Abstract
To explore the critical genes associated with infectious bronchitis virus (IBV) virulence, we compared the genome sequences of virulent YN strain and its attenuated strain aYN. Accumulation of mutations in the S gene and the accessory gene 5a were observed, suggesting a potential role in the loss of viral pathogenicity. Two recombinant IBVs (rIBVs) with replacement of the S gene or 5a with corresponding regions from aYN were rescued to verify this speculation. Embryo mortality time/rate showed that rYN-S-aYN and rYN-5a-aYN had an attenuated phenotype in ovo. Replication assay in ovo and in vitro demonstrated the rIBVs had similar replication patterns with wild-type rIBV. Both rIBVs showed reduced mortality, tissue lesions and tissue virus titers in chicken. In conclusion, S gene and 5a accessory gene are responsible for the attenuation of virulent IBV. Insight into the genes responsible for virus attenuation will facilitate the development of future vaccines against IBV.
Keywords: Infections bronchitis virus, IBV, S gene, 5a gene, Virulence
Highlights
-
•
Genetic differences of pathogenic and attenuated IBV strains were compared.
-
•
Recombinants rYN-S-aYN and rYN-5a-aYN were successfully rescued.
-
•
The recombinants showed an attenuated phenotype both in ovo and in vivo.
-
•
S gene and 5a gene are both responsible in the loss of IBV's pathogenicity.
1. Introduction
Avian infectious bronchitis is a highly contagious disease, caused by infectious bronchitis virus (IBV), that affects poultry production worldwide (Cavanagh, 2007). IBV belongs to the Gammacoronavirus genus, subfamily Coronavirinae, family Coronaviridae, order Nidovirales (Cook et al., 2012). The IBV genome is approximately 27.6 kb in size. All coronaviruses share a similar genome organization with gene 1, the replicase gene, located at the 5ʹ end of the genome and the structural and accessory genes clustered at the 3ʹ end, which produce the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. IBV also possesses two accessory genes, gene 3 and gene 5, and the recently characterized inter region (IR), all of which have been shown to be dispensable for virus replication in cell culture (Bentley et al., 2013; Casais et al., 2005; Hodgson et al., 2006).
The main method of protecting chickens from infectious bronchitis is inoculation with attenuated live vaccines. Commercial infectious bronchitis vaccines are usually developed by multiple passages of a virulent field isolate in specific-pathogen-free (SPF) embryonated eggs until the desired non-pathogenicity and immunogenicity have been achieved (Bande et al., 2015). However, the mutations associated with attenuation of the pathogenicity in chickens remain unknown and are variable leading to the differing efficacies associated with different vaccines. Comparison of the genome sequences of virulent and attenuated avian IBV strain Ark DPI viruses revealed that these viruses are similar and differ only by 21 nucleotides, resulting in 17 amino acids changes. Most of these substitutions were located in the replicase 1a and spike genes (Ammayappan et al., 2009). Philips et al. analyzed the consensus full-length genome sequences of three different IBV serotypes (Ark, GA98, and Mass41) and showed that passage in embryonated eggs, to attenuate the viruses in chickens, resulted in amino acid changes, of which 34.75%–43.66% of all of these amino acid changes occurred in the protein encoded by the nsp 3 gene within a virus type, suggesting the potential role of nsp 3 in the attenuation procedure (Phillips et al., 2012). Huo et al. found that the replicase 1a sequence was truncated by 30 bp in the same region of the gene in all of the passaged viruses (Huo et al., 2016). However, due to the lack of a suitable reverse genetic system for virulent IBV, there is no clear confirmation of the specific gene locus that is directly associated with the attenuation of IBV.
In this study, we used an IBV reverse genetics system based on the virulent YN strain of IBV, to generate recombinant IBVs (rIBVs) in which the S gene or open reading frame (ORF) 5a of the YN genome was replaced with the corresponding regions from the attenuated YN strain (aYN). Successfully rescued viruses were characterized and subsequently investigated for pathogenicity and virulence in ovo and in vivo. The aim of this study was to identify viral components critical for efficient replication and pathogenicity of gamma coronaviruses.
2. Materials and methods
2.1. Viruses and cells
The IBV QX-like strain YN originated from a H120 vaccinated broiler flock with respiratory and renal disease (Feng et al., 2012). The virus was purified and passaged by inoculating 10-day-old SPF embryonated chicken eggs via the allantoic sac route. Attenuated strain aYN were previously propagated in 10-day-old SPF embryonated chicken eggs for 160 passages and preserved at ˗80 °C for further use (Zhao et al., 2015). Vaccinia virus vNotI/tk, CV-1, and D980R, and BHK-21 cells were kindly provided by Dr. Volker Thiel (University of Bern, Switzerland). Vaccinia virus vNotI/tk and vaccinia virus recombinants were propagated, titrated, and purified on monkey kidney fibroblasts (CV-1) by standard procedures (Thiel et al., 2001). All cells were maintained in minimum essential medium containing 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin. Chicken embryonic kidney (CEK) cells were prepared from 18-day embryos and were cultured in Dulbecco's modified Eagle's medium (DMEM) (Hyclone, Logan, UT, USA) supplemented with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin (Tannock et al., 1985).
2.2. Animals and ethics statement
All SPF chickens and SPF embryonated eggs (9-day-old) were purchased from Beijing Boehringer Ingelheim Vital Biotechnology Co. Ltd. (Beijing, China). The protocols of this experiment were performed according to the guidelines of the Animal Welfare and Ethical Censor Committee at China Agricultural University (CAU approval number 19036).
2.3. Sequencing and comparison of the complete genomes of strains YN and aYN
The total RNA was extracted from 200 μl allantoic fluid using the RNA prep Pure Tissue Kit (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions. Twenty-two primer pairs were designed based on the complete published sequences of the IBV genome using Primer Premier software version 5.0 to amplify the complete genome excluding the 5′-terminal segments, 5 primers were used for RACE PCR to amplify 5′- terminal sequence (primer sequences available upon request). cDNA synthesis and PCR were performed as described previously (Zhao et al., 2014). 5′- terminal sequence of genome RNA was amplified using 5′-full race kit (TaKaRa, Japan), cDNA synthesis and PCR were performed according to the manual instruction. The amplified PCR products were analyzed by 1% agarose gel electrophoresis. Nucleotide sequencing was conducted by TsingKe Biological Technology Co. (Beijing, China) using Sanger sequencing with an ABI 3730XL automatic sequencing apparatus. The complete genomes of strains YN and aYN were submitted to the GenBank database under accession numbers JF893452 and MK644086, respectively.
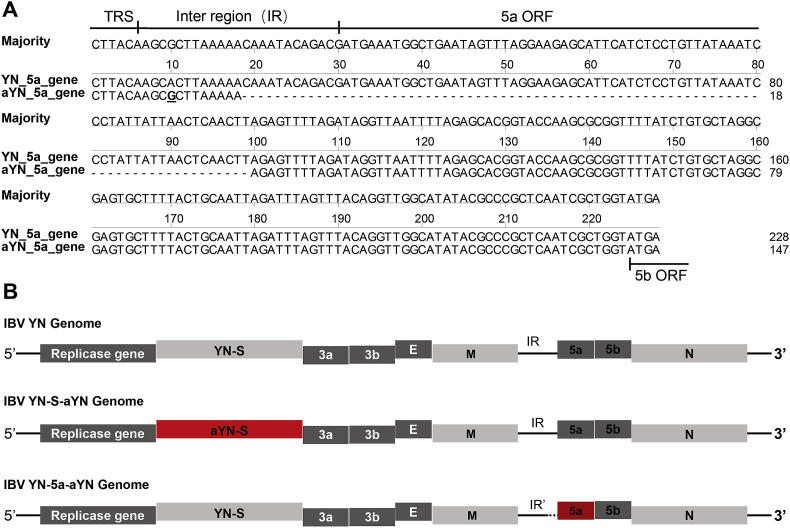
The complete coding sequences of the IBV strains YN and aYN were aligned and analyzed using the ClustalW multiple alignment algorithm in the MegAlign program of the DNASTAR software suite version 3.1 (DNASTAR, Madison, WI, USA). The nucleotide and amino acid (aa) sequence similarities of different gene coding regions were also calculated. Point mutations and deletions were compared between the sequences of strains YN and aYN and are shown in Table 1 and Fig. 1 A.
Table 1.
Differences between the wild-type IBV strain YN and the embryo-attenuated YN strain aYN.
| Locationa | Nucleotide differenceb | Amino acid differencec | Gene region | Location | Nucleotide difference | Amino aciddifference | Gene region |
|---|---|---|---|---|---|---|---|
| 808 | A→C | K→T | 1a | 20659 | A→G | K→R | S |
| 2764 | C→T | P→L | 1a | 20722 | T→C | F→S | S |
| 2892 | G→A | E→K | 1a | 21370 | G→A | R→K | S |
| 3996 | C→T | L→F | 1a | 21543 | C→G | R→G | S |
| 5815 | A→G | N→S | 1a | 22434 | C→T | P→S | S |
| 6466 | C→T | P→L | 1a | 22842 | T→G | Y→D | S |
| 7359 | T→C | W→R | 1a | 23835 | G→T | E→* | S |
| 9553 | A→G | D→G | 1a | 25068 | A→G | T→A | M |
| 14547 | C→T | A→V | 1b | 25140 | G→A | E→K | M |
| 19659 | C→T | S→L | 1b | 25527-25607 | 81 nt deleted | ORF missing | 5a |
| 20455 | T→C | V→A | S | 26519 | G→T | R→L | N |
| 20515 | C→T | T→I | S | 26908 | A→C | T→P | N |
| 20646 | A→G | M→V | S | 26987 | C→T | P→L | N |
According to the IBV wild-type YN strain derived sequence. The entire sequence has been submitted to the GenBank database.
Left nucleotide according to the wild-type YN strain sequence and the right nucleotide from the attenuated YN strain.
Left amino acid according to the wild-type YN strain sequence and the right amino acid from the attenuated YN strain. *indicates the stop codon.
Fig. 1.
Mutations in the 5a ORF of aYN and a schematic overview of the recombinant virus construction. (A) 81 nt deletions in the inter region (IR) between M gene and ORF 5a, and the beginning of ORF 5a. TRS refers to transcription-regulating sequences of the genome. (B) Schematic overview of the recombinant virus construction used in the present study. Original YN sequences are in gray and corresponding regions of aYN are in red. The first two-thirds of the genome is truncated, and the structural and accessory genes are drawn to scale.
2.4. Construction of modified IBV cDNA plasmids and generation of recombinant viruses
All recombinant viruses were generated with a genomic backbone of YN and all nucleotide numbers refer to genome positions in this virus (GenBank number JF893452). Recombinant IBV rYN-S-aYN comprised a direct replacement of the IBV gene S sequence of YN with aYN, beginning at the S gene initiation codon and ending at the S gene. First, two fragments from upstream/downstream of the S gene were amplified with primer pairs: (1) nsp16-NotI-fwd: 5ʹ-CCGCGGTGGCGGCCGCGA TATCTGATATGTATACAG-3ʹ and nsp16-BamHI-rev: 5ʹ-GGAATTGGGGGGATCC CTCTAATCAATAACTTGCC-3ʹ; (2) E-EcoRI-fwd: 5ʹ-GGGCTGCAGGAATTCT GGTTCAAACTCCCGCATCT-3ʹ and E-XhoI-rev: 5ʹ-GGGCCCCCCCTCGAGTTA GCTCCAGGAAGTACTAC-3ʹ. Bold italics show inserted restriction sites EcoRI, XhoI, NotI and BamHI. Fragments were amplified by PCR and cloned into EcoRI/XhoI and NotI/BamHI-digested pGPT-1 step-by-step to generate plasmid pGPT-S-positive. Second, the S gene with mutations was obtained by PCR using aYN strain cDNA as template with the primer pair: nsp16-NotI-fwd and E-XhoI-rev. Fragments were amplified by PCR and cloned into NotI/XhoI-digested pGPT-1 to generate plasmid pGPT-S-negative.
As for the construction of recombinant IBV rYN-5a-aYN, the inter region (IR) between the M and ORF 5a sequences was directly replaced with ORF 5a from the corresponding region of attenuated YN strain. The fragments of IBV genome were modified for the natural deletion of sequence at nucleotides 25527 and 25609, covering the IR sequence and the N-terminal end of ORF 5a. First, two fragments from upstream/downstream of the 5a gene were amplified with primer pairs: (1) 5aR-EcoRI-fwd: 5ʹ-GGGCTGCAGGAATTCATGAATAATAGTAAAGAAAA-3ʹ and 5aR-xhoI-rev: 5ʹ-GGGCCCCCCCTCGAGGTCAGCGGCTGGTCCTGTTC-3ʹ; (2) 5 aL-xbaI-fwd: 5ʹ-GGCGGCCGCTCTAGAGAAACATCTATCGTATGGTG-3ʹ and 5 aL-BamHI-rev: 5ʹ-GAATTGGGGGGATCCCGTCTGTATTTGTTTTTAAG-3ʹ. Bold italics show inserted restriction sites EcoRI, XhoI, XbaI, and BamHI. Fragments were amplified by PCR, and cloned into EcoRI/XhoI and XbaI/BamHI-digested pGPT-1 step-by-step to generate plasmid pGPT-5a-positive. Second, a natural 5a-deleted fragment was obtained by PCR using aYN strain cDNA as template with primer pair: 5 aL-xbaI-fwd and 5aR-xhoI-rev. Fragments were amplified by PCR and cloned into XbaI/XhoI-digested pGPT-1 to generate plasmid pGPT-5a-negative.
Modified regions of the IBV cDNA within plasmids pGPT-S-positive, pGPT-S-negative, pGPT-5a-positive, and pGPT-5a-negative were introduced into the IBV YN full-length cDNA within the vaccinia virus genome by homologous recombination using guanine phosphoribosyltransferase (gpt) selection as described previously (Thiel and Siddell, 2005). In brief, the obtained positive plasmids, pGPT-S-positive and pGPT-5a-positive, were integrated into the vaccinia virus genome by homologous recombination under the positive selection pressure of mycophenolic acid (MPA) in the presence of xanthine and hypoxanthine, to which only recombinant vaccinia viruses with the Ecogpt gene show resistance (Falkner and Moss, 1990). Ecogpt-selected viruses were purified and sequenced to ensure that IBV segment S and 5a has been replaced by the Ecogpt gene in the vaccinia genome respectively. After that, the verified viruses were reconstructed with the obtained negative plasmids, pGPT-S- negative and pGPT-5a-negative, by exerting negative selection pressure on D980R cells due to the presence of 6-thioguanine (Hertzig et al., 2004). IBV full-length RNA transcript was generated in vitro using purified genomic DNA isolated from recombinant vaccinia virus containing the 27.9-kb full-length IBV cDNA, that was digested with NotI and in vitro transcribed by the RiboMAX™ Large Scale RNA Production System--T7 kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. In brief, 10 μg of IBV full-length RNA was used for electroporation of 107 BHK-21 cells. The transfected BHK-21 cells were incubated at 37 °C for 48 h in DMEM supplemented with 10% FBS. After 48 h, the virus-containing medium was collected and inoculated into six 10-day-old SPF embryonated chicken eggs (ECE). After incubation in ECE at 37 °C for 48 h, allantoic fluid from ECE was harvested for further RT-PCR detection and sequencing. The successful rescued viruses were passaged in 10-day-old ECE for ten times. The recombinant viruses after 10 passages in eggs were sequenced to make sure no extra mutation exist.
2.5. In ovo phenotype of rIBVs during passage
The rescued recombinant viruses rYN-S-aYN and rYN-5a-aYN, and wild-type virus rYN were serially passaged 10 times in 10-day-old ECEs by allantoic cavity inoculation. Three eggs were used for each passage per virus. The embryos were candled twice per day to check the death caused by IBV infection. The embryo death time of every egg was recorded in 144 h post inoculation. Embryos that died within 24 h were considered as non-specific deaths and were excluded from the results. Embryos that died after 24 h post inoculation were stored at 4 °C until 144 h to check the embryo curling caused by IBV infection together with the survived embryos. The death time and the curling rate specific for IBV infection was evaluated at each passage to assess the phenotype of the mutants in ECEs.
2.6. In vitro and in ovo growth kinetics of rIBVs
To compare the in vitro replication of the recombinant viruses and wild-type virus rYN on CEK cells, 200 μl of PBS containing 103.0 of the 50% tissue culture infectious dose (TCID50) of rYN-S-aYN, rYN-5a-aYN, or rYN virus were inoculated onto CEK cells in 24-well plates, and the supernatants from three wells from each group were harvested at the time points 6, 12, 24, 36, 48, and 60 h post-inoculation (hpi) for a real-time PCR detection assay for IBV N gene as described before (Zhao et al., 2015). The virus copy numbers for each time point were detected in triplicate and calculated according to a standard curve.
To compare the in ovo replication of the recombinant viruses and wild-type virus rYN in ECEs, 200 μl of PBS containing 102.0 of the 50% egg infectious dose (EID50) of rYN-S-aYN, rYN-5a-aYN, or rYN virus were inoculated into the allantoic cavities of 10-day-old ECEs, and the allantoic fluid of five eggs from each group was harvested at the time points 12, 24, 36, 48, 60, and 72 hpi and pooled for a real-time PCR detection assay, as described above. All of the assays were run in triplicate and calculated according to a standard curve.
2.7. Comparison of the in vivo pathogenicity of rIBVs
One hundred 1-day-old SPF layer chickens were randomly assigned to four groups A, B, C, and D, with each group containing 25 birds being subdivided into 10 and 15 chickens for clinical observation and sampling, respectively. Chickens were inoculated via eye-drop with 0.1 ml containing 105 EID50 of IBV rYN-wt (A), rYN-S-aYN (B), rYN-5a-aYN (C) or no virus as the negative control (D). Chickens were observed daily for 14 days post-challenge (dpc) for clinical signs of ruffled feathers, depression, respiratory signs, diarrhea, and mortality. At 3, 5, 7, 9, and 11 dpc, two birds from each group were euthanized and necropsied. Gross pathology changes were observed and tissue samples from the trachea, kidney, and lung were collected for virus detection by a real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR) assay or histopathological examination. The tracheas of the necropsied chickens were also evaluated for trachea ciliary activity. All surviving chickens were humanely euthanized at the end of the experiment (14 dpc).
2.8. Histopathology test
Tissues collected as described above were fixed in 10% neutral formalin for 24 h, embedded in paraffin, and stained with hematoxylin and eosin (HE) before being observed using standard light microscopy. The lesions were scored according to the severity of the observed lesions in two chickens, three blocks were chosen for each tissue sample per chicken. The indications for the scores were as follows: 0 = no microscopic lesions, 1–3 = mild lesions, 4–6 = moderate lesions, 7–10 = severe and extensive lesions. Mean lesion scores (MLSs) were calculated for each group.
2.9. Inhibition of ciliary activity
For evaluation of tracheal ciliostasis, three sections of the upper, middle, and lower part of the trachea (i.e., nine rings per bird) were analyzed. The rings were placed in a Petri dish containing DMEM with 10% FBS and were then observed under an inverted light microscope at a magnification of 400 × to assess the degree of integrity and preservation of the ciliary movement of the tracheal epithelial cells. For each group, the average ciliostasis score was calculated as previously described (Zhao et al., 2015). A score of 0 was given if the cilia in the complete tracheal section showed movement; a score of 1 was given if the cilia of 75–100% of the tracheal section showed movement; a score of 2 if the cilia of 50–75% of the trachea showed movement; a score of 3 if the cilia of 25–50% of the trachea section showed movement; and a score of 4 if the cilia of less than 25% of the trachea section showed movement or no movement at all. For each group, the average ciliostasis score was calculated.
2.10. IBV detection by real time RT-qPCR
The total RNA of tissues (trachea, kidney, and lung) and allantoic fluid were extracted using the RNA prep Pure Tissue Kit (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions. Reverse transcription was conducted at 37 °C for 1 h, with 3 μg of total RNA, 1 μl of random hexamer primers (500 μg/ml; Promega, Madison, WI, USA) and 0.5 μl of M-MLV reverse transcriptase (200 U/μl; Promega). The cDNA samples were submitted for SYBR Green I real-time RT-qPCR to detect the viral load for IBV 5′UTR gene as described before (Xu et al., 2018). All quantitative PCR reactions were carried out in triplicate and repeated at least twice, and the expression of IBV was calculated according to a standard curve.
2.11. Statistical analysis
Unless otherwise stated, all data are represented as the mean values ± standard deviations obtained from experiments performed at least in triplicate. Viral load qPCR data, mean lesion scores and ciliary activity were statistically evaluated by the one-way or two-way ANOVA test adjusted for post-hoc analysis, followed by Bonferroni's multiple comparison tests. For all tests, the following notations are used to indicate significant differences between groups: *p < 0.05; **p < 0.01; ***p < 0.001. All data were analyzed using GraphPad Prism 6 (GraphPad Software).
3. Results
3.1. Comparison of the complete genome sequences of strains YN and aYN
The complete coding sequences of the YN and aYN strains were aligned and analyzed using the ClustalW multiple alignment algorithm in the MegAlign program of DNASTAR. Comparison of the full-length genome sequences revealed that strain aYN shared 99.57% nucleotide (nt) sequence identity with the parental strain YN: the replicase gene 1 ab (nsp2-16) shared 99.67%–100% nt similarity, the S1 and S2 genes shared 99.45% and 99.84% nt similarity, respectively, and the M and N genes shared 99.41% and 99.76% nt similarity, respectively. The lowest level of similarity appeared in 5a ORF with only 75.25% nt similarity, which was attributable to a 81-nt deletion from nt 25527 to 25609, corresponding to the IR between the M and ORF 5a sequences and the start of the ORF 5a sequence (Fig. 1A). No mutations were found in the 3a, 3b, E, and 5b genes. The nt mutations and deletions are shown in Table 1. Only 10 sense mutations were found in the replicase gene 1 ab, while 10 mutations found in the S gene region mostly occurred within the region from 1 to 400 nt of the S1 gene. Furthermore, a G to T mutation was found at the 3ʹ end of S2 gene, leading to the introduction of a premature termination codon within the S protein, resulting in truncation of the S protein from 1168 to 1159 amino acids. It is noteworthy that the 81 nt deletion in the IR between M and ORF 5a and the beginning of the ORF 5a gene led to the complete lack of 5a accessory protein in strain aYN.
3.2. Design of recombinant IBVs
According to the above results, significant changes in the S and 5a proteins may play a potential role in the loss of viral pathogenicity of strain aYN. To investigate the involvement of these regions in the pathogenicity of IBV, we designed two rIBVs that could be used to assess the replication ability and virulence of IBV. Based on the YN strain of IBV, we designed two rIBVs in which two different IBV genome regions, the S gene and ORF 5a, were replaced with the corresponding regions of attenuated strain aYN (Fig. 1B).
3.3. Virus rescue and the in ovo phenotype of rIBVs during passage
Utilizing the vaccinia recombination based IBV reverse genetics system, we rescued three virus clones, including the original YN strain and recombinant strains of rIBVs rYN-S-aYN and rYN-5a-aYN, in which the S and 5a genes were replaced with the corresponding regions of aYN. Each rescued rIBV was subsequently passaged to P10 in ECEs for further analysis. The successful rescue of infectious rIBVs was confirmed by IBV-induced embryo curling or death within 144 hpi. The time to death was longer for all recombinant rIBVs than for the wild-type YN. Most embryos died within 72 hpi with wild-type rYN, whereas embryos inoculated with rYN-S-aYN or rYN-5a-aYN mostly survived past 96 hpi. Furthermore, the embryo curling rate with all recombinant rIBVs was lower than that with wild-type YN for passages P4–P7 (Table 2 ). These results indicated that rYN-S-aYN and rYN-5a-aYN had an attenuated phenotype in ovo.
Table 2.
Embryonic death time and morality rate of wild-type IBV strain rYN and recombinant strains YN-S-aYN and YN-5a-aYN for each passage.
| Passage | Virus |
|||||
|---|---|---|---|---|---|---|
| rYN |
YN-S-aYN |
YN-5a-aYN |
||||
| Death timea | Curling rate | Death time | Curling rate | Death time | Curling rate | |
| P4 | 96 h × 2b | 100% | 120 h × 1, 144 h × 1 | 100% | 144 h × 2 | 0 |
| P5 | 96 h × 1, 110 h × 2 | 100% | 96 h × 1, 144 h × 1 | 50% | 120 h × 1, 144 h × 1 | 50% |
| P6 | 72 h × 1, 144 h × 2 | 100% | 110 h × 1, 144 h × 2 | 100% | 96 h × 1, 144 h × 2 | 67% |
| P7 | 72 h × 1, 96 h × 1, 144 h × 1 | 100% | 72 h × 1, 110 h × 1, 144 h × 1 | 67% | 96 h × 1, 144 h × 2 | 100% |
| P8 | 144 h × 3 | 100% | 144 h × 2 | 100% | 144 h × 1 | 100% |
| P9 | 72 h × 2 | 100% | 86 h × 1, 96 h × 1 | 100% | 96 h × 1, 144 h × 1 | 100% |
| P10 | 72 h × 2, 110 h × 1 | 100% | 110 h × 1, 120 h × 1, 144 h × 1 | 100% | 144 h × 3 | 100% |
Embryos that died within 24 h were considered as non-specific deaths and were excluded from the results.
*no. means the number of eggs died at the specific timepoint post inoculation within three eggs every passage.
3.4. Growth characteristics of rIBVs following replacement of the S and 5a genes
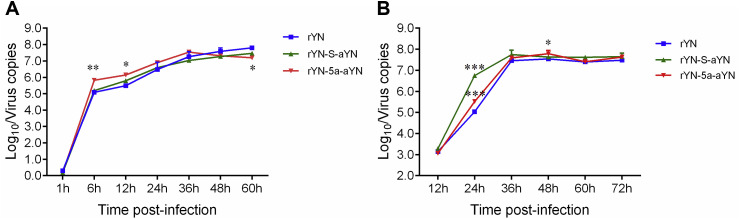
The growth characteristics of the rIBVs, rYN-5a-aYN and rYN-S-aYN, were compared with those of the rescued wild-type virus rYN in vitro and in ovo (Fig. 2 ). Similar replication kinetics were observed for rIBV rYN-S-aYN and rYN in CEK cells. However, peak titers at 6 and 12 hpi for the rIBV rYN-5a-aYN lacking the protein encoded by gene 5a were approximately 1 log10 higher, at 106 virus copies/200 μl, than wild-type rYN strain. Additionally, the replication peak for rYN-5a-aYN was at 36 hpi compared with 60 hpi for the other two rIBVs, suggesting delayed replication or release of rIBV lacking gene 5a. These results suggested that the replacement or deletion of gene 5a had a minor detrimental effect on virus replication in CEK cells. The replication pattern of the two recombinant strains in ECEs was also slightly different to that of the parental IBV strain rYN. At 24 hpi both recombinant strains showed 0.5–1.5 log10 higher copy number than the wild-type rYN strain. While all rIBVs reached a similar peak after 36 hpi, suggesting that replacement of gene 5a did not decrease the replication of recombinant viruses in ECEs.
Fig. 2.
Growth characteristics of IBV-rYNs. (A) Comparison of the replication kinetics of rYNs in CEK cells. The rescued viruses (0.2 ml of 103 TCID50) were inoculated onto CEK cells in 24-well plates, and the supernatants from three wells from each group were harvested at various time-points post-infection for real-time PCR detection of IBV N gene. (B) Comparison of the replication kinetics of rYNs in ovo. The rescued viruses (0.2 ml of 102 EID50) were inoculated into the allantoic cavities of 10-day-old embryonated eggs, and the allantoic fluids of five eggs from each group were harvested at the time points 12, 24, 36, 48, 60, and 72 hpi and pooled for real-time PCR detection of IBV N gene. All of the assays were run in triplicate and calculated according to a standard curve. Data were statistically evaluated by the two-way ANOVA test adjusted for post-hoc analysis, followed by Bonferroni's multiple comparison tests. Statistically significant differences between groups are highlighted; *p < 0.05; **p < 0.01; ***p < 0.001.
3.5. Comparison of the in vivo pathogenicity of rIBVs
Clinical manifestations were observed between 1 and 14 days in chickens challenged with rIBVs. Diseased chicks in the rYN challenged group mainly showed signs of listlessness, huddling, ruffled feathers, increased water intake, and slight watery diarrhea. Of 10 chicks, one bird died at 6 dpc, giving a mortality rate of 10%. No obvious clinical signs or deaths attributable to IBV were observed in the rYN-S-aYN or rYN-5a-aYN challenged groups or the control group. At necropsy, lesions were detected in the respiratory and urinary organs of chickens inoculated with rYN, including punctate hemorrhages and catarrhal exudates in the throat and trachea at 3, 5, and 11 dpc. The lungs of chickens showed congestion and edema at 5 and 9 dpc. The kidneys were swollen with urate deposits in the tubules and ureters at 3, 5, and 9 dpc. The gross lesions in the rYN-S-aYN and rYN-5a-aYN challenged groups were much less pronounced. Only punctate hemorrhages and catarrhal exudates in the throat and trachea were observed during necropsy, while no lesions were observed in the lung and kidney. No gross lesions were observed in any of the birds in the control group (Fig. 3 ).
Fig. 3.
Gross lesions detected in the trachea, lung, and kidney at 9 dpc in chickens experimentally infected with rYN, rYN-S-aYN, rYN-5a-aYN, or the control.
The trachea, lung, and kidney of all groups were also resected for histopathological examination. As shown in Fig. 4 A, the HE staining results were highly consistent with the gross lesion results. In the rYN challenged group, damage to the tracheal epithelium, thickening of the mucosal layer, and cilia-shedding with severe lymphocyte infiltration were observed, whereas only moderate epithelium damage and cilia-shedding were observed in the tracheas of the rYN-S-aYN or rYN-5a-aYN challenged groups. In the rYN challenged group, severe bronchial hemorrhaging was evident in the lungs, with numerous red blood cells in the lumen. Abundant lymphocyte infiltration also existed in the tubulointerstitium region of the kidney. No obvious lesions were observed in the lung or kidney of the rYN-S-aYN or rYN-5a-aYN challenged groups. MLSs also showed that the lesions in rYN-infected group were significantly heavier than those from rYN-S-aYN or rYN-5a-aYN infected groups (Fig. 4B).
Fig. 4.
Histopathologic changes (A) and mean lesion scores (B) in the trachea, lung, and kidney at 9 dpc in chickens experimentally infected with rYN, rYN-S-aYN, rYN-5a-aYN, or the control. Black arrows indicate lesions detected in the tissue. Scale bar = 100 μm. The indications for the scores were as follows: 0 = no microscopic lesions, 1–3 = mild lesions, 4–6 = moderate lesions, 7–10 = severe and extensive lesions (n = 2 × 3). Data were statistically evaluated by one-way ANOVA test adjusted for post-hoc analysis, followed by Bonferroni's multiple comparison tests. Statistical significance was considered as follows: significant at p < 0.05 (*), highly significant at p < 0.01 (**), and very highly significant at p < 0.001 (***).
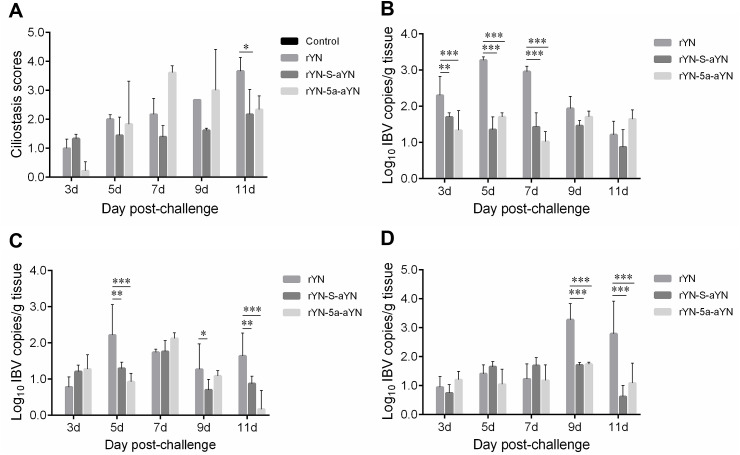
Inhibition of ciliary activity was measured at 3, 5, 7, 9, and 11 dpc in the trachea (Fig. 5 A), and a significant difference was only observed at 11 dpc between the rYN and rYN-S-aYN challenged groups (p < 0.05). No obvious improvement in ciliary activity was observed prior to this between the wild-type and recombinant viruses. Using an SYBR Green I real-time RT-qPCR assay, we detected the viral RNA titers in tissues (trachea, lung, and kidney). The viral load levels in different organs of infected groups are shown in Fig. 5B, C, and D. The viral RNA levels in the tracheas of the rYN-infected group were significantly higher than in both of the recombinant virus-infected groups at 5 and 7 dpc, whereas at 3 dpc, although there was no noticeable difference in the experimental data, the virus copy number in the tracheas of the rYN-infected group was still higher than in the recombinant virus-infected groups (Fig. 5B). In the lungs at 5 dpc, the virus copy number in the rYN-infected group was higher than that in the rYN-S-aYN-infected group and this difference was highly significant (p < 0.01) (Fig. 5C). The number of copies of viral RNA in the kidneys of the rYN-infected group was significantly greater than in the other groups at 9 dpc, and this difference was highly significant (p < 0.001) (Fig. 5D).
Fig. 5.
Trachea ciliostasis scores (A) and viral loads detected in the trachea (B), lung (C), and kidney (D) in chickens experimentally challenged with rYN, rYN-S-aYN, rYN-5a-aYN, and the control. Bars indicate the mean ± SD. Mean ciliostasis scores per group were calculated from ciliostasis scores of 2 euthanized bird at 3, 5, 7, 9, and 11 dpc (9 tracheal rings per individual bird) from each group. Score of 0: the cilia in the complete tracheal section showed movement; score of 1: 75–100% cilia of the tracheal section showed movement; score of 2: 50–75% cilia of the trachea showed movement; score of 3: 25–50% cilia of the trachea section showed movement; score of 4: less than 25% cilia of the trachea section showed movement or no movement at all. For each group, the average ciliostasis score was calculated. Viral load qPCR data comes from 2 euthanized bird at 3, 5, 7, 9, and 11 dpc from each group, includes three technical replicates/animal. Data were statistically evaluated by two-way ANOVA test adjusted for post-hoc analysis, followed by Bonferroni's multiple comparison tests. Statistical significance was considered as follows: significant at p < 0.05 (*), highly significant at p < 0.01 (**), and very highly significant at p < 0.001 (***).
4. Discussion
The avian gamma coronavirus IBV is a highly infectious pathogen of domestic fowl that causes disease in chickens of all ages (Cavanagh, 2005). It replicates primarily in epithelial cells of the respiratory tract causing infectious bronchitis, which is characterized by nasal discharge, snicking, tracheal ciliostasis, and rales in chickens, but is also able to replicate in the epithelial cells of other organs such as the enteric tract, oviducts, and kidneys (Ambali and Jones, 1990; Chen and Itakura, 1996; Maiti et al., 1985). Despite vaccination using both live attenuated and inactivated vaccines, avian IBV is responsible for major economic losses in the poultry industry worldwide as a result of poor weight gain and decreased egg production. The absence of vaccine cross-protection and the frequent emergence of new variant strains complicate the control of IBV (de Wit et al., 2011; Muneer et al., 1987).
Infectious bronchitis is mainly controlled by the use of live attenuated vaccines derived from virulent viruses by multiple serial passages, usually greater than 50 passages, in 10–11-day-old ECEs (Bijlenga et al., 2004; Cavanagh et al., 2007; Gelb and Cloud, 1983). As a consequence of this process, the virus becomes more adapted to the embryo, reflected by more efficient replication and higher pathogenicity for the embryo, with concomitant attenuation in chickens and in some cases loss of immunogenicity. However, the mutations associated with attenuation of pathogenicity in chickens are unknown and variable, leading to the differing efficacies associated with different vaccines. Thus, a new generation of vaccines is needed in which the IBV genome could be precisely modified using reverse genetics.
Reverse genetics systems for IBV, utilizing different construction methods, have been established since 2001, making it possible to explore the use of rIBVs for vaccine development (Britton et al., 2005; Casais et al., 2001; Zhou et al., 2013). Using this system, Casais et al. found that spike protein is a determinant of cell tropism. They constructed a recombinant avian IBV expressing a heterologous spike gene and found that it acquired the same cell tropism phenotype as the donor strain in four different cell types. Hodgson et al. constructed a recombinant IBV strain Beaudette with the S gene of pathogenic M41 strain, and the rIBV remained attenuated but induced protective immunity against strain M41 (Hodgson et al., 2004). Armesto et al. also used the IBV Beaudette strain as a backbone and constructed a recombinant virus containing the replicase gene from strain M41; however, virulence was not restored in this recombinant virus (Armesto et al., 2009; Keep et al., 2018). Shan et al. constructed seven recombinant strains by separately or simultaneously replacing the three hypervariable regions of the S gene of strain Beaudette with the corresponding fragments from a QX-like nephropathogenic isolate. None of these rIBVs showed a significant increase in pathogenicity compared with the Beaudette strain in SPF chickens (Shan et al., 2018). The results presented above were all based on the avirulent Beaudette strain, which may not be an ideal background in which to study the role of IBV-specific mutations in pathogenicity.
In our previous work, we attenuated a virulent QX-type virus, strain YN, in ECEs and the resulting strain, aYN, showed significantly decreased pathogenicity in SPF chickens (Zhao et al., 2015). We compared the complete genome sequences of these two strains and found mutations and deletions distributed across the whole genome. Among these mutations, a mutation cluster region including the introduction of a premature termination codon within the S protein and a serial 81-nt deletion in the 5a accessory gene were most likely to affect the pathogenicity of IBV strain YN. The mutations in the S gene mostly existed in the region from 1 to 400 nt at the start of the S1 gene. This region includes three characterized hypervariable regions possessing high genetic variability that are responsible for the induction of neutralizing and serotype-specific antibodies (Cavanagh et al., 1988; Moore et al., 1997). The role of accessory genes in the attenuation of coronavirus has drawn the attention of researchers in recent years, and the group-specific genes have been proven to have a function in attenuation in natural hosts (Haijema et al., 2004; de Haan et al., 2002; Laconi et al., 2018). To identify the viral components critical for efficient replication and pathogenicity of gamma coronaviruses, in this study, we used an IBV reverse genetics system based on virulent strain YN of IBV to generate rIBVs in which the S gene or ORF 5a of the YN genome were replaced with the corresponding regions from attenuated YN strain aYN. Successfully rescued viruses were characterized and subsequently investigated for pathogenicity and virulence in ovo and in vivo.
Differences in embryo mortality time and curling rate were observed during the passage procedure, with rYN-S-aYN and rYN-5a-aYN showing an attenuated phenotype compared with the wild-type virus rYN. According to the replication kinetic results on CEK cells and in SPF chicken embryos, all of the recombinant mutants showed a higher titer in the first 24 hpi but showed similar growth kinetics after that time, confirming that replacement of the IBV S gene and accessory gene 5a did not affect viral replication in vitro or in ovo. According to previous research, IBV accessory proteins are not essential for viral replication in CEK cells or ECEs (Casais et al., 2005; Hodgson et al., 2006), thus we could conclude that the attenuated phenotype (low embryo mortality and curling rate) of the recombinant virus in ECEs is not due to replication deficiency, but because of the decreased virulence of recombinant virus.
Investigations in 1-day-old SPF chickens provided further evidence of the decrease in virulence of rYN-S-aYN and rYN-5a-aYN. Compared with wild-type rYN, no mortality or clinical signs were observed during the observation period. Gross lesions in the lung and kidney were also negligible, but trachea lesions could still be observed in the rYN-S-aYN and rYN-5a-aYN challenged group. According to histopathological examination, moderate epithelium damage and cilia-shedding were observed in the trachea of the rYN-S-aYN or rYN-5a-aYN challenged groups. The ciliary activity of the rYN-S-aYN or rYN-5a-aYN challenged groups recovered faster than that of the wild-type rYN infected group. Furthermore, the viral loads in the trachea were significantly lower at 5 and 7 dpc. As the primary target organ during IBV infection, the trachea is the most sensitive to IBV infection, followed by the kidney and lung. As demonstrated by previous research, trachea cultures were found to be as sensitive as 9-day-old embryonated eggs in detecting IBV either in pathological material or in serial dilutions (Cook et al., 1976). This may explain the fact that even when the recombinant virus load decreased significantly in the trachea, damage to this organ still occurred and the ciliary activity has not restored in the rYN-S-aYN or rYN-5a-aYN challenged groups. The virus copy numbers and the damage in the lung and kidney of rYN-S-aYN or rYN-5a-aYN challenged groups provided evidence of the reduced pathogenicity of the recombinant viruses.
In conclusion, our results showed that the mutations in S gene and the 5a accessory gene were both accountable for the decreased pathogenicity of virulent IBV, which was not due to a replication deficiency in the recombinant viruses. Furthermore, we demonstrated that the reverse genetics system for IBV based on vaccinia-virus recombination allows for rational modification of virulent IBV strains. This system could be used effectively for studies on gene function and also for the generation of rationally attenuated viruses, as the rescued recombinant viruses proved to be less virulent than the wild-type viruses. Knowledge about the genes responsible for virus attenuation will facilitate the development of future vaccines.
Potential conflicts of interest
The authors have declared no conflict of interest.
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2017YFD0500700). We thank Kate Fox, DPhil, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
References
- Ambali A.G., Jones R.C. Early pathogenesis in chicks of infection with an enterotropic strain of infectious bronchitis virus. Avian Dis. 1990;34:809–817. [PubMed] [Google Scholar]
- Ammayappan A., Upadhyay C., Gelb J.J., Vakharia V.N. Identification of sequence changes responsible for the attenuation of avian infectious bronchitis virus strain Arkansas DPI. Arch. Virol. 2009;154:495–499. doi: 10.1007/s00705-009-0325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armesto M., Cavanagh D., Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bande F., Arshad S.S., Bejo M.H., Moeini H., Omar A.R. Progress and challenges toward the development of vaccines against avian infectious bronchitis. J. Immunol. Res. 2015:1–12. doi: 10.1155/2015/424860. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley K., Keep S.M., Armesto M., Britton P. Identification of a noncanonically transcribed subgenomic mRNA of infectious bronchitis virus and other gammacoronaviruses. J. Virol. 2013;87:2128–2136. doi: 10.1128/JVI.02967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlenga G., Cook J.K., Jr G.J., de Wit J.J. Development and use of the H strain of avian infectious bronchitis virus from The Netherlands as a vaccine: a review. Avian Pathol. 2004;33:550–557. doi: 10.1080/03079450400013154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton P., Evans S., Dove B., Davies M., Casais R., Cavanagh D. Generation of a recombinant avian coronavirus infectious bronchitis virus using transient dominant selection. J. Virol. Methods. 2005;123:203–211. doi: 10.1016/j.jviromet.2004.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Thiel V., Siddell S.G., Cavanagh D., Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 2001;75:12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Davies M., Cavanagh D., Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 2005;79:8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Mockett A.P. Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes. Virus Res. 1988;11:141–150. doi: 10.1016/0168-1702(88)90039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Casais R., Armesto M., Hodgson T., Izadkhasti S., Davies M., Lin F., Tarpey I., Britton P. Manipulation of the infectious bronchitis coronavirus genome for vaccine development and analysis of the accessory proteins. Vaccine. 2007;25:5558–5562. doi: 10.1016/j.vaccine.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.Y., Itakura C. Cytopathology of chick renal epithelial cells experimentally infected with avian infectious bronchitis virus. Avian Pathol. 1996;25:675–690. doi: 10.1080/03079459608419174. [DOI] [PubMed] [Google Scholar]
- Cook J.K., Darbyshire J.H., Peters R.W. The use of chicken tracheal organ cultures for the isolation and assay of avian infectious bronchitis virus. Arch. Virol. 1976;50:109–118. doi: 10.1007/BF01318005. [DOI] [PubMed] [Google Scholar]
- Cook J.K., Jackwood M., Jones R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41:239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- de Haan C.A., Masters P.S., Shen X., Weiss S., Rottier P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J.J., Cook J.K.A., van der Heijden H.M.J.F. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F.G., Moss B. Transient dominant selection of recombinant vaccinia viruses. J. Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J.L., Hu Y.X., Ma Z.J., Yu Q., Zhao J.X., Zhang G.Z. Virulent avian infectious bronchitis virus, People's Republic of China. Emerg. Infect. Dis. 2012;18 doi: 10.3201/eid1812.120552. 1994–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb J.J., Cloud S.S. Effect of serial embryo passage of an Arkansas-type avian infectious bronchitis virus isolate on clinical response, virus recovery, and immunity. Avian Dis. 1983;27:679–687. [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzig T., Scandella E., Schelle B., Ziebuhr J., Siddell S.G., Ludewig B., Thiel V. Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. J. Gen. Virol. 2004;85:1717–1725. doi: 10.1099/vir.0.80044-0. [DOI] [PubMed] [Google Scholar]
- Hodgson T., Casais R., Dove B., Britton P., Cavanagh D. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J. Virol. 2004;78:13804–13811. doi: 10.1128/JVI.78.24.13804-13811.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Britton P., Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006;80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y.F., Huang Q.H., Lu M., Wu J.Q., Lin S.Q., Zhu F., Zhang X.M., Huang Y.Y., Yang S.H., Xu C.T. Attenuation mechanism of virulent infectious bronchitis virus strain with QX genotype by continuous passage in chicken embryos. Vaccine. 2016;34:83–89. doi: 10.1016/j.vaccine.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Keep S., Bickerton E., Armesto M., Britton P. The ADRP domain from a virulent strain of infectious bronchitis virus is not sufficient to confer a pathogenic phenotype to the attenuated Beaudette strain. J. Gen. Virol. 2018;99:1097–1102. doi: 10.1099/jgv.0.001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laconi A., van Beurden S.J., Berends A.J., Krämer-Kühl A., Jansen C.A., Spekreijse D., Chénard G., Philipp H.C., Mundt E., Rottier P.J.M., Hélène Verheije M. Deletion of accessory genes 3a, 3b, 5a or 5b from avian coronavirus infectious bronchitis virus induces an attenuated phenotype both in vitro and in vivo. J. Gen. Virol. 2018;99:1381–1390. doi: 10.1099/jgv.0.001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti N.K., Sharma S.N., Sambyal D.S. Isolation of infectious bronchitis virus from intestine and reproductive organs of laying hens with dropped egg production. Avian Dis. 1985;29:509–513. [PubMed] [Google Scholar]
- Moore K.M., Jackwood M.W., Hilt D.A. Identification of amino acids involved in a serotype and neutralization specific epitope with in the s1 subunit of avian infectious bronchitis virus. Arch. Virol. 1997;142:2249–2256. doi: 10.1007/s007050050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muneer M.A., Newman J.A., Halvorson D.A., Sivanandan V., Coon C.N. Effects of avian infectious bronchitis virus (Arkansas strain) on vaccinated laying chickens. Avian Dis. 1987;31:820–828. [PubMed] [Google Scholar]
- Phillips J.E., Jackwood M.W., Mckinley E.T., Thor S.W., Hilt D.A., Acevedol N.D., Williams S.M., Kissinger J.C., Paterson A.H., Robertson J.S. Changes in nonstructural protein 3 are associated with attenuation in avian coronavirus infectious bronchitis virus. Virus Gene. 2012;44:63–74. doi: 10.1007/s11262-011-0668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D., Fang S., Han Z., Ai H., Zhao W., Chen Y., Jiang L., Liu S. Effects of hypervariable regions in spike protein on pathogenicity, tropism, and serotypes of infectious bronchitis virus. Virus Res. 2018;250:104–113. doi: 10.1016/j.virusres.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock G.A., Bryce D.A., Paul J.A. Evaluation of chicken kidney and chicken embryo kidney cultures for the large-scale growth of attenuated influenza virus master strain A/Ann/Arbor/6/60-ca. Vaccine. 1985;3:333–339. [PubMed] [Google Scholar]
- Thiel V., Siddell S.G. Reverse genetics of coronaviruses using vaccinia virus vectors. Curr. Top. Microbiol. Immunol. 2005;287:199–227. doi: 10.1007/3-540-26765-4_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Xu G., Cheng J.L., Ma S.H., Jia W.F., Yan S.H., Zhang G.Z. Pathogenicity differences between a newly emerged TW-like strain and a prevalent QX-like strain of infectious bronchitis virus. Vet. Microbiol. 2018;227:20–28. doi: 10.1016/j.vetmic.2018.10.019. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Liu X.Y., Cheng J.L., Wu Y.P., Zhang G.Z. Molecular characterization of an infectious bronchitis virus strain isolated from northern China in 2012. Arch. Virol. 2014;159:3457–3461. doi: 10.1007/s00705-014-2213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Cheng J.L., Liu X.Y., Zhao J., Hu Y.X., Zhang G.Z. Safety and efficacy of an attenuated Chinese QX-like infectious bronchitis virus strain as a candidate vaccine. Vet. Microbiol. 2015;180:49–58. doi: 10.1016/j.vetmic.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.S., Zhang Y., Wang H.N., Fan W.Q., Yang X., Zhang A.Y., Zeng F.Y., Zhang Z.K., Cao H.P., Zeng C. Establishment of reverse genetics system for infectious bronchitis virus attenuated vaccine strain H120. Vet. Microbiol. 2013;162:53–61. doi: 10.1016/j.vetmic.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]