Abstract
Circulating molecules that are released into the circulation in response to specific stimuli are considered potential biomarkers for physiological or pathological processes. Their effective usefulness as biomarkers resides in their stability and high availability in all the biological fluids, combined with the limited invasiveness of intervention. Among the circulating molecules, miRNAs represent a novel class of biomarkers as they possess all the required characteristics such as sensitivity, predictivity, specificity, robustness, translatability, and noninvasiveness.
miRNAs are small non-coding RNAs, that act as inhibitors of protein translation, and intervene in the complex network of the post-transcriptional mechanisms finely regulating gene expression.
The emerging role of miRNAs as potential biomarkers for clinical applications (e.g., cancer and cardiovascular diseases diagnosis and prediction, musculoskeletal disease diagnosis and bone fracture risk prediction), however, requires the standardization of miRNA processing, from sample collection and sample storage, to RNA isolation, RNA reverse-transcription, and data analyses. Normalization is one of the most controversial issues related to quantitative Real-Time PCR data analysis since no universally accepted normalization strategies and reference genes exist, even more importantly, for circulating miRNA quantification. As it is widely demonstrated that the choice of different normalization strategies influences the results of gene expression analysis, it is important to select the most appropriate normalizers for each experimental set. This review discloses on the different strategies adopted in RT-qPCR miRNA normalization and the concerning issues to highlight on the need of a universally accepted methodology to make comparable the results produced by different studies.
Keywords: Biomarkers, microRNA, Normalization, Reference genes, RT-qPCR
1. Introduction
MicroRNAs (miRNAs), i.e., small ribonucleotides acting as inhibitors of protein translation, intervene in the complex network of the post-transcriptional mechanisms finely regulating gene expression. Although produced intracellularly and, physiologically, at very low concentrations, they are released into the circulation and, hence, they can be measured as classical biomarkers. However, contrarily to these latter, miRNAs can be considered earlier and more sensitive biomarkers of either a physiological response or a pathological process [1].
Besides their undoubtable usefulness, a number of pre-analytical, analytical, and post-analytical concerns still limits their clinical application. This article is aimed at giving a complete overview of the main available knowledge about the post-analytical processing and data analysis and, particularly, the importance of the normalization strategy for circulating miRNA measurement.
1.1. Micro RNAs: Physiological Meaning and Biomarker Roles
A first hypothesis about regulatory roles for specific RNA molecules was proposed in 1961 by Jacob and Monod [2] but only in 1993 Ambros detected, in Caenorabditis elegans, a small single stranded non-coding RNA, lin-4, with regulatory functions on gene expression and, particularly, on the expression of the lin-14 gene, throughout an antisense interaction [3]. From that time, miRNAs and their regulatory systems have been discovered in all the kingdoms of the living world: prokaryotes [4], protists [5], fungi [6], plants [7] both vertebrates and invertebrates animals [8], and also viruses [9]. The impressive growing number of researches and discoveries in this field has led to the creation of a continuously updated database of miRNAs: www.mirbase.org (release #22, March 2018). At this moment, this database collects over 38.589 miRNA entries (1.800 in humans), providing universal nomenclature, sequences, predicted target genes, and references.
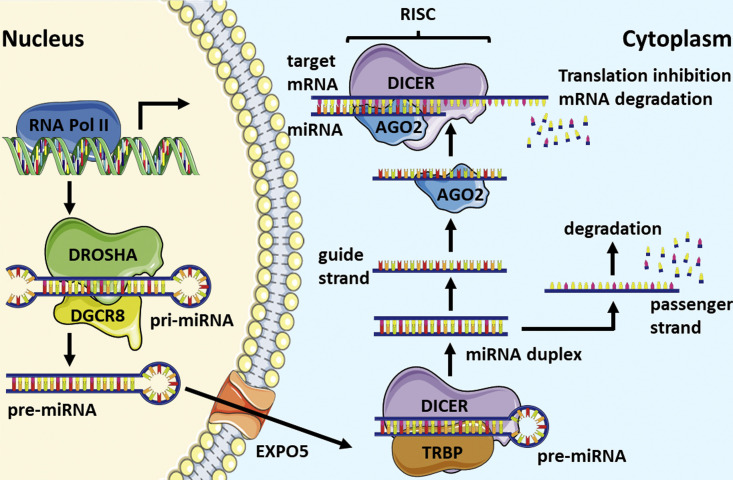
DNA sequences encoding for miRNAs are generally located into non-coding regions, as well as in introns and untranslated regions (UTRs) of protein-encoding genes. As for the canonical coding regions, miRNA-encoding DNA sequences are epigenetically regulated by methylation and deacetylation [10]. miRNA-encoding sequences are transcribed by RNA polymerase II into a pri-miRNA which is cleaved into a pre-miRNA, by the nuclear enzyme Drosha, in order to eliminate the stem loop-flanking sequences. Pre-miRNA is exported by exportin 5 to the cytoplasm where, following the action of the enzyme Dicer, a double strand mature miRNA is released. Although, generally, one of the two strands of the duplex (passenger or star strand) generated by the Dicer is degraded, in some cases also the passenger or star strand acts as a miRNA. The two miRNAs generated by the same transcript are called with either the 3p or 5p suffix depending on the molecular ending of origin of the looped duplex. The mature miRNA is incorporated within the RNA-induced silencing complex (RISC), together with Argonaut (Ago)-2 protein, that targets the mRNAs. The “seed region,” responsible for the mRNA targeting, comprises the nucleotides from position two to seven. The double stranded RNA, raised from the interaction between the miRNA and the target mRNA, is degraded and, hence, the translation is inhibited. Thanks to the nonstringent complementarity and the short length of the seed region, each miRNA targets several (hundreds) mRNAs [11] (Fig. 1 ). The miRNA-dependent regulation of gene expression has a key physiological role as demonstrated by the fact that, in mice, the Dicer-1 knockout is lethal in the early embryo [12], [13] and it is essential in embryological development in humans [14].
Figure 1.
Biogenesis and function of miRNA.
miRNA genes are transcribed from noncoding inter- or intra-genic regions of RNA transcripts. The transcription is mediated by RNA polymerase II and give rise to a long pri-miRNA that is processed by the DROSHA–DGCR8 complex to form a 60 nucleotide-long precursor miRNAs (pre-miRNAs). EXPO5 mediates the export of the pre-miRNA to the cytoplasm where it is further processed by the ribonuclease DICER that trims the pre-miRNAs to form the 18–22 nucleotide-long mature miRNAs. The guide strand is incorporated into RISC involving DICER and AGO2 enzymes to target mRNAs to cause the repression of translation and/or its degradation. This figure was produced using Servier Medical Art, available at https://smart.servier.com/.
While several miRNAs show a cell/tissue-restricted expression profile, many others are more widely expressed. A recent atlas has illustrated the miRNA expression profile in 61 different tissues [15] and, moreover, according to other investigators, miRNAs are present in 12 different body fluids [16]. They are also relatively abundant in serum and plasma and, hence, several researches have investigated their possible use as biomarkers [17]. Indeed, abnormal expression of either a single miRNA or a group of miRNAs has been demonstrated in several disorders including (but not limited to) tumors, cardiovascular diseases, inflammatory/autoimmune diseases [18], [19]. In general, in the case of tumors, miRNAs are downregulated (since they are inhibitors of gene expression) compared to the nontumor counterparts and a few miRNAs can mark a specific neoplasia [20]. Parallel, changes in cell and/or circulating miRNAs mark the cellular response to physiological stimuli such as in the musculoskeletal [21] and myocardial [22] responses to physical activity.
The miRNome has a great diagnostic potential since it may be specific for any disease or any physiological change and, hence, the future implementation of miRNA analysis in routine settings is acknowledged. However, this implementation will require huge efforts in terms of optimization and harmonization of the pre-analytical, analytical, and post-analytical phases [23]. In order to be defined as a biomarker, a molecule might satisfy the following criteria: (1) measurability in a specific clinical setting, (2) validation by multiple studies, (3) direct impact on the medical decision [24]. In the case of miRNAs the criterion of measurability is satisfied: indeed, as stated, they can be detected in situ or in biofluids widening the clinical applicability [25]. On the contrary, validation and impact on medical decision are still affected by several issues mainly derived from the inconsistence of the data among different studies which is in turn due to a lack of standardization of pre-analytical (e.g., choice of sample matrix, samples handling and storage, lifestyle factors [1]), analytical (e.g., low concordance between different platforms [26], platform migration from early discovery to final commercialization [25]), and post-analytical phases (e.g., several data analysis software based on different algorithms, different normalization strategies [25], [27]). A big issue is just represented by the existence of several normalization strategies: miRNA levels in biofluids are expressed as the relative expression values rather than the absolute counts per volume; hence, by using different normalization strategies, different results (sometimes even definitively inconsistent) arise. The comparability of data among different labs, but also among different studies from the same lab, is, thus, seriously limited and this is due to the lack of any consensus over whether a priori defined reference genes might be used to normalize data [25], [27].
1.2. Overview on Present or Future Clinical Applications of miRNAs
Why is it so important to standardize the methods for miRNA determination? There is a huge scientific interest around this class of molecules and enormous research efforts have been spent on their investigation. In order to give an idea about the topicality of this subject it is sufficient to look at PubMed: by trivially typing the term “miRNA,” 70,405 records are displayed of which more than 90% (64,236) have been published since 2010 to today and about 50% (34,733) from 2015 to today. Behind the interest about the definition of their biological role, there is, obviously, the interest around their potential clinical implementation in the next future. This possible implementation is illustrated by some of the examples we report here.
In the field of infectious diseases, for example, the novel oligonucleotide anti-miRNA Miravirsen (anti-hsa-miR-122) was shown to be safe and well tolerated in HCV-infected patients in a phase IIa trials [28] and it is already in phase IIb trials suggesting that miRNA-based treatments may well become a reality; viral vaccines attenuated through incorporation of miRNA target sequences are at the preclinical stage; and miRNA biomarkers of infection hold promise [29].
In the field of metabolic diseases, and specifically in osteoporosis, several miRNAs have been associated with fracture risk and bone mineral density (BMD). Nine miRNAs (hsa-miR-21, hsa-miR-23a, hsa-miR-24, hsa-miR-93, hsa-miR-100, hsa-miR-122a, hsa-miR-124a, hsa-miR-125b, hsa-miR-148a) were found to be upregulated in the serum of patients with osteoporosis while six miRNAs (hsa-miR-21, hsa-miR-23a, hsa-miR-24, hsa-miR-25, hsa-miR-100, hsa-miR-125b) were upregulated in the bone tissue of osteoporotic patients [30]. Among the five miRNAs (hsa-miR-21, hsa-miR-23a, hsa-miR-24, hsa-miR-100, and hsa-miR-125b) upregulated in both serum and bone tissue, hsa-miR-122a-5p, hsa-miR-125b-5p, and hsa-miR-21-5p were identified as markers for osteoporotic fractures and hsa-miR-21-5p, hsa-miR-122-5p, and hsa-miR-125b-5p have been age-independently associated with osteoporotic bone fractures and serum cross-laps (CTx-I) concentrations [31]. hsa-miR-24-3p, hsa-miR-21-5p, hsa-miR-93-5p, hsa-miR-100-5p, and hsa-miR-125-5p have been negatively associated with BMD in osteoporotic patients [32].
The greatest progresses in miRNA-biomarker discovery have been done in oncology and cardiology. In a randomized phase II study of androgen deprivation combined with cixutumumab versus androgen deprivation alone in patients with new metastatic hormone-sensitive prostate cancer, baseline plasma hsa-miR-141, hsa-miR-200a, and hsa-miR-375 levels were associated with baseline circulating tumor cells count and baseline hsa-miR-375 was also associated with the trial endpoint of 28-week prostate-specific antigen (PSA) response [33]. Furthermore, Lin et al. demonstrated, in a phase II study in castration-resistant prostate cancer patients, that higher baseline levels of miRNAs belonging to the hsa-miR-200 family were associated with a shorter overall survival [34]. In a study exploring miRNAs as predictive biomarkers of the efficacy of peptide vaccines alone (phase I) and in combination with chemotherapy (phase II) in colorectal cancer, hsa-miR-125b-1 and hsa-miR-378a have been negatively associated with overall survival [35]. The predictive value of miRNAs in tumor cells and infiltrating immune cells for the efficacy of radiotherapy in combination with either 5-fluorouracil/cisplatin (CDDP-CRTX) or 5-fluorouracil/mitomycin C, in locally advanced head and neck squamous cell carcinoma, was evaluated in a phase III trial highlighting hsa-miR-200b and hsa-miR-155 as potential markers for personalized treatment selection of two standard regimens of chemoradiation [36]. In a large multicenter study investigating the effect of adding bevacizumab to standard chemotherapy in patients diagnosed with epithelial ovarian cancer, low levels of hsa-miR-200b, hsa-miR-1274A (tRNALys5), and hsa-miR-141 were significantly associated with better survival, and the level of hsa-miR-200c seemed predictive of effect of treatment with bevacizumab [37]. In a phase II clinical trial investigating the effects of eribulin in the treatment of metastatic soft tissue sarcoma, 26 specific miRNAs were predictive of response out of the 756 miRNA assessed in paraffin-embedded sections [38]. In their phase II study, Gagez and colleagues found that hsa-miR-125b and hsa-miR-532-3p predicted the efficiency of rituximab-mediated lymphodepletion in chronic lymphocytic leukemia patients [39].
In a clinical trial based on whole-genome miRNA sequencing on RNA extracted from whole blood of 199 patients with non-ST-segment elevation acute coronary syndrome, chronic heart-failure was associated with lower circulating levels of hsa-miR-3135b, hsa-miR-126-5p, hsa-miR-142-5p, and hsa-miR-144-5p, while an increasing acute coronary events risk score was inversely correlated with the levels of hsa-miR-3135b and positively correlated with the levels of hsa-miR-28-3p [40]. On the other hand, Xiao et al. demonstrated that hsa-miR-30d was significantly lower in patients with acute heart failure and this was associated with 1-year all causes mortality in these patients [41].
An important issue concerning the use of miRNAs as biomarkers is represented by their sensitivity and specificity profile in diagnostic applications. Sensitivity and specificity are two key parameters used to define the validity of a biomarker to specifically diagnose a condition, not only for confirming the presence of the disease but also to rule out the disease in healthy subjects. The two parameters are inversely correlated, and plotted as sensitivity versus 1-Specifity in a graph called receiver operating characteristic (ROC) curve, that represents an effective measure of accuracy of a specific biomarker [42]. Several studies focused on the potential clinical role of miRNAs for the diagnosis of a pathology defining the parameters of sensibility and specificity. For example, Ma et al. reported that has-miR-19b-3p and has-miR-29b-3p, selected from a panel of seven peripheral blood mononuclear cells (PBMC)-derived miRNAs and indicated as biomarkers for early detection of non-small cell lung cancer (NSLC), yield a sensitivity and specificity of 72.62% and 82.61%, respectively, in identifying NSLC. Furthermore, these same two miRNAs could be used for the early diagnosis of squamous cell lung carcinoma (SCC), a major type of NSCLC, with 80.00% sensitivity and 89.86% specificity [43]. In a study by Lu et al., plasma derived hsa-miR-10b was identified as a potential biomarker for oral cancer with a sensitivity of 94.40% and a specificity of 80.40% [44]. As for the classical biomarkers, the combination of multiple miRNAs or the combination of a miRNA with protein biomarkers, may increase the sensitivity and specificity of diagnosis and prediction of the studied condition. In supporting this notion, Xiong and co-workers have recently identified a panel of nine miRNAs allowing the early detection of breast cancer with a high diagnostic accuracy with a sensitivity of 98.70% and a specificity of 98.90% [45]. Seeliger et al. identified a set of nine miRNAs (hsa-miR-21, hsa-miR-23a, hsa-miR-24, hsa-miR-93, hsa-miR-100, hsa-miR-122a, hsa-miR-124a, hsa-miR-125b, and hsa-miR-148a) significantly upregulated in serum and six miRNAs (hsa-miR-21, hsa-miR-23a, hsa-miR-24, hsa-miR-25, hsa-miR-100, and hsa-miR-125b) significantly upregulated in bone tissues of patients with osteoporosis fractures compared to patients with non-osteoporotic fractures, with five miRNAs upregulated in both serum and bone tissues. These selected miRNAs could be used as biomarkers for diagnostic purposes for osteoporosis [30]. Combination of miRNA and protein biomarkers were also investigated to improve the differential diagnosis between pancreatic cancer, other gastrointestinal cancer, and benign pancreatic diseases (BPD). Yuan and colleagues demonstrated that the combination of hsa-miR-21, macrophage inhibitory cytokine-1 (MIC-1), and CA19-9 and of hsa-miR-25, MIC-1, and CA19-9, performed better, in terms of specificity, sensitivity and accuracy, in the detection of pancreatic cancer against benign pancreatic diseases compared to CA19-9 alone [46]. Additionally, Cheng et al. tested hsa-miR-141 as a potential biomarker for colorectal cancer and found that high levels of circulating hsa-miR-141 were associated to Stage IV colorectal cancer. The authors demonstrated that hsa-miR-141 combined with carcinoembryonic antigen (CEA), a widely used marker for colorectal cancer, identified additional Stage IV cases in the training cohort, indicating that the combination of the two biomarkers improve the accuracy of the staging of colorectal cancer with distant metastasis; furthermore, high level of plasma hsa-miR-141 alone were associated to poor prognosis [47]. Altogether these examples demonstrate that the use of a panel of miRNAs or the association of specific miRNAs with other classical protein biomarkers significantly increase the accuracy of the diagnosis.
2. RT-qPCR Normalization in Molecular Biology
RT-qPCR (reverse transcription quantitative polymerase chain reaction) is the most widely used method to quantify differences in the expression profile of target genes and it is the technique of choice due to its high sensitivity, specificity, and reproducibility. However, sample collection and preparation, RNA extraction and different reverse-transcription and PCR efficiencies can introduce many technical variables in the experimental steps that can affect the quantification leading to altered results [48]. It is important to choose an accurate normalization method to reduce these errors.
2.1. Reference Genes in RT-qPCR Data Normalization
Data normalization is the mathematical process that rank a set of candidate genes according to their expression stability in a specific sample set and a specific experimental design [49]. Normalization is needed to reduce the analytical variability and to obtain the most reliable and reproducible biological result. Yet, normalization still remains one of the most important and difficult problem associated to qPCR analysis [50]. The gold standard for RT-qPCR data normalization is based on the use of housekeeping (HK) genes, also referred as reference genes. The expression levels of an ideal internal control should remain constant independently of the tissue of origin, the state of disease, stage of cell-cycle, and stage of development and it should also remain constant in all experimental conditions and between experimental groups [51], [52], [53]. Generally, HK genes are constitutive genes involved in the maintenance of cellular functions and are expressed ubiquitously in all cell types under both physiological and pathological conditions [54], [55]. Among the most commonly used human HK genes for RT-qPCR normalization, there are genes encoding for components of the cytoskeleton (β-actin), components of the major histocompatibility complex (β2-microglobulin, B2M), genes involved in the glycolytic pathway (glyceraldehyde-3-phosphate dehydrogenase, GAPDH; phosphoglycerate kinase-1, PGK1), metabolic pathways (hypoxanthine phosphoribosyltransferase, HPRT; hydroxymethylbilane synthase, HMBS), protein folding (cyclophilin A, CypA or PPIA; peptidylprolyl isomerase B, PPIB), synthesis of ribosome units (rRNAs; ribosomal protein large subunit P0, RPLP0) [52], [56], [57]. Importantly, these internal reference genes are subjected to the same preparation steps as the genes of interest, thus co-amplification of these reference genes in the same sample preparation as the target gene allows to minimize the variation due to different amount and quality of RNA obtained during the RNA isolation and to different enzymatic efficiency of reverse transcription and qPCR in each sample [52]. In the latest years, however, it has emerged the evidence that many of these commonly used reference genes are not constantly expressed in every tissue and in every environmental circumstances.
It has been reported that GAPDH and β-actin, two of the most widely used HK genes, vary in the expression according to different tissue types [58], developmental stages [53], cell culturing conditions [59], [60], and cancer types [61]. Schmittgen and co-workers demonstrated that the expression levels of β-actin and GAPDH were affected by culturing condition, showing an up to ninefold increase in the expression levels when cells were cultured in 15% serum compared to starved cells [59]. Also, cell growth under hypoxic conditions was demonstrated to modulate the expression levels of certain commonly used reference genes. Zhong et al. showed that GAPDH, β-actin, and cyclophilin gene expression was affected by hypoxia in normal and tumor cell lines compared to normoxia, whereas the expression levels of the reference gene 28S rRNA were constant in all conditions [60]. Additionally, always in the context of cell culture, β-actin expression was shown to be strongly affected by Matrigel, being inhibited in a dose-dependent way, while in the same conditions 18S rRNA expression was not affected and thus, according to the authors, it represents a suitable reference gene in these specific experimental conditions [62]. GAPDH and β-actin were both found twofold up-regulated in invasive T1C3 melanoma cells compared to noninvasive 1C8 cells, showing that their expression is not stable in cancer [63]. Dydensborg et al. [64] suggested B2M and RPLP0 as the most suitable reference genes for comparing human colon primary carcinoma with normal colon mucosa and for studying differentiation processes of the human intestinal epithelium, respectively, whereas de Kok et al. [61] compared the expression pattern of 13 frequently used reference genes in 80 normal and tumor samples, spanning from colorectal, bladder, skin and other tumors, and identified HPRT as the single best reference gene for normalization in future cancer studies. It has to be considered, however, that HPRT expression levels are relatively low and thus this reference gene is more suitable for normalization of lowly expressed target genes. On the contrary, for highly expressed target genes it is recommended to choose a reference gene with comparable expression levels [52]. Usually β-actin is the normalizer of choice for highly expressed targets, however it has to be considered the variation of its expression according to the experimental settings.
2.2. RT-qPCR for Biomarkers Detection: the Normalization Problem
The use of high-throughput gene expression analysis by RT-qPCR to identify biomarkers and to measure their alteration in either physiological or pathological conditions is increasing. Peripheral blood is a convenient source for the detection of biomarkers. However, gene expression analyses from whole blood is limited by the lack of a standardization in the normalization process. This is particularly difficult as the cellular composition of peripheral blood changes physiologically and even more in pathological conditions such as cancer, infections, autoimmune diseases [65]. When non-homogenous tissue samples composed of different cell types are analyzed, the reference gene(s) of choice has to be stably expressed in all the constituent cell types [52]. Thus, the identification of suitable reference genes in whole blood has to take into account also the different cell populations that may differently express the reference gene of choice. In addition, when analyzing gene expression from peripheral blood for the detection of biomarkers, it has to be discriminated between whole blood and PBMC-derived RNA, which differ in cell populations. PBMCs, which can be isolated from peripheral blood by gradient separation, are mainly represented by T-cells, B-cells, NK cells and monocytes, while whole blood includes also neutrophils, basophils, eosinophils, granulocytes and red blood cells [66]. So far, no common and univocal reference gene exists for normalization of RT-qPCR data from blood samples but several groups tried to give indications on the most fitting reference genes for specific experimental groups. In this contest, Dheda and co-workers [50] analyzed 13 HK genes in PBMC and peripheral whole blood samples derived from either healthy subjects or individuals affected by tuberculosis. The authors identified human acidic ribosomal protein (HuPO) as the most stable reference gene in whole blood samples, with GAPDH, β-actin and HPRT being the most variable (up to 25-fold maximum variability). In PBMC, the most stable genes were HuPO and HPRT while GAPDH, β-actin, B2M and elongation factor 1-α (EF1α) were highly variable (up to 35-fold maximal variability), concluding that for comparing whole blood and PBMC samples from patients affected by pulmonary tuberculosis the most appropriate reference gene was HuPO [50].
To obtain more reliable results, more than one reference gene might be used for normalization when analyzing RT-qPCR data, and the geometric mean of expression of multiple reference genes is preferred to the arithmetic mean [67]. In the last decade, several software have been developed to determine the most stable and reliable reference genes among a subset of genes calculating normalization factors with different statistical algorithm. Among them the most frequently used are geNorm [67], NormFinder [68], and BestKeeper [69]. These algorithms have led to the identification of single reference genes in samples of whole blood or PBMC from subjects affected by chronic fatigue syndrome and healthy controls [66]. After NormFinder and geNorm analysis, the authors identified PPiB as the most stable reference gene in whole blood samples, and RPLP0 as the best reference gene in PBMC. However, PGK1 was the most stable and optimal normalizer to compare results from whole blood and PBMC together. Conversely to what many authors advise, Falkenberg and co-authors demonstrated that the use of a single reference gene is sufficient when the most stable is selected, rather than using multiple reference genes [66]. Similarly, Oturai and colleagues [70] identified the suitable reference genes using different peripheral blood cell populations, such as whole blood cells, PBMC, and PBMC-derived subsets of cells (CD4+ T cells, CD8+ T cells, NK cells, monocytes, B cells, and dendritic cells) obtained from healthy subjects, patients affected by multiple sclerosis (MS), and interferon β (IFNβ)-treated MS patients. Out of eight candidate reference genes, the authors identified ubiquitin-conjugating enzyme E2D2 (UBE2D2) as the best candidate for normalizing samples from control and MS subjects and all cell subsets, while to compare healthy subjects, MS, and IFNβ-treated MS patients, ubiquitin C (UBC) gene was the best normalizer across all the cell subsets. Additionally, they also identified the best combination of two reference genes to normalize either control and MS groups, which were UBE2D2 and HPRT, or controls, MS, and IFNβ-treated MS groups, which were UBC and Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta-polypeptide (YWHAZ) [70]. Noteworthy, reference genes that show a high variability of expression when considered as single normalizer might be optimal in combination with other reference gene(s).
It is clear from these studies that each experimental condition leads to the identification of different reference genes. Reference genes stably expressed in whole blood from subjects affected by a specific pathology may be highly variable in subjects with a different condition. Hence, it is necessary to validate the appropriate reference gene(s) for each specific experimental set, despite how time consuming and cost bearing it might be.
The problem of normalization is gaining interest also in the context of clinical testing of infectious diseases. To correctly diagnose a pathogenic infection, it is crucial to precisely detect the presence of the infectious agent or the corresponding antibodies triggered by the immune response. An incorrect diagnosis leads to wrongly applied therapeutic strategies to the patients as well as an enormous risk in terms of patients life and costs [71]. The implementation of RT-qPCR for reliable quantification of viral loads in clinical specimens requires normalization against defined standards and internal controls. Diagnostic tests are generally based on the titration of the sample on an external standard curve which has been subjected to identical amplification steps. Different sample size can lead to altered readouts. It has been shown that sampling the same volume of blood from HIV positive (HIV+) patients the yields of extracted RNA was significantly higher in HIV+ patients with less advanced immunosuppression (with a count of >200 cells/mL) compared to HIV+ patients with advanced immunosuppression (<200 cells/mL), making impossible the comparison of the two groups based on the processing of the same sample volumes [72]. Thus, diagnostic methods that compare the quantification of viral load to an internal standard/normalizer is needed. This problem was specifically highlighted by Jabs and colleagues, that developed a qPCR-based protocol for the quantification of Epstein–Barr virus (EBV) from PBMCs of transplanted patients as a predictive parameter for post-transplant lymphoproliferative disorders (PTLDs). The authors combined the co-amplification of EBV and genomic C-reactive protein DNA that served as an internal normalizer for the actual amount of viral DNA. They showed that, with the protocol without normalization, the detected EBV copy numbers were within the range of the control group in 4 out of 10 PTLD patients. On the other hand, after normalization, all of the PTLD patients showed higher amount of viral copy number compared to the control group, indicating that by introducing a normalization method for the virus detection the sensitivity and the accuracy of the assay increased [73]. More recently, an alternative method to normalize viral RNA from whole blood samples of cattle infected with Bovine Viral Diarrhea Virus (BVDV) was developed by spiking the blood samples with an external RNA virus, the canine Enteric Coronavirus (CECov), before RNA extraction. Quantification and normalization by RT-qPCR allowed to detect the virus up to 14 days post-challenge while viral detection with the traditional protocol of virus isolation and infection in cell culture was detectable only until 7 days post-challenge [74]. Even though this method was applied in veterinary, the same strategy could be successfully applied to human samples. Altogether, RT-qPCR combined with a normalization strategy would result in a rapid, sensitive, and reliable detection of viral infections.
3. MicroRNAs Normalization
microRNAs are becoming increasingly important as biomarkers but, as stated above, their use in clinical application is limited by the lack of standardized pre-analytical, analytical, and post-analytical procedures [75]. One of the main issue concerning the post-analytical procedure is RT-qPCR data normalization, since there are no universally approved methods. Several normalization methods were developed and used to normalize circulating miRNAs, leading to the impossibility of comparing results from different studies. The most common strategies used to normalize RT-qPCR data are based on exogenous synthetic oligonucleotides, geometrical mean of all the expressed miRNAs, and endogenous miRNAs [76] (Table 1 ).
Table 1.
Examples of Studies that Used Different Strategies for Data Normalization Based on Exogenous or Endogenous miRNAs
| Normalization Method | Authors | Study | Reference Genes | Ref |
|---|---|---|---|---|
| Exogenous | ||||
| Wang et al. | miRNAs in serum from lung cancer patients | Cel-miR-39 | [77] | |
| Yang et al. | miR-20 role in gastric cancer | Cel-miR-39 | [78] | |
| Ho et al. | miR-210 in plasma from pancreatic cancer patients | Cel-miR-54 | [79] | |
| Mitchell et al. | miRNA in serum/plasma from prostatic cancer patients | Cel-miR-39, Cel-miR-54, Cel-miR-238 (averaged) | [80] | |
| Sourvinou et al. | miR-21 from healthy subjects and small cell lung cancer patients | Cel-miR-39, hsa-miR-16 (combination of exogenous and endogenous reference gene) | [81] | |
| Anadol et al. | miRNAs in serum from HIV positive patients | SV40 | [82] | |
| Wang et al. | miRNAs in serum from sepsis and SIRS patients and healthy subjects | mmu-miR-295 (murine miRNA) | [83] | |
| Endogenous | ||||
| Small Nuclear RNAs | Huang et al. | miRNAs in plasma from colorectal neoplasia patients | RNU6B | [84] |
| Benz et al. | miRNAs in plasma from healthy subjects and critically ill and liver fibrosis patients | RNU6B | [85] | |
| Single miRNA | Lawrie et al. | miRNAs in serum from healthy subjects and large B-cell lymphoma patients | hsa-miR-16 | [86] |
| Wong et al. | miR-184 in plasma from squamous cell carcinoma patients | hsa-miR-16 | [87] | |
| Resnick et al. | miRNAs in serum from ovarian carcinoma patients | hsa-miR-142-3p | [88] | |
| Hu et al. | Circulating miRNAs in eight different cancer patients and controls | hsa-miR-1228 | [89] | |
| Tan et al. | miRNAs in serum from primary biliary cirrhosis patients | hsa-miR-24 | [90] | |
| Tan et al. | miRNAs in serum from hepatocellular carcinoma patients | hsa-miR-24 | [91] | |
| Krissansen et al. | miRNAs in serum from inflammatory bowel diseases patients | hsa-miR-484 | [92] | |
| Hao et al. | miRNAs in serum from multiple myeloma patients | hsa-miR-423-5p | [93] | |
| Grassmann et al. | miRNA associated with late stage neovascular age-related macular degeneration | hsa-miR-451 | [94] | |
| Multiple miRNAs | McDermott et al. | Brest cancer study | hsa-miR-16 and hsa-miR-425 (geNorm) | [95] |
| Song et al. | miRNAs in serum from gastric cancer patients | hsa-miR-16 and hsa-miR-93 (geNorm, NormFinder, BestKeeper and Comparative ΔCt) | [96] | |
| Tang et al. | miRNAs in plasma from hepatocellular carcinoma patients | hsa-miR-21 and hsa-miR-106a (geNorm, NormFinder, BestKeeper and Comparative ΔCt) | [97] | |
| Wang et al. | miR-148b-3p in serum from bladder cancer patients | hsa-miR-16a and hsa-miR-193a-5p (geNorm, NormFinder) | [98] | |
| Li et al. | miRNAs in serum from hepatitis B and hepatocellular carcinoma patients | hsa-miR-221, hsa-miR-191, hsa-let -7a, hsa-miR-181a, hsa-miR-181c, hsa-miR-26a (geNorm, NormFinder) | [99] | |
| Sansoni et al. | Fracture risk-associated miRNAs in serum | hsa-miR425-5p and hsa-miR-484 | [21] | |
| Danese et al. | miRNAs from exosome, plasma, and tissue from colorectal adenocarcinoma patients | hsa-miR-1228 and hsa-miR-520d | [100] | |
| Mean of all Expressed Genes | Mestdagh et al. | miRNAs in different tissues: normalization on large amount of data | Global mean | [101] |
Cel-miR, Caenorabditis elegans miRNA; hsa-miR/has-let, Homo sapiens miRNA; SV40, simian virus 40; mmu-miR, Mus musculus miRNA; RNU, small nucleolar RNA.
The choice of reference gene(s) is a crucial aspect in RT-qPCR data normalization in order to avoid erroneous interpretation of data and to obtain the most reliable biological results. The a priori selection of reference gene(s) according to the ones reported in literature represents one method adopted to choose internal control. However, a more precise evaluation is advisable since the expression of a potential reference gene could change under different experimental conditions. Mathematical models, such as geNorm [67], NormFinder [68], BestKeeper [69], and Comparative ΔCt [102] have been developed aimed at identifying the most stable endogenous miRNA(s) to be used as normalizer(s), under a specific experimental conditions.
geNorm is an algorithm that select the most stable internal controls in a group of samples. It ranks the genes on the bases of a gene stability parameter M. M is calculated as the average of the pairwise variation (standard deviation of the logarithmically transformed expression ratio) of each potential reference gene with all the others. Genes with the lower M value during the stepwise exclusion represent the most stable genes [67].
NormFinder is an algorithm that enable the estimation of inter-group and intra-group variations, combining these two variations in a stability value, calculated for each potential reference gene. It ranks genes on their stability value and, moreover, it calculates the accumulate standard deviation (AccSD) that indicates the number of genes to be used for normalization [68].
BestKeeper performs a descriptive statistical analysis of the Cq calculating geometric and arithmetic mean, standard deviation, and coefficient of variance for each analyzed potential reference gene. Furthermore it performs pair-wise correlation analysis of all pairs of candidate genes and calculates the geometric mean of the “best” suited ones to define the optimal reference genes [69].
Comparative ΔCt is a method based on the Ct comparison of pair of genes. On the bases of ΔCt trend of genes (constant or variable) among the analyzed samples it is possible to identify the most stable gene [102].
The development of algorithms as NormFinder, geNorm, BestKeeper, and Comparative ΔCt able to identify the most stable gene(s), in a specific experimental set, has facilitated the delineation of the optimal reference gene(s) to normalize RT-qPCR data. However, the wide heterogeneity of normalization methods and the lack of guidelines for the identification of the reference gene(s) may lead to erroneous interpretation of RT-qPCR data.
Below, the different normalization strategies and the concerning issues are discussed.
3.1. Normalization on Exogenous Reference Genes
Synthetic oligonucleotides (Caenorabditis elegans miRNAs cel-miR-39, cel-miR-54, cel-miR-238, simian virus gene SV40, and the murine mmu-miR-295) are non-human sequences often used to assess the efficiency of RNA processing and the RNA quality. They are added into biological samples at known concentrations before either RNA isolation or RNA reverse-transcription, in order to monitor these processes. In the last few years, since no universally accepted methods to normalize circulating miRNAs exist, these exogenous synthetic oligonucleotides have also been used in miRNA quantification [79], [80], [82], [83]. cel-miR-39 and cel-miR-54 were used as single reference genes to normalize miRNAs. Wang et al. have used cel-miR-39 to quantify serum miRNAs for the early detection of lung cancer [77] while, in another study, Yang et al. used cel-miR-39 as reference gene to determine the potential usefulness of hsa-miR-20 as biomarker in gastric cancer [78]. Also Cel-miR-54 was used for circulating miRNA normalization: Ho et al. used cel-miR-54 to normalize hsa-miR-210 expression levels in plasma from pancreatic cancer patients [79]. In other studies the RT-qPCR data normalization was based on the average of these synthetic oligonucleotides: the average of cel-miR-39, cel-miR-54, and cel-miR-238 was used, for example, to normalize serum/plasma miRNAs levels in prostate cancer patients [80]. Although synthetic oligonucleotides-based normalization strategies have been adopted, their use implies some limitations. Circulating miRNAs and their use as biomarkers are subjected to a set of pre-analytical variables that include the quality and the amount of the starting sample, the collection modality and storage conditions, the efficiency of miRNA isolation and reverse-transcription, that may contribute in altering the total miRNA levels. As synthetic oligonucleotides are added before miRNA isolation and reverse-transcription, they are subjected to the same processing as endogenous miRNA targets regarding to RNA extraction and PCRs. However, other technical variables, generated before those processes, cannot be normalized by synthetic oligonucleotides. To overcome this problem, Sourvinou et al. have adopted the strategy of normalizing on a combination of synthetic oligonucleotides and endogenous genes. cel-miR-39 and hsa-miR-16 were used to quantify hsa-miR-21 in healthy and small cell lung carcinoma patients, demonstrating that differences in miRNA recovery and in cDNA synthesis between samples were compensated [81].
3.2. Normalization on Endogenous Reference Genes
A reference gene should be considered as an optimal normalizer if it is affected by the same variables of the target miRNAs. Endogenous gene(s) hold this prerequisite and several normalization strategies based on endogenous reference genes were developed. Small nucleolar RNAs (RNU), such as RNU6 and RNU6B and the ribosomal RNAs (rRNAs) 5S and 18S have been widely used to normalize circulating miRNAs in RT-qPCR analyses [75], [76]. However, these non-coding RNAs do not reflect the biological properties of miRNAs and several studies have shown their instability and variability in serum and plasma under different experimental conditions. For what concern circulating miRNAs, Huang et al. have investigated the potential role of RNU6B as internal control in plasma miRNA quantification from colorectal neoplasia patients, demonstrating that RNU6B has a lower stability and is less abundant than other miRNAs, such as hsa-miR-16 and hsa-miR-191 [84]. RNU6B was also found variable in serum from healthy volunteers, patients with critical illness and patients with liver fibrosis, indicating that RNU6B is an inappropriate reference gene for normalization of circulating miRNAs [85]. These findings suggest that the use of these small-non coding RNAs is not recommended in RT-qPCR data normalization and that they could introduce biases in miRNAs quantification. Normalization based on endogenous miRNA(s) should represent the most suitable method in RT-qPCR data analyses and strategies based on both a single miRNA and multiple miRNAs have been developed. hsa-miR-16 is one mostly often used reference gene. Lawrie et al. used hsa-miR-16 to normalize miRNAs quantification (hsa-miR-155, hsa-miR-210, and hsa-miR-21) in serum from healthy subjects and large B-cell lymphoma patients [86]. In another study, Wong et al. used hsa-miR-16 to quantify hsa-miR-184 in plasma from tongue squamous cell carcinoma patients [87]. Although in these studies hsa-miR-16 was found to be comparably expressed in serum/plasma of both healthy subjects and cancer patients, other studies highlighted, instead, the high variability of hsa-miR-16 expression in serum/plasma of case and control patients. Analyses on hsa-miR-16 stability revealed that hsa-miR-16 expression was variable in serum from different cancer patients: for example, it was found up-regulated in breast cancer [103] and pancreatic cancer patients [104] compared to controls. Furthermore the use of hsa-miR-16 in miRNA quantification has to be reconsidered since it was found to be susceptible to hemolysis: analyses on plasma miRNAs have demonstrated the up-regulation of hsa-miR-16 in haemolyzed plasma [105]. Therefore, other miRNAs have been evaluated as potential reference genes. In a study on circulating miRNAs in serum from ovarian cancer patients, Resnick et al. used hsa-miR-142-3p to normalize RT-qPCR data [88]. Hu et al. identified hsa-miR-1228 as a potential endogenous reference gene for circulating miRNAs in cancer patients after analyses on its expression stability in patients with eight different cancer types and three control subjects [89]. Elsewhere, hsa-miR-24 was used as reference gene for the quantification of serum miRNAs from primary biliary cirrhosis [90] and hepatocellular carcinoma patients [91], while hsa-miR-484 was used to normalize serum miRNAs in patients with inflammatory bowel diseases [92]. Hao and colleagues used hsa-miR-423-5p to normalize serum miRNAs (hsa-miR-214, hsa-miR-135b, hsa-miR-132, and hsa-miR-92a) in multiple myeloma patients [93], while hsa-miR-451 was used as internal control to quantify miRNAs associated to late stage neovascular age-related macular degeneration [94].
It was demonstrated that the use of multiple reference genes in RT-qPCR data analyses reduces the effects of the analytical variability compared to single gene-based normalization [67], [68]. McDermott et al., in a study on breast cancer patients, have demonstrated that the combination of miR-16 and hsa-miR-425, identified by geNorm as the most stable miRNAs, is more suitable than the use of both these miRNAs alone and RNU6 to normalize RT-qPCR data [95].
Several studies on circulating miRNA have adopted a normalization strategy based on multiple reference genes for miRNA quantification. In a recent study by Danese et al., the combination of hsa-miR-1228 and hsa-miR-520d was identified as the best reference to normalize RT-qPCR miRNA analysis in colorectal adenocarcinoma from exosome, plasma, and tissue samples. These two miRNAs were selected among six potential reference genes (hsa-miR-191, hsa-miR-193a, hsa-miR-345, hsa-miR-520d, and hsa-miR-1228) based on their stability assessed trough both NormFinder and BestKeeper algorithms [100]. Song et al., in a study on serum miRNAs from gastric cancer patients, have identified hsa-miR-16 and hsa-miR-93 as reference genes. In this study the combination of different algorithms (geNorm, NormFinder, BestKeeper and Comparative ΔCt) was employed to identify the best reference genes among 6 pre-selected miRNAs (hsa-let-7a, hsa-miR-16, hsa-miR-93, hsa-miR-103, hsa-miR-192, and hsa-miR-451) and the small nucleolar RNA, RNU6B [96]. A similar approach was adopted by Tang et al. [97]. With the use of RefFinder, a software that integrates geNorm, NormFinder, BestKeeper, and Comparative ΔCt algorithms, they identified hsa-miR-21 and miR-106a as suitable reference genes in plasma of patients with hepatocellular carcinoma, among hsa-let-7a, hsa-miR-21, hsa-miR-106a, hsa-miR-155, hsa-miR219, hsa-miR-221, and hsa-miR-16 [97]. Wang et al. demonstrated that the combination of hsa-miR-16a and hsa-miR-193a-5p gave the most reliable results in miR-148b-3p quantification in serum of bladder cancer patients. These two miRNAs were selected by geNorm and NormFinder for their stability among a set of five potential reference genes: hsa-miR-193a-5p, hsa-miR-16-5p, hsa-miR-191-5p, hsa-let-7d-5p, and RNU6 [98]. geNorm and NormFinder were also employed to investigate on a set of reference genes (5S, U6, hsa-let-7a, hsa-miR-103, hsa-miR-16, hsa-miR-181a, hsa-miR-181c, hsa-miR-191, hsa-miR-22, hsa-miR-221, and hsa-miR-26a) to analyze exosome-associated miRNAs in serum of patients with hepatitis B or hepatocellular carcinoma. The two algorithms have identified the combination of hsa-miR-221, hsa-miR-191, hsa-let-7a, hsa-miR-181a, hsa-miR-181c, and hsa-miR-26a as the optimal reference genes to normalize liver-derived miRNAs [99]. In a recent study on fracture risk-associated miRNA, we used miR-425-5p and miR-484 as reference genes according to a priori analysis [21]. Another endogenous gene-based normalization method is represented by the averaged expression value of all the analyzed miRNAs. This method is generally used in experiments assaying for large numbers of miRNAs and/or in experiments in which an a priori selection of reference genes is absent. Mestdagh et al. compared normalization on mean of all the expressed miRNAs, on stable small RNA controls (RNU24, RNU44, RNU58A, and RNU6B) and on miRNA/small RNA controls resembling the mean expression value, and demonstrated that the mean of all the expressed miRNAs is the most appropriate method to normalize a large amount of data [101].
4. Conclusions
Biomarkers, especially circulating miRNAs, are emerging for their importance in clinical diagnostics. Methods for reliable detection of these molecules are necessary for univocally diagnose pathophysiological conditions. RT-qPCR is an extremely sensitive and accurate method to quantify circulating miRNAs, but a great limitation in the implementation of this technique in clinics is represented by the lack of a standardized normalization methodology. Normalization of RT-qPCR data is the methodology required to reduce the technical bias introduced during the entire experimental process and the choice of the appropriate method is of crucial importance to obtain the most reliable biological result. Several normalization methods have been developed, from the use of exogenous oligonucleotides to the use of different endogenous reference genes. This heterogeneity in normalization strategies with the use of different reference genes leads to an inaccurate quantification of circulating miRNAs and to the possible misinterpretation of results from different studies. Since it is still not possible to define universally accepted reference genes for circulating miRNAs quantification, it is advisable to focus on standardizing the normalization method to select case-specific reference genes. It is now clear that the most corroborated strategy for normalization is to identify and to validate the most stable miRNAs for each experimental setting and to select them as reference genes. Furthermore, in case of large amounts of data, besides the selection of the most stable genes, it is also accepted to use the global mean. Indeed, when a panel of miRNAs is normalized on the most stable genes, all the miRNAs used as reference genes are no longer considerable as targets; the great advantage of normalizing on global mean is to consider all the analyzed miRNAs as target genes.
The drawing up of specific guidelines about the use of the different normalization methods, together with the drawing up of standardized pre-analytical and analytical procedures, may ensure the reproducibility of results and the possibility of comparing miRNA expression levels from different studies, and the use of miRNA in clinical applications.
References
- 1.Lombardi G., et al. Circulating miRNA as fine regulators of the physiological responses to physical activity: pre-analytical warnings for a novel class of biomarkers. Clin. Biochem. 2016;49(18):1331–1339. doi: 10.1016/j.clinbiochem.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 2.Jacob F., Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 3.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Bloch S., et al. Small and smaller-sRNAs and MicroRNAs in the regulation of toxin gene expression in prokaryotic cells: a mini-review. Toxins (Basel) 2017;9(6) doi: 10.3390/toxins9060181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerutti H., Casas-Mollano J.A. On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 2006;50(2):81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang Y., et al. RNA interference in fungi: pathways, functions, and applications. Eukaryot. Cell. 2011;10(9):1148–1155. doi: 10.1128/EC.05109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reinhart B.J., et al. MicroRNAs in plants. Genes Dev. 2002;16(13):1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M., et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 9.Grundhoff A., Sullivan C.S. Virus-encoded microRNAs. Virology. 2011;411(2):325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lujambio A., et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA. 2008;105(36):13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein E., et al. Dicer is essential for mouse development. Nat. Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 13.Murchison E.P., et al. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suh M.R., et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270(2):488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Ludwig N., et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44(8):3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber J.A., et al. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X., et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Chen J., Sen S. MicroRNA as biomarkers and diagnostics. J. Cell. Physiol. 2016;231(1):25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ardekani A.M., Naeini M.M. The role of MicroRNAs in human diseases. Avicenna J. Med. Biotechnol. 2010;2(4):161–179. [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 21.Sansoni V., et al. Effects of repeated sprints training on fracture risk-associated miRNA. Oncotarget. 2018;9(26):18029–18040. doi: 10.18632/oncotarget.24707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X., Platt C., Rosenzweig A. The role of MicroRNAs in the cardiac response to exercise. Cold Spring Harb. Perspect. Med. 2017;7(12) doi: 10.1101/cshperspect.a029850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butz H., Patocs A. Technical aspects related to the analysis of circulating microRNAs. EXS. 2015;106:55–71. doi: 10.1007/978-3-0348-0955-9_3. [DOI] [PubMed] [Google Scholar]
- 24.Morrow D.A., de Lemos J.A. Benchmarks for the assessment of novel cardiovascular biomarkers. Circulation. 2007;115(8):949–952. doi: 10.1161/CIRCULATIONAHA.106.683110. [DOI] [PubMed] [Google Scholar]
- 25.Hackl M., et al. Circulating microRNAs as novel biomarkers for bone diseases - complex signatures for multifactorial diseases? Mol. Cell. Endocrinol. 2016;432:83–95. doi: 10.1016/j.mce.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Mestdagh P., et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 27.Nelson P.T., et al. Technical variables in high-throughput miRNA expression profiling: much work remains to be done. Biochim. Biophys. Acta. 2008;1779(11):758–765. doi: 10.1016/j.bbagrm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Ree M.H., et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014;111:53–59. doi: 10.1016/j.antiviral.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Drury R.E., O'Connor D., Pollard A.J. The clinical application of MicroRNAs in infectious disease. Front. Immunol. 2017;8:1182. doi: 10.3389/fimmu.2017.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeliger C., et al. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. 2014;29(8):1718–1728. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- 31.Panach L., et al. Serum circulating microRNAs as biomarkers of osteoporotic fracture. Calcif. Tissue Int. 2015;97(5):495–505. doi: 10.1007/s00223-015-0036-z. [DOI] [PubMed] [Google Scholar]
- 32.Kelch S., et al. miRNAs in bone tissue correlate to bone mineral density and circulating miRNAs are gender independent in osteoporotic patients. Sci. Rep. 2017;7(1):15861. doi: 10.1038/s41598-017-16113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng H.H., et al. Circulating microRNAs and treatment response in the phase II SWOG S0925 study for patients with new metastatic hormone-sensitive prostate cancer. Prostate. 2018;78(2):121–127. doi: 10.1002/pros.23452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin H.M., et al. Phase 2 study of circulating microRNA biomarkers in castration-resistant prostate cancer. Br. J. Cancer. 2017;116(8):1002–1011. doi: 10.1038/bjc.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka H., et al. miR-125b-1 and miR-378a are predictive biomarkers for the efficacy of vaccine treatment against colorectal cancer. Cancer Sci. 2017;108(11):2229–2238. doi: 10.1111/cas.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hess A.K., et al. MiR-200b and miR-155 as predictive biomarkers for the efficacy of chemoradiation in locally advanced head and neck squamous cell carcinoma. Eur. J. Cancer. 2017;77:3–12. doi: 10.1016/j.ejca.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Halvorsen A.R., et al. Evaluation of prognostic and predictive significance of circulating MicroRNAs in ovarian cancer patients. Dis. Markers. 2017;2017 doi: 10.1155/2017/3098542. 3098542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiemer E.A.C., et al. Identification of microRNA biomarkers for response of advanced soft tissue sarcomas to eribulin: translational results of the EORTC 62052 trial. Eur. J. Cancer. 2017;75:33–40. doi: 10.1016/j.ejca.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 39.Gagez A.L., et al. miR-125b and miR-532-3p predict the efficiency of rituximab-mediated lymphodepletion in chronic lymphocytic leukemia patients. A French Innovative Leukemia Organization study. Haematologica. 2017;102(4):746–754. doi: 10.3324/haematol.2016.153189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang A., et al. Whole blood sequencing reveals circulating microRNA associations with high-risk traits in non-ST-segment elevation acute coronary syndrome. Atherosclerosis. 2017;261:19–25. doi: 10.1016/j.atherosclerosis.2017.03.041. [DOI] [PubMed] [Google Scholar]
- 41.Xiao J., et al. Circulating miR-30d predicts survival in patients with acute heart failure. Cell. Physiol. Biochem. 2017;41(3):865–874. doi: 10.1159/000459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J. Intern. Med. 2013;4(2):627–635. [PMC free article] [PubMed] [Google Scholar]
- 43.Ma J., et al. Differential miRNA expressions in peripheral blood mononuclear cells for diagnosis of lung cancer. Lab. Invest. 2015;95(10):1197–1206. doi: 10.1038/labinvest.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Y.C., et al. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev. Res. (Phila) 2012;5(4):665–674. doi: 10.1158/1940-6207.CAPR-11-0358. [DOI] [PubMed] [Google Scholar]
- 45.Xiong D.D., et al. A nine-miRNA signature as a potential diagnostic marker for breast carcinoma: an integrated study of 1,110 cases. Oncol. Rep. 2017;37(6):3297–3304. doi: 10.3892/or.2017.5600. [DOI] [PubMed] [Google Scholar]
- 46.Yuan W., et al. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget. 2016;7(48):80033–80045. doi: 10.18632/oncotarget.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng H., et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017745. e17745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bustin S.A., Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004;15(3):155–166. [PMC free article] [PubMed] [Google Scholar]
- 49.Bustin S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25(2):169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 50.Dheda K., et al. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37(1):112–114. doi: 10.2144/04371RR03. 116, 118–9. [DOI] [PubMed] [Google Scholar]
- 51.Dheda K., et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005;344(1):141–143. doi: 10.1016/j.ab.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Bar M., Bar D., Lehmann B. Selection and validation of candidate housekeeping genes for studies of human keratinocytes--review and recommendations. J. Invest. Dermatol. 2009;129(3):535–537. doi: 10.1038/jid.2008.428. [DOI] [PubMed] [Google Scholar]
- 53.Mori R., et al. Both beta-actin and GAPDH are useful reference genes for normalization of quantitative RT-PCR in human FFPE tissue samples of prostate cancer. Prostate. 2008;68(14):1555–1560. doi: 10.1002/pros.20815. [DOI] [PubMed] [Google Scholar]
- 54.Zhu J., et al. On the nature of human housekeeping genes. Trends Genet. 2008;24(10):481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Eisenberg E., Levanon E.Y. Human housekeeping genes are compact. Trends Genet. 2003;19(7):362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- 56.Wang T., et al. Selection of suitable housekeeping genes for real-time quantitative PCR in CD4(+) lymphocytes from asthmatics with or without depression. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0048367. e48367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stephens A.S., Stephens S.R., Morrison N.A. Internal control genes for quantitative RT-PCR expression analysis in mouse osteoblasts, osteoclasts and macrophages. BMC Res. Notes. 2011;4:410. doi: 10.1186/1756-0500-4-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Radonic A., et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004;313(4):856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 59.Schmittgen T.D., Zakrajsek B.A. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods. 2000;46(1–2):69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 60.Zhong H., Simons J.W. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem. Biophys. Res. Commun. 1999;259(3):523–526. doi: 10.1006/bbrc.1999.0815. [DOI] [PubMed] [Google Scholar]
- 61.de Kok J.B., et al. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab. Invest. 2005;85(1):154–159. doi: 10.1038/labinvest.3700208. [DOI] [PubMed] [Google Scholar]
- 62.Selvey S., et al. Beta-actin--an unsuitable internal control for RT-PCR. Mol. Cell. Probes. 2001;15(5):307–311. doi: 10.1006/mcpr.2001.0376. [DOI] [PubMed] [Google Scholar]
- 63.Goidin D., et al. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 2001;295(1):17–21. doi: 10.1006/abio.2001.5171. [DOI] [PubMed] [Google Scholar]
- 64.Dydensborg A.B., et al. Normalizing genes for quantitative RT-PCR in differentiating human intestinal epithelial cells and adenocarcinomas of the colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290(5):G1067–G1074. doi: 10.1152/ajpgi.00234.2005. [DOI] [PubMed] [Google Scholar]
- 65.O'Connell G.C., et al. Leukocyte dynamics influence reference gene stability in whole blood: data-driven qRT-PCR normalization is a robust alternative for measurement of transcriptional biomarkers. Lab. Med. 2017;48(4):346–356. doi: 10.1093/labmed/lmx035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Falkenberg V.R., et al. Identification of Phosphoglycerate Kinase 1 (PGK1) as a reference gene for quantitative gene expression measurements in human blood RNA. BMC Res. Notes. 2011;4:324. doi: 10.1186/1756-0500-4-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandesompele J., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andersen C.L., Jensen J.L., Orntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 69.Pfaffl M.W., et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 70.Oturai D.B., et al. Identification of suitable reference genes for peripheral blood mononuclear cell subset studies in multiple sclerosis. Scand. J. Immunol. 2016;83(1):72–80. doi: 10.1111/sji.12391. [DOI] [PubMed] [Google Scholar]
- 71.Souf S. Recent advances in diagnostic testing for viral infections. Biosci. Horiz.: Int. J. Stud. Res. 2016;9 hzw010. [Google Scholar]
- 72.Huggett J., et al. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6(4):279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 73.Jabs W.J., et al. Normalized quantification by real-time PCR of Epstein-Barr virus load in patients at risk for posttransplant lymphoproliferative disorders. J. Clin. Microbiol. 2001;39(2):564–569. doi: 10.1128/JCM.39.2.564-569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Young N.J., et al. Real-time RT-PCR detection of Bovine Viral Diarrhoea virus in whole blood using an external RNA reference. J. Virol. Methods. 2006;138(1–2):218–222. doi: 10.1016/j.jviromet.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reid G., Kirschner M.B., van Zandwijk N. Circulating microRNAs: association with disease and potential use as biomarkers. Crit. Rev. Oncol. Hematol. 2011;80(2):193–208. doi: 10.1016/j.critrevonc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 76.Schwarzenbach H., et al. Data normalization strategies for microRNA quantification. Clin. Chem. 2015;61(11):1333–1342. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang P., et al. Early detection of lung cancer in serum by a panel of microRNA biomarkers. Clin. Lung Cancer. 2015;16(4):313–319 e1. doi: 10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 78.Yang R., et al. Serum miR-20a is a promising biomarker for gastric cancer. Biomed. Rep. 2017;6(4):429–434. doi: 10.3892/br.2017.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ho A.S., et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl. Oncol. 2010;3(2):109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mitchell P.S., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sourvinou I.S., Markou A., Lianidou E.S. Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J. Mol. Diagn. 2013;15(6):827–834. doi: 10.1016/j.jmoldx.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 82.Anadol E., et al. Circulating microRNAs as a marker for liver injury in human immunodeficiency virus patients. Hepatology. 2015;61(1):46–55. doi: 10.1002/hep.27369. [DOI] [PubMed] [Google Scholar]
- 83.Wang J.F., et al. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 2010;394(1):184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 84.Huang Z., et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer. 2010;127(1):118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 85.Benz F., et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp. Mol. Med. 2013;45:e42. doi: 10.1038/emm.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrie C.H., et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 87.Wong T.S., et al. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res. 2008;14(9):2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 88.Resnick K.E., et al. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol. Oncol. 2009;112(1):55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 89.Hu J., et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int. J. Cancer. 2014;135(5):1187–1194. doi: 10.1002/ijc.28757. [DOI] [PubMed] [Google Scholar]
- 90.Tan Y., et al. Serum microRNAs as potential biomarkers of primary biliary cirrhosis. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0111424. e111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan Y., et al. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107986. e107986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krissansen G.W., et al. Overexpression of miR-595 and miR-1246 in the sera of patients with active forms of inflammatory bowel disease. Inflamm. Bowel Dis. 2015;21(3):520–530. doi: 10.1097/MIB.0000000000000285. [DOI] [PubMed] [Google Scholar]
- 93.Hao M., et al. Low serum miR-19a expression as a novel poor prognostic indicator in multiple myeloma. Int. J. Cancer. 2015;136(8):1835–1844. doi: 10.1002/ijc.29199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grassmann F., et al. A circulating microRNA profile is associated with late-stage neovascular age-related macular degeneration. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107461. e107461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McDermott A.M., Kerin M.J., Miller N. Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0083718. e83718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Song J., et al. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig. Dis. Sci. 2012;57(4):897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 97.Tang G., et al. Different normalization strategies might cause inconsistent variation in circulating microRNAs in patients with hepatocellular carcinoma. Med. Sci. Monit. 2015;21:617–624. doi: 10.12659/MSM.891028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L., et al. Identification and validation of reference genes for the detection of serum microRNAs by reverse transcription-quantitative polymerase chain reaction in patients with bladder cancer. Mol. Med. Rep. 2015;12(1):615–622. doi: 10.3892/mmr.2015.3428. [DOI] [PubMed] [Google Scholar]
- 99.Li Y., et al. Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Dis. Markers. 2015;2015 doi: 10.1155/2015/893594. 893594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Danese E., et al. Reference miRNAs for colorectal cancer: analysis and verification of current data. Sci. Rep. 2017;7(1):8413. doi: 10.1038/s41598-017-08784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mestdagh P., et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Silver N., et al. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu Z., et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. 2012;33(4):828–834. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- 104.Liu J., et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int. J. Cancer. 2012;131(3):683–691. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 105.Kirschner M.B., et al. The impact of hemolysis on cell-free microRNA biomarkers. Front. Genet. 2013;4:94. doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]