Abstract
Over 3500 individual water samples, for 131 sampling times, targeting waterborne pathogens/fecal indicator bacteria were collected during a 7-year period from 4 sites along an intermittent stream running through a small livestock pasture system with and without cattle access-to-stream restriction measures. The study assessed the impact of cattle pasturing/riparian zone protection on: pathogen (bacterial, viral, parasite) occurrence, concentrations of fecal indicators, and quantitative microbial risk assessments (QMRA) of the risk of Cryptosporidium, Giardia and Escherichia coli O157:H7 infection in humans. Methodologies were developed to compute QMRA mean risks on the basis of water samples exhibiting potentially human infectious Cryptosporidium and E. coli based on genotyping Crytosporidium, and E. coli O157:H7 presence/absence information paired with enumerated E. coli. All Giardia spp. were considered infectious. No significant pasturing treatment effects were observed among pathogens, with the exception of Campylobacter spp. and E. coli O157:H7. Campylobacter spp. prevalence significantly decreased downstream through pasture treatments and E. coli O157:H7 was observed in a few instances in the middle of the unrestricted pasture. Densities of total coliform, fecal coliform, and E. coli reduced significantly downstream in the restricted pasture system, but not in the unrestricted system. Seasonal and flow conditions were associated with greater indicator bacteria densities, especially in the summer. Norovirus GII was detected at rates of 7–22% of samples for all monitoring sites, and rotavirus in 0–7% of samples for all monitoring sites; pasture treatment trends were not evident, however. Seasonal and stream flow variables (and their interactions) were relatively more important than pasture treatments for initially stratifying pathogen occurrence and higher fecal indicator bacteria densities. Significant positive associations among fecal indicator bacteria and Campylobacter spp. detection were observed. For QMRA, adjusting for the proportion of Cryptosporidium spp. detected that are infectious for humans reduces downstream risk estimates by roughly one order of magnitude. Using QMRA in this manner provides a more refined estimate of beneficial management practice effects on pathogen exposure risks to humans.
Keywords: Beneficial management practice, Riparian zone protection, Public health risks
Highlights
-
•
Over 3000 samples were collected in a stream with/without livestock exclusion.
-
•
Study lasted 7 continuous years, and included a QMRA.
-
•
Viruses/pathogenic bacteria/parasites/coliphages & indicator bacteria were monitored.
-
•
Campylobacter decreased downstream of pasture treatments and Escherichia coli O157:H7 was observed more often in unrestricted pasture.
-
•
QMRA provides a more refined estimate of BMP effects on human health.
1. Introduction
Restricting cattle access to water courses has been shown to reduce nutrient, sediment, and bacteria loads, and improve stream and riparian zone integrity (Line, 2003, Mayer et al., 2007). Yet, how these practices influence the prevalence of zoonotic pathogens and viruses that can result from implementation of stream/riparian zone protection practices is not well documented. Schwarte et al. (2011) reported bovine enterovirus in ∼8–17% of the runoff simulations from bare unrestricted stream access pasture sites under rainfall simulation, while bovine coronavirus and rotavirus group A, and Escherichia coli O157:H7 were rarely detected. Billington et al. (2011) suggest that uncontrolled stock access to waterways results in elevated public health risks from pathogens based on hazard identification, exposure assessment, dose-response analysis, and risk characterisation of stock accessing waterways upstream of drinking water off-takes. Aarons and Gourley (2012) suggest improved riparian zone protection approaches and strategies are needed, and Miller et al. (2010) indicate that not all water quality endpoints are improved with riparian zone fencing.
A complicating factor in the assessment of the water quality benefits imposed by riparian zone protection (a beneficial management practice (BMP)), is that pasturing systems, regardless of whether they are fenced or not, are variably susceptible to livestock, wildlife, and human fecal inputs (Wilkes et al., 2013). Varied fecal inputs have the potential to confound the true nature of BMP impact on, in particular, loading of microorganisms in water and the prevalence and nature of zoonotic pathogens of differing virulence potential to humans.
It is becoming increasingly important to gauge the effectiveness of an agricultural BMP in the context of a breadth of potential services the BMP can provide. Most often, agricultural BMPs that are protective of water quality are evaluated for environmental and production benefits, but their potential human health risk implications are not. Schmidt et al. (2013a) recently demonstrated that quantitative microbial risk assessment (QMRA) could be used to quantify human potential health risks from Campylobacter spp. associated with tile drainage management BMPs imposed en masse at a watershed scale. However, even QMRA approaches must consider true relative and absolute risks in the context of differences in pathogen species, genotype, serovars, strains or lineage occurrence, because not all of these pathogen subtypes necessarily have the same virulence potential to humans (see Afchain et al.'s (2008) accounting of differing strains of Bacillus cereus in QMRA of manufactured food, for example). Parsing out risks on the basis of human pathogenicity better represents true exposure risks in the framework of BMP assessment.
This study evaluated whether restricting cattle access to streams can reduce the prevalence of zoonotic pathogens, parasites, and viruses within a stream discharge and seasonal context. It also assessed the utility of fecal indicator organisms to predict pathogen prevalence. The underpinning study hypothesis is that cattle associated pathogens will decrease or remain constant within the restricted cattle access pasture (RCA), and then increase within a downstream unrestricted cattle access pasture (URCA). A Bayesian QMRA approach was employed to calculate human health risks from a hypothetical recreational exposure to these waters for selected pathogens.
2. Methods and materials
2.1. Study site
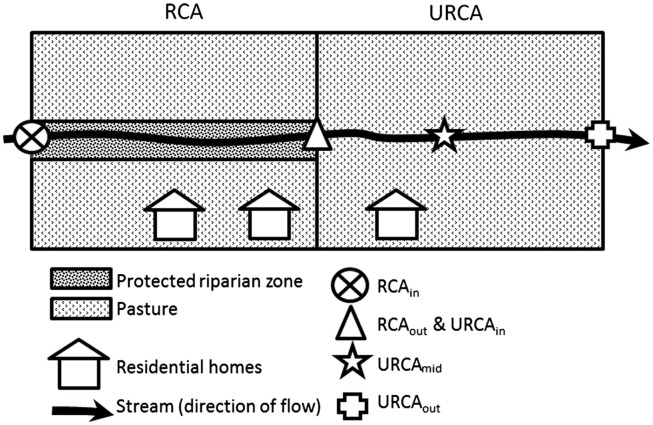
Details of the study site and RCA and URCA are given in Sunohara et al. (2012) (Fig. 1 ). The stream is intermittent (Strahler order = 2) with low and no-flows typically occurring in summer. The stream drains a watershed of roughly 482 ha above the experimental site (above the RCA and URCA area) (Schmidt et al., 2013a). The study framework is an upstream downstream design (USDA, 1996). The RCA area is ∼2 ha in size with a fence buffering the stream from pasturing adult cattle (3–5 m buffer). Stream flow length in the RCA is ∼356 m with a flow gradient of ∼0.002 m m−1. The RCA is situated upstream of the ∼2 ha URCA where adult cattle have unrestricted access to the stream. The URCA possesses a ∼348 m stream flow length and gradient of ∼0.004 m m−1. Holstein cattle densities were maintained at ∼2.5 animals ha−1 in both systems; a density that is not uncommon for pasturing operations in the study region. Cattle were generally allowed to graze from May-June to November-December, beginning in 2005. Adjacent to the RCA and URCA experimental pastures are several residential homes on the southern bank of the stream; small septic system leakages have been observed on occasion. Other prospective fecal inputs along the experimental pastures include house pets, and observed wildlife including small birds, muskrat, vole, turtle, duck, Canada goose, and wild turkey. Fecal inputs upstream of the RCAin could be impacted potentially by a small hobby farm located immediately upstream of RCAin. The farm consists of penned animals of very low and variable seasonal intensity (10–20 goats, 1 donkey, and 2 horses). Yet, it is unclear if there was significant impact of the farm on stream water quality given that many of these livestock were penned in areas where lot drainage to the stream would have been minimal. Liquid swine manures were also applied on fields west of the study area that could have contaminated the roadside ditch that drained near the RCAin sample site in spring and/or fall, but these applications were not frequent.
Fig. 1.
Diagrammatical layout of study site.
2.2. Water sampling, microbiological methods and flow monitoring
Water samples were collected (2004–2010) on a bi-weekly basis beginning April to May until late November (Wilkes et al., 2011). Samples were collected at RCAin (serving as a baseline of water quality input into the restricted cattle access experimental region), RCAout/URCAin (representing the water quality output from the RCA experimental region, and also serving as a baseline of water quality input into the unrestricted cattle access region), at a midpoint of the URCA region (URCAmid), and finally at the output of the URCA region (URCAout) (Fig. 1). Water quality data collected at these sample sites were divided into two groups:
Group 1) Temporally Concurrent Data: data collected concurrently (in synchrony, on the same day) from 2005 to 2010 at RCAin; RCAout/URCAin; and URCAout for bacteria, virus, and coliphage, plus data collected from 2005 to 2010 concurrently at RCAin; URCAout for parasites (due to limited concurrent samples for parasites at output of RCA and input to URCA (RCAout/URCAin) this data was excluded in the temporally concurrent grouping); and Group 2) All Available Data: all data collected at each monitoring site from 2004 to 2010 including URCAmid irrespective of temporal concurrence of sampling (i.e., one site may have more data support than another depending on the microorganism).
Water sampling, hydrological monitoring, and microbiological analysis (pathogenic bacteria, fecal indicator bacteria, coliphage, Giardia and Cryptosporidium (oo)cyst densities) approaches have been provided in other companion papers (Wilkes et al., 2009, Sunohara et al., 2012), with the exception of Aeromonas. Aeromonas (monitoring began in 2009) was processed and enumerated following USEPA Method 1605 (USEPA, 2001) and Havelaar et al. (1987). Cryptosporidium were enumerated, sequenced, and genotyped using phylogenetic analysis (Ruecker et al., 2012).
For group A rotavirus, hepatitis E virus, and norovirus GII, 500 mL of water were processed in accordance with the OPFLP-04 standard method for the recovery and concentration of viruses present in artificially and naturally contaminated water following Health Canada's compendium of analytical methods (Brassard et al., 2005, Brassard et al., 2007; see Supp. Table 1 for detection limits for viruses and other microbiological targets). Negative controls (experimental assay with a non-inoculated water sample as a negative control) were included and processed following the same method applied for virus detection. Viral nucleic acids were extracted using the QIAamp RNA Viral Mini Kit (Qiagen, Mississauga, ON) according to the manufacturer's recommendations. Real-time TaqMan RT-PCR and PCR assays for the detection of norovirus GII and hepatitis E virus, rotavirus, and feline calicivirus (as sample process control) were performed in 25 μL with the 1-step Brillant II QRT-PCR core reagent kit for RNA viruses (Agilent Technologies Canada, Mississauga, ON) and the amplification conditions were carried out according to previously described procedures (Kageyama et al., 2003, Zeng et al., 2008, Ward et al., 2009).
Stream discharge at the RCAin was monitored using a 4150 Area-Velocity Flow Logger (Teledyne Isco, Inc.) and a low-profile area-velocity sensor; an area-velocity calculation was applied to determine stream discharge as described by Sunohara et al. (2012). Due to there being no fixed structure at the RCAout/URCAin and the other downstream sites, we linearly scaled discharge at these sites by applying a proportional amount of discharge based on its contributing area relative to that of RCAin (as the distance between the RCAout and the downstream sites was relatively small); results were verified independently by discharge measurements made by Flow Probe (Global Water Instrumentation, Gold River, CA). Discharge was then grouped based on specific flow conditions measured at the RCAin using percentile classes. Discharge greater than 0.018 m3 s−1 was considered “high flow” (defined as the 75th percentile for discharge measurements), discharge between 0.002 and 0.018 m3 s−1 (defined as discharge between the 25th and 75th percentiles) was considered “low flow,” and discharge <0.002 m3 s−1 (defined as discharge less than the 25th percentile) was considered “no flow.”
2.3. Statistical analysis
Statistical differences in the presence/absence (P/A) of pathogens in water on the basis of sample site and stream flow, and sample site and season, were assessed using Fisher's exact tests. Interactions among site-season-flow characteristics in terms of pathogen P/A were examined in classification mode using CART (Classification and Regression Tree Analysis) (V. 6.6, Salford Systems, CA) (Wilkes et al., 2013). Optimal cross-validated models with a maximum of 3 tree-levels are presented. Site, season, and flow interactions for densities of fecal indicator organism, parasite, and F-coliphage were examined in CART using the least absolute deviation regression tree approach (Wilkes et al., 2011). Differences in the distributions of F-coliphage and fecal indicator bacteria densities within pathogen P/A groupings were assessed by Mann-Whitney U (MWU) tests. Also, F-coliphage and indicator bacteria density thresholds by which pathogen P/A data could be split into similar groups with one binary split were examined using CART in a manner consistent with Wilkes et al. (2009).
2.4. Quantitative microbial risk assessment
It was assumed that all pathogen data from each site are representative of the site-specific variability in pathogen concentrations, and it was assumed that the concentration of total Cryptosporidium oocysts, Giardia cysts, and culturable E. coli at a monitoring site varies temporally according to a gamma distribution. Bayes' theorem and Markov Chain Monte Carlo were used to generate a set of parameter values that is representative of a posterior distribution describing uncertainty in the estimated parameters. For Cryptosporidium and Giardia, the distribution was fit to the raw count and volume data using a probabilistic model that accounts for random measurement errors (Schmidt and Emelko, 2011). Variability in the analytical recovery of the enumeration method was evaluated using matrix spike recovery data (see Supplementary Content, QMRA method).
An assumption in this model is that only a fraction of the (oo)cysts or E. coli colonies that were enumerated are capable of initiating infection in a human host (Table 1 A and B). In the case of Cryptosporidium, site-specific information about the fraction of sequenced oocysts that were either Cryptosporidium parvum or Cryptosporidium hominis (Wilkes et al., 2013) was used to correct for infectivity (disregarding the viability of the oocysts). Uncertainty in this fraction at each site was described using a simple posterior distribution from a hierarchical model (see Supp. Cont.). Infectivity information was not available for Giardia, so all enumerated cysts were assumed infectious. E. coli O157:H7 P/A data were used to adjust total E. coli that were presumed to be infectious, and uncertainty in this fraction was evaluated using Bayes' theorem and Markov Chain Monte Carlo with a uniform prior (see Supp. Cont.).
Table 1.
Pathogenic sequences of Cryptosporidium observed at sample sites for All Available Data (Table A) and the quantity and type of occurrence data used in QMRA for each pathogen (Table B).
| Table A | |||
|---|---|---|---|
| Monitoring site | Total number of Cryptosporidium sequences | Number of pathogenic sequences | Notes |
| RCAin | 76 | 0 | |
| RCAout/URCAin | 9 | 1 | 1 C. hominis |
| URCAout | 89 | 4 | 2 C. parvum; 2 C. hominis |
| * | 13 | 2 | 2 C. parvum |
| * | 7 | 0 | |
| * | 9 | 0 | |
| * | 25 | 0 | |
| * | 63 | 4 | 4 C. hominis |
| * | 68 | 0 | |
| * | 31 | 0 | |
| * | 19 | 0 | |
| * | 88 | 6 | 4 C. parvum; 2 C. hominis |
| * | 48 | 0 | |
| * | 8 | 0 | |
| * | 44 | 0 | |
| * | 51 | 1 | 1 C. parvum |
| * | 9 | 0 | |
| Table B | |||
| Monitoring site | Cryptosporidium/Giardia enumeration dataa | E. coli plating datab | E. coli O157:H7 presence/absence datac |
| RCAin | 40 | 109 | 79 (0) |
| RCAout/URCAin | 8 | 87 | 66 (0) |
| URCAmid | – | 66 | 50 (3) |
| URCAout | 51 | 92 | 75 (0) |
*Other sites in South Nation River Basin not included in this study (see Marti et al., 2013, for reference to other sites).
(Oo)cyst counts and corresponding enumerated volumes.
Reported concentration estimates.
Total number of data (and number of positive results).
Novel dose-response modelling approaches were used herein. For Cryptosporidium, a hierarchical Bayesian model based upon the U.S. EPA's Economic Analysis for the Long Term 2 Enhanced Surface Water Treatment Rule (USEPA, 2005) that included beta-distributed strain-to-strain variation in the exponential dose-response model parameters of six human feeding studies, was used (see Supp. Cont.). The exponential dose-response model for Giardia (based upon the data of Rendtorff (1954)) and the actual beta-Poisson, or hypergeometric, dose-response model for E. coli (based upon the data of DuPont et al. (1971)) were used with Bayesian analysis of parameter uncertainty (Schmidt et al., 2013b).
Because no systematic human exposures (e.g., swimming, drinking water) are known to exist at the study sites, a single hypothetical consumption value of 45 mL was used (representing accidental ingestion during a swim) (Dufour et al., 2006). A second-order risk characterization procedure comparable to that of Schmidt et al. (2013a) was used to separate variability from uncertainty in, 1) the parameters of the gamma distribution describing temporal concentration variability, 2) the fraction of enumerated microorganisms that were infectious, and 3) the parameter(s) of the dose-response model. In this approach, mean risk for a particular set of parameter values was evaluated using numerical integration, and 30,000 such sets of parameters (collectively representing uncertainty in the estimated parameters) were used to evaluate the uncertainty in mean risk. The resulting risk information was summarized using 95% credible intervals for mean risk (illustrating the anticipated probability of infection for the hypothetical exposure and effect of all sources of uncertainty). The posterior probability that the mean risk at one site was greater than that at another was used as the Bayesian analog to significance in a two-tailed hypothesis test. This does not provide a measure of statistical significance (p-value): however, strong support for a substantial difference exists when computed posterior probabilities exceed 0.975, which is the analog of a two-tailed p-value of 0.05 (Schmidt et al., 2013a).
All Bayesian Markov Chain Monte Carlo analyses were performed with the open-source software OpenBUGS (V.3.2.1, rev 781). Three chains with relatively variable initial states were used to assess convergence (often including use of the built-in Gelman-Rubin statistic feature) and history plots were used to evaluate mixing. A burn-in of 5000 iterations was used to ensure that the first recorded iteration was minimally influenced by the initial state of the chain, and 30,000 iterations (usually every fifth of 150,000 iterations) were recorded in most cases as a representative random sample from the posterior distribution of interest.
3. Results
3.1. Prevalence
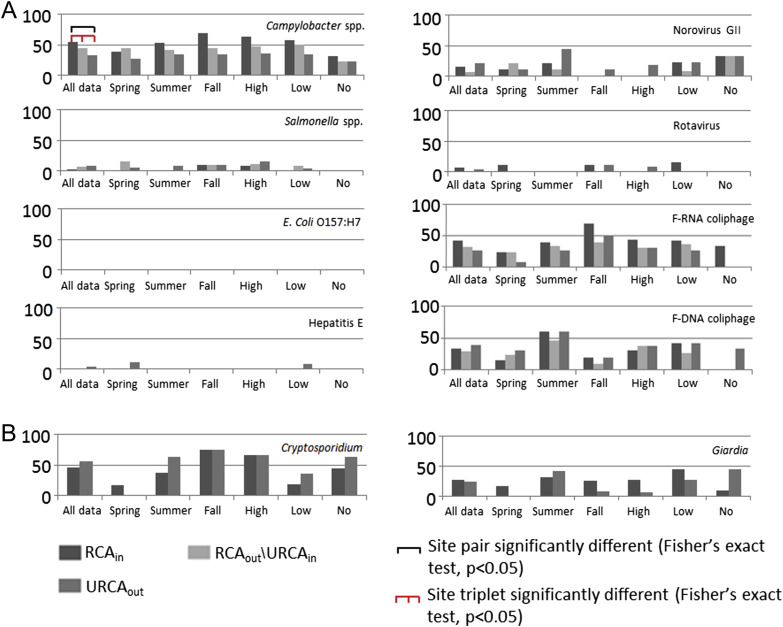
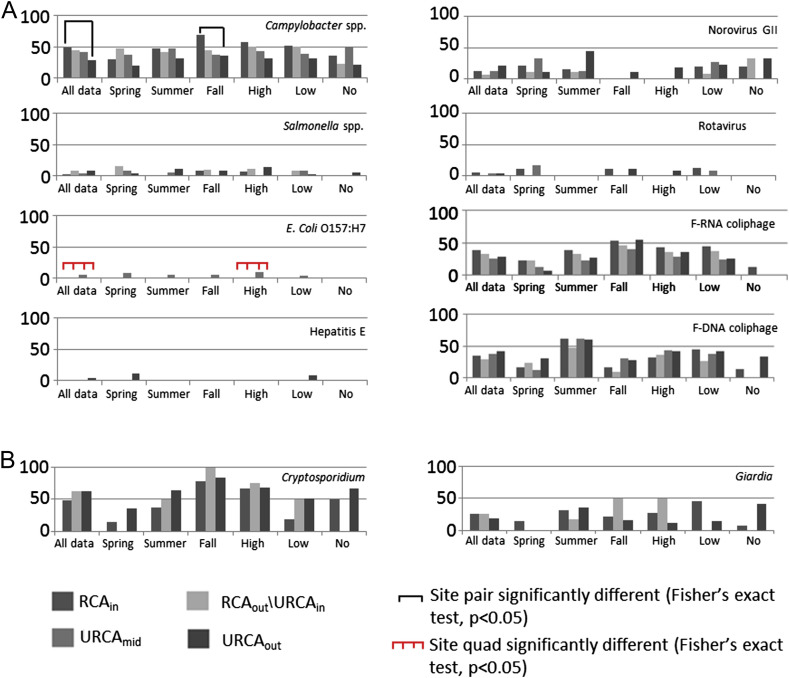
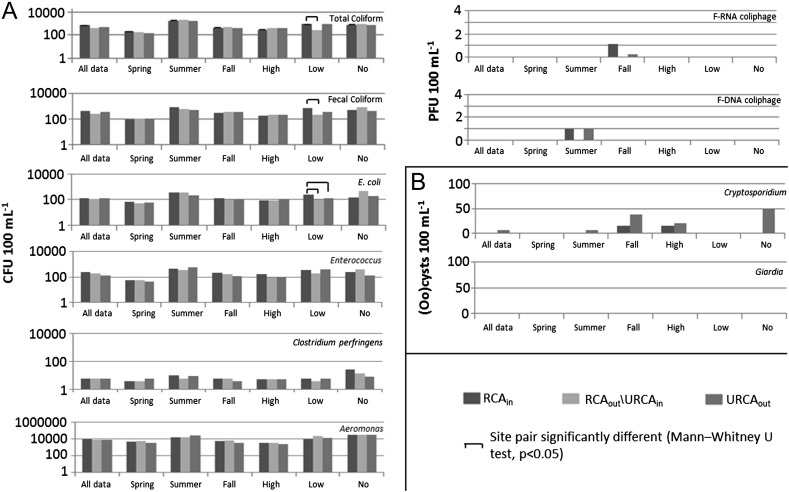
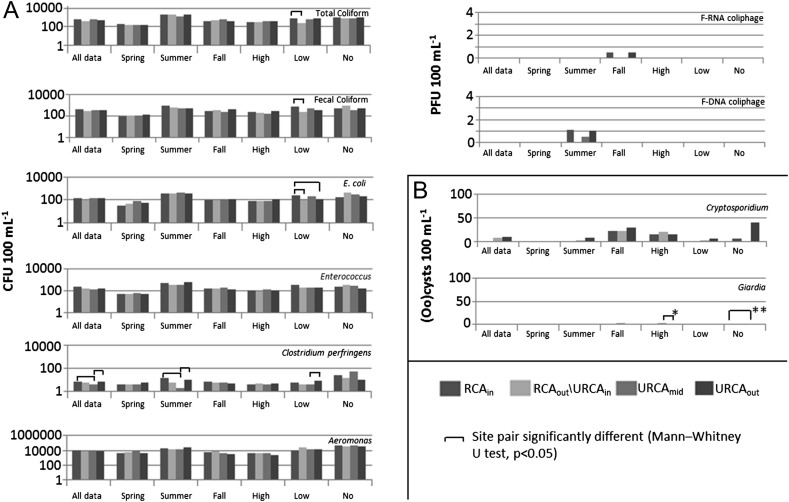
Cryptosporidium oocysts (with sample site prevalence ranging between 46 and 63%) and Campylobacter spp. (29–55%) were the most frequently observed pathogens at the study site for Temporally Concurrent Data (Fig. 2 , ‘all data’) and when All Available Data were considered (Fig. 3 , ‘all data’). Giardia cysts (19–27%) and norovirus GII (7–22%) were observed relatively less frequently on a sample site prevalence basis, and Salmonella spp. (3–8%), rotavirus (0–7%), hepatitis E virus (0–4%), and E. coli O157:H7 (0–6%) were rarely observed in water samples (Figs. 2 and 3, ‘all data’). F-RNA (26–42% samples) and F-DNA (28–41%) coliphage detections were observed from moderately to occasionally. The range of site medians for fecal indicator bacteria and Aeromonas spp. densities (CFU 100 mL−1) were: total coliform (375–710), fecal coliform (290–460), E. coli (106–146), Enterococci (134–240), Aeromonas spp. (7550–9550) and Clostridium perfringens (4–7) (Figs. 4 and 5 , ‘all data’).
Fig. 2.
The percentage (%) of samples under specific site, season, and flow condition positive for detection of microorganisms. Temporally Concurrent Data were used for: (A) the RCAin, RCAout/URCAin, and URCAout for bacteria and viruses, and (B) Cryptosporidium and Giardia which were collected in a temporally concurrent manner at sites RCAin and URCAout only. See Fig. 3 for parasite occurrence at RCAout/URCAin. High flow ≥0.018 m3 s−1; low flow ≥0.002 m3 s−1 and <0.018 m3 s−1, and no flow <0.002 m3 s−1. Black bar represents significant Fisher's exact test p-value (<0.05) from site pair 2 × 2 comparison. Red bar represents significant Fisher's exact test p-value (<0.05) from site triplet 2 × 3 comparison.
Fig. 3.
The percentage (%) of samples under specific site, season, and flow condition positive for detection of microorganisms. Data represented here were based on All Available Data for (A) bacteria and viruses and (B) parasites. High flow ≥0.018 m3 s−1; low flow ≥0.002 m3 s−1 and <0.018 m3 s−1, and no flow <0.002 m3 s−1. Black bar represents significant Fisher's exact test p-value (<0.05) from site pair 2 × 2 comparison. Red bar represents significant Fisher's exact test p-value (<0.05) from site quad 2 × 4 comparison.
Fig. 4.
Median densities of fecal indicator bacteria, Aeromonas, parasites and F-coliphage for samples collected under specific site, season, and flow condition. Temporally Concurrent Data were used for: (A) the RCAin, RCAout/URCAin, and URCAout for bacteria and coliphage, and (B) parasite data which were collected in a temporally concurrent manner at sites RCAin and URCAout only. See Fig. 5 for parasite median density at RCAout/URCAin. High flow ≥ 0.018 m3 s−1; low flow ≥0.002 m3 s−1 and <0.018 m3 s−1, and no flow <0.002 m3 s−1.
Fig. 5.
Median densities of fecal indicator bacteria, Aeromonas, parasites and F-coliphage for samples collected under specific site, season, and flow conditions for All Available Data. (A) bacteria and F-coliphage, (B) parasites. * RCAout/URCAin vs. URCAout; ** RCAin vs. URCAout. High flow 0.018 m3 s−1; low flow 0.002 m3 s−1 and <0.018 m3 s−1, and no flow <0.002 m3 s−1.
3.2. Temporally concurrent data
Unlike the All Available Data datasets, the Temporally Concurrent Data allowed for the evaluation of the stream/riparian zone protection treatment (site) effects on water quality targets. The only significant differences (p-value, Fisher's exact test) in pathogen prevalence for these data occurred for Campylobacter spp. (‘all data’ between RCAin and URCAout sites) (Fig. 2). Here, Campylobacter spp. detections reduced by 11% between the RCAin and RCAout/URCAin, and another 11% between RCAout/URCAin and URCAout.
CART classification tree analysis indicated, overall, no site, season, and flow interactions occurred within tree growing constraints, except for Cryptosporidium (see Supp. Tbl. 2). For Campylobacter spp., CART classified P/A groups by, no flow (presence:absence ratio = 0.35) vs. high AND low flow (0.94). For Salmonella spp., classification was also based on flow: no flow AND low flow (0.03) vs. high flow (0.13). For Cryptosporidium oocyst detects, there was an interaction identified. These were: summer AND fall AND high flow (3.33), and summer AND fall AND no flow AND low flow (0.90), and spring (0.09). For Giardia cyst detection, season was the most important classifier: spring AND fall (0.16) vs. summer (0.58). Too few detects for E. coli O157:H7 precluded CART analysis for this pathogen.
Significant differences among site flow groupings of fecal indicator bacteria were observed, specifically for low flow (p < 0.05) (Fig. 4). For low stream flow, fecal coliform densities were significantly higher at the RCAin relative to RCAout/URCAin, and E. coli densities were significantly higher at RCAin, relative to RCAout\URCAin and URCAout (Fig. 4). For CART regression tree models (Supp. Tbl. 3), for specific microorganisms (total coliforms, fecal coliforms, E. coli, Enterococcus, C. perfringens, Aeromonas, and Cryptosporidium), the data initially subdivided higher and lower density data on the basis of season (except for C. perfringens data which initially split into higher and lower densities by flow and no-flow conditions). Overall, with the exception of C. perfringens and Cryptosporidium (these microorganisms having higher relative densities in fall and to a lesser extent spring), summer, under various flows, was a season that was associated most strongly with higher median densities of the microorganisms in the CART regression tree analysis (Supp. Tbl. 3).
There were some noteworthy data trends, albeit not statistically significant, in the pathogen prevalence data when RCAin, RCAout/URCAin, and URCAout were concurrently monitored. Salmonella spp. tended to increase in prevalence from the RCAin to the URCAout, most markedly at higher flow (Fig. 2). Norovirus GII was, overall, most prevalent in summer and at the URCAout. Rotavirus was most prevalent at the RCAin relative to other sites. Hepatitis E virus occurrence was detected at the URCAout exclusively with spring and low flow being key conditions associated with the occurrence of this virus. F-RNA coliphage occurred most frequently in fall at RCAin. Prevalence of F-DNA coliphage was at or over ∼47% in summer, representing the greatest overall prevalence rate, with RCAin and URCAout having the highest rate during this season. Fall and high flow were conditions where the occurrence of Cryptosporidium was highest, and moreover, prevalence was generally higher at the URCAout relative to the RCAin (Fig. 2b). For Giardia, trends were opposite of that observed for Cryptosporidium, with higher Giardia occurrence in summer and low flow. E. coli O157:H7 was not found during conditions associated with concurrent site sampling despite significant numbers of samples collected.
3.3. All available data
Campylobacter spp. and E. coli O157:H7 detections were significantly different among sites (p < 0.05) (Fig. 3), with significant site differences for Campylobacter spp. for ‘all data’ and ‘fall data’ between RCAin and URCAout (2 × 2, Fisher's exact test), and for E. coli O157:H7 for ‘all data’ and ‘high flow’ (2 × 4 contingency table, Fisher's exact test). E. coli O157:H7 was observed more often at URCAmid (n = 3, out of 50 samples), and was not detected at any other site (Fig. 3). Also, rotavirus and norovirus GII were detected at URCAmid in 4 and 13% of samples, respectively, with equivalent or greater detection rates at URCAout (4 and 22% for rotavirus and norovirus GII, respectively); detection rates for rotavirus and norovirus GII were 6 and 13%, respectively, at RCAin (Fig. 3). Hepatitis E virus was detected at rates of 4% at URCAout for ‘all data’, and was detected in spring (11%) and no flow (8%) there, while no detections were observed upstream.
Trends in indicator bacteria median values (Fig. 5) showed at low flow, there were significant differences in total coliform, fecal coliform, and E. coli densities between RCAin and RCAout/URCAin (with lower median densities at RCAout/URCAin versus RCAin), and similar but non-significant reduction trends between RCAin and RCAout/URCAin for ‘all data’ (Fig. 5). This reduction in densities downstream is further illustrated with significantly lower C. perfringens densities at URCAmid.
F-RNA and F-DNA coliphage densities showed no significant trends. However, F-RNA median densities were greater in the fall only (Fig. 5) at RCAin (0.5 PFU 100 mL−1), decreasing within RCAout/URCAin and URCAmid (0 PFU 100 mL−1), and increasing to 0.5 PFU 100 mL−1 at URCAout. In summer, median densities of F-DNA coliphage were greatest at RCAin (1.1 PFU 100 mL−1), reducing to 0 PFU 100 mL−1 at RCAout/URCAin, and increasing to 0.5 and 1.0 PFU 100 mL−1 at URCAmid and URCAout, respectively.
3.4. Fecal indicator thresholds related to pathogen presence/absence
Relationships between fecal indicator organisms and pathogens can be variable (Yates, 2007, Wilkes et al., 2009); however, they are useful for helping determine conditions and pollution drivers that may impose higher risk for human infection by pathogens, as well as help explain the efficacy of BMPs in reducing surface water impairment by fecal pollution. For this analysis, all available data were used. Distributions of indicator bacteria densities were significantly different in the presence vs. the absence (P.vs.A.) of Campylobacter spp. (MWU tests; p < 0.05), with higher medians in Campylobacter spp. presence (Table 2 ). Distributions of fecal coliforms were significantly different in the P.vs.A. of Cryptosporidium (medians of 485 CFU 100 mL−1 in presence, vs. 320 CFU 100 mL−1 in absence of Cryptosporidium), and distributions of Enterococcus were significantly different in the P.vs.A. of Giardia (medians of 320 CFU 100 mL−1 in presence, versus 144 CFU 100 mL−1 in absence of Giardia). Distributions of Aeromonas spp. were significantly different in Salmonella spp. P.vs.A.; counter intuitively, however, where medians were greater in the absence of Salmonella spp.
Table 2.
Associations between the densities of fecal indicator microorganisms, including Aeromonas, and the occurrence of pathogens in surface water (All Available Data).
| Pathogen | Indicator microorganism (*unit) | Pathogen presence grouping |
Pathogen absence grouping |
Mann-Whitney U P value | ||||
|---|---|---|---|---|---|---|---|---|
| Total number of microorganism samples in presence of pathogen | Median microorganism | Mean rank sum | Total number of microorganism samples in absence of pathogen | Median microorganism | Mean rank sum | |||
| Campylobacter spp. | F-RNA coliphage* | 82 | 0 | 77.6 | 63 | 0 | 67.0 | 0.069 |
| Campylobacter spp. | F-DNA coliphage* | 82 | 0 | 76.6 | 64 | 0 | 69.5 | 0.245 |
| Campylobacter spp. | Total Coliforms** | 109 | 750 | 144.5 | 155 | 400 | 124.0 | 0.032ǂ |
| Campylobacter spp. | Fecal Coliforms** | 103 | 480 | 140.3 | 152 | 340 | 119.6 | 0.028ǂ |
| Campylobacter spp. | E. coli** | 109 | 164 | 144.0 | 155 | 121 | 124.4 | 0.040ǂ |
| Campylobacter spp. | Enterococcus** | 109 | 230 | 145.6 | 155 | 162 | 123.3 | 0.020ǂ |
| Campylobacter spp. | C. perfringens** | 107 | 10 | 144.8 | 154 | 4 | 121.4 | 0.013ǂ |
| Campylobacter spp. | Aeromonas** | 62 | 11900 | 57.4 | 44 | 7300 | 48.0 | 0.125 |
| Cryptosporidium | F-RNA coliphage* | 24 | 1 | 16.9 | 9 | 50 | 17.2 | 0.953 |
| Cryptosporidium | F-DNA coliphage* | 24 | 0 | 16.1 | 9 | 0 | 19.4 | 0.392 |
| Cryptosporidium | Total Coliforms** | 56 | 485 | 53.2 | 45 | 500 | 48.2 | 0.393 |
| Cryptosporidium | Fecal Coliforms** | 56 | 485 | 56.9 | 45 | 320 | 43.6 | 0.023ǂ |
| Cryptosporidium | E. coli** | 56 | 145 | 53.4 | 45 | 122 | 48.0 | 0.353 |
| Cryptosporidium | Enterococcus** | 56 | 164 | 51.5 | 45 | 196 | 50.4 | 0.852 |
| Cryptosporidium | C. perfringens** | 54 | 5 | 53.8 | 45 | 3 | 45.4 | 0.147 |
| Cryptosporidium | Aeromonas** | 5 | 6500 | 4.4 | 2 | 4505 | 3.0 | 0.571 |
| Giardia | F-RNA coliphage* | 6 | 26 | 15.5 | 27 | 0.5 | 17.3 | 0.699 |
| Giardia | F-DNA coliphage* | 6 | 0 | 20.5 | 27 | 0 | 16.2 | 0.348 |
| Giardia | Total Coliforms** | 20 | 675 | 59.6 | 81 | 470 | 48.9 | 0.145 |
| Giardia | Fecal Coliforms** | 20 | 680 | 59.2 | 81 | 370 | 49.0 | 0.165 |
| Giardia | E. coli** | 20 | 186 | 56.3 | 81 | 122 | 49.7 | 0.371 |
| Giardia | Enterococcus** | 20 | 320 | 62.6 | 81 | 144 | 48.1 | 0.047ǂ |
| Giardia | C. perfringens** | 19 | 3 | 49.2 | 80 | 4 | 50.2 | 0.891 |
| Norovirus GII | F-RNA coliphage* | 15 | 0 | 49.0 | 88 | 0 | 52.5 | 0.674 |
| Norovirus GII | F-DNA coliphage* | 15 | 0 | 47.6 | 89 | 0 | 53.3 | 0.503 |
| Norovirus GII | Total Coliforms** | 15 | 290 | 47.1 | 88 | 515 | 52.8 | 0.496 |
| Norovirus GII | Fecal Coliforms** | 13 | 230 | 44.7 | 81 | 370 | 48.0 | 0.689 |
| Norovirus GII | E. coli** | 15 | 130 | 54.4 | 88 | 129 | 51.6 | 0.735 |
| Norovirus GII | Enterococcus** | 15 | 190 | 49.2 | 88 | 200 | 52.5 | 0.694 |
| Norovirus GII | C. perfringens** | 15 | 16 | 46.4 | 88 | 14 | 52.9 | 0.439 |
| Norovirus GII | Aeromonas** | 15 | 11200 | 52.7 | 88 | 9500 | 51.9 | 0.930 |
| Rotavirus | F-RNA coliphage* | 4 | 0 | 41.5 | 99 | 0 | 52.4 | 0.493 |
| Rotavirus | F-DNA coliphage* | 4 | 0 | 36.5 | 100 | 0 | 53.1 | 0.296 |
| Rotavirus | Total Coliforms** | 4 | 495 | 49.3 | 99 | 500 | 52.1 | 0.863 |
| Rotavirus | Fecal Coliforms** | 4 | 505 | 55.0 | 90 | 323 | 47.2 | 0.594 |
| Rotavirus | E. coli** | 4 | 230 | 66.5 | 99 | 130 | 51.4 | 0.340 |
| Rotavirus | Enterococcus** | 4 | 284 | 50.5 | 99 | 200 | 52.1 | 0.928 |
| Rotavirus | C. perfringens** | 4 | 44 | 78.8 | 99 | 12 | 50.9 | 0.068 |
| Rotavirus | Aeromonas** | 4 | 7550 | 49.5 | 99 | 9500 | 52.1 | 0.876 |
| Salmonella spp. | F-RNA coliphage* | 9 | 0 | 49.5 | 136 | 0 | 74.6 | 0.083 |
| Salmonella spp. | F-DNA coliphage* | 9 | 0 | 77.6 | 137 | 0 | 73.2 | 0.773 |
| Salmonella spp. | Total Coliforms** | 15 | 1160 | 144.4 | 250 | 520 | 132.3 | 0.558 |
| Salmonella spp. | Fecal Coliforms** | 13 | 330 | 118.0 | 243 | 408 | 129.1 | 0.602 |
| Salmonella spp. | E. coli** | 15 | 130 | 147.7 | 250 | 133 | 132.1 | 0.448 |
| Salmonella spp. | Enterococcus** | 15 | 110 | 122.1 | 250 | 200 | 133.7 | 0.576 |
| Salmonella spp. | C. perfringens** | 15 | 10 | 153.8 | 247 | 6 | 130.1 | 0.244 |
| Salmonella spp. | Aeromonas** | 8 | 475 | 27.0 | 98 | 9550 | 55.7 | 0.009ǂ |
*PFU per 100 mL−1; **CFU 100 mL−1; ǂ Significant p < 0.05.
CART was able to help quantify threshold fecal indicator and Aeromonas spp. densities that delineated groups of greater detection percentages of targeted pathogens (Supp. Tbl. 4). For approximately 75% (18/24) of the fecal indicator bacteria vs. pathogen classification results that cross-validated, there was greater percentage of pathogen occurrence associated with the > indicator density threshold classification group. For the fecal indicator bacteria vs. bacterial pathogens this value was 75%, for the fecal indicator bacteria vs. parasite results this value was 90%, if only for a few. Coliphage vs. pathogens classification results indicated coliphage was not as specific for discriminating pathogen occurrence as were the fecal indicator bacteria.
3.5. Quantitative microbial risk assessment
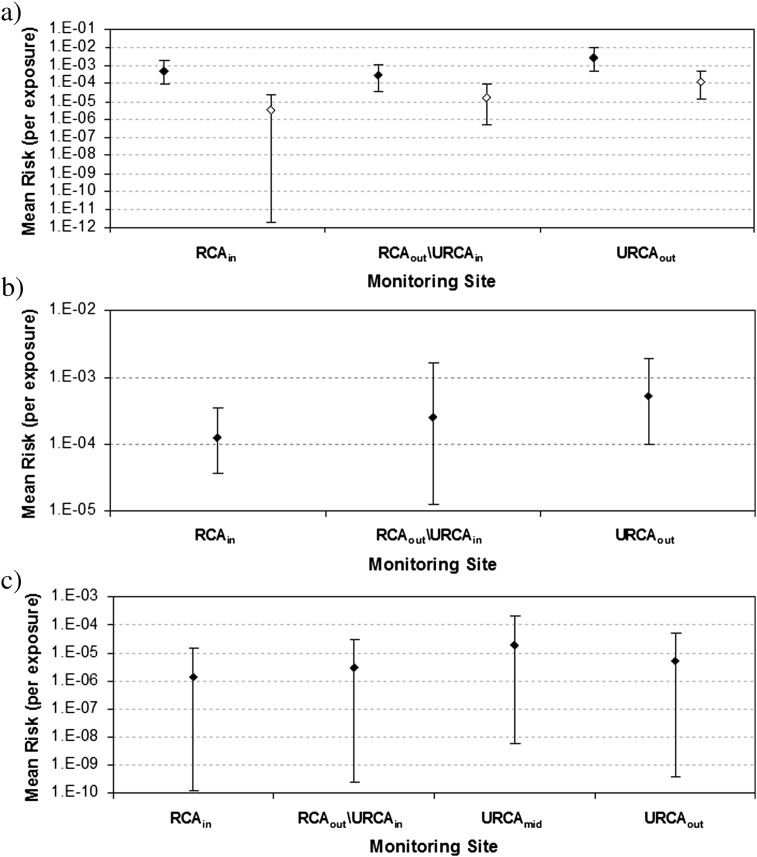
Ninety-five percent credible intervals for mean risk (Fig. 6 ) ranged from 10−4.5 to 10−2 for total Cryptosporidium spp., 10−12 to 10−3 for pathogenic Cryptosporidium spp., 10−5 to 10−2.5 for total Giardia, and 10−10 to 10−3.5 for E. coli O157:H7. Considering only the fraction of the total Cryptosporidium oocysts that are deemed to be pathogenic resulted in mean risks being reduced by approximately an order of magnitude or more.
Fig. 6.
Plots of the uncertainty in the mean risk at each site for a) total Cryptosporidium (black points) and pathogenic Cryptosporidium (white points), b) Giardia, and c) E. coli O157:H7. The points show the posterior mean (of the mean risk) and the error bars show the 95% equal-tailed credible intervals.
Since this risk analysis is comparative rather than predictive, mean risks are compared in a relative way to compute posterior probabilities that the risk at one site is greater than that at another. For total Cryptosporidium, URCAout had greater mean risk than RCAin or RCAout/URCAin with pair-wise posterior probabilities of 0.9993 and 0.9974, respectively. The RCAout/URCAin, however, had lower mean risk than the RCAin with a posterior probability of 0.8531. These results indicated a slight decrease in mean risk from total Cryptosporidium across the RCA followed by an increase in mean risk across the URCA. When only pathogenic Cryptosporidium are considered, the mean risk at the URCAout site is still greater than that of the RCAin and RCAout/URCAin sites (with posterior probabilities of 0.9990 and 0.9750 respectively). The mean risk at the RCAout/URCAin site is also greater than that of the RCAin site with posterior probability 0.8425. These results indicate a slight increase in mean risk from pathogenic Cryptosporidium across the RCA (attributed to a single detection of human source classed C. hominis at RCAout/URCAin) and a further increase across the URCA (attributed to a detection of both C. hominis and C. parvum).
For total Giardia, the mean risk at URCAout is greater than that of the RCAin and RCAout/URCAin (with posterior probabilities of 0.9541 and 0.8658 respectively). The overall average mean risk at the RCAout/URCAin site is greater than that of the RCAin site (Supp. Fig. 1), but the corresponding posterior probability is only 0.3960. This peculiar result is attributable to the relatively small number of available data and corresponding increase in uncertainty. Collectively, these results indicate little change in mean risk from total Giardia across the RCA, followed by an increase in mean risk across the URCA.
In the case of E. coli O157:H7 (for which risk results are also available for URCAmid), URCAmid was found to have greater mean risk than the RCAin, RCAout/URCAin, and URCAout with posterior probabilities of 0.9952, 0.9648, and 0.9189, respectively. This is attributable to the detection of E. coli O157:H7 on three occasions at this site, while it was undetected at all other sites. The mean risk at RCAout/URCAin was greater than that of RCAin with a posterior probability of 0.6735 and lower than that of URCAout with a posterior probability of 0.6180; thus there is a slight increasing E. coli risk trend across both the RCA and URCA. An interesting finding in this risk assessment (unlike Cryptosporidium and Giardia) is that close to two orders of magnitude of uncertainty in the computed mean risk values are attributable to uncertainty in the parameters of the dose–response model (results not shown here). This is attributable to the relatively small number of subjects and range of mean doses in the E. coli dose–response experiment by DuPont et al. (1971).
4. Discussion
It is recognized that water quality surveillance (WHO, 2004, Thomas et al., 2013) can contribute to a better understanding of fecal pollution sources and drivers, and therefore appropriate mitigation measures. Overall, there were not many strong statistically significant trends in pathogen prevalence or densities associated with restricting cattle access to the water course (protecting riparian zone). However, indicator bacteria densities (total coliform, fecal coliform, E. coli) were higher at the RCAin site compared to RCAout/URCAin site (p < 0.05, MWU), illustrating an apparent riparian zone treatment effect. Restricting cattle access did not reduce, statistically speaking, Campylobacter spp. occurrence relative to upstream (site input) prevalence, and surprisingly, Campylobacter spp. occurrence was even lower at the URCAout relative to RCAin. Schmidt et al. (2013a) found that Campylobacter spp. densities were relatively high in these watersheds at RCAin, and upstream of the RCAin, where the only potential for agricultural inputs of Campylobacter spp. would have been related to land applications of manure and subsequent stream inputs via, primarily, subsurface tile drainage. The overall decreasing trend in Campylobacter spp. detection downstream from RCAin to URCAout was most pronounced during fall at higher flows; a time when manure is applied to fields and manure contaminants can be transported off site via tile drainage possibly resulting in overall higher prevalence at RCAin. Sunohara et al. (2012) described the RCA as functioning as an in-line wetland, where loads of many fecal pollution indicators and nutrients were found to decrease significantly from RCAin to RCAout/URCAin and increase again from RCAout/URCAin to URCAout. If the source of Campylobacter spp. was primarily from livestock, the RCA effect may have been formative in terms of reducing the prevalence of this pathogen in water. The light pasturing in the URCA and lack of cattle related manure land applications on fields adjacent to the RCA and URCA may have helped promote the steady downstream decreases in prevalence observed. Moreover, in fall, the pasturing cattle are often trucked back to barns for the winter, which would further reduce potential livestock-based fecal inputs in both pasture treatments. However, Campylobacter spp. has been shown to be associated with the occurrence of avian-sourced Cryptosporidium in the area (Wilkes et al., 2013), which may partially explain site prevalence differences.
The only other noteworthy trend amongst the sites for a pathogen was for E. coli O157:H7, resulting solely from 3 positive detections out of 50 at URCAmid. Infected livestock likely introduced the E. coli serotype (Renter et al., 2003) into the watercourse via fecal contamination in close proximity to the sampling site, given that cattle have direct access to the water course at this site and that there was no detection of this pathogen at URCAout just downstream. When predicting downstream public health risks from exposure to E. coli O157:H7, it was demonstrated that cattle access, most likely, has a direct influence on increasing risk, and although not significant, it is important to differentiate E. coli detected in a surface water system (many of which are not human pathogens) from those that are capable of initiating human infections (like E. coli O157:H7). Unlike Cryptosporidium and Giardia, the breadth of the credible intervals in predicted mean risk shrank by roughly two orders of magnitude when relative mean risks were used instead of mean risks. This suggests that roughly two orders of magnitude of uncertainty are attributable to uncertainty in the parameters of the dose–response model. This illustrates the importance of using relative mean risks to understand a system, given that dose–response data will continue to remain the limiting factor in our ability to more precisely quantify risk.
No significant differences in densities of F-RNA and F-DNA coliphage distribution were observed among the sample sites either, which could be due to, among other things, differential survival rates of subtypes of F- coliphage. Cole et al. (2003) observed seasonal differences in the proportions of F-DNA and F-RNA (higher relative occurrence of F-DNA in summer, complimenting the occurrence results observed in this study), attributing it to survival characteristics, host excretion patterns, or landuse change, and Brion et al. (2002) has observed differential survival rates of subtypes of F-RNA in water. For this study, greatest median densities of F-RNA coliphage occurred in fall while greatest median densities of F-DNA coliphage occurred in spring. Nevertheless, the issue of variable environmental fitness of a microorganism, or strain, can make it additionally difficult to observe the effects of landscape factors on densities (Anderson et al., 2005, van Elsas et al., 2011).
Overall data mining analysis via CART revealed that (Supp. Tbl. 2–3) cattle exclusion measures did not appear to be as important for delineating the occurrence of pathogens, or for parsing of ‘higher’ and ‘lower’ densities of fecal indicator organisms, relative to the more discriminating season and stream flow criteria. This is consistent with findings of Wilkes et al., 2009, Wilkes et al., 2013. Seasonal attributes could be important in the timing and intensity of fecal inputs from various sources, the seasonal growth of filtering vegetation in the riparian zone, and the accumulation of fecal material over time in and along the stream system. Further, season is interrelated with the timing of critical hydrological events for moving contaminants in water, and season is connected to temperature and moisture factors that affect the survivability of pathogens in soil/water environments.
In Ruecker et al. (2012) and Wilkes et al. (2013), wildlife and livestock were dominant genotypes/species of Cryptosporidium observed in surface waters in the study area. Yet many of the wildlife species/genotypes observed were not a known human health risk. Cryptosporidium QMRA results underscore the importance of incorporating the proportion of human pathogenic oocysts (including C. hominis (human host source)) in the final risk estimates, resulting in a reduction in mean risk by at least one order of magnitude. These results, in the context of the detection of norovirus GII and rotavirus (which can be of human origin) at these sites, suggest there could be an influence of human waste in this system (e.g., septic leakage). Regarding Giardia, the lack of additional subtyping data to differentiate between human pathogenic Giardia among all Giardia detected, limits how we interpret the influence of the URCA on the Giardia levels detected at all three sample sites. Based solely on Giardia cyst concentrations, the QMRA analyses illustrate an increasing risk from RCAin to URCAout, although not statistically significant. These QMRA results underscore the importance of understanding the many potential sources of fecal inputs in an open system such as a watershed when interpreting the impact of a land use management practice on public health endpoints.
Norovirus was the most commonly detected virus in surface water in this experiment. Recently Thomas et al. (2013) estimated that Norovirus was the number one ranked foodborne illness in Canada. While extensions in this context cannot be made directly to waterborne exposure, a high norovirus prevalence was observed in this study in relation to other viral targets (greater than ∼40% of samples for URCAout in summer). Perhaps small order streams helped reduce dilution effects. Norovirus genogroup II is one of the currently 5 identified genogroups (GI-GV) of norovirus, for which GII genotypes are the most commonly known for infecting humans (Lindesmith et al., 2008), but some are also found in swine. Norovirus GII and group A rotavirus (found in human, swine, and bovine) tended to be higher at RCAin, decrease in prevalence at RCAout/URCAin, and increase again at the URCAout. These prevalence trends did underscore the significant and documented RCA effect found in the fecal indicator bacteria (Fig. 4) and nutrient and fecal indicator loads given in Sunohara et al. (2012). Liquid swine manure has been applied on fields that can, under high rainfall situations, drain into roadside ditches that could impact the RCAin, as a result, swine inputs cannot be refuted. The contributions from septic systems from homes located along the stream (Sunohara et al., 2012), as well as pets and a few hobby farm animals associated with them, is not clearly explicit from the data reported in this study (outside of the C. hominis observations regarding human source). But these sources could have contributed, along with inhibited dilution effects in this intermittent system, to the higher relative rates of occurrence of this virus in the water course.
A component of this study included a global examination of pathogen associations with indicator bacteria and F- coliphage densities in surface water for the RCA and URCA sites. Assessment of BMP impact on surface water quality within a public health context would currently require monitoring of a suite of pathogens and their densities for different treatments at great monetary and logistical expense. Ideally, fecal indicator organisms could serve as pathogen proxies or surrogates in this capacity (Wilkes et al., 2009). For the pathogenic viruses, there were no strong relationships with F-RNA or F-DNA coliphage density or occurrence which would appear to reduce potential for coliphage data in this small surface water system to serve as proxies for viral pathogens. There were, however, multiple indicator microorganisms that could serve as modest pathogen proxies, especially for Campylobacter spp. and to a lesser extent Cryptosporidium spp., thereby complimenting the results observed in Wilkes et al. (2009). For assessing the effects of pasturing BMPs such as those here, the fact that fecal indicator bacteria demonstrated a ‘low flow’ pasture treatment effect indicates they could potentially serve as adequate, and considerably less expensive organisms to gauge effects of riparian zone protection on net fecal pollution (and potentially serve as a proxy for the occurrence of Campylobacter spp.).
5. Conclusions
Some of the key conclusions that can be drawn from this study include:
-
•
E. coli O157:H7 was infrequently detected, but apparently, light pasturing operations with livestock having the capacity to interact with the water course can increase the likelihood of this pathogen occurring in water, and thereby increase relative mean risks of exposure to this pathogen.
-
•
For the unrestricted and restricted pasture systems, relative exposure risks were ranked in decreasing order as: total Cryptosporidium, total Giardia spp., human pathogenic Cryptosporidium and E. coli O157:H7. Adjusting for the proportion of Cryptosporidium spp. detected that are infectious for humans reduces downstream risk estimates by roughly one order of magnitude. The QMRA indicated that relative risk increased across the RCA and URCA region for E. coli O157:H7; human-pathogenic Cryptosporidium relative risk slightly increased across the RCA due to the occurrence of C. hominis (associated with human and likely of septic origin from homes next to the BMP) at RCAout/URCAin and further increased in the URCA (due to increased occurrence of C. hominis and C. parvum (normally associated with cattle) in tandem at URCAout), and little change in relative risk was observed across the RCA for Giardia, but Giardia relative risk subsequently increased in the URCA. From a relative risk perspective, the fencing BMP did provide improved risk protection for Giardia whereas risks for E. coli and human-pathogenic Cryptosporidium (likely partly due to septic inputs from homes) increased downstream to URCA; the URCA region overall provides an increased risk from background, whereas the RCA region (and the BMP) results varied depending on the targeted pathogen. Using QMRA in this manner provides a more refined estimate of BMP effects on human health.
-
•
The cattle exclusion measures did not have an equal effect on all microorganisms. Campylobacter spp. had high relative prevalence in this agricultural watershed but detections decreased significantly through the restricted pasture and unrestricted pasture exhibiting a pasturing/riparian protection treatment effect (Sunohara et al., 2012). In contrast, Cryptosporidium had high relative prevalence for all sites (both restricted and unrestricted, no reduction) plus subtyping attributes helped to more accurately enumerate human infectivity risks and host specificity (see Ruecker et al., 2012).
-
•
Density distribution differences of total coliform, fecal coliform, E. coli, Enterococcus, and C. perfringens concentrations were associated with Campylobacter spp. detection; greater median densities of indicator organisms were observed in Campylobacter spp. presence. These indicator bacteria show promise as a conservative proxy for this pathogen in agriculturally dominated surface waters.
-
•
Overall, data mining found seasonal and stream flow variables and interactions to be important for delineating pathogen occurrence and higher indicator densities relative to site pasturing treatments. Consistent with other studies, empirical models that may attempt to predict prevalence of pathogen or fecal indicators should at least assess seasonal and flow variable interactions.
-
•
Indicator microorganisms exhibited no statistically significant distribution differences amongst the pasture sites except for total coliform, fecal coliform, and E. coli differences between RCAin and RCAout/URCAin at low flow; where relevant densities for this flow regime reduced from RCAin to RCAout/URCAin demonstrating a riparian zone treatment effect. Seasonal and flow conditions were associated more strongly with greater indicator bacteria densities, especially in the summer. Norovirus GII was detected at rates of 7–22%, and rotavirus 0–7% for monitoring sites, and no statistically significant associations were found with indicator bacteria or F-coliphage densities. It is speculated that at least house septic issues are potential sources of viruses (as supported by occurrence of C. hominis), but other sources (swine) may have contributed as well.
Acknowledgements
This research was funded by the Agriculture and Agri-Food Canada's (AAFC) Agriculture Policy Framework's National Water Quality Surveillance Research Initiative, AAFC's Sustainable Agriculture Environmental Systems (SAGES) program and Watershed Evaluation of Beneficial Management Practice (WEBs) program, and the Alberta Water Research Institute. We also thank the South Nation Conservation Authority for assistance.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.watres.2013.07.041.
Appendix A. Supplementary data
References
- Aarons S.R., Gourley C.J.P. The role of riparian buffer management in reducing off-site impacts from grazed dairy systems. Renew. Agr. Food Syst. 2012;28(1):1–16. [Google Scholar]
- Afchain A.L., Carlin F., Nguyen-The C., Albert I. Improving quantitative exposure assessment by considering genetic diversity of B. cereus in cooked, pasteurised and chilled foods. Int. J. Food Microbiol. 2008;128(1):165–173. doi: 10.1016/j.ijfoodmicro.2008.07.028. [DOI] [PubMed] [Google Scholar]
- Anderson K.L., Whitlock J.E., Harwood V. Persistence and differential survival of fecal indicator bacteria in Subtropical waters and sediments. Appl. Environ. Microbiol. 2005;71:3041–3048. doi: 10.1128/AEM.71.6.3041-3048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington K., Deere D., Ryan U., Stevens D., Davison A. Document produced for Department of Health Victoria by Water Futures Pty Ltd; 2011. Public Health Issues Associated with Stock Accessing Waterways Upstream of Drinking Water Off-takes.http://docs.health.vic.gov.au/docs/doc/Public-health-issues-associated-with-stock-accessing-waterways-upstream-of-drinking-water-off-takes Report. [Google Scholar]
- Brassard J., Seyer K., Houde A., Simard C., Trottier Y.L. Concentration and detection of hepatitis A virus and rotavirus in spring water samples by reverse transcription-PCR. J. Virol. Methods. 2005;123:163–169. doi: 10.1016/j.jviromet.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Brassard J., Simard C., Müller P., Houde A., Trottier Y.L. 2007. Concentration of Hepatitis a Virus and Rotavirus in Spring or Mineral Bottled Water Samples and Their Detection by the Reverse-transcription Polymerase Chain Reaction.http://www.hc-sc.gc.ca/fn-an/res-rech/analy-meth/microbio/volume5/opflp_04-eng.php (accessed 01.10.08.) ed. [Google Scholar]
- Brion G.M., Meschke J.S., Sobsey M.D. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res. 2002;36:2419–2425. doi: 10.1016/s0043-1354(01)00547-4. [DOI] [PubMed] [Google Scholar]
- Cole D., Long S.C., Sobsey M.D. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 2003;69:6507–6514. doi: 10.1128/AEM.69.11.6507-6514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A.P., Evans O., Behymer T.D., Cantú R. Water ingestion during swimming activities in a pool: a pilot study. J. Water Health. 2006;4:425–430. [PubMed] [Google Scholar]
- DuPont H.L., Formal S.B., Hornick R.B., Snyder M.J., Libonati J.P., Sheahan D.G., LaBrec E.H., Kalas J.P. Pathogenesis of Escherichia coli diarrhea. New Engl. J. Med. 1971;285:1–9. doi: 10.1056/NEJM197107012850101. [DOI] [PubMed] [Google Scholar]
- Havelaar A.H., During M., Versteegh J.F.M. Ampicillin-dextrin agar medium for the enumeration of Aeromonas species in water by membrane filtration. J. Appl. Bacteriol. 1987;62:279–287. doi: 10.1111/j.1365-2672.1987.tb02410.x. [DOI] [PubMed] [Google Scholar]
- Kageyama T., Kojima S., Shinohara M., Uchida K., Fukushi S., Hoshino F.B., Takeda N., Katayama K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L., Donaldson E.F., LoBue A.D., Cannon J.L., Zheng D.-P., Vinjé J., Baric R.S. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008;5:0269–0290. doi: 10.1371/journal.pmed.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Line D.E. Changes in a stream's physical and biological conditions following livestock exclusion. Transactions ASAE. 2003;46:287–293. [Google Scholar]
- Marti R., Gannon V.P.J., Jokinen C., Lanthier M., Lapen D.R., Neumann N.F., Ruecker N.J., Scott A., Wilkes G., Zhang Y., Topp E. Quantitative multi-year elucidation of fecal sources of waterborne pathogen contamination in the South Nation River basin using Bacteroidales microbial source tracking markers. Water Res. 2013;47(7):2315–2324. doi: 10.1016/j.watres.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Mayer P.M., Reynolds S.K., McCutchen M.D., Canfield T.J. Meta-analysis of nitrogen removal in riparian buffers. J. Environ. Qual. 2007;36:1172–1180. doi: 10.2134/jeq2006.0462. [DOI] [PubMed] [Google Scholar]
- Miller J.J., Chanasyk D.S., Curtis T., Willms T.C. Influence of streambank fencing on the environmental quality of cattle-excluded pastures. J. Environ. Qual. 2010;39:991–1000. doi: 10.2134/jeq2009.0233. [DOI] [PubMed] [Google Scholar]
- Rendtorff R.C. The experimental transmission of human intestinal protozoan parasites: 2. Giardia lamblia cysts given in capsules. Am. J. Hyg. 1954;59:209–220. doi: 10.1093/oxfordjournals.aje.a119634. [DOI] [PubMed] [Google Scholar]
- Renter D.G., Sargeant J.M., Oberst R.D., Samadpour M. Diversity, frequency, and persistence of Escherichia coli O157 strains from range cattle environments. Appl. Environ. Micro. 2003;69:542–547. doi: 10.1128/AEM.69.1.542-547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruecker N., Matsune J.C., Wilkes G., Lapen D.R., Topp E., Edge T.A., Sensen C.W., Xiao L., Neumann N.F. Molecular and phylogenetic approaches for assessing sources of Cryptosporidium contamination in water. Water Res. 2012;46:5135–5150. doi: 10.1016/j.watres.2012.06.045. [DOI] [PubMed] [Google Scholar]
- Schwarte K.A., Russell J.R., Kovar J.L., Morrical D.G., Einsley S.M., Yoon K.-J., Cornick N.A., Cho Y. Grazing management effects on sediment, phosphorus, and pathogen loading of streams in cool-season grass pastures. J. Environ. Qual. 2011;40(4):1303–1313. doi: 10.2134/jeq2010.0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P.J., Emelko M.B. QMRA and decision-making: are we handling measurement errors associated with pathogen concentration data correctly? Water Res. 2011;45:427–438. doi: 10.1016/j.watres.2010.08.042. [DOI] [PubMed] [Google Scholar]
- Schmidt P.J., Pintar K.D.M., Fazil A.M., Flemming C.A., Lanthier M., Laprade N., Sunohara M., Simhon A., Thomas J.L., Topp E., Wilkes G., Lapen D.R. Using Campylobacter spp. data and Bayesian microbial risk assessment to examine public health risks in agricultural watersheds under tile drainage management. Water Res. 2013;47:3255–3272. doi: 10.1016/j.watres.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Schmidt P.J., Pintar K.D.M., Fazil A.M., Topp E. Harnessing the theoretical foundations of the exponential and beta-Poisson dose-response models to quantify parameter uncertainty using Markov Chain Monte Carlo. Risk Anal. 2013 doi: 10.1111/risa.12006. [DOI] [PubMed] [Google Scholar]
- Sunohara M., Topp E., Wilkes G., Gottschall N., Neumann N., Ruecker N., Jones T.H., Edge E., Marti R., Lapen D.R. Impact of riparian zone protection from cattle on nutrient, bacteria, F-coliphage, Cryptosporidium, and Giardia loading of an intermittent stream. J. Environ. Qual. 2012;41:1–14. doi: 10.2134/jeq2011.0407. [DOI] [PubMed] [Google Scholar]
- Thomas M.K., Murray R., Flockhart L., Pintar K., Pollari F., Fazil A., Nesbitt A., Marshall B. Estimates of the burden of Foodborne illness in Canada for 30 Specified Pathogens and Unspecified Agents, Circa 2006. Foodborne Pathog. Dis. 2013 doi: 10.1089/fpd.2012.1389. http://online.liebertpub.com/doi/full/10.1089/fpd.2012.1389 Online Ahead of Print: May 9, 2013. accessed May, 2013: [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Agriculture (USDA) USDA; Washington, DC: 1996. National Handbook of Water Quality Monitoring. [Google Scholar]
- U.S. Environmental Protection Agency (USEPA) Water Resource Center; Washington, DC: 2001. Method 1605: Aeromonas in Finished Water by Membrane Filtration Using Ampicillin-dextrin Agar with Vancomycin (ADA-v) [Google Scholar]
- U.S. Environmental Protection Agency (USEPA) U.S. Environmental Protection Agency, Office of Water; Washington, DC: 2005. Economic Analysis for the Final Long Term 2 Enhanced Surface Water Treatment Rule; EPA 815-R-06–001. (Appendix N) [Google Scholar]
- van Elsas J.D., Semenov A.V., Costa R., Trevors J.T. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 2011;5:173–183. doi: 10.1038/ismej.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P., Poitras E., Leblanc D., Letellier A., Brassard J., Plante D., Houde A. Comparative analysis of different TaqMan real-time RT-PCR assays for the detection of swine Hepatitis E virus and integration of Feline calicivirus as internal control. J. Appl. Microbiol. 2009;106:1360–1369. doi: 10.1111/j.1365-2672.2008.04104.x. [DOI] [PubMed] [Google Scholar]
- Wilkes G., Edge T., Gannon V., Jokinen C., Lyautey E., Medeiros D., Neumman N., Ruecker N., Topp E., Lapen D.R. Seasonal relationships among indicator bacteria, pathogenic bacteria, Cryptosporidium oocysts, Giardia cysts, and hydrological indices for surface waters within and agricultural landscape. Water Res. 2009;43:2209–2223. doi: 10.1016/j.watres.2009.01.033. [DOI] [PubMed] [Google Scholar]
- Wilkes G., Edge T.A., Gannon V.P.J., Jokinen C., Lyautey E., Neumann N.F., Ruecker N., Scott A., Sunohara M., Topp E., Lapen D.R. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res. 2011;45:5807–5825. doi: 10.1016/j.watres.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Wilkes G., Ruecker N.J., Neumann N.F., Gannon V., Jokinen C., Sunohara M., Topp E., Pintar K.D.M., Lapen D.R. Spatiotemporal analysis of Cryptosporidium species/genotypes and relationships with other zoonotic pathogens in surface water from mixed-use watersheds. Appl. Environ. Micro. 2013 doi: 10.1128/AEM.01924–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organization) In: Waterborne Zoonoses: Identification, Causes and Control. Cotruvo J.A., Dufour A., Rees G., Bartram J., Carr R., Cliver D.O., Craun G.F., Fayer R., Gannon V.P.J., editors. IWA publishing; London: 2004. (for the World Health Organization) [Google Scholar]
- Yates M.V. Classical indicators in the 21st century – far and beyond the coliform. Water Environ. Res. 2007;79:279–286. doi: 10.2175/106143006x123085. [DOI] [PubMed] [Google Scholar]
- Zeng S.-Q., Halkosalo A., Salminen M., Szakal E.D., Puustinen L., Vesikari T. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J. Virol. Meth. 2008;153:238–240. doi: 10.1016/j.jviromet.2008.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.