Abstract
Emerging porcine epidemic diarrhea viruses (PEDVs) have caused large economic losses since 2010, and G2b is the prevalent globally epidemic genotype. Given the fastidious isolation of emerging PEDV in cell culture and difficulties in retaining the isolate infectivity upon further in vitro passage, highly attenuated recombinant vesicular stomatitis virus (rVSVMT) was used as a vector to express the PEDV spike (S) protein, aiming to develop a subunit vaccine against G2b viruses. An S protein with 19 of its cytoplasmic domain amino acids deleted could be incorporated into VSV particles, generating rVSVMT (VSVMT-SΔ19) with high efficiency. Our results suggest that VSVMT-SΔ19 could effectively induce PEDV-specific immunity in pigs via intramuscular, but not intranasal, immunization. Notably, immunizations of sows with VSV MT-SΔ19 provided protective lactogenic immunity against a virulent G2b PEDV challenge in piglets. Consequently, recombinant VSVMT may be a promising platform for preparing a subunit vaccine against PEDV.
Keywords: Vesicular stomatitis virus, Porcine epidemic diarrhea virus, Spike protein, Vaccine
Highlights
-
•
PEDV spike protein can be incorporated into VSV particles.
-
•
VSV-based PEDV vaccine can induce robust PEDV-specific immunities in pigs.
-
•
VSVMT could be a promising platform for developing vaccines against emerging PEDV.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is a coronavirus that was initially identified in 1978 (Pensaert and de Bouck, 1978) and is endemic mainly in Asia. In 2010, a PEDV outbreak emerged in China and then spread to other countries in Asia and North America (Li et al., 2012; Vlasova et al., 2014). The PEDV genome consists of a single-stranded positive-sense RNA molecule. Its structural proteins include the spike (S), membrane (M), envelope (E), and the nucleocapsid (N) proteins. The S protein, which is responsible for virus attachment, receptor binding, cell membrane fusion, and entry, can induce neutralizing antibodies in the host (Lai et al., 2007). A phylogenetic analysis based on S gene alignment indicated that PEDVs can be categorized into two genotypes: genotype 1 (G1) and genotype 2 (G2). These genotypes each consist of two subgroups: G1a and G1b for G1, and G2a and G2b for G2. Isolates of G2 are usually more pathogenic than those of G1 (Lee, 2015). G1 PEDVs include classic strains such as CV777 and DR13. G2a isolates have been found in Asia. Notably, G2b isolates were responsible for the recent pandemic outbreaks in North America and Asia, which were characterized by high mortality in piglets (Huang et al., 2013; Vlasova et al., 2014; Wang et al., 2016). Although inactivated and attenuated vaccines based on the classical CV777 strain have been widely used in China, porcine epidemic diarrhea (PED) is still not controlled effectively (Wang et al., 2016). The low-to-moderate effectiveness of the current CV777-based vaccines may be attributed to antigenic differences between the S protein of the vaccine and those of field epizootic strains (Lee, 2015). Currently, a commercial vaccine against G2b strains remains unavailable. The isolation of PEDV in cell culture has proven fastidious, and a successfully isolated virus may still be incapable of retaining its infectivity upon further in vitro passage (Hofmann and Wyler, 1988; Oka et al., 2014). This laboratory hurdle makes the production of an efficacious vaccine challenging.
The PEDV S protein or a truncated version expressed by recombinant viral vectors has shown potential for use in the development of a subunit vaccine against PEDV (Hain et al., 2016; Yuan et al., 2017). Vesicular stomatitis virus (VSV) is a promising viral vector for expressing foreign antigens, which are capable of potently stimulating host humoral and cellular immune responses (Roberts et al., 1998; Tan et al., 2005). In the present study, a highly attenuated recombinant VSV with triple amino acid mutations in the M protein (ΔM51, V221F, and S226R) (VSVMT) was used to express the S protein of PEDV, with the aim of developing a mucosal vaccine against the predominant epidemic isolates of G2b (Fang et al., 2015). Our data indicate that, when 19 amino acids of its cytoplasmic domain were deleted, the S protein could be efficiently incorporated into VSV particles and lead to the generation of a recombinant VSV (VSVMT-SΔ19) with high efficiency. Our in vivo studies revealed that VSVMT-SΔ19 could effectively induce anti-PEDV immunity in pigs via intramuscular, but not intranasal, immunization. Notably, the administration of VSVMT-SΔ19 in sows stimulated robust passive immunity in neonatal piglets, which could resist a lethal-dose challenge by a virulent homologous PEDV strain. Thus, together with the advantages that VSVMT-SΔ19 can replicate in BHK21 cells at titers above 108 PFU/ml and that BHK21 cells can grow in suspension in disposable bioreactors at cell concentrations of higher than 1 × 107 cells/ml, our findings indicate that recombinant VSVMT could be a promising platform for rapidly developing vaccines against emerging or reemerging epidemic strains of PEDV.
2. Materials and methods
2.1. Cell lines and viruses
Baby hamster kidney cells (BHK-21) (ATCC CCL-10) and African green monkey cells (Vero 81) (ATCC CRL-1587) were grown in DMEM medium supplemented with 10% fetal bovine serum (Gibco, USA). Cells were cultured in humidified air containing 5% CO2 at 37 °C. VSVMT and PEDV CV777 were prepared as previously described (Fang et al., 2012; Hofmann and Wyler, 1988). VSV or PEDV convalescent sera were prepared in infected mice or pigs, respectively.
2.2. Phylogenetic analysis of PEDV emerging isolates
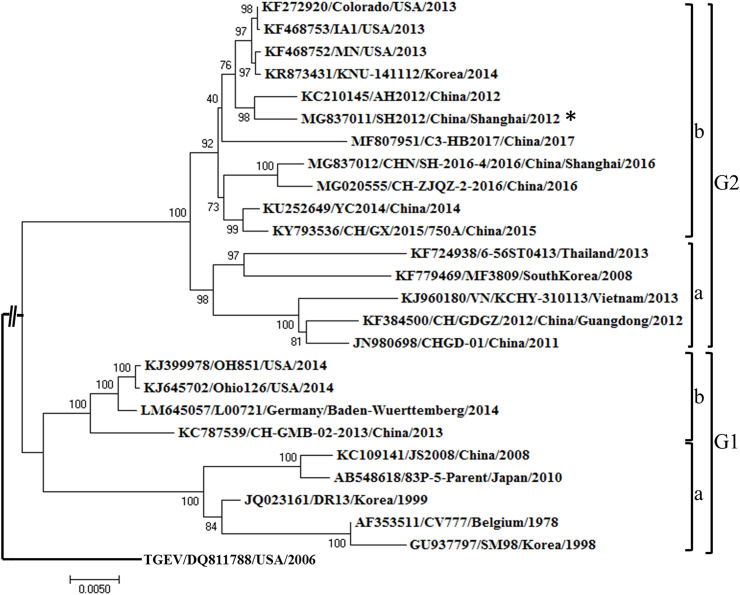
PEDV/CHN/SHANGHAI/2012 (SH 2012) and PEDV/CHN/SHANGHAI/2016 (SH 2016) are two emerging PEDV strains that were isolated in 2012 and 2016, respectively, in Shanghai, China. Their genomes were sequenced and submitted to Genbank (GenBank Nos: MG837011, MG837012). The S genes of these two strains were aligned with the prototype PEDV strain CV777 and other representative emerging PEDV strains, including AH 2012 (reference number: KC210415), Corolado 2013 (reference number: KF272920), and the US-SINDEL strain OH851 (reference number: KJ399978) (Supl. Table 1 ). A phylogenetic tree of these PEDV strains was constructed using the neighbor-joining (NJ) method in MEGA 7.0. The phylogenetic tree was rooted with an out-group TGEV strain (accession number: DQ811785). Bootstrap resampling (1000 replications) was performed and indicated for each node. The scale bars on the resulting tree indicated the nucleotide substitutions per site.
Table 1.
PCR primers for amplifying PEDV S genes with different lengths of cytoplasmic tail sequences.
| Primers Primer sequences |
|---|
| Forward primer P1–F 5′-TAACAGATATCACGCTCGAGATGAAGTCTTTAACCTACTTCTGGT-3′ (XhoI site shown in red) |
| Reverse primers (NheI site shown in red) P2(SFL) 5′-ACATGAAGAATCTGGCTAGCTCATCAGCCGCAGCATCCACAACAACC-3′ |
| P3(SΔ19) 5′-ACATGAAGAATCTGGCTAGCTCATCAGCAGCCGCAGCATCCACAACA-3′ P4(SΔ20) 5′-ACATGAAGAATCTGGCTAGCTCATCAGCAGCAGCCGCAGCATCCACA-3′ |
| P5(SΔ21) 5′-ACATGAAGAATCTGGCTAGCTCATCAACAGCAGCAGCCGCAGCATCC-3′ |
| P6(SΔ22) 5′-ACATGAAGAATCTGGCTAGCTCATCAAGCACAGCAGCAGCCGCAGCA-3′ |
| P7(SΔ23) 5′-ACATGAAGAATCTGGCTAGCTCATCAACAAGCACAGCAGCAGCCGCA-3′ |
| P8(SΔ24) 5′-ACATGAAGAATCTGGCTAGCTCATCAAAAACAAGCACAGCAGCAGCC-3′ P9(SΔ25) 5′-ACATGAAGAATCTGGCTAGCTCATCATGAAAAACAAGCACAGCAGCA-3′ P10(SΔ26) 5′-ACATGAAGAATCTGGCTAGCTCATCAACCTGAAAAACAAGCACAGCA-3′ |
2.3. Recovery of recombinant VSVMT expressing PEDV S protein
The cytoplasmic tail (CT) of PEDV S is composed of 27 amino acids. To construct VSVMT expressing full-length (SFL) or truncated (SΔ#) S protein (VSVMT-S), a sequential deletion of the CT within SH2012 S protein gene was first performed for amino acids (AA) 19 to 26. The PCR primers used for amplifying the S gene or its mutants were shown in Table 1. These mutated genes were inserted into the cloning sites between the G and L genes of the VSV genome using the One Step Seamless Cloning kit (Yeasen, China) with XhoI and NheI restriction enzyme digestion. The resulting plasmids were designated as pVSVMT-SFL, -SΔ19, -SΔ20, -SΔ21, -SΔ22, -SΔ23, -SΔ24, -SΔ25, or -SΔ26.
Recovery of the VSVMT-S virus was performed following the methods by Fang et al. (2012). Briefly, BHK21 cells were infected for 1 h with a recombinant vaccinia virus (vTF7-3) expressing T7 RNA polymerase. The infected cells were then co-transfected with a pVSVMT clone containing the full-length S gene or one of its truncated versions together with the helper plasmids pBS-N, P, and L. After 48 h of transfection, the culture supernatants were collected and filtered into fresh BHK21 cells through a 0.2-μm membrane. These cells were checked daily for infection. If a typical cytopathic effect was observed 2–3 days afterwards, the supernatants were collected, and the viruses were plaque-purified in Vero cells. Individual plaques were isolated, and seed stocks were amplified in BHK21 cells. The rescued viruses were initially confirmed with RT-PCR, and the viral stocks were amplified by passage at multiplicity of infection (MOI) of 0.01 in BHK21 cells. For purifying recombinant VSV, cell culture fluids were clarified by centrifugation at 3000 r/min for 30 min. Viruses were then concentrated via centrifugation at 161,000×g with a 40% (w/v) sucrose cushion for 2 h at 4 °C in a Ti70 rotor (Beckman, USA). The pellet was resuspended in TNC buffer (0.05 M Tris-HCl, 0.15 M NaCl, 15 mM CaCl2 [pH 6.5]). Virions were further purified through a 20–50% (w/v) sucrose gradient by ultracentrifugation at 110,000×g for 1 h at 4 °C in a SW41 rotor (Beckman). The final pellet was resuspended in TNC buffer. Purified viruses were analyzed by western blotting. Typical bands of VSV or PEDV S were detected using convalescent sera from VSV-infected mice or PEDV-infected pigs.
2.4. Replication kinetics of recombinant VSVMT -S
To characterize the replication kinetics of VSVMT-S in vitro, one-step growth curves were set up in BHK21 cells. Briefly, cells in 12-well plates were infected with VSVMT-S or VSVMT at a MOI of 3. After 1 h of absorption, the inoculum was removed, the cells were washed three times with DPBS, and fresh DMEM supplemented with 2% fetal bovine serum was added. The infected cells were incubated at 37 °C, and aliquots of cell culture supernatants were removed in triplicate at 0, 4, 8, 12, 24, and 48 h post-inoculation (hpi). Viral titers in the cellular supernatants were detected by plaque assays as described above, and the mean values were quantified.
2.5. Animal experiments
VSV-based vaccines have been proven to be capable of inducing gut mucosal immunity in mice via the intranasal (IN) route (Wu et al., 2014). To identify the immunogenicity of VSVMT-S in vivo, mice and pigs were inoculated with the virus via the IN or intramuscular (IM) route. All animal experiments were conducted in accordance with the ethical guidelines of Shanghai Jiao Tong University.
2.5.1. Mouse experiments
Specific-pathogen free (SPF) female BALB/c mice (∼20 g body weight) were purchased from Shanghai SLAC Experimental Animal Company (Chinese Academy of Sciences, China). As shown in Table 2 , mice were randomly divided into five groups of five animals each: (1) IN high-dose live VSVMT-S (103 PFU/50 μl); (2) IN low-dose live VSVMT-S (102 PFU/50 μl); (3) IM high-dose binary ethylenimine (BEI)-inactivated VSVMT-S (103 PFU/50 μl); (4) IM low-dose BEI-inactivated VSVMT-S (102 PFU/50 μl); and (5) PBS mock-vaccination group. All animals were primed at day 0 and then boosted once 10 days later. The inactivation of viruses with BEI was performed as described previously (Bahnemann, 1976). In the IM groups, BEI-inactivated VSVMT-S was mixed with Freund's complete adjuvant (Sigma, USA) for priming and with Freund's incomplete adjuvant for booster immunization. Blood samples were collected on day 0 and then every 10 days until 50 days post-vaccination. Animal sera were separated and stored at −70 °C for use in neutralization assays.
Table 2.
Experimental design of mouse and pigs immunization.
| Group | Inoculum | Routes | Immunization dose | Immunization days | |
|---|---|---|---|---|---|
| Mouse | 1 | VSVMT-S | IN | 103 PFU | 0, 10 |
| 2 | VSVMT-S | IN | 102 PFU | 0, 10 | |
| 3 | VSVMT-S | IM | 103 PFU | 0, 10 | |
| 4 | VSVMT-S | IM | 102 PFU | 0, 10 | |
| 5 | PBS | IM | / | 0, 10 | |
| Pig | 1 | VSVMT-S | IN | 107 PFU | 0, 10 |
| 2 | VSVMT-S | IN | 106 PFU | 0, 10 | |
| 3 | VSVMT-S | IM | 107 PFU | 0, 10 | |
| 4 | VSVMT-S | IM | 106 PFU | 0, 10 | |
| 5 | PBS | IM | / | 0, 10 | |
a. IN: intranasal inoculation.
b. IM: intramuscular injection, viruses were mixed with complete or incomplete Freund's adjuvant at ratio of 1:1.
2.5.2. Pig experiments
2.5.2.1. Experiment 1. Immunogenicity assessment in pigs
Pigs were vaccinated with VSVMT-S via the IN or IM route. A total of 15 4-week-old healthy Bama minipigs were purchased from Swine Centre, University of Shanghai Jiao Tong University. All animals were negative for both VSV and PEDV based on a serum neutralizing antibody assay prior to study and were randomly divided into five groups of three animals each: (1) IN high-dose live VSVMT-S (107 PFU/500 μl); (2) IN low-dose live VSVMT-S (106 PFU/500 μl); (3) IM BEI-inactivated high-dose VSVMT-S (107 PFU/500 μl); (4) IM BEI-inactivated low-dose VSVMT-S (106 PFU/500 μl); and (5) PBS mock vaccination group (Table 2). In the IM groups, BEI-inactivated VSVMT-S was also mixed with Freund's complete adjuvant (Sigma) for priming and with Freund's incomplete adjuvant for booster immunization. Blood samples were collected on day 0, and then every 10 days until 50 days post-vaccination. Animal sera were separated and stored at −70 °C for use in additional assays.
2.5.2.2. Experiment 2. Characterization of commercial adjuvants
In this experiment, various commercial adjuvants were tested for use in VSVMT-S vaccine formulation. Nine healthy Bama minipigs (∼5 kg bodyweight) were equally divided into the following three groups: group 1, MONTANIDE™ IMS 1313 VG NPR adjuvant (IMS 1313); group 2, MONTANIDE™ ISA 206 VG adjuvant (ISA 206), and group 3, MONTANIDE™ ISA15A VG adjuvant (ISA15A). The immunization procedure was designed as shown in Table 3 . VSVMT-S was mixed with above adjuvants by following the manufacturer's protocol, and pigs were then inoculated twice (separated by a 10-day interval) via the IM route into two sides of the neck. Pig blood was collected before the primary inoculation and then every 10 days until 50 days post-vaccination. Blood samples were collected through the pig anterior vena cava, and the serum was separated and stored at −70 °C.
Table 3.
Experimental design of pig immunization with various adjuvants.
| Group | Inoculum | Dose | Routes | Immunization adjuvants | Immunization days |
|---|---|---|---|---|---|
| 1 | VSVMT-S | 107 PFU | IM+ | IMS 1313 a | 0, 10 |
| 2 | VSVMT-S | 107 PFU | IM+ | ISA15A b | 0, 10 |
| 3 | VSVMT-S | 107 PFU | IM+ | ISA 206 c | 0, 10 |
IMS 1313: MONTANIDE™ IMS 1313 VG NPR adjuvant.
ISA 206: MONTANIDE™ ISA 206 VG adjuvant.
ISA 15A: MONTANIDE™ ISA15A VG adjuvant.
2.5.2.3. Experiment 3. Sow immunization and piglet challenge
To test lactogenic immunity transferred by pregnant sows to neonatal piglets, three healthy pregnant Bama sows (body weights of ∼70 kg) were intramuscularly vaccinated three times, 14 days apart (42, 28, or 14 days before sow farrowing). Each dose of experimental vaccine contained 108 PFU of the BEI-inactivated VSVMT-S, which was emulsified with a suitable adjuvant. Three sows inoculated with PBS were set up as a mock control. Farrowing was induced at 14 days after the last vaccination. Blood samples were collected from the sows both prior to vaccination and at farrowing. Colostrum samples were collected on the day of sow farrowing. Piglets were allowed to suckle their dams until 5 days after birth, after which litters from the VSVMT-S or mock vaccination groups were orally challenged with the virulent SH2012 isolate at dose of 102 TCID50. In addition, two piglets were randomly selected from each litter of the mock-vaccinated sows and inoculated orally with PBS as a mock challenge control. The clinical signs, diarrhea, morality, and weight of the challenged piglets were monitored daily throughout the study. Rectal swabs were collected daily from each piglet for detecting the presence of viral RNA by RT-qPCR as previously described (Makadiya et al., 2016). Blood samples were collected from piglets on the day of challenge for use in virus neutralization (VN) antibody assays. Animals were monitored for clinical signs until 10 days post-challenge. Clinical significance score (CSS) was determined as previously described (Makadiya et al., 2016), using the following scoring criteria as a measure of diarrhea severity: 0, normal and no diarrhea (mean Ct values of >30); 1, mild and fluidic feces; 2, moderate watery diarrhea; 3, severe watery and projectile diarrhea (mean Ct values of <20); and 4, death.
2.6. Neutralizing antibody detection
Sera from the experimental animals were heat-inactivated at 56 °C for 30 min prior to antibody detection. PEDV-specific neutralizing antibodies in the sera were detected using a VN assay in 96-well microtiter plates as previously described by Makadiya et al. (2016). VSV-specific neutralization antibodies in the animal serum were determined as previously described (Fang et al., 2015). Colostrum or milk from sows were collected and incubated with rennin at a final concentration of 2.5 μg/ml at 37 °C for 30 min. Once solidified, the whey was separated by centrifugation at 8000 rpm for 10 min at 4 °C. The whey was then collected and kept at −70 °C until testing. The presence of PEDV- or VSV-specific neutralizing antibodies in the whey was also determined using a VN assay as previously described (Makadiya et al., 2016).
2.7. Enzyme-linked immunosorbent assay (ELISA)
PEDV-specific IgG in pig serum was detected with an ELISA kit manufactured by National Engineering Research Centre of Veterinary Biologics Corp (Harbin, China). Briefly, 100-fold-diluted serum samples were added into 96-well plates coated with PEDV in duplicate and incubated at 37 °C for 1 h. After the plates were washed three times with 0.05% PBS tween 20, HRP-conjugated anti-pig IgG was added into each well (1:5000), and the plates were incubated at 37 °C for 1 h. After another round of washing, color development was carried out by adding 100 μl of TMB Substrate to each well and incubating the plates for 10 min; after the addition of 100 μl of Stop Solution, the absorbance was measured at 450 nm. The S/P values were calculated as follows: (Test sample value − negative control value)/(positive control value − negative control values). An S/P value of >0.4 was defined as positive.
2.8. Statistics
Statistical analyses were performed using SPSS 19.0 software (Chicago, USA); the exact tests varied depending on the type of experiment. Differences between two groups were assessed using an unpaired two-tailed t-test. Statistical analyses among multiple groups of variance were conducted using a one-way ANOVA, with significant differences between means determined using Duncan's multiple range tests. A p-value of <0.05 was considered statistically significant.
3. Results
3.1. Phylogenetic analysis indicated that epidemic PEDV strain SH2012 belongs to genotype 2b, so its S gene was used to construct a VSV-based PEDV vaccine
Currently, all four PEDV subtypes are endemic in pig farms in China, including vaccine strains (G1a), new variants (G1b), past epidemic strains (G2a), and current dominant epidemic strains (G2b) (Lee, 2015). Given this issue, G2b epidemic or related strains circulating in the field should be employed to develop next generation vaccines. However, a commercial vaccine against G2b strains is still unavailable. SH2012 is a virulent PEDV isolate collected in 2012 in Shanghai, China; this strain caused ∼100% morbidity and >90% mortality in suckling piglets. In our study, a phylogenetic analysis based on the PEDV S gene indicated that SH2012 maps to G2b (Fig. 1 ) and is very closely related to AH 2012, which is regarded as the origin of the PEDV outbreak during 2013–2014 in the USA and Canada (Huang et al., 2013). However, SH2012 has a long genetic distance from the prototype CV777, which was identified as belonging to the G1 genotype (Fig. 1).
Fig. 1.
Phylogenetic analysis of emerging PEDV isolates based on the S gene. The phylogenetic tree, which includes classical strains, global emerging strains, and epidemic strains in China, was constructed by using MEGA 7.0 with the neighbor-joining (NJ) method. Bootstrap resampling (1000 replications) was performed, and bootstrap values are indicated for each node. The TGEV strain (accession number DQ811785) was set as the out-group.
There are at least four neutralizing domains in the PEDV S protein: 1) N-terminal domain (NTD) region (Li et al., 2016); 2) COE domain (residues 499–638) (Chang et al., 2002); 3) S1D region (residues 636–789), which spans the S1–S2 junction region (Sun et al., 2008), and 4) an epitope at the S protein C-terminus (residues 1371–1377) (Cruz et al., 2008). Alignments of full-length S protein amino acid sequences indicated that the similarity between SH2012 and CV777 is 93.2%, and those between SH2012 and AH2012 or Corolado 2013 were each more than 99% (Supl. Fig. 1). The major differences among various strains existed within the S1 region, particularly the NTD domain. Similarities of S1 amino acid sequences among SH 2012, AH 2012, and the US representative strain Colorado 2013 could reach 100%, whereas their similarity to CV777 was only 82.71%, typically with 5 AA insertion at two sites (AA59–62, AA140) and 2 AA deletions at site AA160 in the S protein of G2b strains (SH 2012, AH 2012, or Corolado 2013) in contrast to CV777 (Supl. Fig. 1). There were a total of 11 mutations existing in the COE and S1D regions of SH2012 compared with CV777, of which three were located in S1D and eight in the COE domain. The C-terminal epitope is a highly conserved epitope; not surprisingly, no difference was identified among CV777 and the G2b strains included in our study. The observed differences, especially within the neutralizing epitope(s), may have contributed to the current CV777 vaccine failure in the field (Li et al., 2012). Thus, given the high similarity of the SH2012 S protein to the predominant PEDV G2b strains, it was used to construct a VSV-based PEDV subunit vaccine.
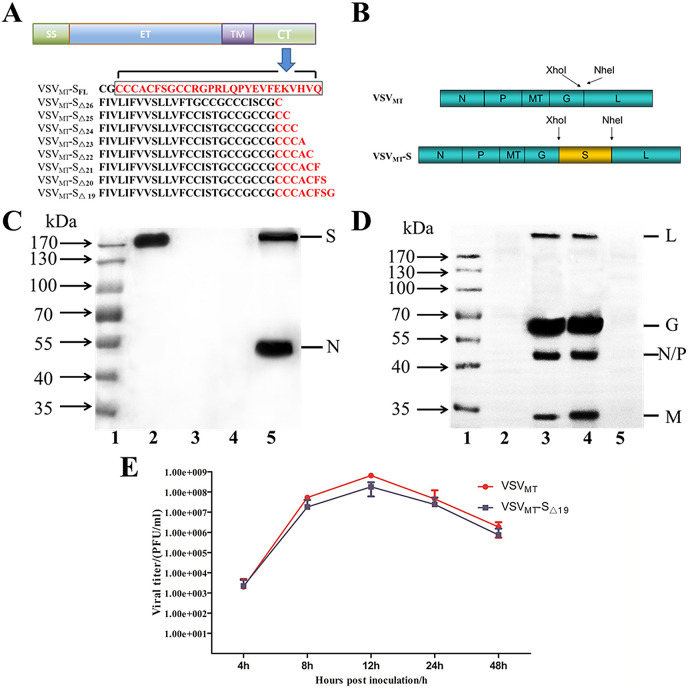
3.2. With 19 amino acids in its cytoplasmic domain deleted, PEDV S protein could be efficiently incorporated into VSVMT particles
Because of its close relatedness with the predominant isolates of G2b in China and other countries, we chose the S protein of SH2012 for use in vaccine development. We initially constructed the rVSV plasmid with the full-length PEDV S gene; however, no replication-competent rVSV was rescued. Next, we generated rVSV plasmids encoding C-terminal-truncated versions of the SH2012 S protein (deletions of AA 19–26) (Fig. 2 A and B). However, only the resulting plasmids, pVSVMT-SΔ19, which encodes the full-length PEDV S protein except for the C-terminal 19 amino acids, was found to be capable of rescuing rVSV. The resulting virus caused typical cytopathic effects in freshly prepared BHK-21 cells and was named as VSVMT-SΔ19. To further confirm the recovery of VSVMT-SΔ19, RNA was extracted from a plaque-purified rVSV clone, and an RT-PCR assay was performed to amplify the S gene, which was identified via DNA sequencing (data not shown). To confirm that the S protein was indeed expressed and incorporated into VSV particles, VSVMT-SΔ19 was purified by ultracentrifugation and then analyzed by western blotting. Using convalescent sera from a PEDV-infected pig as the primary antibody, the ∼200 kDa PEDV S protein was detected in purified VSVMT-SΔ19 but not in VSVMT. In the positive control PEDV strain CV777, in addition to the S protein (∼200 kDa), the N protein (∼55 kDa) was also detected (Fig. 2C). Using convalescent sera from a VSV-infected mouse as the primary antibody, typical bands of VSV structural proteins L, G, N/P, as well as M were detected as previously reported (Roberts et al., 1999) in both VSVMT-SΔ19 and VSVMT but not in CV777 or mock control (Fig. 2D). These results indicated that VSVMT-SΔ19 was successfully rescued, with the PEDV SΔ19 protein incorporated efficiently into VSV particles.
Fig. 2.
Generation of recombinant VSVMTexpressing the PEDV S protein. (A) PEDV S protein structure with different cytoplasmic tail (CT) lengths. SFL, SΔ26, SΔ25, SΔ24, SΔ23, SΔ22, SΔ21, SΔ20, SΔ19 represent the full-length S protein or the S mutants with their CTs truncated from 19 to 26 amino acids, respectively. The remaining CT regions are shown in red. SS: signal peptide sequence, ET: ectodomian, TM: transmembrane domain, CT: cytoplasmic tail. (B) Construction of recombinant VSVMT expressing the PEDV S protein. The parental VSVMT genome encodes the nucleoprotein (N), phosphoprotein (P), glycoprotein (G), RNA polymerase (L), and mutant M protein (MT) that harbors three mutations (M51 deletion, V221F, and S226R). The gene for the PEDV S protein or its truncations were inserted into the cloning sites between the G and L genes of the VSV genome through XhoI and NheI restriction enzyme digestion. (C) VSVMT-SΔ19 identification with PEDV convalescent sera. 1. Protein ladder; 2. Purified VSVMT-SΔ19; 3. Purified VSVMT; 4. Mock control; 5. Purified CV777. (D) VSVMT-SΔ19 identification with VSV convalescent sera. 1. Protein ladder; 2. Purified CV777; 3. Purified VSVMT-SΔ19; 4. Purified VSVMT; 5. Mock control. (E) One-step growth curve of VSVMT and VSVMT-SΔ19 in BHK21 cells. Cells were inoculated with different viruses at MOIs of 3. Plaque titrations from the indicated time points were performed with Vero cells. Data shown are means ± standard deviations.
To characterize the replication kinetics of VSVMT-SΔ19 in vitro, a one-step growth curve of the virus was set up in BHK21 cells at a MOI of 3, with VSVMT as the control. As shown in Fig. 2E, the titers in supernatants of VSVMT-SΔ19-inoculated cells increased post-inoculation, with the titer reaching its peak of 1.8 ± 1.2 × 108 PFU/ml at 12 hpi, then declining. VSVMT also reached its highest titer of 5 ± 1.3 × 108 PFU/ml at 12 hpi. Together, our results showed that VSVMT-SΔ19 could replicate efficiently in vitro, at levels comparable to those of VSVMT.
3.3. VSVMT-SΔ19 IM vaccination simultaneously stimulated PEDV- and VSV-specific humoral immune responses in both pigs and mice
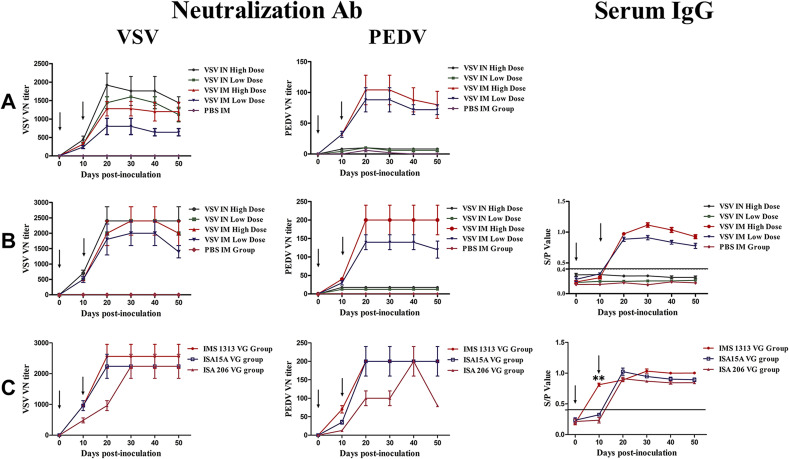
To explore the PEDV-specific immunogenicity of VSVMT-SΔ19 in vivo, mice and pigs were inoculated with the virus through the IN or IM routes, with PBS as the mock vaccination control. As shown in Fig. 3 , no virus-specific antibody was detected in the control mice and pigs. However, when emulsified with Freund's adjuvants, VSVMT-SΔ19 could induce PEDV- and VSV-specific immunity via the IM route in both pigs and mice in a dose- and time-dependent manner. In the IM high-dose mouse group, the PEDV-specific VN titers peaked at 1:100 at 10 days after the second immunization, whereas the VSV-specific VN titers were 1:1200 (Fig. 3A). Notably, although PEDV-specific VN antibodies could not be induced via the IN route by either high or low-dose live VSVMT-SΔ19 in mice, VSV-specific VN antibodies could still be detected in inoculated mice, with peak titers of ∼1:2000 in the high-dose group at 10 days post-booster vaccination (Fig. 3A). The humoral immune responses elicited by VSVMT-SΔ19 in pigs were examined by ELISA and neutralization assay. The pigs vaccinated via the IM route showed an increase in VN antibody titer and serum IgG levels after both the first and second vaccination. In the high-dose group, the PEDV-specific VN antibody titer peaked at 1:250 at 10 days after the second immunization, whereas VSV-specific VN antibody titers peaked at 1:2500 (Fig. 3B). Similarly, the serum IgG level reached its peak at 20 days after the second immunization in the IM high-dose pig group. As with mice, VSV-specific, but not PEDV-specific, antibodies could be detected in pigs immunized with VSVMT-SΔ19 via the IN route (Fig. 3B). Together, these findings indicate that VSVMT-SΔ19 could simultaneously stimulate PEDV- and VSV-specific humoral immune responses in both pigs and mice via the IM but not the IN route.
Fig. 3.
PEDV- and VSV-specific humoral immune responses in VSVMT-SΔ19-immunized animals. Mice and piglets were vaccinated twice, on day 0 and 10, as indicated by arrows. Animal serum samples were collected at day 0 and then every 10 days until 50 days post-vaccination. Virus-neutralizing (VN) antibody and sera IgG were assayed. (A) Mouse VN antibody detection. (B) Pig VN antibody sera IgG detection. (C) Antibody detection in pigs immunized with VSVMT-SΔ19 emulsified with various commercial adjuvants (ISA15A VG, ISA206 VG, and IMS1313VG). The VN titer and sera IgG levels were detected. S/P values from the ELISA test were calculated as follows: (Test sample value − negative control value)/(positive control value − negative control values). An S/P value of >0.4 was defined as positive. Data are presented as means ± SD. Statistical analyses were conducted using a one-way ANOVA, and significant differences between means were determined using Duncan's multiple range tests. **statistically significant at the 0.01 level when the mean of the IMS1313 VG group was compared with the means of the ISA15A VG and ISA206 VG groups.
Although Freund's adjuvants are commonly used for research, they are prohibited for use in the farm industry because of their toxicity. To formulate a clinically useable vaccine with VSVMT-SΔ19, the following three commercial adjuvants were tested: MONTANIDE™ IMS 1313 VG NPR adjuvant (IMS 1313); MONTANIDE™ ISA 206 VG adjuvant (ISA 206), and MONTANIDE™ ISA15A VG adjuvant (ISA15A). IMS1313 is a nanoparticle adjuvant, ISA206 is a Water-Oil-Water emulsion, and ISA15A is an Oil-Water emulsion. Purified VSVMT-SΔ19 was separately mixed with each of these adjuvants. Each pig was intramuscularly inoculated twice, 10 days apart (days 0 and 10), at a dose of 107 PFU. As shown in Fig. 3C, PEDV- and VSV- specific neutralizing antibodies were effectively stimulated in pigs. After the first immunization, the PEDV-specific VN titers in the ISA15A and ISA206 groups were 1:10 and 1:40, respectively, whereas those in the IMS1313 group reached 1:80. The PEDV-specific VN titers in the ISA206 group eventually peaked at 1:200 at 30 days after the second immunization and then declined rapidly; in contrast, the PEDV-specific VN titers in the IMS1313 and ISA15A groups each reached 1:200 and maintained this level until 30 days after the second immunization. Sera IgG levels in pigs vaccinated with VSVMT-SΔ19 emulsified with IMS1313 were positive at only 10 days after the first immunization, with an S/P value that was significantly higher than those of the other adjuvants (p < 0.01) (Fig. 3C). In addition, no adverse reaction was observed at the injection sites when IMS1313 was used as the adjuvant, whereas adverse effects on the skin occurred in some pigs when using ISA206 or ISA15A as adjuvants (data not shown). Together, these results indicated that IMS1313 is suitable for use as an adjuvant for future studies on this vaccine.
3.4. Administration of VSVMT-SΔ19 in pregnant sows provided robust PEDV-specific passive immunity in piglets
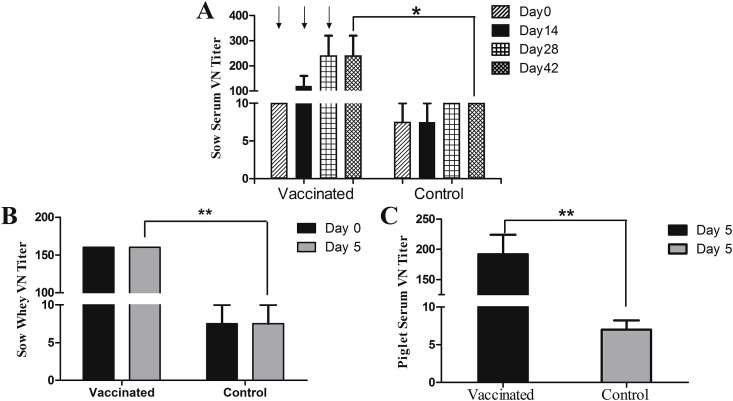
Since 2010, many emerging PEDV isolates have been characterized with high mortality in piglets. Here, we assessed piglets for passive immunity against PEDV transferred by immunized sows. VSVMT-SΔ19, emulsified with IMS1313, was injected into pregnant sows three times (108 PFU each dose) via the IM route, with PBS as the mock control. The humoral immune responses in sows elicited by VSVMT-SΔ19 were examined by neutralization assays of serum or colostrum/milk. The vaccinated sows showed increases in specific serum neutralizing antibody titer after vaccination, which peaked at 1:250 after the second immunization and was maintained at this level until the day of farrowing (Fig. 4 A). The VN titers in whey from colostrum or milk also peaked at 1:150 on the first and fifth day post-farrowing, a level that is significantly higher than the VN titer from control sows (p < 0.01) (Fig. 4B). Sera collected from piglets in the litters of vaccinated sows at 5 days after birth, which was the day of PEDV challenge, had VN titers of ∼1:180; this level was also significantly higher compared with the litters of control sows (p < 0.01) (Fig. 4C).
Fig. 4.
PEDV-specific humoral responses in sows and piglets. Pregnant sows were vaccinated with VSVMT-SΔ19 or PBS three times on days 0, 14, and 28 as indicated by arrows. (A) PEDV-specific virus neutralizing (VN) antibody titers in sera from sows collected at the indicated time points post-inoculation. (B) VN antibody titers in the whey of colostrum or milk collected from sows at the indicated timepoints. (C) VN antibody titers of the sera collected from piglets that suckled vaccinated or control sow milk for 5 days before virulent PEDV challenge. Statistical analyses were performed using an unpaired two-tailed t-test; data are presented as means ± SD. *p < 0.05 and **p < 0.01.
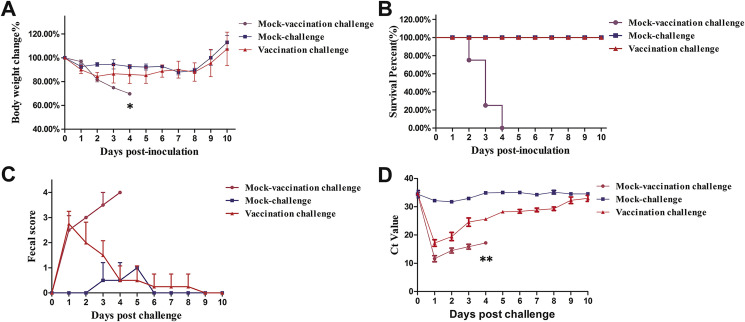
To test the protective potential of immunity transferred by VSVMT-SΔ19-vaccinated sows, piglets delivered by vaccinated and mock-vaccinated sows were orally challenged with virulent SH 2012. In addition, six piglets from mock-vaccinated sows were also inoculated with PBS as the mock-challenge control. All the piglets were monitored daily until 10 days post-challenge (dpc) for clinical symptoms, fecal grade, body weight, and mortality. PEDV RNA was extracted from daily rectal swabs of piglets and assessed by RT-qPCR. As shown in Fig. 5 , following the challenge with SH 2012, the body weights of piglets from mock-vaccination sows dropped ∼30% at 4 dpc, to a level that is significantly lower than that of the other two groups (p < 0.05) (Fig. 5A). Severe watery diarrhea with vomiting could be observed in these animals, with clinical scores reached as high as 4. Virus shedding in their feces was confirmed by RT-qPCR, with the Ct values reaching as low as 12 at 2 dpc and remaining less than 15 at 4 dpc; these levels were also significantly lower than those in the other two groups (Fig. 5D) (p < 0.01). All piglets delivered by mock-vaccination sows died from the virulent PEDV challenge (6/6) (Fig. 5B). In contrast, the piglets delivered by VSVMT-SΔ19-vaccinated sows survived the virulent PEDV challenge, and their body weights started to increase at 6 dpc (Fig. 5A). Not surprisingly, no watery diarrhea was observed in the mock-challenge piglets; their average clinical scores were less than 1, and their Ct values were above 30. Challenged piglets from vaccinated sows had mild diarrhea beginning at 2 dpc, with clinical scores around 2.5. However, at 9 dpc, no watery diarrhea was observed in these animals (Fig. 5C). Additionally, reduced virus shedding was observed in piglets from vaccinated sows following oral challenge, with the mean Ct value increasing from 15 to 30 between 2 and 10 dpc (Fig. 5D). Together, these results indicate that exposure of sows to VSV MT-SΔ19 could provide protective lactogenic immunity to their piglets when challenged with homologous virulent PEDV.
Fig. 5.
Protective efficacy in piglets of passive immunity from sow vaccination. Litters from VSVMT-SΔ19-vaccinated or mock-vaccinated sows were orally challenged with virulent PEDV SH2012 at 5 days of age. The mock-challenge controls, which were orally inoculated with PBS, were born from mock-vaccinated sows. Clinical significance scores were determined as described in the Materials and methods. (A) Piglet body weight changes (%). (B) Survival rate of piglets (%). (C) Piglet fecal score. (D) Virus shedding. PEDV viral RNA was extracted from rectal swabs of challenged or mock-challenged piglets and assessed by real-time RT-PCR. Values are the mean ± standard deviation. Statistical analyses among multiple groups of variance were conducted using a one-way ANOVA, with significant differences between means determined using Duncan's multiple range tests. *p < 0.05 and **p < 0.01 when the mean of the mock vaccination challenge group was compared with the mean of the mock challenge and vaccination challenge groups.
4. Discussion
Vaccination is the fundamental strategy for controlling and eradicating PED; however, cross-protection between the PEDV G1 and G2 genotypes is incomplete (Gerdts and Zakhartchouk, 2017). Traditional vaccines based on the classic G1 genotype strain CV777 cannot effectively control the epidemic of emerging isolates, which predominantly belong to the G2b subtype in Asia and America (Lee, 2015; Wang et al., 2016). Currently, a commercial vaccine against epidemic G2b is still unavailable. Therefore, a G2b strain-based vaccine should be developed.
SH2012 is a virulent PEDV strain that was isolated in 2012 in Shanghai, China. It had a high mortality of almost 100% in infected neonatal piglets and caused a serious clinical disease with symptoms such as watery diarrhea in nursing pigs and sows. An antigenic analysis showed that the major differences among various G1 and G2 genotype PEDV strains were mainly located within the S1 region, especially within the NTD domain. A phylogenetic analysis based on the S gene indicated that SH2012 maps into the G2b subtype. Additionally, an analysis of the S protein amino acid sequences further confirmed its close relationship to other emerging strains of G2b PEDV, including AH 2012, which is considered to be the origin of the PEDV strains that have caused large-scale outbreaks in the USA since 2013 (Huang et al., 2013). Based on these findings, we selected SH2012 as a candidate for use in making a novel PEDV vaccine.
Presently, one of the major hurdles for PEDV vaccine development is that isolating PEDV in cell culture has proven fastidious and time-consuming, and an isolated virus may still be incapable of retaining infectivity upon further in vitro passage. Because of these issues, Gerdts et al. claimed that research on next-generation vaccines, such as subunit and viral-vectored approaches, is critical for the prevention of future outbreaks of emerging coronavirus diseases (Gerdts and Zakhartchouk, 2017). Hain et al. has developed a recombinant parapoxvirus as the vector to express S protein (Hain et al., 2016). Recombinant VSV is a promising vaccine vector for expressing foreign antigens, and has been used as the platform for construction of human vaccines, such as the well-known VSV-based Ebola virus vaccine (Regules et al., 2017). It has been shown that the cytoplasmic tail of coroanvirus spike protein can play key roles for its incorporation into VSV particles (Schwegmann-Wessels et al., 2009). In our study, full-length S gene of SH2012 was initially cloned into genome of highly attenuated VSVMT, with the aim of making mucosal vaccine based on replication-competent VSV for pigs. However, no rVSVMT-SFL recovery was observed. Both PEDV and TGEV belong to Alphacoronavirus genus within the Coronaviridae family. A tyrosine-based motif (YxxI) within cytoplasmic tail of TGEV spike protein can lead to its intracellular retention and thus interfere with its incorporation into VSV particles (Schwegmann-Wessels et al., 2004). Deletion of the YxxI motif results in successful recovery of recombinant VSV pseudotyped with truncated TGEV S (SΔ14) (Schwegmann-Wessels et al., 2009). It is noteworthy that there is a conserved dibasic motif of KxHxx within PEDV S cytoplasmic tail (KVHVQ), which can also lead to retention of S protein to the endoplasmic reticulum–Golgi intermediate compartment (ERGIC) (https://www.ncbi.nlm.nih.gov/pubmed/?term=Shirato%20K%5BAuthor%5D&cauthor=true&cauthor_uid=21840351, Shirato et al., 2011). As shown by Hou et al., the YxxI and KxHxx motifs within PEDV S tail can synergistically regulate its expression on cell surface (Hou, et al., 2019). Consequently, as with TGEV S, the failure of full-length PEDV S incorporation into VSV particles might also be due to its intracellular retention, rather than presence at the plasma membrane, where VSV should be assembled by a budding process. Thus,to remove the YxxI and KxHxx motifs, we generated rVSVMT plasmids expressing S mutants with 19–26 AA of the cytoplasmic domain deleted (Fig. 2A and B). The resulting plasmid pVSVMT-SΔ19, which encodes S except for its C-terminal 19 amino acids, achieved successful rescue, whereas the other constructs (SΔ20-26) failed. The results suggested that, some structural features within the truncated S protein, such as an optimal length of the truncated cytoplasmic tail, might also be essential for its incorporation into VSV particles even though the intracellular retention signals have been removed. However, the detailed mechanism still need clarification in future study. Notably, although inserted with a large-sized foreign protein, VSVMT-SΔ19 still could replicate as efficiently as its parental virus VSVMT in vitro, with the maximum titer exceeding 108 PFU/ml in BHK-21 cells. Together with the advantages that BHK21 cells can grow in suspension in disposable bioreactors at cell concentrations higher than 1 × 107 cells/mL (Genzel, 2015), recombinant VSV could be an ideal platform for conveniently preparing vaccines against emerging or reemerging PEDV. At present, it is still difficult to construct recombinant PEDV using reverse genetics. However, with helps by VSV-based PEDV S protein, epitopes, especially conformational epitopes on S protein might be mapped accurately, even to the level of single amino acid. In addition, the role of S protein in loss of cross-protection between G1 and G2 genotypes isolates might also be identified with the technique.
Foreign antigens delivered by live VSV vector via IN route of immunization have been shown to be capable of inducing mucosal immunity against enteroviruses, like enterovirus coxsackievirus B3 (CVB3), in mice (Wu et al., 2014). However, the induction of mucosal immunity in this manner has not yet been confirmed in a natural host of VSV, such as pigs. Here, we hypothesized that VSVMT-SΔ19 could also stimulate PEDV-specific gut immunity in pigs when administered via the IN route. Our observation of robust VSV-specific humoral immune responses following immunization suggest that the administered VSVMT-SΔ19 replicated in the nasal mucosa. However, to our surprise, PEDV-specific humoral responses could not be detected. The reason for the failure of VSVMT-SΔ19 to stimulate PEDV-specific antibodies when administered via the IN route still needs clarification; it is possible that the S protein was not expressed in tissue cells or not successfully presented to host antigen-presenting cells, thus failing to stimulate the production of PEDV-specific antibodies. Similar results were found in a recombinant parapox virus-based PEDV vaccine, which was reported to stimulate PEDV-specific immunity when administered via the IM route but not the transcutaneous route, which is the natural infection route of parapox virus (Hain et al., 2016). In addition, it was demonstrated that PEDV-specific immune responses could be stimulated effectively in sows immunized via the IM route with VSVMT-expressed PEDV S protein. Notably, among the adjuvants tested in our present study, the nanoparticle adjuvant IMS1313 was the most potent for formulating a VSV-based PEDV vaccine. Neutralization antibody titers, as well as serum IgG, against PEDV were positively detected as early as 10 days post-primary immunization. Furthermore, the passive transfer of antibodies from immunized sows to piglets was observed, as PEDV-specific neutralization antibodies were detected in the serum of piglets born to immunized sows following their ingestion of colostrum and milk.
In summary, our study showed that PEDV S protein with 19 AA within its carboxyl terminal deleted could be expressed and incorporated into VSVMT particles efficiently. Additionally, our VSV-based PEDV subunit vaccine could confer potent passive immunity against PEDV in piglets following the immunization of pregnant sows.
Acknowledgements
The work was funded by the National Natural Sciences Foundation of China (31272562) and the Industry-University Cooperation Project of Minhang District (2016MH250). We thank Dr. Xinchu Zhou for his helps with animal experiments. We also thank Yinjie Zhang, Man Wang and Wen Guan for their technical assistances.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2019.05.009.
Conflicts of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Multiple alignment of S protein dominant neutralizing epitope sequences. Compared with PEDV strain CV777, the amino acid of the new strain has undergone many mutations, and even the polarity of many amino acids has changed. Yellow frame contains the NTD/S0 region; Blue frame contains the COE region; Red frame contains the S1D region; and Orange frame contains the epitope region of residues 1371–1377 AA.
References
- Bahnemann H.G. Inactivation of viruses in serum with binary ethyleneimine. J. Clin. Microbiol. 1976;3:209–210. doi: 10.1128/jcm.3.2.209-210.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.H., Bae J.L., Kang T.J., Kim J., Chung G.H., Lim C.W., Laude H., Yang M.S., Jang Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- Cruz D.J., Kim C.J., Shin H.J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. 2008;132:192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Zhang S., Sun X., Li J., Sun T. Evaluation of attenuated VSVs with mutated M or/and G proteins as vaccine vectors. Vaccine. 2012;30:1313–1321. doi: 10.1016/j.vaccine.2011.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Qi B., Ma Y., Zhou X., Zhang S., Sun T. Assessment of a novel recombinant vesicular stomatitis virus with triple mutations in its matrix protein as a vaccine for pigs. Vaccine. 2015;33:6268–6276. doi: 10.1016/j.vaccine.2015.09.069. [DOI] [PubMed] [Google Scholar]
- Genzel Y. Designing cell lines for viral vaccine production: where do we stand? Biotechnol. J. 2015;10:728–740. doi: 10.1002/biot.201400388. [DOI] [PubMed] [Google Scholar]
- Gerdts V., Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2017;206:45–51. doi: 10.1016/j.vetmic.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain K.S., Joshi L.R., Okda F., Nelson J., Singrey A., Lawson S., Martins M., Pillatzki A., Kutish G.F., Nelson E.A., Flores E.F., Diel D.G. Immunogenicity of a recombinant parapoxvirus expressing the spike protein of Porcine epidemic diarrhea virus. J. Gen. Virol. 2016;97:2719–2731. doi: 10.1099/jgv.0.000586. [DOI] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Meulia T., Gao X., Saif L.J., Wang Q. Deletion of both the tyrosine-based endocytosis signal and the endoplasmic reticulum retrieval signal in the cytoplasmic tail of spike protein attenuates porcine epidemic diarrhea virus in pigs. J. Virol. 2019;93(2) doi: 10.1128/JVI.01758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4 doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.C., Perlman S., Anderson L.J. Coronaviridae. In: Knipe D.M., Howley P.M., Griffin D.E., Martin M.A., Lamb R.A., Roizman B., Straus S.E., editors. Fields Virology. fifth ed. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1306–1336. [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., van Kuppeveld F.J.M., He Q., Rottier P.J.M., Bosch B.-J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016;226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makadiya N., Brownlie R., van den Hurk J., Berube N., Allan B., Gerdts V., Zakhartchouk A. S1 domain of the porcine epidemic diarrhea virus spike protein as a vaccine antigen. Virol. J. 2016;13:57. doi: 10.1186/s12985-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Saif L.J., Marthaler D., Esseili M.A., Meulia T., Lin C.M., Vlasova A.N., Jung K., Zhang Y., Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 2014;173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regules J.A., Beigel J.H., Paolino K.M., Voell J., Castellano A.R., Hu Z., Munoz P., Moon J.E., Ruck R.C., Bennett J.W., Twomey P.S., Gutierrez R.L., Remich S.A., Hack H.R., Wisniewski M.L., Josleyn M.D., Kwilas S.A., Van Deusen N., Mbaya O.T., Zhou Y., Stanley D.A., Jing W., Smith K.S., Shi M., Ledgerwood J.E., Graham B.S., Sullivan N.J., Jagodzinski L.L., Peel S.A., Alimonti J.B., Hooper J.W., Silvera P.M., Martin B.K., Monath T.P., Ramsey W.J., Link C.J., Lane H.C., Michael N.L., Davey R.T., Jr., Thomas S.J., r V.S.V.G.Z.-G.P.S.G. A recombinant vesicular stomatitis virus Ebola vaccine. N. Engl. J. Med. 2017;376:330–341. doi: 10.1056/NEJMoa1414216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Kretzschmar E., Perkins A.S., Forman J., Price R., Buonocore L., Kawaoka Y., Rose J.K. Vaccination with a recombinant vesicular stomatitis virus expressing an influenza virus hemagglutinin provides complete protection from influenza virus challenge. J. Virol. 1998;72:4704–4711. doi: 10.1128/jvi.72.6.4704-4711.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Buonocore L., Price R., Forman J., Rose J.K. Attenuated vesicular stomatitis viruses as vaccine vectors. J. Virol. 1999;73(5):3723–3732. doi: 10.1128/jvi.73.5.3723-3732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Al-Falah M., Escors D., Wang Z., Zimmer G., Deng H., Enjuanes L., Naim H.Y., Herrler G. A novel sorting signal for intracellular localization is present in the S protein of a porcine coronavirus but absent from severe acute respiratory syndrome-associated coronavirus. J. Biol. Chem. 2004;279(42):43661–43666. doi: 10.1074/jbc.M407233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Glende J., Ren X., Qu X., Deng H., Enjuanes L., Herrler G. Comparison of vesicular stomatitis virus pseudotyped with the S proteins from a porcine and a human coronavirus. J. Gen. Virol. 2009;90:1724–1729. doi: 10.1099/vir.0.009704-0. [DOI] [PubMed] [Google Scholar]
- Shirato K., Maejima M., Matsuyama S., Ujike M., Miyazaki A., Takeyama N., Ikeda H., Taguchi F. Mutation in the cytoplasmic retrieval signal of porcine epidemic diarrhea virus spike (S) protein is responsible for enhanced fusion activity. Virus Res. 2011;161(2):188–193. doi: 10.1016/j.virusres.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Feng L., Shi H., Chen J., Cui X., Chen H., Liu S., Tong Y., Wang Y., Tong G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008;131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G.S., McKenna P.M., Koser M.L., McLinden R., Kim J.H., McGettigan J.P., Schnell M.J. Strong cellular and humoral anti-HIV Env immune responses induced by a heterologous rhabdoviral prime-boost approach. Virology. 2005;331:82–93. doi: 10.1016/j.virol.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. vol. 20. North America; 2014. pp. 1620–1628. (Distinct Characteristics and Complex Evolution of PEDV Strains). May 2013-February 2014. Emerging infectious diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Fan X., Yue Y., Xiong S., Dong C. A vesicular stomatitis virus-based mucosal vaccine promotes dendritic cell maturation and elicits preferable immune response against coxsackievirus B3 induced viral myocarditis. Vaccine. 2014;32:3917–3926. doi: 10.1016/j.vaccine.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Lin H., Li B., He K., Fan H. Efficacy and immunogenicity of recombinant swinepox virus expressing the truncated S protein of a novel isolate of porcine epidemic diarrhea virus. Arch. Virol. 2017;162:3779–3789. doi: 10.1007/s00705-017-3548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.