Abstract
The pathogenesis of Type 2 highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) in 10-week old swine in the United States was investigated. rJXwn06, rescued from an infectious clone of Chinese HP-PRRSV, replicated in swine with at least 100-fold increased kinetics over U.S. strain VR-2332. rJXwn06 caused significant weight loss, exacerbated disease due to bacterial sepsis and more severe histopathological lung lesions in pigs exposed to HP-PRRSV than to those infected with VR-2332. Novel findings include identification of bacterial species present, the degree of thymic atrophy seen, and the inclusion of contact animals that highlighted the ability of HP-PRRSV to rapidly transmit between animals. Furthermore, comprehensive detailed cytokine analysis of serum, bronchoalveolar lavage fluid, and tracheobronchial lymph node tissue homogenate revealed a striking elevation in levels of cytokines associated with both innate and adaptive immunity in HP-PRRSV infected swine, and showed that contact swine differed in the degree of cytokine response.
Keywords: HP-PRRSV, JXwn06, VR-2332, US Swine, Pathogenesis, Bacterial, Cytokine Response, Thymic Atrophy, Viral Kinetics
Introduction
In 2006, investigators from several Chinese provinces reported a unique syndrome in growing swine that was highlighted by the predominant clinical signs of high fever, anorexia, listlessness, red discoloration of skin, respiratory distress and very high morbidity and mortality rates (Li et al., 2007, Tian et al., 2007, Tong et al., 2007, Wu et al., 2009, Zhou et al., 2008). Originally known as porcine high fever disease (PHFD), this syndrome spread to Vietnam in 2006 and to Cambodia, Laos, The Philippines, Bhutan, Myanmar, Thailand, South Korea and Russia in later years (An et al., 2011, Feng et al., 2008). Although there was concern that this syndrome may have been caused by a new disease agent, extensive diagnostic testing revealed only known pathogens. One consistent finding was the detection of porcine reproductive and respiratory syndrome virus (PRRSV) with two discontinuous deletions in the replicase polyprotein known as nonstructural protein 2 (nsp2), and two single nucleotide deletions in the 5′ and 3′ untranslated regions (UTRs) (Zhou and Yang, 2010). Experimental infection of Chinese swine with the initial novel PRRSV field isolates reproduced the clinical disease (Li et al., 2007, Tian et al., 2007, Tong et al., 2007, Wu et al., 2009, Zhou et al., 2008), providing strong evidence for its role as the causal agent of PHFD. Since the severity of the clinical disease was greater than expected for a typical PRRSV infection, there remained the chance that an unknown agent in the PRRSV isolates exacerbated the disease symptoms. This question was resolved when PHFD was reproduced in Chinese swine with virus derived from an infectious clone of the JX143 PRRSV isolate (Lv et al., 2008).
The prior studies demonstrated that PRRSV isolates with a common genetic pattern had a causal role in PHFD, leading to the new designation of this viral lineage as highly pathogenic PRRSV (HP-PRRSV). The presence of the unique nsp2 deletion motif was initially thought to contribute to the severity of disease (Tian et al., 2007). However, studies showed that the nsp2 region of HP-PRRSV strain rJXwn06 containing the novel deletion could be replaced with 458 amino acids from low virulence strain HB-1/3.9 to result in a chimeric virus with only a minor delay in mortality (Zhou et al., 2009). There was also concern that there may be some unique aspect that may predispose Asian pigs to a severe outcome, e.g., husbandry practices, endemic infections with other pathogens, climate, or host genetics. Although a number of experiments have associated genetic changes in PRRSV with an attenuation phenotype using point mutations (Grebennikova et al., 2004, Nielsen et al., 2001, Storgaard et al., 1999) or by the construction of chimeric viruses (Ellingson et al., 2010, Kwon et al., 2008, Wang et al., 2008, Zhou et al., 2009), there appears to be no single locus for which mutations confer a predictable change in virulence. Collectively, this area of study suggests the factors that contribute to PRRSV pathogenicity are complex and viral strain-specific.
As part of the single-strand positive-sense RNA virus order Nidovirales, family Arteriviridae, PRRSV genomes can vary between 15 kb to 15.5 kb, consist of at least 10 open reading frames (ORFs) and replicate through a nested set of subgenomic RNAs (van Hemert and Snijder, 2008). Members of this highly variable virus have been grouped into two genotypes, Type 1 (European-like, prototype strain Lelystad) or Type 2 (North American, prototype strain VR-2332), which differ in nucleotide similarity by approximately 45% (Nelsen et al., 1999). Both genotypes have representatives of varying pathogenicity, and intragenic nucleotide sequence variation in each can be as much as 20% (Shi et al., 2010). For this study, we imported a plasmid containing the cDNA copy of the HP-PRRSV strain rJXwn06 genome, a Type 2 PRRSV genome that is 15,321 bases in length and has 89% pairwise nucleotide identity to the prototype genome of VR-2332 (Zhou et al., 2009). To investigate the pathogenesis of HP-PRRSV infection and the potential contribution of climate, host genetics, commensal bacteria, other environmental conditions and husbandry practices to the pathogenicity of HP-PRRSV in U.S. swine, we compared and contrasted the pathogenicity of rescued JXwn06 (rJXwn06) virus to that of VR-2332 in 10-week old swine. We found that this HP-PRRSV strain caused extreme morbidity, as was seen in Asia, but novel to this study, resulted in up to 100x higher abundance of circulating virus when compared to VR-2332, caused extremely exacerbated thymic atrophy such that the thymus was often difficult to discern, and the host response was assessed in comparison to animals infected with strain VR-2332 for the first time by a swine protein array including 5 innate and 5 adaptive cytokines in serum, bronchoalveolar lavage fluid and lymph nodes. Moreover, we completed bacterial speciation and loads after necropsy and specified the degree of weight loss seen. Lastly, we showed that HP-PRRSV readily transmitted to contact swine causing a different pattern of cytokine responses. As a result of our study, the remaining plausible contributing factors to the high virulence seen in Asia were discredited, and thus HP-PRRSV strains pose a serious threat to the U.S. swine industry.
Results
Swine study
Pigs challenged with Chinese HP-PRRSV strain rJXwn06 (Group 2) began exhibiting clinical signs of disease within 2–3 days post exposure (dpe). As a group, the pigs developed fevers, became listless, anorexic, and began shivering and huddling together in a pile. Clinical signs became more severe over the next few days, with pigs rapidly losing body weight ( Fig. 1), becoming dehydrated and weak. Respiratory distress, characterized by dyspnea, tachypnea and coughing was common in all pigs. Erythema of the skin was present in most of the pigs, and several developed cutaneous hemorrhages and cyanotic extremities (blue ears) ( Fig. 2). On 7 dpe, one pig from Group 2 was found dead. Two others were euthanized due to severe weakness and moribund condition on dpe 8 and 9, and one was found dead on dpe 13. On 10 and 11 dpe, clinical signs of disease in some of the HP-PRRSV challenged group began to decrease in severity. While pigs remained huddled in a pile with respiratory signs, approximately half of them were subjectively less listless, would move away when approached and began showing an interest in feed again. During the study, contact pigs displayed similar clinical signs as those challenged with HP-PRRSV. Pigs challenged with VR-2332 (Group 4) were clinically normal until dpe 7, when they began to exhibit slightly increased respiratory rates and became a little less active than the control group. One pig from the VR-2332 group was found dead on dpe 9; however, the cause of death was attributed to gastric dilatation and volvulus, rather than to clinical disease typical of PRRSV strain VR-2332. Control pigs (Group 1) remained clinically normal for the duration of the study.
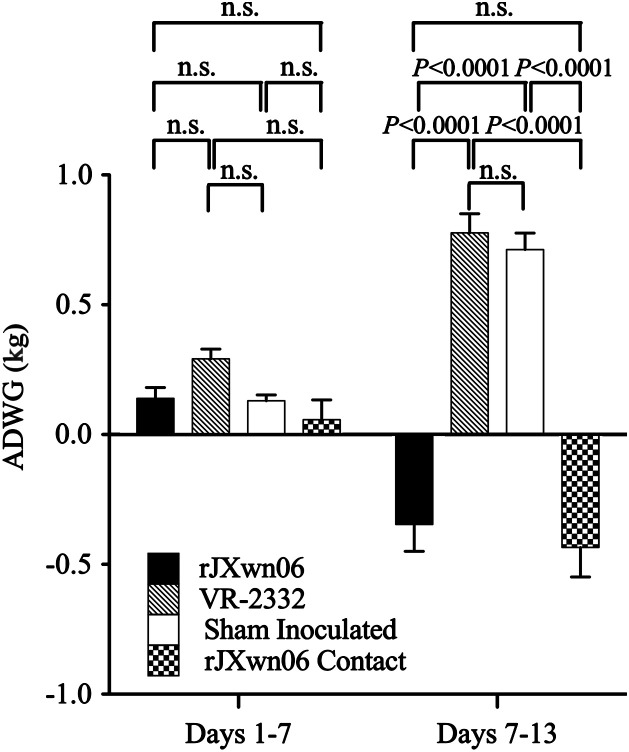
Fig. 1.
Average daily weight gain (ADWG) of swine during the course of the study. Shown are ADWG and associated standard errors for each of the four groups for days −1–7 and days 7–13. Sham inoculated control swine (Group 1) and those challenged with VR-2332 (Group 4) gained weight at an expected rate for their age group. Group 2 swine challenged with HP-PRRSV strain rJXwn06 and their contacts (Group 3) lost body weight during the last week of the experiment. The ADWG for Group 2 on days −1–7 includes only 11 pigs, and on days 7–13 includes only 8 pigs. The body weights for the contact animals were taken on the same day as pigs in the other groups. Significance values are shown.
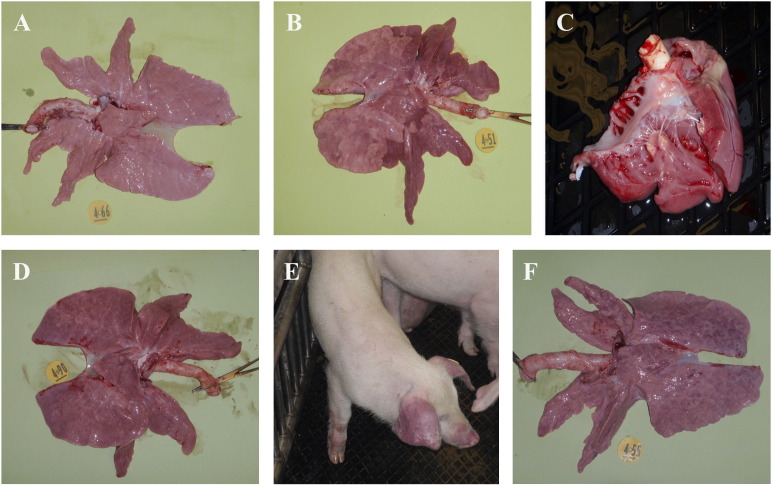
Fig. 2.
Gross pathology. (A) Representative normal lung from a sham inoculated control pig (Group 1). (B) Macroscopic lesions in pigs challenged with HP-PRRSV strain rJXwn06 (Group 2) were severe and include interstitial pneumonia with consolidation, and pulmonary edema. (C) myocardial necrosis. (D) Contact pigs (Group 3) had similar lung lesions. (E) Cyanotic extremities and ears (blue ears) were common as shown in the contact pig. (F) Pigs challenged with VR-2332 (Group 4) had lung lesions characteristic of an uncomplicated PRRSV infection.
Gross pathology
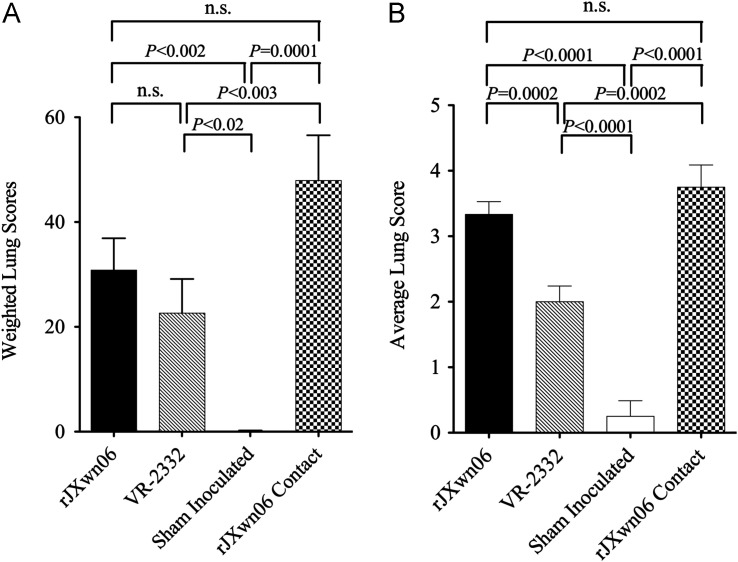
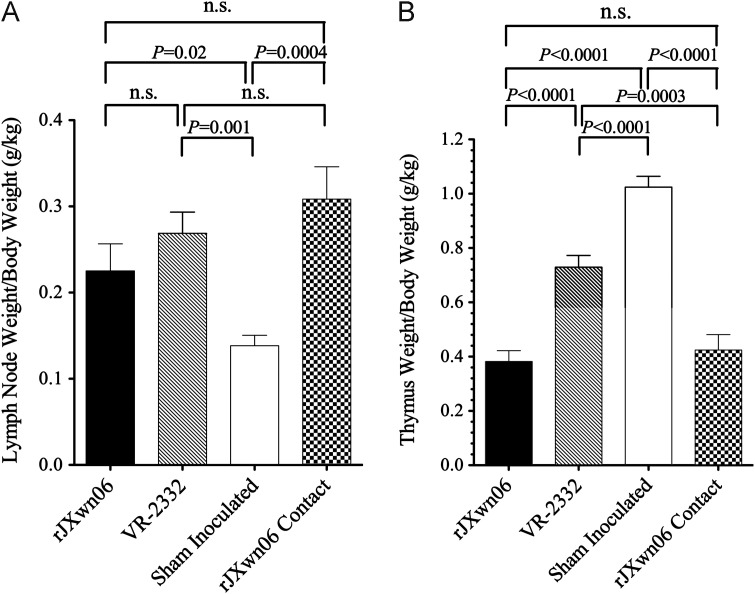
Postmortem examination revealed severe lesions in the HP-PRRSV challenged pigs including: marked interstitial pneumonia, lymphadenopathy and thymic atrophy. Necropsy findings in this group include: pulmonary edema, pleuritis, peritoneal and pericardial effusions, renal petechia, and fibrinous peritonitis that are delineated in Table 1. Lungs and representative lesions are shown in Fig. 2. No pathologic lesions were identified in control pigs (Group 1). Pigs in the VR-2332 challenge group (Group 4) had diffuse interstitial pneumonia, characteristic of an uncomplicated PRRSV infection. Although lung lesions were subjectively more severe in the HP-PRRSV and contact groups, a statistically significant difference in lung scores was only found between the rJXwn06 contact group and the group inoculated with VR-2332 (Groups 4 and 3, P<0.003) ( Fig. 3A). Lymphadenopathy was present in all infected pigs (Groups 2–4), with no statistically significant difference between HP-PRRSV and VR-2332 infected groups ( Fig. 4A). Thymic atrophy was also a common finding in infected pigs and was significantly more severe in HP-PRRSV infected and contact pigs compared to VR-2332 infected pigs (P=0.0003 and P<0.0001; Fig. 4B). Table 2 summarizes aspects of the clinical disease seen in this study.
Table 1.
Macroscopic lesions in treatment groups.
| Organ | Macroscopic lesions | Group 1: Control | Groups 2–3: rJXwn06 challenge and contact | Group 4: VR-2332 challenge pigs |
|---|---|---|---|---|
| Lung | ||||
| Interstitial pneumonia | − | + | + | |
| Pulmonary edema | − | + | − | |
| Infarct/Hemorrhage | − | + | − | |
| Pleuritis | − | + | − | |
| Thymus | − | |||
| Thymic atrophy | − | + | − | |
| Lymph nodes | ||||
| Lymphadenopathy | − | + | + | |
| Hemorrhage, Congestion | − | + | − | |
| Body cavities | ||||
| Hydrothorax, Ascites, Hydropericaridum | − | + | − | |
| Heart | ||||
| Myocardial necrosis/Myocarditis | − | + | − | |
| Hemorrhage | − | + | − | |
| Kidney | ||||
| Petechiae, Hemorrhage | − | + | − | |
| Integument | ||||
| Cyanosis | − | + | − | |
| Periorbital edema | − | + | − | |
| Other | ||||
| Serous atrophy of fat | − | + | − | |
Fig. 3.
Lung scores and associated significance values for all groups. (A) Macroscopic lung scores. (B) Microscopic lung scores.
Fig. 4.
(A) Lymph node weight (LNW) to body weight (BW) ratios showed pronounced lymphadenopathy in all infected animals (Groups 2–4) as compared to control animals (Group 1)(P=0.02 or higher significance), but no statistical significance between PRRSV infected swine. (B) Thymus weight (TW) to BW ratios showed thymic atrophy in all infected animals (Groups 2–4) as compared to control animals (P<0.0001), and statistically significant differences between HP-PRRSV inoculated (P<0.0001) and naturally infected (P=0.0003) animals compared to VR-2332 inoculated swine.
Table 2.
Summary of clinical disease.
| Symptom | Chinese | Contact | VR-2332 | Control |
|---|---|---|---|---|
| Anorexia | +++a | +++ | – | − |
| Listlessness | +++ | +++ | – | − |
| Dyspnea | +++ | +++ | – | − |
| Increased respiration | +++ | +++ | + | − |
| Average weight gain (kg) | −0.79 | +7.23 | +5.31 | |
| % Lung affected | 30.8% | 22.6% | 0.10% | |
| Thymus atrophy (TW/BW) | 0.38 | 0.73 | 1.02 | |
| Lymph nodes (LNW/BW) | 0.23 | 0.27 | 0.14 | |
| Bacterial load—brain, heart & lungs | +++ | +++ | − | − |
| Death | 4/12 | 0/4 | 1b/8 | 0/6 |
Subjective scale where +++ is most severe and − is normal.
Gastric dilatation and volvulus unrelated to PRRSV infection.
Histopathology and immunohistochemistry
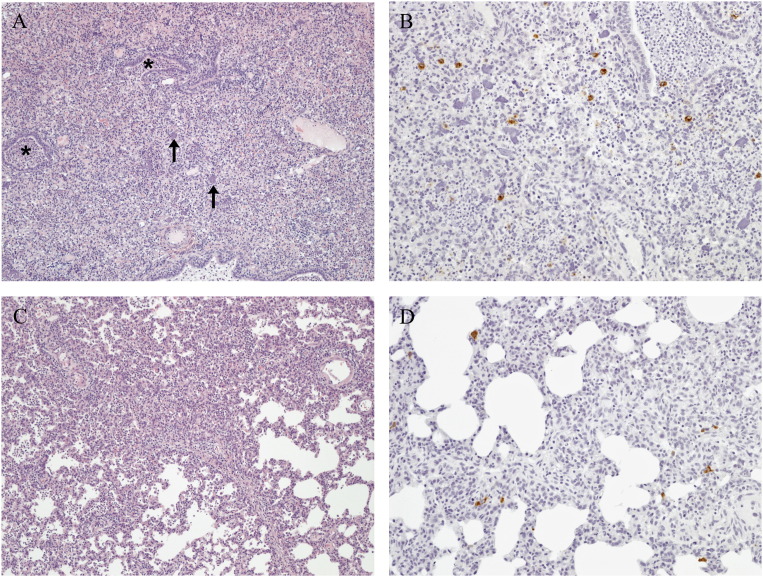
Microscopic lung lesions consisted of histiocytic interstitial pneumonia with increased numbers of macrophages in alveolar septa and lumina ( Fig. 5A, C). Type II pneumocyte hyperplasia was also present. These lesions were seen in the HP- PRRSV challenge group (Group 2), the HP-PRRSV contact pigs (Group 3) and the VR-2332 challenge group (Group 4). Mean interstitial pneumonia lesions were 3.33, 3.75, and 2.00 for Chinese PRRSV challenge group (Group 2), contact Chinese PRRSV pigs (Group 3) and VR-2332 challenge (Group 4), respectively (Fig. 3B). Lesions were significantly (P=0.0002) more severe in the Chinese PRRSV challenge group (Group 2) and contact Chinese PRRSV pigs (Group 3) as compared to the VR-2332 challenge (Group 4). Two sham inoculated control pigs had mild lesions with a score of 1, while the remaining pigs had no lesions. Immunohistochemistry labeling for PRRSV nucleocapsid was positive in 100% of Chinese PRRSV challenge and contact pigs (Groups 2 and 3) and 88% of VR-2332 challenge pigs (Group 4), with mean IHC labeling scores of 2.08, 1.75, and 1.63, respectively (Fig. 5). Positive labeling was present in alveolar macrophages and interstitial macrophages for all infected groups (Fig. 5B and C). No significant difference was present in the amount of PRRSV labeling in the lung among the three treatment groups. Immunohistochemistry labeling for PRRSV in lung tissue was negative in all sham inoculated control pigs.
Fig. 5.
(A) Lung section from a Group 2 pig inoculated with rJXwn06. Severe diffuse histiocytic and necrotizing interstitial pneumonia with a suppurative bronchiolitis (asterisks). Alveolar lumina were filled with alveolar macrophages, neutrophils and necrotic debris; clumps of chromatin (arrows) were present. Alveolar septa were thickened by histiocytes and vessels were cuffed by perivascular macrophages, lymphocytes and plasma cells. 100x. (B) Lung section from a Group 2 HP-PRRSV inoculated pig. Positive cytoplasmic PRRSV labeling was seen in alveolar macrophages, interstitial macrophages and macrophages present in the bronchiole lumen. 200x, Immunoperoxidase-DAB, hematoxylin counterstain. (C) Lung section from a Group 4 pig inoculated with strain VR-2332. Moderate multifocal histiocytic interstitial pneumonia. Alveolar septa were thickened by histiocytes and alveolar lumina contained alveolar macrophages and neutrophils. 100x, HE. (D) Lung section from a Group 4 strain VR-2332 inoculated pig. Positive cytoplasmic PRRSV labeling was seen in alveolar macrophages and interstitial macrophages. 200x, Immunoperoxidase-DAB, hematoxylin counterstain.
Serology
Sera collected at −1, 7, and 13 dpe were tested for PRRSV antibody. Most swine infected with rJXwn06 (Group 2) had seroconverted by 7 dpe (mean S/P=0.51, SD=0.30), and all had seroconverted by 13 dpe (mean S/P=1.14, SD=0.26); however, the rJXwn06 contact animals (Group 3) had not yet seroconverted by 5 dpe (mean S/P=0.00, SD=0.00) but did by 11 dpe (mean S/P=0.99, SD=0.12). VR-2332 infected pigs (Group 4) were seronegative at 7 dpe (mean S/P=0.09, SD=0.12); but positive by 13 dpe (mean S/P=1.05, SD=0.37). The sham inoculated control animals (Group 1) remained negative for PRRSV nucleocapsid antibody throughout the study.
Virus isolation
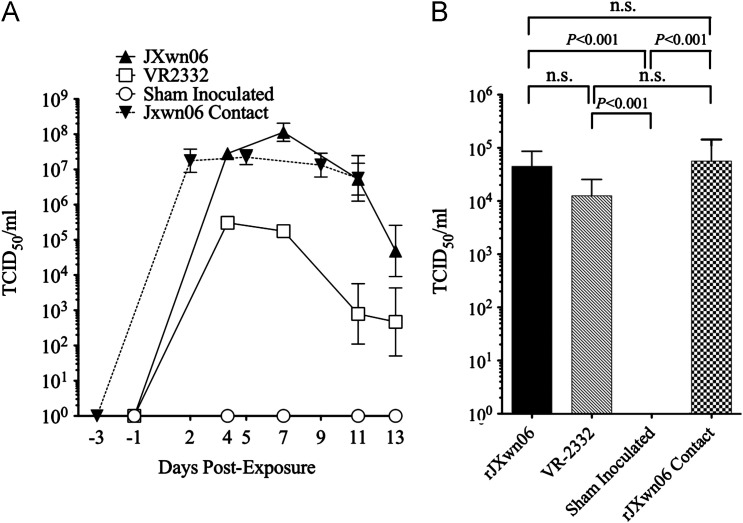
All serum and BALF samples were screened on MARC-145 cells for infectious virus, and VI positive samples were then titered. A significant increase in virus load (100–1000 fold) for the rJXwn06 challenge and rJXwn06 contact pigs was found in the serum when compared to the VR-2332 challenge group ( Fig. 6A). No significant differences were found in serum virus titer between the rJXwn06 challenge and rJXwn06 contact groups at any time points even though the contact pigs were exposed to rJXwn06 infected pigs at 2 dpe. At necropsy, no significant differences in BALF titers were found among the rJXwn06 challenge, rJXwn06 contact, or VR-2332 challenge groups (Fig. 6B). Control serum and BALF samples were VI negative throughout the experiment (Fig. 6A and B).
Fig. 6.
Virus titers expressed as 50% of the tissue culture infective dose per ml (TCID50/ml) of sample. (A) Serum samples during the course of the experiment and (B) Brochoalvealor lavage fluid (BALF) after study termination.
Quantitative real-time RT-PCR (qRT-PCR)
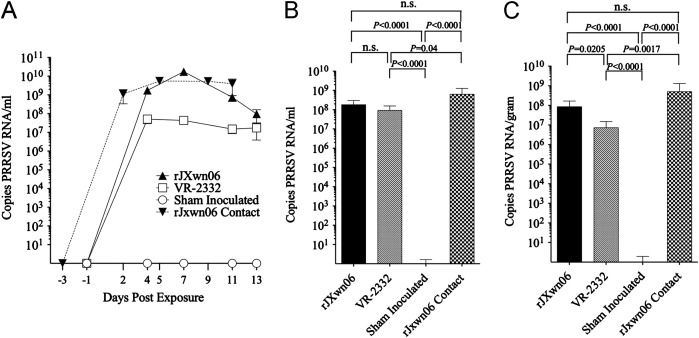
To examine the levels of PRRSV RNA and to strengthen the case for the apparent difference in viral loads between the HP-PRRSV and VR-2332 inoculated animals seen above, qRT-PCR was completed on all serum samples as well as BALF and lymph node tissue ( Fig. 7). The level of viral RNA detected in serum samples mirrors the results obtained by virus isolation on MARC-145 cells (Fig. 7A, P≤0.005 at all dpe). The qRT-PCR results from BALF showed little or no significant difference between the infected groups (Fig. 7B). TBLN samples suggested that swine inoculated with VR-2332 had significantly less viral RNA present than those inoculated (P=0.0205) or infected (P=0.0017) with HP-PRRSV (Fig. 7C). Sham inoculated control serum, BALF, and TBLN tissue samples were viral RNA negative throughout the experiment (Fig. 7A–C).
Fig. 7.
Quantitative RT-PCR (qRT-PCR) results in copies of PRRSV RNA per ml. A. Serum samples. B. BALF. C. Lymph node samples (LN).
Bacterial isolation
Bacteria were isolated from the BALF of 8 of the 12 rJXwn06 inoculated pigs (Group 2) and 2 of the 4 rJXwn06 contact pigs (Group 3) but from only 1 of 8 pigs in each of the sham inoculated control (Group 1) and VR-2332 inoculated (Group 4) groups. In the rJXwn06 challenge group Pasteurella multocida was isolated from four pigs (ranging from 50 to >300 CFU/100 μL), P. multocida (>300 CFU/100 μL) and Actinobacillus suis (>300 CFU/100 μL) were isolated from two pigs, P. multocida, A. suis and Streptococcus suis (approximately 50, 100, and 25 CFU/100 μL, respectively) from one pig, and Staphylococcus aureus and Klebsiella pneumoniae (approximately 150 and 20 CFU/100 μL, respectively) were isolated from one pig. In the contact group, P. multocida and A. suis were isolated from one pig (>300 CFU/100 μL each) and P. multocida, A. suis, and Arcanobacterium pyogenes (approximately 50, >300, and >300 CFU/100 μL, respectively) were isolated from a second pig. S. aureus and Escherichia coli (>300 CFU/100 μL each) were isolated from 1 VR-2332 inoculated pig, and Bordetella bronchiseptica (65 CFU/100 μL) was isolated from 1 sham inoculated control pig.
Effects of PRRSV infection on innate and adaptive immunity cytokine levels
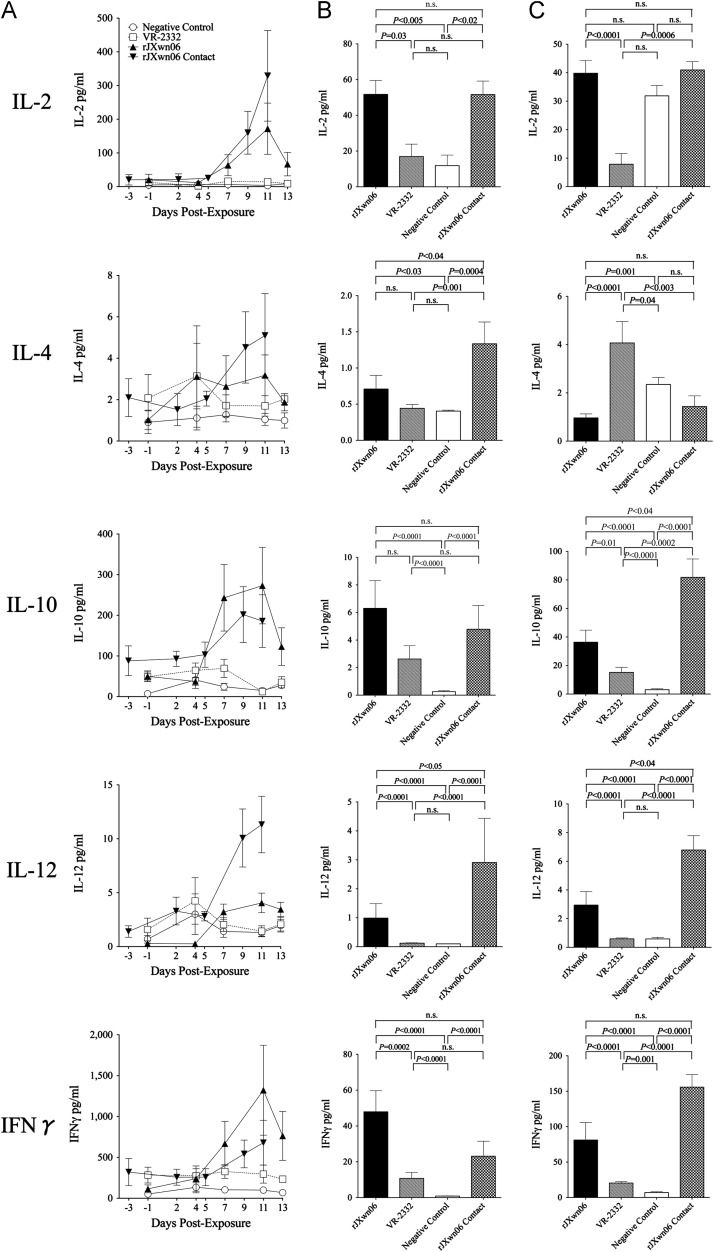
Results on 10 cytokines measured in serum, BALF and TBLN homogenates are presented in Fig. 8, Fig. 9 and the changes in cytokine levels relative to sham-inoculated controls are listed in Table 3. Pigs directly inoculated with rJXwn06 (Group 2) had significantly elevated average serum levels of IFNα, IL-1β, IL-2, IL-10 and IFNγ, ranging from 6 to 11 times the levels detected in sera of sham inoculated controls (Group 1). In pigs inoculated with rJXwn06, serum IFNα levels were significantly elevated at 4 dpe in comparison to both controls and VR-2332 exposed pigs (P<0.05) and remained significantly elevated at 7 dpe versus both groups. Serum levels of IL-10 were elevated by 7 dpe, with IL-2, IL-10, IL-12 and IFNγ all signficantly elevated at 11 dpe compared to both controls and VR-2332 exposed pigs (P<0.05). Compared with sham inoculated controls, contact pigs exposed to rJXwn06 (Group3) developed significant serum level elevations (3 to 89 times greater) in 3 out of 5 innate immunity cytokines measured (IFNα, TNFα and IL-1β) and elevated levels (3 to 21 times greater) for all 5 cytokines measured associated with adaptive immunity (IL-2, IL-4, IL-10, IL-12 and IFNγ). In pigs exposed to rJXwn06 by contact transmission, serum IFNα levels were signifcantly elevated at 2, 5 and 9 dpe in comparison to controls (P<0.05). In addition, contact pigs had significant elevations in IL-1β at 9 and 11 dpe (P<0.05). Contact pigs also had significantly elevated levels of all adaptive immunity cytokines at 9 and 11 dpe (P<0.05) with IL-10 also significantly elevated at 5 dpe. In contrast, none of the 10 cytokines measured had significant elevations in serum levels detected in pigs inoculated with the North American prototype strain VR-2332 (Group 4) when compared with sham inoculated controls.
Fig. 8.
Innate cytokine protein (IFNα, TNFα, IL-1β, IL-6, IL-8) levels in (A) Serum samples at specified days post exposure, (B) Bronchoalveolar lavage (BALF) at necropsy, and (C) Tracheobronchial lymph node (TBLN) at necropsy.
Fig. 9.
Adaptive cytokine protein (IL-2, IL-4, IL-10, IL-12, IFNγ) levels in (A) Serum samples at specified days post exposure, (B) Bronchoalveolar lavage (BALF) at necropsy, and (C) Tracheobronchial lymph node (TBLN) at necropsy.
Table 3.
Changes in cytokine levels in sampled tissues relative to non-challenged control pigs (P-values in comparison to non-challenged controls). Serum values are an area under the curve overall average level measured in samples collected after exposure to virus. Geometric means back-calculated from Log10 means were used to determine relative changes in TBLN and BALF cytokine levels compared to sham-inoculated controls. NS=not significant
| Change Relative to Controls | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cytokine | Serum | P | BALF | P | TBLN | P | ||
| JXwn06 principals | Innate | IFNα | 6.2 X | <0.0001 | 2.4 X | NS | 9.8 X | 0.005 |
| TNFα | 3.9 X | NS | 22.3 X | <0.0001 | 1.0 X | NS | ||
| IL-1β | 9.6 X | 0.04 | 20.1 X | <0.0001 | 2.0 X | 0.002 | ||
| IL-6 | 1.5 X | NS | 3.5 X | NS | 0.9 X | NS | ||
| IL-8 | 0.9 X | NS | 7.4 X | <0.0001 | 3.0 X | 0.001 | ||
| Adaptive | IL-2 | 11.2 X | 0.04 | 4.4 X | 0.005 | 1.2 X | NS | |
| IL-4 | 2.3 X | NS | 1.8 X | <0.03 | 0.4 X | 0.001 | ||
| IL-10 | 6.2 X | 0.002 | 24.1 X | <0.0001 | 11.9 X | <0.0001 | ||
| IL-12 | 1.3 X | NS | 10.1 X | <0.0001 | 5.0 X | <0.0001 | ||
| IFNγ | 7.0 X | 0.01 | 53.5 X | <0.0001 | 11.5 X | <0.0001 | ||
| JXwn06 contacts | Innate | IFNα | 89.4 X | 0.003 | 8.0 X | 0.007 | 37.8 X | 0.0005 |
| TNFα | 3.0 X | 0.02 | 20.6 X | <0.0001 | 2.4 X | 0.03 | ||
| IL-1β | 4.7 X | 0.05 | 4.4 X | 0.02 | 3.3 X | 0.0001 | ||
| IL-6 | 0.2 X | NS | 4.0 X | NS | 1.2 X | NS | ||
| IL-8 | 0.9 X | NS | 7.6 X | <0.0001 | 6.0 X | <0.0001 | ||
| Adaptive | IL-2 | 20.8 X | 0.0007 | 4.3 X | 0.02 | 1.3 X | NS | |
| IL-4 | 3.0 X | 0.04 | 3.3 X | 0.0004 | 0.6 X | NS | ||
| IL-10 | 5.8 X | 0.002 | 18.3 X | <0.0001 | 26.8 X | <0.0001 | ||
| IL-12 | 3.6 X | 0.004 | 29.6 X | <0.0001 | 11.6 X | <0.0001 | ||
| IFNγ | 4.3 X | 0.009 | 25.8 X | <0.0001 | 22.1 X | <0.0001 | ||
| VR2332 principals | Innate | IFNα | 1.4 X | NS | 1.2 X | NS | 1.3 X | NS |
| TNFα | 1.5 X | NS | 2.4 X | NS | 0.9 X | NS | ||
| IL-1β | 1.6 X | NS | 5.0 X | 0.004 | 1.0 X | NS | ||
| IL-6 | 0.04 X | NS | 0.6 X | NS | 0.6 X | NS | ||
| IL-8 | 0.7 X | NS | 3.8 X | <0.0001 | 1.4 X | NS | ||
| Adaptive | IL-2 | 1.6 X | NS | 1.4 X | NS | 0.2 X | 0.0005 | |
| IL-4 | 1.9 X | NS | 1.1 X | NS | 1.7 X | 0.04 | ||
| IL-10 | 2.0 X | NS | 10.1 X | <0.0001 | 5.0 X | <0.0001 | ||
| IL-12 | 1.4 X | NS | 1.2 X | NS | 1.0 X | NS | ||
| IFNγ | 2.8 X | NS | 12.0 X | <0001 | 2.9 X | 0.001 | ||
Similarly, when compared to sham inoculated controls, BALF of swine inoculated with rJXwn06 (Group 2) had significantly elevated (7 to 22 times greater) levels of 3 out of 5 innate immunity cytokines measured (TNFα, IL-1β and IL-8) and elevated levels (2 to 54 times greater) for all 5 cytokines measured associated with adaptive immunity (IL-2, IL-4, IL-10, IL-12 and IFNγ). Compared with sham inoculated controls, contact swine exposed to rJXwn06 (Group 3) had significantly elevated BALF cytokine levels (4 to 21 times greater) in 4 out of 5 innate immunity cytokines measured (IFNα, TNFα and IL-8) and elevated levels (3 to 30 times greater) for all 5 cytokines measured associated with adaptive immunity (IL-2, IL-4, IL-10, IL-12 and IFNγ). In contrast, swine inoculated with VR-2332 (Group 4) had significantly elevated BALF cytokine levels (4 times greater) for only 2 out of 5 innate immunity cytokines measured (IL-1β and IL-8) and elevated levels (10 to 12 times greater) for 2 of 5 cytokines measured associated with adaptive immunity (IL-10 and IFNγ).
Discussion
In this study, we compared the pathogenicity of Chinese HP-PRRSV strain rJXwn06 to North American prototype VR-2332 in U.S. high health swine under controlled conditions. The clinical disease induced in this study by the HP-PRRSV virus was similar to what has been reported in Asia for PHFD and for experimental infections with the wild-type or rescued virus (Li et al., 2007, Lv et al., 2008, Tian et al., 2007, Tong et al., 2007, Wu et al., 2009, Zhou et al., 2009, Zhou et al., 2008). Likewise, the clinical disease induced by the VR-2332 virus was similar to that observed in previous reports (Faaberg et al., 2010, Rossow et al., 1994).
Clinical disease and pathology were much more severe in the rJXwn06 group and their contacts than in the VR-2332 group. Overall, gross pathology lesions in the rJXwn06 challenged and contact groups were much more extensive and not restricted solely to the respiratory tract and lymph nodes as was the case with the VR-2332 challenged pigs. The high occurrence of bacterial co-infections in the rJXwn06 challenge and contact swine likely played a prominent role in the difference in pathology and clinical disease between groups. Bacterial co-infections in pigs naturally infected with PRRSV have been documented, with susceptibility attributed to factors including PRRSV strain differences, host genetics, management practices and environmental factors (Rossow, 1998). In this study, we used swine of high health status in a controlled research environment that were from 2- and 3-way crosses of commercial genetic lines. We believe the incidence and magnitude of bacteria isolated from the rJXwn06 challenge and contact groups when compared to the VR-2332 and control groups suggest the differences in secondary bacterial infection susceptibility are specific to viral strain. In this study, we observed a 30% mortality rate in rJXwn06 infected pigs by 13 dpe, which is less mortality than the original report (Zhou et al., 2009), but similar to other HP-PRRSV strains used (Li et al., 2007, Lv et al., 2008, Zhou et al., 2008). Possible explanations for differences may be the route and dose of inoculation, the age of pigs, the HP-PRRSV strain utilized, and the time course of study. In addition, since the HP-PRRSV lineage may exacerbate subclinical bacterial infections, it is possible different endemic bacterial infections played a role in apparent different mortality rates.
In natural infections with Chinese HP-PRRSV, pulmonary interstitial hyperplasia with hemorrhage and edema is described, which suggests an acute septicemic process due to a secondary bacterial pathogen (Tian et al., 2007, Zhou and Yang, 2010). Upon histopathologic examination of tissues from this study, North American pigs directly inoculated with rJXwn06, as well as contact pigs, had an interstitial pneumonia that was significantly more severe than the VR-2332 inoculated group, which is consistent with the severity of disease reported in China. Although pulmonary lesions were more severe in the rJXwn06 challenge and contact pigs, the amount of antigen labeling was not significantly different from the VR-2332 inoculated swine. Since levels of proinflammatory cytokines, including TNFα, IL-1β and IL-8, were significantly increased in the BALF of rJXwn06-inoculated and contact pigs, the host response to HP-PRRSV may play a role in the augmented lung pathology seen. In addition, the increased incidence of secondary bacterial infections in rJXwn06 challenge and contact pigs may have contributed to increased cytokine production and resultant immunopathology.
There are numerous reports about the interplay of PRRSV with the swine immune system that describe variable responses to infection at the cellular and cytokine level (Miguel et al., 2010, Thanawongnuwech et al., 2004, Thanawongnuwech and Thacker, 2003, Wang et al., 2011). Although a large part of this variability may result from differing methods, challenge viruses, outbred pigs, and experimental designs, there are consistent findings emerging among the studies of increases in levels of selected cytokines associated with both innate and adaptive immunity. Here we report a comprehensive assessment of the effects of PRRSV infection on levels of cytokines critical to innate (IFNα/β, TNFα, IL-1β, IL6 and IL-8) and adaptive (IL-2, IL-4, IL-10, IL-12 and IFNγ) immune systems in serum, BALF and TBLN homogenates. It was demonstrated that infection with a highly pathogenic strain of PRRSV elicited a significant elevation of all adaptive immunity cytokines measured in BALF, as well as a majority of these cytokines in serum and TBLN homogenates of the same groups of pigs. This observation is consistent with previous reports of lymphoid hyperplasia resulting in immune dysregulation in pigs infected with PRRSV (Lemke et al., 2004). Swine infected here with VR-2332 and previous studies with other viral strains found that PRRSV infections are associated with delayed or failed production of detectable serum IFNα levels (Diaz et al., 2005, Gomez-Laguna et al., 2010a, Gomez-Laguna et al., 2010b). Previous studies indicate that PRRSV inhibits IFN-induced JAK/STAT signaling, thus possibly accounting for the delayed or inadequate IFNα response by the host [reviewed in (Sun et al., 2012)]. In contrast, in pigs infected in this study with rJXwn06, we observed significant elevations of serum IFNα as the first cytokine to peak following exposure to rJXwn06 in either the directly challenged or contact pigs, with the zenith occurring 4 and 2 dpe, respectively. However, consistent with a previous study (Gomez-Laguna et al., 2009), elevated serum IFNα had little to no apparent effect on virus clearance as the viremia peaked in the rJXwn06 challenge and contact pigs on dpe 7 and 4, respectively, and remained above the serum virus levels of VR-2332 pigs until the end of the study (11 and 13 dpe). It is well documented that PRRSV infection will increase the susceptibility of swine to co-infection with various bacteria (Brockmeier et al., 2001, Thanawongnuwech et al., 2004, Thanawongnuwech and Thacker, 2003, Thanawongnuwech et al., 1997, Thanawongnuwech et al., 2001, Xu et al., 2010), and based on previous findings with other HP-PRRSV strains (Xu et al., 2010) and our current rJXwn06 findings, it is clear that exposure to HP-PRRSV greatly increases the likelihood of secondary bacterial infection due to commensal or pathogenic organisms typically found in the swine upper respiratory tract. Whether the significant elevations of multiple cytokines that were measured in serum, BALF and TBLN following exposure to rJXwn06 were a direct result of the virus, an indirect effect mediated by the secondary bacterial infections, from extensive host tissue damage, or a combination of all of these events cannot be determined from our experiments. However, the pattern of multiple cytokines being elevated nearly simultaneously in serum (8 of 10 cytokines in rJXwn06 contact pigs and 5 of 10 cytokines in direct inoculated pigs) has not previously been reported and was not detected in VR-2332 infected swine.
Cytokines are a diverse collection of peptides that elicit a wide range of biological responses and are characteristically understood in the context of an immune response wherein inflammation and immunity are carefully orchestrated by sequential secretion of cytokines that coordinate innate and adaptive immune responses. Macrophages and stressed or damaged cells typically initiate a cytokine cascade through secretion of chemokines and proinflammatory cytokines in order to initiate the innate immune response at sites of acute infection or damage. Generally, they act locally at nano- to picogram levels with short half-lives and transient activity. However, inflammatory cytokine cascades classically comprise a sequential appearance and disappearance of proinflammatory cytokines (e.g., TNFα, IL-1 and IL-6) intended to activate immune cells and their recruitment to generate additional cytokines and chemokines. This initial wave of cytokine production is usually followed by anti-inflammatory cytokine production (mainly the IL-10 family) to moderate or down regulate the pro-inflammatory cytokines. Cytokine cascades are therefore usually sequential with transiently detectable levels in peripheral blood; any dysregulation of these cascades can lead to adverse immunopathological responses.
While significant elevations of several cytokine levels in tissues such as BALF and TBLN were expected, near simultaneous elevation of several cytokines in serum was not entirely expected as part of a normal host immune response. Moreover, the levels of cytokines detected were in several instances significantly elevated (usually several times more) over levels detected with the low virulence North American prototype PRRSV strain, VR-2332. Prolonged elevations of serum IFNγ levels have been reported in swine infected with PRRSV, a finding in contrast to serum IFNγ in pigs infected with influenza or respiratory coronavirus, where minimal transient detectable levels are observed (Wesley et al., 2006).
In swine infected with rJXwn06, it appears the normal sequence of cytokine production (e.g., IFNγ, TNFα, and IL-2 often being the first cytokines produced in response to a viral infection, followed by IL-6 and IL-10) leading to effective virus clearance and a normal immune response was dysregulated given that significant elevations of several serum cytokines levels were still evident at 7 to 11 dpe. Two recent studies have reported HP-PRRSV infection in swine results in down regulation of a key toll-like receptor adapter gene, SARM1 (sterile α- and armadillo-motif-containing protein) (Zhou et al., 2011, Zhou et al., 2012). SARM1 normally dampens the proinflammatory immune response by attenuating NF-κB activation and decreasing expression of IL1, IL-8 and TNFα (Carty et al., 2006). Previous studies in pigs infected with strains of HP-PRRSV were reported to have swollen livers and petechial hemorrhages on the kidneys as well as immunohistochemical staining evidence of viral antigen in the liver and kidneys (among other tissues)(Li et al., 2007, Tian et al., 2007). Impaired hepatic and renal function as a consequence of viral or secondary bacterial disease could have a significant effect on clearance of the cytokines detected in serum, and contribute to an apparent severe cytokine release syndrome.
Whether our findings represent a parallel condition in swine to severe cytokine release syndromes or cytokine storms reported in humans that have been attributed to various causes cannot be proven with our data (Descotes and Gouraud, 2008, Tarrant, 2010). However, given PRRSV causes polyclonal B cell activation, autoimmunity, lymphoid hyperplasia and hypergammaglobulinemia in pigs (Lemke et al., 2004), and it modulates multiple intracellular signaling pathways [reviewed in (Sun et al., 2012)], it is not surprising to find an exaggerated immune stimulation in swine infected with a particularly virulent strain of PRRSV. Early studies with cytokine administration to livestock species identified potential toxicities associated with systemic cytokine administration. Interferon-γ was found to have beneficial activity on immune function but was too toxic for practical usage due to febrile responses observed within 24 h of a single dose of 1.25 μg/kg of body weight (Roth and Frank, 1989). Similarly, administration of 166 ng of bovine IL-1β/kg of body weight every 8 h for 4 days caused transient fever, inappetence, increased pulse and respiratory rate, and diuresis (Goff et al., 1992). Blood cytokines have been proposed as biomarkers of in vivo toxicity associated with new drug development (Tarrant, 2010). Under most disease scenarios where there is rapid resolution of the infection by the host immune response, much of the biological activity of cytokines will occur in the locally infected tissues and elevated levels of the cytokines may not be detected in serum.
We cannot definitively state whether the elevated cytokine syndrome reported here contributes to the pathophysiology of the disease caused by this highly pathogenic PRRSV isolate. However, the association between the elevated levels of multiple cytokines and the severe morbidity and high mortality reported is consistent with a multiple cytokine toxicity syndrome in humans associated with various therapeutics (Tarrant, 2010). Adverse reactions ranging from mild-to-moderate flu-like reactions to severe cytokine release syndromes have been observed with many therapeutic proteins in current use in human medicine, and some result in severe and even potentially life-threatening syndromes (reviewed in (Tarrant, 2010)). Macrophage activation syndrome and cytokine storm are different names for two syndromes that share many features including a massive inflammatory response, elevated serum cytokine levels, multiorgan system disease and often death (Behrens et al., 2011). Although these syndromes may be clinically indistinguishable, the cytokines that predominate in each may differ with TNFα being dominant in bacterial sepsis and IFNγ predominate in the macrophage activation syndrome (Behrens et al., 2011). The exact pathophysiology of the systemic toxicity in these syndromes is not fully defined. In pigs infected by natural contact with rJXwn06 inoculated pigs, we detected significant serum elevations in IFNα, TNFα, IL-1β, IL-2, IL-4, IL-10, IL-12 and IFNγ. However, given that PRRSV infects the macrophage cell line in pigs, the elevated serum cytokine levels in pigs infected with HP-PRRSV may represent both conditions (macrophage activation syndrome and cytokine storm) and contribute to the multiorgan damage and high mortality reported here.
Materials and methods
Cells and viruses
MARC-145 cells were cultured in minimum essential medium (MEM, SAFC 56416C) with 10% fetal bovine serum at 37 °C, 5% CO2. Wild-type (wt) Type 2 PRRSV strain VR-2332 (GenBank U87392), passage 6 on MARC-145 cells, was titrated, and used for the swine study. Virus (rescued rJXwn06; rJXwn06) was rescued from a cloned cDNA of Chinese highly pathogenic Type 2 PRRSV strain JXwn06 in MARC-145 cells [pWSK-JXwn; GenBank EF641008, (Zhou et al., 2009)] and passaged 3 times on MARC-145 cells for use in the swine study.
Swine study
The in vivo swine study described here was performed at the National Animal Disease Center under approval from its Animal Care and Use Committee. Thirty-two 10-week-old cross-bred pigs were obtained from a U.S. high-health status herd and were found to be free of PRRSV antibodies by HerdChek ELISA, influenza virus antibodies by NP ELISA, and negative for porcine circovirus type 2 by quantitative real-time PCR (data not shown). One day prior to inoculation, pigs were bled, weighed and randomly assigned to one of four groups. Group 1 (N=8) consisted of sham inoculated control pigs, which received an intranasal 2 ml sham inoculum of MEM on day 0. Group 2 pigs (N=12) were challenged intranasally with 2 ml of 1×106 50% tissue culture infective dose (TCID50)/ml of Chinese PRRSV strain rJXwn06 in animal Biosafety Level 3-Agriculture (BSL3-Ag) housing, where they remained for the duration of the experiment. Group 3 consisted of naïve pigs (N=4) that were placed in contact with Group 2 two days after Group 2 was inoculated (dpe). Group 4 pigs (N=8) were challenged intranasally on day 0 dpe with 2 ml of 1×106 TCID50/ml of Type 2 prototype strain VR-2332. Groups 1 and 4 were housed in separate isolation rooms in an ABSL2 facility.
Clinical and pathological examinations
Clinical monitoring of pigs was performed daily throughout the study. Specifically, observations were made regarding the pig's mental alertness, body condition, appetite, activity level, and clinical signs of respiratory or systemic disease. Serum was collected on −1, 4, 7, 11, and 13 dpe (sera from Group 3 pigs was collected on −3, 2, 5, 9 and 11 dpe), and pigs were weighed on −1, 7 and 13 dpe (Group 3 were weighed on −3, 5 and 11 dpe). Necropsy was scheduled on dpe 13 (dpe 11 for Group 3), or sooner if pigs died or were euthanized due to severe disease. At necropsy, a thorough post-mortem examination was performed, and a complete set of samples was collected for evaluating disease severity. Tracheobronchial lymph nodes (TBLN) and thymic tissue were weighed (LNW and TW, respectively) and compared to respective body weights (BW) to derive a tissue mass index (LNW/BW, TW/BW) measuring the effect of PRRSV infection on organ weight (Mengeling et al., 2003). Upon removal, lungs were examined and extent of macroscopic lung lesions was estimated, as previously described, and reported as a percentage of lungs affected (Halbur et al., 1996). Sections of tissues (lung, tracheobronchial lymph node, trachea, thymus, heart, tonsil, spleen, iliac lymph node, mesenteric lymph nodes, ileum, bone marrow, kidneys, liver, inguinal lymph node, cerebrum, brainstem, cerebellum and ventral midbrain) were collected into 10% neutral-buffered formalin for histopathology and immunohistochemistry. TBLN were collected for RNA extraction and cytokine protein assays. Bronchoalveolar lavage fluid (BALF) was collected after removing whole lungs from pigs and aseptically lavaging with 50 ml antibiotic-free MEM.
Microscopic lesions
Tissues were processed by routine histopathologic procedures and slides were stained with hematoxylin and eosin. A board-certified veterinary pathologist blinded to treatment groups evaluated microscopic lesions. Lung sections were scored on a 5-point scale that accounted for distribution and severity of interstitial pneumonia: 0-No Lesions, 1-Mild, focal to multifocal interstitial pneumonia (<50% of lung section affected), 2-Moderate, multifocal to coalescing (50–75% of lung section affected), 3-Severe, patchy to coalescing and extensive (75–90% of lung section affected), and 4-Severe and diffuse (>90% of lung section affected).
Immunohistochemistry
PRRSV-specific antigen was detected in lung tissues using a previously described immunohistochemical (IHC) method with minor modifications (Halbur et al., 1995). Briefly, tissue sections were deparaffinized and hydrated in distilled water. Slides were quenched in 3% hydrogen peroxide for 10 min, rinsed three times in distilled water and treated in 0.05% protease for 2 min. Slides were then rinsed three times in distilled water. A primary monoclonal antibody (MAb) cocktail of one part 1:800 PRRSV SDOW17 (RTI, Brookings, SD) and one part 1:500 PRRSV SR30 (RTI, Brookings, SD) was applied to the slides and were incubated at room temperature for 1 h. Bound MAbs were stained with peroxidase-labeled anti-mouse IgG followed by chromogen using the DAKO LSAB2-HRP Detection System (DAKO, Carpinteria, CA) according to the manufacturer's instructions. 3,3′-diaminobenzidine (DAB; Vector Laboratories, Burlingame, CA) was applied to the slides for 5 min. The slides were rinsed in deionized water and counterstained with Gill's hematoxylin. PRRSV labeling was graded on a 4 point scale of 0: None, 1: Mild scattered signals (1 to 20 cells in entire section), 2: Moderate scattered signals (less than or equal to 50% of high power fields (hpf) containing immunolabeling) and 3: Abundant scattered signals (greater than 50% of hpf contain labeling and/or there are at least 2–3 groups of 30 cells or more with staining).
Virus isolation
Virus isolation was attempted on all serum and BALF samples as described previously (Faaberg et al., 2010). Those samples that were positive by virus isolation were then titered by serial dilution on MARC-145 cells to determine the quantity of virus present to produce a cytopathic effect in 50% of inoculated tissue culture cells (TCID50).
Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR), as previously described (Faaberg et al., 2010), was used to determine the amount of viral RNA per ml of serum and BALF, and per gram of TBLN homogenized tissue prepared as described below.
Serology
Serum samples were tested on study days −1, 7 and 13 for evidence of seroconversion with the PRRS 2XR enzyme-linked immunosorbent assay (HerdChek ELISA; IDEXX Laboratories). A sample was considered positive for antibodies to PRRSV nucleocapsid protein if the sample-to-positive (S/P) ratio was equal to or greater than 0.4.
Cytokine assays
Levels of TNFα, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70 and IFNγ cytokine levels (pg/ml) were measured in serum, BALF and TBLN samples diluted 1:2 with dilution buffer supplied by the manufacturer using a SearchLight (Aushon BioSystems, Woburn, MA, USA) customized multiplex immunoassay (M-ELISA) following the manufacturer's protocol. M-ELISA samples were assayed in duplicate. The ELISA limits of detection for TNFα, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70 and IFNγ were 0.2, 2.4, 0.78, 0.39, 1.6, 0.098, 0.001, 0.098, and 0.78 pg/ml respectively. Approximately 1 g of TBLN was homogenized in 750 μl of lysis buffer containing 0.5% Triton X-100, 150 mM NaCl, 15 mM Tris, 1 mM CaCl2, and 1 mM MgCl2, pH 7.4, using a tissue homogenizer (Biospec Products, Bartlesville, OK) (Greenberger et al., 1995). Homogenates were incubated on ice for 30 min, then centrifuged at 1258 X g for 10 min. Supernatants were collected, passed through a 0.45-micron filter (Gelman Sciences, Ann Arbor, MI), then stored at −20 °C prior to assessment of cytokine levels.
IFNα enzyme-linked immunosorbent assay (ELISA)
IFNα protein was measured with a porcine IFNα specific ELISA by using F17 monoclonal antibody (MAb) and K9 MAb (R&D Systems Inc.) as previously described (Miller et al., 2009). MAb K9 was conjugated with horseradish peroxidase (HRP) using a peroxidase labeling kit (Roche Molecular Biochemical, Indianapolis, IN). Immulux HB flat-bottomed 96-well plates (Dynex Technology, Chantilly, VA) were coated overnight at 4 °C with F17 at a concentration of 3 μg/plate in coating buffer (100 mM carbonate buffer, pH 9.6, Sigma Inc., St. Louis, MO). After blocking with 1% non-fat dried milk, 0.05% Tween 20 in phosphate buffered saline (PBS) for 1 h at 37 °C, the plates were washed three times with 0.05% Tween 20 in PBS. Samples (50 μl) were added into each well containing 50 μl of 1% non-fat dried milk, 0.05% Tween 20 in PBS and incubated for 2 h at 37 °C. Following three washes, 100 μl of peroxidase conjugated K9 was added to each well. After 1 h incubation, at 37 °C, and three washes, 100 μl of substrate solution, tetramethylbenzidine (KPL Inc., Gaithersburg, MD), was added to each well. After 30 min, the reaction was stopped with tetramethylbenzidine stop solution (KPL Inc., Gaithersburg, MD) and the optical density was measured at 450 nm by an ELISA plate reader. Quantified recombinant porcine IFNα (rIFNα, R&D Systems Inc., Minneapolis, MN) was used as a standard, and IFNα concentrations were calculated based upon a standard curve. One unit/ml of rIFNα is equivalent to 26 pg/ml.
Bacterial assays
Bacterial culture was performed by plating 100 μl of BALF both on a Casman's agar plate supplemented with 0.01% nicotinamide adenine dinucleotide (NAD) and 5% horse serum, and on a 5% sheep's blood agar plate. Both agar plates were incubated for 48 h at 37 °C. Bacterial identification was performed by 16S rRNA-specific PCR and DNA sequencing.
16S rRNA-specific PCR and DNA sequencing
Whole-cell bacterial lysates, used as templates, were prepared by suspending a colony, ∼2 mm in diameter or the equivalent, in 50 μl of sterile water. The mixture was boiled for 10 min, placed on ice until chilled, and centrifuged at 16,000×g for 1 min to pellet cell debris and stored at −20 °C. Supernatant (5 μl) was used as the template in each PCR. The forward primer (5′–AGAGTTTGATCCTGGCTCAG-3′), designated univ16S-3, is homologous to a highly conserved sequence from the 5′ end of the 16S rRNA gene and the reverse primer (5′–GCGGCTGCTGGCACG-3′), designated univ16S-4, is homologous to a highly conserved sequence between the third and fourth variable regions of the 16S rRNA gene. This previously described primer set generates an amplicon of approximately 520 bp (Register and Yersin, 2005). Reactions were carried out in a volume of 50 μl and contained 2 U AmpliTaq polymerase (Applied Biosystems, Foster City, CA), 5 μl 10x buffer II (100 mM Tris–HCl, pH 8.3, 500 mM KCl), 5 μl dimethyl sulfoxide, 1.5 mM MgCl2, 0.5 μM primers, and 200 μM deoxynucleoside triphosphates. An initial denaturation step of 5 min at 95 °C was followed by 35 cycles of 20 s at 94 °C, 30 s at 55 °C, and 3 min at 72 °C, with a final extension step of 10 min at 72 °C. 10 μl of each PCR was analyzed by agarose gel electrophoresis and PCR products were purified with spin columns (Promega, Madison, WI) and sequenced directly by fluorescence-based cycle sequencing with AmpliTaq and BigDye Terminators on an ABI 377 sequencer at the National Animal Disease Center Genomics Unit. Sequences were analyzed using Geneious 5.0 software (Biomatters Ltd, Auckland, New Zealand).
Data analysis
Quantitative virus copy numbers and serum cytokine levels were analyzed using a mixed linear model for repeated measures (Proc Mixed, SAS 9.2 for Windows, SAS Institute, Cary, NC, USA). Linear combinations of the least squares means estimates for each variable were used in a priori contrasts after testing for either a significant (P<0.05) effect of PRRSV challenge strain (VR-2332, rJXwn06, rJXwn06 contacts or sham inoculated controls). Comparisons were made between groups at each time-point using a 5% level of significance (P<0.05) to assess statistical differences. Log10 transformed virus copy numbers and cytokine levels in BALF and TBLN homogenates were analyzed by analysis of variance using a general linear model for unbalanced data (Proc GLM, SAS 9.2 for Windows, SAS Institute, Cary, NC, USA). A 5% level of significance (P<0.05) was used to assess statistical differences. Geometric mean back transformations were made for final data presentation in figures and tables.
Disclaimer
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Acknowledgments
The authors would like to recognize Ann Vorwald, Sarah Anderson, Deb Adolphson and Amanda Burow for their excellent technical assistance, and Jason Huegel, Brian Pottebaum, and Jason Crabtree for their exceptional animal care. We also appreciate the suggestions on manuscript layout and data organization made by Crystal Loving.
References
- An T.Q. Highly pathogenic porcine reproductive and respiratory syndrome virus, Asia. Emerg. Infect. Dis. 2011;17:1782–1784. doi: 10.3201/eid1709.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens E.M. Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J. Clin. Invest. 2011;121:2264–2277. doi: 10.1172/JCI43157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmeier S.L. Effects of intranasal inoculation with Bordetella bronchiseptica, porcine reproductive and respiratory syndrome virus, or a combination of both organisms on subsequent infection with Pasteurella multocida in pigs. Am. J. Vet. Res. 2001;62:521–525. doi: 10.2460/ajvr.2001.62.521. [DOI] [PubMed] [Google Scholar]
- Carty M. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat. Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- Descotes J., Gouraud A. Clinical immunotoxicity of therapeutic proteins. Expert Opin. Drug Metab. Toxicol. 2008;4:1537–1549. doi: 10.1517/17425250802525496. [DOI] [PubMed] [Google Scholar]
- Diaz I. Immune responses of pigs after experimental infection with a European strain of Porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2005;86:1943–1951. doi: 10.1099/vir.0.80959-0. [DOI] [PubMed] [Google Scholar]
- Ellingson J.S. Vaccine efficacy of porcine reproductive and respiratory syndrome virus chimeras. Vaccine. 2010;28:2679–2686. doi: 10.1016/j.vaccine.2009.12.073. [DOI] [PubMed] [Google Scholar]
- Faaberg K.S. In vivo growth of porcine reproductive and respiratory syndrome virus engineered nsp2 deletion mutants. Virus Res. 2010;154:77–85. doi: 10.1016/j.virusres.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Porcine respiratory and reproductive syndrome virus variants, Vietnam and China, 2007. Emerg. Infect. Dis. 2008;14:1774–1776. doi: 10.3201/eid1411.071676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff J.P. Physiological effects of exogenous administration of interleukin-1β in cows. Am. J. Vet. Res. 1992;53:1983–1987. [PubMed] [Google Scholar]
- Gomez-Laguna J. Cytokine expression by macrophages in the lung of pigs infected with the porcine reproductive and respiratory syndrome virus. J. Comp. Pathol. 2010;142:51–60. doi: 10.1016/j.jcpa.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Laguna J. Changes in lymphocyte subsets and cytokines during European porcine reproductive and respiratory syndrome: increased expression of IL-12 and IL-10 and proliferation of CD4(−)CD8(high) Viral Immunol. 2009;22:261–271. doi: 10.1089/vim.2009.0003. [DOI] [PubMed] [Google Scholar]
- Gomez-Laguna J. Acute phase response in porcine reproductive and respiratory syndrome virus infection. Comp. Immunol. Microbiol. Infect. Dis. 2010;33:e51–e58. doi: 10.1016/j.cimid.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Grebennikova T.V. Genomic characterization of virulent, attenuated, and revertant passages of a North American porcine reproductive and respiratory syndrome virus strain. Virology. 2004;321:383–390. doi: 10.1016/j.virol.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Greenberger M.J. Neutralization of IL-10 increases survival in a murine model of Klebsiella pneumonia. J. Immunol. 1995;155:722–729. [PubMed] [Google Scholar]
- Halbur P.G. Immunohistochemical identification of porcine reproductive and respiratory syndrome virus (PRRSV) antigen in the heart and lymphoid system of three-week-old colostrum-deprived pigs. Vet. Pathol. 1995;32:200–204. doi: 10.1177/030098589503200218. [DOI] [PubMed] [Google Scholar]
- Halbur P.G. Comparative pathogenicity of nine US porcine reproductive and respiratory syndrome virus (PRRSV) isolates in a five-week-old cesarean-derived, colostrum-deprived pig model. Journal of veterinary diagnostic investigation: official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc. 1996;8:11–20. doi: 10.1177/104063879600800103. [DOI] [PubMed] [Google Scholar]
- Kwon B. Identification of virulence determinants of porcine reproductive and respiratory syndrome virus through construction of chimeric clones. Virology. 2008;380:371–378. doi: 10.1016/j.virol.2008.07.030. [DOI] [PubMed] [Google Scholar]
- Lemke C.D. Lymphoid hyperplasia resulting in immune dysregulation is caused by porcine reproductive and respiratory syndrome virus infection in neonatal pigs. J. Immunol. 2004;172:1916–1925. doi: 10.4049/jimmunol.172.3.1916. [DOI] [PubMed] [Google Scholar]
- Li Y. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet. J. 2007;174:577–584. doi: 10.1016/j.tvjl.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Lv J. An infectious cDNA clone of a highly pathogenic porcine reproductive and respiratory syndrome virus variant associated with porcine high fever syndrome. J. Gen. Virol. 2008;89:2075–2079. doi: 10.1099/vir.0.2008/001529-0. [DOI] [PubMed] [Google Scholar]
- Mengeling W.L. Strain specificity of the immune response of pigs following vaccination with various strains of porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2003;93:13–24. doi: 10.1016/s0378-1135(02)00427-3. [DOI] [PubMed] [Google Scholar]
- Miguel J.C. Expression of inflammatory cytokines and Toll-like receptors in the brain and respiratory tract of pigs infected with porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2010;135:314–319. doi: 10.1016/j.vetimm.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Miller L.C. Role of Toll-like receptors in activation of porcine alveolar macrophages by porcine reproductive and respiratory syndrome virus. Clinical Vaccine Immunology: CVI. 2009;16:360–365. doi: 10.1128/CVI.00269-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen C.J. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H.S. Reversion of a live porcine reproductive and respiratory syndrome virus vaccine investigated by parallel mutations. J. Gen. Virol. 2001;82:1263–1272. doi: 10.1099/0022-1317-82-6-1263. [DOI] [PubMed] [Google Scholar]
- Register K.B., Yersin A.G. Analytical verification of a PCR assay for identification of Bordetella avium. J. Clin. Microbiol. 2005;43:5567–5573. doi: 10.1128/JCM.43.11.5567-5573.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossow K.D. Porcine reproductive and respiratory syndrome. Vet. Pathol. 1998;35:1–20. doi: 10.1177/030098589803500101. [DOI] [PubMed] [Google Scholar]
- Rossow K.D. Experimental porcine reproductive and respiratory syndrome virus infection in one-, four-, and 10-week-old pigs. J. Vet. Diagn. Invest. 1994;6:3–12. doi: 10.1177/104063879400600102. [DOI] [PubMed] [Google Scholar]
- Roth J.A., Frank D.E. Recombinant bovine interferon-γ as an immunomodulator in dexamethasone-treated and nontreated cattle. J. Interferon Res. 1989;9:143–151. doi: 10.1089/jir.1989.9.143. [DOI] [PubMed] [Google Scholar]
- Shi M. Molecular epidemiology of PRRSV: a phylogenetic perspective. Virus Res. 2010;154:7–17. doi: 10.1016/j.virusres.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Storgaard T. Examination of the selective pressures on a live PRRS vaccine virus. Arch. Virol. 1999;144:2389–2401. doi: 10.1007/s007050050652. [DOI] [PubMed] [Google Scholar]
- Sun Y. Interplay between interferon-mediated innate immunity and porcine reproductive and respiratory syndrome virus. Viruses. 2012;4:424–446. doi: 10.3390/v4040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant J.M. Blood cytokines as biomarkers of in vivo toxicity in preclinical safety assessment: considerations for their use. Toxicological sciences: an official journal of the Society of Toxicology. 2010;117:4–16. doi: 10.1093/toxsci/kfq134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanawongnuwech R. Increased production of proinflammatory cytokines following infection with porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae. Clin. Diagn. Lab. Immunol. 2004;11:901–908. doi: 10.1128/CDLI.11.5.901-908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanawongnuwech R., Thacker E.L. Interleukin-10, interleukin-12, and interferon-gamma levels in the respiratory tract following mycoplasma hyopneumoniae and PRRSV infection in pigs. Viral Immunol. 2003;16:357–367. doi: 10.1089/088282403322396154. [DOI] [PubMed] [Google Scholar]
- Thanawongnuwech R. Effect of porcine reproductive and respiratory syndrome virus (PRRSV) (isolate ATCC VR-2385) infection on bactericidal activity of porcine pulmonary intravascular macrophages (PIMs): in vitro comparisons with pulmonary alveolar macrophages (PAMs) Vet. Immunol. Immunopathol. 1997;59:323–335. doi: 10.1016/s0165-2427(97)00078-0. [DOI] [PubMed] [Google Scholar]
- Thanawongnuwech R. Differential production of proinflammatory cytokines: in vitro PRRSV and Mycoplasma hyopneumoniae co-infection model. Vet. Immunol. Immunopathol. 2001;79:115–127. doi: 10.1016/s0165-2427(01)00243-4. [DOI] [PubMed] [Google Scholar]
- Tian K. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007;2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong G.Z. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg. Infect. Dis. 2007;13:1434–1436. doi: 10.3201/eid1309.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hemert M.J., Snijder E.J. The Arterivirus Replicase. In: Stanley Perlman T.G.a.E.S., editor. The Nidoviruses. ASM Press; Washington, DC: 2008. pp. 83–101. [Google Scholar]
- Wang G. Immune responses in piglets infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2011;142:170–178. doi: 10.1016/j.vetimm.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Wang Y. Attenuation of porcine reproductive and respiratory syndrome virus strain MN184 using chimeric construction with vaccine sequence. Virology. 2008;371:418–429. doi: 10.1016/j.virol.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Wesley R.D. Infection with Porcine reproductive and respiratory syndrome virus stimulates an early gamma interferon response in the serum of pigs. Can. J. Vet. Res. 2006;70:176–182. [PMC free article] [PubMed] [Google Scholar]
- Wu J. Genetic variation and pathogenicity of highly virulent porcine reproductive and respiratory syndrome virus emerging in China. Arch. Virol. 2009;154:1589–1597. doi: 10.1007/s00705-009-0478-6. [DOI] [PubMed] [Google Scholar]
- Xu M. Secondary infection with Streptococcus suis serotype 7 increases the virulence of highly pathogenic porcine reproductive and respiratory syndrome virus in pigs. Virol. J. 2010;7:184. doi: 10.1186/1743-422X-7-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Yang H. Porcine reproductive and respiratory syndrome in China. Virus Res. 2010;154:31–37. doi: 10.1016/j.virusres.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Zhou L. The 30-amino-acid deletion in the nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J. Virol. 2009;83:5156–5167. doi: 10.1128/JVI.02678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. Molecular characterization of transcriptome-wide interactions between highly pathogenic porcine reproductive and respiratory syndrome virus and porcine alveolar macrophages in vivo. Int. J. Biol. Sci. 2011;7:947–959. doi: 10.7150/ijbs.7.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. Molecular characterization of porcine SARM1 and its role in regulating TLRs signaling during highly pathogenic porcine reproductive and respiratory syndrome virus infection in vivo. Dev. Comp. Immunol. 2012 doi: 10.1016/j.dci.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Zhou Y.J. Highly virulent porcine reproductive and respiratory syndrome virus emerged in China. Transbound. Emerg. Dis. 2008;55:152–164. doi: 10.1111/j.1865-1682.2008.01020.x. [DOI] [PubMed] [Google Scholar]