Abstract
The non-structural protein 1 (nsp1) of porcine reproductive and respiratory syndrome virus is partly responsible for inhibition of type I interferon (IFN) response by the infected host. By performing alanine-scanning mutagenesis, we have identified amino acid residues in nsp1α and nsp1β (the proteolytic products of nsp1) that when substituted with alanine(s) exhibited significant relief of IFN-suppression. A mutant virus (16-5A, in which residues 16–20 of nsp1β were substituted with alanines) encoding mutant nsp1β recovered from infectious cDNA clone was shown to be attenuated for growth in vitro and induced significantly higher amount of type I IFN transcripts in infected macrophages. In infected pigs, the 16-5A virus exhibited reduced growth at early times after infection but quickly regained wild type growth properties as a result of substitutions within the mutated sequences. The results indicate a strong selection pressure towards maintaining the IFN-inhibitory property of the virus for successful propagation in pigs.
Keywords: PRRSV, Type-1 interferon inhibition, Innate immunity, Non-structural protein 1α and 1β
Introduction
Porcine reproductive and respiratory syndrome (PRRS) is a significant animal disease affecting swine herds in all pork-producing countries. Two major clinical manifestations of the disease are late term reproductive failure in pregnant sows and respiratory distress in young piglets as well as growing pigs. The annual economic losses as a result of PRRS to the USA swine industry alone has been estimated to be more than $660 million (Miller, 2011). The recent large-scale outbreak of “high fever” disease in China in 2006 with unusually high mortality rate further adds to the significance of this swine pathogen (Tian et al., 2007). The etiological agent of the disease is PRRS virus (PRRSV), an enveloped positive-sense, single-stranded RNA virus with a genome of ∼15 kb. PRRSV belongs to the family Arteriviridae which, along with Coronaviridae and Roniviridae families, form the order Nidovirales (Cavanagh, 1997). Other members of Arteriviridae families include equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV) and simian hemorrhagic fever virus (SHFV). The viral genome encodes 10 open reading frames (ORFs)—ORF1a, ORF1b, ORF2a, ORF2b, ORF3, ORF4, ORF5a, ORF5, ORF6 and ORF7. The ORF1a and ORF1b are translated to generate polyproteins, which are processed by viral proteases to form 14 different non-structural proteins (nsps) (Fang and Snijder, 2010, Snijder and Meulenberg, 1998). Several of the nsps have been identified as integral members of viral replication and transcription machinery while others might be involved in these processes through their interaction with host cell factors (Beura et al., 2011, Fang and Snijder, 2010). Furthermore, the nsps are also likely to regulate viral pathogenesis through their involvement in modulation of host innate immune response.
The type 1 interferon (IFN) constitutes a major player of the host innate immune response system. Viral replication intermediates like double stranded RNA (dsRNA) are sensed by cytoplasmic (RIG-I like helicases) as well as endosomal (Toll-like receptor 3, TLR3) sensors, which trigger a complex signaling cascade (Bowie and Unterholzner, 2008, Kawai and Akira, 2009). These signaling events culminate in activation of several transcription factors including interferon regulatory factor 3 (IRF3), nuclear factor kappa B (NF-κB) and activating transcription factor-2 (ATF-2). These transcription factors coordinately drive expression of type 1 IFN genes. Once secreted, IFNs bind to their cognate receptors on the cell surface and initiate the Janus kinase (JAK)-signal transducers and activators of the transcription (STAT) signaling pathway, which leads to synthesis of IFN-stimulated genes (ISGs). These ISGs then establish the antiviral state. During the course of evolution, viruses have developed numerous strategies to counteract IFN production and signaling pathways to ensure their propagation in the host (Versteeg and Garcia-Sastre, 2010). Infection with PRRSV results in poor type 1 IFN production both in in vitro infected macrophages and infected pigs (Albina et al., 1998, Lee et al., 2004). This low level of IFN induction is a process of active suppression by virus since infection with a strong IFN-inducer transmissible gastroenteritis coronavirus (TGEV) after PRRSV infection could not elicit detectable IFN production (Albina et al., 1998). The nsps of PRRSV inhibit IFN-dependent transcription. Earlier, we reported that five different nsps (nsp1α, nsp1β, nsp2, nsp4 and nsp11) can inhibit IFN-β gene transcription (Beura et al., 2010). The nsp1α and nsp1β protein suppress both IRF3 and NF-κB mediated IFN gene induction (Beura et al., 2010, Chen et al., 2010, Song et al., 2010). The nsp1β also interferes with IFN signaling specifically the JAK-STAT pathway (Chen et al., 2010, Patel et al., 2010). PRRSV nsp2 interferes with NF-κB signaling by deubiquitinating the ubiquitinated IκBα molecule that is important for NF-κB activation (Sun et al., 2010).
After being exposed to PRRSV, the animals develop viremia, which lasts for a month, but the virus can still be detected in certain secondary lymphoid tissue up to 5 months after infection (Allende et al., 2000, Wills et al., 2003). The level of various proinflammatory cytokines, other important components of host innate immune response besides IFN, are low compared to those induced by several other respiratory swine pathogens (Van Reeth et al., 1999, van Reeth and Nauwynck, 2000). The subsequent development of effector components of adaptive immune response e.g., neutralizing antibodies, antigen-specific T-cells are delayed (Lopez and Osorio, 2004). A robust adaptive immune response is dependent on proper priming of the innate immune response. Hence, the initial suboptimal innate response is hypothesized to be responsible for the delayed and defective development of adaptive immune response (Kimman et al., 2009, Murtaugh et al., 2002). Thus, a PRRSV that does not efficiently suppress type 1 IFN induction is predicted to stimulate a strong adaptive immune response culminating in the rapid clearance of PRSSV (Nan et al., 2012).
The objective in this study was to map the domains/residues of PRRSV nsp1α and nsp1β that are responsible for inhibiting IRF3 mediated gene induction. Using alanine-scanning mutagenesis, we have identified such residues in both proteins. We were also able to recover a virus with mutations in the nsp1β protein. Characterization of the nsp1β mutant virus (16-5A) demonstrated that the virus is attenuated for growth and induced higher level of type 1 IFN in vitro. However, the mutation was unstable in vivo and the revertant virus quickly regained the ability to suppress IFN production in infected pigs.
Results
Identification of residues in PCPα domain of PRRSV nsp1α those are important for inhibiting IFN production
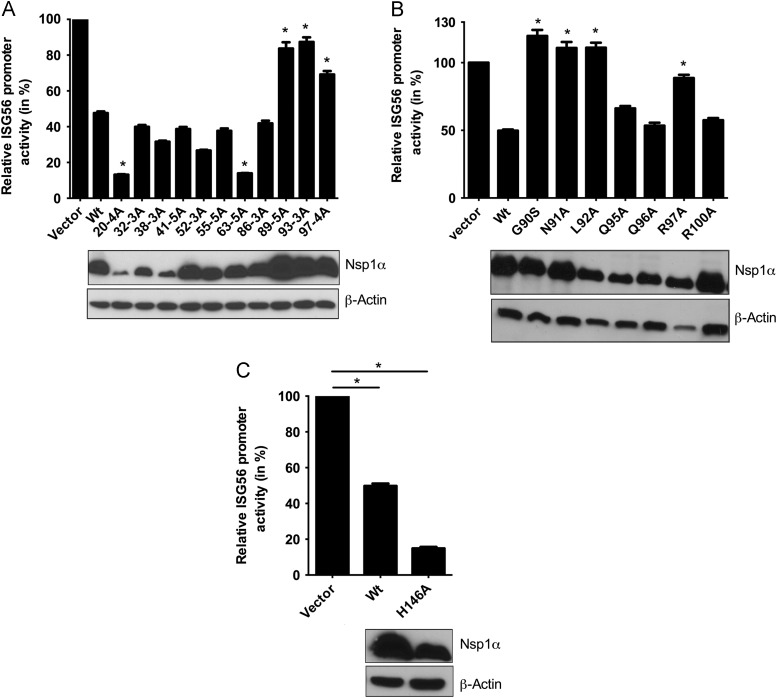
Previously others and we have demonstrated that PRRSV nsp1α is an inhibitor of IFN-β induction (Beura et al., 2010, Chen et al., 2010). The current experiments are intended to map the residues/domains of nsp1α involved in suppressing IFN production. Alanine substitutions (3 aa to 5 aa) were made in both N-terminal zinc finger domain (1–65 aa) and the PCPα domain (66–166 aa) (Sun et al., 2009) of nsp1α. These alanine scanning mutants are designated by the aa residue number of the first aa, followed by the number of residues mutated to alanine e.g., mutant 10-5A has aa residue number 10 to 14 substituted by alanine. It is possible that, these substitutions might affect correct processing of the nsp1α–nsp1β junction by nsp1α protease, which is important for virus viability. To ensure correct processing of this junction, these single amino acid substitutions were made in the background of a full-length nsp1 expression plasmid in which the nsp1β protein has been rendered inactive for ISG56 promoter inhibition. They were first checked for successful cleavage of the nsp1α–nsp1β junction and then subjected to ISG56 reporter assay. The ISG56 reporter vector contains tandem repeats of IFN-stimulated response element (ISRE) promoter elements, which can be activated by IRF3/IRF7. ISG56 reporter assay of these mutants showed that mutants (89-5A, 93-3A, and 97-4A) involving amino acid residues 89 to 100 are relieved of their IFN-suppression property ( Fig. 1A). However residue 96 was not part of these mutants. None of the mutants in the N-terminal zinc finger domain were able to relieve the IFN-suppression. Four single amino acid substitution mutants of nsp1α (G90S, L91A, N92A and R97A) in the 89 aa to 100 aa region showed almost complete relief of the inhibitory effect (Fig. 1B).
Fig. 1.
Mutant nsp1α with reduced ability to antagonize IRF3 mediated gene induction. ISG56 luciferase assay was performed with nsp1α alanine scanning mutant (A), nsp1α single amino acid substitution mutant (B) and nsp1α protease active mutant (C). HEK-293TLR3 cells were co-transfected with indicated nsp1α mutant/wt expression plasmids (1 μg) or empty vector (vector), ISG56-Luciferase plasmid (0.4 μg), and pRLTK (0.01 μg). At 40 h post-transfection, cells were treated with 5 μg/ml dsRNA for 6 h and assayed for luciferase activity. The panel below the bar graph shows the protein expression of respective mutant nsp1α detected using anti-FLAG antibody. Bars represent average (n=3) relative ISG56 promoter activities compared to vector control (converted to 100%). Mutants whose ISG56 promoter activities are significantly different from wt nsp1α are indicated with an asterisk ‘⁎’ (p<0.05). Beta-actin served as loading control.
Since several viral proteases that down regulate IFN production can degrade certain signaling intermediates in the IFN induction pathway (Li et al., 2005a, Li et al., 2005b), it is possible that nsp1α’s IFN suppression activity is mediated through its protease activity. The exact protease active sites of the proteins were determined on the basis of pairwise sequence alignment of the FL12 strain (used in this study) to that of Lelystad strain (Kroese et al., 2008).To examine whether nsp1α proteolytic activity is essential for its IFN-antagonism, we generated mutants containing alanine substitution of nsp1α protease active site residue His146. This mutant did not alter nsp1α's IFN-suppression ability (Fig. 1C). Thus, our mutational studies identified four specific amino acids of PRRSV nsp1α responsible for its IFN-antagonism.
Several amino acid stretches of nsp1β are involved in its IFN-suppression function
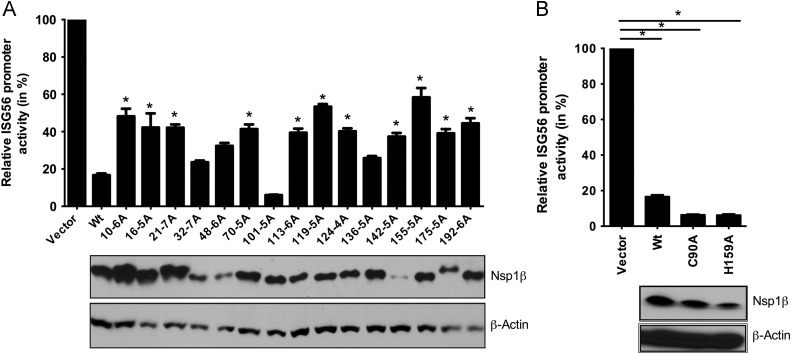
Our subsequent experiments were intended to map the residues/domains of nsp1β involved in suppressing IFN production. Initially, six mutants of nsp1βs were made; each with 30 aa block deletions, and their ability to reduce the IFN-suppression was analyzed by ISG56 reporter assay. To our surprise, all mutants completely relieved the IFN-inhibitory property of wt nsp1β (data not shown). This suggested that the 30 aa block deletions of nsp1β might results in gross deformations in the protein tertiary structure which likely results in nonfunctional protein and thus having no effect on ISG56 promoter. Therefore, we decided to perform alanine-scanning mutagenesis where short amino acid stretches (5–7 aa) of wt nsp1β were replaced by alanine. The crystal structure of PRRSV nsp1β was solved and the protein has been found to possess three distinct domains: the N-terminal nuclease domain (1 aa to 48 aa), the linker domain (49 aa to 84 aa) and the C-terminal cysteine protease domain (85 aa to 181 aa) (Xue et al., 2010). All the three domains were targeted for mutagenesis and most of the residues mutated are well conserved among type II PRRSV strains. The resulting 15 different mutants were then tested for protein expression and their ability to suppress ISG56 promoter activity. There were wide variations in the protein expression level of the different mutant nsp1β relative to wt nsp1β in transient transfection assays, with some mutants exhibiting higher levels of protein accumulation, while others showing lower levels than wt nsp1β ( Fig. 2A bottom). The ISG56 reporter assay results showed variable degree of alleviation in the IFN-suppression effect, with the highest exhibiting 60% relief (Fig. 2A top).
Fig. 2.
Mutant nsp1β with reduced ability to antagonize IRF3 mediated gene induction. ISG56 luciferase assay was performed with nsp1β alanine scanning mutant (A) and nsp1β protease active site mutants (B). HEK-293TLR3 cells were co-transfected with indicated nsp1β mutant expression plasmids (0.5 μg) or empty vector (vector), ISG56-luciferase plasmid (0.4 μg), and pRLTK (0.01 μg). At 40 h post-transfection, cells were treated with 5 μg/ml dsRNA for 6 h and assayed for luciferase activity. The panel below the bar graph shows the protein expression of respective mutant nsp1β detected using anti-FLAG antibody. Bars represent average (n=3) relative ISG56 promoter activities compared to vector control (converted to100%). Mutants whose ISG56 promoter activities are significantly different from wt nsp1β are indicated with an asterisk ‘⁎’ (p<0.05). Beta-actin served as loading control.
The cysteine protease activity of nsp1β is essential for its proteolytic processing of the nsp1β–nsp2 junction, which leads to the release of nsp1β from the polyprotein. To examine whether nsp1β proteolytic activity is essential for its IFN-antagonism, we generated mutants of nsp1β with the individual protease active sites mutated to alanine (C90A and H159A). When tested for their IFN-inhibitory property in an ISG56-luc assay, these mutants showed similar or even stronger suppression of the ISG56 promoter as measured by response to dsRNA stimulation (Fig. 2B). We conclude that the protease active sites of nsp1β are not critical for down regulation of IFN induction. Taken together, we identified several mutant nsp1β with mutations located in all the three domains with reduced ability to suppress induction of an IRF3 dependent promoter in response to dsRNA treatment.
Recovery of PRRSV harboring mutant nsp1β
From the alanine-scanning mutagenesis experiments described above, we identified several mutant nsp1α and nsp1β proteins which exhibit reduced ability to suppress IFN induction in response to dsRNA. Our next objective was to recover viruses encoding these mutant proteins. In the present study, we focused on mutants of nsp1β rather than nsp1α, as the IFN promoter suppression exhibited by nsp1α mutants is less potent than nsp1β. Specifically, we concentrated on the N-terminal nuclease domain of PRRSV nsp1β as certain viral nucleases have been shown to prevent induction of type I IFN (Matzener et al., 2009) in a nuclease dependent manner. Of the five nsp1β nuclease domain mutants (10-6A, 16-5A, 21-7A, 32-7A and 48-6A) generated earlier, the first three were chosen for preparing virus since they not only showed higher alleviation in IFN-inhibition but also have comparable level of protein expression to the wild type (wt) nsp1β. We used standard reverse genetics approach to recover these mutant viruses as described earlier (Kwon et al., 2006, Truong et al., 2004). At 48 h post-electroporation of full length infectious RNA, MARC-145 cells were immunostained for viral nucleocapsid protein (indicator of sg mRNA transcription) as well as viral nsp2/3 protein (indicator of viral replication). Of the three different mutants we attempted for rescue, we were only able to recover one virus containing the nsp1β 16-5A. The other two viruses showed no signs of viral replication and sg RNA synthesis as indicated by absence of nsp2/3 and nucleocapsid protein staining by immunofluorescence (data not shown).
In vitro characterization of 16-5A mutant virus
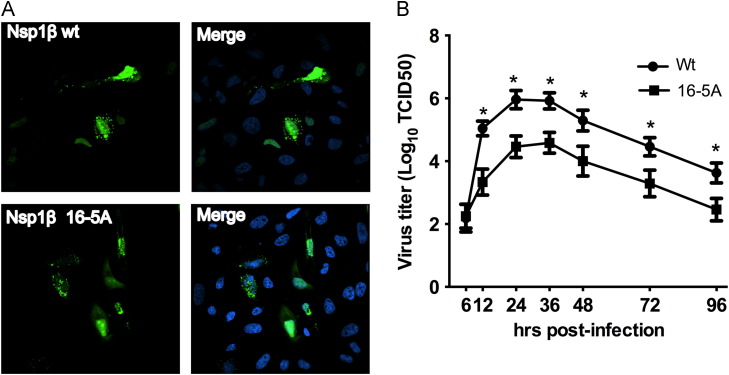
As alteration in protein localization can affect protein function, we checked the localization of nsp1β 16-5A upon transient transfection in HeLa cells ( Fig. 3A). Both wt and mutant nsp1β displayed similar nuclear and cytoplasmic localization as described before (Chen et al., 2010). Next, we compared the in vitro growth characteristics of 16-5A mutant virus with that of the parental wt PRRSV strain (FL12) in monocyte-derived macrophages. Although the multistep growth kinetics of both viruses appeared similar, the mutant virus grew to titers that were on average more than one log lower compared to the wt virus (Fig. 3B). Such reduction in titer was evident at most of the time points examined. Similar growth attenuation was also observed in the permissive monkey kidney cell line MARC-145 (data not shown). Taken together, these studies show that 16-5A mutant virus was growth attenuated in vitro and the mutation did not alter the natural localization of nsp1β.
Fig. 3.
Characterization of 16-5A mutant virusin vitro. (A) Hela cells were transfected with FLAG-tagged wt nsp1β or 16-5A mutant expression plasmid. Nsp1β's (in green) localization was determined 24 h post-transfection by indirect immunofluorescence using anti-FLAG antibody. Position of nucleus is indicated by DAPI (4,6-diamidino-2-phenylindole) (blue) by staining in the merge image (right). (B) Multiple step growth kinetics of 16-5A virus in comparison to parental wt virus. Monocyte-derived macrophages were infected with 0.1 MOI of both viruses and culture supernatant were collected at indicated time post-infection. They were titrated in MARC-145 cells and titers are expressed in Log10TCID50/ml. Error bars indicate SEM values derived from three independent experiments (‘⁎’ indicates p<0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Increased type 1 IFN induction in response to PRRSV 16-5A infection in monocyte-derived macrophages
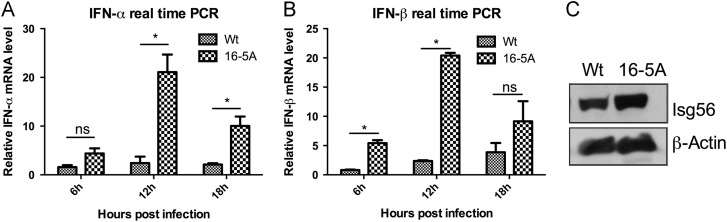
Our next objective was to investigate whether this mutant PRRSV can induce higher levels of type 1 IFNs compared to the wt virus. To this end, monocyte-derived macrophages were mock-infected or infected with 1 MOI of wt PRRSV or 16-5A mutant virus. Total RNA was isolated from the cells and porcine IFN-α and IFN-β mRNA copy numbers were determined by real-time PCR. Both types of IFNs were induced at significantly higher levels in 16-5A virus infected macrophages compared to the wt virus at all the three different time points tested ( Fig. 4A and B). Following IFN synthesis, it up-regulates production of hundreds of ISG and ISG56 is one of them. Therefore, we checked ISG56 protein levels in MARC-145 cells infected with wt or 16-5A mutant virus. A moderate increase in ISG56 level in mutant virus-infected cells compared to wt virus-infected cells (Fig. 4C) was observed. This indicates that the 16-5A mutant virus exhibits reduced ability to suppress IFN induction compared to the wt PRRSV.
Fig. 4.
The 16-5A mutant induces higher level of type I IFN mRNAs. Porcine monocyte- derived macrophages were mock-infected/infected with 1 MOI of 16-5A mutant or wt FL12 virus. Total RNA isolated from cells was reverse transcribed and real time PCR was done for detection of porcine IFN-α (A), IFN-β (B) mRNA. The mRNA copy numbers were calculated after normalization with porcine β-actin copy number and expressed relative to mock control. Bars show average of mRNA copy numbers±SEM from three independent experiments (‘⁎’ indicates p<0.05). (C) MARC-145 cells infected with 16-5A/wt virus and 12 h post-infection were super-infected with sendai virus (SeV) 50 HA (hemagglutinating) Units/ml to induce IFN synthesis for another 12 h. ISG56 protein level was detected by immunoblotting. The β-actin served as loading control.
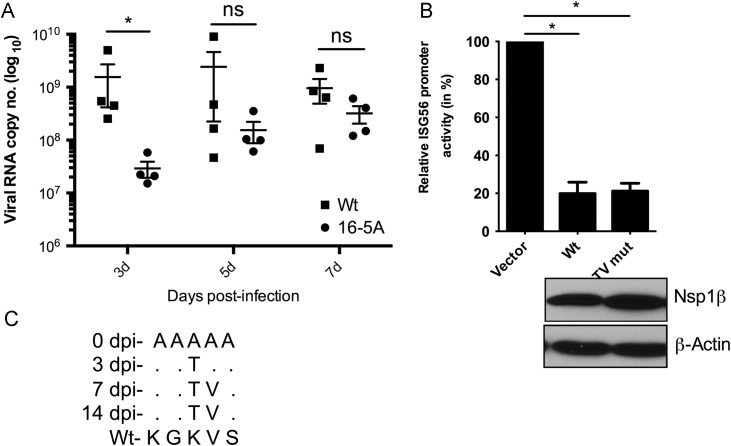
The 16-5A mutant virus exhibits delayed growth characteristics and is unstable in infected pigs
We then pursued in vivo characterization of the 16-5A mutant virus and compared that to the parental virus (wt FL12). Two groups of specific pathogen free young pigs (n=4 each) were infected with either wt or mutant virus. The level of viremia was measured by determining the viral RNA copy numbers in sera at different times post-infection. Animals infected with 16-5A virus showed reduced viremia when compared to wt virus infected animals during early time post-infection (3 dpi) ( Fig. 5A). However, at later times, e.g., 5 dpi and 7 dpi, the difference between the mutant and the wt virus was insignificant. Such rapid recovery in growth of 16-5A mutant virus led us to hypothesize that the mutant virus might have accumulated certain mutations enabling it to attain growth properties similar to the wt PRSSV. To examine this, we sequenced the nsp1β region of virus from serum of 16-5A infected animals at 3 dpi, 7 dpi and 14 dpi. We observed an alanine to threonine substitution at amino acid position 18 of nsp1β at 3 dpi in all 16-5A infected animals (Fig. 5B). These animals also accumulated another mutation at 7 dpi, in which alanine at position 19 reverted to valine, which is the parental amino acid at that position. At 14 dpi, both mutations were still present and no other mutations were observed in nsp1β amino acid sequence. This led us to investigate whether the resulting revertant nsp1β (nsp1β TV mut) could relieve IFN suppression. An ISG56-luciferase based assay with the nsp1β TV mut showed that it has similar IFN-suppression ability as the wt nsp1β and did not offer any relief from suppression unlike 16-5A mutant (Fig. 5C). The results suggest that the rapid growth recovery of the mutant virus was mediated through the mutations in the 18 aa and 19 aa positions of nsp1β 16-5A, which led to the regaining of nsp1β's IFN-suppressive ability.
Fig. 5.
Growth attenuation and instability of 16-5A virusin vivo. (A) The in vivo growth property of 16-5A virus. Wt FL12 or 16-5A mutant viruses were inoculated into growing pigs and serum was collected at indicated days post-infection. After isolation of total RNA from serum, 4 μl of RNA was used in a single step real-time PCR reaction to detect viral RNA copy numbers. The bars represent mean of the viral RNA copy number from 4 different animals in each group. Error bars indicate SEM values. (⁎ indicate p<0.05 and ns is not significant). (B) Mutations in the nsp1β in 16-5A infected pigs. The nsp1β region was amplified by reverse transcription PCR from the serum of 16-5A mutant virus infected pigs at indicated days post-infection (dpi). The amino acid sequences from number 16 to 20 of nsp1β of different dpi were aligned. The 0 dpi indicates the inoculated mutant sequence and dot (.) indicates no change in amino acid. The wt nsp1β sequence is mentioned in bottom for comparison. (C) ISG56 luciferase assay to determine the level of suppression of IRF3 dependent activation by nsp1β wt and nsp1β TVmut. Luciferase assay was performed and the results are presented as mentioned earlier (‘⁎’ indicates p<0.05).
Discussion
Most of the research performed so far in relation to nsp1's anti-IFN activity includes protein overexpression. Such overexpression might lead to atypical expression, unusual localization of protein leading to artifactual functions. In response to such concern, we approached the question of whether nsp1 will act as an IFN-antagonist in the context of virus infection. To this end, the traditional method would call for development of a virus without the IFN antagonistic property followed by checking the relief in IFN suppression. However, since both nsp1α and nsp1β are required for viral sg mRNA synthesis and processing of polyprotein (two crucial steps in viral life cycle), it is impossible to completely remove them from the viral genome (Kroese et al., 2008). Therefore, we performed alanine-scanning mutagenesis to map the residues involved in IFN-suppression. This approach was predicted to have a better chance of recovering mutant viruses that can replicate. We identified several discontinuous stretches of nsp1β encompassing all the three domains as being responsible for the inhibition of IRF3 mediated gene induction. It is possible that different stretches might target distinct steps in the IFN signaling pathway, a circumstance, which could not be distinguished by our end point ISG56 luciferase assay. Interestingly, none of the alanine mutants could provide complete relief in the suppression when expressed individually (Fig. 2A). This suggests that multiple regions of nsp1β are needed for inhibiting IFN production. Since these alanine mutants did not cover the whole nsp1β sequence, the presence of other regions (not included in our assay) with anti-IFN activity cannot be ruled out at this time. Unlike nsp1β, we could more precisely locate (at the single amino acid level) the IFN-antagonism in nsp1α. The three nsp1α mutants G90S, N91A and L92A which are consecutively located exhibited complete relief of inhibition of IRF3 gene induction. All the three amino acids are important for nsp1α homodimer interaction with N91 present on the surface of the protein according the newly described nsp1α crystal structure (Sun et al., 2009). It is therefore possible that nsp1α homodimerization may be a requirement for its IFN inhibitory activity. Both nsp1α and nsp1β are cysteine proteases and their protease activity is essential for correct processing of the viral polyprotein. Mutation of the protease active site residues did not affect their anti-IFN function. This observation is in line with the previous findings that both proteases are only capable of cis-proteolytic activity (cleaving themselves from the viral polyprotein) and once released from the polyprotein, cease at being active as proteases (Sun et al., 2009, Xue et al., 2010). Contrary to this view, Shi et al. (2011) have reported that mutation of the nsp1α protease active site resulted in loss of its anti-IFN property. Such discordance might be explained by the use of the nsp1 full length plasmid in the assay, as mutation of the cys76 (nsp1α protease active residue) in this construct will prevent the processing of mature nsp1α and nsp1β which might be important for IFN inhibition. An alternative explanation would be that the amino acids used for this substitution (serine for cysteine and tyrosine for histidine) might affect the normal protein structure compared to our alanine substitutions. Interestingly, a recent report from our laboratory also implicates some of the same amino acids as found in this study to being involved in interfering with TNFα mRNA synthesis (Subramaniam et al., 2012).
Our efforts to recover viruses with mutations in nsp1β nuclease domain had limited success. We could recover only one mutant virus in nsp1β (16-5A). It is possible that these IFN-antagonist deficient viruses became susceptible to anti-viral action of IFN that is produced by transfection of the in vitro transcript during the virus recovery process. These transcripts possess the 5′ triphosphate group, which is a strong inducer of RIG-I signaling (Schlee et al., 2009). To avoid such endogenous IFN induction, we used BHK-21 cells that have impaired RIG-I signaling (Habjan et al., 2008). However, when we repeated the transfection experiments in BHK-21 cells we could only recover the above-mentioned virus. There was no evidence of genome replication (nsp2/3 immunofluorescence staining) or sg mRNA synthesis (nucleocapsid immunofluorescence staining) in the case of any of the remaining constructs. Besides their role in sg RNA transcription, PRRSV nsp1α and nsp1β are involved in viral polyprotein processing. So to check for the proper protein cleavage, we transferred the nsp1β 10-6A and 21-7A mutations (the non-recoverable viruses) to full-length nsp1. Both the mutants showed reduced processing of nsp1 polyprotein, corroborating that proper nsp1 cleavage is essential for viral replication (data not shown).
Several studies have indicated that IFN antagonist knockout viruses exhibit severe growth defects in vitro and are promptly cleared in vivo by the host IFN response (Richt and Garcia-Sastre, 2009). In this report, we have pursued characterization of one of the nsp1β mutant virus (16-5A). The virus exhibits slow growth kinetics in monocyte-derived macrophages and was able to attain a maximal titer of 104 TCID50/ml. Such growth ability of 16-5A might be due to the residual IFN suppression activity of the virus encoded by four other fully functional IFN antagonistic nsps previously described (Beura et al., 2010). The 16-5A mutant virus showed delayed growth kinetics in vivo but eventually reached titers equivalent to wt PRRSV at 7 dpi. This abnormal in vivo kinetics of growth was due to the reversion of the 16-5A mutation, as the revertant nsp1β obtained from the revertant virus showed equally effective IFN-suppression as wt nsp1β. This gain-of-function mutation most likely contributed to 16-5A virus' ability to attain wt growth. It is possible that a minor species of virus with the mutation(s) may have appeared during our routine passage in cell culture (to prepare stocks for inoculation). However our sequencing control (standard Sanger sequencing) of the inoculant virus did not give any such indication. Nevertheless this indicates a strong selective pressure on these residues during growth inside the host. Interestingly, the first residue to be replaced is alanine at position 18 by a polar residue (threonine) (at 3 dpi). The natural amino acid at this position is lysine, which has been implicated in nsp1β's nuclease activity. Xue et al. (2010) have suggested that lysine18 is responsible for stabilizing the bound nucleic acid in the positively charged pocket formed in the nuclease domain of nsp1β. But presence of threonine at this position in the recovered TV mut virus indicates that the presence of a polar residue might be is essential for a functional nuclease activity, which in turn might be important for efficient in vivo viral replication. Alternatively, the nuclease activity of nsp1β is not required for virus multiplication. Mutation of lysine18 to alanine alone was not sufficient to alleviate nsp1β's IFN-suppression ability (in ISG56 luciferase assay), which suggests that nuclease activity is probably not important for IRF3-dependent IFN modulation (data not shown). It should be noted, however, that a possible role of the nuclease activity in other transcription factor mediated IFN induction cannot be excluded.
PRRSV nsp1's innate immune antagonistic action has attracted considerable attention in recent years resulting in research that has enhanced our understanding of the biological role of this protein. The known mechanisms of IFN-suppression by nsp1α include inhibition of IκB phosphorylation which prevents nuclear translocation of NF-κB (Song et al., 2010). Nsp1β's mode of action includes both inhibition of IRF3 phosphorylation and nuclear translocation, as well as inhibition of STAT1/STAT2 nuclear translocation (Beura et al., 2010, Chen et al., 2010, Patel et al., 2010). In addition, Kim et al. (2010) has shown that expression of nsp1 can target CREB-binding protein (CBP) for proteasomal degradation. However the precise mechanisms regarding how nsp1 achieves these multiple functions need to be identified and will be important for exact mapping of the residues involved in IFN down regulation.
In summary, our results confirm the IFN-inhibitory nature of PRRSV nsp1 in the context of virus infection. Alanine-scanning mutagenesis identified amino acids important for the anti-IFN property of both proteins nsp1α and nsp1β. A mutant virus, which induces significantly higher level of IFN mRNA in vitro quickly, reverted in vivo to acquire the IFN-suppression ability. Future studies should then include a more thorough scanning of the viral proteins to identify residues involved in IFN antagonism. Such mutational studies will likely be especially challenging in case of PRRSV as it would target multiple nsps, most of which are essential for viral replicative cycle. Inclusion of an in vitro replication-transcription assay along with IFN-related assay will help to segregate the IFN-inhibitory domains from replicative domains of the proteins and accelerate the finding of successful candidate mutations. There are ample examples of viruses with rationally designed modifications to remove the anti-IFN property without alteration in their replication potential. Information from such studies can be incorporated to develop effective control strategies against PRRSV (Yoo et al., 2010).
Materials and methods
Cells and viruses
MARC-145 (obtained from Dr. Will Laegreid, USMARC, USDA/ARS) (Kim et al., 1993) and HEK293-TLR3 (Invivogen) cells were maintained in DMEM containing 10% fetal bovine serum and 50 μg/ml of gentamicin (Sigma). Porcine monocyte-derived macrophages were prepared from peripheral blood mononuclear cells as described previously (Loving et al., 2007). Briefly, mononuclear cells were isolated from blood of donor pigs by Ficoll density centrifugation using Lymphocyte separation medium (Mediatech). These cells were allowed to attach to a glass petridish for one day followed by washing with PBS to enrich the monocytes. The attached monocytes were cultured in RPMI 1640 medium supplemented with 10% BVDV-free fetal bovine serum in presence of macrophage colony stimulating factor (5 ng/ml) (Sigma) for 7 day in order to induce their differentiation into macrophages. These monocyte-derived macrophages were then harvested by cell dissociation buffer (Gibco) and seeded with RPMI 1640 and 10% FBS for virus infection.
Infectious clone derived genotype II PRRSV strain FL12 (Truong et al., 2004) was propagated in MARC-145 cells to prepare the virus stocks for this study. The various mutant viruses were also propagated in MARC-145 cells upon recovery. Viruses from passage 3 (P3) were used for most in vitro experiments as well as for animal inoculation. Sendai virus (Cantell strain) was obtained from Charles River Laboratories and was used at 50 haemagglutinating unit/ml concentration for infection of MARC-145 cells.
Plasmids and antibodies
The pIHA-FLAG-nsp1α, pIHA-FLAG-nsp1β, pISG56-luc plasmids have been described earlier (Beura et al., 2010, Subramaniam et al., 2010). The nsp1β block deletion mutants were prepared by overlap extension PCR method. The different alanine substitution mutants have been described (Subramaniam et al., 2012) and were prepared by site-directed mutagenesis by megaprimer PCR method (Sarkar and Sommer, 1990). All the mutant constructs have a N-terminal FLAG tag.
Antibodies used in this study and their sources are as follows—SDOW 17 (anti-PRRSV N monoclonal antibody) (Nelson et al., 1993) was purchased from National Veterinary Services Laboratories (NVSL, Ames, IA). The mouse anti-β-Actin (Sc-47778) was obtained from Santa Cruz Biotech. The anti-Flag M2 (F3165) and rabbit anti-FLAG (F7425) were products of Sigma Aldrich. Secondary antibodies conjugated to HRP were purchased from Kirkegaard and Perry Ltd (KPL) including goat anti-mouse (074-1807) and goat anti-rabbit (214-1516) antibodies. PRRSV nsp1 antibody was developed by immunizing rabbit with the protein and could detect both nsp1α and nsp1β. The anti-ISG56 and anti-nsp2/3 antibodies were generous gifts from Dr. Saumendra Sarkar, U Pittsburg and Dr. Eric J. Snijder, LUMC, The Netherlands, respectively.
Reporter assays
ISG56-luciferase reporter assays were performed as described earlier with minor modifications (Beura et al., 2010). Briefly, HEK293-TLR3 cells were transfected with 0.4 μg of ISG56-luciferase reporter plasmid and 20 ng of pRL-TK (Promega) along with indicated protein expression vectors using TransIT (Mirus Bio) as per manufacturer's protocol. Twenty four hours post-transfection the media was replaced with fresh media. At 40 h post-transfection, cells were either treated with 5 μg/ml of poly (I): poly (C) (GE Health Care) or PBS for 6 h and luciferase assays were performed using Dual Luciferase assay kit from Promega. Then the firefly luciferase activities were expressed as percentages with respect to control after normalizing with renilla luciferase activities (empty vector transfected and dsRNA stimulated cells, were normalized to 100%).
IFN and viral RNA real-time PCR
For IFN real-time PCR, total RNA was isolated from virus infected/mock-infected monocyte-derived macrophages using Trizol (Invitrogen) reagent and was treated with DNase before cDNA synthesis. Then 1 μg of RNA was reverse transcribed using oligo-dT primer and the M-MLV reverse transcription kit (Invitrogen) as per manufacturer's recommendation. Next the cDNA was used for real-time PCR to quantitate the indicated mRNA copy number in a Smartcycler machine (Cepheid) using the Faststart Universal Probe Mastermix (Roche). The cycling conditions used were – initial denaturation −95 °C/10 min, amplification (40 cycles) – 95 °C/30 s and 60 °C/30 s. The swine IFN-α, β and β-actin (internal control) real time PCR was performed using a TaqMan probe based method as described earlier (Beura et al., 2010). IFN-α and IFN-β mRNA levels in virus-infected cells were expressed in copy numbers relative to mock-infected cells as per the method described earlier (Stordeur et al., 2002). The IFN mRNA levels were normalized across the experiments using porcine β-actin as internal control. For measuring viral RNA copy numbers a single step Taqman-based reverse transcription-real time PCR using Hot start-IT Probe one step qRT-PCR master mix (Affymetrix) was designed. Viral RNA was isolated using Qiagen Viral RNA mini kit (Qiagen) from 140 μl of serum from infected pigs following manufacturer's instruction. The isolated RNA (4 μl) was used in the PCR reaction and the RNA copy number was calculated using a standard curve generated with in vitro transcripts. The probe and primers used were—3UTR84P (probe)–TCACCTATTCAATTAGGGCGACCG, 3UTR44F (forward primer)–ATGTGTGGTGAATGGCACTG and 3UTR141R21 (reverse primer)–GCATGGTTCTCGCCAATTAAA. The PCR is targeted to the 3'UTR of type II PRRSV. The cycling conditions for this PCR were—reverse transcription −50 °C/30 min, initial denaturation −95 °C/2 min, amplification 45 cycles (95 °C/15 s and 60 °C/60 s).
Immunoblotting assay
In over-expression experiments, the indicated plasmids were transfected into 293TLR3 cells using Lipofectamine2000 (Invitrogen) or TransIT (Mirus Bio) as per manufacturer's protocol. Then, 36 to 48 h after transfection, the cells were washed twice with PBS and collected for lysate preparation. Lysis was performed in the lysis buffer [20 mM Tris (pH7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate and complete protease inhibitor (Roche)]. After clarification at 13,000 RPM for 10 min at 4 °C, the lysates were resolved in SDS-PAGE followed by transfer onto polyvinylidenedifluoride (PVDF) membrane (Millipore). Then the membranes were blocked in blocking buffer (Tris buffered saline containing 5% nonfat dry milk and 0.1% Tween 20) for 1 h followed by incubation with indicated primary antibodies. After that, membranes were washed and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. Finally the proteins were visualized with an ECL detection system (Pierce).
In vitro transcription, RNA electroporation and mutant virus recovery
Generation of full length PRRSV in vitro transcripts have been described earlier (Kwon et al., 2006, Truong et al., 2004). Briefly the mutant pFL12 full-length plasmids were linearized with AclI restriction enzyme. The digested DNA was phenol: chloroform purified and 1 μg of DNA was used as template in a 20 μl in vitro transcription reaction using mMESSAGEmMACHINE ultra T7 kit (Ambion) as per manufacturer's recommendation. The resulting capped and polyadenylated full length RNA was further purified with phenol: chloroform before being used in the electroporation reaction. MARC-145 cells were electroporated as described previously (Kwon et al., 2006, Truong et al., 2004). After electroporation, cells were seeded in DMEM media containing 10% FBS. Cells were regularly checked for development of cytopathic effect and virus growth was also confirmed by indirect immunofluorescence using anti-N and anti-nsp2/3 antibodies. Upon confirmation of virus growth, supernatants from the plates were collected and clarified. This supernatant was labeled as passage ‘0’ (P0) and was further amplified to prepare stocks for subsequent studies.
Viral growth kinetics
For multiple step growth curve, monocyte-derived macrophages were infected at 0.1 multiplicity of infection (moi) with indicated viruses. Then culture supernatants were collected at indicated time post viral infection and were stored at −80 °C until titration. Titrations of the supernatants were performed in MARC-145 cells and expressed as tissue culture infectious dose 50 per ml (TCID50/ml) as per Spearman and Karber method.
Animal experiments
All animal studies were performed according to the protocols approved by the Institutional Animal Care and Use Committee at the University of Nebraska-Lincoln. Four to six-week old pigs (n=8) were purchased from a specific pathogen-free herd with a certified record of PRRSV absence. Pigs were randomly divided into two groups (n=4) and inoculated with 105.2 TCID50/ml (in total of 2 ml inoculum) of the FL12 (wt PRRSV) or 16-5A virus. The inoculum was divided equally and inoculated through both intramuscular as well as intranasal route. Viremia was measured by real-time PCR in sera collected at 3, 7 and 14 day post-infection (dpi).
Acknowledgments
This research has been supported by grants from USDA NRICGP project No. 2008-00903 (to FAO) and 2009-01654 & 2012-67015-30191 (to AKP). The help of animal care staff in the animal research facility is highly appreciated. We are indebted to Dr. Eric Snijder (LUMC, The Netherlands) for providing necessary reagent. We are also grateful to Dr. Saumendra N. Sarkar (Univ. of Pittsburg) for his suggestions and reagents.
References
- Albina E., Carrat C., Charley B. Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J. Interferon Cytokine Res. 1998;18(7):485–490. doi: 10.1089/jir.1998.18.485. [DOI] [PubMed] [Google Scholar]
- Allende R., Laegreid W.W., Kutish G.F., Galeota J.A., Wills R.W., Osorio F.A. Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J. Virol. 2000;74(22):10834–10837. doi: 10.1128/jvi.74.22.10834-10837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Dinh P.X., Osorio F.A., Pattnaik A.K. Cellular poly(c) binding proteins 1 and 2 interact with porcine reproductive and respiratory syndrome virus nonstructural protein 1beta and support viral replication. J. Virol. 2011;85(24):12939–12949. doi: 10.1128/JVI.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Sarkar S.N., Kwon B., Subramaniam S., Jones C., Pattnaik A.K., Osorio F.A. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 2010;84(3):1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie A.G., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008;8(12):911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142(3):629–633. [PubMed] [Google Scholar]
- Chen Z., Lawson S., Sun Z., Zhou X., Guan X., Christopher-Hennings J., Nelson E.A., Fang Y. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology. 2010;398(1):87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Snijder E.J. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus. Res. 2010;154(1-2):61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M., Penski N., Spiegel M., Weber F. T7 RNA polymerase-dependent and -independent systems for cDNA-based rescue of Rift Valley fever virus. J. Gen. Virol. 2008;89(Pt 9):2157–2166. doi: 10.1099/vir.0.2008/002097-0. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009;21(4):317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S., Kwang J., Yoon I.J., Joo H.S., Frey M.L. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 1993;133(3-4):477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- Kim O., Sun Y., Lai F.W., Song C., Yoo D. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology. 2010;402(2):315–326. doi: 10.1016/j.virol.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimman T.G., Cornelissen L.A., Moormann R.J., Rebel J.M., Stockhofe-Zurwieden N. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine. 2009;27(28):3704–3718. doi: 10.1016/j.vaccine.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Kroese M.V., Zevenhoven-Dobbe J.C., Bos-de Ruijter J.N., Peeters B.P., Meulenberg J.J., Cornelissen L.A., Snijder E.J. The nsp1alpha and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 2008;89(Pt 2):494–499. doi: 10.1099/vir.0.83253-0. [DOI] [PubMed] [Google Scholar]
- Kwon B., Ansari I.H., Osorio F.A., Pattnaik A.K. Infectious clone-derived viruses from virulent and vaccine strains of porcine reproductive and respiratory syndrome virus mimic biological properties of their parental viruses in a pregnant sow model. Vaccine. 2006;24(49-50):7071–7080. doi: 10.1016/j.vaccine.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Lee S.M., Schommer S.K., Kleiboeker S.B. Porcine reproductive and respiratory syndrome virus field isolates differ in in vitro interferon phenotypes. Vet. Immunol. Immunopathol. 2004;102(3):217–231. doi: 10.1016/j.vetimm.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Foy E., Ferreon J.C., Nakamura M., Ferreon A.C., Ikeda M., Ray S.C., Gale M., Jr., Lemon S.M. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Nat. Acad. Sci. U.S.A. 2005;102(8):2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Nat. Acad. Sci. U.S.A. 2005;102(49):17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez O.J., Osorio F.A. Role of neutralizing antibodies in PRRSV protective immunity. Vet. Immunol. Immunopathol. 2004;102(3):155–163. doi: 10.1016/j.vetimm.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Loving C.L., Brockmeier S.L., Sacco R.E. Differential type I interferon activation and susceptibility of dendritic cell populations to porcine arterivirus. Immunology. 2007;120(2):217–229. doi: 10.1111/j.1365-2567.2006.02493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzener P., Magkouras I., Rumenapf T., Peterhans E., Schweizer M. The viral RNase E(rns) prevents IFN type-I triggering by pestiviral single- and double-stranded RNAs. Virus Res. 2009;140(1-2):15–23. doi: 10.1016/j.virusres.2008.10.015. [DOI] [PubMed] [Google Scholar]
- Miller, M. (2011). PRRS Price Tag, $664 million. Porknetwork 〈http://www.porknetwork.com/pork-news/PRRS-price-tag-641-million-127963843.html〉.
- Murtaugh M.P., Xiao Z., Zuckermann F. Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol. 2002;15(4):533–547. doi: 10.1089/088282402320914485. [DOI] [PubMed] [Google Scholar]
- Nan Y., Wang R., Shen M., Faaberg K.S., Samal S.K., Zhang Y.J. Induction of type I interferons by a novel porcine reproductive and respiratory syndrome virus isolate. Virology. 2012;432(2):261–270. doi: 10.1016/j.virol.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson E.A., Christopher-Hennings J., Drew T., Wensvoort G., Collins J.E., Benfield D.A. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 1993;31(12):3184–3189. doi: 10.1128/jcm.31.12.3184-3189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Nan Y., Shen M., Ritthipichai K., Zhu X., Zhang Y.J. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2010;84(21):11045–11055. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richt J.A., Garcia-Sastre A. Attenuated influenza virus vaccines with modified NS1 proteins. Curr. Top Microbiol. Immunol. 2009;333:177–195. doi: 10.1007/978-3-540-92165-3_9. [DOI] [PubMed] [Google Scholar]
- Sarkar G., Sommer S.S. The “megaprimer” method of site-directed mutagenesis. Biotechniques. 1990;8(4):404–407. [PubMed] [Google Scholar]
- Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., Juranek S., Kato H., Kawai T., Poeck H., Fitzgerald K.A., Takeuchi O., Akira S., Tuschl T., Latz E., Ludwig J., Hartmann G. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Zhang G., Wang L., Li X., Zhi Y., Wang F., Fan J., Deng R. The nonstructural protein 1 papain-like cysteine protease was necessary for porcine reproductive and respiratory syndrome virus nonstructural protein 1 to inhibit interferon-beta induction. DNA Cell Biol. 2011 doi: 10.1089/dna.2010.1188. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Meulenberg J.J. The molecular biology of arteriviruses. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Song C., Krell P., Yoo D. Nonstructural protein 1alpha subunit-based inhibition of NF-kappaB activation and suppression of interferon-beta production by porcine reproductive and respiratory syndrome virus. Virology. 2010;407(2):268–280. doi: 10.1016/j.virol.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Stordeur P., Poulin L.F., Craciun L., Zhou L., Schandene L., de Lavareille A., Goriely S., Goldman M. Cytokine mRNA quantification by real-time PCR. J. Immunol. Methods. 2002;259(1-2):55–64. doi: 10.1016/s0022-1759(01)00489-6. [DOI] [PubMed] [Google Scholar]
- Subramaniam S., Beura L.K., Kwon B., Pattnaik A.K., Osorio F.A. Amino acid residues in the non-structural protein 1 of porcine reproductive and respiratory syndrome virus involved in down-regulation of TNF-alpha expression in vitro and attenuation in vivo. Virology. 2012 doi: 10.1016/j.virol.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Subramaniam S., Kwon B., Beura L.K., Kuszynski C.A., Pattnaik A.K., Osorio F.A. Porcine reproductive and respiratory syndrome virus non-structural protein 1 suppresses tumor necrosis factor-alpha promoter activation by inhibiting NF-kappaB and Sp1. Virology. 2010;406(2):270–279. doi: 10.1016/j.virol.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Sun Y., Xue F., Guo Y., Ma M., Hao N., Zhang X.C., Lou Z., Li X., Rao Z. Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha. J. Virol. 2009;83(21):10931–10940. doi: 10.1128/JVI.02579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Chen Z., Lawson S.R., Fang Y. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 2010;84(15):7832–7846. doi: 10.1128/JVI.00217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian K., Yu X., Zhao T., Feng Y., Cao Z., Wang C., Hu Y., Chen X., Hu D., Tian X., Liu D., Zhang S., Deng X., Ding Y., Yang L., Zhang Y., Xiao H., Qiao M., Wang B., Hou L., Wang X., Yang X., Kang L., Sun M., Jin P., Wang S., Kitamura Y., Yan J., Gao G.F. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007;2(6):e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong H.M., Lu Z., Kutish G.F., Galeota J., Osorio F.A., Pattnaik A.K. A highly pathogenic porcine reproductive and respiratory syndrome virus generated from an infectious cDNA clone retains the in vivo virulence and transmissibility properties of the parental virus. Virology. 2004;325(2):308–319. doi: 10.1016/j.virol.2004.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reeth K., Labarque G., Nauwynck H., Pensaert M. Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res. Vet. Sci. 1999;67(1):47–52. doi: 10.1053/rvsc.1998.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Reeth K., Nauwynck H. Proinflammatory cytokines and viral respiratory disease in pigs. Vet. Res. 2000;31(2):187–213. doi: 10.1051/vetres:2000113. [DOI] [PubMed] [Google Scholar]
- Versteeg G.A., Garcia-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr. Opin. Microbiol. 2010;13(4):508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills R.W., Doster A.R., Galeota J.A., Sur J.H., Osorio F.A. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J. Clin. Microbiol. 2003;41(1):58–62. doi: 10.1128/JCM.41.1.58-62.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue F., Sun Y., Yan L., Zhao C., Chen J., Bartlam M., Li X., Lou Z., Rao Z. The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease. J. Virol. 2010;84(13):6461–6471. doi: 10.1128/JVI.00301-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D., Song C., Sun Y., Du Y., Kim O., Liu H.C. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus. Res. 2010;154(1-2):48–60. doi: 10.1016/j.virusres.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]