Abstract
Nitric oxide is an important molecule playing a key role in a broad range of biological process such as neurotransmission, vasodilatation and immune responses. While the anti-microbiological properties of nitric oxide-derived reactive nitrogen intermediates (RNI) such as peroxynitrite, are known, the mechanism of these effects are as yet poorly studied. Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) belongs to the family Coronaviridae, was first identified during 2002-2003. Mortality in SARS patients ranges from between 6 to 55%. We have previously shown that nitric oxide inhibits the replication cycle of SARS-CoV in vitro by an unknown mechanism. In this study, we have further investigated the mechanism of the inhibition process of nitric oxide against SARS-CoV. We found that peroxynitrite, an intermediate product of nitric oxide in solution formed by the reaction of NO with superoxide, has no effect on the replication cycle of SARS-CoV, suggesting that the inhibition is either directly effected by NO or a derivative other than peroxynitrite. Most interestingly, we found that NO inhibits the replication of SARS-CoV by two distinct mechanisms. Firstly, NO or its derivatives cause a reduction in the palmitoylation of nascently expressed spike (S) protein which affects the fusion between the S protein and its cognate receptor, angiotensin converting enzyme 2. Secondly, NO or its derivatives cause a reduction in viral RNA production in the early steps of viral replication, and this could possibly be due to an effect on one or both of the cysteine proteases encoded in Orf1a of SARS-CoV.
Keywords: SARS-CoV, Palmitoylation, Nitric oxide, Spike protein
Introduction
Severe acute respiratory syndrome coronavirus (SARS-CoV) belongs to the family Coronaviridae and was first identified as a novel coronavirus after the first known outbreak in the fall of 2002 in southern part of China (Resh, 2006, Tan et al., 2006). SARS-CoV is a positive-sense RNA virus with a genome of ∼ 30 kb in length that forms a distinct group within the Coronaviridae family. Common to all known coronaviruses are the four structural proteins: spike (S), membrane (M), envelope (E) and nucleocapsid (N) protein (Marra et al., 2003, Rota et al., 2003). The S protein protrudes from the envelope of the virus, which results in its characteristic crown-like appearance. The S protein binds to specific receptors on the cell surface; in the case of SARS-CoV, it is the angiotensin-converting enzyme 2 (ACE2). The S protein has two regions, S1 and S2, where S1 is involved in receptor binding and S2 in membrane fusion (Li et al., 2005, Li et al., 2006).
In order to achieve its functionality, the S protein has to undergo numerous post-translational modifications like glycosylation (Keng et al., 2005). In addition, Petit et al. (2007) recently showed the importance of S-palmitoylation of the endodomain of the spike protein for the mediation of cell fusion. Palmitoylation involves the addition of palmitate, which is a 16-carbon saturated fatty acid, and a wide range of proteins have been shown to carry this specific modification (Resh, 2006). The addition of palmitate can influence the interaction between protein and membrane, protein trafficking, and protein–protein interactions (Resh, 2006, Smotrys and Linder, 2004, Thorp et al., 2006). In S-palmitoylation, the fatty acids are linked through thioester linkages to thiol groups on cysteine residues (Greaves and Chamberlain, 2007, Resh, 2006, Smotrys and Linder, 2004).
Nitric oxide (NO) is involved in a broad range of processes and act as an important signaling molecule between cells (Adler et al., 1997, Zhang et al., 2003). NO is produced in mammalian cells by three enzymes, neuronal (nNOS), endothelial (eNOS) and inducible nitric oxide synthase (iNOS) and these catalyze the oxidation of l-arginine to NO and l-citrulline (Boucher et al., 1999). Inducible nitric oxide in the host cells is commonly elevated during infection by viruses and this can lead to either inhibition or stimulation of viral replication (Adler et al., 1997, Akarid et al., 1995, Akerstrom et al., 2005, Fang, 2004, Klingstrom et al., 2006, Pope et al., 1998, Saxena et al., 2001, Thorp et al., 2006).
In our previous study, we showed that NO inhibits the replication cycle of SARS-CoV (Akerstrom et al., 2005). We clearly showed that the progeny viruses were inhibited in a concentration-dependent manner, using the exogenous NO donor S-nitroso-N-acetyl-penicillamine (SNAP). We also demonstrated that endogenously generated NO can inhibit the replication cycle of SARS-CoV. Another host defence against microbes is superoxide (O2 −) which can be induced when cells are stressed, for example during an infection (Akaike and Maeda, 2000). If NO and O2 − are elevated at the same time, they will react rapidly to form peroxynitrite (ONOO−), which has also been shown to have an inhibitory effect on viruses (Akaike, 2001). Hantaviruses are one group of viruses known to be adversely affected by peroxynitrite (Klingstrom et al., 2006). Hence, it is unclear if the effect of NO on SARS-CoV replication is caused by peroxynitrite or another derivative of nitric oxide. In this study we show that peroxynitrite does not play a significant role in the NO-mediated inhibition of the SARS-CoV replication cycle. Most interestingly, our data suggest that the effect of NO or its derivatives on the replication cycle of SARS-CoV is twofold: Firstly, NO or its derivatives causes a reduction in the palmitoylation of the S protein which results in the inhibition of membrane fusion mediated by the interaction between S protein and the ACE2 receptor. Secondly, NO or its derivatives have an inhibitory effect on the production of viral RNA, which can be observed as early as 3 h post-infection.
Results
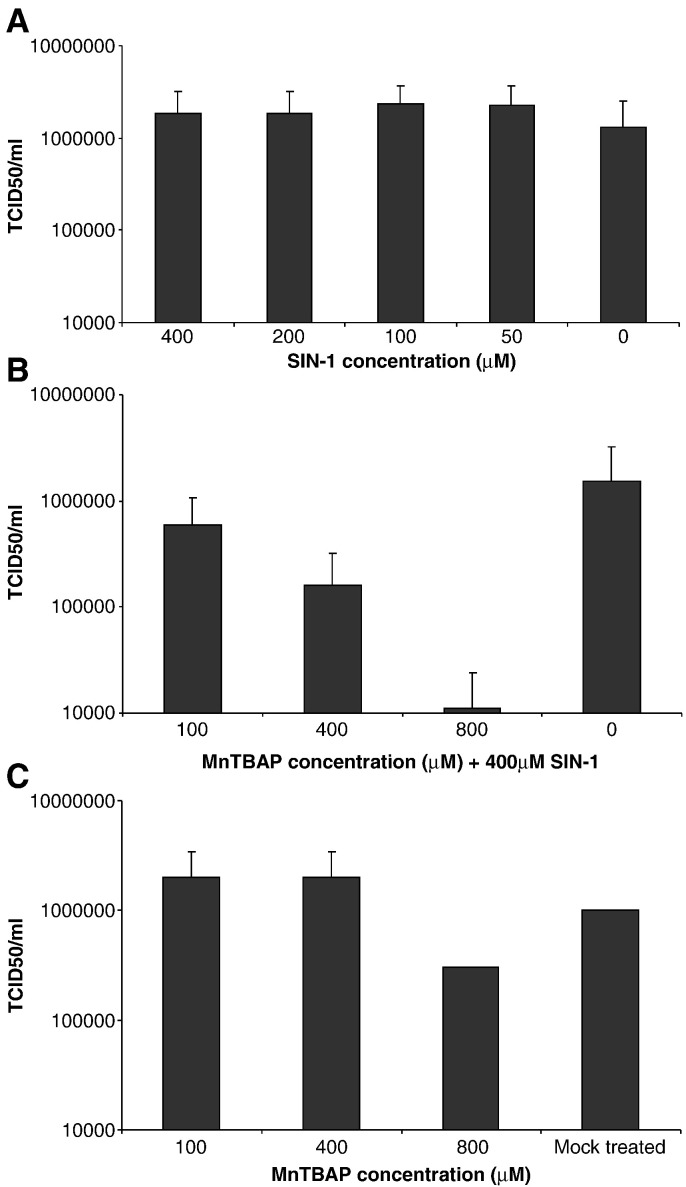
Peroxynitrite has no effect on SARS-CoV
SIN-1, which produces peroxynitrite, was used in conjunction with a superoxide scavenger, MnTBAP, to determine if peroxynitrite was involved in the inhibition of SARS-CoV replication. SARS-CoV infected Vero E6 cells were treated with different concentrations of SIN-1, and it was observed that no significant inhibition of viral replication occurred. This suggested that SIN-1 administration had no effect on the replication cycle of SARS-CoV.
Next, we treated the SARS-CoV infected Vero E6 cells with different concentrations of MnTBAP together with SIN-1. MnTBAP removes the superoxide, resulting in a reduction in the amount of peroxynitrite and an increase in the amount of free NO. The results in Fig. 1B clearly demonstrate that with increasing amount of MnTBAP in presence of SIN-1 (400 μM), there is an inhibition in the production of progeny virus in a concentration-dependent manner. Although a slight reduction on the production of progeny virus was observed when the cells were treated with the highest concentration of MnTBAP used in this study (800 μM, Fig. 1C), the reduction of virus replication in cells treated with both SIN-1 and MnTBAP is significantly higher than those treated with MnTBAP alone. This slight reduction might be explained by the possibility that the use of the superoxide scavenger would have resulted in an increase in intracellular free NO, which could then, directly or through an intermediary compound, exert an inhibitory effect.
Fig. 1.
SIN-1 treatment has no antiviral effect on SARS-CoV infected Vero E6 cells. Vero E6 cells were infected with SARS-CoV at an MOI of 1.0, at 1 hpi cells were treated with different concentrations of SIN-1 and/or MnTBAP. (A) Cells treated with different concentrations of SIN-1. 24 hpi, virus was harvested and titers determined. (B) Cells treated with 400 μM SIN-1 and different concentrations of MnTBAP. Virus was harvested 24 hpi and titers determined. (C) Cells treated with different concentrations of MnTBAP, virus was harvested 24 hpi and titers determined.
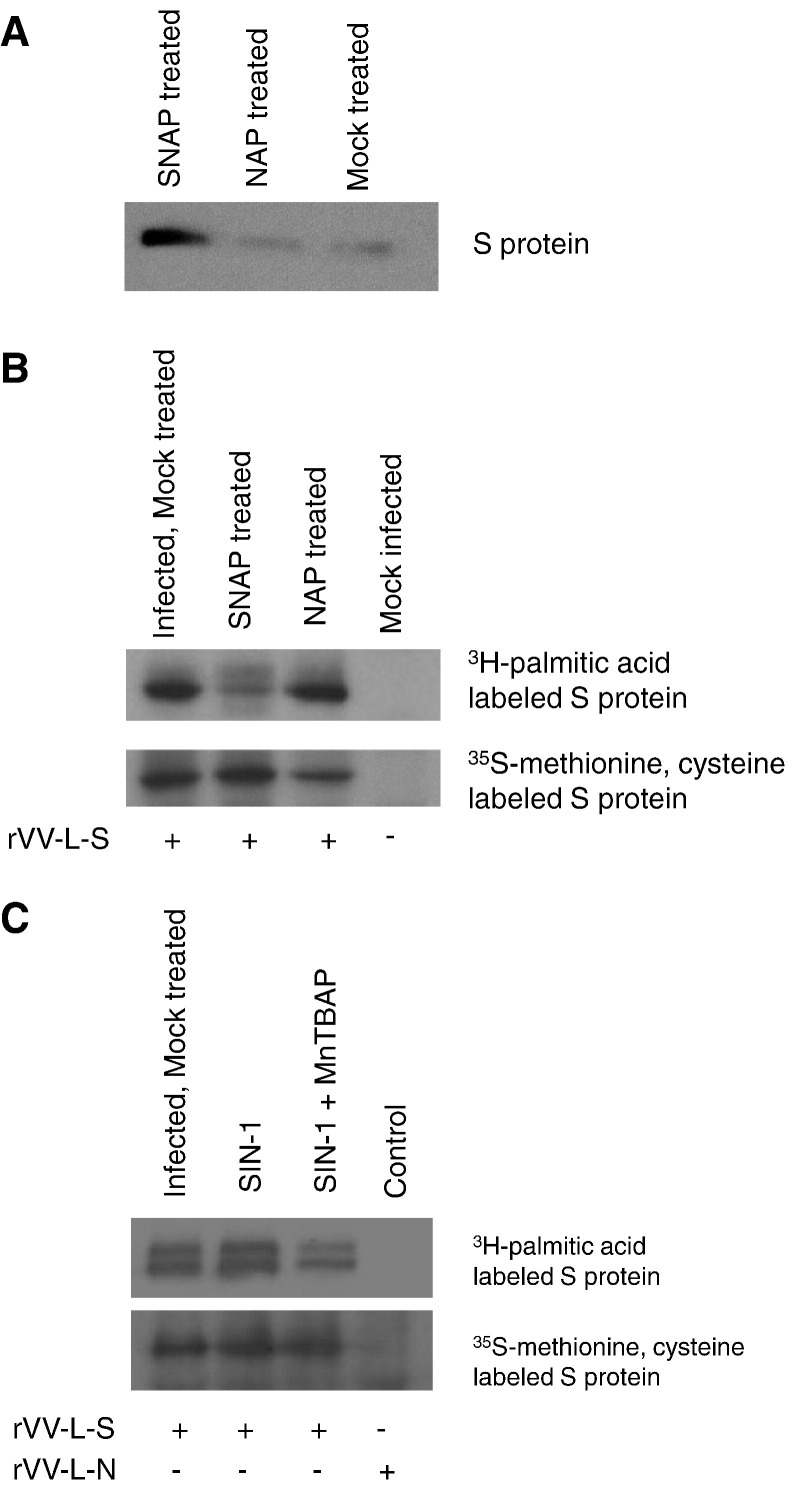
Treatment with NO donor leads to the nitration of the S protein of SARS-CoV and a reduction in its palmitoylation
In order to determine whether nitric oxide or its derivatives were reacting with the SARS-CoV S protein, Vero E6 cells were infected with recombinant vaccinia virus carrying the S gene (rVV-L-S) and treated with 400 μM of SNAP or NAP. Following immunoprecipitation with nitrotyrosine affinity sorbent (Cayman), which specifically binds nitrated proteins, and detection of S by Western blotting, it was clearly demonstrated that S had been nitrated after stimulation with SNAP but not with NAP (Fig. 2A).
Fig. 2.
Effect of SNAP on nitration and palmitoylation of S protein. Vero E6 cells were infected with rVV-L-S at an MOI of 0.1. At 1 hpi cells were treated with 400 μM SNAP or NAP. (A) At 24 h post-infection, the cells were lysed and immunoprecipitation was performed with nitrotyrosine affinity sorbent. Western blot was then performed using rabbit anti-S polyclonal antibody. (B, C) At 24 h post-infection, cells were labelled with 400 μCi of [3H]-palmitic acid for 2 h, or starved with methionine-cysteine (Met-Cys)-free medium for 30 min before being labelled with 22 μCi of [35S]-methionine-cysteine for 2 h. The cells were then lysed and imunoprecipitation was performed with rabbit S polyclonal antibody to detect the amount of radiolabelled S protein.
To determine whether the palmitoylation of S was affected by nitric oxide, cells infected with rVV-L-S were treated with either SNAP, NAP, SIN-1 or SIN-1 and MnTBAP and then labelled with [3H]-palmitic acid. After immunoprecipitation of the S protein with an anti-S rabbit polyclonal antibody, the amount of palmitoylated S protein detected in the SNAP-treated cells was found to be noticeably reduced when compared to the mock-treated or NAP-treated ones (Fig. 2B). Treatment with the peroxynitrite donor SIN-1 did not cause a reduction in the palmitoylation of S (Fig. 2C), confirming other results that showed a lack of effect by peroxynitrite treatment on SARS-CoV titer. On the other hand, if [35S]-methionine-cysteine labelling was used, there was no difference in the amount of total S protein immunoprecipitated for all treatments, indicating that the SNAP and SIN-1 treatments did not affect the expression of S (Figs. 2B, C).
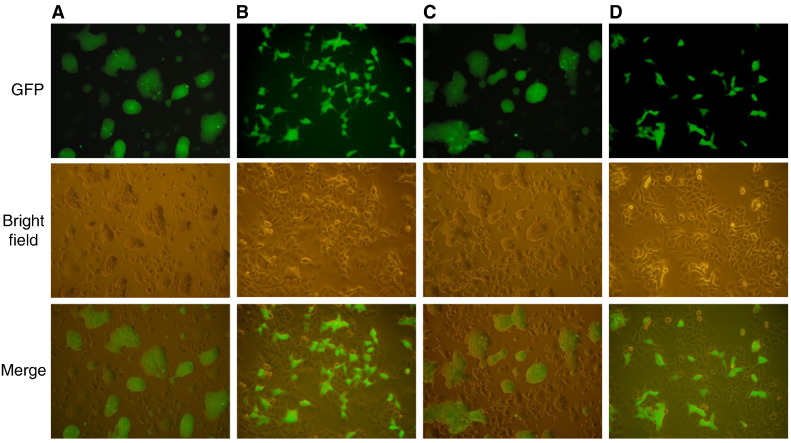
Nitric oxide reduces the cell–cell fusion activity of the S protein
An in vitro assay was used to examine the effect of nitric oxide on the cell–cell fusion activity of the S protein (Lip et al., 2006). In this case, the S-expressing cells were treated with SNAP or NAP before they were mixed with CHO-ACE2 cells stably expressing the ACE2 receptor. The formation of syncytia was observed 6 h later. Cells treated with SNAP showed no fusion (Fig. 3B), whereas cell–cell fusion was clearly visible in the NAP (Fig. 3C) or mock-treated cells (Fig. 3A). As expected, the mock-infected cells showed no fusion as the S protein was not expressed (Fig. 3D).
Fig. 3.
SNAP interferes with cell–cell membrane fusion. A 293T-GFP stable cell line was infected with rVV-L-S and treated with SNAP or NAP 1 h post-infection. At 24 hpi cells were trypsinized and mixed with pre-plated CHO-ACE2 cells. 6 h after mixing the cell lines, syncytium formation was observed. (A) Infected cells, mock-treated (B) Infected cells, treated with 400 μM SNAP (C) Infected cells, treated with 400 μM NAP (D) Mock-infected cells.
SARS-CoV S pseudotyped virus produced in the presence of nitric oxide is less efficient in viral entry
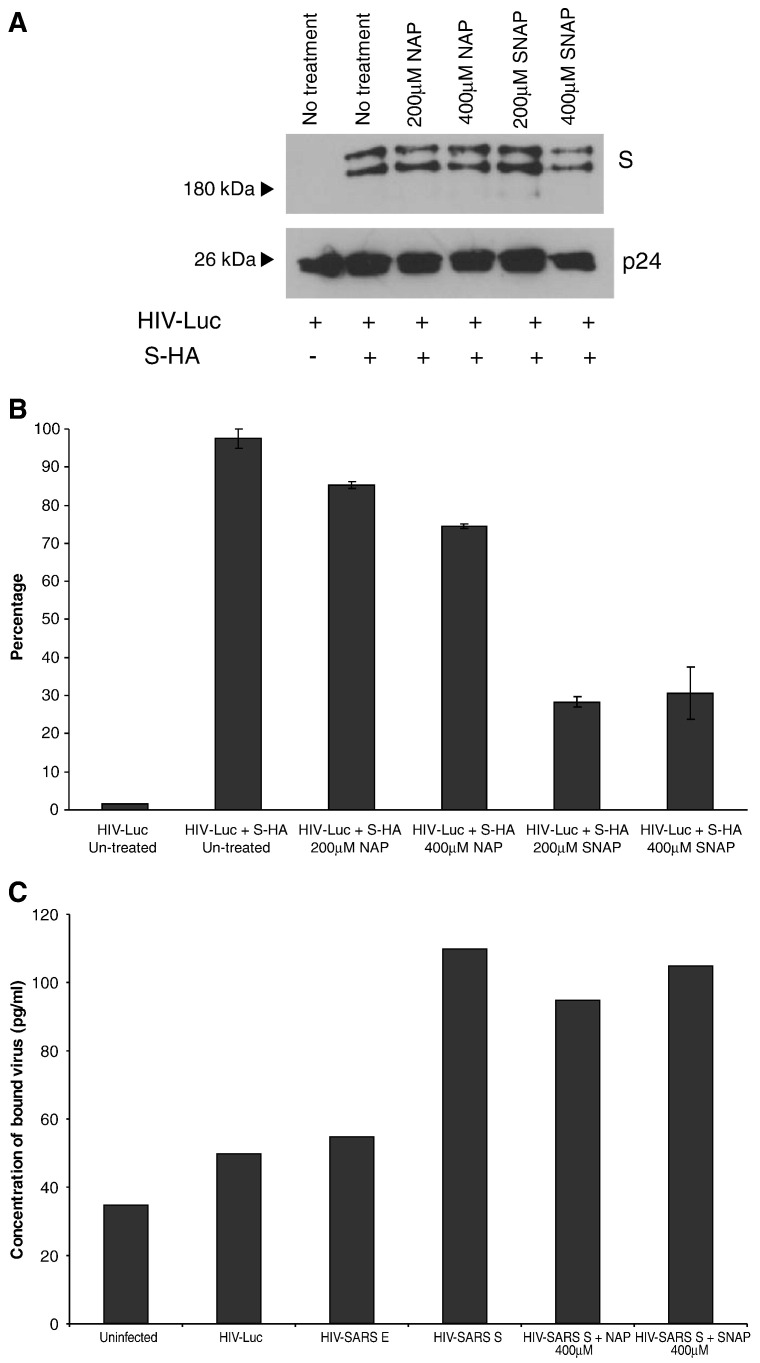
In order to investigate the effect of SNAP on the S-mediated entry of SARS CoV into cells, we used the HIV pseudotyped virus system to produce pseudotyped viruses with S on the surface. Pseudotyped viruses were produced from untreated cells or cells treated with SNAP (200 μM or 400 μM) or NAP (200 μM or 400 μM). SNAP and NAP treatments did not affect the assembly of S on the pseudotyped viruses as Western blot analysis showed that similar amounts of S were incorporated into the pseudotyped viruses (Fig. 4A). Similar to our previous observation (Keng et al., 2005), two forms of S were detected and these correspond to the unglycosylated and glycosylated forms of the protein.
Fig. 4.
SARS-CoV S pseudotyped virus produced in the presence of SNAP is less efficient in viral entry. Pseudotyped viruses produced under different conditions were purified and concentrated using ultra-centrifugation through a 20% sucrose bed. (A) The presence of S and HIV-1 p24 proteins in the purified viruses were determined by Western blot analysis. (B) Purified viruses were used for the transduction of CHO-ACE2 cells and the degrees of viral entry were determined by measuring the luciferase activities. Percentages of infectivity were computed by normalizing the entry of untreated S-bearing pseudotyped viruses to 100%. Means and standard deviation from duplicate readings were shown. (C) Nitric oxide or its derivatives do not exert a noticeable effect on the binding of the SARS S protein to ACE2. Cells were infected with pseudotyped virus bearing the S protein, produced under SNAP or NAP treatment. A p24 ELISA kit was used to measure p24 concentration and concentration of bound virus was derived from a standard graph.
The entry of the pseudotyped virus into CHO-ACE2, which is stably expressing the S receptor ACE2 on the surface of the cells, was reflected by the luciferase activity in the CHO-ACE2 cells at 72 h after infection. As shown in Fig. 4B, the luciferase activity in the cells infected with pseudotyped virus bearing S was significantly higher than those without S as the latter contained no viral envelope protein and could not enter the CHO-ACE2 cells. The reading from the cells infected with pseudotyped virus bearing S was normalized to 100% to calculate percentage of infectivity. Treatment with 200 μM and 400 μM of NAP caused appropriately 15% and 25% reduction in infectivity, respectively. However, treatment with 200 μM and 400 μM of SNAP resulted in a significantly higher reduction, appropriately 70%, in infectivity. In order to exclude any effect on the binding of the S protein to the ACE2 receptor, a p24 ELISA kit (Perkin Elmer) was used to determine the concentration of virus bound to cells infected with the HIV pseudotyped virus which had been produced in cells either treated with 400 μM SNAP or 400 μM NAP. As shown in Fig. 4C, SNAP and NAP treatment had no significant effect on the concentration of bound virus, indicating that the primary effect of SNAP treatment on the S protein occurs post-binding, possibly during entry.
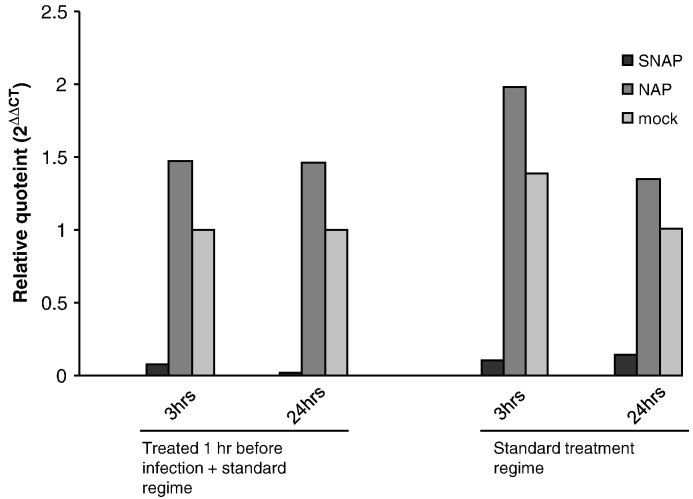
SNAP treatment reduces the production of positive-stranded viral RNA
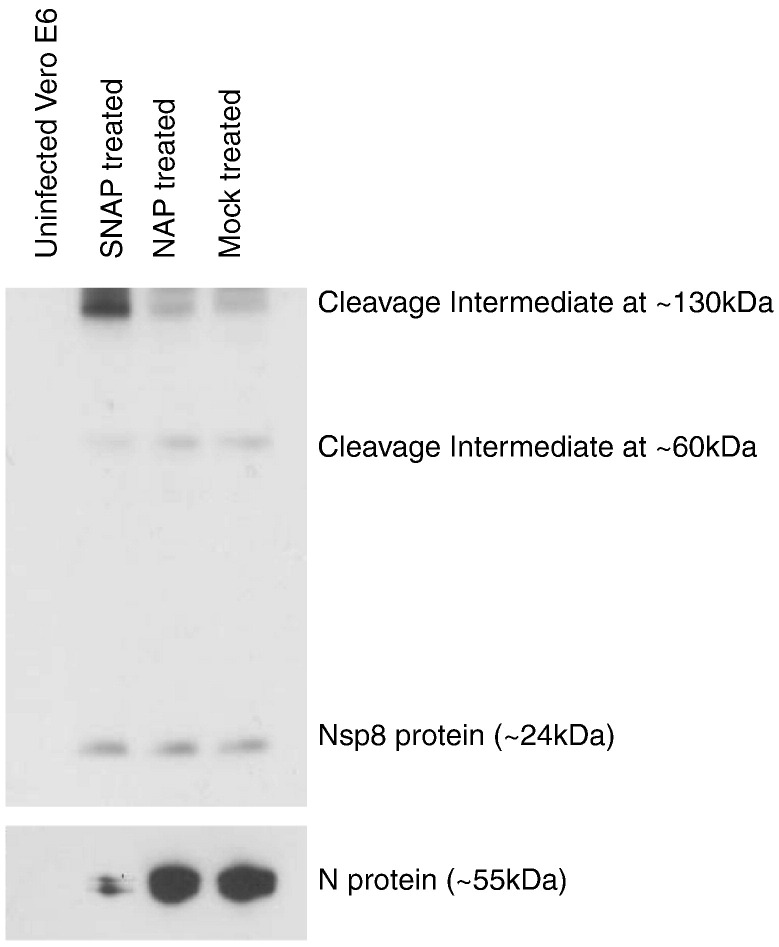
In order to determine if the effect of nitric oxide on SARS-CoV was limited to the post-translational modification of the S protein, we examined the production of viral RNA by realtime PCR analysis. RNA isolated from SARS-CoV infected Vero E6 cells at different time points post-infection in the presence of SNAP, NAP or mock treatment was reverse transcribed and quantitated using primers specific for the N gene of SARS-CoV. An identical amount of each RNA sample was also reverse transcribed and quantitated using primers specific for GAPDH, which served as a housekeeping control. The results showed that SNAP treatment of Vero E6 cells resulted in a reduction in the amount of total positive-stranded viral RNA produced. This effect was observed as early as 3 h post-infection and viral RNA levels remained low at 24 h post-infection (Fig. 5 ). In order to determine if this observed effect could be exaggerated by pretreating Vero E6 cells with nitric oxide, another experiment was run in parallel which included an additional SNAP treatment step an hour before infection. The effect observed was approximately the same as in cells treated post-infection, suggesting that the pretreatment of Vero E6 cells with nitric oxide did not significantly increase the effects of nitric oxide on the production of viral RNA (Fig. 5). The possibility that the pretreatment had an effect on SARS-CoV particles before infection was excluded by titrating SARS-CoV that had been incubated in SNAP-containing media, which showed that the TCID50 of the virus was unchanged (data not shown). It has been previously shown by others that nitric oxide or its derivatives can exert an effect on the activity of cysteine proteases (Cao et al., 2003, Saura et al., 1999). As SARS-CoV encodes two different cysteine proteases, PLpro and 3Cpro, we tried to determine if the effect seen on production of positive/stranded viral RNA might involve a loss or reduction of the function of one or both of these proteases, which serve to cleave the different replicase proteins from the pp1ab polyprotein expressed from Orfs 1a and 1b of the viral genome (Thiel et al., 2003). One simple way to study this effect would be to look at the cleavage of replicase proteins by Western blotting. A previously described monoclonal antibody (Kumar et al., 2007) targeting the nsp8 protein was used against SDS-PAGE-separated lysates from SARS-CoV infected Vero E6 cells which had been treated with either SNAP, NAP or untreated. Surprisingly, there was no significant difference in nsp8 levels at 24 h post-infection (Fig. 6 ). Interestingly though, two other peptide species, detected by the nsp8 antibody in infected cells, showed differences in their respective expression levels in SNAP-treated lysates. These high molecular weight species could possibly represent cleavage intermediates of the ORF1a replicase polyprotein from which nsp8 is cleaved. In order to ascertain that NO was acting in these cells, the same lysates were probed for the SARS N protein using a monoclonal antibody against N (Zymed Laboratories). It was observed that N expression was significantly reduced in the same lysates. This was consistent with earlier work (Akerstrom et al., 2005) and showed that the SNAP treatment was working within the Vero E6 cells.
Fig. 5.
SNAP treatment causes a reduction in the production of positive-stranded viral RNA. Vero E6 cells were either treated with 400 μM SNAP, NAP or mock-treated and infected with SARS-CoV at an MOI of 1. A parallel set of cells were additionally treated an hour before infection. Cells were harvested from all sets of treatments at 3 h and 24 h post infection and subjected to reverse transcription and realtime PCR using primers and probes specific for the N gene of SARS-CoV and GAPDH. Ct values obtained for N in each sample was normalized against GAPDH control values and plotted for each time point and treatment.
Fig. 6.
SNAP treatment causes a difference in observed levels of replicase polyprotein cleavage products. Lysates from either uninfected or SARS-CoV infected Vero E6 cells which had either been treated with SNAP, NAP or mock-treated harvested 24 h post-infection were subjected to Western blot analysis using a monoclonal antibody targeting the nsp8 protein.
Discussion
NO can either inhibit or stimulate viral replication during an infection (Adler et al., 1997, Fang, 2004 Akarid et al., 1995, Akerstrom et al., 2005, Greaves and Chamberlain, 2007, Klingstrom et al., 2006, Pope et al., 1998, Saxena et al., 2001, Thorp et al., 2006). However, there are only a few reports demonstrating the inhibition mechanism of NO. NO has been shown to affect Epstein–Barr virus (EBV) by downregulation of the Zta protein, an immediate early transactivator. The downregulation of Zta helps to maintain latency of the virus. In the same study, it was also shown that apoptosis in B cell lines were l-arginine dependent and that NO can inhibit apoptosis in B cells (Mannick et al., 1994). In a previous study we have shown that NO inhibits the replication cycle of SARS-CoV. The administration of the exogenous NO donor, SNAP, and endogenous cytokine inducers of NO reduced the amount of progeny virus in a concentration-dependent manner. In tissues that are stressed or inflamed, both NO and O2 − are elevated. Although SNAP predominantly increases the NO level in the cell, peroxynitrite may also become elevated after SNAP treatment as NO can react with endogenous O2 − to form peroxynitrite. However, if SIN-1 is used instead, both NO and O2 − are produced simultaneously and peroxynitrite is rapidly formed. Our results show that unlike SNAP, treatment of SARS-CoV infected Vero E6 cells with SIN-1 has no significant effect on the production of progeny virus (Fig. 1A), suggesting that peroxynitrite does not inhibit SARS-CoV replication. Moreover, if a superoxide scavenger (MnTBAP) is used to remove the O2 − and allow more NO to accumulate inside the SIN-1-treated cells, a significant inhibition of the virus replication was observed (Fig. 1B). These results clearly demonstrate that peroxynitrite has no effect on the replication of SARS-CoV and that it is NO, or a derivative of NO, that inhibits the virus.
Palmitoylation is an important post-translational modification of proteins. Petit et al. (2007) have demonstrated that palmitoylation of the SARS-CoV S protein plays a role in S-mediated cell–cell fusion. By substituting select cysteine clusters at the carboxyl terminus of the S protein for alanines, it was shown that palmitoylation of at least two cysteine clusters at the C-terminus was important in membrane fusion that occurred after binding of the S protein to its cognate ACE2 receptor, critical to the infectivity of SARS-CoV. It has also been previously shown that NO or its derivatives have an effect on the palmitoylation of the rat myelin protein (Bizzozero et al., 2001). In agreement with both of these findings, our results in this work indicate that NO or its derivatives reduce the palmitoylation of the S protein and consequently exerts an effect downstream of the S protein binding to ACE2, which is shown by the cell–cell fusion assay. While cell–cell fusion remains an unobserved phenomenon in SARS-CoV infection, in this paper the cell–cell fusion assay has proved useful in examining the interaction of the S protein and its cognate ACE2 receptor. However, the cell–cell fusion assay does not address the question of whether the underpalmitoylation of the S protein affects its expression, folding or incorporation into the mature virus particle. To this end we have created a pseudotyped virus based on HIV which incorporates the S protein and shown that SNAP treatment of cells does not affect the expression or incorporation of the S protein and it is only the infectivity of the pseudovirus, as shown by the luciferase assay, that is affected. It is known that treatment of mammalian cells with NO donors results in at least two different types of post-translational modifications—nitration of tyrosine residues (Ischiropoulos et al., 1992) and S-nitrosylation of cysteine residues (Stamler et al., 1992). We demonstrate that nitric oxide or a derivative compound is interacting with the S protein by means of the tyrosine nitration assay. However, as tyrosine nitration depends on the presence of peroxynitrite (Ischiropoulos et al., 1992), which has been excluded from having an effect on the replication cycle of SARS-CoV in this work, this indicates that either (a) the tyrosine residues which have been nitrated have little or no role to play in the binding between the S protein and ACE2, or (b) that the nitration of these tyrosine residues do not impair their role in the S-protein. The true effect of NO on the binding process might then manifest itself at the cysteine residues on the S protein which may have been palmitoylated, in particular the clusters at the carboxy terminal that have been demonstrated to play a role in cell–cell fusion. But what needs to be determined is whether these residues have simply been underpalmitoylated as a result of an effect on palmitoyl-acyl transferases in the host cell or whether, given that these are cysteine residues, competitively nitrosylated. S-nitrosylation motifs have been previously described as an acid–base motif flanking cysteine residues (Ascenzi et al., 2000, Stamler et al., 1997), and a search of the S-protein amino acid sequence for previously described motifs yielded no results. Further studies will have to be done to answer these questions.
While the palmitoylation of the S protein has been shown to be affected by the administration of SNAP as an NO donor, this is an effect that is seen in the later steps of infection, after the expression of viral proteins. However, nitric oxide and its derivatives have been shown in the case of several micro-organisms to have an effect on the early steps of replication, in both bacterial and viral infections. The work done by several groups has demonstrated that cysteine proteases are particularly susceptible to nitric oxide, as has been described for coxsackieviruses (Saura et al., 1999) and adenoviruses (Cao et al., 2003). The use of realtime PCR directed against positive-stranded RNA species encoded by SARS-CoV showed that SNAP treatment of Vero E6 cells caused a significant decrease in viral RNA production which could be observed as early as 3 h post-infection compared to NAP-treated and mock-treated controls. This observation is interesting as SARS-CoV encodes 2 cysteine proteases, the papain-like protease (PL2pro) and the 3C-like protease(3CLpro) (Thiel et al., 2003). These cleave the pp1ab replicase polyprotein at non-overlapping sites to yield 15 replicase proteins, termed nsp1 to nsp15. PL2pro is predicted to cleave the polyprotein at 3 sites, and the rest of the non-structural proteins have been predicted to be cleaved by 3CLpro. Hence, in order to examine a possible effect of NO on the activity of these proteases, a monoclonal antibody directed against the nsp8 protein was used to look at the relative amounts of cleavage products being formed in SARS-CoV infected Vero E6 cells treated with either SNAP, NAP or in mock-treated cells. If NO or its derivatives exerted a discrete effect on the activity of 3CLpro, one might expect to see a reduction in the amount of nsp8 protein as determined by Western blotting. Surprisingly, no difference was seen in the levels of cleaved nsp8 amongst the different treatments. However, 2 bands of higher molecular weight (of around 60 kDa and 130 kDa) was seen to exhibit altered intensities in SARS-CoV infected cells treated with SNAP. As these bands are specific to infection and are detected by the nsp8 antibody, they might represent cleavage intermediates of the replicase polyprotein. In a 2004 study, Fan and coworkers expressed and purified the SARS-CoV 3CLpro in order to study its activity. Using an HPLC-based peptide cleavage assay, they found that the 11 unique 3CLpro cleavage sites on the pp1ab replicase polyprotein were cleaved with different efficiencies (Fan et al., 2005). In light of this information, the differences seen in the amounts of cleavage products might make more sense to the observer. It could be possible that the effect of NO or its derivatives on 3CLpro might have altered its substrate specificities, resulting in a different pattern of cleavage efficiencies from that previously observed. This might account, then, for the lack of a difference in nsp8 levels while an accumulation is observed for a different intermediate cleavage product which might have had its cleavage efficiency altered more detrimentally. In order to confirm that NO was actually working in these samples, we performed a Western blot using a monoclonal antibody specific to the N protein of SARS-CoV (Fig. 6). It was observed that the expression of the N protein was reduced, in agreement with previous findings (Akerstrom et al., 2005). This showed the intended effect of SNAP treatment and therefore that the observed differences in cleavage product intensities was being observed in cells which were being affected by SNAP treatment.
From the results presented here, we suggest that the effect of nitric oxide on the replication of SARS-CoV is at least twofold: an effect on the production of viral RNA in the early steps of replication and a reduction in the palmitoylation of the S protein later in the replication cycle. What remains to be elucidated is whether both of these effects arise strictly from a direct effect of nitric oxide or its derivatives on viral proteins, or strictly from an effect on host factors which are subverted in the course of infection, or a combination of both.
Materials and methods
Peroxynitrite and superoxide dismutase treatment
Vero E6 cells grown in 24-well plates were infected with SARS-CoV Frankfurt strain 1 at an MOI of 1. At 1 hpi, cells were washed twice with PBS, and medium with or without 3-morpholinosydnonimine hydrochloride (SIN-1; Sigma-Aldrich) was added. 50, 100, 200 or 400 μM of SIN-1 were added 3 times with 4-h intervals. After 24 h, the virus was harvested and titration of progeny virus was titrated out on a 96-well plate containing Vero E6 cells as described previously (Akerstrom et al., 2005). At 48 hpi, the amount of virus (TCID50) was calculated from the cytopathic effect (CPE) induced in cell culture by serial tenfold dilutions of the harvested virus.
Similarly, SARS-CoV infected Vero E6 cells were treated with a superoxide scavenger, Mn(III)tetrakis(4-Benzoic acid)porphyrin chloride (MnTBAP; Calbiochem), in the absence or presence of 400 μM SIN-1. 100, 400 or 800 μM of MnTBAP with or without 400 μM SIN-1 were added 3 times with 4-h intervals. The amount of progeny virus was measured as described above. All SARS-CoV infection work (described here and elsewhere in the manuscript) was carried out at the BSL4 facility at the Swedish Institute for Infectious Disease Control in Solna, Sweden.
Radiolabelled immunoprecipitation
Vero E6 cells grown in 60 mm dishes were infected with recombinant vaccinia virus carrying the S gene (rVV-L-S), at a MOI of 0.1. For mock-infected cells, a recombinant vaccinia virus carrying the N gene (rVV-L-N) was used. Both rVV-L-S and rVV-L-N are kind gifts from Baxter Vaccine (Orth/Donau, Austria) and have been previously described (Lip et al., 2006). At 1 h post-infection, the cells were treated with 400 μM of SNAP (Sigma, St. Louis, Mo.), 400 μM N-acetylpenicillamine (NAP; Sigma), 400 μM of SIN-1 (Sigma, St. Louis, Mo) or 400 μM of SIN-1 and 400 μM of MnTBAP (Calbiochem). Treatments were repeated 4 times with 2-h intervals. At 24 h post-infection, cells were labelled with 400 μCi of [3H]-palmitic acid (Perkin Elmer, USA) for 2 h or starved with methionine-cysteine (Met-Cys)-free medium for 30 min before being labelled with 22 μCi of [35S]-methionine-cysteine for 2 h. The labelled cells were washed 3 times with cold PBS and lysed in 1 ml TES lysis buffer (20 mM Tris–HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 2 mM PMSF). Cell debris was removed from the lysates by centrifugation at 13,000 rpm for 10 min. Immunoprecipitation was carried out by adding rabbit anti-SΔ10 antibody (Keng et al., 2005) to the lysates and rotating at 4 °C for 1 h, followed by overnight incubation with protein A-Sepharose beads (Roche Diagnostics). The beads were washed 3 times with lysis buffer and boiled in 20 μl of 2 × Laemmli's SDS loading buffer. Samples were separated in a 7.5% SDS-PAGE gel, fixed for 30 min using fixing solution (10% acetic acid, 45% methanol), and treated with Amplify Fluorographic Reagent (Amersham Biosciences) according to protocol. Visualization was done by autoradiography.
Cell–cell membrane fusion assay
An in vitro cell fusion assay that can be used to study S-mediated membrane fusion has been previously developed (Lip et al., 2006). Briefly a 293T cell-line stably expressing GFP was infected with rVV-L-S in order to express the S protein and then treated with SNAP or NAP as described above. Another stable cell line stably expressing ACE2, CHO-ACE2, was plated in a 12-well plate in DMEM containing 1 mg/ml porcine trypsin (JRH Biosciences Inc.). Infected 293T-GFP cells were mixed with the CHO-ACE2 cells in a 1:1 ratio per well and syncytium formation was observed 6 h later.
Pseudotyped virus entry assay
To produce HIV pseudotyped virus bearing the SARS-CoV S protein, 9 μg of HIV-1 luciferase reporter vector pNL4.3.Luc.R-E- (HIV-luc) and 4.5 μg of SARS-CoV S protein expression plasmid (pXJ3′-S-HA) were co-transfected into 293T cells in a 100-mm dish using Lipofectamine2000 reagent (Invitrogen) following manufacturer's instructions. For the negative control, the cells were transfected with pNL4-3.Luc.R-E-alone. The construction of the plasmids has been described previously (Connor et al., 1995, He et al., 1995, Tan et al., 2004). The virus-containing medium was harvested after 72 h of incubation and centrifuged at 2000 rpm for 5 min to remove any cell debris. Virus in the supernatant was subsequently concentrated through a 20% sucrose cushion for 3 h at 30,000 rpm and 4 °C by using an SW41 rotor, and subsequently resuspended in DMEM. In order to normalize the amount of pseudotyped virus used, 10 μl of each sample was used to measure p24 content using a p24 ELISA kit (Perkin Elmer), according to the manufacturer's protocol. Absorbance readings were made at 492 nm in a plate reader (Tecan) and sample concentrations were determined from a standard graph. Appropriate volumes corresponding to 16 ng of pseudotyped virus were then used to infect CHO-ACE2 cells (Chou et al., 2005). At 72 hpi, supernatant was removed and the cells were washed and harvested in trypsin and washed again in PBS. Cell pellet was lysed using the Promega lysis buffer and luciferase activity was determined using a luciferase assay kit, following manufacturer's instructions (Promega). Luciferase readings were measured using a Veritus microplate luminometer (Turner Biosystems).
Pseudotyped virus binding assay
HIV pseudotyped virus bearing the SARS-S protein was produced as described in the pseudotyped virus entry assay (see above). Virus-containing medium was harvested after 24 h of incubation and centrifuged at 3000 rpm for 5 min to remove any cell debris. 10 μl of the medium were diluted 20 × using DMEM and used to measure virus concentration using a P24 ELISA kit (Perkin Elmer), according to the manufacturer's protocol. Virus concentration was determined from a standard graph, and 80 ng of virus was used for infection of CHO-ACE2 cells. CHO-ACE2 cells were pre-incubated in DMEM with 1% BSA at 4 °C for 1 h prior to infection at 4 °C for 2 h. After infection, the cells were washed 4 × using 1 × PBS with 1% BSA and harvested in DMEM, freeze–thawed six times and sonicated for 2 min with 20-s intervals. Samples were centrifuged at 13,000 rpm to remove any cell debris and 200 μl of supernatant was used to determine P24 content using the P24 ELISA kit. The concentration of bound virus was then determined using the standard graph.
Western blot and detection of nitrated SARS-CoV S protein
Vero E6 cells grown in 60 mm dishes were infected with rVV-L-S and rVV-L-N and treated with 400 μM SNAP or NAP as described above. After 24 h, the cells were lysed (50 mM Tris [pH 8], 150 mM NaCl, 0.5% NP40, 0.5% deoxycholic acid, 0.005% SDS, 1 mM PMSF) and the concentration of total protein in the cell lysates was determined using Coomassie Plus reagent (Pierce). Appropriate volumes yielding 500 μg of total protein from each sample were added to 20 μl of Nitrotyrosine Affinity Sorbent (Cayman Chemical) after washing the beads three times in PBS. The samples were left for rotation at 4 °C over night. Next day, the beads were washed 3 times with lysis buffer, resuspended in 20 μl of Laemmli's SDS loading buffer and boiled for 10 min at 100 °C. Samples were separated on a 7.5% SDS-PAGE gel, transferred to a nitrocellulose membrane, and blocked for 1 h in room temperature in 5% non fat dry milk in PBS with 0.1% Tween 20. Membrane was incubated overnight in 5% non-fat dry milk with rabbit anti-SΔ10 antibody (diluted 1:6000) at 4 °C. The next day, the membrane was incubated with HRP-conjugated secondary antibody (goat anti-rabbit antibody diluted 1:2000) for 1 h at room temperature and washed in PBS-Tween. Detection was performed with SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Quantification of viral RNA
Vero E6 cells were grown in 24-well plates and infected with SARS-CoV at an MOI of 1. Cells were treated with SNAP, NAP or mock-treated either 1 h before infection and 1 h, 3 h or 6 h post-infection or only 1 h, 3 h, 6 h post-infection without pretreatment. At 24 h post-infection, cells were harvested in Trizol reagent (Invitrogen) and total RNA was extracted using an RNeasy mini kit (Qiagen). Viral RNA was quantitated using a Nanodrop spectrophotometer and equal amounts of total RNA were added to a reverse transcription reaction using either of the following gene-specific primers: SARSNRTREV-5′ TTATGCCTGAGTTGAATCAGCAGAA 3′; GAPDHRTREV-5′ AGCCTTCTCCATGCTGGTGAAGAC 3′. Equal amounts of each reverse transcription reaction were then added to a portion of LightCycler Taqman Master reaction mix (Roche) along with either of the following primer pairs: SARSNCCORFP-5′ TGCCTCTGCATTCTTTGGA 3′, SARSNCCORRP-5′-TAAGTCAGCCATGTTCCCG 3′, GAPDHF-5′ GAAGATGGTGATGGGATTTC 3′, GAPDHR-5′ GAAGGTGAAGGTCGGAGT 3′ and the following probes: SARSNCCORP1-5′ FAM (6-carboxy-flourescein)-, tgtgacttccatgccaatgcgtg-TAMRA (6-carboxy-tetrylrhodamine quencher) 3′ or GAPDHP-5′FAM-,CAAGCTTCCCGTTCTCAGCC-TAMRA 3′. Cycle threshold values obtained for the SARS N gene were normalized against those for GAPDH to obtain relative quotients (2ΔΔCt) which were then plotted for SNAP, NAP and mock treatments.
Acknowledgments
We thank Baxter Vaccine (Orth/Donau, Austria) for sharing their proprietary vaccinia virus expression system, and S. Shen (Institute of Molecular and Cell Biology, Singapore) for technical assistance. The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3.Luc.R-E- from Dr. Nathaniel Landau.
References
- Ascenzi P., Colasanti M., Persichini T., Muolo M., Polticelli F., Venturini G., Bordo D., Bolognesi M. Re-evaluation of amino acid sequence and structural consensus rules for cysteine-nitric oxide reactivity. Biol. Chem. 2000;381(7):623–627. doi: 10.1515/BC.2000.081. [DOI] [PubMed] [Google Scholar]
- Adler H., Beland J.L., Del-Pan N.C., Kobzik L., Brewer J.P., Martin T.R., Rimm I.J. Suppression of herpes simplex virus type 1 (HSV-1)-induced pneumonia in mice by inhibition of inducible nitric oxide synthase (iNOS, NOS2) J. Exp. Med. 1997;185:1533–1540. doi: 10.1084/jem.185.9.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike T. Role of free radicals in viral pathogenesis and mutation. Rev. Med. Virol. 2001;11:87–101. doi: 10.1002/rmv.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike T., Maeda H. Nitric oxide and virus infection. Immunology. 2000;101:300–308. doi: 10.1046/j.1365-2567.2000.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akarid K., Sinet M., Desforges B., Gougerot-Pocidalo M.A. Inhibitory effect of nitric oxide on the replication of a murine retrovirus in vitro and in vivo. J. Virol. 1995;69:7001–7005. doi: 10.1128/jvi.69.11.7001-7005.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerstrom S., Mousavi-Jazi M., Klingstrom J., Leijon M., Lundkvist A., Mirazimi A. Nitric oxide inhibits the replication cycle of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1966–1969. doi: 10.1128/JVI.79.3.1966-1969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzozero O.A., Bixler H., Parkhani J., Pastuszyn A. Nitric oxide reduces the palmitoylation of rat myelin proteolipid protein by an indirect mechanism. Neurochem. Res. 2001;26:1127–1137. doi: 10.1023/a:1012370822754. [DOI] [PubMed] [Google Scholar]
- Boucher J.L., Moali C., Tenu J.P. Nitric oxide biosynthesis, nitric oxide synthase inhibitors and arginase competition for l-arginine utilization. Cell Mol. Life Sci. 1999;55:1015–1028. doi: 10.1007/s000180050352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Baniecki M.L., McGrath W.J., Bao C., Deming C.B., Rade J.J., Lowenstein C.J., Mangel W.F. Nitric oxide inhibits the adenovirus proteinase in vitro and viral infectivity in vivo. FASEB J. 2003;17(15):2345–2346. doi: 10.1096/fj.03-0396fje. [DOI] [PubMed] [Google Scholar]
- Chou C.-F., Shen S., Tan Y.-J., Fielding B.C., Tan T.H.P., Fu J.-L., Xu X., Lim S.G., Hong W. A novel cell-based binding assay system reconstituting interaction between SARS-CoV S protein and its cellular receptor. J. Virol. Methods. 2005;123:41–48. doi: 10.1016/j.jviromet.2004.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor R.I., Chen B.K., Choe S., Landau N.R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Fan K., Ma L., Han X., Liang H., Wei P., Liu Y., Lai L. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2005;329(3):934–940. doi: 10.1016/j.bbrc.2005.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F.C. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- Greaves J., Chamberlain L.H. Palmitoylation-dependent protein sorting. J. Cell Biol. 2007;176:249–254. doi: 10.1083/jcb.200610151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Choe S., Walker R., Di Marzio P., Morgan D.O., Landau N.R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischiropoulos H., Zhu L., Chen J., Tsai M., Martin J.C., Smith C.D., Beckman J.S. Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch. Biochem. Biophys. 1992;298(2):431–437. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- Keng C.T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H., Chou C.F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.J. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingstrom J., Akerstrom S., Hardestam J., Stoltz M., Simon M., Falk K.I., Mirazimi A., Rottenberg M., Lundkvist A. Nitric oxide and peroxynitrite have different antiviral effects against hantavirus replication and free mature virions. Eur. J. Immunol. 2006;36:2649–2657. doi: 10.1002/eji.200535587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Gunalan V., Liu B., Chow V.T., Druce J., Birch C., Catton M., Fielding B.C., Tan Y.J., Lal S.K. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366(2):293–303. doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li F., Berardi M., Li W., Farzan M., Dormitzer P.R., Harrison S.C. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 2006;80:6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lip K.M., Shen S., Yang X., Keng C.T., Zhang A., Oh H.L., Li Z.H., Hwang L.A., Chou C.F., Fielding B.C., Tan T.H., Mayrhofer J., Falkner F.G., Fu J., Lim S.G., Hong W., Tan Y.J. Monoclonal antibodies targeting the HR2 domain and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. J. Virol. 2006;80:941–950. doi: 10.1128/JVI.80.2.941-950.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick J.B., Asano K., Izumi K., Kieff E., Stamler J.S. Nitric oxide produced by human B lymphocytes inhibits apoptosis and Epstein-Barr virus reactivation. Cell. 1994;79:1137–1146. doi: 10.1016/0092-8674(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Petit C.M., Chouljenko V.N., Iyer A., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion. Virology. 2007;360:264–274. doi: 10.1016/j.virol.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope M., Marsden P.A., Cole E., Sloan S., Fung L.S., Ning Q., Ding J.W., Leibowitz J.L., Phillips M.J., Levy G.A. Resistance to murine hepatitis virus strain 3 is dependent on production of nitric oxide. J. Virol. 1998;72:7084–7090. doi: 10.1128/jvi.72.9.7084-7090.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh M.D. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE. 2006;2006:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Saura M., Zaragoza C., McMillan A., Quick R.A., Hohenadl C., Lowenstein J.M., Lowenstein C.J. An antiviral mechanism of nitric oxide: inhibition of a viral protease. Immunity. 1999;10:21–28. doi: 10.1016/S1074-7613(00)80003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S.K., Mathur A., Srivastava R.C. Induction of nitric oxide synthase during Japanese encephalitis virus infection: evidence of protective role. Arch. Biochem. Biophys. 2001;391:1–7. doi: 10.1006/abbi.2001.2360. [DOI] [PubMed] [Google Scholar]
- Smotrys J.E., Linder M.E. Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- Stamler J.S., Simon D.I., Osborne J.A., Mullins M.E., Jaraki O., Michel T., Singel D.J., Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. U. S. A. 1992;89(1):444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler J.S., Toone E.J., Lipton S.A., Sucher N.J. (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 1997;18(5):691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Tan Y.J., Teng E., Shen S., Tan T.H.P., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel SARS coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Lim S.G., Hong W. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antiviral Res. 2006;72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Thorp E.B., Boscarino J.A., Logan H.L., Goletz J.T., Gallagher T.M. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 2006;80:1280–1289. doi: 10.1128/JVI.80.3.1280-1289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Kuncewicz T., Yu Z.Y., Zou L., Xu X., Kone B.C. Protein–protein interactions involving inducible nitric oxide synthase. Acta Physiol. Scand. 2003;179:137–142. doi: 10.1046/j.1365-201X.2003.01119.x. [DOI] [PubMed] [Google Scholar]