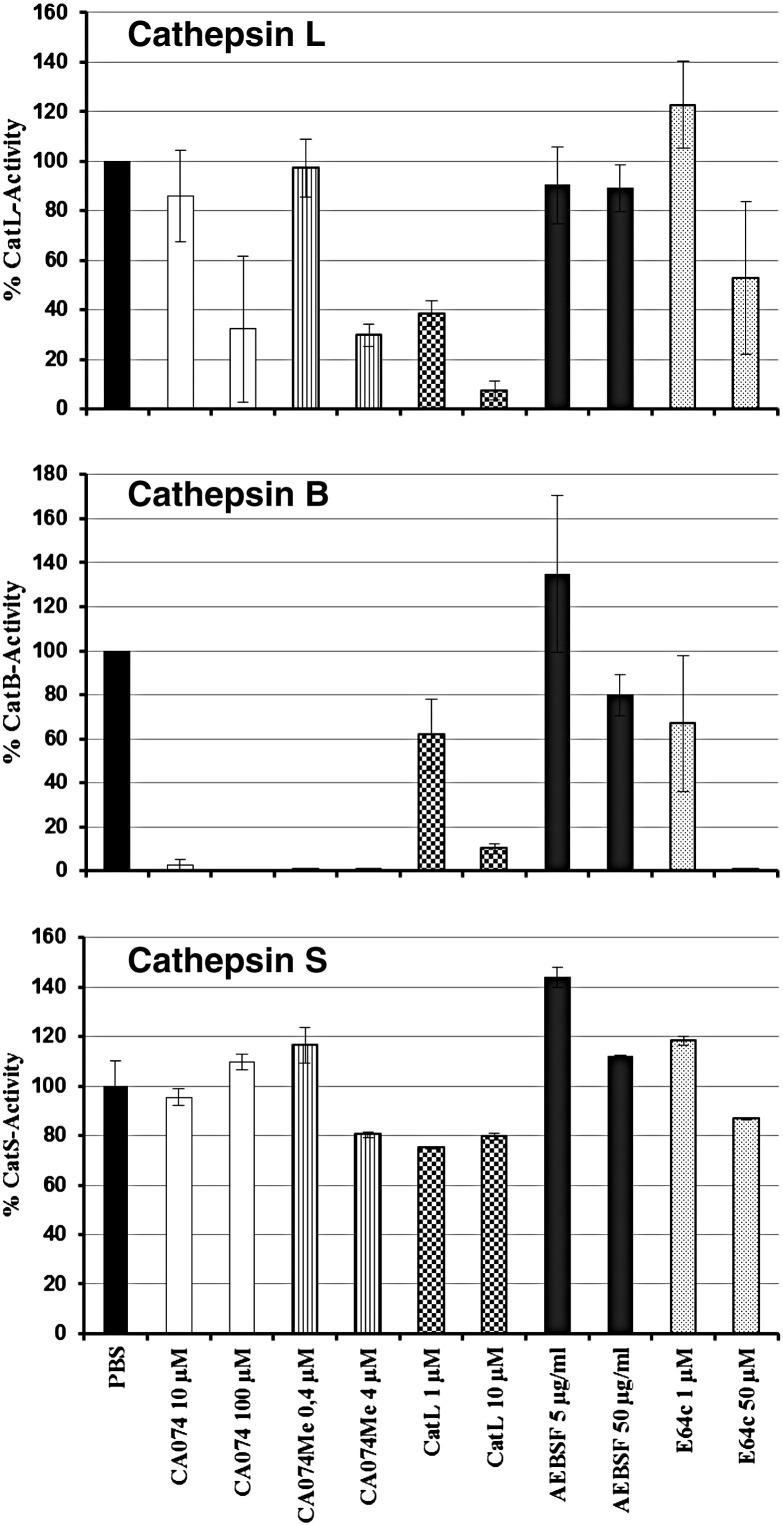
In order to assess the proteolytic activation of EBOV-GP by cathepsins B and L, we selected a panel of previously characterized, commercially available inhibitors (Table 1 ). For several of these inhibitors, we determined their activity against cathepsins B, L and S in 293T cells, employing inhibitor concentrations subsequently used for infection experiments. CA074, a cathepsin B inhibitor (Sigma), potently blocked cathepsin B activity, as expected, but at a concentration of 100 μM also reduced cathepsin L activity by about 70% (Fig. 1 ). CA074Me, a CA074 derivative with high membrane permeability (Calbiochem), exhibited activity similar to CA074 at the concentrations tested. Notably, CA074Me was previously shown to inhibit cellular entry of ecotropic murine leukemia virus via inhibition of cathepsin L, demonstrating that the modest activity of CA074Me against cathepsin L (Fig. 1) can be sufficient to exert antiviral effects (Yoshii et al., 2009). Cathepsin L inhibitor III, an irreversible inhibitor of cathepsin L (Merck), was highly active against cathepsin L but, unexpectedly, also inhibited cathepsin B with similar efficiency (Fig. 1). AEBSF, a serine protease inhibitor (Roth), displayed no activity against cathepsins B and L. E64c, a cysteine protease inhibitor (Sigma), fully inhibited cathepsin B and reduced cathepsin L activity by half when applied at a concentration of 50 μM (Fig. 1). None of the inhibitors tested were active against cathepsin S, indicating a substantial level of selectivity, although inhibition of other cysteine cathepsins, like cathepsins H, K and O, has not been tested. Finally, leupeptin, a well characterized inhibitor of serine and cysteine proteases (Aoyagi et al., 1969), and MDL 28170, an efficient inhibitor of cathepsin B activity (Mehdi, 1991), cathepsin L activity (Simmons et al., 2005) and SARS-S driven host cell entry (Simmons et al., 2005), were also included in the panel.
Table 1.
Specificity and commercial source of protease inhibitors used in the present study.
| Inhibitor | Distributor | Protease specificity⁎ | CatL | CatB | CatS |
|---|---|---|---|---|---|
| Leupeptin | Sigma | Cysteine-, serine- proteases | n.d. | n.d. | n.d. |
| AEBSF | Roth | Serine-proteases | − | − | − |
| E64c | Sigma | Cysteine proteases | (+) | ++ | − |
| MDL28170 | Sigma | Calpain I and II | n.d. | n.d. | n.d. |
| CA074 | Sigma | Cathepsin B | (+) | ++ | − |
| CA074Me | Calbiochem | Cathepsin B | (+) | ++ | − |
| Cathepsin L inhibitor III | Merck | Cathepsin L | + | (+) | − |
⁎According to distributor, ++ = more than 100-fold inhibition at highest concentration tested, + = 10–100 fold inhibition at highest concentration tested, (+) = 2–10 fold inhibition at highest concentration tested, − = no inhibition, n.d., not done.
Fig. 1.
Inhibition of cathepsin B, L and S activity by protease inhibitors. 293T cells were incubated with the indicated inhibitors and cathepsin activity was determined 8 h later using commercially available kits. The cathepsin B, L and S activity in cells treated with protease inhibitors is shown relative to that measured upon treatment of cells with PBS, which was set as 100%. Results for cathepsin B and L activity represent the average of three independent experiments. Error bars indicate standard error of the mean. The results of a representative experiment performed in duplicates are shown for cathepsin S and were confirmed in a separate experiment. Error bars indicate standard deviation (SD). CatB, cathepsin B; CatL, cathepsin L; CatS, cathepsin S.
The glycoproteins of all Ebola virus species depend on cathepsin B and L activity for host cell entry
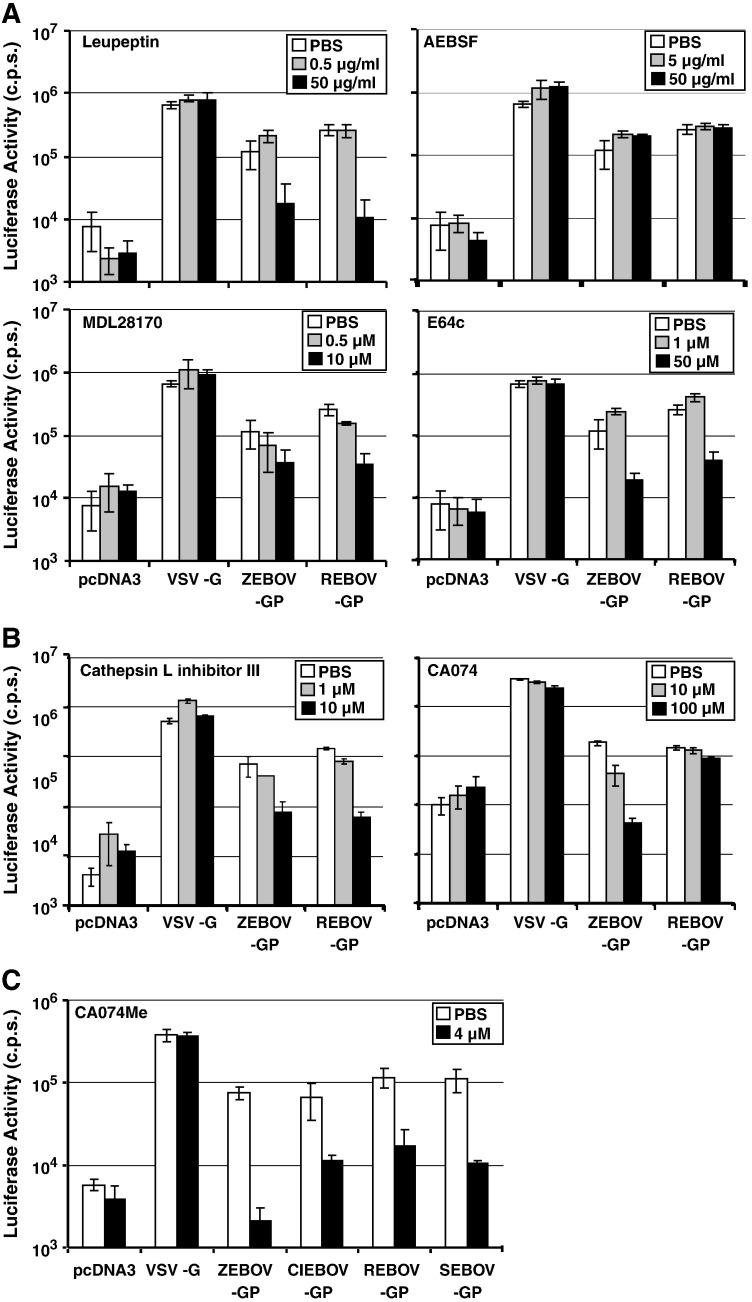
Cathepsins B and L were previously shown to activate ZEBOV-GP for host cell entry (Chandran et al., 2005). In order to address if the GPs of different EBOVs are activated differentially by host cell proteases, we compared activation of GP derived from ZEBOV and REBOV, which exhibit differential pathogenicity in humans (Feldmann and Geisbert, 2011). For this, we employed infectivity-normalized HIV-derived vectors pseudotyped with ZEBOV- and REBOV-GP for infection of 293T target cells that were either pretreated with protease inhibitor (Fig. 2 , gray and black bars) or inhibitor solvent as control (white bars). Particles bearing no GP (Fig. 2, pcDNA3) or the G-protein of vesicular stomatitis virus (VSV-G) were used as controls. AEBSF treatment of target cells had no effect on entry of all pseudotypes tested (Fig. 2A, upper panel, right), while leupeptin inhibited ZEBOV- and REBOV-GP-dependent entry (Fig. 2A, upper panel, left), indicating that cysteine but not serine proteases are involved in EBOV-GP activation. MDL28170 and E64c both inhibited ZEBOV- and REBOV-GP-driven entry, in agreement with the concept that cathepsins B/L can activate EBOV-GP (Fig. 2A, lower panel). However, the relative importance of cathepsins B and L for activation of these glycoproteins might differ, since cathepsin L inhibitor III (Table 1), which displayed the highest activity against cathepsin L of all compounds tested, inhibited cell entry driven by both ZEBOV- and REBOV-GP, while the largely cathepsin B-specific inhibitor CA074 (Table 1) was active only against ZEBOV-GP (Fig. 2B). Finally, CA074Me, which reduced cathepsin B activity by about 99.9% and cathepsin L activity by about 70% at a concentration of 4 μM (Fig. 1), inhibited entry driven by the GPs of all EBOV species, although with different efficiencies (Fig. 2C), suggesting that cathepsin B and potentially also cathepsin L dependence for host cell entry is conserved between EBOV-GPs. In sum, these results indicate that efficient cellular entry driven by the GPs of the EBOV species depends on the activity of cathepsins B and/or L, while the relative contribution of both enzymes to GP activation might vary between the EBOV species.
Fig. 2.
Entry driven by the glycoproteins of the four Ebola virus species depends on cathepsin activity. A. 293T target cells were preincubated with the indicated concentrations of cathepsin B and L inhibitors (MDL28170, E64c), a serine protease inhibitor (AEBSF) and an inhibitor blocking cysteine and serine proteases (leupeptin). Subsequently, cells were infected with pseudotypes bearing the indicated GPs, normalized for equal infectivity. Luciferase activity in cell lysates was measured at 72 h post infection. B. The experiment was carried out as described in (A), using two different concentrations of cathepsin L inhibitor III and cathepsin B inhibitor CA074. C. The experiment was carried out as described in (A) but cells were incubated with the cathepsin B inhibitor CA074Me. The results of representative experiments performed in triplicates are shown in A–C; error bars indicate SD. Similar results were obtained in three separate experiments.
Evidence that the Marburg virus glycoprotein can be activated by proteases other than cathepsins B and L
A previous study indicated that replication competent MARV might not depend on cathepsin B/L activity for host cell entry (Sanchez, 2007). Cellular entry of lentiviral vectors pseudotyped with MARV-GP showed variable dependence on cathepsin B and L activity: CA074Me, when applied at a concentration of 4 μM, modestly suppressed MARV-GP-driven entry into 293T cells (3.3- to 6.2-fold inhibition, data not shown) in two experiments but had no appreciable effect in two separate experiments (0- to 1.5-fold inhibition, Fig. 3 and data not shown). These results suggest that MARV-GP is indeed less dependent on cathepsin B/L activity for entry into 293T cells when compared to EBOV-GP. However, leupeptin but not AEBSF robustly and invariably suppressed host cell entry driven by MARV-GP (Fig. 3), demonstrating that MARV-GP like EBOV-GP depends on activation by host cell proteases for infectious entry, and the nature of the responsible protease(s) remains to be defined.
Fig. 3.
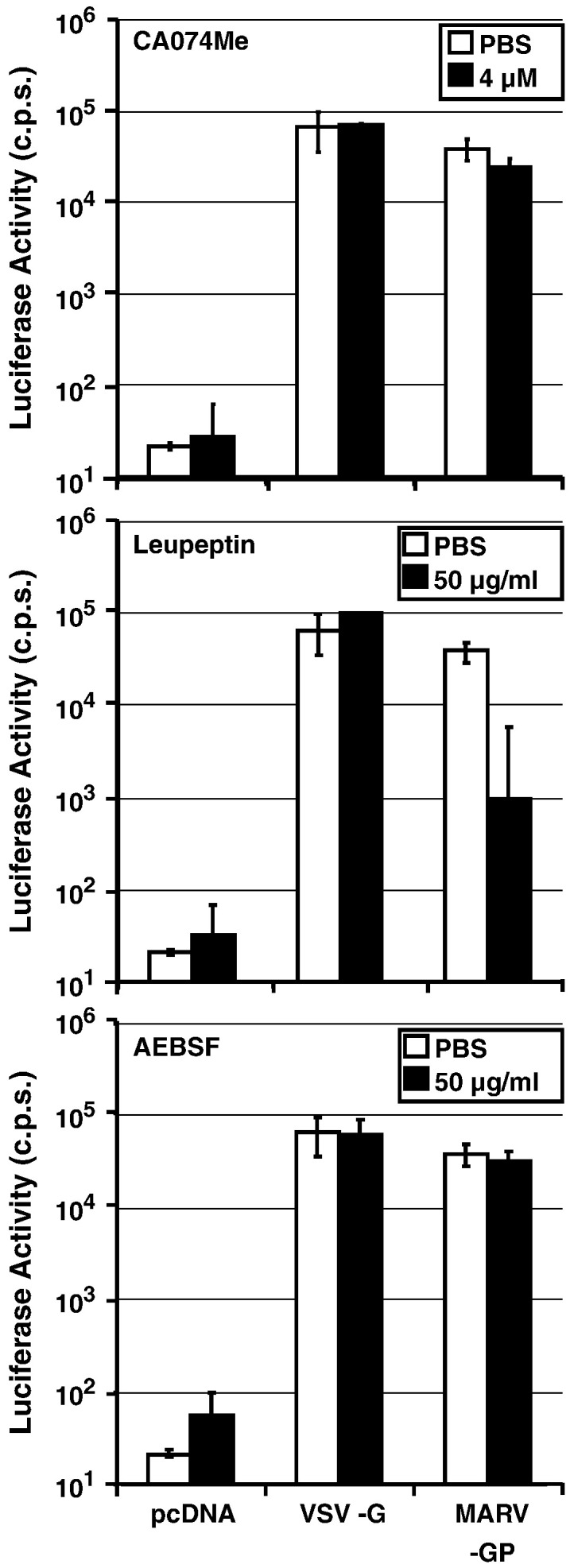
Cellular entry mediated by Marburg virus glycoprotein is largely cathepsin-independent. 293T target cells were preincubated with the indicated concentrations of protease inhibitors leupeptin, AEBSF and CA074Me. Subsequently, cells were infected with pseudotypes bearing the indicated GPs, normalized for equal infectivity and luciferase activity in cell lysates was measured at 72 h post infection. The results ± SD of a representative experiment performed in triplicates are shown and were confirmed in at least two separate experiments.
Type II transmembrane serine proteases cannot substitute for cathepsins B/L in Ebolavirus glycoprotein-driven host cell entry
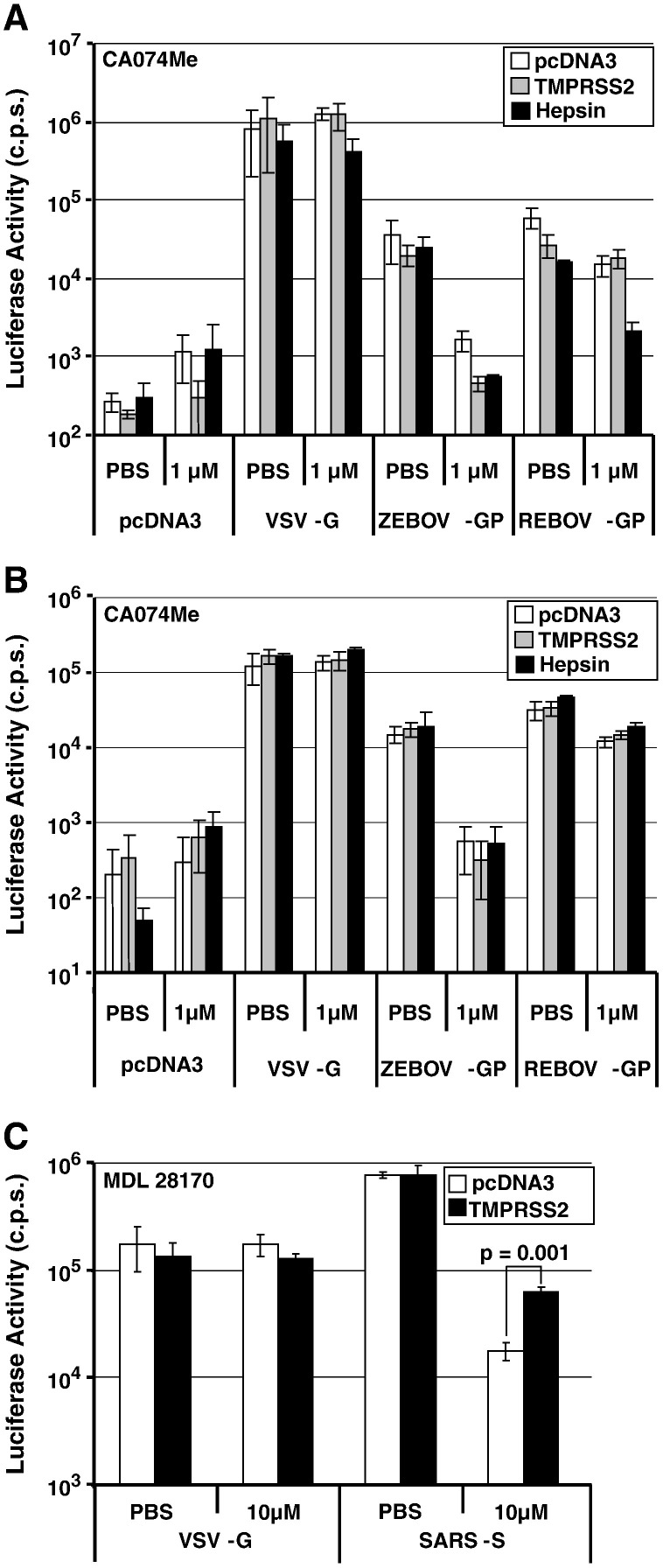
The S-protein of SARS-CoV is also activated by cathepsins B and L (Simmons et al., 2005). However, the dependence on cathepsin activity is markedly reduced if target cells express the type II transmembrane serine protease TMPRSS2 (Glowacka et al., 2011, Matsuyama et al., 2010, Shulla et al., 2011). We next addressed if TMPRSS2 and the related protease hepsin modulate cathepsin-dependence of EBOV-GP for host cell entry. For this, we coexpressed EBOV-GPs with a C-terminal V5 tag and TMPRSS2 or hepsin in 293T cells and analyzed GP cleavage by Western blot. All GPs were cleaved in the absence of TTSP expression (Fig. 4 ), in agreement with the previously published finding that GP is proteolytically processed by furin (Volchkov et al., 1998), an ubiquitously expressed pre-protein convertase active in the 293T cell line used for GP expression in the present study (Bush et al., 2001, Yano et al., 2009). However, cleavage efficiency differed between the GPs analyzed: ZEBOV- and SEBOV-GP were efficiently cleaved while cleavage of CIEBOV- and REBOV-GP was inefficient (Fig. 4). Notably, the inefficient cleavage of REBOV-GP was rescued by hepsin coexpression, and TMPRSS2 coexpression allowed efficient cleavage of both REBOV- and CIEBOV-GP. Thus, hepsin and TMPRSS2 might activate REBOV- and/or CIEBOV-GP for host cell entry. However, coexpression of neither protease in virus producing cells (Fig. 5 A) or in viral target cells (Fig. 5B) allowed efficient GP-driven entry into cells pretreated with a cathepsin B/L inhibitor, while TMPRSS2 was able to partially rescue blockade of SARS-S-driven infection (Fig. 5C), as expected (Glowacka et al., 2011, Matsuyama et al., 2010, Shulla et al., 2011). These results suggest that TMPRSS2 and hepsin cannot functionally replace cathepsin B/L activity in EBOV target cells.
Fig. 4.
Proteolytic processing of the Ebola virus glycoprotein by TMPRSS2 and hepsin. Lysates of 293T cells coexpressing the indicated proteases and EBOV-GPs or empty vector (pCAGGS) were analyzed for GP cleavage by Western blot using a V5-tag specific antibody. Detection of ß-actin served as a loading control. The results are representative for at least three independent experiments.
Fig. 5.
TMPRSS2 and hepsin do not facilitate cathepsin-independent, Ebola virus glycoprotein-dependent host cell entry. A. Pseudotypes were generated in 293T cells expressing the indicated GPs in combination with the indicated proteases. Released pseudoparticles were normalized for equal content of p24-antigen and used to infect mock-transfected 293T cells pretreated with PBS or PBS containing 1 μM CA074Me. At 8 h post infection, medium was replaced, and luciferase activity was measured in cell lysates at 72 h post infection. B. Pseudotypes bearing the indicated GPs were normalized for comparable infectivity prior to infection of PBS or CA074Me treated 293T cells expressing the indicated proteases. C. Infectivity-normalized SARS-S pseudotypes were used to infect PBS or MDL 28170 treated 293T-ACE2 target cells expressing TMPRSS2 or no protease. The results of representative experiments performed in triplicates are shown in A–C; error bars indicate SD. Similar results were obtained in at least two separate experiments. For testing of statistical significance, a two-tailed student's t-test was employed.
Cathepsin B/L activity is essential for efficient Ebola but not Marburg virus glycoprotein-driven entry into macrophages
Previous studies on the role of cathepsins in the proteolytic activation of EBOV-GP largely focused on cell lines (Chandran et al., 2005, Schornberg et al., 2006), while the importance of these enzymes for viral entry into macrophages, a key target cell in filovirus infection, has not been examined. Incubation of macrophages with CA074Me and leupeptin inhibited entry driven by ZEBOV- and REBOV-GP but not VSV-G (Fig. 6 ). In contrast, only leupeptin but not CA074Me efficiently reduced MARV-GP-dependent infection (Fig. 6) and AEBSF had no effect (data not shown), indicating that MARV might employ other cellular proteases in addition to or instead of cathepsins B and L for entry into macrophages.
Fig. 6.
Macrophage infection driven by Zaire and Reston ebolavirus as well as Marburg virus glycoproteins is cathepsin-dependent. A. Macrophages were preincubated with 4 μM CA074Me before infection with pseudotypes bearing the indicated GPs, normalized for equal infectivity. Luciferase activity in cell lysates was measured at 72 h post infection. B. Macrophages were preincubated with the indicated concentrations of leupeptin prior to infection with ZEBOV-GP or MARV-GP pseudoparticles. C. The experiment was carried out as described in (B) but the cells were treated with the indicated concentrations CA074Me prior to infection. The results of representative experiments performed in triplicates are shown in A–C; error bars indicate SD. Similar results were obtained in two independent experiments.
Discussion
Many viruses which cause disease in humans, including HIV, influenza virus, and SARS-CoV, acquire infectivity only upon proteolytic activation of their glycoproteins (Bertram et al., 2010b, Harrison, 2008). Whether the same holds true for EBOV has long been unclear, since cleavage at a furin consensus site in EBOV-GP was found to be dispensable for infectivity (Neumann et al., 2002, Neumann et al., 2007). Recent findings demonstrated that the endo-/lysosomal cysteine proteases cathepsin L and particularly cathepsin B can activate ZEBOV-GP (Chandran et al., 2005, Schornberg et al., 2006), indicating that proteolytic activation of EBOV occurs upon viral uptake into host cells. Interestingly, proteolysis of GP by cathepsins B/L seems to be required for engagement of the endosomal membrane protein Niemann–Pick C1, which is essential for infectious filovirus entry (Carette et al., 2011, Cote et al., 2011). Here, we show that the GPs of all EBOV subspecies depend on cathepsin B/L activity for viral entry into 293T cells, although to different degrees. Similarly, EBOV-GP-driven infection of macrophages required cathepsin B/L activity. In contrast, MARV-GP-driven entry was largely cathepsin B/L-independent and was promoted by so far uncharacterized host cell proteases. Finally, TMPRSS2 and hepsin, members of the TTSP family known to activate SARS-CoV and influenza viruses (Böttcher et al., 2006, Glowacka et al., 2011, Matsuyama et al., 2010, Shulla et al., 2011), were able to cleave EBOV-GP but were unable to support GP-dependent, cathepsin B/L-independent host cell entry.
Previous studies reported that the GPs of the four EBOV subspecies differ in their interactions with host cells; for instance, ZEBOV- but not REBOV-GP was found to interact efficiently with the dendritic cell lectin hMGL (Takada et al., 2004) and expression of ZEBOV-GP but not REBOV was shown to interfere with the integrity of the blood vessel endothelium (Yang et al., 2000). Our results demonstrate that the GPs of all four EBOV species depend on cathepsin B/L-activity for host cell entry. Sensitivity of ZEBOV-GP to inhibition by CA074Me was highest of all GPs tested, suggesting that ZEBOV-GP might be particularly dependent on activation by cathepsins B/L. It needs to be noted that CA074Me was more than 200-fold more active against cathepsin B compared to cathepsin L at the highest concentration used (4 μM), and the inhibitory effects observed in infection experiments might thus mainly stem from cathepsin B blockade. The dependence on the individual activities of cathepsin B and cathepsin L might vary between ZEBOV- and REBOV-GP, with the latter being insensitive to inhibition by CA074 but sensitive to blockade by cathepsin L inhibitor III. The reasons for these differences are at present unknown, but might relate to differences in the cathepsin B/L target sites. Cathepsin L cleavage sites in ZEBOV-GP have recently been proposed (Hood et al., 2010) and their sequences indeed differ between ZEBOV- and REBOV-GP. Moreover, Wong and colleagues selected EBOV-GP variants with amino acid changes, which allow entry into cells treated with a cathepsin B inhibitor, and some of the respective amino acids are not conserved between EBOV species (Wong et al., 2010). Alternatively, some GPs might be intrinsically more amenable to proteolysis than others and might thus exhibit a different requirement for cathepsin activity.
Entry of MARV-GP-bearing viruses into 293T cells and macrophages was largely resistant to the cathepsin B/L inhibitor CA074Me. These results are in agreement with a previous study showing that inhibition of cathepsin B/L efficiently blocked ZEBOV but not MARV entry into Vero E6 cells, although inhibition of viral replication was observed (Sanchez, 2007). In contrast, Huang and colleagues reported that engineered expression of cathepsin L in murine cathepsin L knock-out cells increased MARV-GP-mediated entry (Huang et al., 2006). The reasons for these discrepant results are at present unclear. Our observations suggest that MARV does either not require proteolytic activation of its GP for entry or that MARV and EBOV rely on different host cell proteases to ensure their activation. The finding that leupeptin, an inhibitor of cysteine and serine proteases, blocked MARV-GP-driven entry indicates that the latter is the case, but the responsible protease(s) remain(s) to be defined.
Many TTSPs play an important role in cancer development (Choi et al., 2009). Recent studies demonstrate that several of these proteins might also promote viral infections by facilitating activation of viral glycoproteins. Thus, TMPRSS2 was shown to activate influenza virus (Böttcher et al., 2006), human metapneumovirus (Shirogane et al., 2008) and SARS-CoV (Glowacka et al., 2011, Matsuyama et al., 2010, Shulla et al., 2011). The latter finding is remarkable, because it has previously been reported that SARS-CoV relies on cathepsins B and L for its activation (Simmons et al., 2005). The observation that cathepsin B/L-dependence is abrogated by TMPRSS2 suggests that virions are activated by TMPRSS2 during contact with the plasma membrane of target cells and then reach endo-/lysosomes, which contain cathepsins B and L, in an already activated from. Our results show that TMPRSS2 and the related protease hepsin can cleave REBOV- and/or CIEBOV-GP, potentially at the same site as furin. ZEBOV-GP and SEBOV-GP are presumably also cleaved by TMPRSS2 but cleavage could not be demonstrated because of the efficient processing of these proteins by endogenous furin in 293T cells (Wool-Lewis and Bates, 1999). However, cleavage by TMPRSS2 and hepsin did not result in GP activation and cathepsin B/L-independent cellular entry, in agreement with previous findings indicating that the furin cleavage site is dispensable for viral spread and pathogenicity (Neumann et al., 2002, Neumann et al., 2007). Thus, unlike the situation noted for SARS-CoV, TTSPs are unlikely to rescue EBOV from the antiviral effect of therapeutically applied cathepsin B/L-inhibitors.
Macrophages are key targets of EBOV in experimentally infected animals and humans and infection is believed to result in the uncontrolled release of cytokines, a hallmark of EBOV disease (Feldmann and Geisbert, 2011, Hartman et al., 2010). ZEBOV- and REBOV-GP-driven entry into macrophages was dependent on cathepsin B/L activity. These results suggest that cathepsin B/L inhibitors could display antiviral activity in EBOV infected individuals. However, a recent study suggested that ZEBOV-GP-driven entry into dendritic cells, which are targets of ZEBOV (Geisbert et al., 2003, Mahanty et al., 2003), requires cathepsin B but not L activity (Martinez et al., 2010). These findings, combined with the present observation that blockade of EBOV-GP-dependent entry by cathepsin inhibitors is frequently incomplete suggest that efficacy of inhibitors needs to be improved compared to presently available compounds in order to achieve a profound antiviral effect and such efforts are ongoing (Shah et al., 2010). In sum, cathepsins B and L are required for efficient EBOV- but not MARV-GP-driven infection of cell lines and macrophages.
Materials and methods
Cell culture
293T cells were maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS; Biochrom) and 1% penicillin/streptomycin. 293T-ACE2 (293T-232 cells, (Glowacka et al., 2010)) cells were cultured similar to 293T with additional 0.1% Zeocin (Invitrogen). Primary human monocyte-derived macrophages were cultured in monocyte differentiation medium (X-VIVO 10 medium (Lonza) supplemented with 1% human fibrin-depleted plasma and 1% penicillin/streptomycin). All cells were grown in a humidified atmosphere at 37 °C and 5% CO2.
Generation of primary human monocyte-derived macrophages
For primary human monocyte-derived macrophage preparation, the buffy coat of thrombocyte-depleted blood was collected upon Ficoll density gradient centrifugation. Buffy coats were washed three times by resuspension in ice-cold PBS and centrifugation at 2000, 1500 and 1200 rpm. Cells were resuspended in 40 ml monocyte adhesion medium (RPMI 1640 (PAA) supplemented with 7.5% human fibrin-depleted plasma and 1% penicillin/streptomycin) and seeded in two 15-cm tissue-culture dishes. After 45–90 min incubation non-adherent cells were removed by washing the cells seven times with 20 ml 37 °C warm PBS. The adherent cells were incubated in 20 ml monocyte adhesion medium overnight. The next day, the cells were washed with PBS, detached with StemPro Accutase (Invitrogen) and seeded into 96-well plates. Monocytes were differentiated into macrophages by culture in monocyte differentiation medium for seven days.
Plasmids
Expression plasmids encoding the glycoproteins of the EBOV species Zaire ebolavirus, Sudan ebolavirus, Cote d'Ivoire ebolavirus and Reston ebolavirus with or without a C-terminal V5-tag as well as MARV, SARS-coronavirus and vesicular stomatitis virus (VSV) were described previously (Hofmann et al., 2004, Kühl et al., 2011a, Kühl et al., 2011b, Marzi et al., 2006). Expression plasmids for proteases TMPRSS2 and hepsin were also described previously (Bertram et al., 2010a, Chaipan et al., 2009, Shirogane et al., 2008). For generation of human immunodeficiency virus (HIV) pseudotypes, plasmid pNL4-3 E–R–Luc was employed (Connor et al., 1995).
Western blotting
For detection of TMPRSS2- and hepsin-mediated cleavage of EBOV-GP, 293T cells seeded in 6-well plates were cotransfected with 5 μg of EBOV-GP and 1 μg protease expression plasmids employing calcium phosphate and transfection medium was replaced by fresh culture medium at 8 h post transfection. Cells were lysed at 48 h post transfection, proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes. The V5-tag was detected by using a mouse anti-V5 antibody (Invitrogen). As loading control, β-actin was detected employing a mouse monoclonal antibody (Sigma).
Production of pseudotypes
For generation of HIV-1 NL4-3-based pseudotypes carrying the different EBOV-GPs, SARS-S, VSV-G or no glycoprotein as control, cells were calcium phosphate-cotransfected with 12 μg DNA at a 1:1 ratio of pNL4-3 E–R–Luc and glycoprotein expression plasmid or empty plasmid in T25 tissue culture flasks. In some experiments, additionally 0.25 μg protease expression plasmids were cotransfected. Medium was changed at 16 h post transfection. At 48 h post transfection, supernatants were harvested, passed through 0.45 μm pore-size filters and concentrated using VivaSpin columns (Sartorius). Virus stocks were either normalized for equal p24-content by ELISA (Advanced Bioscience Laboratories) or comparable infectivity for 293T cells by luciferase assay (Promega).
Infection assay
For infection experiments, 293T cells or 293T-ACE2 cells were seeded in 96-well plates at a density of 5 × 104 cells/well. Monocytes were seeded and differentiated in 96-well plates at a density of 7.5 × 104 cells/well, and infected the latest at day 10 after addition of differentiation medium. Cells were infected with 50 μl of p24-antigen- or infectivity-normalized virus. 293T cells and macrophages were infected by spin-oculation (O'Doherty et al., 2000) at 2000 rpm and 4 °C for 2 h. Medium was exchanged 8 h post infection and infectivity was measured by determining luciferase activity in cell lysates at 72 h post infection using a commercially available kit (Promega).
Determination of protease inhibitor activity and specificity
For determination of inhibitor specificity, 293T cells were seeded and treated with protease inhibitors as described below for infection experiments. After 8 h cells were washed with PBS and lysed. Cathepsin B, L (Merck) and S (AnaSpec) activity in cell lysates was measured using commercially available kits according to the manufacturer's instructions.
Protease inhibitor experiments
Protease inhibitors CA074Me (Calbiochem), AEBSF (Roth), cathepsin L inhibitor III (Merck), MDL28170, E64c or leupeptin (Sigma) were diluted in PBS and used in the indicated concentrations. For protease inhibition, target cells were pretreated with the indicated inhibitors for 60 min and infected with infectivity-normalized pseudotypes in the presence of inhibitor. The inhibitor containing medium was replaced by fresh culture medium without inhibitor at 8 h post infection.
Acknowledgments
We thank T.F. Schulz for support and BMBF (grant 01KI 1005C, S.B. and I.G.), DFG (SFB 900, C.K.) and HBRS/Center for Infection Biology/DEWIN (K.G.) for funding.
References
- Aoyagi T., Takeuchi T., Matsuzaki A., Kawamura K., Kondo S. Leupeptins, new protease inhibitors from Actinomycetes. J. Antibiot. (Tokyo) 1969;22:283–286. doi: 10.7164/antibiotics.22.283. [DOI] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Blazejewska P., Soilleux E., Allen P., Danisch S., Steffen I., Choi S.Y., Park Y., Schneider H., Schughart K., Pöhlmann S. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 2010;84:10016–10025. doi: 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Steffen I., Kühl A., Pöhlmann S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev. Med. Virol. 2010;20:298–310. doi: 10.1002/rmv.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher E., Matrosovich T., Beyerle M., Klenk H.D., Garten W., Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush G., diSibio G., Miyamoto A., Denault J.B., Leduc R., Weinmaster G. Ligand-induced signaling in the absence of furin processing of Notch1. Dev. Biol. 2001;229:494–502. doi: 10.1006/dbio.2000.9992. [DOI] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., Dal Cin P., Dye J.M., Whelan S.P., Chandran K., Brummelkamp T.R. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaipan C., Kobasa D., Bertram S., Glowacka I., Steffen I., Tsegaye T.S., Takeda M., Bugge T.H., Kim S., Park Y., Marzi A., Pöhlmann S. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 2009;83:3200–3211. doi: 10.1128/JVI.02205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.Y., Bertram S., Glowacka I., Park Y.W., Pöhlmann S. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol. Med. 2009;15:303–312. doi: 10.1016/j.molmed.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Connor R.I., Chen B.K., Choe S., Landau N.R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Cote M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Small molecule inhibitors reveal Niemann–Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H., Geisbert T.W. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.W., Hensley L.E., Larsen T., Young H.A., Reed D.S., Geisbert J.B., Scott D.P., Kagan E., Jahrling P.B., Davis K.J. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 2003;163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., Simmons G., Hofmann H., Kuri T., Weber F., Eichler J., Drosten C., Pöhlmann S. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 2010;84:1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pöhlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenberger S., Bosch V., Angliker H., Shaw E., Klenk H.D., Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A.L., Towner J.S., Nichol S.T. Ebola and Marburg hemorrhagic fever. Clin. Lab Med. 2010;30:161–177. doi: 10.1016/j.cll.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M., Kuate S., Uberla K., Niedrig M., Pöhlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood C.L., Abraham J., Boyington J.C., Leung K., Kwong P.D., Nabel G.J. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J. Virol. 2010;84:2972–2982. doi: 10.1128/JVI.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bosch B.J., Li W., Farzan M., Rottier P.M., Choe H. SARS-CoV, but not HCoV-NL63, utilizes cathepsins to infect cells: viral entry. Adv. Exp. Med. Biol. 2006;581:335–338. doi: 10.1007/978-0-387-33012-9_60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R.L., Simmons G., Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl A., Banning C., Marzi A., Votteler J., Steffen I., Bertram S., Glowacka I., Konrad A., Sturzl M., Guo J.T., Schubert U., Feldmann H., Behrens G., Schindler M., Pöhlmann S. The Ebola virus glycoprotein and HIV-1 Vpu employ different strategies to counteract the antiviral factor tetherin. J. Infect. Dis. 2011;204(Suppl 3):S850–S860. doi: 10.1093/infdis/jir378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl A., Hoffmann M., Muller M.A., Munster V.J., Gnirss K., Kiene M., Tsegaye T.S., Behrens G., Herrler G., Feldmann H., Drosten C., Pöhlmann S. Comparative analysis of Ebola virus glycoprotein interactions with human and bat cells. J. Infect. Dis. 2011;204(Suppl 3):S840–S849. doi: 10.1093/infdis/jir306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty S., Hutchinson K., Agarwal S., McRae M., Rollin P.E., Pulendran B. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J. Immunol. 2003;170:2797–2801. doi: 10.4049/jimmunol.170.6.2797. [DOI] [PubMed] [Google Scholar]
- Martinez O., Johnson J., Manicassamy B., Rong L., Olinger G.G., Hensley L.E., Basler C.F. Zaire Ebola virus entry into human dendritic cells is insensitive to cathepsin L inhibition. Cell. Microbiol. 2010;12:148–157. doi: 10.1111/j.1462-5822.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A., Akhavan A., Simmons G., Gramberg T., Hofmann H., Bates P., Lingappa V.R., Pöhlmann S. The signal peptide of the ebolavirus glycoprotein influences interaction with the cellular lectins DC-SIGN and DC-SIGNR. J. Virol. 2006;80:6305–6317. doi: 10.1128/JVI.02545-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune J.M., Rabin L.B., Feinberg M.B., Lieberman M., Kosek K.J.C., Reyes G.R., Weissmann I.L. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- Mehdi S. Cell-penetrating inhibitors of calpain. Trends Biochem. Sci. 1991;16:150–153. doi: 10.1016/0968-0004(91)90058-4. [DOI] [PubMed] [Google Scholar]
- Neumann G., Feldmann H., Watanabe S., Lukashevich I., Kawaoka Y. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J. Virol. 2002;76:406–410. doi: 10.1128/JVI.76.1.406-410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Geisbert T.W., Ebihara H., Geisbert J.B., Daddario-DiCaprio K.M., Feldmann H., Kawaoka Y. Proteolytic processing of the Ebola virus glycoprotein is not critical for Ebola virus replication in nonhuman primates. J. Virol. 2007;81:2995–2998. doi: 10.1128/JVI.02486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty U., Swiggard W.J., Malim M.H. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. Analysis of filovirus entry into vero e6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J. Infect. Dis. 2007;196(Suppl 2):S251–S258. doi: 10.1086/520597. [DOI] [PubMed] [Google Scholar]
- Schornberg K., Matsuyama S., Kabsch K., Delos S., Bouton A., White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K.L., Shoemaker C.J., Dube D., Abshire M.Y., Delos S.E., Bouton A.H., White J.M. Alpha5beta1-integrin controls ebolavirus entry by regulating endosomal cathepsins. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8003–8008. doi: 10.1073/pnas.0807578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.P., Wang T., Kaletsky R.L., Myers M.C., Purvis J.E., Jing H., Huryn D.M., Greenbaum D.C., Smith A.B., III, Bates P., Diamond S.L. A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol. Pharmacol. 2010;78:319–324. doi: 10.1124/mol.110.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirogane Y., Takeda M., Iwasaki M., Ishiguro N., Takeuchi H., Nakatsu Y., Tahara M., Kikuta H., Yanagi Y. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 2008;82:8942–8946. doi: 10.1128/JVI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Watanabe S., Ito H., Okazaki K., Kida H., Kawaoka Y. Downregulation of beta1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology. 2000;278:20–26. doi: 10.1006/viro.2000.0601. [DOI] [PubMed] [Google Scholar]
- Takada A., Fujioka K., Tsuiji M., Morikawa A., Higashi N., Ebihara H., Kobasa D., Feldmann H., Irimura T., Kawaoka Y. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J. Virol. 2004;78:2943–2947. doi: 10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J.S., Sealy T.K., Khristova M.L., Albarino C.G., Conlan S., Reeder S.A., Quan P.L., Lipkin W.I., Downing R., Tappero J.W., Okware S., Lutwama J., Bakamutumaho B., Kayiwa J., Comer J.A., Rollin P.E., Ksiazek T.G., Nichol S.T. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS.Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volchkov V.E., Feldmann H., Volchkova V.A., Klenk H.D. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.C., Sandesara R.G., Mulherkar N., Whelan S.P., Chandran K. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 2010;84:163–175. doi: 10.1128/JVI.01832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wool-Lewis R.J., Bates P. Endoproteolytic processing of the ebola virus envelope glycoprotein: cleavage is not required for function. J. Virol. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Duckers H.J., Sullivan N.J., Sanchez A., Nabel E.G., Nabel G.J. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000;6:886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]
- Yano H., Torkin R., Martin L.A., Chao M.V., Teng K.K. Proneurotrophin-3 is a neuronal apoptotic ligand: evidence for retrograde-directed cell killing. J. Neurosci. 2009;29:14790–14802. doi: 10.1523/JNEUROSCI.2059-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii H., Kamiyama H., Minematsu K., Goto K., Mizota T., Oishi K., Katunuma N., Yamamoto N., Kubo Y. Cathepsin L is required for ecotropic murine leukemia virus infection in NIH3T3 cells. Virology. 2009;394:227–234. doi: 10.1016/j.virol.2009.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]