Abstract
Swine enteric alphacoronavirus (SeACoV), also known as swine acute diarrhea syndrome coronavirus (SADS-CoV), belongs to the species Rhinolophus bat coronavirus HKU2. Herein, we report on the primary characterization of SeACoV in vitro. Four antibodies against the SeACoV spike, membrane, nucleocapsid and nonstructural protein 3 capable of reacting with viral antigens in SeACoV-infected Vero cells were generated. We established a DNA-launched SeACoV infectious clone based on the cell adapted passage-10 virus and rescued the recombinant virus with a unique genetic marker in cultured cells. Six subgenomic mRNAs containing the leader-body junction sites, including a bicistronic mRNA encoding the accessory NS7a and NS7b genes, were experimentally identified in SeACoV-infected cells. Cellular ultrastructural changes induced by SeACoV infection were visualized by electron microscopy. The availability of the SeACoV infectious clone and a panel of antibodies against different viral proteins will facilitate further studies on understanding the molecular mechanisms of SeACoV replication and pathogenesis.
Keywords: Swine enteric alphacoronavirus (SeACoV), Viral antibodies, Infectious clone, Subgenomic mRNAs, Electron microscopy (EM)
Highlights
-
•
Generation of four antibodies to distinct SeACoV protein for detection of SeACoV infection.
-
•
Development of a DNA-launched reverse genetics system for SeACoV.
-
•
Recombinant SeACoV with a genetic marker had similar growth kinetics to the parental virus.
-
•
Identification of all SeACoV subgenomic mRNAs containing the leader-body junction sites.
-
•
SeACoV infection induces cellular ultrastructural changes.
1. Introduction
Swine enteric alphacoronavirus (SeACoV), also known as swine acute diarrhea syndrome coronavirus (SADS-CoV), is a novel porcine enteric coronavirus that causes acute vomiting and watery diarrhea in piglets (Gong et al., 2017; Pan et al., 2017; Zhou et al., 2018). This emerging virus was first isolated from clinically sick animals in commercial swine herds at Guangdong province, China during February–May 2017. The mortality rate in less than 5 days old piglets was over 90%, whereas it dropped to 5% in piglets older than 8 days (Zhou et al., 2018). The clinical samples examined by polymerase chain reaction (PCR) or reverse transcription PCR (RT-PCR) during laboratory investigation were negative for the other swine coronaviruses such as porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), porcine deltacoronavirus (PDCoV) and porcine hemagglutinating encephalomyelitis virus (PHEV), as well as the other known viral pathogens (Pan et al., 2017). Isolation of the pathogen in African green monkey Vero cells resulted in the discovery of SeACoV (Pan et al., 2017), which belongs to the species Rhinolophus bat coronavirus HKU2 identified in the same region a decade earlier (Lau et al., 2007). A retrospective study indicated that the virus had emerged in Guangdong since August 2016 (Zhou et al., 2019). The isolated virus was infectious to pigs and cause mild or severe diarrhea symptom when inoculated orally into conventional newborn piglets (Pan et al., 2017; Xu et al., 2019; Zhou et al., 2018). Nevertheless, as SeACoV fulfilled the premises of Koch's Postulates, this was regarded to be the etiologic agent of the epidemic.
Like other CoVs, SeACoV is a single-stranded and positive-sense RNA virus in the genus alphacoronavirus (α-CoVs) of the subfamily Coronavirinae of the family Coronaviridae. Its genome is approximately 27.2 kb in size with the gene order of 5’-ORF1a/1b (ORF1ab)-Spike (S)-ORF3-Envelope (E)-Membrane (M)-Nucleocapsid (N)-NS7a/NS7b-3’. SeACoV shared 95% nucleotide (nt) sequence identity with the bat CoV HKU2 strains and 96–98% nt identity with the HKU2-derived bat SADS-related coronavirus (SADSr-CoV) strains at the complete genome level (Pan et al., 2017; Zhou et al., 2018). Interestingly, SeACoV and other HKU2-related α-CoVs possess the unique S genes closely related to the betacoronavirus (β-CoV), in a manner similar to those by rodent and Asian house shrew α-CoVs (Tsoleridis et al., 2019; Wang et al., 2015, 2017b), suggesting the occurrence of ancient recombination events between α-CoV and β-CoV (Lau et al., 2007; Pan et al., 2017).
The CoV genome harbors a few genus-specific accessory genes within the 3’-part genomic region encoding the four structural proteins (S-E-M-N). It is found that SeACoV contains a putative open reading frame (ORF), NS7a, and a downstream NS7b ORF (overlapped with NS7a) after the N gene at the 3’-end genome (Lau et al., 2007; Pan et al., 2017). The NS7a is shared by the HKU2 and SeACoV strains, whereas NS7b is only present in the SeACoV genome (Zhou et al., 2018). Many of CoV accessory proteins play some important roles in immune modulation and viral pathogenesis (Liu et al., 2014). For examples, the severe acute respiratory syndrome coronavirus (SARS-CoV) ORF-3a was found to induce necrotic cell death, lysosomal damage and caspase-1 activation, which largely contribute to the clinical manifestations of SARS-CoV infection (Yue et al., 2018). In addition, SARS-CoV ORF6 and ORF7b may also be also associated with the virulence. In another newly emerged swine CoV, PDCoV, its accessory NS6 protein has been reported to counteract host innate antiviral immune response by inhibiting IFN-β production that interacts with RIG-I/MDA5 (Fang et al., 2018). Whether the predicted NS7a and NS7b of SeACoV encode functional accessory proteins remain to be confirmed experimentally.
Discovery of SeACoV, largely dissimilar to PEDV, TGEV and PDCoV, challenges to the prospects of detection, prevention and control of diarrheal pathogens in swine (Wang et al., 2019). It is pivotal to undertake comprehensive investigations on the basic genetics of this emerged enteric CoV since very little is known about the molecular virology of SeACoV. The purpose of this study was to develop SeACoV-specific antibodies to distinct viral protein as the research tools used to investigate the basic characteristics of SeACoV infection in vitro. We also aimed to develop a DNA-launched reverse genetics system for SeACoV that will be useful for future studies.
2. Results and discussion
2.1. Polyclonal antibodies against four recombinant SeACoV proteins can react with viral antigens in SeACoV-infected cells
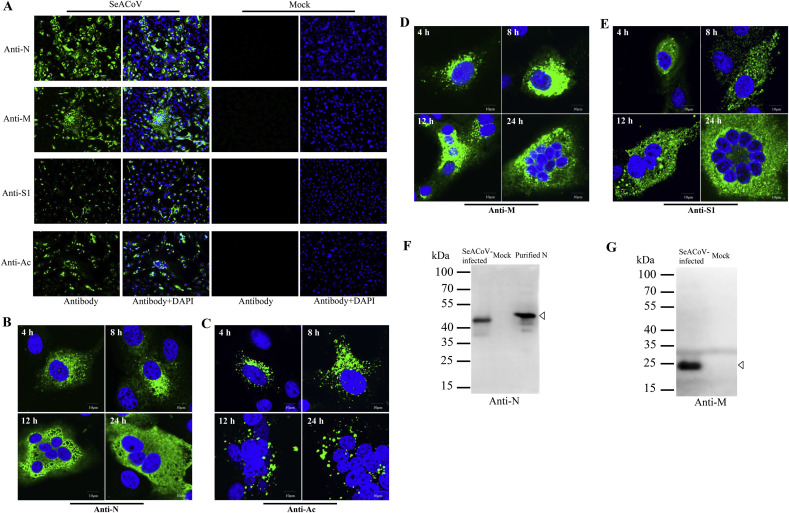
Four SeACoV specific polyclonal antibodies (pAbs) against distinct viral protein antigens were generated and validated. Two viral genes, SeACoV N and the nonstructural protein 3 (Nsp3) acidic domain (Ac) of ORF1a, were expressed as soluble products in the bacteria; the SeACoV spike subunit 1 (S1) was expressed in insect cells, secreting into the cultured medium. Purified recombinant SeACoV proteins (N, S1 and Ac) and an antigenic peptide corresponding to the last 14 amino acids (aa) at the carboxyl terminus of the M protein were used to immunize rabbits, respectively, generating four polyclonal sera that were then used to detect viral proteins on SeACoV-infected Vero cells. Immunofluorescence assay (IFA) conducted at 48 h post-infection (hpi) using respective pAb showed that the four viral antigens (N, M, S1 or Ac) were each expressed in the cytoplasm of the infected cells, with the anti-N and anti-M pAbs displaying the higher fluorescence intensity (Fig. 1 A). In contrast, mock-infected controls did not show any positive IFA signals (Fig. 1A).
Fig. 1.
Characterization of the four anti-SeACoV polyclonal antibodies (pAbs). (A) Immunofluorescence assay (IFA) results at 48 h post-infection (hpi) in SeACoV-infected or mock-infected Vero cells with an anti-N-pAb, an anti-M-pAb, an anti-S1-pAb and an anti-Ac-pAb, respectively (magnification = 200×). Alexa Fluor 488-conjugated goat anti-rabbit IgG (green) was used as the secondary antibody in the IFA. Antibody staining merged with nuclear staining using DAPI (blue) is also shown. (B–E) Time course analysis of N, Ac, M or S1 detection using an Olympus confocal microscope. Vero cells infected with SeACoV were fixed at 4, 8, 12, and 24 hpi, and labeled with four pAbs, respectively. Bar = 10 μm. (F) Western blot analysis using cell lysates of SeACoV-infected or mock-infected Vero cells with an anti-N pAb. The purified N protein expressed in E.coli was used as the control. (G) Western blot analysis using cell lysates of SeACoV-infected or mock-infected Vero cells with an anti-peptide pAb specific to M. Open arrowheads indicate the detected N or M protein.
To determine the intracellular localization and the timing of the viral protein expression with higher magnification, time course analysis of confocal image was performed. Vero cells infected with SeACoV were fixed at 4, 8, 12, and 24 hpi, and labeled with four pAb, respectively. Perinuclear and cytoplasmic foci were detected by anti-N staining at 4 and 8 hpi, and were distributed throughout the cytoplasm at 12 and 24 hpi, probably reflecting that N protein is associated with sites of viral RNA replication in early infection phase (Verheije et al., 2010) and assembled into virions subsequently (Fig. 1B). Anti-Ac (Nsp3) staining also resulted in detection of perinuclear foci at four time points, indicating localization to the viral replication-transcription complexes (Fig. 1C), which was similar to the pattern of Nsp3 antibody observed in SARS-CoV-infected Vero cells (Prentice et al., 2004). Confocal microscopy detected discrete cytoplasmic fluorescence signal throughout the cytoplasm with anti-M (Fig. 1D) and anti-S1 (Fig. 1E) as early as 4 hpi. Diffuse and more intense fluorescence was observed over time, demonstrating the process of virus assembly by incorporation of M and S proteins into virus particles.
The anti-N pAb recognized a single band of 42 kDa in the lysate of SeACoV-infected cells but not in control cells at 48 hpi by western blot analysis (Fig. 1F). The molecular size was consistent with the deduced aa sequence of the N protein but was a little less than the purified products expressed in the bacteria (Fig. 1F). Expression of the M protein with the predicted 25-KDa molecular size was also detected by using anti-M pAb in SeACoV-infected cells (Fig. 1G). The reactivity of anti-S1 or anti-Ac was less distinct as seen by western blot analysis (data not shown). Therefore, all the four SeACoV pAbs can be used for specific detection of SeACoV infection in the cultured cell by IFA staining, and the anti-N and anti-M pAbs can also be used particularly in western blot analysis. The antibodies are available to the research community upon request.
2.2. Rescue of recombinant SeACoV from a SeACoV full-length cDNA clone in Vero cells
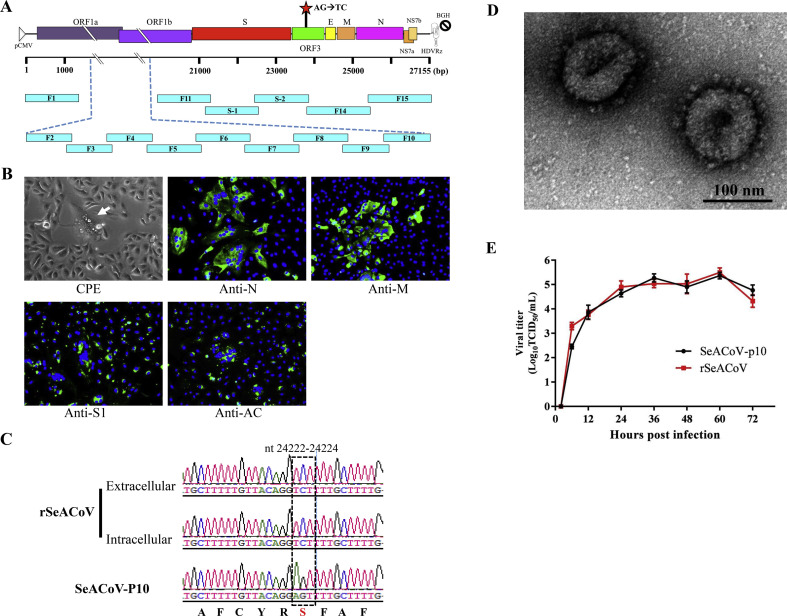
Genetic manipulation of viral genomes and dissection of the structural and functional relationships of viral genes depend on the development of powerful reverse genetics systems. Thus far, the RNA polymerases II-based DNA-launched reverse genetics system using a bacterial artificial chromosome (BAC) as the backbone vector has been applied to rescue of multiple CoVs (Almazan et al., 2014). Basically, homogenous RNA transcripts are generated from transfected full-length cDNA clone in permissive cells to launch virus life cycle. Recently, our lab has just developed a novel and efficient method to assemble a full-length cDNA clone of measles virus (~16 kb) by using the GeneArt™ High-Order Genetic Assembly System, without the need for restriction endonucleases, which was used to rescue recombinant measles virus and the derived vaccine candidates (Wang et al., 2018). We employed this strategy successfully to assemble the 27.2-kb SeACoV genomic cDNA from the passage-10 virus (“SeACoV-p10”) by a single step ligation of 15 overlapping fragments into a BAC expression vector, resulting in a full-length cDNA clone of SeACoV named pSEA (Fig. 2 A). The SeACoV genomic cDNA cassette on pSEA was engineered with a cytomegalovirus (CMV) promoter and a hepatitis delta virus ribozyme (HDVRz) followed by a bovine growth hormone polyadenylation and termination sequences (BGH) at both termini, respectively. In addition, two silent mutations (A24222T and G24223C) in ORF3 were introduced in pSEA as a genetic marker to distinguish the parental virus SeACoV-p10 (Fig. 2A).
Fig. 2.
Construction and rescue of a full-length cDNA clone of SeACoV-p10. (A) Organization of the SeACoV genome structure and location of two unique nucleotide changes (nt 24222–24223; red star) in ORF3 gene is shown. The numbers under the scale bar indicate distances from the 5’ end. Fifteen DNA fragments, represented by turquoise bars, were amplified by PCR from the respective plasmid DNA clones containing the consensus SeACoV-p10 genomic sequence and assembled into a full-length cDNA clone, pSEA, by using the GeneArt™ High-Order Genetic Assembly System (Invitrogen). The names of each fragment are indicated. A cytomegalovirus promoter (pCMV; open triangle) was engineered at the 5’ end of the genomic cDNA, whereas a hepatitis delta virus ribozyme (HDVRz) followed by a bovine growth hormone polyadenylation and termination sequences (BGH; indicated by a “stop” symbol) were engineered at the 3’ end. (B) Rescue of SeACoV in Vero cells by co-transfection with the plasmids pSEA and pRK-N. The supernatants from the transfected cells were passaged onto fresh Vero cells, which were subsequently examined for CPE by direct observation (the first panel). Detection of expression of four SeACoV proteins using the available four pAbs was conducted at 48 hpi by IFA (the rest panels). Magnification = 200×. (C) Sequencing results of the region containing the genetic marker (AG→TC; boxed by dashed lines) by RT-PCR amplification of extracellular and intracellular viral RNA extracted from rSeACoV in comparison with that of SeACoV-p10. The corresponding amino acids are shown below. (D) Electron microscope image of the purified rSeACoV virions (by ultracentrifugation) using phosphotungstic acid negative staining. Bar = 100 nm. (E) Comparison of growth kinetics between rSeACoV and the parental SeACoV-p10 in Vero cells. Cells were infected in triplicate with virus at a MOI = 0.1. Cells were harvested at 2, 6, 12, 24, 36, 48, 60 and 72 hpi, and virus titers (TCID50/ml) were determined in triplicate on Vero cells.
BHK-21 cells were co-transfected with pSEA and a helper plasmid expressing the N protein (pRK-N) in order to recover the infectious SeACoV. Supernatants from transfected BHK-21 cells were inoculated onto fresh Vero cells at 2–3 days post-transfection. SeACoV-induced cytopathic effects (CPE) were visualized at 48 hpi in inoculated Vero cells; viral antigens were detected by IFA using anti-N, anti-M, anti-S1 or anti-Ac to stain cells, confirming the successful recovery of recombinant SeACoV (rSeACoV; Fig. 2B). A region containing the marker from extracellular and intracellular samples of extracted viral RNA was amplified and sequenced to determine the retention of the genetic markers in the rescued viruses. The two introduced mutations (TC) were still present in both samples, confirming that the rescued virus originated from the clone pSEA (Fig. 2C). There were no other mutations detected in genomic RNA of rSeACoV by genome re-sequencing.
We further assessed the morphology of the purified rSeACoV virions via ultracentrifugation followed by EM observation. The virus particles measured 100–120 nm in diameter with surface projections (Fig. 2D), consistent with our previous report of SeACoV isolation in Vero cells (Pan et al., 2017). The comparative growth kinetics of rSeACoV and the parental SeACoV-p10 were analyzed by infection of Vero cells with the respective virus at the same multiplicity of infection (MOI) of 0.1. The infectious virus titers were determined at different time points post-infection (2, 6, 12, 24, 36, 48, 60 and 72 hpi). The result showed that rSeACoV had the growth kinetics similar to the parental SeACoV-p10 (Fig. 2E). Of note, the maximal rates of SeACoV-p10 or rSeACoV production were from 6 to 12 hpi, suggesting that the exponential release of virus occurred before 6 hpi, which was consistent with detection of N, M, S and Ac expression as early as 4 hpi (Fig. 1B–E). The single-cycle growth of SeACoV in Vero cells is hence similar to those of mouse hepatitis virus (MHV), SARS-CoV and PDCoV, taking approximately 4–6 h (Prentice et al., 2004; Qin et al., 2019). These data collectively demonstrated that rSeACoV and its parental virus share the same virological features. To our knowledge, this is the first study describing a SeACoV/SADS-CoV infectious clone. Previous studies on CoV reverse genetics have shown that CoV accessory genes such as ORF3 [in TGEV (Sola et al., 2003), SARS-CoV (Yount et al., 2005), PEDV (Ji et al., 2018) or human CoV NL63 (Donaldson et al., 2008)] and the gene 7 [in TGEV (Ortego et al., 2003)] are dispensable for propagation in vitro. The corresponding genes, ORF3 and NS7a, are also present in the SeACoV genome; therefore, we will aim to generate reporter virus expressing luciferase or green fluorescent protein by replacement of ORF3 or NS7a with the reporter gene in future studies.
2.3. Identification of the leader-body junctions for all predicted subgenomic mRNAs of SeACoV
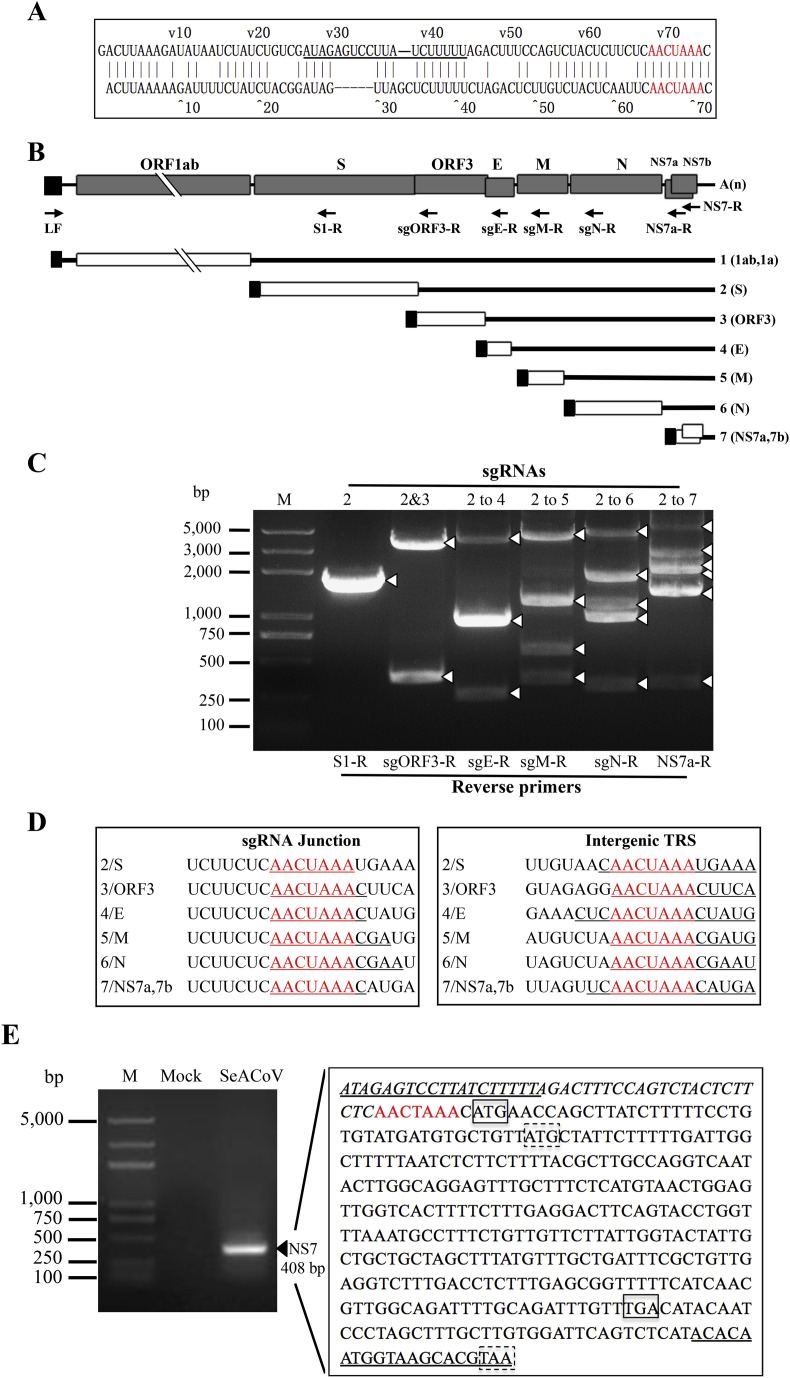
Coronaviruses can produce multiple sgRNAs are produced by discontinuous transcription. Each sgRNA contains a short 5’ leader sequence derived from the 5’-end of the genome and a body sequence from the 3’-poly (A) stretching to a position in the upstream of each ORF encoding a structural or accessory protein (Sola et al., 2015). The fusion site of the leader and body sequence in each sgRNA is termed transcription regulatory sequence (TRS). The SeACoV leader sequence of 75 nt from the 5’-end to the leader TRS was proposed according to the previous report (Lau et al., 2007); it was compared with that of another swine α-CoV, PEDV, indicating an identical leader TRS sequence (AACTAAA) shared by these two α-CoVs (Huang et al., 2013) (Fig. 3 A). The existence of all predicted subgenomic mRNAs (sgRNA; mRNA 2 to mRNA 7) for the expression of S, ORF3, E, M, N and NS7a was investigated further (Fig. 3B).
Fig. 3.
Identification of the leader-body junctions for all SeACoV subgenomic mRNAs. (A) Alignment of the leader sequences between SeACoV (upper line) and PEDV (lower line; US-MN strain, GenBank accession no. KF4687752). The sequence of the leader primer LF is underlined. Each of the leader transcription regulatory sequence (TRS) is marked in red. (B) Genomic and subgenomic organizations of SeACoV. The 5’-leader region is represented by a solid box, and the 3’-poly (A) tail is depicted by A(n). Positions of forward (LF) and reverse primers (S1-R, sgORF3-R, sgE-R, sgM-R, sgN-R and NS7a-R/NS7-R) used for PCR amplification of distinct subgenomic mRNAs (sgRNAs) are indicated by arrows under the genome. The seven small black boxes at the 5’ ends of the genomic RNA (gRNA) and sgRNAs depict the common leader sequence. Genomic and subgenomic RNA numbers (1 for gRNA and 2 to 7 for sgRNAs) are also indicated. (C) Detection of SeACoV sgRNAs by RT-PCR. Different combinations of the forward primer (LF) and one of the six reverse primers (indicated below each lane) were used. The bands representing RT-PCR products of specific SeACoV sgRNAs are marked with white arrowheads. The numbers above each lane represent specific sgRNAs amplified with the corresponding reverse primers. Lane M: DNA markers. (D) Leader-body fusion sites of sgRNAs and their corresponding intergenic sequences. The TRS is indicated in red. The junction sequences in sgRNAs (left panel) and the body sequences fused with the 5’ leader (right panel) are underlined. (E) Detection of the unique SeACoV NS7 sgRNA (arrowhead) by RT-PCR (left panel) and further determination of the sequence containing the leader-body junction site (right panel). The leader sequence is italicized, with the forward primer LF underlined. The body TRS is shown in red. The putative start and stop codons of NS7a and NS7b are boxed by solid and dashed lines, respectively. The binding site for the reverse primer NS7-R used in RT-PCR is also underlined.
The leader-body junctions and surrounding regions of all of the putative sgRNAs were amplified by RT-PCR. Each of the combination of the forward primer (LF) and one of the six reverse primers (S1-R, sgORF3-R, sgE-R, sgM-R, sgN-R and NS7a-R) amplified at least one major band of the expected size by agarose gel electrophoresis analysis (Fig. 3C). The appearance of multiple PCR bands was in line with what was expected, since except for the primers LF and S1-R, the other primer combinations could produce larger PCR fragments that correspond to the upstream-larger sgRNAs. For examples, the primer sgN-R, intended to amplify the leader-body fusion site of mRNA 6, could also amplify those of mRNAs 2 to 5, resulting in detection of five bands (Fig. 3C). Sequencing of individual PCR fragments confirmed that the leader-body junction sequences of sgRNAs are identical to the conserved core elements in the intergenic TRS (Fig. 3D).
We also noticed that both ORFs of NS7a and NS7b are connected with a body TRS in the upstream, implying a bicistronic mRNA encoding NS7a and NS7b (Fig. 3E). Since amplification with the reverse primer NS7a-R could not cover the entire NS7b, we next determined whether a potential NS7b sgRNA is present using the leader primer LF and a new reverse primer NS7-R corresponding to the 3’-end of ORF7b by RT-PCR. A single band of approximately 400-bp was amplified by optimizing the PCR condition and detected by agarose gel electrophoresis analysis; the other smaller bands were not found (Fig. 3E). Sequence analysis revealed that the TRS for this bicistronic sgRNA NS7 was exactly AACUAAA and one nt upstream of the AUG start codon of NS7a, which is consistent with the prediction (Fig. 3E). We further expressed and purified the complete NS7a or NS7b gene in the bacteria. Both products were found in the inclusion bodies. However, the resulting anti-NS7a or anti-NS7b pAb did not react with any antigens in SeACoV-infected cells by IFA and western blot analysis (data not shown) in contrast to the four working SeACoV pAbs. This suggests that NS7a and NS7b are either, not highly antigenic or the denatured antigens used to generate pAbs destroy the native protein structure. Development of monoclonal antibodies against NS7a and NS7b used for experimental validation of the existence of two expression products at the protein level is underway.
2.4. Ultrastructural changes in cells infected with SeACoV
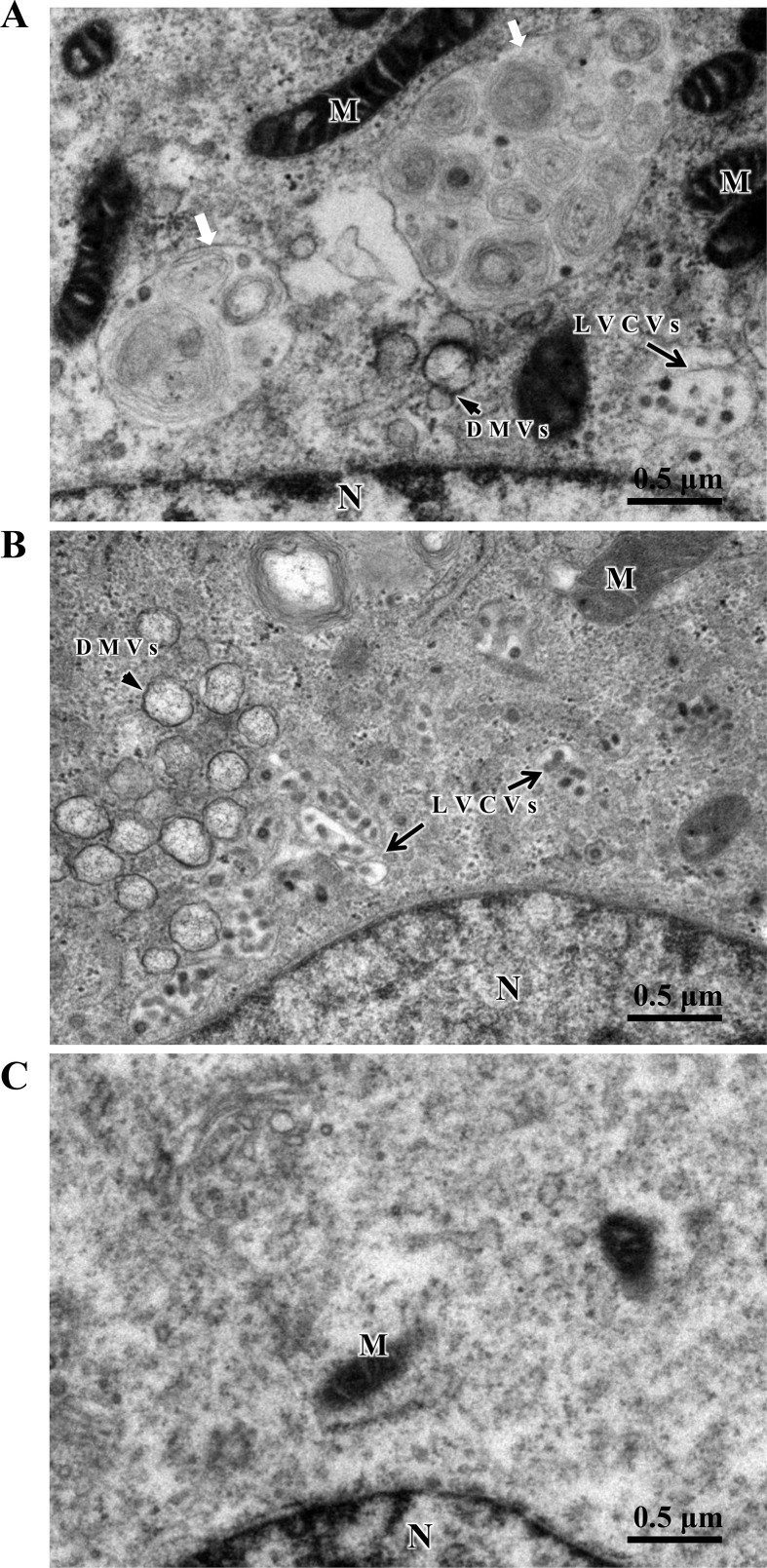
A number of studies on ultrastructural characterization of CoV-infected cells in vitro have demonstrated the presence of altered membrane architectures such as the double-membrane vesicles (DMVs), the large virion-containing vacuoles (LVCVs) and the phagosome-like vacuoles during CoV replication and morphogenesis (Goldsmith et al., 2004; Gosert et al., 2002; Qin et al., 2019; Salanueva et al., 1999; V'Kovski et al., 2015). DMVs are membrane structures where viral genomic RNA is recognized by the host cell machinery and translated into non-structural proteins (ORF1ab), assembling into viral replication–transcription complexes (Gosert et al., 2002), whereas LVCVs are large circular organelles that are thought to originate from Golgi compartments expanding to accommodate numerous precursor virions (Ulasli et al., 2010). The other type of membrane structure usually seen is phagosome-like vacuoles or lysosomes containing endoplasmic reticulum (ER), small vesicles, damaged mitochondrion and other vesicles. These conserved structures were also observed directly under an electron microscope (EM) in SeACoV-infected Vero cells (Fig. 4 A; 24 hpi) but not in uninfected cells (Fig. 4C). Of note, time course analysis of Nsp3 detection in Fig. 1C likely indicated corresponding locations of the DMVs.
Fig. 4.
EM observation of in vitro SeACoV infection in comparison with PEDV infection. Vero cells were infected with SeACoV (A) or PEDV (B) or mock-infected (C) at 24 hpi. CoV-induced cellular ultrastructural changes, including double-membrane vesicles (DMVs) (arrowheads), large virion-containing vacuoles (LVCVs) (black arrows), and phagosome-like vacuoles or lysosomes containing endoplasmic reticulum and other vesicles (white arrows) were visible in the cytoplasm. N: Nucleus. M: mitochondria. Bar = 0.5 μm.
Since infection of Vero cells with either SeACoV or PEDV resulted in indistinguishably cytopathic phenotype, i.e., syncytia formation (Pan et al., 2017), the ultrastructural changes in PEDV-infected Vero cells (at the same MOI of 0.1) were examined under EM for comparison of possibly morphological differences. Interestingly, PEDV appeared to induce a higher number of DMVs and LVCVs in large clusters surrounding the nucleus at 24 hpi and thereafter (Fig. 4B). A previous study on qualitative and quantitative ultrastructural analysis of membrane rearrangements induced by MHV proposed that CoV RNA synthesis is dictated by the number of DMVs, whereas an increasing production of viral particles is accommodated by LVCVs from expanding of ER-Golgi intermediate compartment (ERGIC)/Golgi compartments (Ulasli et al., 2010). It will be interesting to investigate whether synthesis of PEDV/SeACoV RNA and assembly of PEDV/SeACoV virions are correlated with the level of ultrastructural changes in the future.
3. Conclusions
In summary, we generated rabbit antisera against four of the SeACoV structural and nonstructural proteins and validated their reactivity and use of time course analysis of viral protein expression in SeACoV-infected Vero cells. Furthermore, we established a DNA-launched reverse genetics system for SeACoV and rescued the recombinant virus with a unique genetic marker in cultured cells. Recombinant SeACoV had similar growth kinetics to the parental virus. The single-cycle growth of SeACoV in Vero cells was determined to take approximately 4–6 h. By RT-PCR analysis, we experimentally identified all proposed SeACoV sgRNAs containing the leader-body junction sites. Among six sgRNAs, a bicistronic mRNA 7 was utilized by the accessory NS7a and NS7b genes. Finally, we characterized the cellular ultrastructural changes induced by SeACoV infection in vitro. Our study develops essential research tools and establishes the basic characteristics of SeACoV that will facilitate future studies on understanding the molecular mechanisms of SeACoV replication and pathogenicity.
4. Materials and methods
4.1. Cell lines and virus stocks
A monkey kidney cell line Vero (ATCC CCL-81) and a baby hamster kidney fibroblast cell line, BHK-21 (ATCC CCL-10) were grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics at 37 °C, respectively. The SeACoV isolate CH/GD-01/2017 at the passage 10 (p10) used in this study (Pan et al., 2017) was cultured in Vero cells. The virus titers were determined by endpoint dilutions as 50% tissue culture infective dose (TCID50) on Vero cells. The control virus PEDV (ZJU/G2/2013 strain; GenBank accession no. KU558701) was also cultured in Vero cells as described earlier (Ji et al., 2018; Qin et al., 2017).
4.2. Transmission electron microscopy (TEM)
Vero cells infected by the SeACoV or PEDV (at 24 h postinoculation, hpi) were fixed with 2.5% glutaraldehyde in phosphate buffer (0.1 M, pH 7.0) and 1% OsO4 in phosphate. Ultrathin sections were prepared as described previously (Qin et al., 2019), stained by uranyl acetate and alkaline lead citrate for 5–10 min, and observed using a Hitachi Model H-7650 TEM.
4.3. Generation of SeACoV polyclonal antibodies
Polyclonal antibodies (pAb) against the spike subunit 1 (anti-S1), membrane (anti-M), nucleocapsid (anti-N) and the nonstructural protein 3 (nsp3) acidic domain (anti-Ac) of SeACoV were produced in rabbits. For generation of anti-M pAb, prediction of transmembrane helices of the SeACoV M protein was first performed using the TMpred software (https://embnet.vital-it.ch/software/TMPRED_form.html). The M protein antigenic peptide was predicted as “CSDNLTENDRLLHLV”, and synthesized by Hua-An Biotechnology Co., Ltd (Hangzhou, China). This peptide was purified and used to immunize two New Zealand white rabbits and antiserum was harvested at 55 days post-immunization (DPI). Anti-S1, anti-N and anti-Ac pAbs of SeACoV were prepared in-house. Briefly, full-length N (1128 nt, 379 aa, ~42 kDa) or Ac (435 nt, 145 aa, ~16 kDa) of SeACoV were expressed with a six-histidine tag in Escherichia coli according to methods described previously (Huang et al., 2011), whereas SeACoV-S1 (1638 nt, 174 aa, ~62 kDa) with a six-histidine tag was expressed by baculovirus system in SF9 insect cells as described previously (Wang et al., 2017a). The purified proteins were used to immunize rabbits, and antisera were harvested at 55 DPI, respectively.
4.4. Analysis of the leader-body junctions of SeACoV subgenomic mRNAs
Total RNA from SeACoV-infected Vero cell was extracted using Trizol reagent (Invitrogen) and then reverse-transcribed with a SuperScript II reverse transcriptase (Invitrogen) using oligo-dT (Promega) as the reverse primer according to the manufacturer's instructions. The forward primer LF (5’-ATAGAGTCCTTATCTTTTT-3’) and six gene specific reverse primers, S1-R (5’-CAATGGCATTTCTGTGTACCTCTC-3’), sgORF3-R (5’-AGTAATCTGCTTACAACAGC-3’), sgE-R (5’-AGACATTAATTATGGGGCAT-3’), sgM-R (5’-GTTCGCGTTCTGCGATAAAG-3’), sgN-R (5’-ATCTGCGTGAGGACCAGTAC-3’), NS7a-R (5’-AATCTGCAAAATCTGCCAAC-3’), were designed for amplification of all SeACoV subgenomic mRNAs (Fig. 3A) from the obtained cDNA with a Taq DNA polymerase (Transgen, Beijing, China) in a total volume of 50 μl by PCR. The PCR condition was set at 35 cycles of 94 °C for 30 s, 50 °C for 30 s, 72 °C for 3 min with an initial denaturing of the template DNA at 94 °C for 3 min and a final extension at 72 °C for 5 min. The resulting PCR fragments were analyzed on a 1% agarose gel (Fig. 3B) and then subcloned into a pEASY-T1 vector (Transgen, Beijing, China) followed by Sanger sequencing. For amplification of the subgenomic mRNA 7 containing the entire NS7a/NS7b, the reverse primer NS7-R (5’-TTACGTGCTTACCATTGTGT-3’) was used, and the PCR extension time was shortened to 45 s. Analysis of DNA sequences was performed using the Lasergene Package (DNASTAR Inc., Madison, WI).
4.5. Construction of a DNA-launched SeACoV full-length cDNA clone
The expression vector, designated as pSB2μ, used to construct a full-length SeACoV cDNA clone, was based on a BAC backbone vector pSMART-BAC-BamHI (CopyRight v2.0 BAC Cloning Kits, Lucigen). This pSMART-BAC vector was modified to insert a yeast replication origin (2μ) from the plasmid pYES2 (Invitrogen), a cytomegalovirus (CMV) promoter from the plasmid pcDNA3 (Invitrogen), a hepatitis delta virus ribozyme (HDVRz) sequence from a PRRSV (porcine reproductive and respiratory syndrome virus) infectious clone pTri-53Rz-PGXG (Huang et al., 2009), and a bovine growth hormone (BGH) polyadenylation and terminator from the plasmid pcDNA3 (Invitrogen) by several rounds of amplification and “In-fusion” PCR according to our previous publication (Wang et al., 2018). The primer sequences and approaches used in the PCR assays are available upon request.
The full-length consensus sequence of SeACoV-p10 (27,155 nt) was determined as described previously (Pan et al., 2017). Briefly, a total of 15 overlapping fragments covering the entire genome was amplified by RT-PCR using the Q5 High-Fidelity 2×Master Mix (New England Biolabs, USA). PCR products were purified and cloned into a pEASY-Blunt vector (Transgen, Beijing, China). For each amplicon, five individual clones were sequenced to validate the consensus sequence.
To create a 2-nt genetic marker on the ORF3 gene of the infectious clone, two point mutations, A to T, and G to C at nucleotide positions 24222–24223, corresponding to the SeACoV-p10 genome, were generated on the fragment S-2 by fusion PCR (Fig. 2A). Subsequently, all 14 fragments identical to the consensus sequence together with the mutated S-2 fragment were re-amplified from each clone with primers listed in Table 1 . It was then assembled into the expression vector (pSB2μ) between the CMV promoter and the HDVRz+BGH element, using the GeneArt™ High-Order Genetic Assembly System according to the manufacturer's manual, to create a DNA-launched SeACoV full-length cDNA clone, pSEA (Fig. 2). The plasmid pSEA is available to the research community upon request. The sequence encoding the full-length SeACoV nucleocapsid gene was amplified and inserted into a pRK5 eukaryotic expression vector containing a FLAG-tag at its C terminus to construct pRK-N-FLAG as a helper plasmid for rescuing the infectious clone.
Table 1.
Primers used for amplifications of the SeACoV genomic fragments.
| Primer ID | Sequence (5’ to 3’) | Positiona | Fragment |
|---|---|---|---|
| F1–F | ATAAGCAGAGCTCGTTTAGTGAACCGT GACTTAAAGATATAA | 1–15 | F1 |
| F1-R | GTCATCACAGAGGGCAGTAAAGC | 1702–1724 | |
| F2–F2 | GCATTCAGTGTTGTTGACGGC | 1605–1625 | F2 |
| R2-R2 | GCACCGCTAAGTTCTTCGAAG | 3710–3730 | |
| F3–F | GATGTTGCACATTGTTTAGAGGTA | 3624–3647 | F3 |
| F3-R | CGAACTTGTTCCACAAATCCTCC | 5423–5445 | |
| F4–F2 | CAATTGCTGGGTTAATGCGAC | 5354–5374 | F4 |
| F4-R2 | AAATGCCTTATGCAAAGCACC | 7224–7244 | |
| F5–F | GTTTATCTCTCACAACTTCTGTGT | 7143–7166 | F5 |
| F5-R | ATTGATAAGACGCTCATAAGAAC | 8905–8927 | |
| F6–F2 | GCCATGGTGGTTGCTTACAT | 8750–8769 | F6 |
| F6-R2 | GCACAACATTGGCACACTTAAG | 10878–10899 | |
| F7–F | GTCCTTTTGACTCTGTATTACTTAG | 10783–10807 | F7 |
| F7-R | TTTGTTATACATGGACTGCTCGT | 12651–12673 | |
| F8–F | TAAGCATGATGCCTTCTTTGTTATT | 12598–12622 | F8 |
| F8-R | TTTGAACCGAGAACCATAGCAGC | 14309–14331 | |
| F9–F | TCCTAAATGTGATAGAGCTATGCCT | 14266–14290 | F9 |
| F9-R | AATAATACGTGAGCATCTGTCTA | 16188–16210 | |
| F10–F2 | GTGGCAAATCACATTGTGTT | 16065–16084 | F10 |
| F10-R2 | ACCATTAACGCCTTCTAGTG | 18204–18223 | |
| F11–F3 | GTGCCTATTTTGGAACTGTAATG | 18118–18140 | F11 |
| F11-R3 | CATAATAGTGGAATTGCGCC | 20274–20293 | |
| S-1-F1 | CGCTATGGCTGTTAAGATTACCG | 20071–20093 | S-1 |
| S-1-R1 | CAATGGCATTTCTGTGTACCTCTC | 22165–22188 | |
| S-2-F1 | GCTAGTTACGCACCTAATGACACC | 22021–22044 | S-2 |
| S-2-R1 | CATTAGGGTCAAGTTTAGCAGCTC | 23926–23949 | |
| F14–F | TTTTGCTAATGTCATTGCCGTTTCA | 23403–23427 | F14 |
| F14-R | GGCGACAGTCACAAATTGCGGTA | 25276–25298 | |
| F15–F | ATGGCATCAGAATTGCTACTGGTGT | 25237–25261 | F15 |
| F15-R | TTTTTTTTTTTTTTTTTTTTTTTTTTTGTGTATCACTGTCAA | 27141–27182 |
Positions correspond to the SeACoV CH/GD-01/2017/P2 strain (GenBank accession no. MF370205). Nucleotides overlapping with the vector (pSB2μ) sequences in primers F1 and F15 are underlined.
4.6. Transfection and rescue of recombinant SeACoV
The plasmid pSEA was purified from the E. coli DH10B strain using QIAprep Miniprep Kit (Qiagen) and quantified by a NanoDrop spectrophotometry. BHK-21 cells were seeded at 2×105 per well of a six-well plate and grown until 60–70% confluence before transfection. One microgram each of pSEA and pRK-N-FLAG were co-transfected into the cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer's protocol. Transfected cells were cultured for 2–3 days. The supernatant was collected and passaged onto fresh Vero cells on 12-well plates and cultured for 3 days before the detection of viral protein expression by IFA. The recombinant SeACoV rescued from the pSEA infectious clone was named rSeACoV. The rSeACoV titers were determined by endpoint dilutions as TCID50. Viral particles in the supernatants from rSeACoV-infected cell cultures were negatively stained and examined under TEM. A 1.5-kb DNA fragment harboring the introduced mutations in the ORF3 gene was amplified by RT-PCR using primers TF21 (5’-TACTGGATGTTGTGGCATGT-3’) and TR21 (5’-TTCCACTTAAAATCGTCAGA-3’). The amplicons were sequenced to affirm that rSeACoV contained the desired mutations.
4.7. Immunofluorescence assay (IFA) and western blot analysis
SeACoV-infected or rSeACoV-infected cells were washed twice with PBS, fixed with 4% paraformaldehyde in PBS for 20 min and then permeabilized with 0.5% Triton X-100 for 10 min. Anti-N, anti-M, anti-S1 or anti-Ac pAb, each at a 1:1000 dilution in PBS, was added over the cells and incubated for 1 h at 37 °C. Cells were then washed thrice with PBS and Alexa Fluor 488-labeled goat anti-rabbit IgG (Thermo Fisher Scientific) at a 1:1000 dilution was then added. After 30 min of incubation at 37 °C, the cells were again washed thrice with PBS followed by 4',6-diamidino-2-phenylindole (DAPI) staining, and were visualized under a fluorescence microscope (DMI3000B, Leica, Germany). For time course analysis of detection of N, M, S1 or Ac, fluorescent images were obtained with a confocal laser scanning microscope (Fluoviewver FV1000-IX81; Olympus, Japan).
For western blot analysis, SeACoV-infected cells were lysed in lysis buffer (25 mM Tris-HCl, 200 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 25 mM β-glycerophosphate, 1% NP40, and protease cocktail [Biotool, Houston, TX]). Samples were resolved on SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membrane that was subsequently blocked with Tris-buffered saline (TBS) containing 3% bovine serum albumin (BSA) overnight at 4 °C. Proteins were detected using the anti-N pAb or anti-M pAb at 1:1000 dilution followed by incubation with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:5000 dilution; Thermo Fisher Scientific).
4.8. Nucleotide sequence accession number
The consensus sequence of SeACoV-p10 used for construction of the infectious clone has been deposited in GenBank under accession no. MK977618.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFD0500102), the National Natural Science Foundation of China (31872488), and the Fundamental Research Funds for the Central Universities of China (2019FZA6014). We thank the staff in the Shared Experimental Platform for Core Instruments, College of Animal Science, Zhejiang University for assistance with analysis of confocal microscopy.
References
- Almazan F., Sola I., Zuniga S., Marquez-Jurado S., Morales L., Becares M., Enjuanes L. Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 2014;189:262–270. doi: 10.1016/j.virusres.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Yount B., Sims A.C., Burkett S., Pickles R.J., Baric R.S. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J. Virol. 2008;82:11948–11957. doi: 10.1128/JVI.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Ren J., Hong Y., Liu X., Zhao Y., Wang D., Peng G., Xiao S. Porcine deltacoronavirus accessory protein NS6 antagonizes interferon beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J. Virol. 2018;92 doi: 10.1128/JVI.00712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L., Li J., Zhou Q., Xu Z., Chen L., Zhang Y., Xue C., Wen Z., Cao Y. A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 2017;23 doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4 doi: 10.1128/mBio.00737-13. e00737-00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Fang Y., Meng X.J. Identification and characterization of a porcine monocytic cell line supporting porcine reproductive and respiratory syndrome virus (PRRSV) replication and progeny virion production by using an improved DNA-launched PRRSV reverse genetics system. Virus Res. 2009;145:1–8. doi: 10.1016/j.virusres.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Huang Y.W., Harrall K.K., Dryman B.A., Beach N.M., Kenney S.P., Opriessnig T., Vaughn E.M., Roof M.B., Meng X.J. Expression of the putative ORF1 capsid protein of Torque teno sus virus 2 (TTSuV2) and development of Western blot and ELISA serodiagnostic assays: correlation between TTSuV2 viral load and IgG antibody level in pigs. Virus Res. 2011;158:79–88. doi: 10.1016/j.virusres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Ji C.M., Wang B., Zhou J., Huang Y.W. Aminopeptidase-N-independent entry of porcine epidemic diarrhea virus into Vero or porcine small intestine epithelial cells. Virology. 2018;517:16–23. doi: 10.1016/j.virol.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Wang M., Lam C.S., Xu H., Guo R., Chan K.H., Zheng B.J., Yuen K.Y. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K.L., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Sola I., Almazan F., Ceriani J.E., Riquelme C., Balasch M., Plana J., Enjuanes L. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology. 2003;308:13–22. doi: 10.1016/S0042-6822(02)00096-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y.L., Wang L., Wang D., Song Y., Zhang X., Huang Y.W. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Du E.Z., Luo W.T., Yang Y.L., Zhang Y.Q., Wang B., Huang Y.W. Characteristics of the life cycle of porcine deltacoronavirus (PDCoV) in vitro: replication kinetics, cellular ultrastructure and virion morphology, and evidence of inducing autophagy. Viruses-Basel. 2019;11:455. doi: 10.3390/v11050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Li H., Wang J.W., Wang B., Xie R.H., Xu H., Zhao L.Y., Li L., Pan Y., Song Y., Huang Y.W. Genetic and pathogenic characterization of a novel reassortant mammalian orthoreovirus 3 (MRV3) from a diarrheic piglet and seroepidemiological survey of MRV3 in diarrheic pigs from east China. Vet. Microbiol. 2017;208:126–136. doi: 10.1016/j.vetmic.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanueva I.J., Carrascosa J.L., Risco C. Structural maturation of the transmissible gastroenteritis coronavirus. J. Virol. 1999;73:7952–7964. doi: 10.1128/jvi.73.10.7952-7964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazan F., Zuniga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Alonso S., Zuniga S., Balasch M., Plana-Duran J., Enjuanes L. Engineering the transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity. J. Virol. 2003;77:4357–4369. doi: 10.1128/JVI.77.7.4357-4369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoleridis T., Chappell J.G., Onianwa O., Marston D.A., Fooks A.R., Monchatre-Leroy E., Umhang G., Muller M.A., Drexler J.F., Drosten C., Tarlinton R.E., McClure C.P., Holmes E.C., Ball J.K. Shared common ancestry of rodent alphacoronaviruses sampled globally. Viruses. 2019;11 doi: 10.3390/v11020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulasli M., Verheije M.H., de Haan C.A., Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010;12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P., Al-Mulla H., Thiel V., Neuman B.W. New insights on the role of paired membrane structures in coronavirus replication. Virus Res. 2015;202:33–40. doi: 10.1016/j.virusres.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Hagemeijer M.C., Ulasli M., Reggiori F., Rottier P.J., Masters P.S., de Haan C.A. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84:11575–11579. doi: 10.1128/JVI.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lei X., Qin P., Zhao P., Wang B., Wang Y., Li Y., Jin H., Li L., Huang Y.W. Development and application of real-time RT-PCR and S1 protein-based indirect ELISA for porcine deltacoronavirus. Sheng wu gong cheng xue bao = Chin. J. Biotechnol. 2017;33:1265–1275. doi: 10.13345/j.cjb.170119. [DOI] [PubMed] [Google Scholar]
- Wang Q., Vlasova A.N., Kenney S.P., Saif L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X.D., Guo W.P., Zhou R.H., Wang M.R., Wang C.Q., Ge S., Mei S.H., Li M.H., Shi M., Holmes E.C., Zhang Y.Z. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X.D., Liao Y., Guan X.Q., Guo W.P., Xing J.G., Holmes E.C., Zhang Y.Z. Discovery of a highly divergent coronavirus in the asian house shrew from China illuminates the origin of the alphacoronaviruses. J. Virol. 2017;91 doi: 10.1128/JVI.00764-17. e00764-00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu R., Lu M., Yang Y., Zhou D., Hao X., Zhou D., Wang B., Li J., Huang Y.W., Zhao Z. Enhancement of safety and immunogenicity of the Chinese Hu191 measles virus vaccine by alteration of the S-adenosylmethionine (SAM) binding site in the large polymerase protein. Virology. 2018;518:210–220. doi: 10.1016/j.virol.2018.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.C., Zhang Y., Gong L., Huang L.C., Lin Y., Qin J.R., Du Y.P., Zhou Q.F., Xue C.Y., Cao Y.C. Isolation and characterization of a highly pathogenic strain of Porcine enteric alphacoronavirus causing watery diarrhoea and high mortality in newborn piglets. Transbound Emerg. Dis. 2019;66:119–130. doi: 10.1111/tbed.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Nabar N.R., Shi C.S., Kamenyeva O., Xiao X., Hwang I.Y., Wang M., Kehrl J.H. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Sun Y., Lan T., Wu R.T., Chen J.W., Wu Z.X., Xie Q.M., Zhang X.B., Ma J.Y. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound Emerg. Dis. 2019;66:687–695. doi: 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]