Abstract
The coronaviruses, porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), and porcine deltacoronavirus (PDCoV) represent important sources of neonatal diarrhea on pig farms. The requirement for aminopeptidase N (APN) as a receptor for TGEV, but not for PEDV, is well established. In this study, the biological relevance of APN as a receptor for PDCoV was tested by using CRISPR/Cas9 to knockout the APN gene, ANPEP, in pigs. Porcine alveolar macrophages (PAMs) from ANPEP knockout (KO) pigs showed resistance to PDCoV infection. However, lung fibroblast-like cells, derived from the ANPEP KO PAM cultures, supported PDCoV infection to high levels. The results suggest that APN is a receptor for PDCoV in PAMs but is not necessary for infection of lung-derived fibroblast cells. The infection of the ANPEP KO pigs with PDCoV further confirmed that APN is dispensable as a receptor for PDCoV.
Keywords: Aminopeptidase N, Porcine deltacoronavirus, APN, CRISPR, ANPEP, CD13
Highlights
-
•
APN is a receptor for PDCoV in PAMS, but not fibroblast cells.
-
•
ANPEP KO pigs are not resistant to PDCoV.
1. Introduction
Coronaviruses belong to the family Coronaviridae, order Nidovirales, and are important pathogens of humans and animals. Coronaviruses are divided into four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. Infection of neonatal pigs with the alphacoronaviruses, transmissible gastroenteritis virus (TGEV) or porcine epidemic diarrhea virus (PEDV), results in mal-absorptive diarrhea, which can lead to dehydration and death (Madson et al., 2016; Saif et al., 2012). The first outbreak of PEDV on U.S. farms in 2013 resulted in the death of nearly 7 million pigs or about 10% of U.S. pig production for that year (Stevenson et al., 2013). In 2014, a diarrhea-causing porcine deltacoronavirus (PDCoV) was isolated from five pig farms in Ohio (Wang et al., 2014). The M and N gene sequences were 99% identical to another PDCoV, HKU15, from China. Since 2014, PDCoV has rapidly spread throughout pig-producing regions in the U.S.
To date, four host proteins, aminopeptidase N (APN), angiotensin-converting enzyme 2 (ACE2), dipeptidyl peptidase 4 (DPP4), and carcinoembryonic antigen-related cell adhesion.
molecule 1 (CEACAM1), have been described to function as coronavirus receptors (Delmas et al., 1992; Li et al., 2003; Raj et al., 2013; Dveksler et al., 1991). Porcine APN (pAPN), a 963 amino acid, type II membrane metallopeptidase, participates in the removal of N-terminal amino acids from protein substrates during digestion. Delmas et al. (1994) were the first to characterize an APN peptide sequence, located between amino acids 717 and 813, as required for infection of cells with TGEV. The corresponding receptor region on TGEV is located on the S1 subunit C-terminal domain (S1-CTD) of the TGEV spike (S) protein (Godet et al., 1994).
Recent studies have focused on the potential role of APN as a receptor for PDCoV. The data have yielded three different observations and conclusions. In 2018, Wang et al. showed that cell lines, such as Vero and BHK-21, were resistant to infection with PDCoV and TGEV. Permissiveness was acquired after transfection of both cell lines with a plasmid expressing a pAPN cDNA. PDCoV and TGEV growth curves on the transfected cells showed a yield greater than 5 log10 TCID50/ml. The authors concluded that pAPN is a functional receptor for PDCoV. Also, in 2018, Zhu et al., reported similar results. However, HeLa cells, which were pAPN-negative, and resistant to TGEV, supported PDCoV infection. Furthermore, a knockout of pAPN expression in the porcine IPI-21 cell line was prepared by using CRISPR to inactivate ANPEP, the gene which codes for APN. The ANPEP KO IPI-21 cells were completely resistant to TGEV, but retained permissiveness for PDCoV. The authors concluded that “pAPN is likely not a critical functional receptor for PDCoV, although it is involved in PDCoV infection”. The third outcome related to pAPN and PDCoV is found in Li et al. (2018), who evaluated the permissiveness of ANPEP knockout (KO) ST cells to infection with TGEV and PDCoV. ST cells lacking pAPN were completely resistant to TGEV but retained a small capacity to support PDCoV infection. The authors concluded that PDCoV utilizes pAPN as a primary receptor for virus attachment, but the presence of a second co-receptor contributes to the permissiveness of cells for infection. Furthermore, the PDCoV co-receptor can retain function independent of pAPN.
In this study, we investigated the role of pAPN as a receptor for PDCoV by evaluating the permissiveness of different cell populations derived from the lungs of ANPEP KO and wild-type (WT) pigs. Porcine alveolar macrophages from ANPEP KO pigs were resistant to PDCoV and TGEV. However, lung fibroblast-like cells, which appeared following the outgrowth of ANPEP KO PAM cultures, were susceptible to PDCoV but remained resistance to TGEV. Furthermore, ANPEP KO pigs supported PDCoV infection. The results support a role for pAPN as a receptor for PDCoV, but the presence of a second, unknown receptor or factor can substitute for pAPN function.
2. Results
2.1. Permissiveness of cells from ANPEP KO and WT pigs for PDCoV infection
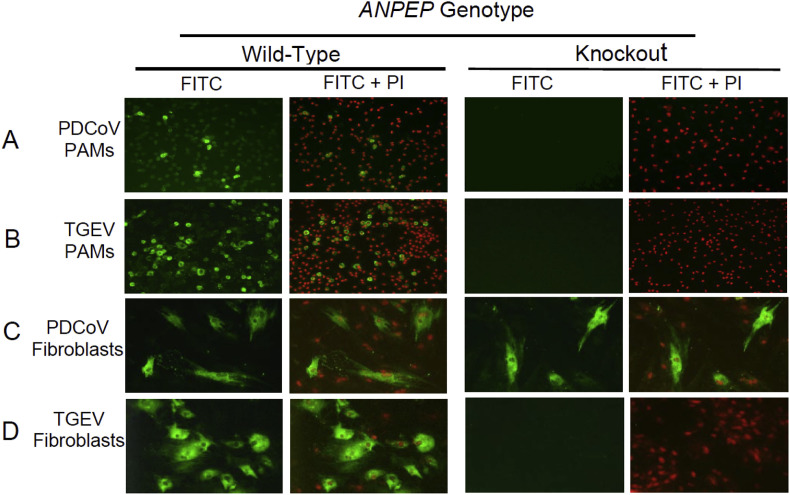
Porcine alveolar macrophages (PAMs) from ANPEP KO and WT pigs were used to evaluate the permissiveness of cells for infection with PDCoV and TGEV. As shown in figure panels 1A and 1B, the ANPEP WT PAMs were permissive for infection with PDCoV and TGEV, while no infected TGEV or PDCoV cells were detected in PAMs from the ANPEP KO pigs. The results showed that PAMs from pigs lacking a functional ANPEP gene are resistant to TGEV and PDCoV infection.
Fig. 1.
Coronavirus infection of porcine alveolar macrophages (PAMs) and lung-derived fibroblast-like cells from ANPEP wild-type (WT) and knockout (KO) pigs. (A, C) PAMs and lung fibroblasts were infected with PDCoV at an MOI of 0.1; the infected cells were fixed and stained with AlexaFluor488-labeled anti-PDCoV antibody and nuclei were counterstained with propidium iodide (PI) at two days after infection. (B, D) PAMs and lung fibroblasts were infected with TGEV at an MOI of 1; the infected cells were fixed and stained with AlexaFluor488-labeled anti-TGEV antibody and nuclei were counterstained with PI at 48 h post-infection. Representative pictures are shown from experiments performed on cells derived from three WT and three KO pigs.
The long-term culture of PAM cultures typically results in the outgrowth of a minor population of lung mesenchymal stem cells (MSCs), which exhibit a fibroblast-like morphology (Khatri et al., 2015). By two weeks, the PAM cultures were completely overgrown with fibroblast-like cells along with the disappearance of macrophages. The fibroblast-like cells from the WT and KO pigs were passaged at least two times and then infected with PDCoV or TGEV. The fibroblast-like cells derived from the ANPEP WT pigs were permissive for both TGEV and PDCoV (Fig. 1 C and D). However, the ANPEP KO fibroblast-like cell cultures showed no evidence of TGEV infection, but showed several PDCoV-infected cells, all possessing a fibroblast-like morphology. These data confirmed the requirement of APN for the permissiveness of the fibroblast cells to TGEV; however, the absence of APN had no effect on infection of fibroblast-like cells with PDCoV.
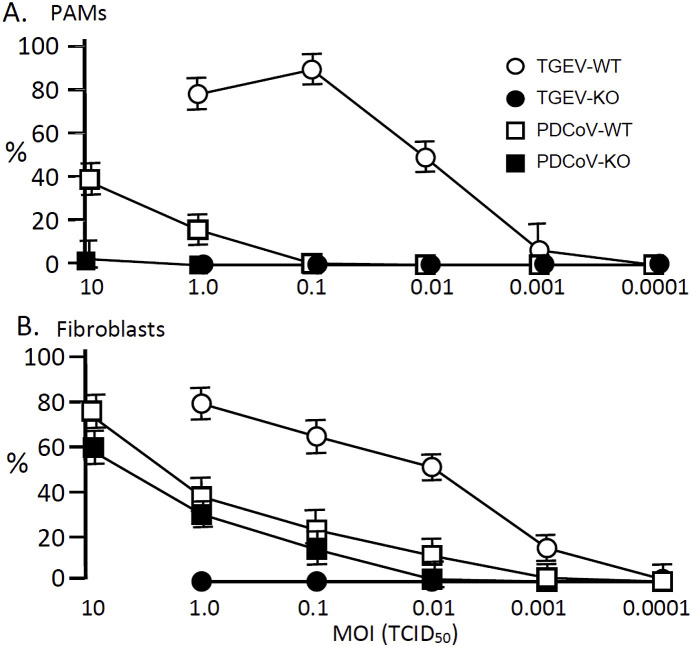
The permissiveness of ANPEP WT and KO PAMs and fibroblast-like cells for TGEV and PDCoV infection was also evaluated by determining percent virus antigen-positive cells after infection with different MOIs of virus. The results for WT PAMs are shown in Fig. 2 A. An MOI = 1 produced 20% and 80% antigen-positive cells for PDCoV and TGEV, respectively. The corresponding virus dilution endpoints were 0.0001 and 0.1 MOI. Increasing the MOI to 10 increased the percent PDCoV antigen-positive WT PAMs to 40%. For the KO PAMs, an MOI of 1 showed no TGEV or PDCoV antigen-positive cells. However, increasing the MOI to 10 resulted in 2% PDCoV antigen-positive PAMs.
Fig. 2.
PDCoV and TGEV infection of PAMs and lung-derived fibroblast-like cells. (A) ANPEP WT and KO PAMs were infected with different MOIs of TGEV or PDCoV. After 24 h from infection, the cells were stained and fixed with AlexaFluor488-labeled anti-TGEV or anti-PDCoV antibodies and the percent antigen-positive cells recorded. (B) ANPEP WT and KO lung-derived fibroblast-like cells were infected with TGEV or PDCoV at an MOI of 0.0001–10. After 24 h from infection, the cells were stained and fixed with AlexaFluor488-labeled anti-TGEV or anti-PDCoV antibodies and the percent antigen-positive cells determined. Results are shown as the mean and standard deviation of results from three ANPEP WT and three KO pigs.
Results for the infection of the WT fibroblast-like cells, presented in Fig. 2B, showed that an MOI = 1 produced 80% TGEV and 40% PDCoV antigen-positive cells. The dilution endpoints were 0.0001 MOI for TGEV and 0.001 MOI for PDCoV. For the KO fibroblast-like cells, TGEV infection was not detected; whereas, PDCoV infection resulted in 35% antigen-positive cells at an MOI = 1 with an endpoint of 0.01.
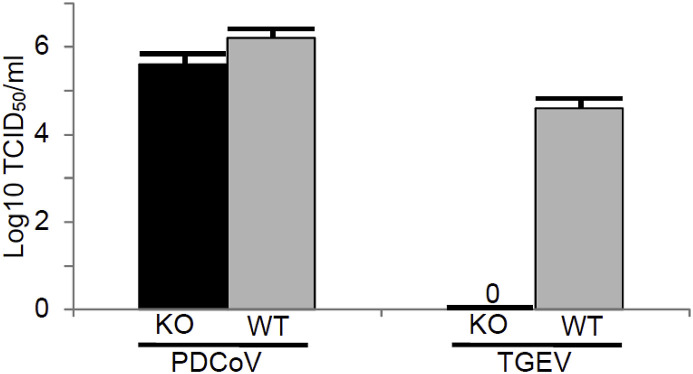
The results for virus yields on ANPEP WT and KO fibroblast-like cells are shown in Fig. 3 . At an MOI = 1, PDCoV grew to near equal levels on fibroblast-like cells derived from the ANPEP WT and KO pigs; whereas, TGEV could only be propagated on fibroblast-like cells derived from the ANPEP WT pigs.
Fig. 3.
Yields of PDCoV and TGEV on fibroblast-like cells. Fibroblast-like cells were infected with an MOI = 1. Virus concentration was measured by titration of virus on ST cells at two days after infection.
2.2. PDCoV infection of ANPEP WT and KO pigs
Previous work from our lab showed that ANPEP KO pigs are completely resistant to infection with TGEV (Whitworth et al., 2018). We tested the ability of PDCoV to infect ANPEP KO pigs. The results for virus infection are summarized in Table 1 . All pigs were positive for PDCoV nucleic acid at one day after infection. On the second day, two ANPEP WT and three KO pigs remained RT-PCR-positive. By three days after infection, only one pig, an ANPEP KO pig, was positive for PDCoV nucleic acid. By four days after infection, all pigs were negative for virus nucleic acid in feces. Serum samples obtained at three days after infection were negative for virus nucleic acid (data not shown). At four days after infection, one WT and one KO pigs were removed from the study and necropsied. RT-PCR amplification of PDCoV nucleic acid in intestines and mesenteric lymph nodes showed that both pigs were negative for PDCoV. The presence of PDCoV infection was also assessed by virus-specific neutralizing activity measured in serum at the termination of the study, 14 days after infection. The results, summarized in Table 1, showed that two of the remaining six pigs seroconverted; one WT and one KO pig. Clinical signs, such as diarrhea and fever, were not detected in any of the infected WT or KO pigs. The absence of clinical signs is likely a result of the older-age pigs used in the infection study. Together, the RT-PCR and serological results showed that ANPEP KO pigs are not resistant to infection with PDCoV.
Table 1.
RT-PCR for PDCoV nucleic acid in feces and virus neutralizing activity in seruma.
| Pig | ANPEP Genotype | RT-PCR |
Virus Neutralization | |||
|---|---|---|---|---|---|---|
| Day after infection | ||||||
| 1 | 2 | 3 | 4 | |||
| 18 | Wild-Type | 38.1 | >40 | >40 | >40 | 1/64 b |
| 19 | Wild-Type | 35.2 | 37.3 | >40 | >40 | <16 |
| 22 | Wild-Type | 36.6 | 38.1 | >40 | >40 | ND c |
| 20 | Knockout | 37.1 | 36.0 | >40 | >40 | 1/64 |
| 21 | Knockout | 35.1 | 37.1 | >40 | >40 | <16 |
| 23 | Knockout | 32.5 | 32.5 | 36.3 | >40 | ND |
Ct > 40, negative for detectable quantities of PDCoV nucleic acid. Virus neutralization assays were performed at 14 days after infection. A tite <1/16 was considered negative for detectable neutralizing activity. All pigs at day 0 were negative for neutralizing activity.
Neutralizing activity was measured on day 10 after infection.
ND, Not Done.
3. Discussion
The results from this study showed no PDCoV infection of PAMs obtained from ANPEP KO pigs (see Fig. 1A), which supports the observations of Wang et al. (2018), who concluded that APN is a receptor for PDCoV. In contrast, the PDCoV-positive infection results for fibroblast-like cells derived from the ANPEP KO cultures support the observations of Zhu et al. (2018), who concluded that APN is not a functional receptor for PDCoV (see Fig. 1C). When taken together, our data support the observations of Li et al. (2018) who concluded that there are APN-dependent and APN-independent receptors for PDCoV. We hypothesize that PAMs possess only the APN receptor, while the lung-derived fibroblast-like cells possess both receptors. However, since the APN-independent receptors for PDCoV have not been identified, we currently cannot easily prove this hypothesis and it remains to be elucidated in future studies.
Ultimately, the importance of pAPN as a receptor for PDCoV is determined by the permissiveness of ANPEP KO pigs for infection. The results in Table 1, showing positive results for the presence of PDCoV nucleic acid in the feces of infected ANPEP KO pigs confirms the results obtained following the infection of KO fibroblast-like cells. These data support the notion that APN is not required for PDCoV infection of pigs. However, the conclusion that ANPEP KO pigs retain permissiveness for infection with PDCoV comes with a few caveats. For example, all fecal samples on day 1 after infection were positive for PDCoV nucleic acid and by 3 days, the virus was only detected in one KO pig. Intestinal tissues from the two pigs tested, one WT and one KO, were negative for PDCoV. The positive results obtained for feces can be explained by the presence of environmental contamination caused by the residual inoculum used for infection. However, support for a productive infection of the KO pigs comes from the seroconversion of one KO pig at 14 days after infection (see Table 1). A second caveat comes from the transient nature of PDCoV infection in the WT and KO pigs used in this study, which made it difficult to determine if APN plays a role in the disease process. Previous studies show that neonatal piglets are more susceptible than weaned pigs to enteric virus infection (Thomas et al., 2015; Jung et al., 2015). The relationship of pig age and PDCoV infection outcome has not been studied in detail; however, it is speculated that PDCoV infection may be age-dependent, similar to PEDV, with more severe outcomes in younger piglets. In this study, 28-33 day-old ANPEP KO and WT pigs were used for PDCoV inoculation. In future studies, a more sensitive neonatal pig infection model can be used which may yield a more accurate determination regarding the role of APN in PDCoV infection of its natural host.
4. Materials and methods
4.1. ANPEP KO pigs, viruses and cells
The ANPEP-edited pigs used in this study were derived by breeding founder animals created using direct zygote injection of CRISPR/Cas9 along with two CRISPR guides directed at exon 2 of ANPEP (Whitworth et al., 2017). Along with sequencing of the ANPEP gene, the APN phenotype for each knockout allele was confirmed by the absence of CD13 expression in intestines and by the resistance of ANPEP KO pigs to infection with TGEV (Whitworth et al., 2018).
The PDCoV isolate, South Dakota, was used for the infection of cells and pigs (Vitosh-Sillman et al., 2016). PDCoV stocks were prepared on ST cells maintained in MEM supplemented with 7% FBS, Pen-Strep (80 Units/ml and 80 μg/ml, respectively), 3 μg/ml Fungizone, 25 mM HEPES MEM, and 0.2 μg/ml L-1-Tosylamide-2-phenylethyl chloromethyl ketone (TPCK) trypsin, as described in Chen et al. (2015). After 2 h, the media was replaced with MEM-FBS with antibiotics. TGEV, Purdue strain, was obtained from Iowa State University. TGEV was maintained in the same media without TPCK. Cells were maintained at 37°C and 5% CO2. After 48 h, the virus was harvested and titrated on ST cells. Serial 1:10 dilutions of virus in triplicate were performed on a 96-well plate containing ST cells. Twenty-four h later, the cells were fixed for 10 min in 80% acetone and air-dried. For detection of PDCoV, cells were stained with anti-N protein mAb SD110-121 (kindly provided by Steve Lawson at South Dakota State University), which was diluted 1:500 in PBS with 5% goat serum (PBS-GS). Detection of TGEV was performed using anti-FIPV3-70 mAb (Custom Monoclonals International, USA), diluted 1:700 in PBS-GS. After 1 h incubation with antibody at 37o C, the cells were washed with PBS and bound antibody detected with AlexaFluor488-labeled goat anti-mouse IgG (Cat. No. A-11001, Invitrogen) diluted 1:400 in PBS-GS. After 1 h incubation at 37 °C, the plates were washed with PBS, nuclei counterstained with propidium iodide (PI), and cells viewed under a fluorescence microscope. The 50% tissue culture infectious dose (TCID50/ml) was calculated according to the method of Reed and Muench (1938).
The recovery of porcine alveolar macrophages is described in Wells et al. (2017). Lungs were removed from euthanized pigs and lavaged by pouring 100 ml of cold PBS into the trachea. PAMs were sedimented by centrifugation at 1200×g for 10 min at 4°C and cells re-suspended and washed once in cold sterile PBS. The final cell pellet was re-suspended in freezing medium containing 45% RPMI 1640 with antibiotics, 45% FBS, 10% dimethylsulfoxide (DMSO) and stored in liquid nitrogen until use. For the infection, approximately 104 PAMs were added to each well of a 96 well plate and incubated overnight at 37oC in 5% CO2. The cells were gently washed to remove non-adherent cells. Serial 1:10 dilutions of virus in media were added to wells in triplicate. After an overnight incubation, the cells were washed with PBS and fixed for 10 min in 80% acetone. Presence of either PDCoV or TGEV in the infected PAMs was confirmed as described above.
For enrichment of lung mesenchymal stem cells, PAMs were cultured in RPMI-1640 supplemented with 10% FBS, l-glutamine, and antibiotics. The continuous maintenance of PAMs for two weeks resulted in the overgrowth of cultures by fibroblast-like cells, which comprise only about 1% of the lung lavage material. The confluent cells were removed by trypsinization and passaged at least 2 times. The infection of fibroblast-like cells was carried out as described for the PAM cultures.
4.2. Infection of pigs
All experiments involving animals and viruses were performed in accordance with the Federation of Animal Science Societies Guide for the Care and Use of Agricultural Animals in Research and Teaching, the USDA Animal Welfare Act and Animal Welfare Regulations; and after approval by the Kansas State University and University of Missouri institutional animal care and institutional biosafety committees. Three KO piglets were obtained from an ANPEP −/− sow mated with an ANPEP −/− boar. The ANPEP KO genotype of each piglet was confirmed by PCR and DNA sequencing. To avoid unnecessary stress, the weaned piglets were transported from the University of Missouri rearing facility to Kansas State University at three weeks of age. Three ANPEP WT piglets, obtained from a separate mating, were included as positive infection controls.
At the time of virus infection, the KO and WT pigs were 28 and 33 days old, respectively. Pigs were infected with 6.2 log10 TCID50 of PDCoV administered as a single oral dose in 20 ml of culture medium. A 5 cm tube was attached to the end of the inoculating syringe to allow the virus to flow down the back of the throat. Throughout the study, the infected WT and KO pigs were housed in the same pen to allow for continuous pig-to-pig contact. Caretakers and researchers were blind as to the genotype of each pig. Fecal swabs were collected daily from each pig beginning one day prior to infection until the end of the study. Each swab was placed in a 15 ml conical tube containing 1 ml of MEM with Pen-Strep and Fungizone. The tube was vortexed briefly to mix the swab contents, aliquoted into 1.5 ml cryovial storage tubes, and stored at −80 °C. Blood for serum was collected into serum separator tubes at 0, 3, 7, 10 and 14 days after infection.
4.3. PDCoV RT-PCR
Total RNA was extracted from fecal and serum samples using a MagMAXTM-96 Total RNA Isolation Kit (InvitrogenTM) according to the manufacturer's instructions. RT-PCR was performed using a commercial kit, EZ-PED/TGENatur/PDCoV MPX 1.0 (Tetracore) on a CFX-96 real-time PCR system (Bio-Rad). Results were reported as CT.
4.4. Measurement of virus neutralizing activity
Serum samples were serially diluted 1:2, starting at a dilution of 1:16 in 100 μl of MEM supplemented with 7% FBS, antibiotics and 0.2 μg/ml TPCK trypsin. One hundred microliters of diluted serum were mixed with 100 μl of PDCoV South Dakota an 800 TCID50 virus in 100 μl media 800 TCID50/100 μl on a 96 well plate and incubated 1 h at 37°C. Samples were transferred to a 96-well plate of confluent ST cells and incubated for 24 h at 37°C, 5% CO2. Infected cells were detected by IFA using an anti-PDCoV mAb. The neutralizing antibody titer was reported as the highest serum dilution at which 100% of virus infection was inhibited.
Author contribution
Ana Stoian, Methodology, Validation, Formal analysis, Investigation, Writing, Editing.
Raymond R.R. Rowland, Conceptualization, Methodology, Formal analysis, Writing, Supervision, Funding acquisition.
Vlad Petrovan, Methodology, Investigation.
Maureen Sheahan, Methodology, Validation, Formal analysis.
Melissa S. Samuel, Methodology, Investigation.
Kristin M. Whitworth, Conceptualization, Writing.
Kevin D. Wells, Conceptualization, Writing, Supervision.
Jianqiang Zhang, Methodology, Validation, Formal analysis, Investigation.
Benjamin Beaton, Conceptualization, Supervision.
Mark Cigan, Conceptualization, Supervision, Funding acquisition.
Randall S. Prather, Conceptualization, Writing, Supervision, Funding acquisition.
Acknowledgements
The work was supported by Genus plc and Food for the 21st Century at the University of Missouri. The authors acknowledge many people, including undergraduate students who made many of the experiments possible.
References
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L'Haridon R., Vogel L.K., Sjostrom H., Noren O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Kut E., Sjostrom H., Noren O., Laude H. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase-N that is distinct from the enzymatic site. J. Virol. 1994;68:5216–5224. doi: 10.1128/jvi.68.8.5216-5224.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dveksler G.S., Pensiero M.N., Cardellichio C.B., Williams R.K., Jiang G.S., Holmes K.V., Dieffenbach C.W. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68:8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Annamalai T., Lu Z., Saif L.J. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 2015;178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri M., O'Brien T.D., Chattha K.S., Saif L.J. Porcine lung mesenchymal stromal cells possess differentiation and immunoregulatory properties. Stem Cell Res. Ther. 2015;6:222. doi: 10.1186/s13287-015-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Kenney S.P., Widjaja I., Jung K., Alhamo M.A., van Dieren B., van Kuppeveld F.J.M., Saif L.J., Bosch B.J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. 2018;115:E5135–E5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426 doi: 10.1038/nature02145. 450–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson D.M., Arruda P.H., Magstadt D.R., Burrough E.R., Hoang H., Sun D., Bower L.P., Bhandari M., Gauger P.C., Stevenson G.W., Wilberts B.L., Wang C., Zhang J., Yoon K.J. Characterization of porcine epidemic diarrhea virus isolate US/Iowa/18984/2013, infection in 1-day-old cesarean-derived colostrum-deprived piglets. Vet. Pathol. 2016;53:44–52. doi: 10.1177/0300985815591080. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.L., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hygiene. 1938;27:493–497. [Google Scholar]
- Saif L.J., Pensaert M.B., Sestak K., Yeo S., Jung K. Coronaviruses. In: Zimmerman J.J., Karriker L.A., Ramirez A., Schwartz K.J., Stevenson G.W., editors. Diseases of Swine. tenth ed. Wiley; New York: 2012. pp. 501–524. [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Investig. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Thomas J.T., Chen Q., Gauger P.C., Gimenez-Lirola L.G., Sinha A., Harmon K.M., Madson D.M., Burrough E.R., Magstadt D.R., Salzbrenner H.M., Welch M.W., Yoon K.J., Zimmerman J.J., Zhang J. Effect of porcine epidemic diarrhea virus infectious doses on infection outcomes in naive conventional neonatal and weaned pigs. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitosh-Sillman S., Loy J.D., Brodersen B., Kelling C., Doster A., Topliff C., Nelson E., Bai J., Schirtzinger E., Poulsen E., Meadors B., Anderson J., Hause B., Anderson G., Hesse R. Experimental infection of conventional nursing pigs and their dams with Porcine deltacoronavirus. J. Vet. Diagn. Investig. 2016;28:486–497. doi: 10.1177/1040638716654200. [DOI] [PubMed] [Google Scholar]
- Wang B., Liu Y., Ji C.M., Yang Y.L., Liang Q.Z., Zhao P., Xu L.D., Lei X.M., Luo W.T., Qin P. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J. Virol. 2018;92 doi: 10.1128/JVI.00318-18. e00318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA. Emerg. Infect. Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells K.D., Bardot R., Whitworth K.M., Trible B.R., Fang Y., Mileham A., Kerrigan M.A., Samuel M.S., Prather R.S., Rowland R.R.R. Replacement of porcine CD163 scavenger receptor cysteine-rich domain 5 with a CD163-like homolog confers resistance of pigs to genotype 1 but not genotype 2 porcine reproductive and respiratory syndrome virus. J. Virol. 2017;91 doi: 10.1128/JVI.01521-16. e01521-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth K.M., Benne J.A., Spate L.D., Murphy S.L., Samuel M.S., Murphy C.N., Richt J.A., Walters E., Prather R.S., Wells K.D. Zygote injection of CRISPR/Cas9 RNA successfully modifies the target gene without delaying blastocyst development or altering the sex ratio in pigs. Transgenic Res. 2017;26:97–107. doi: 10.1007/s11248-016-9989-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth K.M., Rowland R.R.R., Petrovan V., Sheahan M., Cino-Ozuna A.G., Fang Y., Hesse R., Mileham A., Samuel M.S., Wells K.D., Prather R.S. Resistance to coronavirus infection in amino peptidase N-deficient pigs. Transgenic Res. 2018;28:21–32. doi: 10.1007/s11248-018-0100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Liu S., Wang X., Luo Z., Shi Y., Wang D., Peng G., Chen H., Fang L., Xiao S. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection. Emerg. Microb. Infect. 2018;7:65. doi: 10.1038/s41426-018-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]