Abstract
During viral entry, enveloped viruses require the fusion of their lipid envelope with host cell membranes. For coronaviruses, this critical step is governed by the virally-encoded spike (S) protein, a class I viral fusion protein that has several unique features. Coronavirus entry is unusual in that it is often biphasic in nature, and can occur at or near the cell surface or in late endosomes. Recent advances in structural, biochemical and molecular biology of the coronavirus S protein has shed light on the intricacies of coronavirus entry, in particular the molecular triggers of coronavirus S-mediated membrane fusion. Furthermore, characterization of the coronavirus fusion peptide (FP), the segment of the fusion protein that inserts to a target lipid bilayer during membrane fusion, has revealed its particular attributes which imparts some of the unusual properties of the S protein, such as Ca2+-dependency. These unusual characteristics can explain at least in part the biphasic nature of coronavirus entry. In this review, using severe acute respiratory syndrome coronavirus (SARS-CoV) as model virus, we give an overview of advances in research on the coronavirus fusion peptide with an emphasis on its role and properties within the biological context of host cell entry.
Keywords: Coronavirus, SARS, Spike protein, Virus entry, Endosomes, Calcium, Fusion peptide
Highlights
-
•
SARS-CoV as model for studying the coronavirus (CoV) fusion peptide (FP).
-
•
Evidence based on functional and biophysical analyses that reveal the region downstream of the coronavirus spike protein S2′ cleavage as the bona fide FP that forms an extended “fusion platform”.
-
•
Evidence for a direct role for calcium cations in mediating membrane fusion of SARS-CoV.
-
•
Unusual features of the CoV FP that set it apart from other class I fusion peptides.
-
•
The biphasic nature of SARS-CoV cellular entry pathways can be explained by calcium-dependency of fusion.
1. Introduction
Coronaviruses are a diverse group of single-stranded plus-sense RNA viruses belonging to the Coronaviridae family and Nidovirales order (de Groot et al., 2012). They infect a wide array of mammalian and avian species, including bats, and have a propensity for interspecies jumping and zoonotic transmission as exemplified by severe acute respiratory syndrome coronavirus (SARS-CoV) and more recently by Middle East respiratory syndrome coronavirus (MERS-CoV) (Graham et al., 2013, Woo et al., 2009). As coronaviruses possess an envelope, membrane fusion with host cell membranes is a required and critical step in the replication cycle allowing for delivery of genomic RNA into the cytoplasm, which eventually leads to the start of replication. This critical early step is being viewed as an attractive target for therapeutic interventions (White et al., 2008).
Virus entry constitutes a series of interactions between a virion and its host cell occurring early in the viral life cycle (Boulant et al., 2015, Grove and Marsh, 2011, Hofmann and Pöhlmann, 2004, Marsh and Helenius, 2006). For an enveloped virus, these steps allow the virus to (i) bind to a target host cell, typically via interactions with cellular receptors, (ii) fuse its envelope with a cellular membrane, either at the plasma membrane or through the endocytic pathway, and (iii) deliver its genetic material inside the cell. Virus entry is a finely regulated process, often requiring a specific sequence of interactions (binding to receptors and/or co-receptors), triggers or cues (pH, proteolytic activation), and cellular processes such as endocytosis for successful delivery of viral genomic nucleic acids (White and Whittaker, 2016). Interestingly, coronaviruses display a large degree of plasticity regarding the entry pathways they use, which can occur at the plasma membrane or through the endocytic pathway (Belouzard et al., 2012, Matsuyama et al., 2005, Nash and Buchmeier, 1997). The mechanisms by which coronaviruses enter cells depends on the strain and species considered, along with tissue and cell-type specificities (e.g. receptor and protease availability, local microenvironment).
For enveloped viruses, a critical player in the entry process is the viral fusion protein as it mediates the membrane fusion reaction (Chernomordik and Kozlov, 2008, Colman and Lawrence, 2003, Harrison, 2008, White et al., 2008, White and Whittaker, 2016). While the process of merging two distinct lipid bilayers into a single one is a thermodynamically favorable reaction, it is associated with a high kinetic barrier (Harrison, 2008). As such, and because viral fusion proteins facilitate this process, they can be viewed as catalysts for the membrane fusion reaction. This has been very well studied both structurally and functionally with the influenza hemagglutinin (HA) fusion protein (Harrison, 2015). After attachment of HA to sialic acids cellular receptors the virion is internalized through the endocytic pathway. Because of endosomal acidification, increased H+ ion concentration within the endosome forces HA to undergo major conformational changes, which allows exposure of the fusion peptide and its insertion into target cellular membrane. This brings the viral and endosomal membrane in close proximity. Further conformational changes of several HA trimers allow merging of the outermost lipid leaflets forming an intermediate structure called the hemifusion stalk. This transient structure collapses into an expanding fusion pore allowing release of viral genetic material in the cytoplasm.
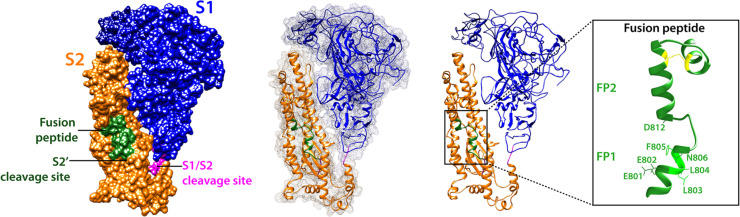
In the case of coronaviruses, viral entry into target cells is performed by the spike (S) envelope glycoprotein, which mediates both host cell receptor binding and membrane fusion. The S protein is classified as a class I viral fusion protein (Bosch et al., 2003), which includes the prototypical influenza virus hemagglutinin (HA) and retrovirus envelope (env) proteins (White et al., 2008). Class I viral fusion proteins form trimers and each monomer can often be divided into two domains, a receptor-binding domain (e.g. HA1 and gp120 for influenza virus and HIV, respectively), and a fusion domain (HA2 and gp40). Fusion domains are enriched in alpha-helices and contain regions called heptad repeats (HR) which are repetitive heptapeptides containing some hydrophobic residues and which are involved in the refolding process and coiling of central helices during membrane fusion. After membrane fusion has occurred, class I fusion proteins adopt a compact conformation, with a well-defined coiled-coil structure called a 6-helix bundle or 6HB (Belouzard et al., 2012, White et al., 2008). Importantly, the fusion domain contains a short segment, the fusion peptide (Epand, 2003, Lai et al., 2005, Tamm and Han, 2000, Tamm et al., 2002), typically composed of 15–25-amino acids, generally hydrophobic in nature, which becomes anchored to a target membrane when the fusion protein adopts the pre-hairpin conformation. The fusion peptide plays an essential role in mediating the membrane fusion reaction as it directly interacts with lipid bilayers, enabling to disrupt and connect two apposing membranes. Class I fusion proteins are often proteolytically activated or primed for fusion by host cell proteases at a specific cleavage site that usually forms the boundary between the receptor-binding and fusion domains (White and Whittaker, 2016). In the case of influenza HA and HIV env proteins, the cleavage event releases the fusion peptide as it is located at the N-terminal end of the fusion domain. While coronavirus S proteins possess salient features of class I fusion proteins, such as being a type I membrane proteins organized in trimers, possessing heptad repeats regions (HR1 and HR2), and requiring proteolytic cleavage for activation, they differ in several key aspects. The S proteins form substantially larger trimers, with S monomer size in the range of 1200–1400 amino acids (~180–200 kDa) compared to ~500 aa for influenza HA and ~800 aa for HIV. Until recently structural data on the coronavirus S was limited, due mostly to the difficulty of obtaining X-ray crystallographic structures of intact ectodomains of S. However, recent advances in cryo-electron microscopy (cryo-EM) have allowed the determination of the structure covering the majority of the ectodomain of several coronavirus S proteins in their pre-fusion conformation such as those of the murine hepatitis virus (MHV) (Walls et al., 2016a), human coronavirus HCoV-NL63 (Walls et al., 2016b), HCoV-HKU1 (Kirchdoerfer et al., 2016), MERS-CoV and SARS-CoV (Gui et al., 2017, Yuan et al., 2017). These efforts and studies constitute a huge step forward in the field as they uncovered the complexity of coronavirus S proteins and their highly glycosylated nature. The S protein can be divided into the S1 receptor-binding subunit and S2 fusion domain, usually separated by a cleavage site (S1/S2). However, coronavirus S proteins possess an additional cleavage site located within S2 and called (Belouzard et al., 2009, Belouzard et al., 2012, Millet and Whittaker, 2014). Not only are coronaviruses S proteins unusual for harboring multiple cleavage sites, they are also activated by a wide variety of host cell proteases (Millet and Whittaker, 2015), spanning different families such as cathepsins (Simmons et al., 2005), trypsin-like serine proteases such as members of the transmembrane serine protease (TTSP) family (Bertram et al., 2013, Gierer et al., 2013, Glowacka et al., 2011, Matsuyama et al., 2010), and the furin-like proprotein convertases (PCs) (Burkard et al., 2014, Millet and Whittaker, 2014). While the location of the fusion peptide has been debated, most recent data appears to argue that the segment immediately downstream of S2′ cleavage site is the bona fide fusion peptide. In the cryo-electron microscopy structures of coronavirus S, the fusion peptide segment appears to be exposed at the surface of the protein in the pre-fusion state ( Fig. 1), another unique characteristic setting S proteins apart from other class I viral fusion proteins like the influenza virus HA. While the fusion peptides of coronaviruses are not as well characterized as the ones from other prototypical class I fusion proteins like influenza HA or HIV env, evidence has been steadily accumulating for the identification of the bona fide fusion peptide.
Fig. 1.
Structural model of the SARS-CoV spike in pre-fusion uncleaved and monomeric form, based on pdb 3JCL (MHV spike). SARS-CoV S is colored with the S1 domain in blue and the S2 domain in orange, and is shown in in three different representations: space-filling, surface mesh and cartoon. The fusion peptide/predicted neutralizing epitope is shown green. The two proteolytic cleavage sites are shown in magenta, with S1/S2 exposed and the fusion peptide-proximal S2′ site protected. An enlarged cartoon depiction of the fusion peptide region is shown with key negatively charged and hydrophobic residues indicated.
In this review, we use SARS-CoV as model for studying the mechanism of coronavirus-membrane fusion. We aim to give an overview of the entry pathway of SARS-CoV and connect this to recent advances in our understanding of the coronavirus spike fusion peptide, highlighting its unique features as well as putting these findings back to their biological context and the apparently biphasic nature of coronavirus entry. An underlying theme that emerges from these studies is that the flexible characteristic of coronavirus entry pathways is in some ways imparted by the unique and special features of the coronavirus fusion peptide.
2. SARS-CoV mediated entry into cells and the activation of membrane fusion
As with many viruses, SARS-CoV entry into cells is dictated by the presence of its receptor. During the 2003–2004 SARS epidemic, a functional viral receptor for SARS-CoV was rapidly identified using a biochemical approach, where purified S1 was shown to bind human angiotensin-converting enzyme (ACE2) (Li et al., 2003). The SARS-CoV/ACE2 interaction has allowed much to be discovered regarding SARS-CoV tropism changes (Hulswit et al., 2016) and is considered and essential part of the virus entry mechanism, along with additional and binding factors, such as CD209L (Jeffers et al., 2004).
While ACE2 is an essential factor in all cell types, subsequent entry events are more cell-type dependent. In typical cell culture systems, and following receptor engagement, SARS-CoV is taken up by both clathrin and non-clathrin pathways (Inoue et al., 2007, Wang et al., 2008), with entry blocked by inhibitors of endosomal acidification and endosomal proteases such as cathepsin L (Simmons et al., 2005, Simmons et al., 2004). However, treatment of cells with trypsin or trypsin-like proteases (TTSPs) can also result in entry and spike-mediated fusion in a pH-independent manner (Bertram et al., 2011, Matsuyama et al., 2010, Matsuyama et al., 2005, Simmons et al., 2004). These findings show that in addition to ACE2, host cell proteases are key activators of SARS-CoV entry, and while the two identified activation sites (S1/S2 and S2′) are critical, the process of protease activation is quite complex, possibly depending heavily on cell type (Reinke et al., 2017). Overall, it is believed that the TTSP-mediated pathway is the relevant pathway in primary respiratory epithelia, the site of SARS-CoV infection in vivo (Shulla et al., 2011). These findings also suggest that low pH in itself is not a driving force for the necessary conformational events needed for coronavirus fusion, and that, as suggested by Matsumaya and Taguchi for MHV-2 (Matsuyama and Taguchi, 2009), additional factors are involved. Related to this, single particle studies of entry, using feline coronavirus as a model system, have shown that effects of pH on the rate constant of coronavirus fusion are negligible (Costello et al., 2013), with pH-dependency of entry possibly linked more to the enzymatic activity of the activating protease. One feature of SARS-CoV is that in cell culture, entry only begins after a lag time of 30 min (Mingo et al., 2015), suggesting that substantial endosomal maturation is necessary in the endosomal route of entry. The finding that wild-type MHV and feline coronavirus infection of HeLa cells also depends heavily on endosomal maturation, including components such as the VPS/HOPS complex involved in late endosome-lysosome trafficking, also suggests that endosome maturation (but not endosomal acidification per se) is a key player in infection across the Coronaviridae (Burkard et al., 2014). An overarching model, based primarily on studies with MERS-CoV, is that coronaviruses can enter cells either following fusion at or close to the cell surface, the so-called “early” pathway, or following prolonged endocytic trafficking, the “late” pathway (Earnest et al., 2017).
3. Fusion peptides form the core of viral fusion protein machinery
From the perspective of membrane fusion, viral fusion peptides are perhaps the most critical region of virus envelope glycoproteins as they directly interact and disrupt target host cell membranes. Thus, the corresponding regions have been extensively studied in model viral fusion proteins. While there are no strict definitions for what constitutes a viral fusion peptide, a number of criteria have to be met in order to designate a segment of viral fusion protein a bona fide fusion peptide. Often, the region corresponding to an FP is composed of hydrophobic amino acids, particularly enriched in glycine (G) and alanine (A) residues. It is important to note that fusion peptides can contain a few charged residues as well as bulky hydrophobic residues such as tryptophans (W). Another feature of fusion peptides is that they correspond to regions which are extremely sensitive to point mutations in the context of full-length fusion protein, i.e. that a single residue substitution within an FP often results in loss of fusion activity. Related to this characteristic is that although viral fusion protein sequences tend to vary greatly amongst different viral families, they are extremely well conserved within a given family. The capacity of a synthetic peptide to induce lipid-mixing and fusion of liposomes is another criterion that can help in identifying regions hypothesized to be FPs. Furthermore, the study of the effects of such peptides on lipid bilayer ordering using powerful spectroscopy techniques such as electron spin resonance (ESR) has also been successfully used to characterize other cellular and viral FPs such as those of influenza virus and HIV (Ge and Freed, 2009, Ge and Freed, 2011, Lai and Freed, 2014, Lai and Freed, 2015, Pinello et al., 2017).
Within each major classes of viral fusion proteins (I, II, and III) the associated fusion peptides share basic characteristics. Class I fusion peptides are usually enriched in alanine and/or glycine residues, and can either be “N-terminal” (i.e. located immediately downstream of the activating cleavage site) or “internal” depending on their positions respective to the cleavage site (Apellaniz et al., 2014). For class II and III fusion proteins, the fusion peptides are not released by proteolytic cleavage but instead, form so-called internal “fusion loops” which can be bipartite as in the case of class III fusion proteins (e.g. vesicular stomatitis virus glycoprotein G) (Apellaniz et al., 2014, Sun et al., 2008, White et al., 2008). It is important to highlight that it is the anchoring of the fusion peptides to apposing target membranes that allows for the formation of a unique biological structure: two lipid bilayers connected by fusion proteins.
4. Locating the coronavirus S fusion peptide
Identifying and locating the coronavirus fusion peptide has been most extensively performed using SARS-CoV spike (S) as a model. Initial work, using a peptide library derived from the SARS-CoV S protein, showed that three regions (R1, R2, and R3) within the S2 domain displayed membrane-interacting properties in experiments measuring vesicle membrane leakage (Guillen et al., 2005). R1 (aa 858–886) was found located upstream of HR1, R2 was identified between HR1 and HR2 (aa 1077–1092) and R3 (aa 1190–1202) was found to be situated proximal to the transmembrane region. Mutations within the region spanning residues 852–883, which approximately corresponds to the above-mentioned R1 region, were found to decrease cell-cell fusion in S-expression mediated syncytia formation assays (Petit et al., 2005). Another group identified two regions using the Wimley and White interfacial hydrophobicity analysis approach. They defined these regions as WW-I (aa 770–778) and WW-II (aa 864–886) with the latter almost matching the R1 region identified by Guillen and colleagues. WW-I was found to strongly partition into large unilaminar vesicles (LUV) and be most important for fusion. Subsequently, a model for how different membranotropic segments identified within SARS-CoV S2 would function has been proposed in which the putative fusion peptide (for consistency, this FP is referred to here as FP770–788) located N-terminal of the HR1 region (aa 770–788) is followed by an “internal” FP or IFP873–888 (aa 873–888) and another segment located upstream of the transmembrane domain (PTM1185–1202, aa 1185–1202) (Guillen et al., 2008, Guillén et al., 2008). In this model, all three segments work sequentially and in concert: the FP first inserts into target membranes, the IFP would then facilitate the hemifusion process by interacting with the apposing membrane and the interaction of the PTM with target lipid bilayer after further conformational changes of spike would also facilitate fusion pore expansion. More recently, structural characterization of these regions has been undertaken (Basso et al., 2016, Mahajan and Bhattacharjya, 2015). Mahajan and Bhattacharjya have used nuclear magnetic resonance (NMR) spectroscopy to gain insight into the structures adopted by isolated peptides corresponding to the above-mentioned regions FP770–788, IFP873–888, and PTM1185–1202 regions. They have determined the atomic-resolution structure of these peptides in presence of dodecylphosphocholine (DPC) detergent micelles by solution NMR (Mahajan and Bhattacharjya, 2015). Basso and colleagues have used electron spin resonance (ESR) as well as differential scanning calorimetry (DSC) to probe the interactions of the FP and IFP peptides with lipid bilayers (Basso et al., 2016).
These initial studies have identified regions of S2 with the capacity to interact with membranes. They also suggest that several regions within S2 could act in a concerted fashion to mediate the membrane fusion process. Further studies have been conducted taking a more functional approach and considering other criteria that define fusion peptides, such as proximity to a cleavage site and sequence conservation (Madu et al., 2009a, Madu et al., 2009b). The starting point for these studies was the identification of a second cleavage site in the SARS-CoV S S2 domain called S2′ (at R797 residue) (Belouzard et al., 2009). Authors found that the SARS-CoV S protein was proteolytically activated sequentially at the S1/S2 and S2′ sites, and that the latter event was critical for activating membrane fusion. Using these findings as a basis for identifying the SARS-CoV fusion peptide, additional work has shown that the segment located immediately downstream of the SARS-CoV S2′ cleavage site, 798SFIEDLLFNKVTLADAGF815, displayed features of a fusion peptide. In addition to being proximal to the S2′ cleavage site, the sequence of this region was found to be remarkably conserved within the Coronaviridae, with the 800IEDLLF805 motif showing minimal divergence. Interestingly, the highly conserved 803LLF805 correspond to the beginning of a major antigenic determinant of SARS-CoV S protein (aa 803–828), capable of eliciting the generation of neutralizing antibodies (Zhang et al., 2004). Furthermore, mutagenesis analysis of the S2′-proximal 798–815 segment in the context of full-length protein demonstrated its importance in mediating membrane fusion. Subsequent structural analysis of the isolated peptide by circular dichroism (CD) spectral analysis and vesicle lipid mixing assays confirmed the role of the segment as a fusion peptide (Madu et al., 2009b). More recently this domain has been subjected to extensive characterization by electron spin resonance (ESR) spectroscopy (along with CD spectroscopy) to show membrane ordering and the critical involvement of the LLF motif (Lai et al., 2017). Another mutagenesis study based on cell-cell fusion and pseudovirion infectivity assays has also found that a pair of highly conserved cysteines (C822 and C833) and the flanking residues D830 and L831 located C-terminally to the 798–815 region played a critical role for fusion (Madu et al., 2009a). The small sub-domain formed by the cysteine pair form a loop structure, a situation that is reminiscent to the avian sarcoma/leukosis virus subtype A in which a pair of cysteines flank the internal fusion peptide of the Env glycoprotein (Delos et al., 2008).
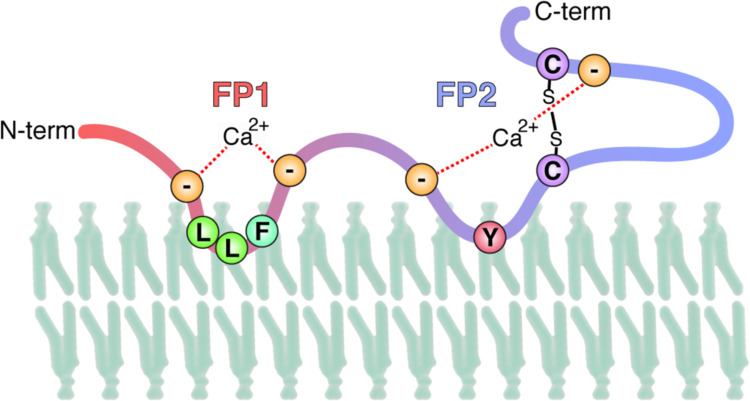
ESR is a powerful tool enabling to analyze membrane-ordering effects of fusion peptides with lipid bilayers. Using this technique the two distinct domains downstream of S2′ have recently been characterized. The 22 amino acids region immediately after the S2′ cleavage site (798SFIEDLLFNKVTLADAGFMKQY818) were termed FP1 and induced significant membrane ordering. The work also showed that the subsequent 21 amino acid, disulfide-bonded, domain (816KQYGECLGDINARDLICAQKF835), termed FP2, also displayed membrane-ordering properties. Furthermore, these effects of the two domains were dependent on Ca2+ ions, a situation similar to the calcium-dependency of fusion observed for the Rubella virus E1 envelope glycoprotein (Dube et al., 2016, Dube et al., 2014). A specific role for calcium in binding to both the FP1 and FP2 domains of SARS-CoV S was reinforced by isothermal titration calorimetry (ITC) (Lai et al., 2017). Overall these data suggest that for coronaviruses the fusion peptide is complex, possibly forming an extended FP1-FP2 “platform” with a conserved LLF core, as well as other regions of S2 previously identified as membrane-interacting ( Fig. 2), and suggesting that Ca2+ ions bind to conserved negatively charged residues in the fusion platform, to comprise the missing factor required to trigger membrane fusion.
Fig. 2.
Predicted model of the CoV fusion peptide region, along with its interaction with the lipid bilayer. Conserved negatively charged and hydrophobic residues and a proposed location of Ca2+ ions are shown.
5. The coronavirus fusion peptide in the context of the cell biology of virus entry
The notion that SARS-CoV fusion peptide structure and function in vitro is controlled by Ca2+ ions suggests that a similar situation occurs in vivo. Indeed, SARS-CoV entry is heavily dependent on the Ca2+ concentration of the media in which cells are grown, and is inhibited by calcium chelators such as BAPTA-AM, which act in endosomes (Lai et al., 2017). It is also interesting to note that amiodarone, a drug that blocks endosomal/lysosomal calcium channels, also inhibits SARS-CoV entry, after endosomal uptake (Stadler et al., 2008). The inhibition by amiodarone was originally attributed to Ca2+-dependent defects in endosome trafficking, but may also act more directly in the membrane fusion process.
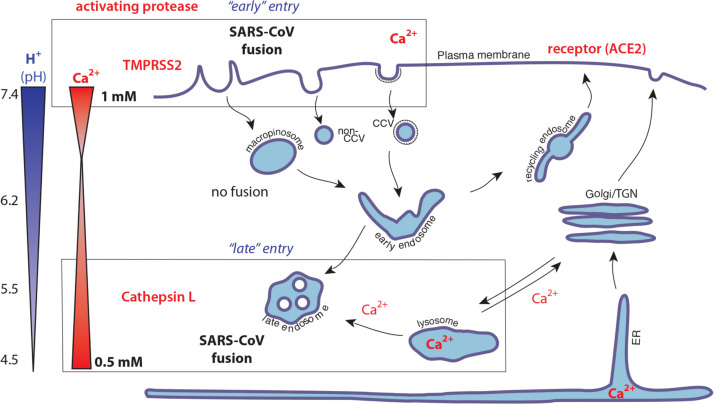
In general, the role of Ca2+ and other ions in the context of endosomal trafficking is poorly understood. While extracellular calcium is high (approximately 1 mM), levels drop rapidly in the lumen of newly formed endocytic vesicles, due to the action of efflux pumps (Huotari and Helenius, 2011, Luzio et al., 2007, Scott and Gruenberg, 2010). However, lysosomes can act as a calcium store via their interaction with major calcium stores in the endoplasmic reticulum, with levels as high as 0.5 mM found in lysosomes. During the later stages of endosome maturation, lysosome-late endosome fusion mediated by the HOPs complex can also result in substantial levels of Ca2+ in endosomal compartments. Such trafficking pathways may therefore lead to the availability of Ca2+ in two distinct points in the coronavirus entry pathway. First, at the cell surface where TTPS-mediated cleavage can activate fusion, and second, in “mature” endocytic vesicles where cathepsins can activate fusion.
Overall, we propose a model of SARS-CoV and for coronavirus entry in general, which integrates the various triggers and activators of the viral spike protein as follows: 1) Protease cleavage at S1/S2, which provides a preliminary priming step; 2) Receptor engagement; 3) Protease cleavage at S2′ to expose the viral fusion peptide (FP1-2); 4) Ionic changes to promote conformational changes and fusion peptide insertion. These ionic changes are not exclusively driven by H+ ions, and we argue that Ca2+ ions are necessary to promote fusion peptide insertion into the lipid bilayer. Our model encompasses both the available in vitro and in vivo data, with entry occurring from the extracellular space (TTSP-driven) – the “early’ pathway, or following substantial endosome maturation (cathepsin-driven) – the “late pathway”. Variations on this pathway may occur for some coronaviruses in the situations where furin-like proteases activate viruses in an intermediate endosomal compartment such as the early endosome (Burkard et al., 2014, Millet and Whittaker, 2014). A cartoon diagram of a basic entry model for SARS-CoV is presented in Fig. 3.
Fig. 3.
Cartoon model of SARS-CoV entry into cells. In this model, fusion can take place in two Ca2+-containing compartments corresponding to the observed “early” and “late” entry pathways, with cleavage occurring by distinct activating proteases following receptor engagement.
Acknowledgments
We thank Ruth Collins, Susan Daniel and members of the Whittaker and Daniel labs for helpful discussions. This work was supported by the National Institutes of Health Grants R21 AI1076258 and R21 AI111085.
References
- Apellaniz B., Huarte N., Largo E., Nieva J.L. The three lives of viral fusion peptides. Chem. Phys. Lipids. 2014;181:40–55. doi: 10.1016/j.chemphyslip.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso L.G., Vicente E.F., Crusca E., Jr., Cilli E.M., Costa-Filho A.J. SARS-CoV fusion peptides induce membrane surface ordering and curvature. Sci. Rep. 2016;6:37131. doi: 10.1038/srep37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H., Thiel V., Pöhlmann S. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87:6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C., Soilleux E.J., Jahn O., Steffen I., Pöhlmann S. Cleavage and activation of the SARS-coronavirus spike-protein by human airway trypsin-like protease. J. Virol. 2011;85:13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S., Stanifer M., Lozach P.Y. Dynamics of virus-receptor interactions in virus binding, signaling, and endocytosis. Viruses. 2015;7:2794–2815. doi: 10.3390/v7062747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J.M., Bosch B.J., de Haan C.A.M. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman P.M., Lawrence M.C. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- Costello D.A., Millet J.K., Hsia C.Y., Whittaker G.R., Daniel S. Single particle assay of coronavirus membrane fusion with proteinaceous receptor-embedded supported bilayers. Biomaterials. 2013;34:7895–7904. doi: 10.1016/j.biomaterials.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Cowley J.A., Enjuanes L., Faaberg K.S., Perlman S., Rottier P.J.M., Snijder E.J., Ziebuhr J., Gorbalenya A.E. Order - Nidovirales. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy. Elsevier; San Diego: 2012. pp. 784–794. [Google Scholar]
- Delos S.E., Brecher M.B., Chen Z., Melder D.C., Federspiel M.J., White J.M. Cysteines flanking the internal fusion peptide are required for the avian sarcoma/leukosis virus glycoprotein to mediate the lipid mixing stage of fusion with high efficiency. J. Virol. 2008;82:3131–3134. doi: 10.1128/JVI.02266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M., Etienne L., Fels M., Kielian M. Calcium-dependent rubella virus fusion occurs in early endosomes. J. Virol. 2016;90:6303–6313. doi: 10.1128/JVI.00634-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M., Rey F.A., Kielian M. Rubella virus: first calcium-requiring viral fusion protein. PLoS Pathog. 2014;10:e1004530. doi: 10.1371/journal.ppat.1004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest J.T., Hantak M.P., Li K., McCray P.B., Jr., Perlman S., Gallagher T. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog. 2017;13:e1006546. doi: 10.1371/journal.ppat.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand R.M. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta (BBA) - Biomembr. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- Ge M., Freed J.H. Fusion peptide from influenza hemagglutinin increases membrane surface order: an electron-spin resonance study. Biophys. J. 2009;96:4925–4934. doi: 10.1016/j.bpj.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge M., Freed J.H. Two conserved residues are important for inducing highly ordered membrane domains by the transmembrane domain of influenza hemagglutinin. Biophys. J. 2011;100:90–97. doi: 10.1016/j.bpj.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kramer-Kuhl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von Hahn T., Simmons G., Hofmann H., Pöhlmann S. The spike-protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2 and is targeted by neutralizing antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pöhlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J., Marsh M. The cell biology of receptor-mediated virus entry. J. Cell Biol. 2011;195:1071–1082. doi: 10.1083/jcb.201108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen J., Kinnunen P.K.J., Villalain J. Membrane insertion of the three main membranotropic sequences from SARS-CoV S2 glycoprotein. Biochim. Biophys. Acta (BBA) - Biomembr. 2008;1778:2765–2774. doi: 10.1016/j.bbamem.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen J., Perez-Berna A.J., Moreno M.R., Villalain J. Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16/18-mer peptide scan: implications for the viral fusion mechanism. J. Virol. 2005;79:1743–1752. doi: 10.1128/JVI.79.3.1743-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén J., Pérez-Berná A.J., Moreno M.R., Villalaín J. A Second SARS-CoV S2 Glycoprotein Internal Membrane-Active Peptide. Biophysical Characterization and Membrane Interaction. Biochemistry. 2008;47:8214–8224. doi: 10.1021/bi800814q. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pöhlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12:466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J., Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., Demartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.L., Freed J.H. HIV gp41 fusion peptide increases membrane ordering in a cholesterol-dependent fashion. Biophys. J. 2014;106:172–181. doi: 10.1016/j.bpj.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.L., Freed J.H. The Interaction between Influenza HA Fusion Peptide and Transmembrane Domain Affects Membrane Structure. Biophys. J. 2015;109:2523–2536. doi: 10.1016/j.bpj.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.L., Li Y., Tamm L.K. Wiley-VCH Verlag GmbH & Co. KGaA; 2005. Interplay of Proteins and Lipids in Virus Entry by Membrane Fusion, Protein–Lipid Interactions; pp. 279–303. [Google Scholar]
- Lai A.L., Millet J.K., Daniel S., Freed J.H., Whittaker G.R. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium-dependent manner. J. Mol. Biol. 2017;429:3875–3892. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio J.P., Bright N.A., Pryor P.R. The role of calcium and other ions in sorting and delivery in the late endocytic pathway. Biochem. Soc. Trans. 2007;35:1088–1091. doi: 10.1042/BST0351088. [DOI] [PubMed] [Google Scholar]
- Madu I.G., Belouzard S., Whittaker G.R. SARS-coronavirus spike S2 domain flanked by cysteine residues C822 and C833 is important for activation of membrane fusion. Virology. 2009;393:265–271. doi: 10.1016/j.virol.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M., Bhattacharjya S. NMR structures and localization of the potential fusion peptides and the pre-transmembrane region of SARS-CoV: implications in membrane fusion. Biochim. Biophys. Acta. 2015;1848:721–730. doi: 10.1016/j.bbamem.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh M., Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Taguchi F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J. Virol. 2009;83:11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. 2005;102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingo R.M., Simmons J.A., Shoemaker C.J., Nelson E.A., Schornberg K.L., D'Souza R.S., Casanova J.E., White J.M. Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J. Virol. 2015;89:2931–2943. doi: 10.1128/JVI.03398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash T.C., Buchmeier M.J. Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology. 1997;233:1–8. doi: 10.1006/viro.1997.8609. [DOI] [PubMed] [Google Scholar]
- Petit C.M., Melancon J.M., Chouljenko V.N., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology. 2005;341:215–230. doi: 10.1016/j.virol.2005.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello J.F., Lai A.L., Millet J.K., Cassidy-Hanley D., Freed J.H., Clark T.G. Structure-function studies link class II viral fusogens with the ancestral gamete fusion protein HAP2. Curr. Biol.: CB. 2017;27:651–660. doi: 10.1016/j.cub.2017.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke L.M., Spiegel M., Plegge T., Hartleib A., Nehlmeier I., Gierer S., Hoffmann M., Hofmann-Winkler H., Winkler M., Pohlmann S. Different residues in the SARS-CoV spike protein determine cleavage and activation by the host cell protease TMPRSS2. PLoS One. 2017;12:e0179177. doi: 10.1371/journal.pone.0179177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.C., Gruenberg J. Ion flux and the function of endosomes and lysosomes: pH is just the start. Bioessays. 2010;33:103–110. doi: 10.1002/bies.201000108. [DOI] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler K., Ha H.R., Ciminale V., Spirli C., Saletti G., Schiavon M., Bruttomesso D., Bigler L., Follath F., Pettenazzo A., Baritussio A. Amiodarone alters late endosomes and inhibits SARS coronavirus infection at a post-endosomal level. Am. J. Respir. Cell Mol. Biol. 2008;39:142–149. doi: 10.1165/rcmb.2007-0217OC. [DOI] [PubMed] [Google Scholar]
- Sun X., Belouzard S., Whittaker G.R. Molecular architecture of the bipartite fusion loops of vesicular stomatitis virus glycoprotein G, a class III viral fusion protein. J. Biol. Chem. 2008;283:6418–6427. doi: 10.1074/jbc.M708955200. [DOI] [PubMed] [Google Scholar]
- Tamm L.K., Han X. Viral fusion peptides: a tool set to disrupt and connect biological membranes. Biosci. Rep. 2000;20:501. doi: 10.1023/a:1010406920417. [DOI] [PubMed] [Google Scholar]
- Tamm L.K., Han X., Li Y., Lai A.L. Structure and function of membrane fusion peptides. Biopolymers. 2002;66:249–260. doi: 10.1002/bip.10261. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.-J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., DiMaio F., Bosch B.-J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Whittaker G.R. Fusion of enveloped viruses in endosomes. Traffic. 2016;17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang G., Li J., Nie Y., Shi X., Lian G., Wang W., Yin X., Zhao Y., Qu X., Ding M., Deng H. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 2004;78:6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]