Abstract
Priming of the viral glycoprotein (GP) by the cellular proteases cathepsin B and L (CatB, CatL) is believed to be essential for cell entry of filoviruses. However, pseudotyping systems that predominantly produce non-filamentous particles have frequently been used to prove this concept. Here, we report that GP-mediated entry of retroviral-, rhabdoviral and filoviral particles depends on CatB/CatL activity and that this effect is cell line-independent. Moreover, we show that the human cell line Calu-3, which expresses low amounts of CatL, is largely resistant to entry driven by diverse filovirus GPs. Finally, we demonstrate that Calu-3 cell entry mediated by certain filovirus GPs can be rescued upon directed expression of CatL or DC-SIGN. Our results identify Calu-3 cells as largely resistant to filovirus GP-driven entry and demonstrate that entry is limited at the stage of virion attachment and GP priming.
Keywords: Filovirus, Ebola virus, Marburg virus, Glycoprotein, Host cell entry, Cathepsin L, DC-SIGN
1. Introduction
The family Filoviridae comprises three genera, Ebolavirus, Marburgvirus and Cuevavirus. To date, there are five species of ebolaviruses according to the ICTV (International Committee on Taxonomy of Viruses), Zaire ebolavirus (only member: Ebola virus [EBOV]) Sudan ebolavirus (only member: Sudan virus [SUDV]), Bundibugyo ebolavirus (only member: Bundibugyo virus [BDBV]), Taï Forest ebolavirus (only member: Taï Forest virus [TAFV]), Reston ebolavirus (only member: Reston virus, [RESTV]), one species of marburgvirus, Marburg marburgvirus (two members: Marburg virus [MARV] and Ravn virus [RAVV]) and one species of cuevavirus, Lloviu cuevavirus (only member: Lloviu virus [LLOV]) (Kuhn, 2017). There is compelling evidence that all members of the three genera circulate in bats, which are believed to serve as their natural reservoir, and that zoonotic transmissions of most of these viruses to humans can induce severe disease (Mandl et al., 2018; Olival and Hayman, 2014). Multiple filovirus outbreaks in Africa were recorded during the last 52 years and usually entailed less than 500 cases (Centers for Disease Control and Prevention, 2018). In contrast, an outbreak of Ebola virus disease (EVD) in West Africa in 2013 resulted in an EVD epidemic with more than 10.000 deaths and secondary transmission events in Europe and the US (Lo et al., 2017). Further, a recent outbreak of EVD has started in the Democratic Republic of Congo, causing 543 confirmed cases and 309 deaths (WHO, 2018), and is still ongoing (as of January 2019). These examples show that filovirus infection constitutes a serious health threat that is not limited to African countries in which most filoviruses are endemic. Despite promising results of clinical trials, neither vaccines nor antiviral agents approved by regulatory agencies are currently available to combat filovirus infection. Therefore, the development of antiviral strategies remains a research focus and host cell factors required by diverse filoviruses for spread but dispensable for host cell survival are potential drug targets.
The filovirus glycoprotein (GP) mediates viral entry into target cells. It is a highly N- and O-glycosylated type I transmembrane protein and it is the sole viral protein incorporated into the viral envelope (Martin et al., 2016). Host cell entry of filoviruses is best studied for EBOV and we will therefore focus the subsequent discussion on factors important for EBOV GP-driven entry. To initiate infection, EBOV particles first attach to target cells, which can be facilitated by GP interactions with attachment factors like the C-type lectin DC-SIGN (Alvarez et al., 2002; Simmons et al., 2003). In addition or alternatively, attachment can be facilitated in a GP-independent fashion: Phosphatidylserine residues on the viral envelope can be recognized by TIM (T-Cell Immunoglobulin And Mucin Domain-Containing) proteins (Jemielity et al., 2013; Kondratowicz et al., 2011; Moller-Tank et al., 2013) or, via an adaptor, by tyro 3 kinases, including Axl, which are located on the cell surface (Shimojima et al., 2006). After attachment, virions are taken up into cells by macropinocytosis (Quinn et al., 2009; Saeed et al., 2010) and are trafficked into late endosomes, where GP is processed by the host cell cysteine proteases cathepsin B and L (CatB, CatL) (Chandran et al., 2005; Kaletsky et al., 2007; Schornberg et al., 2006), which are known to also process several other viral glycoproteins (Pager et al., 2006; Simmons et al., 2005). Proteolytically processed GP (also termed primed GP) then interacts with the cholesterol transporter Niemann-Pick C1 (NPC1) (Carette et al., 2011; Cote et al., 2011) and, upon low pH exposure and a poorly defined additional stimulus (Bale et al., 2011; Brecher et al., 2012; Schornberg et al., 2006) mediates the fusion of viral and late endosomal membranes, thereby allowing the release of the viral genetic information in the host cell cytoplasm. Priming of GP by CatB and to a lesser degree CatL is essential for host cell entry and results frequently obtained with surrogate systems indicate that entry driven by the GPs of all filoviruses might depend on CatB/CatL activity (Gnirss et al., 2012; Maruyama et al., 2014; Misasi et al., 2012). However, the relative dependence varies, with RESTV showing generally modest CatB/CatL dependence and MARV exhibiting pronounced CatL (Misasi et al., 2012) but not CatB dependence (Gnirss et al., 2012; Misasi et al., 2012). Moreover, EBOV entry into dendritic cells was reported to be CatB but not CatL dependent (Martinez et al., 2010). Notably, a study by Marzi and colleagues conducted with authentic viruses suggested that EBOV and BDBV entry into cultured cells is CatB dependent while entry of several other ebolaviruses is not (Marzi et al., 2012). Moreover, the study showed that lack of CatB or CatL expression is compatible with full viral spread and pathogenesis in the rodent host (Marzi et al., 2012). The discrepancies between these results and the CatB/CatL dependence reported by several other studies are incompletely understood. However, one can speculate that particle shape - filoviruses are filamentous while bullet-shaped or spherical particles are frequently used to model filovirus entry - and target cell type may play a role.
Here, we show that EBOV-GP-driven entry is CatB/CatL dependent irrespective of the surrogate system and target cell type. Moreover, we demonstrate that Calu-3 cells express very low levels of endogenous CatL, in keeping with a previous study (Park et al., 2016), and are largely refractory to entry driven by ebola-, marburg- and cuevavirus GPs. Finally, we show that entry into Calu-3 cells can be rescued by directed expression of DC-SIGN or CatL, suggesting that attachment and GP priming can limit entry into these cells.
2. Results
2.1. EBOV-GP-driven entry into cell lines requires CatB/CatL activity irrespective of the surrogate system and the target cell type
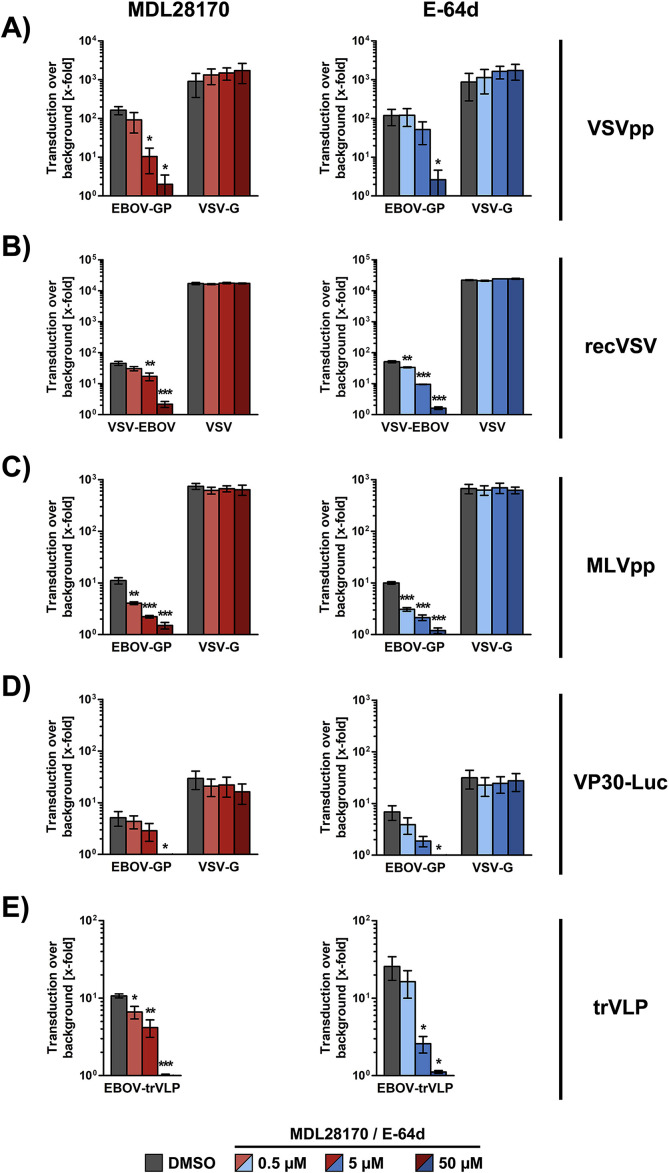
We first examined whether retroviral (mainly spherical), rhabdoviral (mainly bullet-shaped) and filoviral (mainly filamentous) particles differ in their CatB/CatL-dependence for host cell entry. For this, we used retro- and rhabdoviral vectors previously used to analyze EBOV-GP-mediated host cell entry (Takada et al., 1997; Wool-Lewis and Bates, 1998) as well as replication competent vesicular stomatitis virus (VSV) encoding EBOV-GP (Gonzalez-Hernandez et al., 2018). Moreover, we employed two systems that depend exclusively on filovirus proteins. First, we produced EBOV-like particles harboring luciferase by directed expression of all viral proteins from plasmids, including a VP30 version that is fused to firefly luciferase (Dietzel et al., 2017). Second, we employed the EBOV trVLP system in which a EBOV minigenome is packaged into filoviral particles due to expression of the missing open reading frames (EBOV-NP, -VP30, - VP35 and - L) in trans (Schmidt et al., 2018; Watt et al., 2014). While the VP30-Luc system allows measuring a single delivery of VP30-luciferase into cells, the trVLP system allows studying entry and spread of filovirus-like particles. Finally, we employed the endosomal cysteine protease inhibitors E-64d and MDL28170 to block CatB/CatL activity in target cells. These inhibitors were previously shown to interfere with CatB/CatL activity (Brana et al., 1999; Gewies and Grimm, 2003; Hoffmann et al., 2016; Simmons et al., 2005) and were previously used to inhibit EBOV-GP-driven entry (Gnirss et al., 2012; Kaletsky et al., 2007).
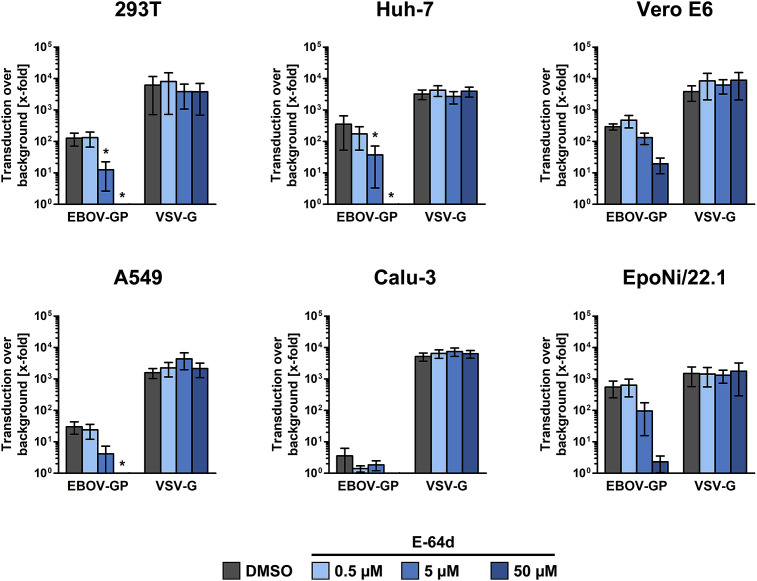
Analysis of GP-driven entry into 293T cells revealed that entry was blocked by E-64d and MDL28170 in a concentration-dependent manner, irrespective of whether GP was presented on retro-, rhabdo- or filovirus particles (Fig. 1 ). In contrast, entry driven by the glycoprotein (G) of vesicular stomatitis virus (VSV) was not affected by E-64d and MDL28170, as expected. Moreover, analysis of entry of EBOV-GP-bearing rhabdoviral particles into six different cell lines of human (accidental host; 293T, Huh-7, A549 and Calu-3), non-human primate (accidental host; Vero E6) and fruit bat (suspected reservoir host; EpoNi/22.1) origin revealed that entry was sensitive to inhibition by E-64d irrespective of the target cell type, although it should be stated that entry into Calu-3 was very close to background (Fig. 2 ). Collectively, our results show that, under the experimental conditions chosen, EBOV-GP-driven entry was CatB/CatL-dependent irrespective of the surrogate system and target cell line. Moreover, our results identify Calu-3 cells as one of the very few cell lines that are largely resistant to GP-mediated entry.
Fig. 1.
EBOV-GP-driven entry into 293T cells requires CatB/CatL activity irrespective of the transduction system. 293T cells were seeded in 96-well plates and pre-incubated with DMSO or the indicated concentrations of E-64d or MDL28170 and then transduced with equal volumes of (A) single-cycle VSV particles (VSVpp) pseudotyped with the indicated glycoproteins or no glycoprotein as negative control; (B) replication-competent VSV or VSV-chimera encoding EBOV-GP instead of VSV-G at a MOI of 0.1; (C) MLV particles pseudotyped with the indicated glycoproteins or no glycoprotein; (D) EBOV-like particles containing VP30-luciferase and bearing EBOV-GP, VSV-G or no glycoprotein; (E) EBOV-like particles generated in the trVLP system. Luciferase activities in cell lysates were determined at 24 h post transduction as indicator of transduction efficiency. In panels A–D, transduction mediated by EBOV-GP and VSV-G is shown relative to transduction mediated by control particles bearing no glycoprotein, which was set as 1. In panel E, transduction by particles produced in the absence of the viral polymerase, L, were set as 1. The average of three independent experiments is shown in each panel. Error bars indicate standard error of the mean (SEM). One-way ANOVA (ANalysis Of VAriance) with Bonferroni post-test analysis was performed to test statistical significance (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001).
Fig. 2.
EBOV-GP-driven entry is CatB/CatL-dependent irrespective of the target cell line. Equal volumes of VSVpp bearing the indicated glycoproteins or bearing no glycoprotein were used for transduction of 293T, Huh-7, A549, Calu-3, Vero E6, and EpoNi/22.1 cells pre-incubated for 3 h with E-64d in the indicated concentrations or DMSO as control. At 24 h post transduction, luciferase activities in cell lysates were quantified as indicator of transduction efficiency. Transduction mediated by EBOV-GP and VSV-G is shown relative to transduction mediated by control particles bearing no glycoprotein, which was set as 1. The average of four independent experiments conducted with separate pseudotype preparations is shown. Error bars indicate SEM. One-way ANOVA with Bonferroni post-test analysis was performed to test statistical significance (*, p ≤ 0.05).
2.2. Calu-3 cells are largely resistant to entry driven by filoviral but not arena-, paramyxo- and rhabdoviral glycoproteins
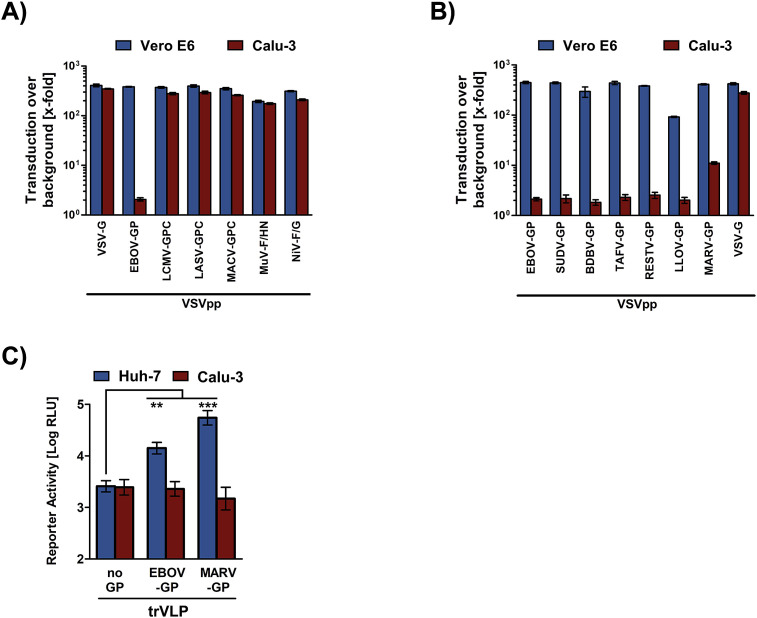
The inefficient EBOV-GP-driven entry into Calu-3 cells raised the question whether this phenotype was specific for EBOV-GP or could also be observed for other viral glycoproteins. In order to answer this question, we investigated whether glycoproteins from viruses of diverse viral families also fail to mediate efficient Calu-3 entry or whether this effect is rather specific for EBOV-GP. We found that glycoproteins of arena- (Lassa virus [LASV], lymphocytic choriomeningitis virus [LCMV] and Machupo virus [MACV]), paramyxo- (Mumps virus [MuV] and Nipah virus [NiV]) and rhabdoviruses (VSV) mediated entry into Calu-3 cells with high efficiency and to roughly the same levels as measured for Vero E6 cells (Fig. 3 A). EBOV-GP-driven entry into Vero E6 cells was highly efficient and comparable to that measured for the other glycoproteins tested, while entry into Calu-3 cells was severely attenuated (∼200-fold compared to Vero E6 cells) (Fig. 3A). In order to investigate whether Calu-3 cells are largely refractory to entry driven by filovirus GPs in general, we performed the same experiment but tested GPs from viruses of all known filovirus species. All GPs tested mediated efficient entry into control Vero E6 cells, with entry driven by LLOV-GP being slightly less efficient than that observed for the other GPs (Fig. 3B). In contrast, Calu-3 cell entry was severely attenuated for all filovirus GPs with MARV-GP-driven entry being most efficient and ranging ∼10-fold over background (Fig. 3B). Finally, use of the trVLP system showed that Calu-3 cells were non-susceptible to EBOV-GP-mediated transduction even if GP was presented in the context of EBOV-like particles, although entry into Huh-7 cells was robust, and comparable results were obtained for MARV-GP (Fig. 3C). These results reveal that Calu-3 cells are poorly susceptible to entry driven by filoviral glycoproteins but are readily susceptible to entry driven by arena-, paramyxo- and rhabdoviral glycoproteins.
Fig. 3.
Calu-3 cells are largely resistant to entry driven by filoviral but not arena-, paramyxo-and rhabdoviral glycoproteins. Vero E6 and Calu-3 cells were transduced with equal volumes of VSV particles pseudotyped with the glycoproteins from diverse viruses (A) or pseudotyped with filovirus glycoproteins (B). Particles bearing no glycoprotein served as negative control. At 24 h post transduction, luciferase activities in cell lysates were quantified as indicator of transduction efficiency. Transduction mediated by the different glycoproteins is shown relative to transduction mediated by control particles bearing no glycoprotein, which was set as 1. The average of three independent experiments is shown. Error bars indicate SEM. (C) Huh-7 and Calu-3 cells were inoculated with trVLPs encoding nano-luciferase for 6 h, washed three times and incubated for 48 h. Thereafter, the luciferase activity (given as relative luminescence units, RLU) in cell lysates was measured. The averages of four (Huh-7) and six (Calu-3) independent experiments are shown, error bars indicate SEM. Two-way ANOVA with Sidak posttest analysis was performed to test statistical significance (**, p ≤ 0.01; ***, p ≤ 0.001).
2.3. Directed expression of DC-SIGN or CatL can rescue Calu-3 cells entry driven by several but not all filovirus GPs
A previous study found that Calu-3 cells express very low amounts of endogenous CatL (Park et al., 2016). Quantitative RT-PCR analysis confirmed the presence of low amounts of CatL transcripts as compared to 293T (∼183-fold higher levels than Calu-3) and Huh-7 cells (∼264-fold higher levels than Calu-3) (Fig. 4 A). In contrast, CatB transcript numbers in Calu-3 cells were comparable or only slightly lower than those in 293T and Huh7 cells (∼3- and 13-fold), respectively (Fig. 4A). Therefore, we next asked whether the low endogenous CatL levels in Calu-3 cells were responsible for inefficient GP-mediated cell entry. In parallel, we examined whether directed expression of other cellular factors known to promote EBOV entry could rescue entry efficiency. To address these questions, we transduced Calu-3 cells with vectors encoding CatB, CatL, Axl, DC-SIGN and NPC1, selected transduced cells via antibiotics and then examined protein expression by immunoblot. These analyses revealed robust expression of CatB, CatL, DC-SIGN, NPC1 and Axl, indicating that transduction and selection were successful (Fig. 4B).
Fig. 4.
Directed expression of CatL and DC-SIGN can rescue filovirus GP-driven entry into Calu-3 cells. (A) CatB, CatL and β-actin mRNA expression in 293T, Huh-7 and Calu-3 cells was quantified by quantitative RT-PCR. CatB and CatL copy numbers were plotted in relation to β-actin copy numbers for each cell line. Shown is a single representative experiment with triplicate samples, error bars indicate standard deviation (SD). The results were confirmed in a second independent experiment. (B) Calu-3 cells engineered to stably express CatB, CatL, DC-SIGN, NPC1, and Axl with a cMYC antigenic tag were subjected to immunoblot analysis using an anti-cMYC antibody. Detection of β-actin expression served as loading control. Similar results were obtained in a separate experiment. (C) Equal volumes of VSVpp bearing the indicated filovirus glycoproteins were used to transduce Calu-3 cells stably expressing the indicated entry and attachment factors. At 24 h post transduction, luciferase activities in cell lysates were quantified as indicator of transduction efficiency. Transduction driven by the different GPs in Calu-3 cells stably expressing the indicated cellular factors is shown relative to transduction of Calu-3 control cells which was set as 1. The average of five independent experiments is shown. Error bars indicate SEM. (D) Calu-3 control cells and Calu-3 CatL-cMYC cells were incubated for 3 h with DMSO or 50 μM of MDL28170 and transduced with equal volumes of VSV pseudotyped with MARV-GP, VSV-G or bearing no glycoprotein. (E) Calu-3 control cells and Calu-3 DC-SIGN-cMYC cells were pre-incubated for 1 h with 20 μg/ml of anti-DC-SIGN monoclonal antibody or an isotype matched control antibody and then transduced with equal volumes of VSV particles bearing the indicated glycoproteins or bearing no glycoprotein. For panels (D) and (E) luciferase activity was quantified at 24 h post transduction. Entry into Calu-3 CatL and Calu-3 DC-SIGN cells is shown relative to entry into Calu-3 control cells, which was set as 1. The average of at least five independent experiments is shown. Error bars indicate SEM. One-way ANOVA with Bonferroni post-test analysis was performed to test statistical significance (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001).
Next, we determined whether the directed expression of the entry factors stated above modulated entry driven by EBOV-GP and other filovirus GPs. All particles examined were readily able to transduce Vero E6 control cells with high efficiency, whereas entry into Calu-3 cells was poor (Fig. 3, Fig. 4C). In comparison, directed expression of CatL in Calu-3 cells increased transduction driven by MARV-GP more than 50-fold (Fig. 4C). In contrast, a roughly 2–6-fold increase was observed for EBOV-GP-, SUDV-GP-, TAFV-GP- and RESTV-GP-driven entry and only a 1.3-fold enhancement was observed for entry driven by BDBV-GP and LLOV-GP. Moreover, directed expression of DC-SIGN in Calu-3 cells augmented entry driven by EBOV-GP, SUDV-GP and MARV-GP by at least 15-fold. For particles bearing RESTV-GP or TAFV-GP a ∼2–4-fold increase of entry efficiency was observed, whereas the increase of entry driven by BDBV-GP and LLOV-GP was less than 2-fold (Fig. 4C). Finally, expression of the other entry factors did not modulate transduction mediated by any of the filovirus GPs and none of the factors had an impact on transduction mediated by VSV-G, as expected (Fig. 4C). These results indicate that filovirus GP-driven entry into Calu-3 cells is limited at the stage of attachment and priming and that the severity of these limitations varies among the GPs of different filovirus species.
2.4. Augmentation of filovirus GP-driven entry into Calu-3 cells via directed expression of CatL or DC-SIGN can be blocked by a cathepsin L inhibitor and a DC-SIGN antibody, respectively
We next sought to provide formal proof that the increased filovirus GP-driven entry into Calu-3 cells expressing DC-SIGN and CatL as compared to Calu-3 control cells was a direct consequence of the expression of these entry factors. For this, we investigated whether entry augmentation could be blocked by the CatL inhibitor MDL28170 and an antibody directed against DC-SIGN. We selected particles bearing MARV-GP for these experiments, since MARV-GP-driven entry into CatL and DC-SIGN expressing cells was markedly more efficient than entry into Calu-3 control cells. Directed CatL expression in Calu-3 cells increased the efficiency of MARV-GP-driven entry as expected and pre-treatment with MDL28170 abolished this effect (Fig. 4D). Similarly, directed expression of DC-SIGN augmented entry, confirming our results discussed above, and augmentation was partially abrogated by pretreatment of target cells with a monoclonal antibody directed against the ectodomain of DC-SIGN (Fig. 4E). In contrast, entry of control particles bearing VSV-G was not sensitive to DC-SIGN antibody or MDL28170. These results indicate that directed expression of CatL or DC-SIGN augments MARV-GP-driven Calu-3 cell entry through direct effects.
3. Discussion
Controversial results regarding the requirement of CatB/CatL activity for filovirus entry have been reported (Chandran et al., 2005; Marzi et al., 2012) and differences in the experimental systems used to study GP-driven entry might account, at least in part, for the discrepant observations. Our study shows that EBOV-GP-driven entry into different cell lines depends on CatB/CatL activity and indicates that entry is CatB/CatL dependent irrespective of the surrogate system used to analyze GP. Moreover, we identify Calu-3 cells as largely resistant to entry driven by EBOV-GP and other filovirus GPs and found that entry can be restored by promoting attachment and/or CatL-dependent GP priming.
CatB/CatL were identified as GP processing enzymes essential for EBOV entry in a study that relied on both surrogate systems and infectious EBOV (Chandran et al., 2005). Moreover, subsequent analyses performed with surrogate models and/or authentic filoviruses confirmed CatB/CatL dependence of EBOV-GP-driven entry but also provided evidence that CatB/CatL dependence of entry driven by other filoviral GPs was variable (Chandran et al., 2005; Gnirss et al., 2012; Martinez et al., 2010; Misasi et al., 2012; Sanchez, 2007). For instance, directed expression of CatB in CatB/CatL double knockout cells increased EBOV-GP- and TAFV-GP-driven entry while directed expression of CatL had only a moderate effect. Moreover, expression of both proteases only moderately increased SUDV-GP-driven entry. In contrast, directed expression of either CatB or CatL markedly increased RESTV-GP-driven entry and expression of CatL but not CatB efficiently promoted MARV-GP-driven entry (Misasi et al., 2012). The general requirement for CatB and/or CatL activity for entry driven by most ebola- and marburgvirus glycoproteins demonstrated by these studies was contrasted by a report by Marzi and colleagues, who showed that Vero cell infection by EBOV and to some degree BDBV depends on CatB but not CatL activity while entry of TAFV, SUDV and RESTV is independent of CatB/CatL activities (Marzi et al., 2012). Finally, this study demonstrated that knockout of CatB or CatL does not interfere with EBOV spread and pathogenesis in mice (Marzi et al., 2012). The reason for the discrepancy between these results is largely unclear and differences in the particles used to study GP-driven entry and in the target cells might be accountable.
The present study shows that the nature of the surrogate system used to study EBOV-GP-driven entry does not have a prominent impact on CatB/CatL dependence. Thus, 293T cell entry of retroviral particles, rhabdoviral particles and filoviral particles was inhibited by E-64d and MDL28170 in a dose-dependent fashion. In this context, it should be stated that particles produced using the trVLP system should adequately mimic virions produced in EBOV infected cells, since particle production in the trVLP system exclusively depends on viral proteins and all proteins encoded by EBOV are expressed in the particle producing cells (Watt et al., 2014), which can release filamentous particles (Wang et al., 2018). These results suggest that particle shape might not be a major determinant of CatB/CatL dependence of EBOV-GP-driven entry. However, particle production in the surrogate systems studied is likely not uniform and it can at present not be excluded that for instance a fraction of particles produced in the trVLP system is spherical or bullet shaped and that only the cell entry of these particles is CatB/CatL-dependent.
Analysis of lung- (A549, Calu-3), liver- (Huh-7) and kidney-derived human cell lines showed that CatB/CatL dependence was independent of the target cell type. Similarly, CatB/CatL dependence was not impacted by the donor species, since EBOV-GP-driven entry into human, non-human primate and fruit bat cell lines was CatB/CatL-dependent, confirming a previous study (Hoffmann et al., 2016). However, these experiments revealed that Calu-3 cell entry driven by EBOV-GP and all other filovirus GPs studied was inefficient. This finding was remarkable for two reasons: First, EBOV exhibits a broad cell tropism and EBOV-GP is known to mediate entry into a diverse panel of cell lines, with only lymphoid cells being refractory for incompletely understood reasons (Dube et al., 2010; Wool-Lewis and Bates, 1998; Yang et al., 1998). Second, Calu-3 cells have previously been shown to express very low levels of endogenous CatL, which were incompatible with CatL-dependent entry of MERS-CoV into these cells (Park et al., 2016). This finding raised the question whether the low endogenous CatL expression might also limit EBOV-GP-driven entry into these cells.
We confirmed low CatL expression in Calu-3 cells and equipped these cells with an expression cassette for CatL in order to unravel whether CatL expression limits EBOV-GP dependent entry. In parallel, we examined whether directed expression of the filovirus entry factors NPC1, Axl and DC-SIGN increased entry efficiency, which would suggest that attachment (DC-SIGN, Axl) or receptor engagement (NPC1) limits entry into Calu-3 cells. Finally, cells were equipped with CatB as control. We found that both directed expression of DC-SIGN and CatL can promote Calu-3 cell entry driven by filoviral GPs. However, the entry promoting effect of these factors was dependent on the viral GP studied. Expression of DC-SIGN, a calcium-dependent lectin that binds to glycans on GP and promotes viral attachment to cells (Alvarez et al., 2002; Simmons et al., 2003), markedly increased entry driven by EBOV-GP, SUDV-GP and MARV-GP. Moreover, DC-SIGN expression moderately augmented entry driven by RESTV-GP and TAFV-GP, suggesting that viral attachment limits Calu-3 cell entry driven by most if not all GPs tested. In comparison, directed expression of CatL markedly increased MARV-GP- and moderately augmented EBOV-GP-, SUDV-GP-, TAFV-GP- and RESTV-GP-driven entry. These findings are largely in keeping with a previous report suggesting a pronounced contribution of CatL to RESTV- and MARV-GP-driven entry (Misasi et al., 2012) and demonstrate that priming can also limit GP-driven entry into Calu-3 cells. Notably, entry driven by LLOV- and BDBV-GP was not appreciably rescued upon CatL or DC-SIGN expression, although previous studies showed that entry driven by these GPs depends on CatL activity (LLOV) and is augmented by DC-SIGN expression (LLOV, BDBV) (Maruyama et al., 2014; Marzi et al., 2006). The reasons for this effect are at present unknown. However, one can speculate that both attachment and priming limit LLOV and BDBV entry into Calu-3 cells, suggesting that coexpression of DC-SIGN and CatL might render the cells susceptible, a possibility that remains to be examined. Finally, it is noteworthy that entry driven by MARV-GP was efficiently and comparably augmented upon directed expression of DC-SIGN or CatL. One explanation could be that attachment and priming are operative at low levels in these cells, in keeping with the low but detectable MARV-GP-driven entry into untreated Calu-3 cells, and that boosting either one of the two processes is sufficient to allow for robust MARV-GP-driven entry. In this context, it is noteworthy that a functional link between lectin-mediated augmentation of viral entry and CatB/L mediated GP priming has previously been suggested (Matsuno et al., 2010). One could thus speculate that binding of MARV-GP to DC-SIGN might reduce CatL-dependence of viral entry, potentially by routing particles into a particular uptake pathway or by stabilizing a GP conformation in which the CatL cleavage site is more exposed.
Collectively, our results show that low levels of endogenous CatL can limit susceptibility of cells to filovirus GP-driven entry and may thus constitute a determinant of filovirus cell tropism.
4. Materials and methods
4.1. Cell culture
Vero E6 (African green monkey kidney cells), 293T (human kidney cells), Huh-7 (human liver cells), A549 (human lung adenocarcinoma cells), Calu-3 (human bronchial adenocarcinoma cells) and EpoNi/22.1 (Buettikofer's epauletted fruit bat kidney cells) were incubated in a humidified atmosphere at 37 °C and 5% CO2. Vero E6, 293T, EpoNi/22.1, and Huh7 cell lines were maintained in Dulbecco's modified Eagle medium (DMEM, PAN-Biotech), A549 cells were grown in DMEM: Nutrient Mixture F-12 + GlutaMax medium (Gibco) and Calu-3 cells were maintained in minimum essential medium (MEM, Gibco). All media were supplemented with 10% fetal bovine serum (Biochrom) and 100 U/ml of penicillin and 0.1 mg/ml of streptomycin (PAN-Biotech). Calu-3 cells stably expressing cMYC-tagged CatB (GenBank: XM_017013097.2), CatL (GenBank: XM_005251716.4), DC-SIGN (GenBank: AB527563.1), NPC1 (GenBank: AB971140.1), and Axl (GenBank: BC032229.1) were generated using retroviral transduction and vector pQCXIP. For further cultivation, cells were maintained in MEM supplemented with 0.5 μg/ml of puromycin, 10% fetal bovine serum, and 1% of penicillin-streptomycin. All cells were detached by either resuspension in fresh culture medium (293T cells) or by the use of trypsin/EDTA (PAN-Biotech) for further subcultivation and seeding.
4.2. Viruses
We employed a previously described recombinant VSV (recVSV) containing a dual reporter consisting of eGFP and firefly luciferase (fLuc) that is located between the open reading frames for VSV-G and VSV-L(Gonzalez-Hernandez et al., 2018). Similarly, a chimeric VSV was used in which the genetic information for VSV-G was replaced by that of EBOV-GP (Gonzalez-Hernandez et al., 2018). All viruses were propagated on Vero E6 cells.
4.3. Plasmids
Expression plasmids encoding for the envelope glycoproteins of EBOV, BDBV, TAFV, RESTV, SUDV, MARV, LLOV, NiV, MuV, LASV, LCMV, MACV and VSV have been described previously (Hoffmann et al., 2016; Wrensch et al., 2014; Krüger et al., 2015). The plasmids required for the trVLP system (Watt et al., 2014) and VP30-luciferase containing particles have also been described before (Hoffmann et al., 2017). For the generation of CatB and CatL encoding retroviral vectors, the respective open reading frames were amplified by PCR from cDNA prepared from A549 cells and inserted into the vector pQCXIP using NotI and BamHI restriction enzymes. The same strategy was applied for generation of NPC1, DC-SIGN and Axl encoding vectors, and cDNA prepared from Huh-7 cells (NPC1) and previously described expressions plasmids (DC-SIGN, Axl (Pohlmann et al., 2001; Watt et al., 2014),) were used as PCR template. The integrity of all PCR amplified sequences was confirmed by automated sequence analysis.
4.4. Production of rhabdoviral pseudotypes
The pseudotypes were generated and used for transduction as described previously (Hoffmann et al., 2016). Briefly, 293T cells were seeded in 6-well plates and transfected using calcium phosphate precipitation with plasmids encoding the viral glycoproteins under study or empty plasmid (pCAGGS) as a negative control. At 18 h post transfection, the cells were inoculated at a multiplicity of infection (MOI) of 3 with a replication-deficient VSV, in which the ORF for VSV-G had been exchanged by two separate ORFs for eGFP and firefly luciferase (fLuc) (Berger Rentsch and Zimmer, 2011) (kindly provided by G. Zimmer). After 1 h incubation at 37 °C, the cells were washed with PBS and incubated for 1 h with a 1:1,000 dilution of I1 (an anti-VSV-G mouse hybridoma supernatant from CRL-2700; American Type Culture Collection) to neutralize residual input virus. Finally, fresh culture medium was added to the cells. At 18–20 h post transduction, supernatants were collected, clarified from cell debris by centrifugation at 4,700×g for 10 min, aliquoted, and stored at −80 °C until use.
4.5. Production of MLV pseudotypes
Transduction vectors based on murine leukemia virus (MLV, MLVpp) were generated according to an established protocol (Wrensch et al., 2014) with slight modifications. First, 293T cells grown in T-25 flasks were transfected with MLV-Gag/Pol (4 μg), MLV-Luc (6 μg) (Wrensch et al., 2014)and expression plasmid for the respective glycoprotein (4 μg). At 16 h post transfection, the cell culture supernatant was replaced by fresh culture medium. 48 h later, supernatants were collected and clarified from debris by centrifugation (4,000×g, 10 min).
4.6. Production of EBOV-like particles
Generation of EBOV-like particles containing VP30-Luc or the tetracistronic transcription and replication competent EBOV minigenome (trVLP) was performed according to previously published protocols (Hoffmann et al., 2017, 2019; Watt et al., 2014):
Briefly, for production of VP30-Luc particles, 293T cells were grown in T-75 cell culture flasks and transfected with expression plasmids for EBOV-NP (1 μg), EBOV-VP35 (1 μg), EBOV-L (8.3 μg), EBOV-VP40 (2 μg), EBOV-GP or VSV-G (2 μg), and EBOV-VP30-Luc (12.5 μg). At 16 h post transfection, the cell culture supernatant was replaced by fresh culture medium. 24 h later, supernatants were collected, clarified from debris by centrifugation (4,000×g, 10 min) and subsequently concentrated employing Vivaspin concentrator columns (molecular mass cutoff = 30,000 kDa, Sartorius). Finally, volumes were equilibrated to 500 μl by addition of DMEM.
For production of EBOV trVLPs, 293T cells were grown in 6-well plates and transfected with expression plasmids for EBOV-NP (125 ng), EBOV-VP35 (125 ng), EBOV-VP30 (75 ng) EBOV-L (1 μg), p4cis-vRNA-RLuc (250 ng) and T7-polymerase (250 ng). At 16 h post transfection, the cell culture supernatant was replaced by fresh culture medium. 48 h later, supernatants were collected and clarified from debris by centrifugation (4,000×g, 10 min). trVLP assays with Calu-3 and Huh-7 target cells were essentially performed as described above, except that the target cells were not pretransfected with support plasmids. Moreover, the reporter gene was exchanged against nano-luciferase, and the EBOV-GP ORF was either exchanged against MARV-GP or the ORF was completely deleted. 200 μl cleared supernatant was used to infect 90%–100% confluent target cells in 96-well format. Infection was allowed to proceed for 6 h, and then cells were washed three times and incubated for another 48 h, after which nano-luciferase activity was measured.
4.7. Transduction/infection of cell lines and its inhibition with CatB and CatL inhibitors
For transduction and infection, target cells were seeded in 96-well plates. At 24 h after seeding, medium was removed and cells were inoculated with equal volumes of transduction vectors (VSVpp, MLVpp, VP30-luc particles or trVLPs) or recVSV (MOI of 0.1). If required, cells were previously treated with inhibitor or antibody (see below). In case of transduction of 293T cells using EBOV-like particles based on the trVLP system, target cells were transfected with expression plasmids for EBOV-NP, -VP30, -VP35, -L and DC-SIGN prior (24 h) to transduction. In order to quantify transduction/infection efficiencies, firefly (fLuc, VSVpp, recVSV, MLVpp, VP30-Luc particles) or Renilla luciferase (rLuc, trVLP particles) activities in cell lysates were measured 24 (VSVpp, recVSV, VP30-Luc) or 48 h (MLVpp, trVLP) post transduction. For this, cell culture medium was aspirated, 50 μL of cell culture lysis reagent (Promega) was added and cells incubated for 30 min at room temperature. Lysates were then transferred to a white, opaque-walled 96-well plate (Thermo Fisher Scientific), luciferin (fLuc; Beetle-Juice, PJK) or coelenterazine (rLuc; Sigma-Aldrich) was added and luciferase activity was measured in a microplate reader (Hidex Sense Microplate Reader) using the PlateReader Software (version 0.5.41.0, Hidex).
4.8. Treatment of target cells with cathepsin inhibitors or anti-DC-SIGN antibody
To investigate entry inhibition by MDL28170 (Tocris), E-64d (Tocris) or DC-SIGN antibody (120,526, NIH-AIDS Research and Reference Reagent Program), the inhibitors or medium was added to target cells and the cells were incubated for 3 h (MDL28170/E-64d) or 1 h (DC-SIGN antibody) before transduction/infection was performed.
4.9. Quantification of CatB/CatL mRNA expression
Total RNA from 293T, Huh-7, and Calu-3 cell lines was extracted using the RNeasy Mini Kit (Qiagen) following the manufacturer instructions. Afterwards, cDNA was produced using the SuperScript III First Strand Synthesis System (Invitrogen) with random hexamers. To determine the copy numbers CatB/CatL mRNA expression, quantitative PCR was performed using the QuantiTect SYBR Green PCR Kit (Qiagen), 1 μl of cDNA as template and the following primers: Cathepsin B 5′-TACAGCCCGACCTACAAACA-3′, 5′-CCATGATGTCCTTCTCGCTA-3’; Cathepsin L 5′-GCAGGTCATGAGTCCTTCCT-3′, 5′-CTTTACGTAGCCACCCATGC-3’; β-actin 5′-GGCTCCCAGCACAATGAAGA-3′, 5′-GGAGCCGCCGATCCA-3’. As standard, serial dilutions of expression plasmids for CatB, CatL, and β-actin were subjected to PCR analysis. Ct values were determined using the Rotor-Gene Q device along with the Rotor-Gene Q software (Qiagen) and used to calculate their respective copy numbers. Further, copy numbers for CatB and CatL were normalized against that of β-actin.
4.10. Immunoblotting
To detect expression of entry and attachment factors in stably transduced Calu-3 cell lines, the cells were lysed using 200 μL 2x sodium dodecyl sulfate (SDS)-containing lysis buffer (50 mM Tris (pH 6.8), 10% glycerol, 2% SDS, 5% β-mercaptoethanol, 0.1% bromophenol blue, 1 mM EDTA), and boiled for 15 min at 95 °C. Subsequently, samples were separated by SDS-PAGE and blotted onto nitrocellulose membranes (GE Healthcare Life Sciences; 0.2 μm). Afterwards, membranes were blocked for 30 min in PBS containing 5% milk powder and 0.1% Tween 20. Finally, expression of cMYC tagged CatL, CatB, DC-SIGN, NPC1 and Axl was determined using undiluted supernatants of a hybridoma cell line that secretes anti-MYC antibody (9E10). Bound antibodies were detected using a horseradish peroxidase linked anti-mouse antibody (Dianova) at a 1:5,000 dilution. Signals were detected using the ChemoCam imaging system along with the ChemoStarProfessional software (Intas).
4.11. Statistical analysis
To test for statistical significance, one-way or two-way analysis of variance (ANOVA) with Bonferroni or Sidak posttest, respectively, was performed using the GraphPad Prism software version 7.03 (GraphPad Software).
Acknowledgements
The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: DC-SIGN antibody 120526 (# 6886). We thank M. A. Müller and C. Drosten for EpoNi/22.1 cells. M. González-Hernández was supported by the Deutscher Akademischer Austauschdienst (DAAD) country-related cooperation program with Mexico (CONACYT, Consejo Nacional de Ciencia y Tecnología) (stipend). Funding for Andreas Müller and Thomas Hoenen was provided in part by the German Federal Ministry of Food and Agriculture (BMEL) based on the decision of the Parliament of the Federal Republic of Germany through the Federal Office for Agriculture and Food (BLE).
References
- Alvarez C.P., Lasala F., Carrillo J., Muniz O., Corbi A.L., Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S., Liu T., Li S., Wang Y., Abelson D., Fusco M., Woods V.L., Jr., Saphire E.O. Ebola virus glycoprotein needs an additional trigger, beyond proteolytic priming for membrane fusion. PLoS Neglected Trop. Dis. 2011;5 doi: 10.1371/journal.pntd.0001395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Rentsch M., Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brana C., Benham C.D., Sundstrom L.E. Calpain activation and inhibition in organotypic rat hippocampal slice cultures deprived of oxygen and glucose. Eur. J. Neurosci. 1999;11:2375–2384. doi: 10.1046/j.1460-9568.1999.00653.x. [DOI] [PubMed] [Google Scholar]
- Brecher M., Schornberg K.L., Delos S.E., Fusco M.L., Saphire E.O., White J.M. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J. Virol. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., Dal Cin P., Dye J.M., Whelan S.P., Chandran K., Brummelkamp T.R. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention U. 2018. 40 Years of Ebola Virus Disease Around the World. [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel E., Schudt G., Krahling V., Matrosovich M., Becker S. Functional characterization of adaptive mutations during the west african ebola virus outbreak. J. Virol. 2017;91 doi: 10.1128/JVI.01913-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube D., Schornberg K.L., Shoemaker C.J., Delos S.E., Stantchev T.S., Clouse K.A., Broder C.C., White J.M. Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16637–16642. doi: 10.1073/pnas.1008509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewies A., Grimm S. Cathepsin-B and cathepsin-L expression levels do not correlate with sensitivity of tumour cells to TNF-alpha-mediated apoptosis. Br. J. Canc. 2003;89:1574–1580. doi: 10.1038/sj.bjc.6601297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirss K., Kuhl A., Karsten C., Glowacka I., Bertram S., Kaup F., Hofmann H., Pöhlmann S. Cathepsins B and L activate Ebola but not Marburg virus glycoproteins for efficient entry into cell lines and macrophages independent of TMPRSS2 expression. Virology. 2012;424:3–10. doi: 10.1016/j.virol.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hernandez M., Hoffmann M., Brinkmann C., Nehls J., Winkler M., Schindler M., Pöhlmann S. A GXXXA motif in the transmembrane domain of the ebola virus glycoprotein is required for tetherin antagonism. J. Virol. 2018;92 doi: 10.1128/JVI.00403-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Crone L., Dietzel E., Paijo J., Gonzalez-Hernandez M., Nehlmeier I., Kalinke U., Becker S., Pöhlmann S. A polymorphism within the internal fusion loop of the ebola virus glycoprotein modulates host cell entry. J. Virol. 2017;91 doi: 10.1128/JVI.00177-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Gonzalez Hernandez M., Berger E., Marzi A., Pöhlmann S. The glycoproteins of all filovirus species use the same host factors for entry into bat and human cells but entry efficiency is species dependent. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Nehlmeier I., Brinkmann C., Krahling V., Behner L., Moldenhauer A.S., Kruger N., Nehls J., Schindler M., Hoenen T., Maisner A., Becker S., Pöhlmann S. Tetherin inhibits Nipah virus but not Ebola virus replication in fruit bat cells. J. Virol. 2019;93(3) doi: 10.1128/JVI.01821-18. pii: e01821-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity S., Wang J.J., Chan Y.K., Ahmed A.A., Li W., Monahan S., Bu X., Farzan M., Freeman G.J., Umetsu D.T., Dekruyff R.H., Choe H. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R.L., Simmons G., Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratowicz A.S., Lennemann N.J., Sinn P.L., Davey R.A., Hunt C.L., Moller-Tank S., Meyerholz D.K., Rennert P., Mullins R.F., Brindley M., Sandersfeld L.M., Quinn K., Weller M., McCray P.B., Jr., Chiorini J., Maury W. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for zaire ebolavirus and lake victoria marburgvirus. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger N., Hoffmann M., Drexler J.F., Müller M.A., Corman V.M., Sauder C., Rubin S., He B., Orvell C., Drosten C., Herrler G. Functional properties and genetic relatedness of the fusion and hemagglutinin-neuraminidase proteins of a mumps virus-like bat virus. J. Virol. 2015;89:4539–4548. doi: 10.1128/JVI.03693-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H. Guide to the correct use of filoviral nomenclature. Curr. Top. Microbiol. Immunol. 2017;411:447–460. doi: 10.1007/82_2017_7. [DOI] [PubMed] [Google Scholar]
- Lo T.Q., Marston B.J., Dahl B.A., De Cock K.M. Ebola: anatomy of an epidemic. Annu. Rev. Med. 2017;68:359–370. doi: 10.1146/annurev-med-052915-015604. [DOI] [PubMed] [Google Scholar]
- Mandl J.N., Schneider C., Schneider D.S., Baker M.L. Going to bat(s) for studies of disease tolerance. Front. Immunol. 2018;9:2112. doi: 10.3389/fimmu.2018.02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B., Hoenen T., Canard B., Decroly E. Filovirus proteins for antiviral drug discovery: a structure/function analysis of surface glycoproteins and virus entry. Antivir. Res. 2016;135:1–14. doi: 10.1016/j.antiviral.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O., Johnson J., Manicassamy B., Rong L., Olinger G.G., Hensley L.E., Basler C.F. Zaire Ebola virus entry into human dendritic cells is insensitive to cathepsin L inhibition. Cell Microbiol. 2010;12:148–157. doi: 10.1111/j.1462-5822.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama J., Miyamoto H., Kajihara M., Ogawa H., Maeda K., Sakoda Y., Yoshida R., Takada A. Characterization of the envelope glycoprotein of a novel filovirus, lloviu virus. J. Virol. 2014;88:99–109. doi: 10.1128/JVI.02265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A., Reinheckel T., Feldmann H. Cathepsin B & L are not required for ebola virus replication. PLoS Neglected Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A., Wegele A., Pöhlmann S. Modulation of virion incorporation of Ebolavirus glycoprotein: effects on attachment, cellular entry and neutralization. Virology. 2006;352:345–356. doi: 10.1016/j.virol.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Matsuno K., Kishida N., Usami K., Igarashi M., Yoshida R., Nakayama E., Shimojima M., Feldmann H., Irimura T., Kawaoka Y., Takada A. Different potential of C-type lectin-mediated entry between Marburg virus strains. J. Virol. 2010;84:5140–5147. doi: 10.1128/JVI.02021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi J., Chandran K., Yang J.Y., Considine B., Filone C.M., Cote M., Sullivan N., Fabozzi G., Hensley L., Cunningham J. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J. Virol. 2012;86:3284–3292. doi: 10.1128/JVI.06346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Tank S., Kondratowicz A.S., Davey R.A., Rennert P.D., Maury W. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J. Virol. 2013;87:8327–8341. doi: 10.1128/JVI.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J., Hayman D.T. Filoviruses in bats: current knowledge and future directions. Viruses. 2014;6:1759–1788. doi: 10.3390/v6041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pager C.T., Craft W.W., Jr., Patch J., Dutch R.E. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006;346:251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Jr., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöhlmann S., Baribaud F., Lee B., Leslie G.J., Sanchez M.D., Hiebenthal-Millow K., Munch J., Kirchhoff F., Doms R.W. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 2001;75:4664–4672. doi: 10.1128/JVI.75.10.4664-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn K., Brindley M.A., Weller M.L., Kaludov N., Kondratowicz A., Hunt C.L., Sinn P.L., McCray P.B., Jr., Stein C.S., Davidson B.L., Flick R., Mandell R., Staplin W., Maury W., Chiorini J.A. Rho GTPases modulate entry of Ebola virus and vesicular stomatitis virus pseudotyped vectors. J. Virol. 2009;83:10176–10186. doi: 10.1128/JVI.00422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M.F., Kolokoltsov A.A., Albrecht T., Davey R.A. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. Analysis of filovirus entry into vero e6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J. Infect. Dis. 2007;196(Suppl. 2):S251–S258. doi: 10.1086/520597. [DOI] [PubMed] [Google Scholar]
- Schmidt M.L., Tews B.A., Groseth A., Hoenen T. Generation and optimization of a green fluorescent protein-expressing transcription and replication-competent virus-like particle system for ebola virus. J. Infect. Dis. 2018;218:S360–S364. doi: 10.1093/infdis/jiy405. [DOI] [PubMed] [Google Scholar]
- Schornberg K., Matsuyama S., Kabsch K., Delos S., Bouton A., White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M., Takada A., Ebihara H., Neumann G., Fujioka K., Irimura T., Jones S., Feldmann H., Kawaoka Y. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J. Virol. 2006;80:10109–10116. doi: 10.1128/JVI.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Grogan C.C., Vandenberghe L.H., Baribaud F., Whitbeck J.C., Burke E., Buchmeier M.J., Soilleux E.J., Riley J.L., Doms R.W., Bates P., Pohlmann S. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology. 2003;305:115–123. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- Takada A., Robison C., Goto H., Sanchez A., Murti K.G., Whitt M.A., Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li J., Fu Y., Zhao Z., Zhang C., Li N., Li J., Cheng H., Jin X., Lu B., Guo Z., Qian J., Liu L. A rapid screen for host-encoded miRNAs with inhibitory effects against ebola virus using a transcription- and replication-competent virus-like particle system. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19051488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt A., Moukambi F., Banadyga L., Groseth A., Callison J., Herwig A., Ebihara H., Feldmann H., Hoenen T. A novel life cycle modeling system for Ebola virus shows a genome length-dependent role of VP24 in virus infectivity. J. Virol. 2014;88:10511–10524. doi: 10.1128/JVI.01272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2018. Ebola Situation Reports: Democratic Republic of the Congo. [Google Scholar]
- Wool-Lewis R.J., Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J. Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrensch F., Winkler M., Pöhlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6:3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Delgado R., Xu L., Todd R.F., Nabel E.G., Sanchez A., Nabel G.J. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science. 1998;279:1034–1037. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]