Abstract
The Middle East respiratory syndrome coronavirus (MERS-CoV) closely resembled severe acute respiratory syndrome coronavirus (SARS-CoV) in disease manifestation as rapidly progressive acute pneumonia with multi-organ dysfunction. Using monocyte-derived-dendritic cells (Mo-DCs), we discovered fundamental discrepancies in the outcome of MERS‐CoV‐ and SARS-CoV-infection. First, MERS-CoV productively infected Mo-DCs while SARS-CoV-infection was abortive. Second, MERS-CoV induced significantly higher levels of IFN-γ, IP-10, IL-12, and RANTES expression than SARS-CoV. Third, MERS-CoV-infection induced higher surface expression of MHC class II (HLA-DR) and the co-stimulatory molecule CD86 than SARS-CoV-infection. Overall, our data suggests that the dendritic cell can serve as an important target of viral replication and a vehicle for dissemination. MERS-CoV-infection in DCs results in the production of a rich combination of cytokines and chemokines, and modulates innate immune response differently from that of SARS-CoV-infection. Our findings may help to explain the apparent discrepancy in the pathogenicity between MERS-CoV and SARS-CoV.
Keywords: MERS-CoV, SARS-CoV, Viral replication, Pathogenesis, Cytokine and chemokine response, Antigen-presentation
Highlights
-
•
MERS-CoV productively infected Mo-DCs while SARS-CoV-infection was abortive.
-
•
MERS-CoV induced higher levels of IFN-γ, IP-10, IL-12, and RANTES than SARS-CoV.
-
•
MERS-CoV induced higher surface expression of MHC II and CD86 than SARS-CoV.
Introduction
The novel Middle East respiratory syndrome coronavirus (MERS-CoV), previously also known as human coronavirus (HCoV) EMC, was first identified in a patient who died from severe acute pneumonia and renal failure in Bisha, Saudi Arabia, in September 2012 (Chan et al., 2013b, Chan et al., 2012, de Groot et al., 2013, Zaki et al., 2012). Although a zoonotic source has been suspected in bats and camels, the definitive and intermediate animal reservoirs have so far not been confirmed, which may have partly contributed to the continuing spread of the epidemic (Annan et al., 2013, Chan et al., 2013c, Chan et al., 2013, Lau et al., 2013, Reusken et al., 2013, Woo et al., 2007, Woo et al., 2012). As of February 7th, 2014, a total of 182 laboratory-confirmed cases of MERS-CoV infection with 79 fatalities have been reported in the Middle East, Europe, and Africa (WHO, 2014). The evolving outbreak has raised global concern of a SARS-like epidemic particularly because of their comparably protean clinical manifestations involving both the respiratory tract and extrapulmonary tissues, and the unusually high crude fatality rate among infected patients (Assiri et al., 2013b, Chan et al., 2012, Guery et al., 2013, Memish et al., 2013). These clinical observations corroborated with several key experimental findings on the high pathogenicity of MERS-CoV. First, MERS-CoV replicated more rapidly and showed a much broader tissue tropism in-vitro than any other coronaviruses associated with human infections including SARS-CoV (Chan et al., 2013a). Second, using human lung epithelial cell line as a model, it was shown that MERS-CoV induced a massive dysregulation of the host transcriptome, which may prevent the host from mounting an optimal immune response (Josset et al., 2013). Third, the utilization of dipeptidyl peptidase 4 (DPP4) instead of angiotensin-converting enzyme 2 (ACE2; SARS-CoV and HCoV-NL63), aminopeptidase N (HCoV-229E), and 9-O-acetylated sialic acid (HCoV-OC43) as a functional receptor by MERS-CoV may also account for the differences in the spectrum of organ involvement between MERS and other human coronavirus infections (Du et al., 2013, Raj et al., 2013). Finally, there is currently no specific anti-MERS-CoV treatment proven to be effective in clinical trials, though mycophenolic acid, interferons, ribavirin, cyclosporin A, and an HR2 peptide have demonstrated in-vitro activities (Chan et al., 2013d; de Wilde et al., 2013; Falzarano et al., 2013; Lu et al., 2014).
Despite these initial clinical and laboratory correlations, the virulence of MERS-CoV has remained controversial especially after a recent study which reported that most of the patients who required hospitalization or died from MERS were elderlies with comorbidities, specifically diabetes mellitus, hypertension, and chronic renal, heart and lung diseases (Assiri et al., 2013b). In contrast, healthy children and adults might be asymptomatic or develop mild infection detected only during contact tracing (Assiri et al., 2013b, Chan et al., 2013b). Furthermore, many MERS patients developed extrapulmonary manifestations including renal impairment, hepatic dysfunction, gastrointestinal symptoms, coagulopathy, cytopenias, and pericarditis (Assiri et al., 2013a, Chan et al., 2013b). The mechanisms by which these extrapulmonary tissues are involved in MERS are incompletely understood. MERS-CoV RNA was detected in mononuclear cells and stellate cells in the mediastinal lymph nodes of infected rhesus macaques, hinting the possibility of macrophage or dendritic cell involvement (de Wit et al., 2013). More in-depth studies on the pathogenesis of MERS-CoV are urgently needed to ascertain its virulence and pathogenetic mechanisms for more accurate prediction of its clinical behavior and better design of therapeutic options. Given the ability of MERS-CoV to infect different human immune cell lines including monocytes and T lymphocytes, we postulated that it might also be able to infect dendritic cells (DCs), which are potent antigen presenting cells (APCs) with instrumental roles in linking the innate and adaptive immune systems. Although previous literatures suggested that SARS-CoV was unable to establish a productive infection in DCs, we suspected that MERS-CoV might be able to infect DCs in a more efficient manner due to the wide distribution of its cellular receptor DPP4 in human cells including activated leukocytes (Raj et al., 2013).
DCs are key players of the innate immune system. As perhaps the most potent APCs, DCs are capable of entering peripheral tissues, taking up antigens, migrating to lymphoid tissues, and activating helper T cells (Banchereau and Steinman, 1998, Steinman and Banchereau, 2007). In this capacity, DCs play unique roles in bridging the innate and the adaptive immunity and therefore become important targets for microbial invasion. For instance, HIV has evolved a number of mechanisms to exploit the function of DCs for its own benefit, including escaping from the immune surveillance and facilitating cell-to-cell dissemination (Wu and KewalRamani, 2006).
DCs are susceptible to SARS-CoV infection. However, as suggested by a number of studies, the infection was either abortive (Law et al., 2005, Tseng et al., 2005, Ziegler et al., 2005) or at low level (Spiegel et al., 2006) and caused no adverse effect to cell viability. Infection of DCs by SARS-CoV failed to trigger a strong type I interferon (IFN) response but resulted in an up-regulation of inflammatory cytokines and chemokines (Law et al., 2005, Tseng et al., 2005). Functional activation of DCs by SARS-CoV has been analyzed but the result was inconclusive as both activation (Spiegel et al., 2006, Tseng et al., 2005) and absence of activation (Ziegler et al., 2005) have been proposed. In the current study, we used monocyte-derived DCs (Mo-DCs) as a system to recapitulate MERS-CoV infection in DCs and we compared the results with that of SARS-CoV-infected Mo-DCs. Our results revealed fundamental differences in the replication kinetics of MERS-CoV and SARS-CoV in infected Mo-DCs. In addition, we also demonstrated characteristic changes in the pattern of cytokine/chemokine expression as well as antigen-presenting function of infected Mo-DCs.
Results
Monocyte-derived-dendritic cells (Mo-DCs) were susceptible to MERS-CoV infection
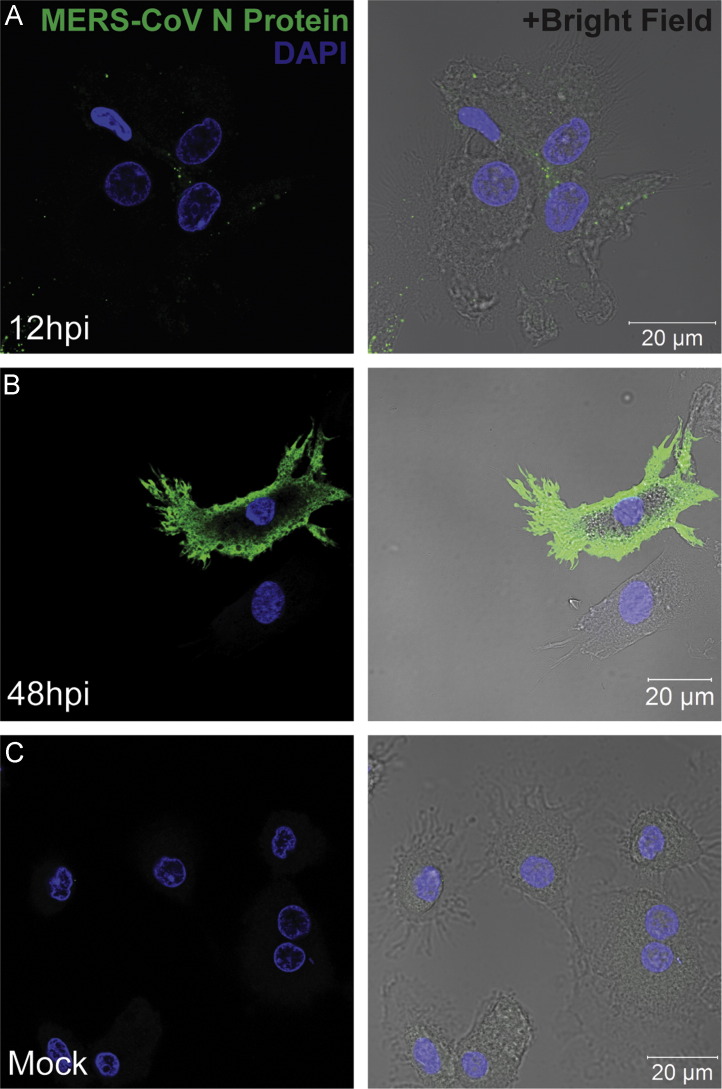
To determine whether primary human DCs are susceptible to MERS-CoV infection, we infected Mo-DCs with MERS-CoV and examined the expression of MERS-CoV nucleoprotein (NP) at different time points post inoculation. Our data revealed that MERS-CoV NP expression could be detected at 12 hours post infection (hpi) ( Fig. 1A). The signal for NP appeared to be puncta-like and distributed in the cytoplasm of infected cells. At 48 hpi, the signal for NP was dramatically enhanced and was detected throughout the cytoplasm of infected cells (Fig. 1B). Mock-infected cells (Fig. 1C) as well as cells treated with pre-immune sera (data not shown) both failed to display any signal for NP. Our immunostaining study revealed that Mo-DCs were susceptible to MERS-CoV infection. The enhanced expression of NP at 48 hpi versus 12 hpi suggested that MERS-CoV not only infected Mo-DCs but also continued viral transcription and translation in these infected cells. In addition, the percent of Mo-DCs infected by MERS-CoV was assessed by flow cytometry and was determined to be 12.6%±1.2% at 48 hpi (data not shown).
Fig. 1.
Detection of MERS-CoV nucleocapsid protein (NP) in MERS-CoV-infected Mo-DCs with immunofluorescence staining. Mo-DCs were seeded on glass coverslips in 24-well plates at a density of 1×105 cells per well and infected with MERS-CoV at 2 TCID50/cell for 1 h. Infected Mo-DCs were fixed at 12 hpi (A) and 48 hpi (B) and processed for immunostaining. MERS-CoV NP was detected with guinea pig anti-NP antibody followed by FITC-conjugated rabbit-anti-guinea pig IgG. The slides were mounted with a mounting buffer that contained DAPI and examined with a Carl Zeiss LSM 710 microscope. Mock-infected Mo-DCs that were fixed at 48 hpi are shown in panel (C).
MERS-CoV infection in Mo-DCs was productive
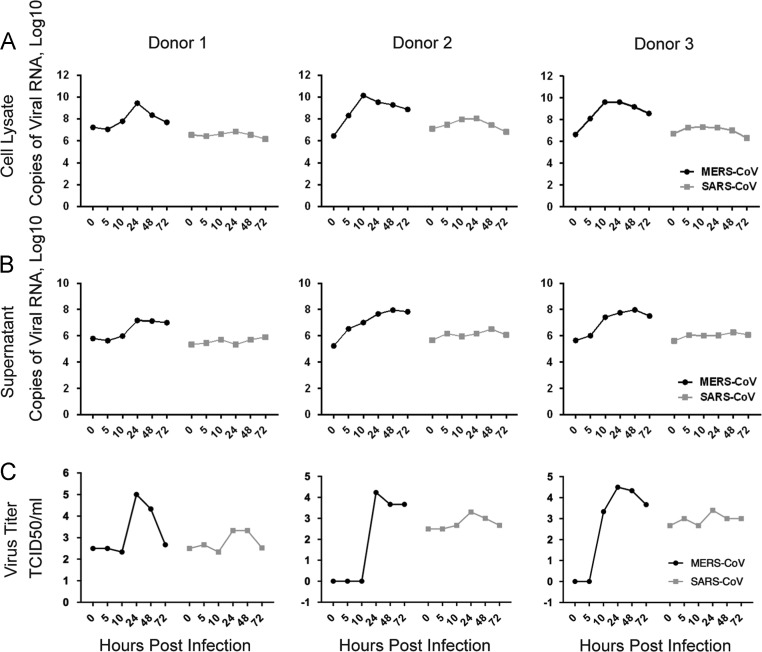
With the immunofluorescence study described above, we illustrated that MERS-CoV was capable of infecting DCs with efficient transcription and translation of the viral genome, which was supported by the substantial increase in cellular NP expression (Fig. 1). To obtain a more comprehensive picture of the kinetics of MERS-CoV infection in Mo-DCs, we infected Mo-DCs with MERS-CoV and compared the results with that of SARS-CoV-infected cells. Importantly, a 2-to-4 log increase in viral RNA in both cell lysates ( Fig. 2A) and supernatants (Fig. 2B) of MERS-CoV-infected Mo-DCs was detected in samples from all donors. In stark contrast, little or no increase in viral RNA was detected in the cell lysates or supernatants of SARS-CoV-infected Mo-DCs, which agreed with previous reports that SARS-CoV could infect but was unable to propagate in Mo-DCs. We further assessed the infectivity of viral particles released from infected Mo-DCs with TCID50 assays. Our results demonstrated that while low levels of infectious particles were detected from the supernatants of SARS-CoV-infected Mo-DCs, MERS-CoV-infected Mo-DCs consistently released a considerable amount of infectious particles with a peak at 24 hpi (Fig. 2C). Overall, our data suggested that MERS-CoV could establish a productive infection in Mo-DCs.
Fig. 2.
Replication kinetics of MERS-CoV and SARS-CoV in infected Mo-DCs. Mo-DCs were infected with MERS-CoV or SARS-CoV at 2 TCID50 per cell for 1 h. Supernatants and cell lysates were harvested at the indicated time points and processed for RNA extraction, reverse transcription, and quantitative PCR. The copy number of the positive-sense viral genomic RNA are illustrated for cell lysate (A) and supernatant (B). The number of infectious particles was quantified with TCID50 assays (C). Data from three representative experiments are shown.
MERS-CoV triggered stronger cytokine and chemokine response than SARS-CoV in Mo-DCs
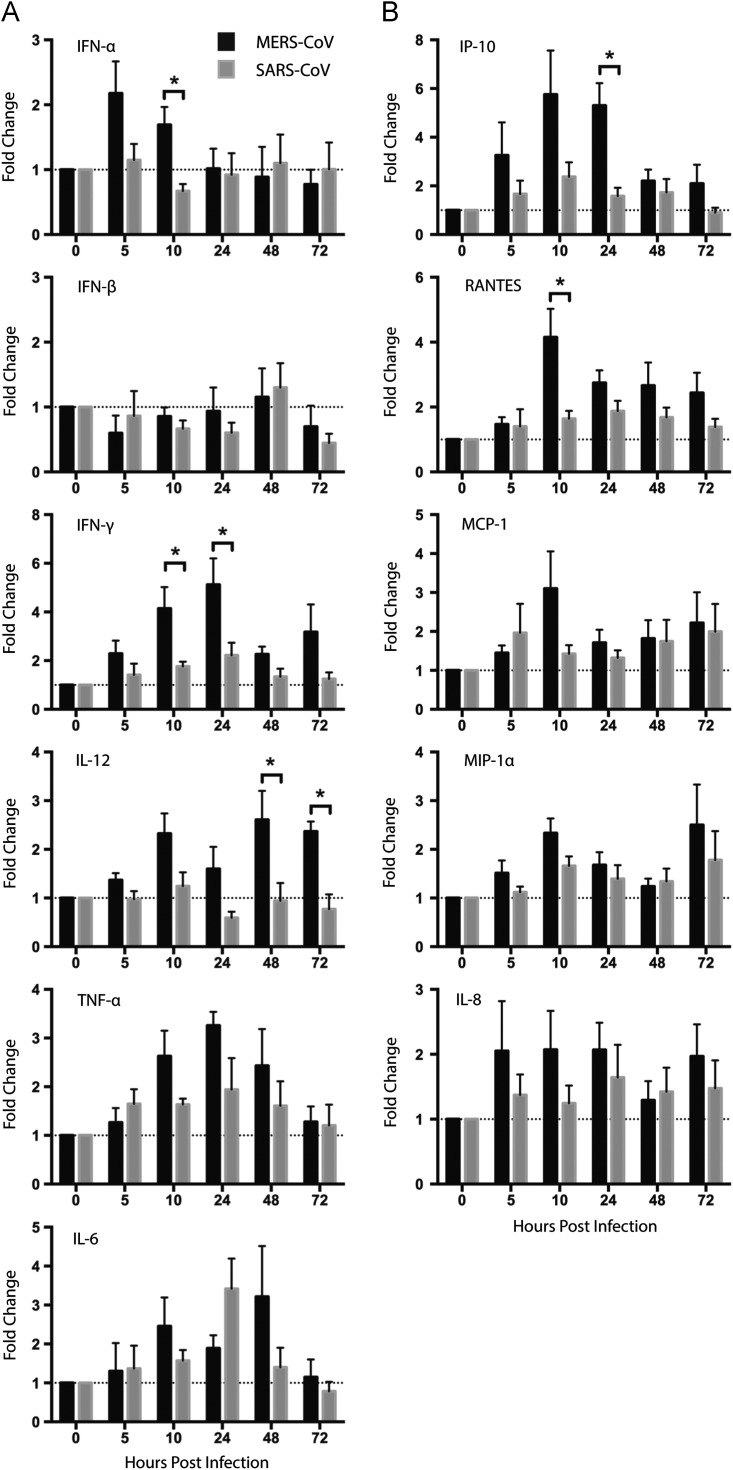
DCs are among the first line of innate immune response and are capable of producing a large number of cytokines and chemokines upon microbial challenge (Banchereau and Steinman, 1998). We examined the ability of MERS-CoV to trigger cytokine and chemokine response in Mo-DCs and compared the results with that of SARS-CoV-infected Mo-DCs. Mo-DCs were infected with MERS-CoV or SARS-CoV at 2 TCID50 per cell for one hour and cell lysates were harvested at 0, 5, 10, 24, 48, and 72 hpi. The expression of cytokine and chemokine genes was quantified with RT-qPCR and normalized first with GAPDH and then with the gene expression from the mock-infected cells. Our data recapitulated the lack of IFN-α and IFN-β induction in SARS-CoV-infected Mo-DCs. At the same time, while MERS-CoV similarly failed to trigger the expression of IFN-β, it induced an up-regulation of IFN-α at the early time points. As shown in Fig. 3A, SARS-CoV marginally up-regulated the expression of IFN-γ and did not induce an up-regulation of IL-12. On the other hand, MERS-CoV induced significantly higher expression levels of IFN-γ and IL-12 than SARS-CoV. Pro-inflammatory cytokine TNF-α and IL-6 were similarly up-regulated in MERS-CoV-and SARS-CoV-infected Mo-DCs. The peak fold inductions for TNF-α and IL-6 were documented between 10 hpi and 48 hpi and were at comparable levels for MERS-CoV and SARS-CoV (Fig. 3A).
Fig. 3.
Quantitations of cytokine and chemokine induction in MERS-CoV- or SARS-CoV-infected Mo-DCs. Mo-DCs were infected with MERS-CoV or SARS-CoV at 2 TCID50 per cell for 1 h. At the indicated time points post infection, cells were lysed for RNA extraction, RT and qPCR to detect the mRNA expression level of antiviral and proinflammatory cytokines (A) as well as a number of representative chemokines (B). Results were normalized to GAPDH and presented as fold change in gene expression in relation to mock-infected Mo-DCs. Data represented the mean and SEM from at least five donors. Statistical analysis was performed between MERS-CoV- and SARS-CoV-infected Mo-DCs for all time points in all panels using Student׳s t test. p<0.05 was considered to be statistically significant and is indicated by asterisk marks.
In parallel, we quantified the expression levels of a number of representative inflammatory and chemotactic chemokines, including IP-10, IL-8, MCP-1, MIP-1α and RANTES. The degree of induction by MERS-CoV and SARS-CoV on IL-8, MCP-1, and MIP-1α were subtle with comparable levels. In contrast, MERS-CoV induced significantly higher levels of IP-10 and RANTES than SARS-CoV in Mo-DCs (Fig. 3B).
MERS-CoV induced higher surface expression of antigen-presentation and co-stimulatory molecules than SARS-CoV in Mo-DCs
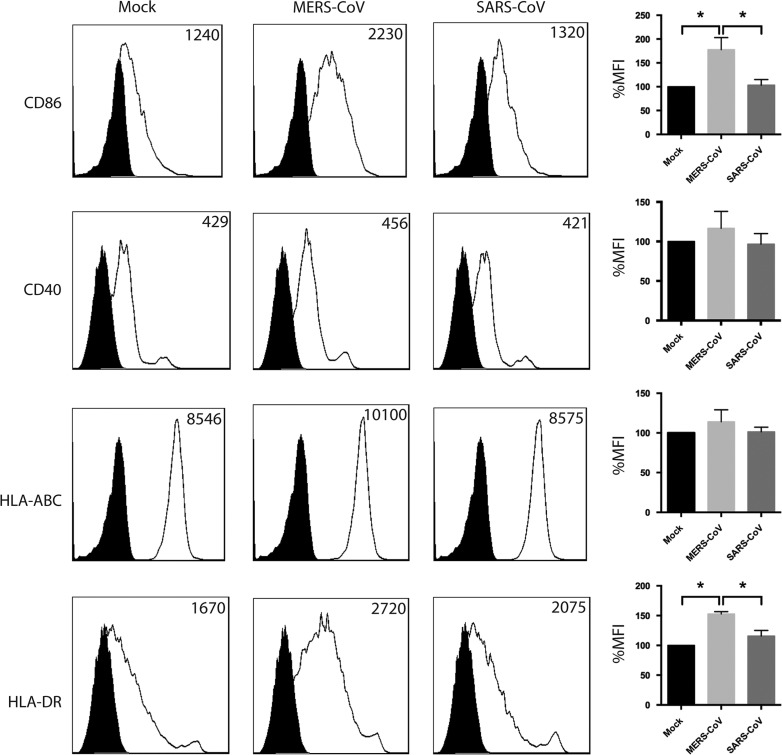
Dendritic cells are professional antigen presenting cells that play critical roles in bridging the innate and the adaptive immunity. Upon activation by foreign pathogens, DCs prime naive T cells and trigger antigen-specific T cell responses. We assessed the degree of activation of Mo-DCs upon MERS-CoV or SARS-CoV challenge by measuring the surface expression of MHC class I (HLA-ABC), MHC class II (HLA-DR), and co-stimulatory molecules (CD40, CD86). Mo-DCs were infected with MERS-CoV or SARS-CoV at 2 TCID50 per cell for one hour. At 48 hpi, the cells were detached, fixed, immunolabeled for surface makers for co-stimulation (CD40, CD86), MHC Class I (HLA-ABC), MHC Class II (HLA-DR), and analyzed with flow cytometry. As shown in Fig. 4, while the surface expression of CD40 and MHC Class I (HLA-ABC) were not significantly higher in MERS-CoV-infected Mo-DCs compared with that of mock-infected or SARS-CoV-infected Mo-DCs, the surface expression of CD86 and MHC Class II (HLA-DR) were significantly increased in MERS-CoV-infected Mo-DCs. Summaries of the change in the mean fluorescence intensity were illustrated in the right panels. Intriguingly, little change in surface expression of MHC and co-stimulatory molecules were observed in SARS-CoV-infected Mo-DCs (Fig. 4). Taken together, our result demonstrated that MERS-CoV was more capable of inducing Mo-DC activation than SARS-CoV.
Fig. 4.
Surface expression of antigen-presentation and co-stimulatory molecules in Mo-DCs infected with MERS-CoV or SARS-CoV. Mo-DCs were infected with MERS-CoV or SARS-CoV at 2 TCID50 per cell for 1 h. At 48 hpi, cells were detached, fixed, and labeled for specific cellular surface markers. The expression of surface markers was assessed by flow cytometry using a BD FACSCanto II flow cytometer and the data was analyzed using FlowJo. The numbers in each panel represent mean fluorescent intensity. The filled curve represents the staining of isotype controls. Results in the right panels represent mean %MFI and SEM summarized from four independent experiments. Statistical analysis was performed between all groups using Student׳s t test. p<0.05 was considered to be statistically significant and is indicated by asterisk marks.
Discussion
Previous studies suggested that SARS-CoV could infect Mo-DCs. However, there was a marked reduction in viral genome copy over time and a notable absence of infectious progeny from the inoculated cells, which indicated that the infection, if any, was at low level (Spiegel et al., 2006) or was abortive (Law et al., 2005, Tseng et al., 2005). The absence of SARS-CoV NP expression in Mo-DCs suggested that the viral replication cycle was incomplete (Ziegler et al., 2005). In our current study, we demonstrated conclusively that MERS-CoV could productively infect Mo-DCs. First, the immunofluorescence study showed a progressive increase in NP signal in infected Mo-DCs over time (Fig. 1). Second, the positive-strand viral genome of MERS-CoV increased by 2-to-4 logs in the supernatants and cell lysates of infected Mo-DCs (Fig. 2A and B). Third, TCID50 assays performed using supernatants from MERS-CoV-infected Mo-DCs demonstrated that MERS-CoV produced a substantial amount of infectious virus particles with a peak production time at around 24 hpi (Fig. 2C). Clinically, patients who contracted MERS-CoV developed coagulopathy, multi-organ dysfunction and hematological changes including lymphopenia, neutrophilia and thrombocytopenia. The virus could be detected not only in the respiratory tract, but also in the blood (Guery et al., 2013), urine (Drosten et al., 2013), and stool (Drosten et al., 2013) of infected individuals, which strongly argued for a systemic dissemination of the virus. In this regard, MERS-CoV-infected DCs and monocyte-derived macrophages (MDMs) (Zhou et al., 2013) could potentially serve as vehicles of dissemination and facilitate the spread of the virus to the lymph nodes, where they interact with T cells, and further worsen the cytokine/chemokine storm, mimicking the scenario in SARS (Gu et al., 2005).
Currently, the exact reason of why MERS-CoV but not SARS-CoV could productively infect DCs is unclear. Differential infection outcome with MERS-CoV and SARS-CoV may be partially attributed to the difference in the level of receptor expression. ACE2 (Li et al., 2003) and DPP4 (Raj et al., 2013) are the functional receptors of SARS-CoV and MERS-CoV in humans, respectively. Previous reports have suggested that ACE2 is not expressed in monocytes or DCs (Law et al., 2005). A separate group suggested that ACE2 could be detected by Western Blots in MDMs and DCs but surface expression of ACE2 could not be detected by flow cytometry (Tseng et al., 2005). Our own analysis by RT-qPCR showed that while Mo-DCs express high level of DPP4, the expression level of ACE2 is marginal and is close to the detection limit (unpublished data). In addition, SARS-CoV uses DC-SIGN and L-SIGN as alternative receptors (Han et al., 2007, Jeffers et al., 2004). DCs are known to express abundant levels of C-type lectins, which facilitates their antigen-uptake (Figdor et al., 2002). For instance, DCs internalize and sequester HIV particles through DC-SIGN, which is exploited by HIV to facilitate efficient cell-to-cell trans-infection (Geijtenbeek et al., 2000). A similar mechanism is also utilized by hepatitis C virus in trans-infection of liver cells (Cormier et al., 2004). Therefore, it is possible that while MERS-CoV infects DCs with virus production, SARS-CoV is primarily sequestered by DCs. Further investigation on this possibility will be necessary.
DCs are important stimulators of T cells and B cells for specific immune responses. In addition, upon encountering pathogens, DCs are capable of releasing a large number of cytokines and chemokines. Infection of Mo-DCs by SARS-CoV did not induce up-regulation of anti-viral interferons, IFN-α and IFN-β, but resulted in the up-regulation of other pro-inflammatory cytokines and chemokines (Law et al., 2005, Tseng et al., 2005). Recent research demonstrated that MERS-CoV failed to trigger anti-viral interferon or pro-inflammatory innate immune response in primary human airway epithelial cells and in cultured cells (Chan et al., 2013, Zielecki et al., 2013). In the current study, we reported that MERS-CoV induced no IFN-β and marginal IFN-α expression in infected Mo-DCs. MERS-CoV induced comparable levels of pro-inflammatory cytokine IL-6 and TNF-α compared to SARS-CoV. In parallel, the expression level of chemokine IL-8, MCP-1, and MIP-1α was subtle and was not significantly different between MERS-CoV- and SARS-CoV-infected Mo-DCs. Most importantly, MERS-CoV triggered a significantly higher expression level of IFN-γ, IL-12, IP-10, and RANTES, compared to SARS-CoV-infected Mo-DCs (Fig. 3). In SARS-CoV-infected patients, viral replication was limited to the first two weeks after symptom onset, without continued widespread replication after this period (Nicholls et al., 2006). On the other hand, a number of cytokines and chemokines including but not limited to IFN-γ, IL-12, and IP-10 were found to be highly up-regulated in SARS-CoV-infected patients even after viral clearance (Huang et al., 2005, Jiang et al., 2005, Wong et al., 2004). The resulting immune dysregulation including an IFN-γ mediated cytokine storm might have accounted for a large proportion of SARS-associated mortality (Huang et al., 2005). In the current study, we showed that MERS-CoV induced significantly higher expression level of IFN-γ, IL-12, IP-10, and RANTES in infected Mo-DCs, compared to SARS-CoV. While IP-10 and RANTES both recruit T cells, IL-12 mediates the differentiation of naive T cells into Th1 cells and stimulates the production of IFN-γ from T cells, representing a positive feedback loop. Our data raised the possibility of an IFN-γ-induced, Th1 cell-mediated immunity and hyperactive inflammatory response in MERS-CoV-infected patients causing severe diseases before host adaptive immune response could be mounted.
In the capacity of professional antigen-presenting cells, DCs capture and process both endogenous and exogenous antigens and present the processed antigens through MHC class I or MHC class II molecules. During DC maturation, antigen capturing, processing, and presentation are accompanied with an increase in the surface expression of co-stimulatory molecules, including CD40, CD80, and CD86, which are critical for T cell signaling and activation. Previous studies performed using SARS-CoV-infected Mo-DCs indicated that while the expression of MHC class I was comparable in infected and mock-infected cells, the expression of MHC class II and co-stimulatory molecules (CD40, CD80 and CD86) were up-regulated (Spiegel et al., 2006, Tseng et al., 2005), although conflicting results existed (Ziegler et al., 2005). In this study we demonstrated that MERS-CoV induced higher surface expression of MHC class II (HLA-DR), and co-stimulatory molecule CD86 than SARS-CoV in infected Mo-DCs (Fig. 4). Although only around 13% of Mo-DCs were infected by MERS-CoV, it appeared that CD86 and HLA-DR were up-regulated in a much larger proportion of cells, which could be a result of paracrine signaling from the infected Mo-DCs to the un-infected cells since the expressions of a number of key cytokines and chemokines were up-regulated in MERS-CoV-infected Mo-DCs. As a result of the up-regulated MHC class II (HLA-DR) and co-stimulation molecule CD86, the robust activation of T cells may contribute to an exaggerated T cell function, which may lead to more destruction of infected pulmonary tissues when adaptive immune response is mounted later in the course of illness. Alternatively, activated T cells also produce high levels of inflammatory cytokines that may potentially aggravate the cytokine cascade.
Taken together, our study demonstrated that MERS-CoV could productively infect DCs, thereby giving itself an extra edge in systemic dissemination over SARS-CoV. At the same time, MERS-CoV induced higher expression of IFN-γ and IFN-γ-related cytokines and chemokines in infected DCs than SARS-CoV with comparable levels of TNF-α and IL-6 mRNA expression. In addition, antigen-presentation and co-stimulatory molecules are higher up-regulated in MERS-CoV-infected DCs than SARS-CoV-infected DCs. Overall, our study may help to confirm the high pathogenicity of MERS-CoV and explain the protean clinical manifestation seen in MERS. Further investigation on the underlying mechanisms will aid in the development of effective therapy for MERS.
Materials and methods
Preparation of Mo-DCs
Healthy volunteer blood samples were collected from Hong Kong Red Cross Blood Transfusion Service according to a protocol approved by the Institutional Review Board of the University of Hong Kong. Monocyte preparation and differentiation were performed according to a well established protocol we described previously (Chu et al., 2012). In brief, peripheral blood mononuclear cells (PBMCs) were obtained from buffy coats after Ficoll centrifugation and were allowed to adhere to plastic surface coated with poly-d-lysine (Sigma Aldrich, Saint Louis, MO, USA) for 1 h. Adherent monocytes were then differentiated into Mo-DCs according to established protocols. Specially, the cells were treated with RPMI-1640 media supplemented with 10% FBS, 100 ug/ml streptomycin, 100 U/ml penicillin, 2 mM glutamine, 1% sodium pyruvate, 1% non-essential amino acids, 10 ng/ml recombinant human granulocyte macrophage colony-stimulating factor (GM-CSF), and 10 ng/ml recombinant human interleukin-4 (IL-4) (R&D Systems, Minneapolis, MN, USA) for differentiation into Mo-DCs. The Mo-DCs were differentiated for 8 days before used for infection, during which the media was replaced every 2 days.
Virus
SARS-CoV was cultured in FRhK-4 cells in serum free DMEM supplemented with penicillin and streptomycin. MERS-CoV was kindly provided by Dr. Fouchier׳s laboratory (Zaki et al., 2012) and cultured in Vero cells in the same medium. Two to three days post infection, supernatants were collected, aliquoted, and stored at −80 °C. 50% tissue culture infective dose (TCID50) assays were performed to quantify viral titers.
Virus Infections
For infections, Mo-DCs were plated in 24-well plates and infected with MERS-CoV or SARS-CoV at 2 TCID50/cell for one hour at 37 °C. Supernatants and cell lysates were harvested at 0, 5, 10, 24, 48, and 72 hpi. For immunostaining, Mo-DCs were plated in 24-well plates on glass cover slips. The cells were infected with MERS-CoV at 2 TCID50/cell. At 12 and 48 hpi, the cells were fixed with 4% paraformaldehyde and underwent immunostaining procedures.
Quantification of viral and cellular RNA transcript by RT-qPCR
Cellular RNA extraction, reverse transcription and quantitative PCR were performed as we described previously (Zhou et al., 2011). Supernatants from infected cells were extracted with the PureLink Viral RNA/DNA mini kit (Life Technologies). Reverse primers against the NP gene of MERS-CoV and SARS-CoV were used to retrieve cDNA complementary to the positive strand of viral genomes and Oligo(dT) was used to reverse transcribe cellular cDNAs. The levels of cellular gene expression were normalized to GAPDH and presented as fold change in gene expression of infected Mo-DCs relative to that of mock-infected Mo-DCs. Specific primer sequences are available upon request.
Immunofluorescence staining
Mo-DCs cultured on glass cover slips were infected with MERS-CoV at 2 TCID50 per cell for 1 h at 37 °C. At 12 and 48 hpi, cells were fixed in 4% paraformaldehyde, permeabilized, blocked, and immunostained for MERS-CoV NP. MERS-CoV NP was detected using guinea pig anti-NP antibody as we previously described (Chan et al., 2013a, Zhou et al., 2013), followed by FITC-conjugated rabbit-anti-guinea pig IgG (Life Technologies). Slides were mounted with ProLong Gold antifade reagent with DAPI (Life Technologies) and examined with a Carl Zeiss LSM 710 microscope.
Flow cytometry
Mo-DCs were infected with MERS-CoV or SARS-CoV at 2 TCID50 per cell for 1 h at 37 °C. At 48 hpi, infected cells were detached with 10 mM EDTA, fixed in 4% paraformaldehyde, and immunolabeled with specific cell surface markers. APC-anti-human CD40, APC-anti-human CD86, APC- anti-human HLA-ABC, APC-anti-human HLA-DR, and APC-IgG isotype control were ordered from Biolegend (SanDiego, CA, USA). For infectivity assessment, infected Mo-DCs were fixed, permeabilized, and immunolabeled first with guinea pig anti-MERS-CoV NP antibody as we previously described (Chan et al., 2013a, Zhou et al., 2013), followed by FITC-conjugated rabbit anti-guinea pig IgG (Life Technologies). The flow cytometry was performed using a BD FACSCanto II flow cytometer (BD Biosciences, San Jose, CA, USA) and data was analyzed using FlowJo (Tree Star, Inc).
Statistical analysis
Experimental results represented mean and SEM from at least three different donors. Statistical comparison between the groups was performed by Student׳s t-test using GraphPad Prism 6. Differences were considered statistically significant when p<0.05.
Financial support
This work was supported by The Providence Foundation Limited in memory of the late Dr Lui Hac Minh, and the HKSAR Health and Medical Research Fund (HMRF).
Acknowledgments
We thank Dr. Ron Fouchier for providing us the MERS-CoV isolate. We acknowledge the Core Imaging Facility, Li Ka Shing Faculty of Medicine, the University of Hong Kong, for facilitation of the study.
References
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K., Lina P.H., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human Betacoronavirus 2c EMC/2012-related Viruses in Bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F., Makhdoom H.Q., Zumla A.I., Memish Z.A. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., Madani H., Alhakeem R., Al-Tawfiq J.A., Cotten M., Watson S.J., Kellam P., Zumla A.I., Memish Z.A. Hospital Outbreak of Middle East Respiratory Syndrome Coronavirus. N. Engl. J. Med. 2013;369(5):407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Chan K.H., Choi G.K., To K.K., Tse H., Cai J.P., Yeung M.L., Cheng V.C., Chen H., Che X.Y., Lau S.K., Woo P.C., Yuen K.Y. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013;207:1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Lau S.K., Woo P.C. The emerging novel Middle East respiratory syndrome coronavirus: The "knowns" and "unknowns". J. Formos. Med. Assoc.=Taiwan yi zhi. 2013;112:372–381. doi: 10.1016/j.jfma.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Li K.S, To K.K., Cheng V.C., Chen H., Yuen K.Y. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J. Infect. 2012;65:477–489. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., To K.K., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013 doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Chan K.H., Kao R.Y., To K.K., Zheng B.J., Li C.P., Li P.T., Dai J., Mok F.K., Chen H., Hayden F.G., Yuen K.Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.H., Chan J.F., Tse H., Chen H., Lau C.C., Cai J.P., Tsang A.K., Xiao X., To K.K., Lau S.K., Woo P.C., Zheng B.J., Wang M., Yuen K.Y. Cross-reactive antibodies in convalescent SARS patients׳ sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J. Infect. 2013;67:130–140. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R.W., Chan M.C., Agnihothram S., Chan L.L., Kuok D.I., Fong J.H., Guan Y., Poon L.L., Baric R.S., Nicholls J.M., Peiris J.S. Tropism and innate immune responses of the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 2013 doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Wang J.J., Qi M., Yoon J.J., Chen X., Wen X., Hammonds J., Ding L., Spearman P. Tetherin/BST-2 is essential for the formation of the intracellular virus-containing compartment in HIV-infected macrophages. Cell Host Microbe. 2012;12:360–372. doi: 10.1016/j.chom.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier E.G., Durso R.J., Tsamis F., Boussemart L., Manix C., Olson W.C., Gardner J.P., Dragic T. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. USA. 2004;101:14067–14072. doi: 10.1073/pnas.0405695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W., Posthuma C.C., van der Meer Y., Barcena M., Haagmans B.L., Snijder E.J., van den Hoogen B.G. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Rasmussen A.L., Falzarano D., Bushmaker T., Feldmann F., Brining D.L., Fischer E.R., Martellaro C., Okumura A., Chang J., Scott D., Benecke A.G., Katze M.G., Feldmann H., Munster V.J. Vol. 110. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques; pp. 16598–16603. (Proc. Natl. Acad. Sci. USA). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Seilmaier M., Corman V.M., Hartmann W., Scheible G., Sack S., Guggemos W., Kallies R., Muth D., Junglen S., Muller M.A., Haas W., Guberina H., Rohnisch T., Schmid-Wendtner M., Aldabbagh S., Dittmer U., Gold H., Graf P., Bonin F., Rambaut A., Wendtner C.M. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 2013;13(9):745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Kou Z., Ma C., Sun S., Poon V.K., Lu L., Wang L., Debnath A.K., Zheng B.J., Zhou Y., Jiang S. Identification of a receptor-binding domain in the s protein of the novel human coronavirus middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 2013;87:9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Martellaro C., Callison J., Munster V.J., Feldmann H. Inhibition of novel beta coronavirus replication by a combination of interferon-alpha2b and ribavirin. Sci. Rep. 2013;3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figdor C.G., van Kooyk Y., Adema G.J. C-type lectin receptors on dendritic cells and Langerhans cells. Nat. Rev. Immunol. 2002;2:77–84. doi: 10.1038/nri723. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B., Kwon D.S., Torensma R., van Vliet S.J., van Duijnhoven G.C., Middel J., Cornelissen I.L., Nottet H.S., KewalRamani V.N., Littman D.R., Figdor C.G., van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z., Zhuang H., Wu B., Zhong H., Shao H., Fang W., Gao D., Pei F., Li X., He Z., Xu D., Shi X., Anderson V.M., Leong A.S. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guery B., Poissy J., el Mansouf L., Sejourne C., Ettahar N., Lemaire X., Vuotto F., Goffard A., Behillil S., Enouf V., Caro V., Mailles A., Che D., Manuguerra J.C., Mathieu D., Fontanet A., van der Werf S., group M.E.-C.s. Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.P., Lohani M., Cho M.W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007;81:12029–12039. doi: 10.1128/JVI.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.J., Su I.J., Theron M., Wu Y.C., Lai S.K., Liu C.C., Lei H.Y. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., Demartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Xu J., Zhou C., Wu Z., Zhong S., Liu J., Luo W., Chen T., Qin Q., Deng P. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am. J. Respir Crit. Care Med. 2005;171:850–857. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- Josset L., Menachery V.D., Gralinski L.E., Agnihothram S., Sova P., Carter V.S., Yount B.L., Graham R.L., Baric R.S., Katze M.G. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4:e00165–00113. doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Li K.S., Tsang A.K., Lam C.S., Ahmed S., Chen H., Chan K.H., Woo P.C., Yuen K.Y. Genetic characterization of Betacoronavirus Lineage C viruses in bats reveals marked sequence divergence in the spike protein of Pipistrellus bat Coronavirus HKU5 in Japanese Pipistrelle: implications for the origin of the novel middle east respiratory syndrome coronavirus. J. Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law H.K., Cheung C.Y., Ng H.Y., Sia S.F., Chan Y.O., Luk W., Nicholls J.M., Peiris J.S., Lau Y.L. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhiu Y., Chan K.H., Qin L., Li Y., Wang Q., Chan J.F., Du L., Yu F., Ma C., Ye S., Yuen K.Y., Zhang R., Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Zumla A.I., Assiri A. Middle East respiratory syndrome coronavirus infections in health care workers. N. Engl. J. Med. 2013;369:884–886. doi: 10.1056/NEJMc1308698. [DOI] [PubMed] [Google Scholar]
- Nicholls J.M., Butany J., Poon L.L., Chan K.H., Beh S.L., Poutanen S., Peiris J.S., Wong M. Time course and cellular localization of SARS-CoV nucleoprotein and RNA in lungs from fatal cases of SARS. PLoS Med. 2006;3:e27. doi: 10.1371/journal.pmed.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Vries L.S., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortazar-Schmidt C., Drosten C., Koopmans M.P. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013 doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Schneider K., Weber F., Weidmann M., Hufert F.T. Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J. Gener. Virol. 2006;87:1953–1960. doi: 10.1099/vir.0.81624-0. [DOI] [PubMed] [Google Scholar]
- Steinman R.M., Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- Tseng C.T., Perrone L.A., Zhu H., Makino S., Peters C.J. Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. J. Immunol. 2005;174:7977–7985. doi: 10.4049/jimmunol.174.12.7977. [DOI] [PubMed] [Google Scholar]
- WHO, Middle East respiratory syndrome coronavirus (MERS-CoV) - update of Feb 7 2014 〈http://www.who.int/csr/don/2014_02_07mers/en/index.html〉
- Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clini. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Li L.S., Tsang KS, Yuen KY AK. Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg. Microbes Infect. 2012:1. doi: 10.1038/emi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Wang M., Lau S.K., Xu H., Poon R.W., Guo R., Wong B.H., Gao K., Tsoi H.W., Huang Y., Li K.S., Lam C.S., Chan K.H., Zheng B.J., Yuen K.Y. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., KewalRamani V.N. Dendritic-cell interactions with HIV: infection and viral dissemination. Nat. Rev. Immunol. 2006;6:859–868. doi: 10.1038/nri1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhou J., Chu H., Li C., Wong B.H., Cheng Z.S., Poon V.K., Sun T., Lau C.C., Wong K.K., Chan J.Y., Chan J.F., To K.K., Chan K.H., Zheng B.J., Yuen K.Y. Active replication of middle east respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2013 doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Tan T., Tian Y., Zheng B., Ou J.H., Huang E.J., Yen T.S. Kruppel-like factor 15 activates hepatitis B virus gene expression and replication. Hepatology. 2011;54:109–121. doi: 10.1002/hep.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler T., Matikainen S., Ronkko E., Osterlund P., Sillanpaa M., Siren J., Fagerlund R., Immonen M., Melen K., Julkunen I. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J. Virol. 2005;79:13800–13805. doi: 10.1128/JVI.79.21.13800-13805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielecki F., Weber M., Eickmann M., Spiegelberg L., Zaki A.M., Matrosovich M., Becker S., Weber F. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J. Virol. 2013;87:5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]