Abstract
Recently it has been reported that a cathepsin B inhibitor, CA-074Me, attenuates ecotropic murine leukemia virus (Eco-MLV) infection in NIH3T3 cells, suggesting that cathepsin B is required for the Eco-MLV infection. However, cathepsin B activity was negative or extremely low in NIH3T3 cells. How did CA-074Me attenuate the Eco-MLV infection? The CA-074Me treatment of NIH3T3 cells inhibited cathepsin L activity, and a cathepsin L specific inhibitor, CLIK148, attenuated the Eco-MLV vector infection. These results indicate that the suppression of cathepsin L activity by CA-074Me induces the inhibition of Eco-MLV infection, suggesting that cathepsin L is required for the Eco-MLV infection in NIH3T3 cells. The CA-074Me treatment inhibited the Eco-MLV infection in human cells expressing the exogenous mouse ecotropic receptor and endogenous cathepsins B and L, but the CLIK148 treatment did not, showing that only the cathepsin L suppression by CLIK148 is not enough to prevent the Eco-MLV infection in cells expressing both of cathepsins B and L, and CA-074Me inhibits the Eco-MLV infection by suppressing both of cathepsins B and L. These results suggest that either cathepsin B or L is sufficient for the Eco-MLV infection.
Keywords: Cathepsin B, Cathepsin L, Ecotropic murine leukemia virus
Introduction
Ecotropic murine leukemia viruses (Eco-MLVs) recognize cationic amino acid transporter 1 (CAT1) as the entry receptor (Albritton et al., 1989). After the receptor recognition, the Eco-MLV particles are internalized into endosomes, and then the virus core enters into host cytoplasm by fusion between viral envelope and host cell endosomal membrane. The membrane fusion mediated by the Eco-MLV envelope glycoprotein (Env) is activated by endosome acidification (Katen et al., 2001, McClure et al., 1990).
The Eco-MLV Env protein has the 16-amino acid peptide (R peptide) at the C-terminal tail that inhibits the membrane fusion reaction. The R peptide is cleaved during virion maturation to achieve viral entry by the membrane fusion (Kubo and Amanuma, 2003, Kubo et al., 2007, Ragheb and Anderson, 1994, Rein et al., 1994). The R peptide-truncated Env protein induces syncytia in susceptible cells, but the R peptide-containing Env protein does not. However, the R peptide-containing Env protein successfully induces syncytia in rat XC and fu-1 cells (Jones and Risser, 1993, Wong et al., 1977). We have previously reported that treatment of XC cells with tunicamycin, an N-linked glycosylation inhibitor, significantly suppresses the syncytium formation of XC cells by the R peptide-containing Env protein, but does not that by the R peptide-truncated Env protein, suggesting that an unknown glycosylated cellular protein is involved in the XC cell-specific syncytium formation by the R peptide-containing Env protein (Kubo, Ishimoto, and Amanuma, 2003). Because XC and fu-1 cells were both derived from muscle tissues, it was speculated that the cellular factor is also associated with syncytium formation in myogenesis.
In order to identify a key molecule for the XC cell-specific syncytium formation by the R peptide-containing Env protein, we analyzed expressions of several glycosylated proteins in XC and NIH3T3 cells that are involved in myogenesis. Cathepsin B is a glycosylated protein, and its expression is upregulated during early myoblast fusion and is downregulated in myotubes. It has been reported that cathepsin B-deficient myoblast cells undergo differentiation with impaired cell fusion (Gogos et al., 1996), and a cathepsin B inhibitor, CA-074Me, inhibits L6 myoblast differentiation (Jane et al., 2002). We found that cathepsin B was expressed in XC cells, but was undetectable in NIH3T3 cells. However, it has been reported that CA-074Me suppresses the Eco-MLV infection in NIH3T3 cells (Kumar et al., 2007). In our experiments, the compound also inhibited the Eco-MLV vector infection in NIH3T3 cells. How did the cathepsin B inhibitor, CA-074Me, inhibit the Eco-MLV infection in the cathepsin B-undetectable NIH3T3 cells?
There are many evidences showing that cathepsin proteases B and L both participate in viral infections, including Ebola virus, paramyxovirus, coronavirus, and reovirus (Chandran et al., 2005, Ebert et al., 2002, Pager and Dutch, 2005, Qiu et al., 2006, Simmons et al., 2005). In addition, CA-074Me inhibits both of cathepsins B and L (Alain et al., 2007, Montaser et al., 2002). Therefore, cathepsin L might be involved in the Eco-MLV infection in cathepsin B-undetectable NIH3T3 cells. To assess this possibility, we analyzed effects of a cathepsin L specific inhibitor, CLIK148, on the Eco-MLV infection. We found in this study that CLIK148 significantly inhibits the Eco-MLV infection in the cathepsin B-undetectable NIH3T3 cells, but does not in cells expressing both of cathepsins B and L, showing that cathepsin L is required for the Eco-MLV infection in the cathepsin B-undetectable NIH3T3 cells, but not in cathepsin B-expressing cells. These results show that either cathepsin B or L is sufficient for the Eco-MLV infection.
Results
Cathepsin B inhibitor, CA-074Me, inhibits Eco-MLV infection
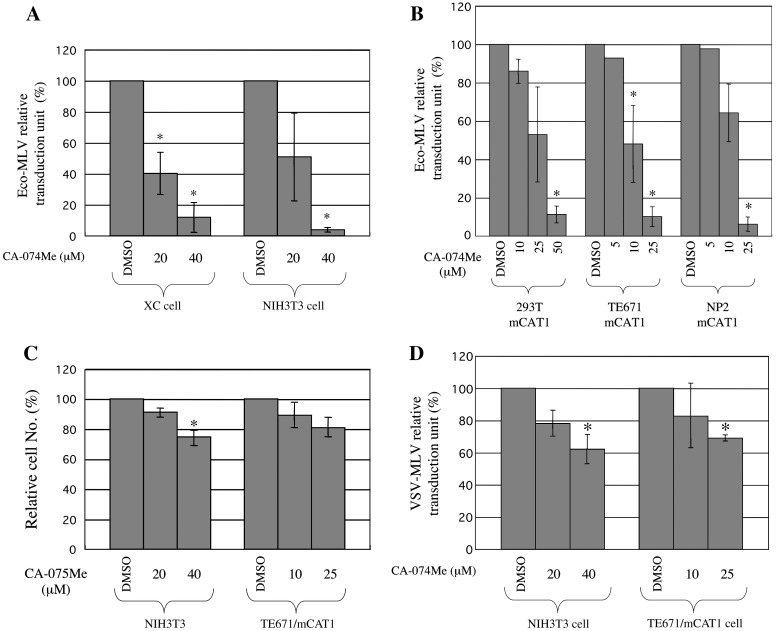
It has been reported that a cathepsin B inhibitor, CA-074Me, attenuates the Eco-MLV infection (Kumar et al., 2007). To confirm this result, we analyzed the effects of CA-074Me on the Eco-MLV vector infection in mouse NIH3T3, rat XC, and human cells (293T, TE671, and NP2) expressing mouse CAT1 (mCAT1). These cells were pretreated with CA-074Me for 5 h, and were inoculated with the Eco-MLV vector having the LacZ gene as a marker. As already reported (Kumar et al., 2007), the CA-074Me treatment attenuated the Eco-MLV vector transduction in a dose-dependent manner in all cell lines examined (Figs. 1 A and B), but did not significantly affect cell viability (Fig. 1C). The inhibitory effect of CA-074Me on the Eco-MLV vector infection was statistically significant by ANOVA and Tukey's test in all the cell lines (p < 0.05). The CA-074Me treatment moderately inhibited VSV-G-pseudotyped MLV vector transduction (Fig. 1D) as already reported (Kumar et al., 2007), but the inhibitory effect on the VSV vector transduction was lower than that on the Eco-MLV vector transduction (Figs. 1A and B). These results suggest that cathepsin B is required for the Eco-MLV vector infection.
Fig. 1.
A cathepsin B inhibitor, CA-074Me, inhibits Eco-MLV vector infection. (A) Transduction titers of the Eco-MLV vector were measured in CA-074Me-pretreated XC and NIH3T3 cells. (B) Transduction titers of the Eco-MLV vector were measured in CA-074Me-pretreated human cells expressing mCAT1. Relative values to titers in cells treated with equal volume of DMSO are indicated. (C) Numbers of viable cells were counted, and relative values to the numbers of viable cells in DMSO-treated cells are indicated. (D) Transduction titers of VSV-G-pseudotyped MLV vector were measured in CA-074Me-pretreated NIH3T3 and TE671/mCAT1 cells. Relative values to titers in DMSO-treated cells were indicated. These experiments were repeated three times. Error bars indicate standard deviations. Asterisks indicate significant differences compared with titers in DMSO-treated cells.
CA-074Me is a membrane-permeable form of CA-074. Treatment of NIH3T3 and XC cells with the original CA-074 at 40 μM did not affect the Eco-MLV vector infection (data not shown). This result indicates that secreted cathepsin B is not involved in the Eco-MLV infection.
Cathepsin B is undetectable in NIH3T3 cells
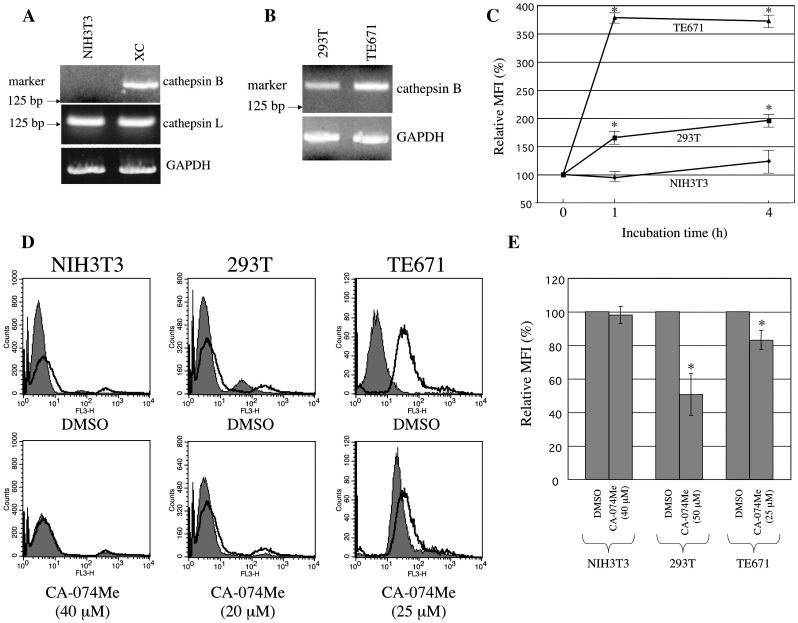
To assess whether cathepsin B is expressed in mouse NIH3T3, rat XC, human 293T, and human TE671 cells, cathepsin B mRNA expression was assessed by RT-PCR. A predicted size of PCR product (about 190 bp) was detected in XC, 293T, and TE671 cells, but was not in NIH3T3 cells (Figs. 2 A and B). These PCR conditions amplify the 3′ cathepsin B protease domain sequence. Even if a cathepsin B mRNA variant lacking this region is expressed, the product does not have protease activity. The cathepsin B mRNA level of TE671 cells was higher than that of 293T cells. Cathepsin L mRNA was detected in NIH3T3 and XC cells by RT-PCR. These results indicate that NIH3T3 cells express cathepsin L mRNA, but not the cathepsin B mRNA.
Fig. 2.
Expression of cathepsin B in target cells. (A) Expressions of mouse cathepsin B (upper panel) and cathepsin L (middle panel) mRNAs in NIH3T3 and XC cells were analyzed by RT-PCR. (B) Expression of human cathepsin B mRNA in human 293T and TE671 cells was analyzed by RT-PCR. A Hind III-digested product of the λ phage DNA was used as molecular size marker (left side of the panels). (C) Cathepsin B activity in living cells was analyzed by incubation of cells with the cathepsin B detection reagent for indicated times. Means of fluorescence intensities (MFIs) of the stained cells were measured by a flow cytometer. Relative values to MFIs of cells incubated with the cathepsin B detection reagent for 0 h are indicated. (D and E) Cathepsin B activities of the CA-074Me-treated cells were measured by the cathepsin B detection reagent. The treated cells were incubated with the cathepsin B detection reagent for 1 h. Histograms are indicated in panel (D). Grey areas in upper panels indicate cells treated with DMSO and unstained with the cathepsin B detection reagent. Open areas indicate cells treated with DMSO and stained with the cathepsin B detection reagent. Grey areas in lower panels indicate cells treated with CA-074Me and stained with the cathepsin detection reagent. Relative values to MFI of DMSO-treated cells are also indicated in panel (E). These experiments were repeated three times. Error bars indicate standard deviations. Asterisks indicate significant differences compared with MFI of the control cells.
To confirm that the cathepsin B is not expressed in NIH3T3 cells, cathepsin B activity was measured using the cathepsin B detection reagent. When the reagent is digested by cathepsin B in cells, the digested product generates red fluorescence. Fluorescence strength of 293T cells was increased by incubation with the cathepsin B detection reagent, but that of NIH3T3 cells was not (Fig. 2C). This result indicates that cathepsin B activity is negative or extremely low in NIH3T3 cells. Cathepsin B activity in TE671 cells was much higher than 293T cells (Figs. 2C and D) consistent with the result of cathepsin B mRNA detection by RT-PCR (Fig. 2B).
The CA-074Me treatment reduced the fluorescence strength of 293T and TE671 cells stained with the cathepsin B detection reagent, indicating that CA-074Me indeed inhibits cathepsin B activity in 293T and TE671 cells (Figs. 2D and E). Cathepsin B activity was more effectively inhibited by CA-074Me in 293T cells than in TE671 cells, because 293T cells were treated with CA-074Me at higher concentration (50 μM) than TE671 cells (25 μM). The fluorescence strength of NIH3T3 cells stained with the cathepsin B detection reagent was not reduced by the CA-074Me treatment (40 μM), supporting that cathepsin B activity is negative in NIH3T3 cells, although the CA-074Me treatment at the same concentration significantly reduced the Eco-MLV vector transduction (Fig. 1A). These results suggest that CA-074Me does not attenuate the Eco-MLV infection by suppressing cathepsin B activity.
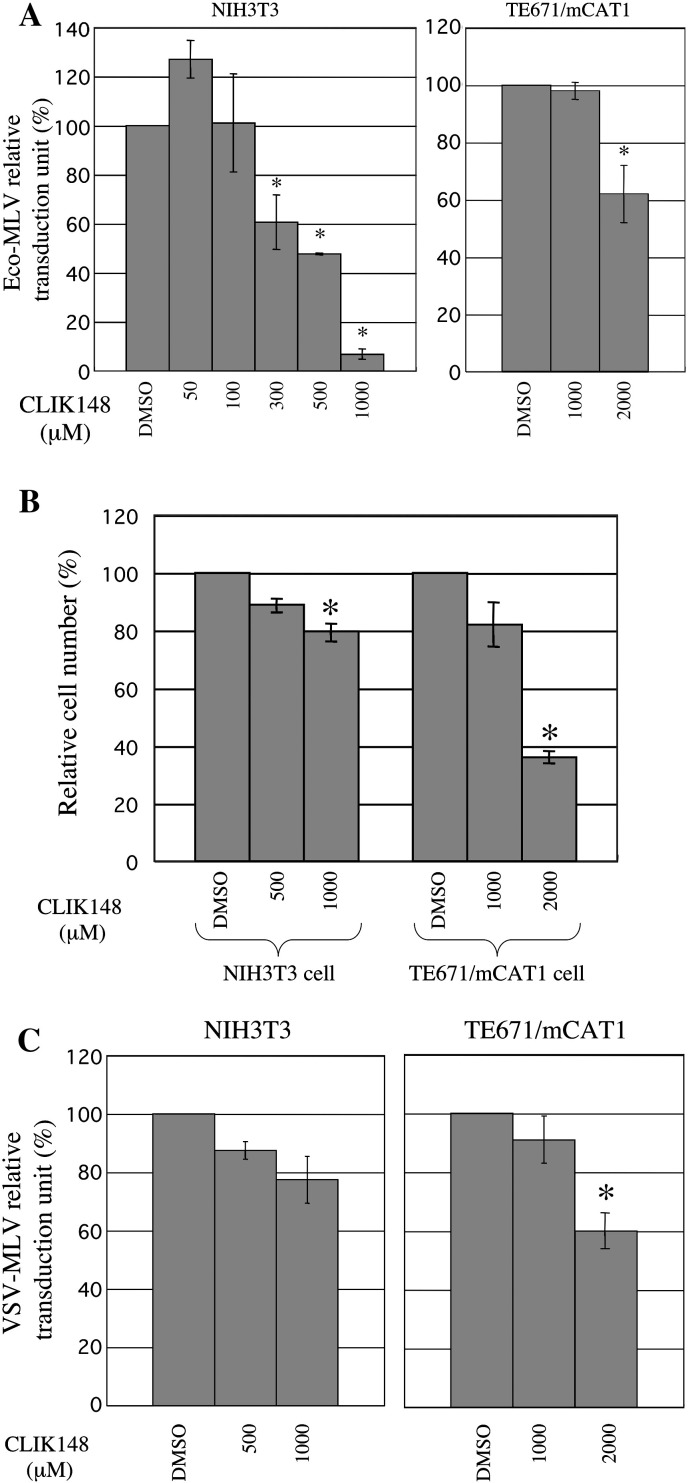
Cathepsin L specific inhibitor, CLIK148, attenuates Eco-MLV infection in NIH3T3 cells
It has been reported that CA-074Me inhibits both of cathepsins B and L (Alain et al., 2007, Montaser et al., 2002). Therefore, inhibition of cathepsin L activity by CA-074Me could attenuate Eco-MLV vector infection in NIH3T3 cells. To assess this possibility, we analyzed effects of a cathepsin L specific inhibitor, CLIK148 (Tsuge et al., 1999), on the Eco-MLV vector infection. NIH3T3 cells were pretreated with CLIK148 for 5 h, and inoculated with the Eco-MLV vector. The CLIK148 treatment significantly suppressed the Eco-MLV vector transduction in dose-dependent manner in NIH3T3 cells (Fig. 3A, left panel). The inhibitory effect of CLIK148 on the Eco-MLV vector infection in NIH3T3 cells was statistically significant by ANOVA (p < 0.05). The treatment of NIH3T3 cells with CLIK148 did not significantly suppressed cell viability (Fig. 3B, left panel) and VSV-G-pseudotyped MLV vector infection (Fig. 3C, left panel). These results indicate that cathepsin L is required for the Eco-MLV infection in cathepsin B-negative NIH3T3 cells.
Fig. 3.
Involvement of cathepsin L in Eco-MLV vector infection. (A) NIH3T3 and mCAT1-expressing TE671 (TE671/mCAT1) cells were pretreated with a cathepsin L inhibitor, CLIK148. Transduction titers of the Eco-MLV vector were measured in the treated cells. Relative values to titers in cells treated with equal volume of DMSO are indicated. (B) Numbers of viable cells were measured, and relative values to numbers of viable cells in DMSO-treated cells are indicated. (C) Transduction titers of VSV-G-pseudotyped MLV vector in the pretreated cells were measured. Relative values to titers in DMSO-treated cells are indicated. These experiments were repeated three times. Error bars indicate standard deviations. Asterisks indicate significant differences compared with values in DMSO-treated cells.
In contrast to NIH3T3 cells, human TE671 cells expressed both of cathepsins B and L analyzed by the cathepsin detection reagents (Figs. 2 and 4 ). The CLIK148 treatment at 1000 μM did not attenuate the Eco-MLV vector infection in mCAT1-expressing TE671 cells (TE671/mCAT1) (Fig. 3A, right panel). The CLIK148 treatment of TE671/mCAT1 cells at 2000 μM moderately inhibited the Eco-MLV (Fig. 3A, right panel) and VSV-G vector (Fig. 3C, right panel) infections, and additionally induced cell growth suppression (Fig. 3B, right panel). The cell growth suppression by the CLIK148 treatment (2000 μM) should result in the attenuation of MLV vector infection independently of envelope proteins. Therefore, we could not conclude that cathepsin L is required for the Eco-MLV infection in TE671 cells expressing both of cathepsins B and L.
Fig. 4.
Effects of CLIK148 on cathepsin B and L activities in NIH3T3 and TE671 cells. (A) NIH3T3 and TE671 cells were incubated with the cathepsin L detection reagent for 0 or 1 h, and MFIs of the cells were measured by a flow cytometer. Relative values to MFIs of cells incubated for 0 h are indicated. (B and C) Cathepsin L activities of CA-074Me- or CLIK148-treated cells were measured by the cathepsin L detection reagent. Histograms are indicated in panel (B). Closed areas indicate cells treated with DMSO and unstained with the cathepsin L detection reagent. Open areas indicate cells treated with DMSO and stained with the reagent. Grey areas indicate cells treated with CLIK148 (middle panel) or CA-074Me (lower panel) and stained with the reagent. Relative values to MFIs of DMSO-treated cells are also indicated in panel (C). (D) Cathepsin B activity of CLIK148-treated cells was measured by the cathepsin B detection reagent. Relative values to MFIs of DMSO-treated cells are indicated in the left panel. Histogram was indicated in the right panel. Closed area indicates cells treated with DMSO and stained with the cathepsin B detection reagent. Red line indicates cells treated with CLIK148 and stained with the reagent. These experiments were repeated three times. Error bars indicate standard deviations. Asterisks indicate significant differences compared to values in the control cells.
Cathepsin L activity in NIH3T3 cells was higher than that in TE671 cells (Fig. 4A). The CLIK148 treatment at 1000 μM indeed inhibited cathepsin L activity in NIH3T3 and TE671 cells analyzed by the cathepsin L detection reagent (Figs. 4B and C). The CA-074Me treatment of NIH3T3 and TE671 cells also inhibited cathepsin L activity, as already reported (Montaser, Lalmanach, and Mach, 2002). The CLIK148 treatment of TE671 cells did not affect cathepsin B activity (Fig. 4D) (Katunuma et al., 1999). Therefore, it is suggested that the CA-074Me treatment attenuates the Eco-MLV infection by inhibiting cathepsin L activity in NIH3T3 cells, and by inhibiting both of cathepsin B and L in TE671 cells.
The above results indicate that cathepsin L is required for the Eco-MLV infection in cathepsin B-undetectable NIH3T3 cells, but not in cathepsin B-expressing TE671 cells, suggesting that either cathepsin B or L is sufficient for the Eco-MLV infection. To confirm this result, NIH3T3 cells were transduced by an MLV vector encoding a mouse cathepsin B. However, cathepsin B-expressing NIH3T3 cells were not obtained (data not shown), perhaps because of cathepsin B-induced apoptosis (Conus and Simon, 2008, Guicciardi et al., 2001).
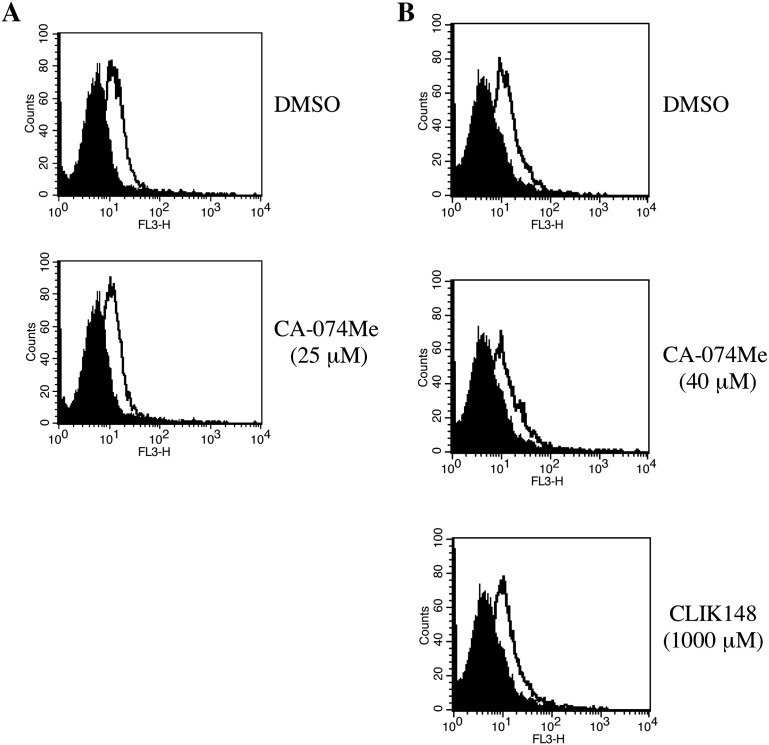
Cathepsin inhibitors do not affect Eco-MLV virion binding to target cells
We assessed whether the CA-074Me and CLIK148 treatments affect the Eco-MLV vector binding to the target cells (Yoshii et al., 2008). The CA-074Me treatment (25 μM) did not affect the vector binding to the TE671/mCAT1 cells (Fig. 5A). Similarly the CA-074Me (40 μM) and CLIK148 (1000 μM) treatments of NIH3T3 cells did not affect the MLV vector binding (Fig. 5B). These results indicate that the CA-074Me and CLIK148 treatments did not attenuate the Eco-MLV infection by suppressing the Eco-MLV vector binding to the target cells.
Fig. 5.
Cathepsin inhibitors do not attenuate virus–cell interaction. TE671/mCAT1 (A) were treated with DMSO (upper panel) or CA-074Me (25 μM) (lower panel). NIH3T3 cells (B) were treated with DMSO (upper panel), CA-074Me (40 μM) (middle panel), or CLIK148 (1000 μM) (lower panel). Black area indicates cells suspended with culture supernatant of TELCeB6 cells that do not express any Env proteins. Open area indicates cells suspended with culture supernatant of the Eco-MLV Env-expressing TELCeB6 cells.
Discussion
Although cathepsin B activity was negative or extremely low in NIH3T3 cells, the cathepsin B inhibitor, CA-074Me, significantly suppressed the Eco-MLV vector infection. The CA-074Me treatment inhibited cathepsin L activity as already reported (Alain et al., 2007, Montaser et al., 2002). A cathepsin L specific inhibitor, CLIK148, attenuated the Eco-MLV infection in NIH3T3 cells. CLIK148 did not inhibit cathepsin B activity. These results indicate that cathepsin L is required for the Eco-MLV infection in cathepsin B-undetectable NIH3T3 cells. In contrast, the CLIK148 treatment did not inhibit the Eco-MLV infection in TE671/mCAT1 cells expressing both of cathepsins B and L, indicating that cathepsin L is not required for the Eco-MLV infection in cells expressing both of cathepsins B and L. Therefore, it is suggested that either cathepsin B or L is sufficient for the Eco-MLV infection.
The CA-074Me treatment of NIH3T3 cells at 40 μM induced 90% reduction in the Eco-MLV vector infection. The CLIK148 treatment at much higher concentration (1000 μM) induced similar reduction in the infection. CA-074Me is a membrane-permeable form of CA-074. The CA-074 treatment at the same concentration (40 μM) did not affect the Eco-MLV vector infection. Because CLIK148 is not membrane-permeable like CA-074, much higher concentration of CLIK148 was needed to attenuate the Eco-MLV infection than membrane-permeable CA-074Me.
The CA-074Me treatment of NIH3T3 and TE671 cells at 40 and 25 μM, respectively, resulted in 90% reduction in the Eco-MLV vector infection. NIH3T3 cells needed higher concentration of CA-074Me to attenuate the Eco-MLV infection than TE671 cells. Cathepsin B activity was undetectable in NIH3T3 cells, but cathepsin L activity in NIH3T3 cells was higher than that in TE671 cells. Because CA-074Me attenuates the Eco-MLV infection by inhibiting both of cathepsins B and L, CA-074Me concentration required to attenuate the Eco-MLV infection should be affected by total activities of cathepsins B and L.
The CLIK148 treatment moderately reduced cathepsin L activity to 50% of that in DMSO-treated cells. The CA-074Me treatment slightly reduced cathepsin B activity to 80% of that in DMSO-treated cells. In contrast, these treatments induced significant reduction (90%) in the Eco-MLV vector infection. When undiluted Eco-MLV vector solution was inoculated into target cells, about 2 × 106 infected cells were detected in DMSO-treated cells per culture dish. Because 2 × 105 infected cells were still detected in the inhibitor-treated cells, it is not suspicious that cathepsin B activity of the target cells correlates to susceptibility to the Eco-MLV infection.
The CLIK148 treatment of NIH3T3 cells at 50 μM rather slightly increased the Eco-MLV vector infection. A part of the Eco-MLV vector particles could be degraded by cathepsin L in NIH3T3 cells expressing cathepsin L at high level. The CLIK148 treatment at 50 μM could inhibit the degradation of vector particles but could not attenuate the Eco-MLV infection. Therefore, the CLIK148 treatment at low concentration could increase the Eco-MLV infection.
The CA-074Me treatment at 40 μM and the CLIK148 treatment at 1000 μM both induced 90% reduction in the Eco-MLV vector infection in NIH3T3 cells, whereas the CLIK148 treatment more efficiently inhibited cathepsin L activity in NIH3T3 cells than the CA-074Me treatment did. It seems to be inconsistent that the CA-074Me treatment less efficiently inhibits cathepsin L activity than the CLIK148 treatment but these treatments attenuate the Eco-MLV infection at similar extent. Therefore, CA-074Me might inhibit other proteases that are involved in the Eco-MLV infection.
Cathepsin B activity was negative or extremely low in NIH3T3 cells. It has been reported that cathepsin B is required for reovirus-mediated oncolysis, and transformed NIH3T3 cells are susceptible to the reovirus-mediated oncolysis, but normal NIH3T3 cells are not (Alain et al., 2007). Cathepsin B expression is significantly (more than 50 times) elevated by transformation of NIH3T3 cells (Chambers et al., 1992, He et al., 2004). These results suggest that cathepsin B activity in normal NIH3T3 cells is negative or much lower than that in transformed NIH3T3 cells, consistent with our result.
Cathepsin B is involved in syncytium formation during myogenesis (Gogos et al., 1996, Jane et al., 2002), and is expressed in XC cells in which the R peptide-containing Env protein can induce syncytium formation (Jones and Risser, 1993, Kubo et al., 2003). As mentioned above, cathepsin B expression is significantly activated in transformed NIH3T3 cells (Chambers et al., 1992, He et al., 2004). It has been reported that transformed NIH3T3 cells are highly susceptible to the Eco-MLV particle-induced syncytium formation compared to control NIH3T3 cells (Wilson, Marsh, and Eiden, 1992). The activation of cathepsin B by the cellular transformation could confer NIH3T3 cells more susceptible to the Eco-MLV particle-induced syncytium formation. However, cathepsin B is not a determinant molecule for the XC cell-specific syncytium formation induced by the R peptide-containing Env protein, because the R peptide-containing Env protein could not induce syncytia in transformed NIH3T3, 293T/mCAT1, and TE671/mCAT1 cells expressing cathepsin B (data not shown). Further study is needed to understand the mechanism by which the R peptide-containing Env protein induces syncytia in XC cells.
In conclusion, cathepsin L is required for the Eco-MLV infection in cathepsin B-undetectable NIH3T3 cells, but is not in cathepsin B-expressing cells. CA-074Me attenuated the Eco-MLV infection by inhibiting both of cathepsins B and L. These results suggest that either cathepsin B or L is sufficient for the Eco-MLV infection. This study presents the novel finding that cathepsin B is not the only protease involved in the Eco-MLV infection, and cathepsin L also participates in the infection.
Materials and methods
Cells
Human 293T (Pear et al., 1993), TE671, TELCeB6 (Cosset et al., 1995), NP2 (Soda et al., 1999), mouse NIH3T3, and rat XC (Kubo, Ishimoto, and Amanuma, 2003) cells were cultured in Dulbecco's modified Eagle's medium (Wako, Osaka, Japan) supplemented with 8% fetal bovine serum (Biosource, Rockville, MD) at 37 °C in 5% CO2. Friend Eco-MLV vector-producing cells were constructed by stable transfection of TELCeB6 cells with the Friend MLV Env expression plasmid (Kubo et al., 2004). Ecotropic MLV receptor-expressing 293T, TE671, and NP2 cells were constructed by a mCAT1-encoding MLV vector as already reported (Kubo, Ishimoto, and Amanuma, 2003).
Semi-quantitative RT-PCR of cathepsin mRNAs
Total RNA samples were isolated by the TRIzol Reagent (Invitrogen, Carlsbad, CA). First strand cDNA was synthesized by a reverse-transcriptase (RT) using random primers (TaKaRa, Otsu, Japa). Polymerase chain reaction (PCR) was performed to measure mRNA levels of cathepsins B and L using the LA Taq DNA polymerase (TaKaRa). Nucleotide sequences of PCR primers for mouse cathepsin B mRNA detection are 5′-GTA TAC AAG CAT GAA GCC GGT-3′ and 5′-TCA AGT CCC AGC AGA TTA-3′. Nucleotide sequences of PCR primers for human cathepsin B mRNA detection are 5′-GTG TAC CAA CAC GTC ACC GGA-3′ and 5′-CAG GCC CAC GGC AGA TTA-3′. The PCR products were subjected to NuSieve agarose gel electrophoresis (TaKaRa) for separation of low molecular weight DNA fragments. Nucleotide sequences of PCR primers for mouse cathepsin L mRNA detection are 5′-ACT ATGAAC CCA ACT GTA GCA-3′ and 5′-TCA ATT CAC GAT AGG ATA GCT-3′. GAPDH mRNA levels were also measured by RT-PCR as controls (Kubo et al., 2008). The PCR products of cathepsin L and GAPDH were subjected to standard agarose gel electrophoresis.
Transduction assay
Target cells (2 × 105) were plated onto a 6-cm culture dish, and cultured for 24 h. The cells were treated with CA-074Me (Sigma, St. Louis, MO) or CLIK148 (Tsuge et al., 1999) for 5 h. The cells were washed to remove the inhibitor. VSV-G-pseudotyped MLV vector was constructed by transient transfection of TELCeB6 cells by a VSV-G expression plasmid. Culture supernatants of the vector-producing cells were inoculated into the treated cells in the presence of polybrene (Sigma) (4 μg/ml). When undiluted culture supernatant of the Eco-MLV vector-producing cells was inoculated, numbers of infected cells were too many to count. Therefore, the culture supernatant was diluted 1000 times with fresh medium and inoculated into target cells. Two days after inoculation, the cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Wako). Numbers of blue cells were counted in randomly selected 10 microscopic fields per dish. Usually 30–100 infected cells were detected in a microscopic field. Total numbers of infected cells in 10 microscopic fields were compared.
Cell viability
Target cells were treated with CA-074Me or CLIK148 for 5 h, and were cultured for additional 5 h in a fresh medium. The cells were collected and treated with trypan blue. Numbers of unstained cells were counted using a counting chamber to estimate cell viability.
Cathepsin activity measurement in living cells
Cells were stained with the cathepsin B or L detection reagent (Cell Technology, Minneapolis, MN). The reagent utilizes fluorophore cresyl violet that is bi-substituted by an amide linkage to a peptide that contains a cathepsin B or L target cleavage sequence. In this form, the cresyl violet leaving group is non-fluorescent. Following cleavage at the amide linkage site by the cathepsin protease B or L in living cells, the mono and non-substituted cresyl violet fluorophores generate red fluorescence. The stained cells were subjected to a flow cytometer (BD Biosciences, San Jose, CA) to measure fluorescence strength of the cells.
MLV vector binding assay
A MLV vector binding assay was performed as reported previously (Lavillette et al., 2000, Yoshii et al., 2008). Cells (1 × 106) were incubated with the Eco-MLV vector solution for 1 h at 4 °C, and unbound vector was removed by two washes with PBS. Vector–cell complexes were incubated sequentially at 4 °C with goat anti-MLV SU antiserum and then with PE-conjugated anti-goat IgG (Jackson Laboratories). Fluorescence intensity of the cells was analyzed by a flow cytometer (BD Biosciences).
Statistical analysis
Differences between two groups of data were determined by Student's t-test. Additionally, the effects of cathepsin inhibitors on the Eco-MLV vector infection were evaluated using analysis of variance (ANOVA) and Tukey's test. Statistical significance was set at p < 0.05 for all tests. The statistical analysis was performed using the SPSS v15.0 software (SPSS Japan, Tokyo, Japan).
Acknowledgments
We thank Dr. L. Chang for VSV-G expression plasmid. The plasmid was provided through the AIDS Research and Reference Reagent Program, NIAID, NIH. We also thank Dr. T. Matsuyama (Nagasaki University) and Dr. H. Sato (National Institute of Infectious Diseases) for discussions. This work was partially supported by the NEKKEN Research Found 2008 of Nagasaki University.
References
- Alain T., Kim T.S., Lun X., Liacini A., Schiff L.A., Senger D.L., Forsyth P.A. Proteolytic disassembly is a critical determinant for reovirus oncolysis. Mol. Ther. 2007;15(8):1512–1521. doi: 10.1038/sj.mt.6300207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albritton L.M., Tseng L., Scadden D., Cunningham J.M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57(4):659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- Chambers A.F., Colella R., Denhardt D.T., Wilson S.M. Increased expression of cathepsins L and B and decreased activity of their inhibitors in metastatic, ras-transformed NIH 3T3 cells. Mol. Carcinog. 1992;5(3):238–245. doi: 10.1002/mc.2940050311. [DOI] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308(5728):1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conus S., Simon H.U. Cathepsins: key modulators of cell death and inflammatory responses. Biochem. Pharmacol. 2008;76(11):1374–1382. doi: 10.1016/j.bcp.2008.07.041. [DOI] [PubMed] [Google Scholar]
- Cosset F.L., Takeuchi Y., Battini J.L., Weiss R.A., Collins M.K. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 1995;69(12):7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D.H., Deussing J., Peters C., Dermody T.S. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 2002;277(27):24609–24617. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- Gogos J.A., Thompson R., Lowry W., Sloane B.F., Weintraub H., Horwitz M. Gene trapping in differentiating cell lines: regulation of the lysosomal protease cathepsin B in skeletal myoblast growth and fusion. J. Cell Biol. 1996;134(4):837–847. doi: 10.1083/jcb.134.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guicciardi M.E., Miyoshi H., Bronk S.F., Gores G.J. Cathepsin B knockout mice are resistant to tumor necrosis factor-alpha-mediated hepatocyte apoptosis and liver injury: implications for therapeutic applications. Am. J. Pathol. 2001;159(6):2045–2054. doi: 10.1016/s0002-9440(10)63056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.H., Li J.J., Xie Y.H., Tang Y.T., Yao G.F., Qin W.X., Wan D.F., Gu J.R. Altered gene expression profiles of NIH3T3 cells regulated by human lung cancer associated gene CT120. Cell Res. 2004;14(6):487–496. doi: 10.1038/sj.cr.7290252. [DOI] [PubMed] [Google Scholar]
- Jane D.T., Morvay L.C., Allen F., Sloane B.F., Dufresne M.J. Selective inhibition of cathepsin B with cell-permeable CA074Me negatively affects L6 rat myoblast differentiation. Biochem. Cell Biol. 2002;80(4):457–465. doi: 10.1139/o02-134. [DOI] [PubMed] [Google Scholar]
- Jones J.S., Risser R. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J. Virol. 1993;67(1):67–74. doi: 10.1128/jvi.67.1.67-74.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katen L.J., Januszeski M.M., Anderson W.F., Hasenkrug K.J., Evans L.H. Infectious entry by amphotropic as well as ecotropic murine leukemia viruses occurs through an endocytic pathway. J. Virol. 2001;75(11):5018–5026. doi: 10.1128/JVI.75.11.5018-5026.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunuma N., Murata E., Kakegawa H., Matsui A., Tsuzuki H., Tsuge H., Turk D., Turk V., Fukushima M., Tada Y., Asao T. Structure based development of novel specific inhibitors for cathepsin L and cathepsin S in vitro and in vivo. FEBS Lett. 1999;458(1):6–10. doi: 10.1016/s0014-5793(99)01107-2. [DOI] [PubMed] [Google Scholar]
- Kubo Y., Amanuma H. Mutational analysis of the R peptide cleavage site of Moloney murine leukaemia virus envelope protein. J. Gen. Virol. 2003;84(Pt 8):2253–2257. doi: 10.1099/vir.0.19126-0. [DOI] [PubMed] [Google Scholar]
- Kubo Y., Ishimoto A., Amanuma H. N-Linked glycosylation is required for XC cell-specific syncytium formation by the R peptide-containing envelope protein of ecotropic murine leukemia viruses. J. Virol. 2003;77(13):7510–7516. doi: 10.1128/JVI.77.13.7510-7516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y., Ishimoto A., Ono T., Yoshii H., Tominaga C., Mitani C., Amanuma H., Yamamoto N. Determinant for the inhibition of ecotropic murine leukemia virus infection by N-linked glycosylation of the rat receptor. Virology. 2004;330(1):82–91. doi: 10.1016/j.virol.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Kubo Y., Tominaga C., Yoshii H., Kamiyama H., Mitani C., Amanuma H., Yamamoto N. Characterization of R peptide of murine leukemia virus envelope glycoproteins in syncytium formation and entry. Arch. Virol. 2007;152(12):2169–2182. doi: 10.1007/s00705-007-1054-6. [DOI] [PubMed] [Google Scholar]
- Kubo Y., Yoshii H., Kamiyama H., Tominaga C., Tanaka Y., Sato H., Yamamoto N. Ezrin, Radixin, and Moesin (ERM) proteins function as pleiotropic regulators of human immunodeficiency virus type 1 infection. Virology. 2008;375(1):130–140. doi: 10.1016/j.virol.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Kumar P., Nachagari D., Fields C., Franks J., Albritton L.M. Host cell cathepsins potentiate Moloney murine leukemia virus infection. J. Virol. 2007;81(19):10506–10514. doi: 10.1128/JVI.02853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavillette D., Ruggieri A., Russell S.J., Cosset F.L. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 2000;74(1):295–304. doi: 10.1128/jvi.74.1.295-304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure M.O., Sommerfelt M.A., Marsh M., Weiss R.A. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 1990;71(Pt 4):767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- Montaser M., Lalmanach G., Mach L. CA-074, but not its methyl ester CA-074Me, is a selective inhibitor of cathepsin B within living cells. Biol. Chem. 2002;383(7–8):1305–1308. doi: 10.1515/BC.2002.147. [DOI] [PubMed] [Google Scholar]
- Pager C.T., Dutch R.E. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 2005;79(20):12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear W.S., Nolan G.P., Scott M.L., Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad Sci. U.S.A. 1993;90(18):8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu C., Das Sarma J., Bates P., Weiss S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006;80(12):5768–5776. doi: 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragheb J.A., Anderson W.F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 1994;68(5):3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A., Mirro J., Haynes J.G., Ernst S.M., Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 1994;68(3):1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad Sci. U.S.A. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda Y., Shimizu N., Jinno A., Liu H.Y., Kanbe K., Kitamura T., Hoshino H. Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem. Biophys. Res. Commun. 1999;258(2):313–321. doi: 10.1006/bbrc.1999.0633. [DOI] [PubMed] [Google Scholar]
- Tsuge H., Nishimura T., Tada Y., Asao T., Turk D., Turk V., Katunuma N. Inhibition mechanism of cathepsin L-specific inhibitors based on the crystal structure of papain–CLIK148 complex. Biochem. Biophys. Res. Commun. 1999;266(2):411–416. doi: 10.1006/bbrc.1999.1830. [DOI] [PubMed] [Google Scholar]
- Wilson C.A., Marsh J.W., Eiden M.V. The requirements for viral entry differ from those for virally induced syncytium formation in NIH 3T3/DTras cells exposed to Moloney murine leukemia virus. J. Virol. 1992;66(12):7262–7269. doi: 10.1128/jvi.66.12.7262-7269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P.K., Yuen P.H., Kaufman S.J. Induction of syncytia by Moloney murine leukemia virus in myoblasts defective in differentiation. J. Virol. 1977;21(1):319–327. doi: 10.1128/jvi.21.1.319-327.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii H., Kamiyama H., Amanuma H., Oishi K., Yamamoto N., Kubo Y. Mechanisms underlying glycosylation-mediated loss of ecotropic receptor function in murine MDTF cells and implications for receptor evolution. J. Gen. Virol. 2008;89(Pt 1):297–305. doi: 10.1099/vir.0.83430-0. [DOI] [PubMed] [Google Scholar]