Abstract
Transcriptomics has the potential to discover new RNA virus genomes by sequencing total intracellular RNA pools. In this study, we have searched publicly available transcriptomes for sequences similar to viruses of the Nidovirales order. We report two potential nidovirus genomes, a highly divergent 35.9 kb likely complete genome from the California sea hare Aplysia californica, which we assign to a nidovirus named Aplysia abyssovirus 1 (AAbV), and a coronavirus-like 22.3 kb partial genome from the ornamented pygmy frog Microhyla fissipes, which we assign to a nidovirus named Microhyla alphaletovirus 1 (MLeV). AAbV was shown to encode a functional main proteinase, and a translational readthrough signal. Phylogenetic analysis suggested that AAbV represents a new family, proposed here as Abyssoviridae. MLeV represents a sister group to the other known coronaviruses. The importance of MLeV and AAbV for understanding nidovirus evolution, and the origin of terrestrial nidoviruses are discussed.
Keywords: Nidovirales, Transcriptome, Virus discovery, Proteinase, Protease, Protein expression, Translation, Readthrough
Graphical abstract
1. Introduction
Until recently, discovery of new RNA viruses proceeded slowly in a mostly hypothesis-driven manner while searching for an agent of a disease, and using antibody cross-reactivity or enough conserved motifs for successful amplification by reverse transcriptase polymerase chain reaction. With improvements in RNA transcriptome sequencing and homology-based search methods, it is now possible to capture the complete infecting RNA virome of an organism by deep-sequencing total intracellular RNA pools (Miranda et al., 2016, Shi et al., 2018, Shi et al., 2016).
The new sequencing methods have brought a great change to the Nidovirales, an order that includes viruses with complex replicase polyproteins and the largest known RNA genomes (Lauber et al., 2013). This order previously contained four family-level groups, the Coronaviridae which infect birds and mammals including humans, the Arteriviridae which infect non-human mammals, the Mesoniviridae which infect arthropods, and the Roniviridae which infect crustaceans (Lauber et al., 2013). However, recent papers (Lauck et al., 2015, O’Dea et al., 2016, Saberi et al., 2018, Shi et al., 2018, Shi et al., 2016, Tokarz et al., 2015, Vasilakis et al., 2014, Wahl-Jensen et al., 2016) and our results (see below) have added to within-family diversity and revealed several highly divergent nido-like viruses which the Nidovirales Study Group proposed, pending ICTV ratification, to form four new virus families within the Nidovirales (Gorbalenya et al., 2017a).
In this report we describe the discovery and characterization of one of the nidoviruses prototyping a new family along with another putative nidovirus. We used BLAST searches to scan the publicly available transcriptomes and expressed sequence tag libraries available at the US National Center for Biotechnology Information, and revealed two novel nido-like virus sequences from the frog Microhyla fissipes developmental transcriptome (Zhao et al., 2016) and from several transcriptome studies dealing with the marine gastropod Aplysia californica (Fiedler et al., 2010, Heyland et al., 2011, Moroz et al., 2006). We describe the bioinformatics of the new virus-like sequences, and demonstrate that the Aplysia virus-like sequence encodes a functional proteinase, and a translational termination-suppression signal. Implications for nidovirus evolution and the origin of nidovirus structural proteins are discussed.
2. Results
2.1. Virus discovery
Recent studies have identified a wide variety of virus-like sequences in intracellular RNA pools, but few new members of the Nidovirales have been reported compared to groups such as the Picornavirales. In order to determine whether additional lineages of nido-like viruses might be present, tBLASTn (Altschul et al., 1990) was used to search the transcriptome shotgun assembly (TSA) and expressed sequence tag (EST) databases for sequences encoding proteins similar to the main proteinase (Mpro), polymerase and helicase, or complete pp1b regions of the nidovirus strains Infectious bronchitis virus, Gill-associated virus, White bream virus, Cavally virus and Wobbly possum disease virus. The tBLASTn results were checked by using BLASTx to compare each result to the non-redundant protein database, and results that matched back to any member of the Nidovirales were selected for further analysis. This led to the discovery of a 35.9 kb transcript and 243 other fragments from the California sea hare, Aplysia californica, and a 22.3 kb transcript from Microhyla fissipes, known as the ornamented pygmy frog. Putative virus transcripts were then compared to DNA sequences from the same organisms by nucleotide BLAST, and no evidence of either virus was found. Together, these tests suggest that both nidovirus-like transcripts most likely come from RNA viruses associated with host transcriptomes.
2.2. Phylogenetic analysis
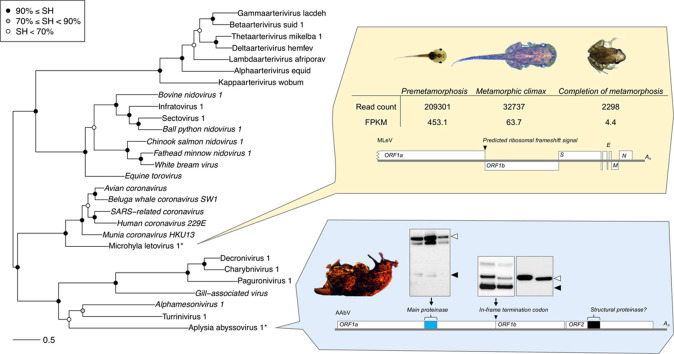
Phylogenetic analysis was performed by IQ Tree 1.5.5 (Nguyen et al., 2015) using five protein domains universally conserved in known and proposed nidoviruses plus the virus-like sequences described in this study (see below). The produced maximum-likelihood tree was mid-point rooted to reveal two strongly-supported super-clades, consisting of four strongly-supported major clades corresponding to arteri-like viruses, toro-like viruses, corona-like viruses, and invertebrate nidoviruses ( Fig. 1). A Bayesian rooted tree (not shown) was also constructed using the same viral sequences, and it yielded the same four major clades, but with weaker support values on some branches and a basal position of the arteri-like major clade. Together these results suggest that the novel virus-like sequences likely represent distantly related members of the Nidovirales, but the tree branch uncertainty also demonstrates the limitations of these phylogenetic approaches in dealing with the extreme diversity of the sparsely sampled nido-like viruses.
Fig. 1.
Nidovirus phylogeny reconstructed based on concatenated MSA of five replicative domains universally conserved in nidoviruses. SH-aLRT branch support values are depicted by shaded circles. Species names that are not currently recognized by ICTV are written in plain font. Asterisks designate viruses described in this study.
The virus-like sequence from Aplysia californica formed a relatively long and moderately supported branch that clustered with other invertebrate nidoviruses, forming a sister group to a clade consisting of the Mesoniviridae and a recently discovered nidovirus from the marine snail Turritella, TurrNV. The virus-like sequence from Microhyla fissipes clustered with strong support as a sister group to the known Coronavirinae. We named these putative viruses Aplysia abyssovirus (AAbV) and Microhyla letovirus (MLeV), respectively.
While we were expressing viral proteins to biologically validate the new sequences and preparing this manuscript, a second manuscript appeared on BioRxiv (Debat, 2018) from Humberto Debat who was describing the same Aplysia virus from the same source material, posted April 24th, 2018, where it is called Aplysia californica nido-like virus. That report covers the tissue tropism and age-dependent prevalence of the Aplysia virus thoroughly, so in this manuscript we will focus on bioinformatics analysis and biological validation of this virus. It is our opinion that the name Aplysia californica nido-like virus should be regarded as an alternate name to Aplysia abyssovirus.
2.3. Naming and etymology
After assigning AAbV and MLeV to nidoviruses by the above bioinformatics analysis, the genome sequences were submitted to the Nidovirus Study Group (NSG) of the International Committee on the Taxonomy of Viruses (ICTV) for their accommodation in the nidovirus taxonomy; BN, senior author of this manuscript, is a member of the NSG and AAG assisted NSG with analysis of these viruses. Classification of these and other viruses were described in several taxonomic proposals that were made publicly available in the pending proposals section of ICTV on June 23rd, 2017, revised on November 26th, 2017 (Gorbalenya et al., 2017b, Gorbalenya et al., 2017a; Ziebuhr et al., 2017) and August 12, 2018. They were approved by the ICTV Executive Committee in July 2018 and will be placed for ratification by ICTV in 2018. Throughout this report, we will follow the taxa naming and taxonomy from the pending ICTV taxonomic proposals cited above, which we interpret to establish priority in discovering and naming these viruses and establishing the respective taxa.
The etymology of the name abyssovirus is from the word abyss, a reference to the aquatic environment where Aplysia lives, to the Sumerian god of watery depths Abzu, and to its discovery in an RNA transcriptome obtained by “deep” sequencing technology. Based on relatively low amino acid identity to the other families in the Nidovirales, it is our opinion that AAbV prototypes a new nidovirus family, which was confirmed in the analysis described in the pending proposal. The NSG has also accepted our proposal to name the new family Abyssoviridae, the new genus Alphaabyssovirus and the new species Aplysia abyssovirus 1.
The etymology of the name letovirus is in reference to the source of the virus in frogs, and their connection to the mythological Leto, daughter of the titans Coeus and Phoebe. In the story, Leto turned some inhospitable peasants into frogs after they stirred up the mud at the bottom of a pool so that she could not drink from it. Based on the low sequence identity but high conservation of domains found in the Coronavirinae, it is our opinion that MLeV is a member of a sister group to all known coronaviruses, but still within the Coronavirinae. Based on our input, the NGS named the new genus Alphaletovirus in the pending proposal.
2.4. AAbV genome and subgenome sequences and their potential expression
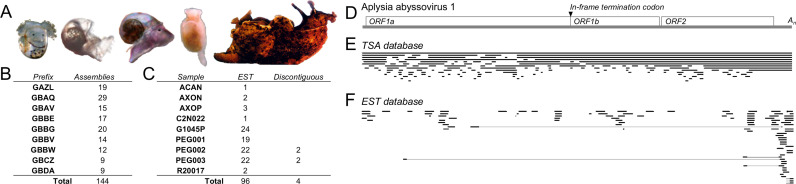
The host of AAbV is shown in Fig. 2A. The virus was recovered from a variety of adult tissues, and from several developmental stages of the host organism, as described elsewhere (Debat, 2018). Fragments of AAbV were detected in 9 TSA and 9 EST databases, compiled over several years by three labs working in Florida and the UK (Fig. 2B-C).
Fig. 2.
Sequence coverage of AAbV in public NCBI libraries. (A) Examples of the host organism Aplysia californica at swimming veliger, settled, metamorphic, juvenile and adult developmental stages (images not to scale, adapted from Heyland et al. (2011) and Moroz et al. (2006)). Summary of distinct sequence assemblies and reads in the TSA (B) and EST (C) matching AAbV for which the nucleotide BLAST E value was 2 × 10–70 or smaller. (D) Map of AAbV, showing the location of the replicase polyprotein genes (ORF1a, ORF1b), structural polyprotein gene (ORF2) and poly-adenosine tail (An). The position of sequences from the TSA (E) and EST (F) databases matching AAbV is shown.
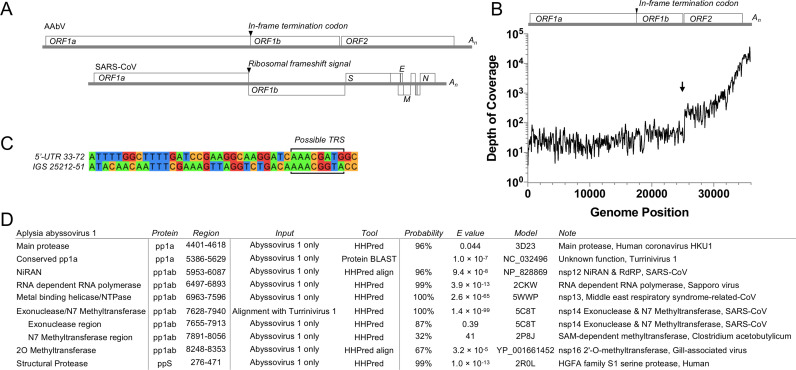
The AAbV genome is represented in its longest and most complete available form by the transcriptome shotgun assembly sequence GBBW01007738 which represents a reverse-complementary genomic sequence. Remarkably, the organization of the AAbV genome has several features typical for viruses of the Alphavirus genus of the Togaviridae family (King et al., 2012) that could be contrasted with those conserved in the nidoviruses. They include: a) two in-frame open reading frames (ORFs; ORF1a and ORF1b) of the replicase gene that are separated by a stop codon rather than overlapping and including a nidovirus-like ribosomal frameshift signal in the overlap, and b) a single structural polyprotein gene (ORF2) rather than several ORFs encoding structural proteins. The 35913 nt long AAbV genome has a 74 nt 5′-untranslated region, a 964 nt 3′-untranslated region, and a short poly-A tail (Fig. 2D). Despite these alphavirus-like features, BLASTx analysis confirmed that the AAbV replicase polyprotein clusters with the Nidovirales, as depicted in Fig. 1. Each part of the genome is represented in 3–20 independent sequences from the TSA and EST databases available at www.ncbi.nlm.nih.gov as of November 26th, 2017 (Fig. 2E-F). The AAbV genome ( Fig. 3A) is the second-largest currently reported RNA virus genome, behind a new 41.1 kb planarian nidovirus described in a BioRxiv manuscript (Saberi et al., 2018).
Fig. 3.
Coding capacity, depth of coverage and bioinformatics of AAbV. (A) Genome and coding capacity of AAbV and SARS-CoV are shown to scale. (B) Total depth of coverage based on a sample of 672017 aligned spots matching AAbV from Aplysia californica RNA sequence read archives including SRR385787, SRR385788, SRR385792, SRR385793, SRR385795, SRR385800, SRR385802 and SRR385815. The putative start site of a viral subgenomic RNA species is marked with an arrow. (C) Alignment of the 5′-untranslated region and the intergenic sequence between the pp1b and pp2 genes showing a potential transcription-regulatory sequence (boxed). (D) Bioinformatic assignment of domains in AAbV. Sequence(s) used for prediction (Input) were either AAbV alone or a multiple sequence alignment containing AAbV and TurrNV. Probability score from HHPred and E value from HHPred or BLAST are shown. Accession numbers are given for sequences or protein structures identified as a match for an AAbV domain (Model).
The sequence of the genomic 5′-terminus is supported by the five assemblies (GBBW01007738, GAZL01021275, GBDA01037198, GBCZ01030948, and GBCZ01030949) that end within one nucleotide of each other. The EST sequence EB188990 contains the same sequence with an additional 5′-GGCTCGAG-3′ that may represent part of the 5′-terminal region missing from GBBW01007738. However, we prefer to side with the preponderance of sequence data and consider GBBW01007738 the most complete AAbV genome available until further biological evidence emerges.
The sequence of the 3′-terminus is supported by 6 TSA sequence assemblies and 1 EST sequence that all end within one nucleotide of each other. Every part of the genome is represented in at least three TSA sequence assemblies. Genome coverage is more abundant at the 3′-end, which could be evidence of 3′-coterminal subgenomic RNA species, or could be a result of the method used to prepare cDNA.
Genetic variation among these sequences is as follows. There are four short EST sequences which appear to join different discontinuous regions of the genome together, but the joins occur at different positions in the middle of genes and cannot be explained by nidovirus-like discontinuous transcription. These oddly joined sequence fragments likely represent either defective RNA species (Furuya et al., 1993), or artifacts of the EST preparation process. Two sequence assemblies differed from the others, with A replacing G at nucleotide 1627, and in another assembly A replacing the consensus G at position 28005, both of which could be attributed to natural mutations or the actions of host cytidine deaminase on the viral minus strand. There is also some variation in the preserved poly-A tail sequences, presumably from the difficulty of accurately reading long stretches of a single nucleotide.
In order to test whether there was support for AAbV subgenomic RNA species in the raw sequence data, individual sequence reads were mapped to the AAbV genome using Bowtie 2.3.4.1 (Langmead and Salzberg, 2012) and SAMtools 1.9 (Li et al., 2009). There was no a noticeable change in read depth at the junction between ORF1a and ORF1b, but there was a sudden increase of about seven-fold in read depth immediately before the start of ORF2 (Fig. 3B), suggesting that ORF2 may be expressed from a subgenomic mRNA produced in relative abundance compared to the genomic RNA, as would be expected for a member of the Nidovirales. Numerous low-frequency AAbV sequence variants were identified in the raw sequence data, but none were consistent across all datasets, and no indels were consistently present within 1000 nucleotides of the start of ORF2. This was interpreted to indicate that either the viral subgenomic mRNA did not contain the expected nidovirus-like leader-body structure, or that any potential 5′-terminal leader sequences were not captured in the raw data.
Nidoviruses express their structural and accessory proteins via a set of 3′-coterminal nested subgenomic RNAs, which are produced by discontinuous transcription on the genomic template. In this process, the polymerase is thought to pause at transcription-regulatory sequences located upstream of each gene, occasionally resulting in a template switch to homologous transcription-regulatory sequence in the viral 5′-untranslated region to produce negative-stranded RNAs of subgenomic size (Sola et al., 2015). The longest sequence match between the 5′-untranslated region and intergenic sequence of AAbV is shown in Fig. 3C. It consists of six of eight identical nucleotides, which could form eight base pairs with a reverse-complementary viral minus strand due to the possibility of both A-U and G-U wobble base pairing. However, none of the available TSA or EST sequences showed direct evidence of a subgenomic RNA species, such as a consistently-spliced transcript, or a large number of sequence reads that stop at the putative transcription-regulatory sequence. This sequence AAACGATG or AAACGGTA needs to be investigated further to determine whether it functions as a transcription-regulatory sequence for viral subgenomic RNA production.
Together these data suggest that the AAbV genome is reasonably complete, robust, and represents a novel and exceptionally large nido-like virus. It has the unusual genome organization which is nonetheless consistent with the canonical nidovirus features of large replicase polyproteins 1a and 1ab, pp1a and pp1ab, respectively. They are expressed via a translational readthrough rather than frameshift mechanism, while potential structural protein genes are presumably expressed from a single subgenomic RNA to produce structural polyprotein pp2.
2.5. AAbV protein bioinformatics
To annotate the functional protein domains encoded in the AAbV genome, a series of bioinformatics tools were used. Wherever possible, we have followed the convention of SARS-associated coronavirus (SARS-CoV) species in naming domains and polyprotein processing products (Ref?). When run against the PDB database, HHPred (Söding et al., 2005) predicts function based on structure. For domains like the polymerase where a nidovirus structure is not yet available, HHPred can sometimes detect a match to a homologous protein, such as the picornavirus polymerase.
HHPred produced confident predictions for a coronavirus-like Mpro (Anand et al., 2002) in pp1a (Fig. 3D). In pp1b HHPred identified a picornavirus-like RNA-dependent RNA polymerase (RdRp (te Velthuis et al., 2009)), nsp13 metal-binding helicase (Deng et al., 2014, Ivanov et al., 2004), nidovirus-specific nsp14 exonuclease (ExoN (Ma et al., 2015)) and nsp14 N7 methyltransferase (N7 MTase (Chen et al., 2009; Ma et al., 2015)). In pp2, HHPred identified a chymotrypsin-like serine proteinase (Birktoft and Blow, 1972), a feature analogous to the alphavirus capsid proteinase (Melancont and Garoff, 1987), but until now predicted in only one nidovirus, TurrNV. We have termed this the structural proteinase (Spro).
Where HHPred was unable to annotate a region, a protein BLAST search was carried out to identify likely homologs among other known nidoviruses. When a match was found, both proteins were aligned using Clustal Omega (Sievers et al., 2011), and the multiple sequence alignment was used in HHPred. The most consistent matches to AAbV were from TurrNV. This identified a larger region and a more confident match to the coronavirus nsp14 ExoN-N7 MTase.
Protein BLAST was used to map the AAbV nidovirus RdRp-associated nucleotidyl transferase (NiRAN) and nsp16 2O-MTase domains to homologous domains from other nidoviruses. The corresponding regions of AAbV and the top protein BLAST match were then submitted to HHPred in align mode, which uses predicted structure and primary sequence data to compare proteins. This led to confident identifications of the NiRAN and a match for the divergent but functional 2O MTase domain of Gill-associated virus (Zeng et al., 2016). One other uncharacterized domain was also identified in both AAbV and TurrNV by protein BLAST, in the position where the coronavirus conserved replication accessory proteins nsp7–10 were expected (Fig. 3D). However, there was not enough similarity between the AAbV-TurrNV conserved domain and other nidovirus domains to confidently assign a function to this region.
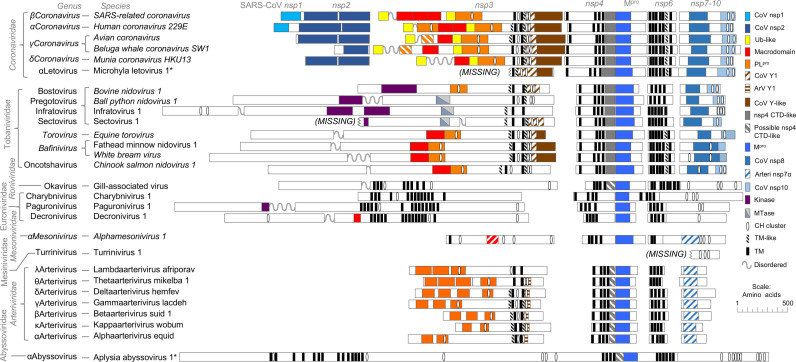
We also looked for transmembrane regions which are typically clustered in three regions in nidovirus pp1a. Domain-level maps of new and known nidoviruses pp1a and pp1b are shown in Figs. 4 and 5A, respectively. Nidoviruses typically have a cluster of an even number of transmembrane helices near the midpoint of pp1a, equivalent to nsp3 of SARS coronavirus. Nidoviruses also have two other clusters of 2–8 transmembrane helices flanking the Mpro domain from both sides.
Fig. 4.
Comparison of predicted domain-level organization in polyprotein 1a of new viruses to previously described nidoviruses. Gaps have been introduced so to align predicted homologous domains. Virus naming and taxonomy conventions follow the ICTV proposals in which MLeV and AAbV were first described (Gorbalenya et al., 2017b, Gorbalenya et al., 2017a, Ziebuhr et al., 2017). New viruses are marked with stars, accepted taxonomic ranks are italicized and proposed taxonomic ranks are not italicized. Polyprotein processing products from SARS-CoV are shown at top. Domains are colored to indicate predicted similarity to SARS-CoV nsp1 (CoV nsp1), SARS-CoV nsp2 (nsp2-like), ubiquitin (Ub-like), macrodomains, papain-like proteinase (PLpro), first section of the coronavirus Y domain (CoV Y1), first section of the arterivirus Y domain (ArV Y1) coronavirus-specific Y domain-like (CoV Y-like), carboxyl-terminal domain of coronavirus nsp4 (nsp4 CTD-like), region with PSIPRED predicted structural similarity to nsp4 CTD, main proteinase (Mpro), SARS-CoV nsp8-like (CoV nsp8), Equine arteritis virus nsp7α (ArV nsp7α), SARS-CoV nsp10 (CoV nsp10), protein kinase-like (Kinase), RNA methyltransferase (Mtase), potential metal ion-binding clusters with 4 cysteine or histidine residues in a 20 amino acid window (CH-cluster), transmembrane helices, hydrophobic transmembrane-like regions that may not span the membrane by analogy to coronavirus nsp4 and nsp6 (TM-like) and disordered regions (Unstructured).
Fig. 5.
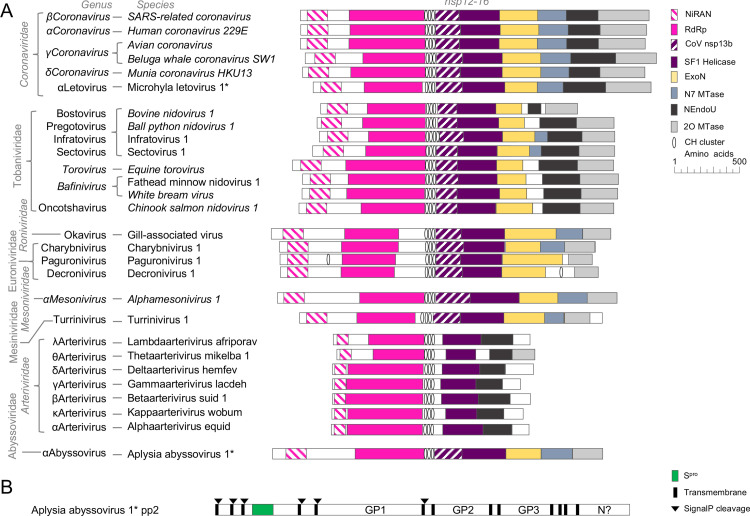
Comparison of predicted domain-level organization in polyprotein 1b of new viruses to previously described nidoviruses. (A) Domains include the nidovirus RdRp-associated nucleotidyl transferase (NiRAN), RdRp, potential metal ion binding clusters with four cysteine or histidine residues in a window of 20 amino acids (CH cluster), homologs of the domain of unknown function in the middle of coronavirus nsp13 (CoV nsp13b), superfamily 1 helicase (SF1 Helicase), nidovirus-specific exonuclease (ExoN) and uridylate-specific endonuclease (NEndoU), RNA cap N7 methyltransferase (N7 MTase) and RNA cap 2′-O-methyltransferase (2O MTase). (B) Domains of pp2 include the structural protease (Spro), putative glycoproteins GP1, GP2 and GP3, and a nucleoprotein-like domain (N?), TMHMM-predicted transmembrane domains and SignalP-predicted signal peptidase cleavage sites.
AAbV is also missing some common but not universally conserved nidovirus domains. AAbV does not appear to encode a homolog of the uridylate-specific nidovirus endonuclease (NendoU), nor is there enough un-annotated protein sequence in pp1b to accommodate an NendoU. This result is in line with the lack of this domain in other invertebrate nidoviruses (Nga et al., 2011). We were also not able to corroborate the prediction (Debat, 2018) of a papain-like proteinase domain situated among the predicted transmembrane regions of the first transmembrane cluster, or of a potential S-like domain of the structural polyprotein.
The pp2 gene of AAbV encodes a putative structural polyprotein of 3224 amino acids. HHPred and BLAST were not able to detect matches for any domains except Spro in AAbV pp2. TMHMM (Krogh et al., 2001) predicted 13 transmembrane helices in pp2, which were generally arranged in pairs with large intervening domains, which we have tentatively named Spro, predicted surface glycoproteins GP1–3 and a possible nucleoprotein (Fig. 5B). Included in pp2 are additional smaller domains that have not been named yet, pending a better understanding of pp2 proteolytic processing. SignalP (Petersen et al., 2011) predicted an initial signal peptide at the extreme amino terminus, but after removing the predicted signal peptide and re-running the prediction with the “N-terminal truncation of input sequence” parameter set to zero, a total of six potential signal peptidase cleavage sites were detected. The identification of the nucleoprotein-like domain is based on a resemblance to the N proteins of Bovine torovirus and Alphamesonivirus 1, and to the carboxyl-terminal half of the SARS-CoV N. The features the AAbV N-like protein shares with N of other established nidoviruses are an initial glycine-rich region that may be flexibly disordered, followed by a lysine and arginine-rich region from amino acid 2869–2913 that could facilitate RNA binding, followed by a domain predicted by PSIPRED (Buchan et al., 2013) to contain a secondary structure profile similar to that of the Equine arteritis virus N and the SARS-CoV N carboxyl-terminal domain. We did not find strong evidence to support the analysis of Debat (Debat, 2018) predicting a spike-like fold in GP3, but we concur with Debat in noticing that GP2 (and we would add, GP3) have a protein secondary structure profile that resembles an alphavirus E1 protein and the E1-like protein of TurrNV.
One previous report (Prince, 2003) had noted virus-like particles described as resembling intracellular alphavirus virions, that were widespread in transmission electron micrographs of Aplysia californica tissue, which would seem to be consistent with the alphavirus-like organization of the structural polyprotein and apparent E1 homology. However, further testing is necessary to confirm whether those virus-like particles are related to AAbV.
2.6. AAbV proteinases
When identifying viruses through bioinformatics, there is a risk that the sequences are either mis-assembled, contain errors, or are artifacts of the sequencing and sequence assembly processes. We tested the function of some AAbV protein features to determine if any was biologically functional, as a way to better assess whether the AAbV genome represented a replicating virus encoding functional parts.
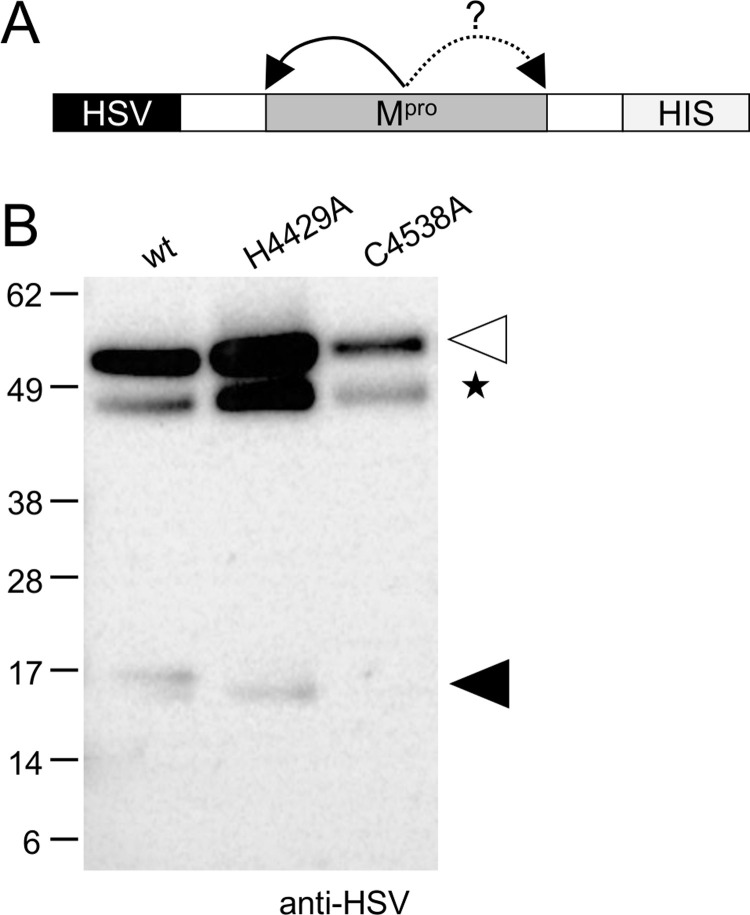
The AAbV Mpro and Spro plus surrounding regions up to the nearest preceding and following predicted transmembrane helix were cloned into pTriEx 1.1 and expressed with an amino-terminal herpes simplex virus epitope (HSV) tag, and a carboxyl-terminal poly-histidine (HIS) tag. Expressions were carried out by in vitro coupled T7 transcription and rabbit reticulocyte lysate translation. Mpro cleavage at an amino-terminal site was detected by the presence of an approximately 16 kDa HSV-tagged fragment ( Fig. 6), which would be expected if Mpro cleavage occurred in the vicinity of amino acid 4375, located near the start of the region of Mpro homology at amino acid 4401 (Fig. 3D). Spro was expressed, but did not produce any detectable cleavage products in the same assay (data not shown). From this we concluded that AAbV Mpro appeared to have proteinase activity in the context of our expression construct, while our Spro construct did not. Further work will be needed to determine whether the failure of the putative Spro to cleave was a result of the construct boundaries, assay conditions, lack of an appropriate substrate, or errors in the protein sequence.
Fig. 6.
Investigation of proteinase activity of AAbV Mpro. The AAbV main proteinase (Mpro; A-B) and surrounding regions were expressed as HSV and HIS-tagged constructs as shown in panel A. A white triangle marks the expected size of the 52.5 kDa uncleaved Mpro constructs. Black triangles mark the size of approximately 16 kDa amino-terminal cleavage products. Non-specific bands that were also present in control lanes are indicated with a star.
To further characterize the activity of AAbV Mpro, alanine-scanning mutations were made to amino acids that appeared to match the catalytic cysteine and histidine residues of other coronavirus main proteinases. Mutation of the putative catalytic histidine H4429 did not strongly reduce proteolytic processing, while mutation of the cysteine C4538 blocked proteinase activity (Fig. 6). These data demonstrate that AAbV encodes at least one functional proteinase, but further work is needed to determine the cleavage specificity and map proteolytic processing by the AAbV Mpro.
2.7. AAbV pp1ab expression
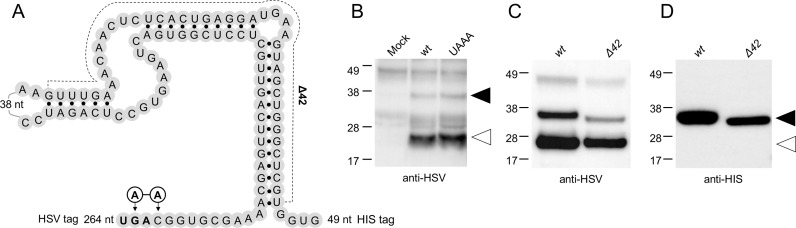
Another unusual feature of AAbV was the presence of an in-frame stop codon separating the pp1a and pp1b genes, rather than the expected ribosomal frameshift signal found in most other nidoviruses. We note that an in-frame stop codon separates the putative pp1a and pp1b of the molluscan nidovirus Tunninivirus 1, which was phylogenetically grouped with AAbV and Alphamesonivirus 1 (Fig. 1). This suggested that AAbV may use a translational termination-suppression signal as a way to control expression of the pp1b region. Termination-suppression signals are found in several other viruses including alphaviruses and some retroviruses, and typically consist of a UAG or UGA stop codon followed by an RNA secondary structure element, and the efficiency of suppression normally depends on the stop codon, the nucleotides immediately following the stop codon, and the free energy of the RNA secondary structure element (Feng et al., 1992). The pp1a gene of AAbV ends in a UGA stop codon, and the region that follows was predicted by Mfold (Zuker, 2003) to be capable of forming several related RNA secondary structure elements, of which the most consistently predicted is shown in Fig. 7A. A potential pseudoknot-like conformation in the same region is shown by Debat (Debat, 2018).
Fig. 7.
Mutational analysis of the termination-suppression signal (TSS) at the ORF1a/b junction. (A) Schematic view of the TSS expression construct and introduced HSV and HIS tags, showing only predicted RNA secondary structures that were consistent in the best six models generated by Mfold. Mutations around the stop codon (bold, producing the UAAA construct) or removing one side of the predicted stem-loops (Δ42) are shown. (B-D) Western blots showing translation of mutant TSS expression constructs in a coupled T7 polymerase rabbit reticulocyte lysate expression system. Blots were probed with anti-HSV (B, D) to detect both 25 kDa terminated and 32–35 kDa readthrough products, or with anti-HIS (C) to detect only readthrough products.
To investigate protein expression at the pp1a-pp1b region, nucleotides 17255–17707 were cloned into pTriex 1.1 with amino-terminal HSV and carboxyl-terminal HIS tags. This construct would allow detection and quantification of the 25 kDa proteins that stopped at the natural UGA stop codon that would have an HSV tag only, and 35 kDa readthrough products that would have both HSV and HIS tags. Expression of this construct produced the expected 25 kDa termination product and 35 kDa readthrough product (Fig. 7B-D). Based on densitometry analysis (not shown), it was estimated that 25–30% of translation events resulted in readthrough.
The choice of stop codon and elements of the two codons that follow have been shown to affect the efficiency of translational termination (Cridge et al., 2018, Skuzeski et al., 1991). To further investigate the AAbV termination-suppression signal, constructs were made in which the region around the pp1a stop codon was perturbed from the wild-type UGAC, predicted to produce near optimal termination, to UAAA, predicted to produce much less than optimal termination. In another construct, 42 nucleotides predicted to form one side of the predicted RNA stem-loops were deleted (Δ42; Fig. 7A). Mutation of the AAbV pp1a stop codon had little effect on readthrough efficiency (Fig. 7B), but deletion of 42 nucleotides predicted to be involved in RNA secondary structures appeared to decrease readthrough, and led to a smaller readthrough product as predicted. Together these results indicate that the pp1b region of AAbV is probably expressed by readthrough of a UGA stop codon, mediated by a functional termination-suppression signal that is dependent on sequences following the stop codon.
2.8. MLeV genome
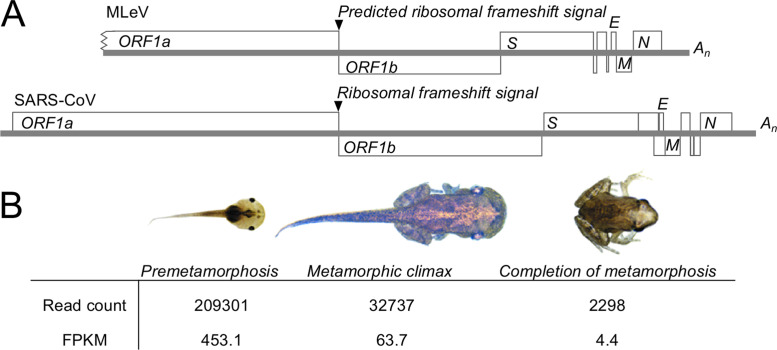
Microhyla letovirus is represented by a single assembly (accession number GECV01031551) of 22304 nucleotides that potentially encodes a partial corona-like virus from near the end of a protein equivalent to SARS-CoV nsp3 to the 3′-end ( Fig. 8A). No other matches for this sequence were found in the TSA or EST databases by nucleotide BLAST. The host organism of MLeV is shown in Fig. 8B. Mapping single sequence reads onto the genome revealed a strong age dependence of MLeV detection. The number of fragments per kilobase of transcript per million mapped reads decreased by seven-fold from pre-metamorphosis to metamorphic climax, then decreased again by fourteen-fold from metamorphic climax to completion of metamorphosis. Further testing was done by reverse transcriptase polymerase chain reaction using MLeV-specific primers on the same population of adult frogs later in the year, but all the adult material tested was negative for MLeV (LZ, personal communication).
Fig. 8.
Coding capacity and prevalence of MLeV (A) Schematic representation of the coding capacity of MLeV compared to SARS-CoV, showing the similarities in genome organization. (B) Prevalence of MLeV transcripts in Microhyla fissipes by age, by total number of reads and fragments per kilobase of transcript per million mapped reads (FPKM).
The MLeV genome is missing the 5′-end of the genome, including a 5′-untranslated region and sequences corresponding to coronavirus nsp1, nsp2 and part of nsp3. The size of the missing part of the genome can be estimated at 1500–4000 nucleotides based on comparison to complete genomes from the relatively small deltacoronaviruses or the relatively large alphacoronaviruses. The MLeV genome contains a 572 nucleotide 3′-untranslated region and an 18-nucleotide poly-adenosine tail.
The genome organization of MLeV was similar to that of coronaviruses, with a predicted -1 ribosomal frameshift signal. Usually, a programmed -1 ribosomal frameshift signal consists of three elements: a slippery sequence that is most commonly UUUAAAC in coronaviruses, a stop codon for the upstream coding region, and a strong RNA secondary structure or pseudoknot. MLeV encodes a potential slippery sequence at nucleotide 6085 (UUUAAAC) followed immediately by a UAA stop codon for pp1a. The region following the putative frameshift signal was predicted by Mfold to adopt a stem-loop conformation which may be part of an RNA pseudoknot (not shown), but further biological characterization is needed to determine the boundaries of the frameshifting region and test its frameshifting efficiency.
The 3′-end of the MLeV genome contains six ORFs that could encode proteins of 50 or more amino acids, which presumably include the viral structural proteins. Five of the six 3′-end ORFs are preceded by a sequence UCUAAHA (where H is any nucleotide except G), that resembles the UCUAAAC transcription regulatory sequence of the coronavirus mouse hepatitis virus. These candidate transcription-regulatory sequences start 6–66 nucleotides before the AUG start codon of the next ORF. Without the 5′-end or any evidence of viral subgenomic RNAs, it is not possible to be certain how the 3′-end ORFs are expressed, but these repeated sequences are evidence that MLeV may express its structural proteins from subgenomic RNAs in the manner of coronaviruses. Unfortunately, the original RNA sample that was used for Microhyla fissipes transcriptomic analysis was completely consumed, and could not be further tested by RT-PCR.
The first of these downstream ORFs encodes a large S-like protein of 1526 amino acids with an amino-terminal signal peptide predicted by SignalP and a carboxyl-terminal transmembrane region predicted by TMHMM. The second and third ORFs appear to encode a unique single-pass transmembrane protein of 55 amino acids (ORF 2b) and a unique soluble 157 (ORF 3) amino acid protein, respectively, which are likely strain-specific accessory proteins. The fourth ORF encodes an E-like protein of 77 amino acids, with an amino-terminal predicted transmembrane region followed by a potential amphipathic helix predicted by Amphipaseek (Sapay et al., 2006). The fifth ORF encodes a 241 amino acid long three-pass transmembrane protein that resembles the coronavirus M protein, and the sixth ORF encodes a putative N protein of 459 amino acids. Together, these 3′-ORFs appear to encode a complete coronavirus functional repertoire, and are present in the same order found on all other currently known coronavirus genomes (Neuman and Buchmeier, 2016). The start codons of the putative S and M ORFs appear to overlap with the stop codons of preceding ORFs, indicating a relatively compact genome.
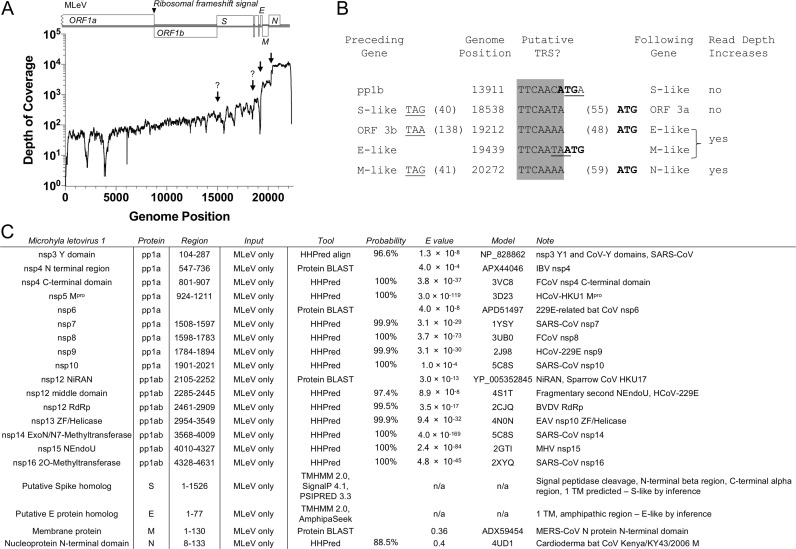
To test whether there was support for MLeV subgenomic RNA species in the raw sequence data, individual sequence reads were mapped to the MLeV genome using the same method used for AAbV above ( Fig. 9A). There was not a noticeable change in read depth at the junction between ORFs 1a and 1b of MLeV, suggesting that polyprotein 1b is expressed by a translational rather than transcriptional mechanism. However, there were two sudden increases of about eight-fold in read depth immediately before the start of the N ORF and near the beginning of the adjacent E and M ORFs (Fig. 9B). Expected increases in read depth before the putative S gene and the largest putative accessory gene were not detected. As with AAbV, many low-frequency sequence variants were detected in the raw sequence data, but no indels were consistently present in the region surrounding the putative transcription-regulatory sequences. These data suggest that at least the M and N genes of MLeV are expressed via subgenomic mRNAs.
Fig. 9.
Depth of coverage and bioinformatics of MLeV. (A) Total depth of coverage is based on 275503 aligned spots matching MLeV from Microhyla fissipes RNA sequence read archives SRR2418812, SRR2418623 and SRR2418554. The putative start sites of a viral subgenomic RNA species are marked with an arrow. Potential subgenomic RNA start sites not marked by a sharp rise in read depth are indicated with question marks. (B) Positions and usage of putative transcription-regulatory sequences. Termination codons from the preceding gene are underlined, initiation codons of the following gene are in bold. (C) Bioinformatic assignment of domains in MLeV.
2.9. MLeV protein bioinformatics
In the pp1a region, HHPred detected matches for conserved coronavirus domains including the carboxyl-terminal domain of coronavirus nsp4, Mpro, nsp7, nsp8, nsp9 and nsp10 (Fig. 8C). In the pp1b region, HHPred detected matches for a picornavirus-like RdRp, the nsp13 metal-binding helicase, the nsp14 ExoN-N7 MTase, the nsp15 NEndoU, and the nsp16 2O MTase. In the structural protein region, HHPred detected a match for the amino-terminal domain of coronavirus N in the putative MLeV N protein.
As with AAbV, we then widened our search to include conserved coronavirus domains that do not yet have known protein structures. This led to a match for the carboxyl-terminal region of nsp3, amino-terminal region of nsp4, nsp6, the nsp12 NiRAN domain, and a match between coronavirus M and the proposed MLeV M protein. Neither the proposed MLeV S nor E protein could be further corroborated by bioinformatics tools. Together, this indicated that MLeV appears to encode a complete set of conserved coronavirus-like proteins from the carboxyl-terminal region of nsp3 through the end of the genome.
3. Discussion and conclusions
With the addition of MLeV, AAbV and a host of other recently-published highly divergent nidoviruses, the field of nidovirus evolution is due for a revision, which will require a detailed approach and that will fit best in another study. However, a few tentative conclusions can be drawn from these new viruses.
Firstly, the new viruses confirm that the region of pp1a up to the SARS-CoV nsp4 equivalent, which seems to contain a variety of anti-host countermeasures in the viruses where this region has been studied (Neuman et al., 2014), is highly variable and does not appear to contain any universally-conserved domains. As previously noted (Lauber et al., 2013), this part of the genome appears to have the most genetic flexibility, even within viral genera, and likely has great relevance to those studying interactions between viruses and innate immunity (Bailey-Elkin et al., 2014, Lokugamage et al., 2015, Mielech et al., 2014). It is worth noting that the region preceding the Mpro in AAbV is over 13 kb – larger than most other complete RNA virus genomes.
Secondly, two elements of genome architecture seem to be conserved throughout the Nidovirales: a Mpro flanked by multi-pass transmembrane regions, and the block containing NiRAN-RNA polymerase-metal binding-Helicase. Knowledge of these apparent nidovirus genetic synapomorphies should make it possible to design searches to detect even more divergent nido-like viruses in transcriptomes.
Thirdly, the NendoU domain appears to be found only in viruses infecting vertebrate animals, and is lacking in every known nidovirus-like genome from an invertebrate host. This suggests that the function of NendoU may have evolved as a countermeasure to conserved metazoan viral RNA recognition machinery involved in innate immunity (Lokugamage et al., 2015).
Fourthly, while most currently known nidovirus species are associated with terrestrial hosts, the greatest phylogenetic diversity of nidoviruses is now associated with hosts that live in aquatic environments. Since terrestrial metazoan transcriptomes are relatively well-sampled in comparison to aquatic and particularly marine metazoa, we would predict this trend is likely to continue. Of the eight proposed nidovirus families shown in Fig. 4, Fig. 5, four contain only viruses associated with aquatic hosts, two (Arteriviridae (Shi et al., 2018) and the proposed Tobaniviridae) are found in a mix of strictly aquatic and strictly terrestrial animals, and two (Coronaviridae, Mesoniviridae) are in part associated with hosts such as mosquitoes and frogs that have an obligate aquatic larval phase. Taken together, this data suggests that it may be useful to consider potential routes of interspecies transmission between marine, freshwater and terrestrial hosts in future studies of nidovirus evolution, as more data becomes available.
Lastly, the structural protein repertoire of nidoviruses appears to be quite broad compared to other known virus orders. There do not appear to be any conserved nidovirus structural proteins with the possible exception of the nucleoprotein (discussed elsewhere (Neuman and Buchmeier, 2016)), and even that homology can only be regarded as hypothetical until more structures of putative nucleoproteins are solved. A tentative categorization of nidovirus structural proteins, based on size, predicted transmembrane regions, and predicted protein secondary structure is shown in Fig. 10. If correct, this would indicate that nidoviruses have a diverse set of structural proteins that includes a variety of possibly unrelated spike-like proteins plus components shared with Orthomyxoviridae (HA and HE), Togaviridae (E1 and the E3 structural serine proteinase), Flaviviridae (the capsid RNAse). This structural repertoire appears to be variously expressed from subgenomic RNAs encoding a single gene (as proposed for MLeV), giant polyproteins such as that of AAbV, and a mix of intermediate-sized polyproteins and single genes, as in the Roniviridae. Taken together, these observations suggest that structural proteins are widely shared and exchanged among RNA viruses, and that conserved elements of the replicase will be more useful than structural proteins for anyone trying to construct trees that connect viruses at taxonomic ranks above the family level.
Fig. 10.
Speculative annotation of nidovirus structural proteins. Where structures or functions were not known, proteins were categorized according to general PSIPRED secondary structure profile. Marked domains include coronavirus spike protein homologs (Spike) and structurally similar regions (β-α), alphavirus E1 homologs (E1) and structurally similar regions (βαβ), coronavirus envelope-like proteins (E-like), coronavirus membrane proteins (M-like) and structurally similar proteins (β), potential nucleoprotein (N-like), chymotrypsin-like structural proteinase (Spro), similar to the bovine viral diarrhea virus structural RNAse (BVDV RNAse), proteins related to influenza A virus hemagglutinin (HA) or torovirus hemagglutinin-esterase (HE), other viral surface glycoproteins (GP-like), domains of no known function (Unknown), SignalP-predicted signal peptidase cleavage sites (SP cleavage), and potential sites cleaved by unknown proteinases by analogy to other nidovirus structural proteins.
4. Materials and methods
4.1. Phylogeny
Nidovirus phylogeny was reconstructed based on MSA of concatenated Mpro, NiRAN, RdRp, CH cluster and SF1 Helicase conserved cores (3417–3905, 5441–5866, 6095–7291, 7340–7504, 7781–8545 nt of the Equine arteritis virus genome X53459.3), prepared with the help of Viralis platform (Gorbalenya et al., 2010). Representatives of 28 nidovirus species (Supplementary table 1) delineated in recent ICTV proposals (Brinton et al., 2017, Gorbalenya et al., 2017b, Gorbalenya et al., 2017a, Ziebuhr et al., 2017) were used. Phylogeny was reconstructed by IQ Tree 1.5.5 using a partition model where the evolutionary model for each of the five domains was selected by ModelFinder (Chernomor et al., 2016, Kalyaanamoorthy et al., 2017, Nguyen et al., 2015). To estimate branch support, Shimodaira-Hasegawa-like approximate likelihood ratio test (SH-aLRT) with 1000 replicates was conducted. The tree was midpoint rooted and visualized with the help of R packages APE 3.5 and phangorn 2.0.4 (Paradis et al., 2004; R Development Core Team, 2011; Schliep, 2011).
4.2. Protein assays
Nucleotides 12926–14176 containing the AAbV Mpro and flanking regions extending to the preceding and following predicted transmembrane regions was produced as a synthetic GeneArt Strings DNA fragment (Invitrogen). This was used as the template in a 50 µl PCR reaction using primers Aby_IF_MP_F (CCCCGAGGATCTCGAGTTGCGAATGATTTTGTCTACC) and Aby_IF_MP_R (GATGGTGGTGCTCGAGACACAGACAACACAACAAAAA) with 1x Phusion High Fidelty PCR Mastermix (Thermo Fisher Scientific). The 1283 bp PCR product was gel extracted using a QIAquick gel extraction kit (Qiagen) and cloned into pTriEx1.1 (Novagen/Merck) linearised with XhoI using In-Fusion HD cloning reagents (Clontech). 2 µl of the In-Fusion reaction was transformed into Stellar chemically competent cells as per the manufacturers protocol (Clontech) and selected on LB agar containing 100 μg/mL ampicillin. The final construct with a T7 RNA polymerase promoter and in-frame amino-terminal HSV and carboxyl-terminal HIS tags was verified by Sanger sequencing (Source Bioscience) of plasmid DNA purified using a QIAquick spin miniprep kit (Qiagen). Site-directed mutagenesis was carried out using the Quikchange II (Agilent) reagents and protocol. Protein expression was carried out in a 50 µl reaction volume using 0.5 µg of plasmid DNA with the TnT® Quick Coupled Transcription/Translation System (Promega) reagents and protocol. In vitro transcription and translation was carried out for 1 h.
Samples containing expressed proteins were mixed with an equal volume of 2× SDS PAGE loading buffer containing 100 mM Tris-HCL pH6.8, 4% w/v SDS, 20% w/v glycerol, 0.2% bromophenol blue, 2% β- mercaptoethanol. Samples were boiled at 100 °C for 10 min, collected by gentle centrifugation, and loaded in Mini-PROTEAN precast polyacrylamide gels (BioRad). After electrophoresis, proteins were blotted to PVDF membranes for 80 min at 150 mA using a Trans-Blot Turbo (BioRad). Membranes were blocked overnight at 4 °C with 5% (w/v) non-fat milk powder in TBST (50 mM Tris, 150 mM NaCl, 0.1% Tween 20, pH 7.5). Membranes were then washed three times for 5 min each on a rocking platform at 25 rpm with TBST buffer before addition unconjugated rabbit anti-HIS tag monoclonal antibody (Abcam) or unconjugated rabbit anti-HSV tag monoclonal antibody (Abcam) for 1 h. Membranes were again washed three times for 5 min each with TBST buffer before addition of horseradish peroxidase-conjugated goat anti-rabbit secondary antibody for 1 h. For detection, ChemiFast chemiluminescent reagent (Syngene) was used to detect bound secondary antibody. Samples were visualized using a Syngene Chemi XL G:Box gel documentation system. Gel images were cropped and brightness and contrast of images was adjusted using GIMP software (GIMP team).
The region from the pp1a-pp1b junction containing the putative termination-suppression signal of AAbV, nucleotides 17255–17707, was PCR amplified from a synthetic GeneArt Strings fragment (Invitrogen) using primers Aby_IF_SS_F (CCCCGAGGATCTCGAGGAGTCTTGTCGTGTGAAGT) and Aby_IF_SS_R (GATGGTGGTGCTCGAGAGGATTAATCCGTCTGTCAA). The predicted Spro-containing region of AAbV, nucleotides 25918–27183, was PCR amplified from a synthetic GeneArt Strings fragment (Invitrogen) using primers Aby_IF_TryP_R (GATGGTGGTGCTCGAGCGGTTTGTTCGCATACAGA) and Aby_IF_TryP_R (GATGGTGGTGCTCGAGCGGTTTGTTCGCATACAGA). Both the Spro and putative pp1a-pp1b termination-suppression signal products were cloned, expressed and detected in the same way as AAbV Mpro.
4.3. Microhyla prevalence
Data for the MLeV prevalence study comes from a published report (Zhao et al., 2016). Briefly, nine tadpoles were sacrificed, using three individuals from each of the three developmental stages as independent biological replicates. One microgram of mRNA of each stage sample was sequenced on an Illumina HiSeq. 2000 platform by NovoGene (Beijing), and paired-end reads were generated.
Acknowledgements
A.A.G. is a Ph.D. student with Alexander E. Gorbalenya (A.E.G.) and her work and resources she used were partially supported by European Union Horizon2020 EVAg 653316 grant and Leiden University Medical Center, United Kingdom MoBiLe Program to A.E.G. She thanks Igor A. Sidorov, Dmitry V. Samborskiy, and A.E.G. for help with the dataset used in her analysis. The work of L.Z., G.S. and J.J. was supported by a key project from Chinese Academy of Sciences (KJZD-EW-L13) and the National Natural Science Foundation of China (No. 31471964). K.B. was supported by a studentship from the Ministry of Education in Saudi Arabia (S13280). K.B. thanks Ian M. Jones of the University of Reading for assistance in planning and carrying out protein expression and detection studies.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2018.08.010.
Appendix A. Supplementary material
Supplementary material
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Elkin B.A., Knaap R.C.M., Johnson G.G., Dalebout T.J., Ninaber D.K., Van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the middle east respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birktoft J.J., Blow D.M. Structure of crystalline α-chymotrypsin. V. The atomic structure of tosyl-α-chymotrypsin at 2 Å resolution. J. Mol. Biol. 1972;68:187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Brinton, M.A., Gulyaeva, A., Balasuriya, U.B.R., Dunowska, M., Faaberg, K.S., Goldberg, T., Leung, F.-C., Nauwynck, H.J., Snijder, E.J., Stadejek, T., Gorbalenya, A.E., 2017. ICTV Pending proposal 2017.012S. Expansion of the rank structure of the family Arteriviridae and renaming its taxa.
- Buchan D.W.A., Minneci F., Nugent T.C.O., Bryson K., Jones D.T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013:41. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomor O., Von Haeseler A., Minh B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridge A.G., Crowe-Mcauliffe C., Mathew S.F., Tate W.P. Eukaryotic translational termination efficiency is influenced by the 3′ nucleotides within the ribosomal mRNA channel. Nucleic Acids Res. 2018;46:1927–1944. doi: 10.1093/nar/gkx1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debat H.J. Expanding the size limit of RNA viruses: Evidence of a novel divergent nidovirus in California sea hare, with a ~ 35.9 kb virus genome. bioRxiv. 2018 [Google Scholar]
- Deng Z., Lehmann K.C., Li X., Feng C., Wang G., Zhang Q., Qi X., Yu L., Zhang X., Feng W., Wu W., Gong P., Tao Y., Posthuma C.C., Snijder E.J., Gorbalenya A.E., Chen Z. Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase. Nucleic Acids Res. 2014;42:3464–3477. doi: 10.1093/nar/gkt1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.X., Yuan H., Rein A., Levin J.G. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J. Virol. 1992;66:5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler T.J., Hudder A., McKay S.J., Shivkumar S., Capo T.R., Schmale M.C., Walsh P.J. The transcriptome of the early life history stages of the California Sea Hare Aplysia californica. Comp. Biochem. Physiol. Part D Genom. Proteom. 2010;5:165–170. doi: 10.1016/j.cbd.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T., Macnaughton T.B., La Monica N., Lai M.M.C. Natural evolution of coronavirus defective-interfering rna involves rna recombination. Virology. 1993;194:408–413. doi: 10.1006/viro.1993.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A.E., Brinton, M.A., Cowley, J., de Groot, R., Gulyaeva, A., Lauber, C., Neuman, B.W., Ziebuhr, J., 2017a. ICTV Pending Proposal 2017.015S. Reorganization and expansion of the order Nidovirales at the family and sub-order ranks.
- Gorbalenya, A.E., Brinton, M.A., Cowley, J., de Groot, R., Gulyaeva, A., Lauber, C., Neuman, B.W., Ziebuhr, J., 2017b. ICTV Pending Proposal 2017.014S. Establishing taxa at the ranks of subfamily, genus, sub-genus and species in six families of invertebrate nidoviruses.
- Gorbalenya A.E., Lieutaud P., Harris M.R., Coutard B., Canard B., Kleywegt G.J., Kravchenko A.A., Samborskiy D.V., Sidorov I.A., Leontovich A.M., Jones T.A. Practical application of bioinformatics by the multidisciplinary VIZIER consortium. Antivir. Res. 2010 doi: 10.1016/j.antiviral.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyland A., Vue Z., Voolstra C.R., Medina M., Moroz L.L. Developmental transcriptome of Aplysia californica’. J. Exp. Zool. Part B Mol. Dev. Evol. 2011;316(B):113–134. doi: 10.1002/jez.b.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J. Virus Taxonomy. 2012. Togaviridae; pp. 1103–1110. [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Goeman J.J., de Parquet M.C., Thi Nga P., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013:9. doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M., Alkhovsky S.V., Bào Y., Bailey A.L., Shevtsova Z.V., Shchetinin A.M., Vishnevskaya T.V., Lackemeyer M.G., Postnikova E., Mazur S., Wada J., Radoshitzky S.R., Friedrich T.C., Lapin B.A., Deriabin P.G., Jahrling P.B., Goldberg T.L., O’Connor D.H., Kuhn J.H. Historical outbreaks of simian hemorrhagic fever in captive macaques were caused by distinct arteriviruses. J. Virol. 2015;89:8082–8087. doi: 10.1128/JVI.01046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Data G.P., Sam T., Subgroup, 1000 Genome Project Data Processing The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Narayanan K., Nakagawa K., Terasaki K., Ramirez S.I., Tseng C.-T.K., Makino S. Middle east respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of Cytoplasmic Origin. J. Virol. 2015;89:10970–10981. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. USA. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancont P., Garoff H. Processing of the Semliki forest virus structural polyprotein: role of the capsid protease. J. Virol. 1987;61:1301–1309. doi: 10.1128/jvi.61.5.1301-1309.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Chen Y., Mesecar A.D., Baker S.C. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.A., Culley A.I., Schvarcz C.R., Steward G.F. RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 2016 doi: 10.1111/1462-2920.13291. [DOI] [PubMed] [Google Scholar]
- Moroz L.L., Edwards J.R., Puthanveettil S.V., Kohn A.B., Ha T., Heyland A., Knudsen B., Sahni A., Yu F., Liu L., Jezzini S., Lovell P., Iannucculli W., Chen M., Nguyen T., Sheng H., Shaw R., Kalachikov S., Panchin Y.V., Farmerie W., Russo J.J., Ju J., Kandel E.R. Neuronal transcriptome of aplysia: neuronal compartments and circuitry. Cell. 2006;127:1453–1467. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Buchmeier M.J. Supramolecular architecture of the coronavirus particle. Adv. Virus Res. 2016:1–27. doi: 10.1016/bs.aivir.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Chamberlain P., Bowden F., Joseph J. Atlas of coronavirus replicase structure. Virus Res. 2014;194:49–66. doi: 10.1016/j.virusres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T., Parquet MdC., Lauber C., Parida M., Nabeshima T., Yu F. Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes. PLoS Pathog. 2011;7(9):e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dea M.A., Jackson B., Jackson C., Xavier P., Warren K. Discovery and partial genomic characterisation of a novel nidovirus associated with respiratory disease in wild shingleback lizards (Tiliqua rugosa) PLoS One. 2016:11. doi: 10.1371/journal.pone.0165209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., Von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011 doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Prince J.S. A presumptive alphavirus in the gastropod mollusc, Aplysia californica. Bull. Mar. Sci. 2003;73:673–677. [Google Scholar]
- R Development Core Team, R., 2011. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing. 〈 10.1007/978-3-540-74686-7〉. [DOI]
- Saberi A., Gulyaeva A.A., Brubacher J., Newmark P.A., Gorbalenya A. A planarian nidovirus expands the limits of RNA genome size. bioRxiv. 2018 doi: 10.1371/journal.ppat.1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapay N., Guermeur Y., Deléage G. Prediction of amphipathic in-plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinform. 2006:7. doi: 10.1186/1471-2105-7-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep K.P. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Lin X.D., Chen X., Tian J.H., Chen L.J., Li K., Wang W., Eden J.S., Shen J.J., Liu L., Holmes E.C., Zhang Y.Z. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- Shi M., Lin X.D., Tian J.H., Chen L.J., Chen X., Li C.X., Qin X.C., Li J., Cao J.P., Eden J.S., Buchmann J., Wang W., Xu J., Holmes E.C., Zhang Y.Z. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011:7. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuzeski J.M., Nichols L.M., Gesteland R.F., Atkins J.F. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol. 1991;218:365–373. doi: 10.1016/0022-2836(91)90718-L. [DOI] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005:33. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J.W., Arnold J.J., Cameron C.E., van den Worm S.H.E., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2009;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R., Sameroff S., Hesse R.A., Hause B.M., Desai A., Jain K., Ian Lipkin W. Discovery of a novel nidovirus in cattle with respiratory disease. J. Gen. Virol. 2015;96:2188–2193. doi: 10.1099/vir.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N., Guzman H., Firth C., Forrester N.L., Widen S.G., Wood T.G., Rossi S.L., Ghedin E., Popov V., Blasdell K.R., Walker P.J., Tesh R.B. Mesoniviruses are mosquito-specific viruses with extensive geographic distribution and host range. Virol. J. 2014:11. doi: 10.1186/1743-422X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl-Jensen V., Johnson J.C., Lauck M., Weinfurter J.T., Moncla L.H., Weiler A.M., Charlier O., Rojas O., Byrum R., Ragland D.R., Huzella L., Zommer E., Cohen M., Bernbaum J.G., Caì Y., Sanford H.B., Mazur S., Johnson R.F., Qin J., Palacios G.F., Bailey A.L., Jahrling P.B., Goldberg T.L., O’Connor D.H., Friedrich T.C., Kuhn J.H. Divergent simian arteriviruses cause simian hemorrhagic fever of differing severities in macaques. MBio. 2016:7. doi: 10.1128/mBio.02009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Wu A., Wang Y., Xu S., Tang Y., Jin X., Wang S., Qin L., Sun Y., Fan C., Snijder E.J., Neuman B.W., Chen Y., Ahola T., Guo D. Identification and characterization of a ribose 2′-O-methyltransferase encoded by the ronivirus branch of nidovirales. J. Virol. 2016;90:6675–6685. doi: 10.1128/JVI.00658-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Liu L., Wang S., Wang H., Jiang J. Transcriptome profiles of metamorphosis in the ornamented pygmy frog Microhyla fissipes clarify the functions of thyroid hormone receptors in metamorphosis. Sci. Rep. 2016:6. doi: 10.1038/srep27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr, J., Baric, R.S., Baker, S., de Groot, R.J., Drosten, C., Gulyaeva, A., Haagmans, B.L., Neuman, B.W., Perlman, S., Poon, L.L.M., Sola, I., Gorbalenya, A.E., 2017. ICTV Pending Proposal 2017.013S. Reorganization of the family Coronaviridae into two families, Coronaviridae (including the current subfamily Coronavirinae and the new subfamily Letovirinae) and the new family Tobaniviridae (accommodating the current subf).
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material