Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is known to interfere with the signaling of type I interferons (IFNs). Here we found PRRSV A2MC2 induced type I IFNs in cultured cells. A2MC2 replication in MARC-145 cells resulted in the synthesis of IFN-α2 protein, transcript elevation of the IFN-stimulated genes ISG15 and ISG56, and the proteins of the signal transducer and activator of transcription 2 (STAT2) and ISG56. A2MC2 infection of primary porcine pulmonary alveolar macrophages (PAMs) also led to the elevation of the two proteins, but had little cytopathic effect. Furthermore, A2MC2 infection of MARC-145 or PAM cells had no detectable inhibitory effect on the ability of IFN-α to induce an antiviral response. Sequencing analysis indicated that A2MC2 was closely related to VR-2332 and Ingelvac PRRS MLV with an identity of 99.8% at the nucleotide level. The identification of this IFN-inducing PRRSV isolate may be beneficial for vaccine development against PRRS.
Keywords: Interferon induction, Porcine reproductive and respiratory syndrome virus (PRRSV), IFN, IFN signaling
Highlights
► We discovered a PRRSV isolate A2MC2 that induces synthesis of type I interferons. ► A2MC2 induces IFN production in both MARC-145 cells and PAMs. ► A2MC2 is a Type 2 PRRSV closely related to VR-2332 and MLV strains. ► Like MLV strain, A2MC2 infection of PAMs leads to undetectable adverse effect.
Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV) is a positive-sense single-stranded RNA virus belonging to the family Arteriviridae (Faaberg et al., 2011, Meulenberg, 2000). It causes an economically important disease, resulting in an estimated $560 million loss per year to the swine industry in the United States (Neumann et al., 2005). PRRSV is known to inhibit the synthesis of type I interferons (IFNs) in infected pigs (Albina et al., 1998, Buddaert et al., 1998). IFNs are not detectable in the lungs of pigs, in which PRRSV actively replicates. PRRSV-infected pigs develop delayed and low titer neutralizing antibodies (Labarque et al., 2000) and weak cell-mediated immune responses (Xiao et al., 2004). Suppression of innate immunity may be an important contributing factor to PRRSV modulation of host immune responses.
PRRSV can be propagated in vitro in an epithelial-derived monkey kidney cell line, MARC-145 (Kim et al., 1993), and in primary cultures of porcine pulmonary alveolar macrophages (PAMs). PAMs are the main target cells for PRRSV during its acute infection of pigs (Rossow et al., 1995). PRRSV infection of PAM and MARC-145 cells in vitro leads to a very low expression of interferon-α (IFN-α) for viral strains studied to date (Albina et al., 1998, Lee et al., 2004, Miller et al., 2004).
Type I IFNs, such as IFN-α and -β, are critical to innate immunity against viral infection and contribute to the modulation of adaptive immunity (Takaoka and Yanai, 2006). The innate immune system is activated after cellular pattern recognition receptors (PRR) sense pathogen associated molecular patterns (PAMPs) of invading pathogens. Host PRRs for RNA viruses include Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs). Activation of the TLR or RLR pathways eventually leads to the secretion of type I IFNs. The binding of type I IFNs to their receptors activates the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway, which induces expression of IFN-stimulated genes (ISGs) and results in the establishment of an antiviral state (Darnell et al., 1994, Schindler and Darnell, 1995, Stark et al., 1998).
Some PRRSV strains suppress IFN-β expression in MARC-145 cells and PRRSV non-structural proteins (nsp) 1, 2, 4, and 11 inhibit IFN induction when over-expressed (Beura et al., 2010, Chen et al., 2010, Kim et al., 2010). PRRSV can also inhibit IFN downstream signaling and expression of ISGs in both MARC-145 and PAM cells (Patel et al., 2010). The nuclear translocation of STAT1/STAT2/IRF9 heterotrimers was blocked in PRRSV-infected cells, while the IFN-induced phosphorylation of STAT1 and STAT2 was not affected.
Many efforts to control PRRS have been attempted, but few are successful because of the antigenic and genomic diversity among PRRSV isolates, and persistence of the virus in infected herds. Attenuated live virus vaccines have been commercially available for years; however, outbreaks of PRRS resulting from viral strains nearly identical in sequence to the vaccine strain have been reported (Botner et al., 1997, Opriessnig et al., 2002). Outbreaks of atypical or acute PRRS in vaccinated pigs have raised serious concern about the efficacy and safety of the current vaccines (Madsen et al., 1998, Meng, 2000).
In the present study, we discovered a PRRSV isolate that induced production of type I IFNs in both MARC-145 and PAM cells and appeared to have an undetectable inhibitory effect on the ability of IFN-α to induce an antiviral response. Its infection of PAMs led to an undetectable cytopathic effect. Characterization of this unique isolate will be beneficial to the study of PRRSV-cell interactions and the development of improved vaccines.
Results
Detection of antiviral activity in cell culture supernatant from A2MC2-infected MARC-145 cells
In studying PRRSV interference of IFN signaling, we discovered one PRRSV cell culture isolate that did not inhibit IFN signaling but induced antiviral activity in MARC-145 cells. After plaque purification of this isolate three times, one plaque was named A2MC2.
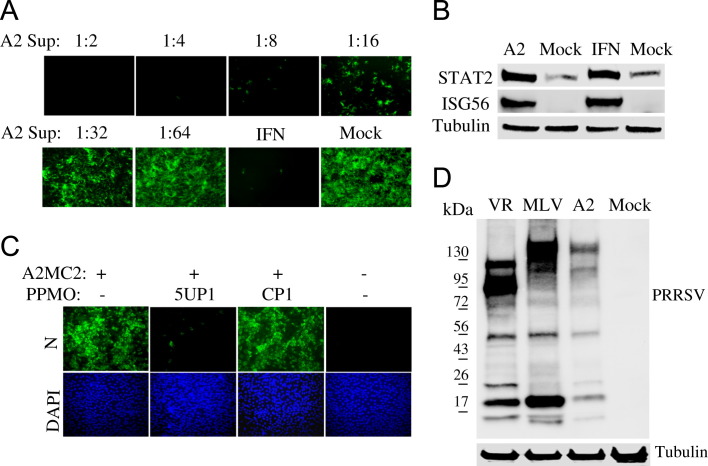
Vero cells are not susceptible to PRRSV infection and were used as an indicator cell line for our studies. NDV-GFP is sensitive to type I IFNs, so pre-treatment of Vero cells with IFN-α inhibited NDV-GFP replication and was included as an assay control. Treatment of Vero cells with dilutions of A2MC2-infected MARC-145 cell culture supernatant reduced the number of NDV-GFP-positive cells ( Fig. 1A), which indicated that the NDV replication was inhibited. This result suggested the existence of type I IFNs in the culture supernatant of the A2MC2-infected cells.
Fig. 1.
Detection of antiviral activity in cell culture supernatant from A2MC2-infected MARC-145 cells. (A) Inhibition of NDV-GFP replication in Vero cells. Vero cells were treated with dilutions of cell culture supernatant of A2MC2-infected MARC-145 cells. The Vero cells were inoculated with NDV-GFP 12 h after the treatment, and observed under fluorescence microscopy at 24 h post-infection. Treatment of the cells with IFN-α at a final concentration of 1000 U/ml was included as a positive control. (B) Elevation of STAT2 and ISG56 proteins in Vero cells after treatment with the supernatant from A2MC2-infected MARC-145 cells detected by Western blot analysis. Vero cells treated with IFN-α and mock-treated were included as positive and negative controls, respectively. Blotting with β-tubulin antibody was done to normalize protein loading. (C) Inhibition of A2MC2 replication in MARC-145 cells by PRRSV-specific peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO) 5UP1. A scrambled control PPMO CP1 was included as a negative control. An indirect immunofluorescence assay with PRRSV N-specific monoclonal antibody was conducted. The bottom panel of images shows the nuclear DNA stained with 4′,6-19 diamidino-2-phenylindole (DAPI). (D) Detection of PRRSV proteins in whole cell lysate of A2MC2-infected cells (A2 lane) by Western blotting with pig antiserum. Cell lysate samples from PRRSV VR-2385-infected (VR lane) or MLV-infected cells were included as positive controls. Molecular weight markers are illustrated on the left.
To further confirm that the antiviral activity was due to interferons, Western blot analysis was conducted to assess the protein levels of interferon-stimulated gene 56 (ISG56) and STAT2, in Vero cells. Blotting results showed that both ISG56 and STAT2 were elevated after treatment with the cell culture supernatant from A2MC2-infected MARC-145 cells (Fig. 1B). The levels of the proteins were similar to those of cells treated with 1000 U/ml IFN-α. These results suggested that A2MC2 induced synthesis of type I IFNs in MARC-145 cells.
PRRSV is known to inhibit production of type I IFNs. To confirm that A2MC2 is a genuine PRRSV isolate, an inhibition assay was performed by using antisense peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO), 5UP1, which inhibits replication of PRRSV in MARC-145 cells in a sequence-specific manner (Zhang et al., 2006). A scrambled control PPMO CP1 was included as a negative control. An indirect immunofluorescence assay showed that 5UP1 blocked A2MC2 replication in MARC-145 cells (Fig. 1C), while CP1 had no effect. The lysate of A2MC2-infected MARC-145 cells was used for Western blot analysis with pig antiserum against PRRSV. Lysate samples of VR-2385- and MLV-infected cells were included as controls. Blotting results showed that A2MC2-infected cells had a band profile similar to MLV, though the bands were at a weaker intensity (Fig. 1D). The difference in band pattern between VR-2385 and MLV is likely because there is a deletion in nsp2 of VR-2385 (Ni et al., 2011). These results confirmed that A2MC2 was a strain of PRRSV. Genotyping of this isolate was then performed.
Genotyping of PRRSV A2MC2 strain
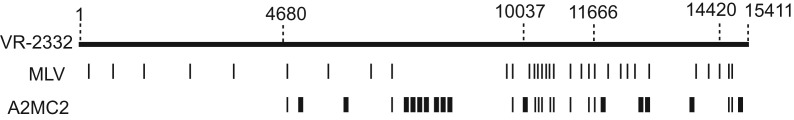
RT-PCR was conducted for the whole A2MC2 RNA genome. Sequencing of the cDNA was done and sequence analysis showed that it closely resembles Ingelvac PRRS MLV (GenBank ID: AF066183) and VR-2332 (GenBank ID: U87392), strains of genotype 2 PRRSV, at identity of 99.8%. There were a total of 28 nucleotide (nt) variations when compared to VR-2332, resulting in 14 amino acid changes (Supplementary Table 1). The nucleotide variations are scattered from nt 4681 to the end of the genome ( Fig. 2). The first 4680 nucleotides are identical to VR-2332. There were a total of 34 nucleotide variations when compared to strain MLV, resulting in 19 different amino acids. Compared to both VR-2332 and MLV, A2MC2 has 15 unique nucleotides scattered from nt 4681 to the end of the genome (Fig. 2). Ten of the unique changes are between nt 4681 and nt 10037 of the A2MC2 genome. The sequence from nt 11667 to 14420 of A2MC2 is the same as VR-2332 except 4 unique nucleotide variations. The sequence from nt 14421 to the end of the A2MC2 genome is the same as MLV except 1 unique nucleotide variation.
Fig. 2.
Illustration of sequence variation of A2MC2 in comparison to VR-2332 and MLV. The top line indicates the genomic sequence of VR-2332 and the numbers above the line indicate nucleotide positions in the genome. The nucleotide variations of MLV in comparison with VR-2332 are indicated by narrow vertical bars. The nucleotide variations of A2MC2 in comparison with VR-2332 are indicated by both narrow and wide vertical bars, among which the narrow vertical bars indicates they are the same as MLV and the wide bars indicates they are unique for A2MC2 sequence.
At the amino acid level, the A2MC2 differences when compared to VR-2332 were located in nsp3, nsp7, nsp8, nsp10, nsp11, nsp12, GP3, and M; and the variations from MLV sequence were located in nsp1β, nsp2, nsp8, nsp10, nsp11, nsp12, GP2, GP3, GP5, and M (Supplementary Table 1). Six unique amino acid changes occurred in A2MC2 when compared to VR-2332 and MLV: threonine to serine in nsp8, serine to alanine and proline to leucine in nsp10, serine to glycine in nsp12, methionine to valine, and isoleucine to valine in GP3 ( Table 1). Nsp10 is a RNA helicase that unwinds dsRNA (Bautista et al., 2002), while the functions of nsp8 and nsp12 are unknown. GP3 is a glycoprotein found in PRRSV virions as a minor structural component (de Lima et al., 2009). The genomic sequence of A2MC2 when compared to both that of VR-2332 and MLV indicated that A2MC2 was possibly a chimera derived from these two strains.
Table 1.
Nucleotide variations in A2MC2 sequence leading to unique amino acid changes compared with both MLV and VR-2332a.
| Positionb | Nucleotidec |
Amino acidd |
Proteine | ||||
|---|---|---|---|---|---|---|---|
| A2MC2 | MLV | VR-2332 | A2MC2 | MLV | VR-2332 | ||
| 7621 | T | A | A | S | T | T | nsp8 |
| 9627 | G | T | T | A | S | S | nsp10 |
| 9655 | T | C | C | L | P | P | nsp10 |
| 12012 | G | A | A | G | S | S | nsp12 |
| 12972 | G | A | A | V | M | M | GP3 |
| 12975 | G | A | A | V | I | I | GP3 |
GenBank accession numbers: VR-2332 (GenBank ID: U87392), MLV (GenBank ID: AF066183), and A2MC2 (GenBank ID: JQ087873).
Nucleotide positions are indicated on left column based on VR-2332 sequence.
Nucleotides at the indicated positions are listed.
Amino acids derived from the codon of indicated nucleotides are listed.
Proteins corresponding to the amino acids derived from the codon of indicated nucleotide positions are listed on the right column.
Growth properties of A2MC2 in MARC-145 and PAM cells
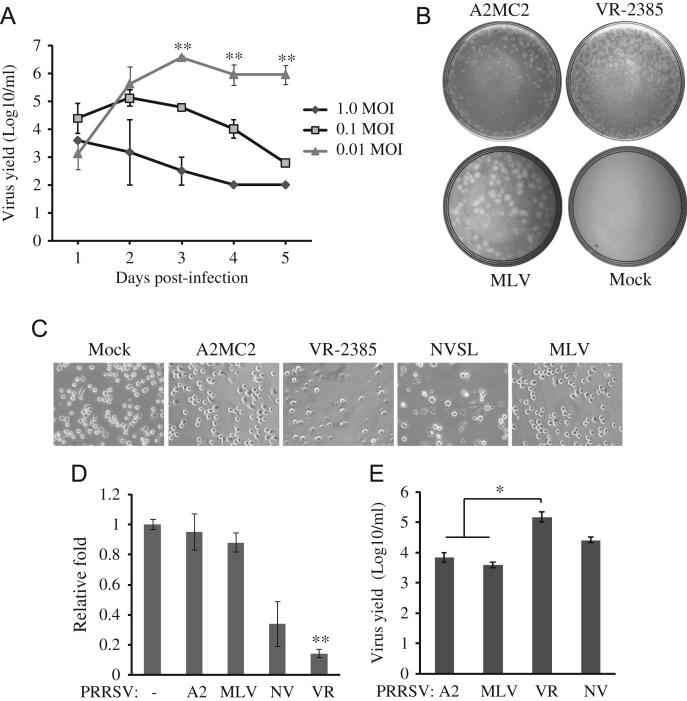
To determine the growth properties of A2MC2 in MARC-145 cells, a multi-step growth curve analysis was conducted. The cells were inoculated at a MOI of 0.01, 0.1 and 1 TCID50 per cell, respectively. Cell culture supernatant samples were collected daily for five days after the inoculation and titrated for virus yield. The cells inoculated with 0.01 TCID50 per cell had the highest virus yield, 106.67 TCID50 per ml on day 3, while the cells inoculated with 1 TCID50 per cell had the lowest yield, lower than 103 ( Fig. 3A). The virus yields of cells with 0.01 TCID50 increased from 103 on day 1 to 106.67 on day 3, and remained at 106 on day 5. The virus yield of cells with 1 TCID50 per cell decreased from 103.5 on day 1 to 102.5 on day 5. The virus yields of cells with 0.01 TCID50 on day 3, 4 and 5 were significantly higher than those from cells with 0.01 and 1 TCID50.
Fig. 3.
Growth properties of A2MC2 in MARC-145 and PAM cells. (A) Multi-step growth curve of A2MC2 in MARC-145 cells. The cells were inoculated with A2MC2 virus at a multiplicity of infection of 0.01, 0.1 or 1 TCID50 per cell. Virus yields at different time points after inoculation were titrated by an immunofluorescence assay. Error bars represent variation of three repeated experiments. Significant differences in virus yields between cells with 0.01 TCID50 and the rest two other groups are denoted by “**”, which indicate P-value of<0.01. (B) Plaque assay completed in MARC-145 cells. The cells were infected with diluted A2MC2, VR-2385 or MLV and overlaid with agarose. A plate of mock-infected cells was included as a negative control. Plaques were revealed at 4 dpi and photographed for comparison. (C) Cytopathic effect in PRRSV-infected PAMs. PAM cells were inoculated with PRRSV and at 20 hpi, observed using bright field microscopy. Mock-infected cells were included for comparison. PRRSV VR-2385 and NVSL led to cell death and lysis, while A2MC2 and MLV had little cytopathic effect. (D) Cell viability assay of PAMs. PRRSV-infected PAMs were assayed at 20 hpi with CellTiter-Glo kit (Promega). Relative fold cell viability in comparison with uninfected PAMs were plotted. Only VR-2385-infected cells had significantly lower viability (denoted by “**”, indicating P<0.01) viability than uninfected PAMs. A2: A2MC2, NV: NVSL, VR: VR-2385. (E) Virus yield titrated using MARC-145 cells. Cell culture supernatant samples from PRRSV-infected PAMs at 24 hpi were titrated in MARC-145 cells by IFA. Median tissue culture infectious dose per ml is shown. Error bars represent variation of three repeated experiments. The virus yields of A2MC2 and MLV were significantly lower (denoted by “*”, indicating P<0.05) than VR-2385.
To further characterize the growth properties of A2MC2, plaque assays were conducted in comparison with VR-2385 and MLV strains. MARC-145 cells were inoculated with A2MC2, VR-2385, and MLV, respectively. Plaques were observed on 4 dpi after neutral red staining. The A2MC2-infected cells resulted in a small plaque morphology at around less than 1 mm in diameter, similar to VR-2385, while cells infected with MLV revealed a plaque morphology at around 5 mm in diameter, which was at least 5 times larger than those of A2MC2 (Fig. 3B). This result indicated that A2MC2 replication in MARC-145 cells was different from that of MLV.
To test if A2MC2 caused cytopathic effects (CPE) after infection of PAM cells, as it does in MARC-145 cells, we inoculated primary PAMs with PRRSV at the MOI of 0.05 TCID50 per cell and observed the cells at 20 hpi under bright field microscopy. A2MC2 infection of PAMs caused no observable CPE, while VR-2385 and NVSL led to cell death (Fig. 3C). A2MC2-infected cells appeared similar to MLV-infected or uninfected PAM cells in morphology. A cell viability assay was conducted to assess the relative viability level between treatments. A2MC2-infected PAM cells showed a similar viability rate as was seen in uninfected cells, as did MLV infection, while VR-2385 significantly reduced viability to 0.14-fold (Fig. 3D). This result was consistent with the CPE observed under bright field microscopy.
To determine virus yield in PAMs, cell culture supernatant was collected at 24 hpi and titrated in MARC-145 cells by IFA. The virus yields of A2MC2, MLV, VR-2385, and NVSL were 103.8, 103.6, 105.2, and 104.4 TCID50 per ml, respectively (Fig. 3E). The results showed that viral yields of A2MC2 and MLV were significantly lower than VR-2385 (P<0.05), but did not vary much from the viral yield of NVSL.
A2MC2 replication induces strong expression of STAT2 and ISG56 in MARC-145 cells
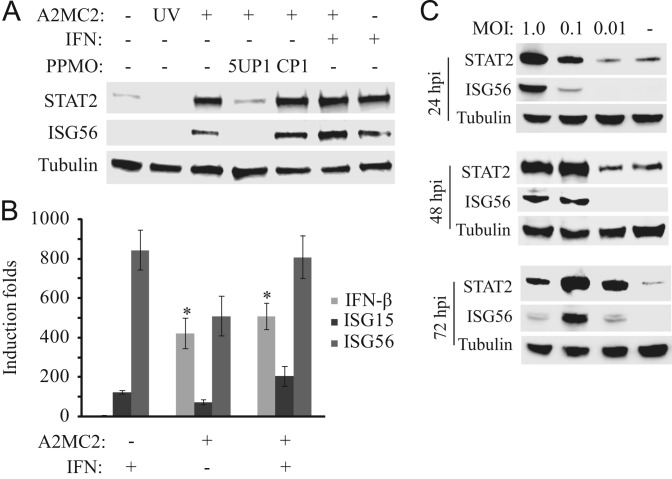
To determine if A2MC2 replication induces IFN-stimulated genes in MARC-145 cells, the cells were infected with the virus at 1 TCID50 per cell and harvested at 24 h post-infection (hpi). Western blot analysis showed that the levels of STAT2 and ISG56 in MARC-145 cells were remarkably elevated after A2MC2 infection ( Fig. 4A). Treatment of A2MC2-infected cells with PPMO 5UP1 abolished the elevation, which indicated that the inhibition of A2MC2 removed the stimulation. UV-inactivated A2MC2 failed to induce the elevation of these two proteins, indicating that the elevation was A2MC2 replication-dependent.
Fig. 4.
A2MC2 replication induces elevated expression of IFN-stimulated genes in MARC-145 cells. (A) Elevation of STAT2 and ISG56 detected by Western blotting. The cells were infected with A2MC2 or UV-inactivated virus at 1 TCID50 per cell, followed by treatment with PPMO 5UP1 to inhibit A2MC2 replication, and at 24 hpi, treated with or without IFN-α. Cell lysate from uninfected cells was included as a control. (B) Elevation of IFN-β, ISG15 and ISG56 expression detected by real-time PCR. Treatment of the cells with IFN-α was included as a control. Relative induction folds in comparison with mock-treated cells are shown. Error bars represent variation between repeated experiments. Significant differences between A2MC2-infected cells and the uninfected cells are denoted by “*”, which indicates a P-value of<0.05. (C) Kinetics of STAT2 and ISG56 expression in MARC-145 cells infected with different MOIs of A2MC2. The cells were harvested at 24, 48 and 72 hpi for Western blot amalyses. Samples of uninfected cell lysates were included as controls.
Real-time RT-PCR was conducted to detect the transcripts of IFN-β, ISG15, and ISG56 in MARC-145 cells. Compared to mock-treated control wells, A2MC2 infection at 1 TCID50 per cell induced 422-, 73-, and 509-fold RNA elevations of IFN-β, ISG15, and ISG56, respectively (Fig. 4B). The IFN-β transcript in A2MC2-infected cells was 105-fold higher than that of IFN-α-treated PRRSV-negative cells. The average levels of ISG15 and ISG56 transcripts in A2MC2-infected cells without external IFN-α were 1.68- and 1.6-fold, respectively, lower than those in IFN-α-treated PRRSV-negative cells. The differences of ISG15 and ISG56 between the A2MC2-infected and the IFN-a-treated uninfected cells were statistically insignificant. Addition of IFN-α to A2MC2-infected cells did not lead to a significant increase in expression of these three genes, compared to A2MC2-infected cells without external IFN-α.
As different MOI led to variable virus yields in MARC-145 cells, the protein levels of STAT2 and ISG56 in the cells after infection with different MOIs were assessed. Western blotting showed that the greater the MOI that was used to infect MARC-145 cells, the higher the level of STAT2 and ISG56 at 24 hpi detected (Fig. 4C). By 48 hpi, the cells with 0.1 TCID50 had similar levels of these two proteins to 1 TCID50. By 72 hpi, the cells with 1 TCID50 had the lowest level of these two proteins. The results indicated that the high MOI inoculum was able to induce early synthesis of the two proteins, while a low MOI led to delayed induction.
Comparison with other PRRSV strains on IFN production in MARC-145 cells
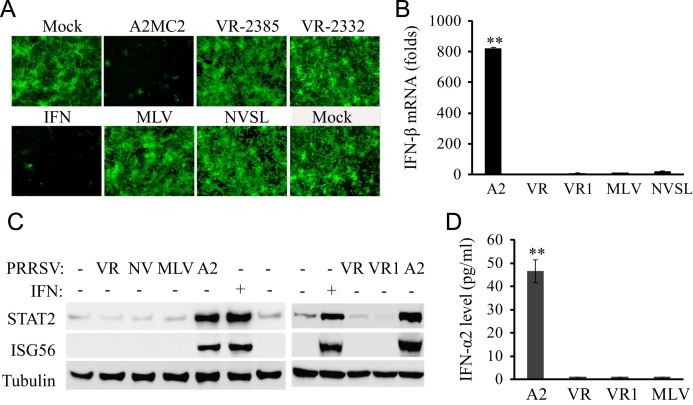
To compare A2MC2 with other PRRSV strains in regards to IFN induction, MARC-145 cells were inoculated with A2MC2, VR-2385, NVSL, MLV, and VR-2332 at 1 TCID50 per cell, separately. These strains were selected because they are frequently used in this laboratory, because VR-2385 (Halbur et al., 1995, Meng et al., 1994), VR-2332 (Benfield et al., 1992), and NVSL (Kwon et al., 2006) are PRRSV strains of varying virulence in experimentally infected pigs, and Ingelvac PRRS MLV is a licensed modified live vaccine strain. Cell culture supernatant samples were collected at 24 hpi and used to treat Vero cells for the IFN bioassay. Supernatants from A2MC2-infected cells inhibited NDV-GFP replication in Vero cells, while supernatant samples from MARC-145 cells infected with VR-2385, VR-2332, NVSL or MLV had no effect on NDV-GFP propagation in Vero cells ( Fig. 5A).
Fig. 5.
Comparison of A2MC2 to other PRRSV strains in IFN production using MARC-145 cells. (A) IFN bioassay in Vero cells. Cell culture supernatants from MARC-145 cells infected with an MOI of 1 TCID50 each of PRRSV strains A2MC2, VR-2385, VR-2332, MLV, or NVSL, respectively, were collected at 36 hpi. Vero cells were treated with 1:4 dilution of the respective supernatants for 12 h, and then infected with NDV-GFP. Fluorescence microscopy was conducted at 24 hpi. Treatment with IFN-α was included as a positive control. (B) IFN-β expression in MARC-145 cells detected by real-time RT-PCR. The cells were infected with PRRSV and harvested for detection of IFN-β transcript. Relative fold of induction in comparison with uninfected cells are shown. Error bars represent variation of three repeated experiments. Significant difference between A2MC2 and the rest of the samples is denoted by “**”, which indicates P<0.01. A2: A2MC2; VR: VR-2385; VR1: VR-2332. (C) STAT2 and ISG56 protein level in MARC-145 cells detected by Western blotting. Treatment of uninfected cells with IFN-α was included as a positive control. A2: A2MC2; VR: VR-2385; NV: NVSL; VR1: VR-2332. (D). IFN-α2 level in culture supernatants of MARC-145 cells infected with A2MC2, VR-2385, VR-2332, and MLV, respectively. ELISA analyses were conducted to quantify the IFN-α2 levels and concentrations were calculated on the basis of a standard curve. The significant difference between A2MC2 and the rest of the samples is denoted by “**”, which indicates P<0.01.
Real-time RT-PCR analysis showed that A2MC2 induced an 820-fold elevation of IFN-β transcripts in MARC-145 cells, significantly higher than that induced by VR-2385, VR-2332, NVSL, or MLV (Fig. 5B). Western blot analysis showed that VR-2385, VR-2332, NVSL, and MLV infection had no effect on STAT2 and ISG56 protein level, while A2MC2 infection led to a higher amount of the two proteins (Fig. 5C).
ELISA was done to detect IFN level in culture supernatants of MARC-145 cells infected with A2MC2, VR-2385, VR-2332, or MLV, respectively. Due to paucity of ELISA kits for type I IFNs of monkeys, only the level of monkey IFN-α2 was quantified. The level of IFN-α2 in culture supernatant of A2MC2-infected cells was 46.6 pg/ml and significantly higher than the supernatants of MARC-145 cells infected with VR-2385, VR-2332 or MLV (Fig. 5D). These results indicated that A2MC2 induced synthesis of type I IFNs in MARC-145 cells, while the other four PRRSV strains inhibited IFN induction.
Kinetics of IFN-β expression in A2MC2-infected MARC-145 cells
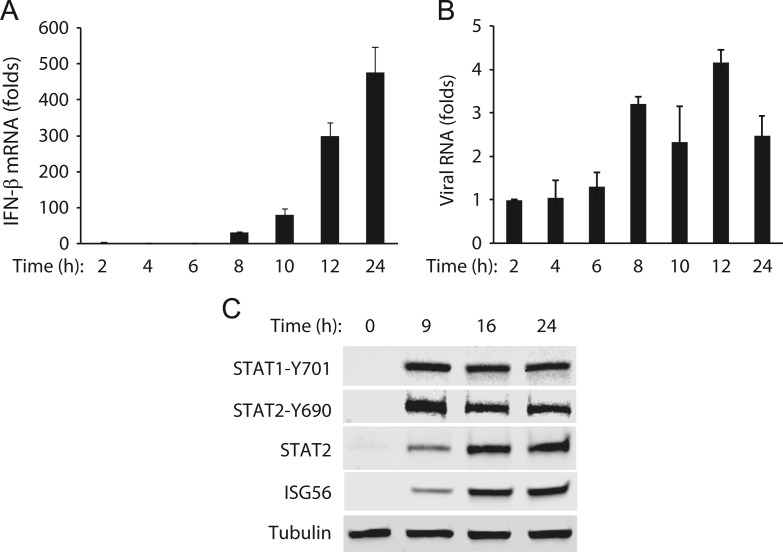
To further examine the expression of IFN-β in A2MC2-infected MARC-145 cells, the cells were harvested at 2, 4, 6, 8, 10, 12, and 24 hpi for RNA isolation and real-time RT-PCR. The IFN-β mRNA increased from 2-fold at 2 hpi to 474-fold at 24 hpi ( Fig. 6A). The large increase of IFN-β transcripts started at 8 hpi. This result indicates that IFN-β expression increased concurrently with A2MC2 replication. Viral RNAs at these time points were detected by real-time RT-PCR. The results showed that the viral RNAs detected at 8, 10, 12 and 24 hpi were 3.2-, 2.3-, 4.2-, and 2.5-fold, respectively, higher than 2 hpi (Fig. 6B). The relatively small increase in the viral RNA level is consistent with the result of the multi-step growth curve showing limited virus replication in cells inoculated with 1 TCID50 per cell.
Fig. 6.
Time-course kinetics of IFN-β expression and activation of the JAK–STAT signaling pathway in A2MC2-infected MARC-145 cells. (A). Time-course kinetics of IFN-β expression. The cells were infected with A2MC2 at 1 TCID50 per cell and harvested at 2, 4, 6, 8, 10, 12, and 24 hpi for real-time RT-PCR detection of IFN-β transcript. Relative fold of induction in comparison with uninfected cells are shown. Error bars represent variation between repeated experiments. (B) Viral RNA levels detected by real-time RT-PCR. Relative fold of viral RNA in comparison with that detected at 2 hpi are shown. (C) Activation of the JAK-STAT signaling pathway. The cells were infected with A2MC2 at an MOI of 1 TCID50 per cell and harvested at 0, 9, 16, and 24 hpi for Western blot analysis of phosphorylated STAT1 (STAT1-Y701) and STAT2 (STAT2-Y690), whole STAT2, and ISG56.
The data above showed that A2MC2 induced expression of type I IFNs. We were interested in the status of JAK-STAT signaling pathway in A2MC2-infected cells. STAT1 and STAT2 proteins are key players in JAK-STAT signaling, a pathway activated by type I IFNs (Darnell et al., 1994, Schindler and Darnell, 1995, Stark et al., 1998). Phosphorylation of STAT1 and STAT2 is an early step in the pathway after IFNs bind to their receptors. To determine if A2MC2-induced IFNs resulted in the activation of these two proteins, we tested the phosphorylation status of STAT1 and STAT2 in MARC-145 cells at 0, 9, 16 and 24 hpi. The selection of 9 hpi was based on the speculation that after increase of IFN-β transcript at 8 hpi, phosphorylation of STAT1 and STAT2 would be detected. The inclusion of 16 and 24 hpi was based on the speculation that along with the increase of IFN-β expression, phosphorylation of STAT1 and STAT2 would continue. The result showed that the levels of phosphorylated STAT1 at tyrosine 701 (STAT1-Y701) and STAT2 at tyrosine 690 (STAT2-Y690) were greatly increased at 9 hpi (Fig. 6C), indicating A2MC2-induced IFNs led to the activation of STAT1 and STAT2. Moreover, the total STAT2 and ISG56 were detectable at 9 hpi and increased at 16 and 24 hpi (Fig. 6C). This result suggested that A2MC2-induced IFNs resulted in the activation of JAK–STAT signaling, which then led to the increased expression of STAT2 and ISG56.
A2MC2 induces elevated expression of ISGs in primary porcine alveolar macrophages
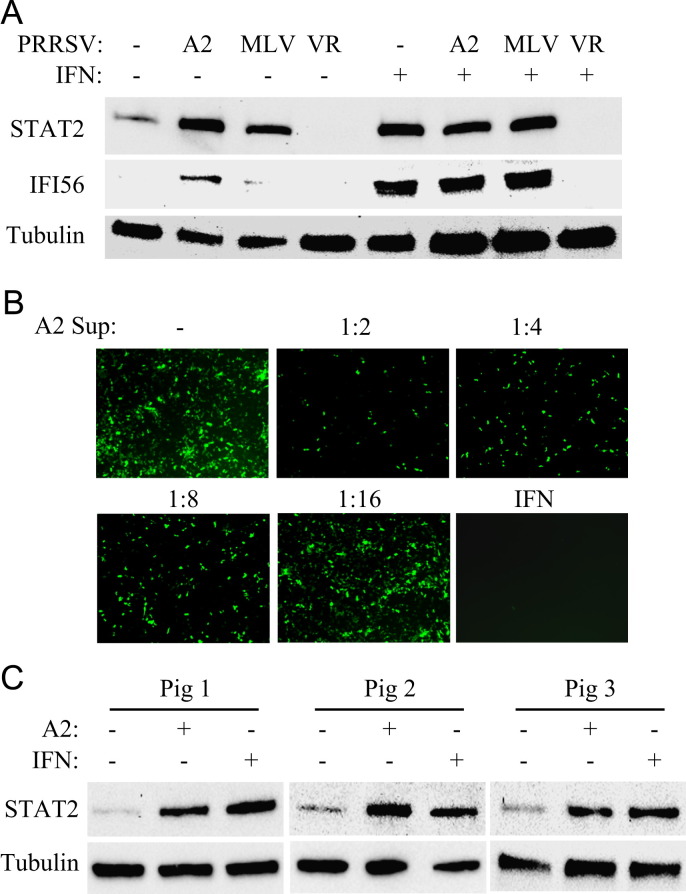
PAMs are the main target cells for PRRSV infection in vivo. To determine the effect of A2MC2 on IFN synthesis in PAM cells, PAMs were infected with A2MC2 and harvested for Western blot analyses at 20 hpi. Infections of PAMs with VR-2385 and MLV were included as controls. Compared to uninfected cells, A2MC2 infection resulted in the elevation of STAT2 and IFI56 (equivalent to ISG56 in primates), while VR-2385 led to no change of these two proteins, and MLV led to a slight elevation of STAT2 ( Fig. 7A). To test the effect of these virus strains on IFN signaling, IFN-α was added to PAMs at 12 hpi. PAMs infected with A2MC2 and MLV had elevated STAT2 and IFI56 to a similar level of uninfected cells treated with external IFN-α, while VR-2385-infected cells had no increase in STAT2 and IFI56 levels (Fig. 7A). This result indicated that A2MC2 and MLV had an undetectable effect on IFN downstream signaling while VR-2385 inhibited the IFN activation in PAMs.
Fig. 7.
A2MC2 induces expression of IFN-stimulated genes in primary porcine pulmonary alveolar macrophages (PAMs). (A) STAT2 and IFI56 detected by Western blotting. PAMs were infected with PRRSV VR-2385, A2MC2, and MLV, and at 12 hpi, treated with or without IFN-α. The cells were harvested at 20 hpi for Western blotting. Cell lysate samples from uninfected PAMs with or without IFN treatment were included as controls. (B) IFN bioassay in CRL2843 cells. Supernatant from A2MC2-infected PAMs was diluted and added to the CRL2843 cells 12 h before NDV-GFP inoculation. The cells were observed 24 h after NDV-GFP inoculation. Treatment of the cells with swine IFN-α at a final concentration of 1000 U/ml was included as a positive control. (C) A2MC2 induces elevation of STAT2 in PAM cells from different piglets. PAMs from three piglets were inoculated with A2MC2 at 0.05 TCID50 per cell, respectively, and incubated for 20 h. Cell lysate samples from IFN-α-treated PAM cells were included as positive controls. Cell lysate samples from non-infected cells were included as negative controls in the Western blot analyses.
The IFN bioassay was conducted to assess IFNs in culture supernatant of A2MC2-infected PAMs. CRL2843 cells are immortalized porcine alveolar macrophages that are not susceptible to PRRSV infection (Weingartl et al., 2002). The cells were treated with dilutions of the supernatant from A2MC2-infected PAMs and, on the next day, inoculated with NDV-GFP. The supernatant dilutions up to 1:8 inhibited NDV-GFP replication, compared with mock-infected cells (Fig. 7B). This result indicated that the culture supernatant of A2MC2-infected PAMs contained interferons.
To determine if the induction of IFNs by A2MC2 is not limited to PAMs from one piglet, PAMs from three other piglets were inoculated with A2MC2 and incubated for 20 h. Western blot analysis showed that A2MC2 infection induced the elevation of STAT2 in PAM cells from all three other piglets (Fig. 7C). The level of STAT2 in A2MC2-infected PAMs was similar to that in PAMs treated with external IFN-α.
Discussion
Many PRRSV strains are known to inhibit the production of type I IFNs in cultured cells and in infected pigs. A2MC2 induces the synthesis of interferons in MARC-145 and primary PAM cells, and will be useful in studying PRRSV interference with host innate immune responses. In this study, the induction of IFNs by A2MC2 was examined from several aspects. First, the culture supernatant from A2MC2-infected MARC-145 cells protected Vero cells from NDV infection. Vero cells are defective in IFN production and not susceptible to PRRSV. Pretreatment of the cells with dilutions of A2MC2-derived supernatant induced an antiviral response in Vero cells that inhibited the replication of NDV. The result was corroborated by elevation of STAT2 and ISG56, two genes stimulated by IFN signaling in Vero cells.
Second, the elevated expression of STAT2 and ISG56 was shown in A2MC2-infected MARC-145 cells. Virus replication was essential for the elevation of the two proteins because UV-inactivated virus and PPMO inhibition of PRRSV replication did not lead to elevation of these host proteins. The results suggested that viral replication in the cytoplasm stimulated PRRs, leading to IFN synthesis. The elevation of the transcripts of ISG15 and ISG56 further confirmed the observation. This result also ruled out the possibility of contamination by other swine pathogens because PPMO-mediated inhibition of A2MC2 led to an absence of IFN induction. Other common swine pathogens, such as porcine respiratory coronavirus (PRCV) or swine influenza virus (SwIV), can induce synthesis of a high level bioactive IFN-α (Van Reeth et al., 1999). Interestingly, A2MC2 infection at different MOIs induced variable levels of ISG expression. The higher the MOI, the earlier A2MC2 induced ISG elevation. The lower the MOI, the later the virus-induced ISG expression and the lower level of induction. This result provided an explanation as to why an MOI of 1 TCID50 per cell led to a lower virus yield than MOIs of 0.1 and 0.01 TCID50. It indicates that the inoculum of 0.01 TCID50 per cell resulted in limited initial virus replication, leading to a weaker stimulation on cellular PRRs, which allowed the virus to complete its replication cycles. On the other hand, the inoculum at 1 TCID50 per cell stimulated a more robust response in regards to PRRs and IFN synthesis, which in turn inhibited virus spread to neighboring cells or continued replication.
Third, several other PRRSV strains including the vaccine strain MLV were shown to inhibit IFN production. IFN-α2 was detected in culture supernatant from A2MC2-infected cells, but not in the samples from cells infected with VR-2332, VR-2385, or MLV. It was possible that there were other subtypes of type I IFNs in the supernatant from A2MC2-infected cells, as IFN-β mRNA level significantly increased. We further tested IFN production in A2MC2 infection of PAM cells. The STAT2 and IFI56 were elevated in A2MC2-infected cells, but not in VR-2385-infected cells. The A2MC2 induction of IFNs was pig-independent as PAMs isolated from three other pigs had similar elevations of STAT2 after A2MC2 infection. Infection of PAMs with MLV also led to a slight elevation in STAT2 level, which indicates that MLV might induce STAT2 albeit at much lower levels than A2MC2.
Analysis of the cDNA sequence of the A2MC2 genome showed that it was highly homologous to both VR-2332 and MLV. This result indicated that A2MC2 might be a chimera of VR-2332 and MLV strains. Based on sequence analysis, we reasoned that the first 4.6 kb and the fragment from nt 11966 to 14420 were derived from VR-2332; fragments of nt 10697 to 11666 and nt 14421 to the end of the genome were possibly of MLV origin; and the fragment of nt 4681 to 10037 was derived from either one of them, but with mutations as 10 unique changes are located in this fragment. Compared to both VR-2332 and MLV, A2MC2 possessed 6 unique amino acids distributed in nsp8, nsp10, and nsp12, and GP3. Based on this sequence comparison, future experiments on virus mutagenesis using reverse genetic technology will be conducted to pinpoint the A2MC2 sequences that are involved in the loss of inhibition of IFN production.
Both A2MC2 and MLV have an undetectable effect on the ability of IFN-α to induce an antiviral response, as their infection of PAM cells did not affect expression of STAT2 and IFI56 activated by external IFN-α. This result is consistent with a previous report that virulent strain VR-2385 inhibits IFN signaling while MLV does not (Patel et al., 2010). The plaque morphology of A2MC2 was much smaller than that of MLV, which indicated that MLV replicated with faster kinetics than A2MC2 in MARC-145 cells. In addition, A2MC2 infection did not lead to any observable cytopathic effect in PAM cells, and A2MC2-infected cells were of similar viability as mock-infected or MLV-infected cells. These features indicated that like MLV, A2MC2 might be less suitable to replicate in PAMs.
The well-characterized cellular PRRs for IFN production during virus infection include the TLR and RLR pathways. Activation of either the TLR or the RLR pathway leads to the phosphorylation of interferon regulatory factor 3 (IRF-3), followed by its nuclear translocation and transcriptional activation of type I IFNs. We tested if A2MC2 replication led to IRF-3 phosphorylation using polyinosinic-polycytidylic acid (poly(I:C)), a synthetic analog of double-stranded RNA (dsRNA), as a positive control. The phosphorylated IRF-3 was detected in the poly(I:C)-treated cells but undetectable in cells infected with A2MC2 (data not shown). The result that A2MC2 infection led to increase of IFN-β transcript from 8 hpi to less than 1000-fold at 24 hpi (Fig. 6) indicated that A2MC2 replication might lead to a low level of IRF-3 signaling for an extended period. The reason for this conjecture is that poly(I:C) treatment led to over a 10,000-fold induction of IFN-β transcript with a peak at 6 h after treatment (data not shown). The continuous detection of phosphorylation of both STAT1 and STAT2 in A2MC2-infected MARC-145 cells from 9 to 24 hpi supports this speculation, while stimulation with external IFN-α leads to transient increase of phosphorylation of STAT1 that was reduced below the detection level after 2.5 h treatment (Patel et al., 2010).
In conclusion, we have discovered a strain of PRRSV, A2MC2, that induced IFN production in both MARC-145 and PAM cells while other tested PRRSV strains inhibited IFN induction. Specifically, A2MC2 induced type I IFNs and led to an elevation of IFN-stimulated genes. Full genome sequence analysis showed that A2MC2 was closely related to VR-2332 and MLV. The identification of A2MC2 as an IFN-inducer may be beneficial for studying PRRSV interference in IFN signaling, and developing improved vaccines for protective immunity against PRRS.
Materials and methods
Cells and viruses
MARC-145 (Kim et al., 1993) and Vero cells (ATCC CCL-81) were grown in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Immortalized porcine macrophages (CRL2843) were cultured in RPMI1640 medium supplemented with 10% FBS. Primary PAM cells were prepared from bronchoalveolar lavage of 4-week-old PRRSV-negative piglets. The preparation and subsequent culture of PAMs in RPMI1640 culture medium were conducted, as previously described (Patel et al., 2008). PRRSV strains A2MC2, VR-2385, NVSL 97-7895, and Ingelvac PRRS MLV were used to inoculate MARC-145 cells at 1 multiplicity of infection (MOI). Virus titers were determined in MARC-145 cells for the median tissue culture infectious dose (TCID50), as previously described (Zhang et al., 2006). Avirulent LaSota Newcastle disease virus carrying the gene of green fluorescence protein (NDV-GFP) was propagated in Vero cells, as previously described (Kim and Samal, 2010).
Virus inactivation was conducted with a UV cross-linker (Spectrolinker XL-1500, Agilent Technologies, Santa Clara, CA) at 1200 mJ/cm2 for two 10-min pulses at 1-min interval. The inactivation was confirmed by the absence of virus replication in MARC-145 cells at 72 h post-infection (hpi) as assessed by immunofluorescence assay (IFA).
For interferon stimulation, universal type I IFN-α (R&D Systems, Minneapolis, MN) was added to the cultured cells at a final concentration of 1000 U/ml. The cells were harvested at indicated time points for further analysis.
Interferon bioassay
Vero cells were seeded into cell culture plates, incubated overnight, and, on the next day, treated with culture supernatant from PRRSV-infected MARC-145 cells. The cells were infected with LaSota NDV-GFP 12 h after the treatment. Fluorescence microscopy was conducted 24 h after infection to observe GFP-positive cells.
Immunofluorescence assay (IFA)
An IFA was carried out as previously reported with an N-specific monoclonal antibody EF11 to detect PRRSV N-proteins in MARC-145 cells on coverglass slips (Zhang et al., 1998). Specific reactions between EF11 and the N-protein were detected with goat anti-mouse IgG-fluorescein isothiocyanate (FITC) conjugate (Sigma, St. Louis, MO). The coverglass was mounted onto slides using SlowFade Gold antifade reagent containing 4′6-diamidino-2-phenylindole (DAPI) (Life Technologies Corporation, Carlsbad, CA) and observed under fluorescent microscopy.
Western blot analysis
Cell lysate samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis as described previously (Zhang et al., 2007). Briefly, separated proteins from SDS-PAGE were transferred onto a nitrocellulose membrane and probed with antibodies against STAT2 (Santa Cruz Biotechnology, Santa Cruz, CA), β-tubulin (Sigma), phospho-STAT2 (STAT2-Y690) (Santa Cruz Biotechnology), phospho-STAT1 (STAT1-Y701) (Millipore, Billerica, MA), and ISG56 (Thermo Fisher Scientific, Rockford, IL). The chemiluminescent signal was recorded digitally by Quantity One Program, Version 4.6, in a ChemiDoc XRS imaging system (Bio-Rad Laboratories, Hercules, CA). Pig antiserum against PRRSV NVSL strain (NVSL, Ames, IA) was used to detect PRRSV proteins in lysate of PRRSV-infected cells (Patel et al., 2010).
RNA isolation, reverse transcription, and real-time PCR
Total RNA was isolated from MARC-145 and PAM cells with TRIzol® Reagent (Life Technologies) following the manufacturer's instructions. Reverse transcription and real-time PCR were conducted as previously described (Patel et al., 2008, Patel et al., 2009). Transcripts of ribosomal protein L32 (RPL32) were also amplified from the samples of PAM and MARC-145 cells and used to normalize the total input RNA. Primers used in this study to conduct reverse transcription and real-time RT-PCR were previously described (Patel et al., 2010). Relative transcript levels were quantified by the 2−ΔΔCT method (Livak and Schmittgen, 2001) and shown as a relative fold of change in comparison with mock-treated control.
Cell viability assay
Viability of PAMs was determined with CellTiter-Glo Cell Viability Assay (Promega). Briefly, CellTiter-Glo reagent was added to cells in a 96-well plate and the luminescence signal was measured with VICTOR3™ Multilabel Counter (Perkin-Elmer Life and Analytical Sciences, Wellesley, MA). Relative percentages of luminescence intensity were calculated by comparison to controls.
Plaque assay
Plaque assays were done with modifications from a previously described protocol (O'Reilly et al., 1992). MARC-145 cells were seeded into 35 mm culture dishes and incubated overnight. PRRSV was diluted in a ten-fold series and added to the cells. The virus inoculum was removed 2 h after inoculation and replaced with 0.5% agarose overlay containing complete growth medium. The cells were stained with another layer of agarose overlay containing neutral red at 50 μg/ml 4 days after inoculation. Plaques were observed after overnight incubation.
Quantifying IFN level by ELISA
Culture supernatant samples of MARC-145 cells infected with A2MC2, VR-2385, VR-2332, or MLV at a MOI of 1 TCID50 per cell were collected at 24 hpi. Detection of IFN-α2 in each sample was done by using VeriKine™ Cynomolgus/Rhesus Interferon-Alpha Serum ELISA kit (PBL InterferonSource, Piscataway, NJ) according to the manufacturer's instruction. The concentration of IFN-α2 in the samples was calculated on the basis of a standard curve prepared from supplied IFN-α2 in the kit.
Sequencing
A2MC2 genomic RNA was isolated from cell culture supernatant with TRIzol LS reagent (Life Technologies). Reverse transcription of the viral RNA with primers 32nsp12R1 (5′-TCAATTCAGGCCTAAAGTTG-3′) and P6-7-R (5′-CGCCCTAATTGAATAGGTGACTT-3′) was done with Maxima reverse transcriptase (Thermo Fisher Scientific). PCR amplification was done with Phusion high-fidelity DNA polymerase (New England Biolab, Ipswich, MA). 5′-RACE (rapid amplification of cDNA ends) and 3′-RACE of the A2MC2 genome were done as previously described (Sambrook and Russell, 2001). Sequencing of the PCR products was performed with ABI Prism 3130 Genetic Analyzer (Life Technologies). Sequence assembly and analysis was done with LaserGene Core Suite (DNASTAR Inc., Madison, WI). The GenBank accession number of the cDNA sequence of the A2MC2 genome is JQ087873.
Statistical analysis
Differences in indicators between treatment samples, such as cellular RNA level between the groups in the presence or absence of PRRSV infection, were assessed by the Student t-test. A two-tailed P-value of less than 0.05 was considered significant.
Acknowledgments
We are grateful to Dr. Joseph F. Urban at Human Nutrition Research Center, USDA, Beltsville, MD for his gift of the lung lavage of piglets. We thank Sa Xiao at University of Maryland, College Park, MD for his advice and discussion in sequencing of A2MC2 cDNA. Y. Nan and R. Wang were partially supported by China Scholarship Council. M. Shen was partially supported by Shandong Bureau of Education, China. This project was supported by National Pork Board, USA.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2012.05.015.
Appendix A. Supporting information
Supplementary material
References
- Albina E., Carrat C., Charley B. Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J. Interferon Cytokine Res. 1998;18:485–490. doi: 10.1089/jir.1998.18.485. [DOI] [PubMed] [Google Scholar]
- Bautista E.M., Faaberg K.S., Mickelson D., McGruder E.D. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology. 2002;298:258–270. doi: 10.1006/viro.2002.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfield D.A., Nelson E., Collins J.E., Harris L., Goyal S.M., Robison D., Christianson W.T., Morrison R.B., Gorcyca D., Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332) J. Vet. Diagn. Invest. 1992;4:127–133. doi: 10.1177/104063879200400202. [DOI] [PubMed] [Google Scholar]
- Beura L.K., Sarkar S.N., Kwon B., Subramaniam S., Jones C., Pattnaik A.K., Osorio F.A. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 2010;84:1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botner A., Strandbygaard B., Sorensen K.J., Have P., Madsen K.G., Madsen E.S., Alexandersen S. Appearance of acute PRRS-like symptoms in sow herds after vaccination with a modified live PRRS vaccine. Vet. Rec. 1997;141:497–499. doi: 10.1136/vr.141.19.497. [DOI] [PubMed] [Google Scholar]
- Buddaert W., Van Reeth K., Pensaert M. In vivo and in vitro interferon (IFN) studies with the porcine reproductive and respiratory syndrome virus (PRRSV) Adv. Exp. Med. Biol. 1998;440:461–467. doi: 10.1007/978-1-4615-5331-1_59. [DOI] [PubMed] [Google Scholar]
- Chen Z., Lawson S., Sun Z., Zhou X., Guan X., Christopher-Hennings J., Nelson E.A., Fang Y. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology. 2010;398:87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E., Jr., Kerr I.M., Stark G.R. Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Sci. (New York, N.Y.) 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- de Lima M., Ansari I.H., Das P.B., Ku B.J., Martinez-Lobo F.J., Pattnaik A.K., Osorio F.A. GP3 is a structural component of the PRRSV type II (US) virion. Virology. 2009;390:31–36. doi: 10.1016/j.virol.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Faaberg K.S., Balasuriya U.B., Brinton M.A., Gorbalenya A.E., Leung F.C.-C., Nauwynck H., Snijder E.J., Stadejek T., Yang H., Yoo. D. Family arteriviridae. In: King A.M.Q., editor. Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; London: 2011. [Google Scholar]
- Halbur P.G., Paul P.S., Frey M.L., Landgraf J., Eernisse K., Meng X.J., Lum M.A., Andrews J.J., Rathje J.A. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet. Pathol. 1995;32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Kwang J., Yoon I.J., Joo H.S., Frey M.L. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 1993;133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- Kim, O., Sun, Y., Lai, F.W., Song, C., Yoo, D., 2010. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology 402, 315-326. [DOI] [PMC free article] [PubMed]
- Kim S.H., Samal S.K. Role of untranslated regions in regulation of gene expression, replication, and pathogenicity of Newcastle disease virus expressing green fluorescent protein. J. Virol. 2010;84:2629–2634. doi: 10.1128/JVI.02049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon B., Ansari I.H., Osorio F.A., Pattnaik A.K. Infectious clone-derived viruses from virulent and vaccine strains of porcine reproductive and respiratory syndrome virus mimic biological properties of their parental viruses in a pregnant sow model. Vaccine. 2006;24:7071–7080. doi: 10.1016/j.vaccine.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Labarque G.G., Nauwynck H.J., Van Reeth K., Pensaert M.B. Effect of cellular changes and onset of humoral immunity on the replication of porcine reproductive and respiratory syndrome virus in the lungs of pigs. J. Gen. Virol. 2000;81:1327–1334. doi: 10.1099/0022-1317-81-5-1327. [DOI] [PubMed] [Google Scholar]
- Lee S.M., Schommer S.K., Kleiboeker S.B. Porcine reproductive and respiratory syndrome virus field isolates differ in in vitro interferon phenotypes. Vet. Immunol. Immunopathol. 2004;102:217–231. doi: 10.1016/j.vetimm.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego, CA) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Madsen K.G., Hansen C.M., Madsen E.S., Strandbygaard B., Botner A., Sorensen K.J. Sequence analysis of porcine reproductive and respiratory syndrome virus of the American type collected from Danish swine herds. Arch. Virol. 1998;143:1683–1700. doi: 10.1007/s007050050409. [DOI] [PubMed] [Google Scholar]
- Meng X.J. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 2000;74:309–329. doi: 10.1016/S0378-1135(00)00196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.J., Paul P.S., Halbur P.G. Molecular cloning and nucleotide sequencing of the 3′ terminal genomic RNA of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 1994;75:1795–1801. doi: 10.1099/0022-1317-75-7-1795. [DOI] [PubMed] [Google Scholar]
- Meulenberg J.J. PRRSV, the virus. Vet. Res. 2000;31:11–21. doi: 10.1051/vetres:2000103. [DOI] [PubMed] [Google Scholar]
- Miller L.C., Laegreid W.W., Bono J.L., Chitko-McKown C.G., Fox J.M. Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch. Virol. 2004;149:2453–2463. doi: 10.1007/s00705-004-0377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann E.J., Kliebenstein J.B., Johnson C.D., Mabry J.W., Bush E.J., Seitzinger A.H., Green A.L., Zimmerman J.J. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 2005;227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- Ni Y.Y., Huang Y.W., Cao D., Opriessnig T., Meng X.J. Establishment of a DNA-launched infectious clone for a highly pneumovirulent strain of type 2 porcine reproductive and respiratory syndrome virus: identification and in vitro and in vivo characterization of a large spontaneous deletion in the nsp2 region. Virus Res. 2011;160:264–273. doi: 10.1016/j.virusres.2011.06.027. [DOI] [PubMed] [Google Scholar]
- O'Reilly D.R., Miller L.K., Luckow V.A. 2nd ed. W.H. Freeman and Company; New York: 1992. Baculovirus Expression Vectors: A laboratory manual. [Google Scholar]
- Opriessnig T., Halbur P.G., Yoon K.J., Pogranichniy R.M., Harmon K.M., Evans R., Key K.F., Pallares F.J., Thomas P., Meng X.J. Comparison of molecular and biological characteristics of a modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine (ingelvac PRRS MLV), the parent strain of the vaccine (ATCC VR2332), ATCC VR2385, and two recent field isolates of PRRSV. J. Virol. 2002;76:11837–11844. doi: 10.1128/JVI.76.23.11837-11844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Nan Y., Shen M., Ritthipichai K., Zhu X., Zhang Y.J. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2010;84:11045–11055. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Opriessnig T., Stein D.A., Halbur P.G., Meng X.J., Iversen P.L., Zhang Y.J. Peptide-conjugated morpholino oligomers inhibit porcine reproductive and respiratory syndrome virus replication. Antiviral Res. 2008;77:95–107. doi: 10.1016/j.antiviral.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Stein D.A., Zhang Y.J. Morpholino oligomer-mediated protection of porcine pulmonary alveolar macrophages from arterivirus-induced cell death. Antiviral Ther. 2009;14:899–909. doi: 10.3851/IMP1409. [DOI] [PubMed] [Google Scholar]
- Rossow K.D., Collins J.E., Goyal S.M., Nelson E.A., Christopher Hennings J., Benfield D.A. Pathogenesis of porcine reproductive and respiratory syndrome virus infection in gnotobiotic pigs. Vet. Pathol. 1995;32:361–373. doi: 10.1177/030098589503200404. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. Third ed. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular Cloning: A Laboratory Manual. pp. 8.54–65. [Google Scholar]
- Schindler C., Darnell J.E., Jr. Transcriptional responses to polypeptide ligands: the JAK–STAT pathway. Ann. Rev. Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr I.M., Williams B.R., Silverman R.H., Schreiber R.D. How cells respond to interferons. Ann. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Takaoka A., Yanai H. Interferon signalling network in innate defence. Cell. Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- Van Reeth K., Labarque G., Nauwynck H., Pensaert M. Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res. Vet. Sci. 1999;67:47–52. doi: 10.1053/rvsc.1998.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H.M., Sabara M., Pasick J., van Moorlehem E., Babiuk L. Continuous porcine cell lines developed from alveolar macrophages: partial characterization and virus susceptibility. J. Virol. Methods. 2002;104:203–216. doi: 10.1016/S0166-0934(02)00085-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Batista L., Dee S., Halbur P., Murtaugh M.P. The level of virus-specific T-cell and macrophage recruitment in porcine reproductive and respiratory syndrome virus infection in pigs is independent of virus load. J. Virol. 2004;78:5923–5933. doi: 10.1128/JVI.78.11.5923-5933.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sharma R.D., Paul P.S. Monoclonal antibodies against conformationally dependent epitopes on porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 1998;63:125–136. doi: 10.1016/s0378-1135(98)00231-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y.J., Stein D.A., Fan S.M., Wang K.Y., Kroeker A.D., Meng X.J., Iversen P.L., Matson D.O. Suppression of porcine reproductive and respiratory syndrome virus replication by morpholino antisense oligomers. Vet. Microbiol. 2006;117:117–129. doi: 10.1016/j.vetmic.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.J., Wang K.Y., Stein D.A., Patel D., Watkins R., Moulton H.M., Iversen P.L., Matson D.O. Inhibition of replication and transcription activator and latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus by morpholino oligomers. Antiviral Res. 2007;73:12–23. doi: 10.1016/j.antiviral.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material