Abstract
Avian reovirus non-structural protein muNS is partially cleaved in infected chicken embryo fibroblast cells to produce a 55-kDa carboxyterminal protein, termed muNSC, and a 17-kDa aminoterminal polypeptide, designated muNSN. In this study we demonstrate that muNS processing is catalyzed by a caspase 3-like protease activated during the course of avian reovirus infection. The cleavage site was mapped by site directed mutagenesis between residues Asp-154 and Ala-155 of the muNS sequence. Although muNS and muNSC, but not muNSN, are able to form inclusions when expressed individually in transfected cells, only muNS is able to recruit specific ARV proteins to these structures. Furthermore, muNSC associates with ARV factories more weakly than muNS, sigmaNS and lambdaA. Finally, the inhibition of caspase activity in ARV-infected cells does not diminish ARV gene expression and replication, but drastically reduces muNS processing and the release and dissemination of progeny viral particles.
Keywords: Avian reovirus, muNS, Apoptosis, Caspase, Proteolytic processing, Viroplasms, Virus spread
Highlights
-
•
Avian reovirus-triggered apoptosis promotes cleavage of the viral nonstructural muNS protein.
-
•
muNS processing is catalyzed by a caspase 3-like protease activated during avian reovirus infection.
-
•
Cleavage occurs between residues Asp-154 and Ala-155 of the muNS sequence.
-
•
Avian reovirus-induced apoptosis enhances the release and dissemination of progeny viral particles.
Introduction
Avian reoviruses (ARVs) are important pathogens that cause great economic losses in the poultry industry. Infection by these viruses has been associated with a variety of disease conditions, including viral arthritis, chronic respiratory diseases, and malabsorption syndrome (Jones, 2000, van der Heide, 2000). ARVs are members of the Orthoreovirus genus, one of the 12 genera of the Reoviridae family. They are non-enveloped fusogenic viruses whose particles contain 10 double-stranded RNA genome segments enclosed within two concentric protein shells (Benavente and Martinez-Costas, 2007). Viral genomic segments can be separated electrophoretically into three different size classes, named L (Large; three segments), M (Medium; three segments) and S (Small; four segments). The ARV genome expresses at least 8 structural and four non-structural proteins (Bodelon et al., 2001), but the protein repertoire of ARV is increased to at least 12 structural proteins and six non-structural proteins by post-translational cleavage of some viral proteins (Busch et al., 2011, Ji et al., 2010, Varela. et al., 1996).
Genome replication and assembly of ARVs takes place in distinctive cytoplasmic globular inclusions called viral factories where viral components concentrate to increase the efficiency of these processes (Benavente and Martinez-Costas, 2006, Touris-Otero et al., 2004a). The framework of the factories is thought to be formed by the non-structural muNS protein, because it is the only viral protein that accumulates in factory-like inclusions when expressed individually in transfected cells (Touris-Otero et al., 2004b). Furthermore, the fact that viral core protein lambdaA and the non-structural protein sigmaNS redistribute to inclusions when individually co-expressed with muNS in transfected cells suggests that muNS is able to recruit these proteins to viral factories of infected cells (Touris-Otero et al., 2004a).
Similar to many other viruses, ARVs induce apoptotic cell death of infected cells, and the activation of the intracellular apoptotic program takes place during an early stage of the ARV life cycle (Labrada et al., 2002). Apoptosis is triggered by ARV in the absence of viral gene expression, but is no longer induced when intracellular viral uncoating is blocked, suggesting both that apoptosis does not depend on viral protein synthesis and that it is triggered from within the infected cell by viral products generated after intraendosomal uncoating of parental reovirions (Labrada et al., 2002). A previous report revealed that the M3 gene of ARV expresses three muNS isoforms in infected cells and that the two smaller isoforms originate by a specific post-translational cleavage near the amino terminus of muNS. This cleavage produces a 55-kDa carboxyterminal protein, termed muNSC, and a complementary 17-kDa aminoterminal polypeptide, designated muNSN. Cleavage of muNS occurs with ~30% efficiency, so precursor muNS and its cleavage products are all present in ARV-infected cells (Busch et al., 2011). In this study we show that muNS processing is indirectly promoted by ARV infection through activation of an effector caspase that cleaves muNS between amino acid residues 154 and 155.
Results
Cleavage of ARV muNS is promoted by ARV infection
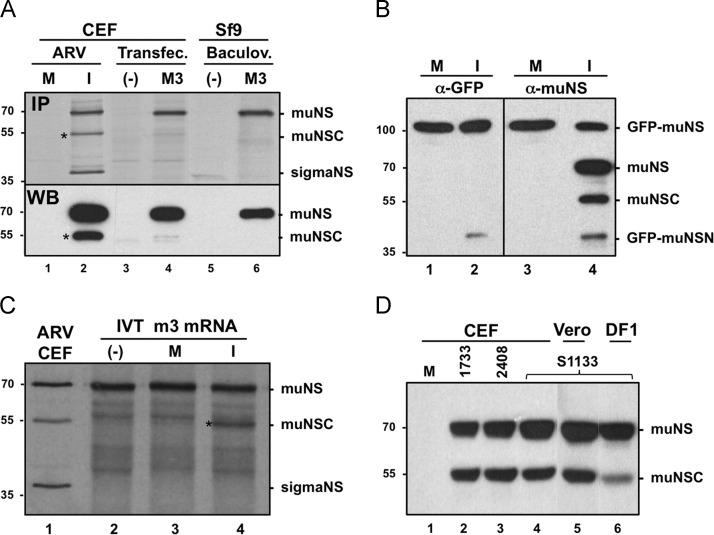
We have previously shown that ARV muNS is cleaved in ARV-infected cells to produce the aminoterminal peptide muNSN and the carboxyterminal protein muNSC (Busch, et al., 2011). To assess whether muNS cleavage is promoted by avian reovirus proteins and/or by changes induced in the host during infection, we compared muNS-to-muNSC conversion in ARV-infected cells with that in transfected cells and in insect cells infected with a muNS-expressing recombinant baculovirus ( Fig. 1A). For this, extracts from CEF monolayers either mock-infected (lane 1) or infected with ARV S1133 (lane 2), from CEF monolayers transfected with either empty pCINeo plasmid (lane 3) or pCINeo–muNS plasmid (lane 4), and from Sf9 cells infected with either wild-type baculovirus (lane 5) or recombinant baculovirus AcNPV–S1133–muNS (lane 6) were subjected to immunoprecipitation (upper panel) and immunoblot (lower panel) analyses. The results revealed that while a significant amount of muNSC was generated in ARV-infected cells (its position is marked with an asterisk at the left of lane 2), this protein was hardly detected in transfected cells (lane 4) or in insect cells infected with a muNS-expressing recombinant baculovirus (lane 6). These data strongly suggest that intracellular processing of muNS is promoted by ARV infection.
Fig. 1.
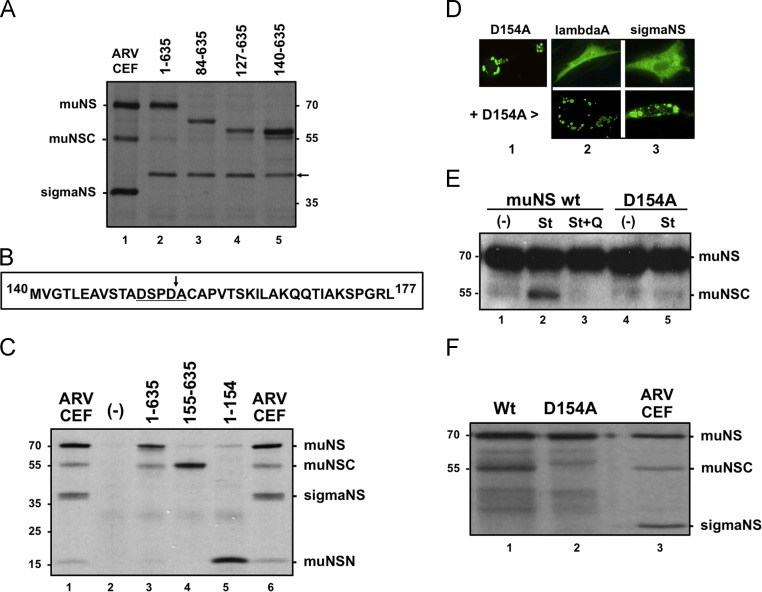
Proteolytic processing of muNS occurs in ARV-infected cells, but not in transfected or baculovirus-infected cells. (A) CEF monolayers are either mock-infected (lane 1) or infected with 10 PFU/cell of ARV S1133 (lane 2) for 16 h. The same cells are transfected with either an empty pCINeo3.1 plasmid (lane 3) or with pCINeo-muNS plasmid (lane 4) for 24 h. Sf9 insect cells are infected with either wild-type baculovirus (lane 5) or with recombinant baculovirus AcNPV-S1133-muNS (lane 6) for two days. The cells in the upper panel are incubated for 1 h with 100 μCi/ml of [35S]amino acids, then lysed with RIPA buffer and immunoprecipitated with muNS-specific antiserum. An immunoblot analysis of nonradiolabeled samples is shown in the lower panel. (B) CEF monolayers are transfected with GFP-muNS for 10 h and then mock-infected (lanes 1 and 3) or infected with 10 PFU/cell of ARV S1133 for 16 h (lanes 2 and 4). The cells are then lysed in RIPA buffer and analyzed by Western blot with polyclonal antibodies against both GFP (lanes 1 and 2) and muNS (lanes 3 and 4). (C) 35S-radiolabeled in-vitro-synthesized muNS (lane 2) is incubated for 2 h at 37 °C with extracts from mock-infected cells (lane 3) or from ARV-infected cells (lane 4). These samples, as well as an immunoprecipitated extract of ARV-infected CEF (lane 1), are analyzed by 10% SDS-PAGE and autoradiography. Positions of protein markers are indicated on the left, and those of nonstructural ARV proteins on the right. The position of the muNSC band is marked with an asterisk. (D) Western blot analysis of extracts from CEF monolayers either mock-infected (lane 1) or infected for 16 h with 10 PFU/cell of the ARV isolates 1733 (lane 2), 2408 (lane 3), and S1133 (lane 4). In lane 5 the analysis is performed with an extract of Vero cells infected for 24 h with 50 PFU of ARV S1133, and in lane 6 with an extract of DF-1 cells infected for 16 h with 10 PFU of ARV S1133. The membranes in lanes 1–4 and 6 are exposed for 5 s and the one in lane 5 for 30 s.
Two additional experiments were subsequently performed to confirm this suggestion. In the first one, a plasmid expressing GFP fused to the amino terminus of muNS (GFP-muNS) was lipofected into CEF monolayers and 10 h later the monolayers were mock-infected or infected with ARV S1133 for 16 h. Extracts from these cells were analyzed by Western blotting using antibodies specific for both muNS and GFP. The results shown in Fig. 1B reveal that while cleavage products derived from GFP-muNS are not detected in mock-infected cells (lanes 1 and 3), a ~45-kDa polypeptide that is recognized by antibodies against both GFP and muNS is present in ARV-infected cells (lanes 2 and 4). Its electrophoretic mobility and its capacity to interact with the two antibodies suggest that this polypeptide is GFP–muNSN. In the second confirmation experiment (Fig. 1C), 35S-labeled in-vitro-generated muNS (lane 2) was incubated with extracts obtained from either mock-infected cells (lane 3) or ARV-infected cells (lane 4), and the resulting samples, as well as radiolabeled extracts from ARV-infected CEF that had been immunoprecipitated against muNS (lane 1), were analyzed by SDS-PAGE and autoradiography. The results revealed that muNS-to-muNSC conversion took place in the sample incubated with extracts from ARV-infected cells (lane 4), but not in the one incubated with extracts from mock-infected cells (lane 3). Taken together, these results indicate that muNS cleavage is promoted by ARV infection.
ARV-triggered apoptosis causes muNS processing
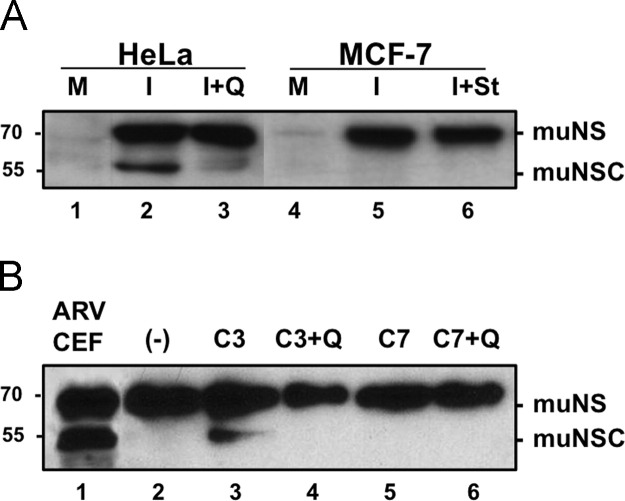
The muNS protein expressed by three different ARV isolates is processed to a similar extent in infected CEF cells (Fig. 1D, lanes 2–4), indicating that muNS processing is not a unique property of the ARV S1133 isolate, but a more general property of ARVs. To assess whether muNS cleavage is influenced by cell-type specific factors we examined muNS-to-muNSC conversion in cells other than CEF. The results revealed that partial cleavage of ARV muNS also took place in ARV-infected monkey Vero cells (Fig. 1D, lane 5) and in human HeLa cells (Fig. 5A, lane 2). Surprisingly, the load of muNSC is highly reduced following infection with ARV S1133 of the CEF-derived avian cell line DF-1 (Fig. 1D, lane 6), suggesting that muNS processing is controlled by cell factors.
Fig. 5.
Identification of the caspase that catalyzes muNS cleavage. (A) Semiconfluent monolayers of HeLa (lanes 1–3) and MCF-7 (lanes 4–6) cells are mock-infected (lanes 1 and 4) or infected with 50 PFU/cell of ARV 1733 (lanes 2, 3, 5 and 6). The cells in lane 3 are incubated with 10 μM Q-VD-OPh from the onset of the infection and the cells in lane 6 are incubated with 0.5 μM staurosporine during the last 6 h of infection. At 16 hpi the cells are lysed in RIPA buffer and the resulting cell extracts are analyzed by Western blotting with muNS-specific antiserum. (B) 1 μg of recombinant muNS purified from baculovirus-infected insect cells (lane 2) is incubated with 1 unit of either caspase 3 (lanes 3 and 4) or caspase 7 (lanes 5 and 6) for 4 h at 37 °C, in the presence (lanes 4 and 6) or absence (lanes 3 and 5) of 10 μM Q-VD-OPh. These samples, as well as an extract of ARV-infected CEF (lane 1), are subjected to Western blot analysis using polyclonal anti-muNS serum. The positions of protein markers are shown on the left and those of nonstructural ARV proteins on the right.
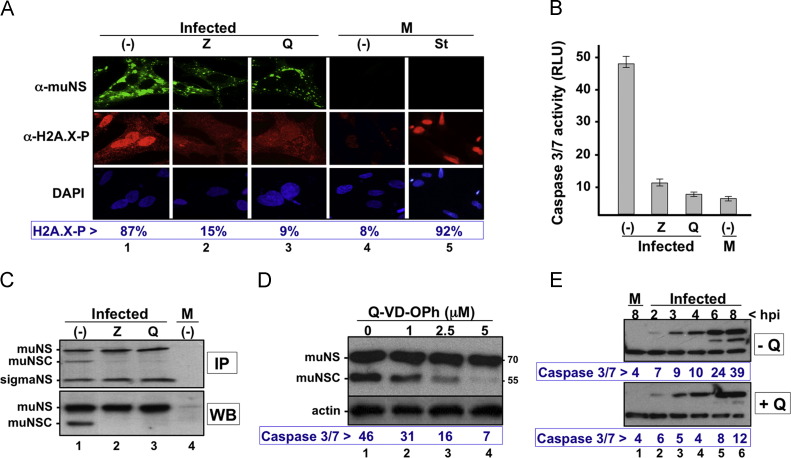
The immortal DF-1 cell line was established spontaneously from Line 0 endogenous-virus negative embryos and had been widely used for the propagation of various avian viruses (Bacon et al., 2000). A genome-wide transcription profile of DF-1 cells and a comparison of their global gene expression with that of primary CEF revealed that DF-1 cells are characterized by enhanced molecular mechanisms for cell cycle progression and proliferation, suppressing cell death pathways, downregulation of caspase 3, altered cellular morphogenesis, and accelerated capacity for molecule transport (Kong et al., 2011). The observation that reduced caspase activity of DF1 cells is accompanied by diminished muNS processing when ARV infects these cells raised the interesting possibility that caspase activation and muNS cleavage are interlinked events. This hypothesis is reinforced by our finding that muNS is not processed when expressed in baculovirus-infected cells (Fig. 1A), probably because baculoviruses express anti-apoptotic factors that prevent caspase activation (Clem, 2007, Clem et al., 1991), although this awaits experimental confirmation. To explore whether there was a connection between caspase activity and muNS processing we first investigated the effect that two broad-spectrum caspase inhibitors, Z-VAD-FMK and Q-VD-OPh, exert on these processes when added to ARV-infected cells at the onset of infection. The effect of these inhibitors on the apoptotic state of infected cells was monitored by two different approaches. In the first one, we used immunofluorescence analysis of histone H2AX phosphorylation to determine the percentage of cells containing damaged DNA (Cook et al., 2009, Yuan et al., 2010). For this, the cells were fixed, stained with a monoclonal antibody against phosphorylated H2AX and counterstained with DAPI. Polyclonal antiserum against the nonstructural muNS protein was also used to visualize ARV-infected cells ( Fig. 2A). Quantification of the stained cells revealed that, similar to staurosporine treatment (lane 5), ARV infection caused a 10 fold increase in the percentage of cells containing phosphorylated H2AX (compare lanes 1 and 4). However, this increase was highly attenuated when the infected cells were incubated in the presence of either of the two caspase inhibitors (lanes 2 and 3). In the second approach, we used the luminiscent Caspase-Glo 3/7 Assay kit (Promega) for measuring caspase-3/7 activities. The cells were lysed in Caspase-Glo 3/7 substrate and protease activity was measured as relative light units (RLU). The results shown at the bottom of Fig. 2B indicate that infection of CEF cells with ARV induces a 10 fold increase in caspase activity, but this increase is significantly reduced when the infected cells are incubated in the presence of either of the two pancaspase inhibitors.
Fig. 2.
Effect of apoptosis inhibitors on muNS processing. (A) Effect of apoptotic inhibitors on DNA damage. CEF monolayers are infected with 10 PFU/cell of ARV in the absence (lane 1) or presence of either 100 μM Z-VAD-FMK (Z; lane 2) or 10 μM Q-VD-OPh (Q; lane 3). Mock-infected cells either untreated (lane 4) or incubated for 6 h in the presence of 0.5 μM staurosporine (lane 5) are used as control samples. The cells are analyzed by indirect immunofluorescence with rabbit polyclonal antibodies against muNS (upper row; green), with a monoclonal antibody against phosphorylated H2AX (middle row; red), and nuclei are stained with DAPI (lower row; blue). The percentage of red-stained nuclei shown at the bottom of the figure is the mean of three independent experiments, and 100 cells were counted for each experiment. (B) Effect of the same concentrations of the two apoptotic inhibitors on caspase 3/7 activity. Caspase activity is determined with the Caspase-Glo® 3/7 Assay kit (Promega), as described in Methods section, and expressed as arbitrary RLU units. Each value is the mean of three independent experiments. (C) Effect of the two apoptotic inhibitors on muNS processing in ARV-infected CEF. Processing is monitored by both immunoprecipitation (IP; upper panel) and Western blotting (WB; lower panel) using muNS-specific antiserum. The sample in lane 1 is run on the same gel, but an internal lane is removed. (D) Effect of Q-VD-OPh on muNS processing and caspase 3/7 activity. ARV-infected CEF monolayers are incubated from the onset of the infection with the concentrations of the pancaspase inhibitor shown on top. At 16 hpi the cells are lysed and the resulting extracts are assayed for caspase 3/7 activity and muNS processing. Processing of muNS is monitored by Western blotting with polyclonal antibodies against muNS and actin. The values of caspase activity shown at the bottom of the figure are the mean of three independent experiments. (E) Time course of muNS processing and caspase 3/7 activity in ARV-infected CEF cells. CEF cell monolayers are infected with 10 PFU/cell of ARV, either in the absence (top blot) or presence (bottom blot) of 10 μM Q-VD-OPh. The cells are lysed at the infection times indicated at the top of the figure. The extracts are subsequently processed for both immunoblotting and caspase 3/7 activity, as for Fig. 2D.
Once demonstrated that the two inhibitors were highly efficient in preventing ARV-induced caspase activation, we next examined their capacity to prevent muNS processing in ARV-infected cells. Both immunoblotting and immunoprecipitation analyses of extracts from ARV-infected CEF revealed that muNS-to-muNSC conversion was dramatically reduced when the cells were incubated in the presence of either of the two inhibitors (Fig. 2C, compare lane 1 with lanes 2 and 3). In subsequent experiments we exclusively used the inhibitor Q-VD-OPh because it blocks apoptosis at very low nontoxic concentrations, and also because it inhibits caspase activity, but not cathepsin activity (Caserta et al., 2003, Kuželová et al., 2011). The results shown in Fig. 2D reveal that the ability of Q-VD-OPh to inhibit both apoptosis and muNS processing in ARV-infected CEF is dose-dependent, and further show that the load of muNSC is drastically reduced at a Q-VD-OPh concentration as low as 5 μM. A time course of muNS processing and caspase activation performed in ARV-infected cells in the presence or absence of Q-VD-OPh (Fig. 2E) revealed that even though the presence of muNS was first detected at 3 hpi, muNSC was not detected until 6 hpi, coinciding with a large increase in caspase 3/7 activity. Furthermore, both muNS processing and an increase in caspase activity were not observed when Q-VD-OPh was present during the infection (Fig. 2E, +Q). Taken together, these results strongly suggest that there is a correlation between ARV-induced apoptosis and muNS processing.
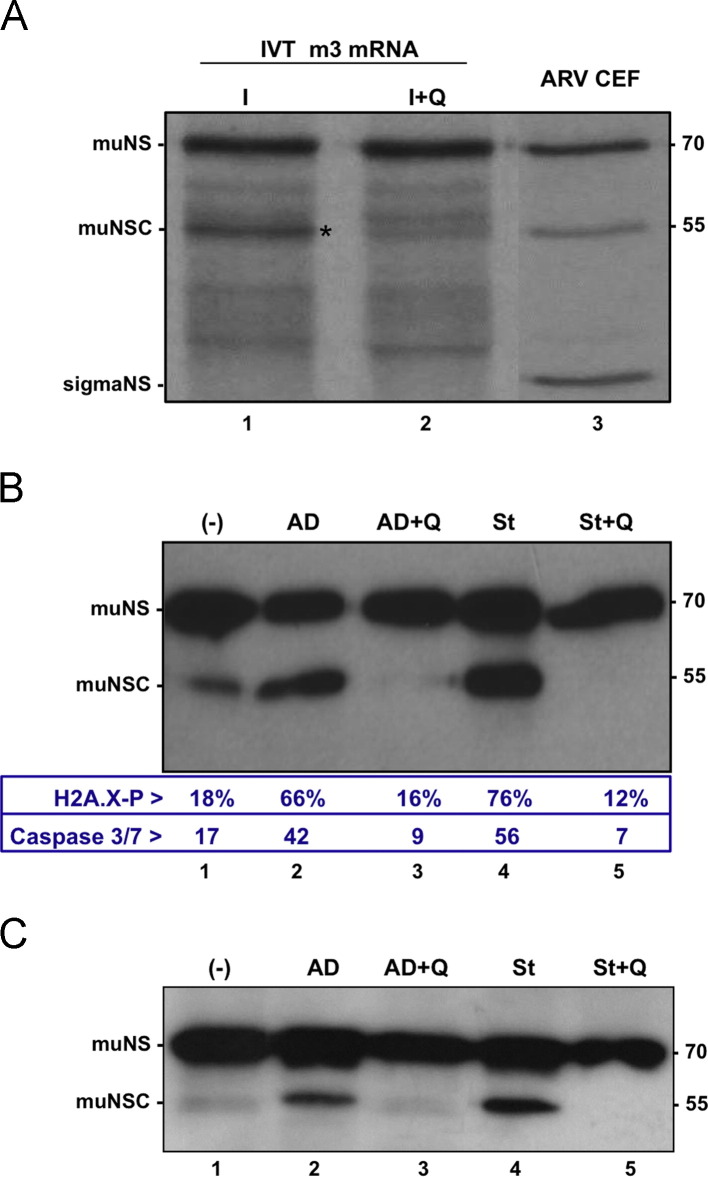
To confirm this suggestion additional experiments were performed. We first observed that the capacity of extracts of ARV-infected cells to cleave in-vitro-synthesized muNS was largely abolished when the infected cells were incubated in the presence of Q-VD-OPh ( Fig. 3A, compare lanes 1 and 2). In addition, the reduced ability of ARV to induce muNS processing in DF-1 cells was significantly enhanced when the cells were incubated during the last 6 h of infection with the pro-apoptotic agents actinomycin D and staurosporine (Fig. 3B, compare lane 1 with lanes 2 and 4), but the ability of these compounds to enhance muNS processing was blocked in the presence of the apoptotic inhibitor Q-VD-OPh (Fig. 3B, lanes 3 and 5). Finally, these pro-apoptotic agents were also effective in promoting muNS processing in transfected cells (Fig. 3C). Our findings that caspase inhibitors prevent muNS processing in ARV-infected cells and that apoptosis enhancers promote muNS cleavage in both transfected cells and ARV-infected DF1 cells indicate that muNS cleavage is catalyzed by an effector caspase activated during ARV infection.
Fig. 3.
Effect of apoptosis inhibitors/enhancers on muNS processing. (A) Effect of Q-VD-OPh on muNS processing. 35S-radiolabeled in-vitro-synthesized muNS is incubated for 4 h at 37 °C with extracts from ARV-infected cells that have been incubated (lane 2) or not (lane 1) with 10 μM Q-VD-OPh from the onset of the infection. These samples, as well as an extract of ARV-infected CEF immunoprecipitated against muNS (lane 3), are analyzed by 10% SDS-PAGE and autoradiography. (B) Effect of apoptotic enhancers and inhibitors on muNS processing and apoptosis in DF1 cells. DF-1 cell monolayers are infected with 10 PFU/cell of ARV S1133. The cells in lanes 3 and 5 are treated at the onset of the infection with 10 μM Q-VD-OPh. The cells in lanes 2 and 3 are treated from 10 to 16 hpi with 1 μg/ml actinomycin D, and in lanes 4 and 5 with 0.5 μM staurosporine. Caspase 3/7 activity, DNA damage and muNS processing are determined at 16 hpi as for Fig. 2A–C. (C) Effect of apoptotic enhancers and inhibitors on muNS processing in transfected cells. CEF monolayers are transfected with the pCINeo-muNS plasmid and 24 h later the cells are incubated for 6 h with the same compounds as for Fig. 3B. The cells are then lysed and the resulting extracts subjected to Western blot analysis with anti-muNS serum. The positions of nonstructural viral proteins are indicated on the left and that of protein markers on the right.
Mapping the muNS cleavage site
In a first attempt to map the muNS cleavage site we compared the electrophoretic mobility of muNSC with that of several muNS amino-terminal truncations whose translation initiates at different internal methionine codons ( Fig. 4A). Plasmids expressing these truncations (lanes 3–5) and full-length muNS (lane 2) were transfected into CEF and the transfected cells, as well as ARV-infected CEF (lane 1), were incubated with [35S]amino acids at 24 h after transfection or 16 hpi. The cells were lysed with RIPA buffer, the extracts were immunoprecipitated with muNS-specific antiserum and the resulting samples were analyzed by SDS-PAGE and autoradiography. The results reveal that muNSC migrates slightly faster than the muNS truncation 140–635, suggesting that the cleavage site should be located several residues downstream of Met-140 (Fig. 4A). A similar conclusion has been recently reached when comparing the electrophoretic mobility of muNSC with that of in-vitro-translated muNS truncations (Busch et al., 2011). A survey of the deduced amino acid sequence downstream of Met-140 for an appropriate caspase cleavage motif revealed the presence of the sequence 151DSPD↓A155 (Fig. 4B, underlined), which is a canonical cleavage motif for caspases 3 and 7 (DXXD↓Y; “X” indicates any amino acid; “Y” indicates G, A, T, S or N) (Thornberry et al., 1997, Timmer and Salvesen, 2007). To verify that this putative caspase recognition sequence is a bona fide cleavage site we first examined whether the electrophoretic mobility of muNS, muNSC and muNSN matched with that of transiently expressed polypeptides comprising muNS residues 1–635, 155–365 and 1–154, respectively (Fig. 4C). ARV-infected CEF (lanes 1 and 6), as well as cells lipofected with empty plasmid (lane 2) or with plasmids expressing the muNS sequences depicted on top of Fig. 4C (lanes 3–5), are radiolabeled with [35S]amino acids and lysed in RIPA buffer. The resulting extracts were immunoprecipitated with polyclonal anti-muNS serum and the imunoprecipitated proteins were resolved on an electrophoresis gel system (8% tricine–SDS-PAGE gel) specifically designed to resolve small peptides (Schägger, 2006). The results reveal that the muNS, muNSC and muNSN proteins present in ARV-infected cells (Fig. 4C, lane 1) have the same electrophoretic mobility as that of the muNS-derived recombinant polypeptides comprising residues 1–635, 155–635 and 1–154, respectively (Fig. 4C, lanes 3–5). It should be however noted that partial conversion of muNS to muNSC took place in transfected CEF cells when the cell monolayers were kept in serum-free medium for more than 5 h during lipofection (Fig. 4C, lane 3). This is probably due to the fact that apoptosis is induced when the cells are incubated for long periods of time in serum-free medium (not shown).
Fig. 4.
Mapping the muNS cleavage site. (A) CEF monolayers are infected with ARV S1133 (lane 1) or lipofected with plasmids expressing muNS versions comprising the residues shown on top of lanes 2–5. The cells are incubated with [35S]amino acids for 1 h at 16 hpi or for 2 h at 24 h after transfection, and then lysed with RIPA buffer. The resulting extracts are immunoprecipitated with muNS-specific antiserum and the samples are analyzed by 8% SDS-PAGE and autoradiography. The position of a non-specific protein band in transfected cells is marked with an arrow. (B) Deduced amino acid sequence of the ARV S1133 muNS region comprising residues 140–177. A potential consensus cleavage sequence for caspase 3/7 is underlined and the putative cleavage site is marked with an arrow. (C) CEF monolayers are infected with ARV S1133 (lanes 1 and 6) or transfected with empty pCINeo plasmid (lane 2) and with plasmids expressing muNS versions comprising the amino acid residues shown on top of lanes 3–5. The cells are radiolabeled with [35S]amino acids for 1 h at 16 hpi or for 5 h at 24 h after transfection, lysed with RIPA buffer and the extracts are immunoprecipitated with muNS-specific antiserum. Immunoprecipitated proteins are resolved on an 8% tricine–SDS-PAGE gel and protein bands are visualized by autoradiography. (D) The D154A muNS mutant (lane 1) and the ARV proteins shown on top (lanes 2 and 3) are transiently expressed in CEF cells, either individually (top row) or in combination with D154A (bottom row). At 24 h after transfection the cells are subjected to immunofluorescence analysis using, as primary antibodies, polyclonal antiserum against muNS (lane 1), ARV cores (lane 2), and sigmaNS (lane 3). (E) CEF monolayers are transfected with plasmids pCINeo-muNS (lanes 1–3) and pCINeo-D154A (lanes 4–5), and at 18 h post-transfection the cells in lanes 2, 3 and 5 are incubated for 6 h in the presence of 0.5 μM staurosporine, and the cells in lane 3 are also supplemented with 10 μM Q-VD-OPh. The cells are then lysed and subjected to Western blot analysis with anti-muNS serum. (F) 35S-radiolabeled in-vitro-synthesized muNS (lane 1) or its D154A point mutant (lane 2) is incubated for 4 h at 37 °C with extracts from ARV-infected cells. These samples, as well as an extract of ARV-infected CEF immunoprecipitated against muNS (lane 3), are analyzed by 10% SDS-PAGE and autoradiography. The positions of protein markers are indicated on the left and those of ARV nonstructural proteins on the right.
To confirm that muNS is cleaved after Asp-154 we next mutated this residue to Ala and investigated abrogation of cleavage. To rule out the possibility that a function of the protein other than proteolysis could be modified by the mutation, we examined the capacity of the D154A mutant to form inclusions and to recruit the viral proteins lambdaA and sigmaNS to these structures. The immunofluorescent images shown in Fig. 4D reveal that the single site mutant D154A maintains the ability of wild-type muNS to form inclusions (lane 1) and to recruit the two ARV proteins to these structures (compare top and bottom rows of lanes 2 and 3). These observations suggest that the D-to-A mutation does not significantly alter the spatial conformation and activity of the nonstructural viral protein. We next compared the capacity of staurosporine to promote processing of both wild-type muNS and its D154A mutant in transfected cells. The results revealed that staurosporine promoted the cleavage of wild-type muNS, but not of the mutant D154A, in transfected cells (Fig. 4E). Similarly, extracts of ARV-infected CEF promoted the cleavage of wild-type muNS, but not of the D154A mutant (Fig. 4F). Taken together, these observations indicate that ARV muNS is cleaved by caspases between muNS residues Asp-154 and Ala-155.
muNS cleavage is catalyzed by a caspase 3-like protease
The results presented so far indicate both that muNS cleavage is catalyzed by a caspase and that caspase 3 and/or caspase 7 are activated in ARV infected cells. Our observation that muNS cleavage takes place in ARV-infected cells of both avian and mammalian origin indicates that muNS is a substrate for avian and mammalian caspases. To assess the role of caspase 3 in muNS processing we compared muNS-to muNSC conversion in ARV-infected HeLa and MCF-7 human cells. HeLa is a caspase 3-competent cell line, whereas MCF-7 is a breast cancer cell line that does not express a functional caspase 3 due to a 125 bp deletion in exon 3 of the caspase 3 gene (Jänicke, 2009). The results of the immunoblot analysis shown in Fig. 5A reveal that muNS-to-muNSC conversion takes place in ARV-infected HeLa cells (lane 2) and that this conversion is prevented by the pancaspase inhibitor Q-VD-OPh (lane 3). However, muNS-to-muNSC conversion did not occur in ARV-infected MCF-7 cells (lane 5), even when these cells were treated for the last 6 h of infection with pro-apoptotic staurosporine (lane 6). This result suggests that caspase 3 or a protease activated by caspase 3 is directly involved in the cleavage of muNS.
We next examined whether a recombinant muNS protein purified from baculovirus-infected insect cells is a substrate for active caspases 3 and 7 of human origin available commercially. The samples obtained when muNS was incubated with these caspases, either in the presence or absence of Q-VD-OPh, as well as an extract of ARV-infected CEF, were subjected to Western blot analysis with muNS-specific antiserum. The results shown in Fig. 5B reveal that partial muNS-to-muNSC conversion occur in the samples incubated with caspase 3, and that this conversion is prevented by the pancaspase inhibitor Q-VD-OPh. Although the result shown in lane 5 of Fig. 5B appears to suggest that muNS is not a substrate for caspase 7, a faint band with the same electrophoretic mobility as muNSC, which disappears in the presence of Q-VD-OPh, is detected in some caspase 7 incubations after long exposures of the membrane (not shown).
Distinctive properties of muNS isoforms
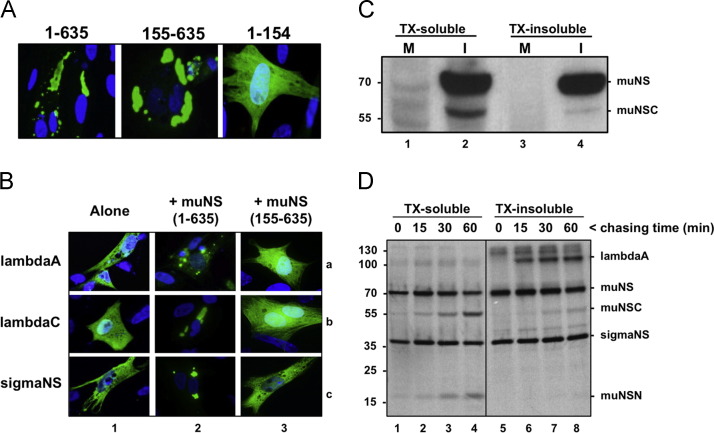
We first analyzed the capacity of three muNS isoforms to form inclusions and to recruit other viral proteins to these structures. The pictures shown in Fig. 6A reveal that both muNS and muNSC, but not muNSN, collect into globular inclusions when expressed individually in transfected cells. Furthermore, all muNS truncations containing the intercoil region (residues 477–542) including muNSC, but not HA-tagged muNSN, collected into inclusions when co-expressed with muNS (Brandariz-Nuñez et al., 2010a). However, these results should be taken with caution, since the attached tag could modify the properties of the proteins. On the other hand, the immunofluorescent images presented in Fig. 6B show that muNS, but not muNSC, is able to recruit lambdaA and sigmaNS to inclusions (compare lanes 2 and 3). This result suggests that the portion of muNS corresponding to muNSN contains sequences required for the interaction with each of the two ARV proteins.
Fig. 6.
Properties of muNS, muNSC and muNSN. (A) Plasmids expressing muNS versions comprising the amino acid residues shown on top are lipofected into CEF monolayers, and at 20 h post-transfection the cells are subjected to immunofluorescence analysis with muNS-specific antiserum as primary antibody (green), and counterstained with DAPI (blue). (B) Plasmids expressing the ARV proteins depicted on the left are transfected into CEF monolayers either individually (lane 1) or together with plasmids expressing full-length muNS (lane 2) or muNS(155–635). At 20 h post-transfection the cells are processed for immunofluorescence using as primary antibodies polyclonal antisera against ARV cores (top row) and sigmaNS (bottom row), and counterstained with DAPI (blue). (C) CEF monolayers are mock-infected (lanes 1 and 3) or infected with 10 PFU/cell of ARV S1133 (lanes 2 and 4) for 16 h. The cells are then lysed with TX-buffer, incubated for 10 min on ice and the TX-soluble fraction is removed (lanes 1 and 2). The plate-attached fraction is then solubilized with RIPA buffer (TX-insoluble fraction; lanes 3 and 4). The two fractions are analyzed by Western blotting with muNS-specific antiserum. (D) ARV-infected cells are labeled for 10 min with [35S]amino acids and then incubated in non-radioactive medium for the indicated chasing times. The cells are subsequently processed as for Fig. 2C and the resulting TX-soluble and -insoluble fractions are immunoprecipitated with muNS-specific antiserum. Immunoprecipitated proteins are resolved by electrophoresis on an 8% tricine-SDS-PAGE gel and visualized by autoradiography. Positions of protein markers are indicated on the left and those of ARV proteins on the right.
We next examined the strength with which muNS and muNSC associate with ARV factories. For this we compared the capacity of Triton-X-100-containing buffer (TX) to extract muNS and muNSC from ARV-infected cells. This buffer has been previously used to discriminate between soluble and cytoskeleton-associated mammalian reovirus proteins, as well as soluble and inclusion-associated ARV proteins (Mora et al., 1987, Touris-Otero et al., 2004a). In the first approach, mock-infected (Fig. 6C, lanes 1 and 3) and ARV-infected CEF (Fig. 6C, lanes 2 and 4) were lysed with the Triton-X-100-containing buffer and the extracts resolved into TX-soluble (lanes 1 and 2) and TX-insoluble (lanes 3 and 4) fractions. The resulting samples were subjected to Western blot analysis with muNS-specific antiserum. The results revealed that while similar amounts of muNS were present in the TX-soluble and -insoluble fractions, most muNSC was extracted by the TX buffer. In the second approach, ARV-infected cells were pulsed with [35S]amino acids for 10 min, then chased in nonradioactive medium for different time periods, and finally fractionated into TX-soluble and -insoluble fractions. The samples were immunoprecipitated with muNS-specific antiserum and analyzed by 8% tricine–SDS-PAGE and autoradiography (Fig. 6D). The results revealed that newly-synthesized muNS and sigmaNS distributed between the two fractions, with a slightly higher proportion in the TX-insoluble fraction. In contrast, most muNSC and almost all muNSN were extracted by the TX buffer at all chasing times. The reliability of the extracting procedure was evidenced by the finding that the structural protein lambdaA coimmunoprecipitated with muNS exclusively in the TX-insoluble fraction at all chasing times (lanes 6–8). These findings indicate that muNS, sigmaNS and lambdaA associate with ARV factories more tightly than muNSC, despite that muNSC, but not lambdaA or sigmaNS, collects into inclusions when expressed individually in transfected cells.
Effect of caspase inhibition on ARV replication and spread
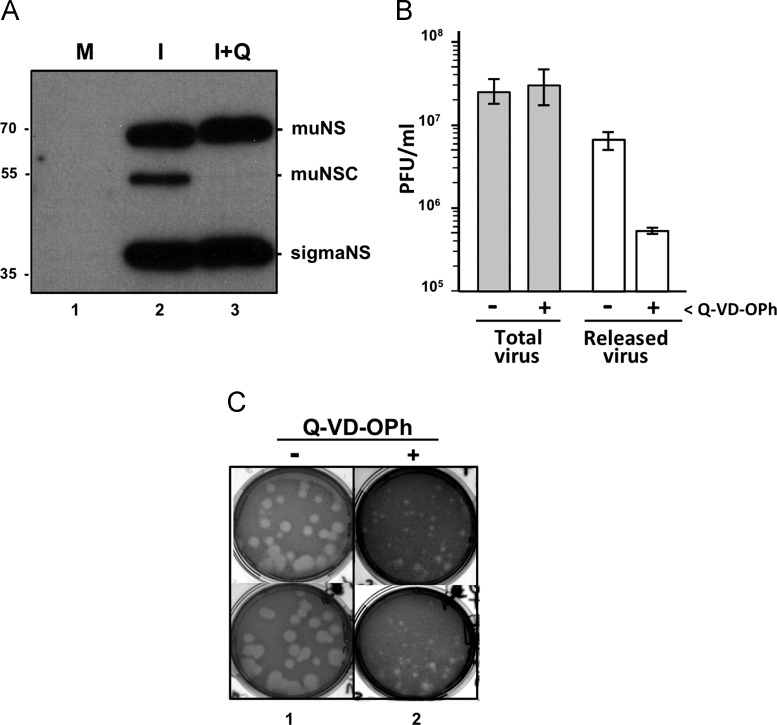
It is of interest to determine the role that muNS processing plays on ARV replication. In the absence of an established reverse genetics system for ARV that would allow us to generate a recombinant virus that expresses an uncleavable muNS protein, like D154A, we examined the effect of Q-VD-OPh on ARV replication in CEF cells, since this compound had been shown to block both apoptosis and muNS processing. The Western blot shown in Fig. 7A reveals that although the presence of the inhibitor from the onset of the infection completely blocks muNS processing, it does not significantly alter the intracellular concentration of the ARV nonstructural protein sigmaNS. This result indicates that apoptosis and muNS processing do not have significant effects on ARV gene expression and on stages of the virus life cycle prior to gene expression. Furthermore, the inhibitor did not significantly change the production of infective ARV particles, as determined by a virus plaque assay (Fig. 7B, total virus), indicating that both apoptosis and muNS processing are not required for the production of infective progeny in cultured cells.
Fig. 7.
Effect of Q-VD-OPh on ARV replication and dissemination. (A) CEF monolayers are mock-infected (M) or infected with 10 PFU/cell of ARV S1133 (I), in the presence (+Q) or absence of 10 μM Q-VD-OPh. The cells are processed for immunoblotting at 16 hpi and the membranes are probed with rabbit polyclonal antibodies against both muNS and sigmaNS. The positions of protein markers are indicated on the left and those of the non-structural ARV proteins on the right. (B) CEF monolayers are infected with 0.1 PFU/cell of ARV S1133, and the production of both total virus (intracellular+culture medium; filled bars) and released virus (cultured medium; open bars) is determined after 24 h of infection by plaque assay on CEF monolayers. (C) Plaque pictures obtained by titrating a stock of ARV S1133 on CEF monolayers in the absence (lane 1) or presence (lane 2) of 10 μM Q-VD-OPh. The pictures in lane 1 correspond to a 106 dilution and the ones in lane 2 to 105 dilution of the same virus stock. Plaques of two different plaque assays are shown.
We next investigated the effect of Q-VD-OPh on ARV release by titrating the infectious viral particles released into the incubation medium. The results showed that virus release was drastically reduced when the infected cells were incubated in the presence of Q-VD-OPh (Fig. 7B, released virus), suggesting that ARV-induced apoptosis enhances dissemination of progeny reovirions from infected cells. This suggestion was further supported by our finding that the presence of Q-VD-OPh during plaque assay of an ARV stock, where the agar overlay limits virus dissemination to neighboring cells, caused a significant reduction in the size of the viral plaques (Fig. 7C), suggesting that the inhibitor reduces cell-to-cell viral spread over the different rounds of replication, release and re-infection that occur during plaque formation.
Discussion
Activation of programmed cell death is one of the first protective defences setup by the host cell to restrict viral amplification and spread. When triggered early in infection apoptosis may limit both the time and the cellular machinery available for virus replication. Therefore, it is not surprising that many viruses try to overcome the apoptotic threat by using a battery of different strategies and by expressing a variety of antiapoptotic products aimed to block or delay the induction of apoptosis (Galluzzi et al., 2010). However, other viruses induce apoptosis actively at late stage of infection to facilitate virus release and spread with limited induction of inflamatory and immune host responses (Teodoro and Branton, 1997). The results shown in Fig. 2E confirm previous findings from our laboratory that ARV infection triggers apoptosis in cultured cells at an early stage of the viral life cycle (Labrada et al., 2002), yet the results of this study demonstrate that early apoptosis triggering does not have adverse effects on either ARV gene expression or infectious progeny production. These apparently conflicting results can be reconciled considering that ARV has a short replication time (Benavente and Martinez-Costas, 2006), which would allow the virus to proceed its life cycle successfully until completion and to reach a satisfactory intracellular production of progeny viral particles before the infected cell dies or is severely damaged by the execution of apoptosis.
A growing number of viruses have been documented to take advantage of apoptosis induction and caspase activation to promote their own replication. For instance, viruses as diverse as mammalian reovirus, human astrovirus, measles virus, porcine circovirus 2, African swine fever virus, bovine herpesvirus 1, coronavirus, influenza virus and the Moscow strain of Ectromelia virus, all have been reported to usurp apoptosis for facilitating the release of progeny viral particles from infected cells (Best, 2008, Galluzzi et al., 2010). The results of the present study suggest that ARV should be included in that list, since this virus activates rather than suppress caspases and this activation is necessary for efficient virus release and spread. On the other hand, many apoptosis-inducing viruses express proteins that are processed by caspases to generate cleavage products with novel properties, ranging from apoptosis downregulation to enhancement or attenuation of viral replication (Richard and Tulasne, 2012). Still, the functional consequences of many protein caspase cleavages remain elusive so far, yet the conservation of caspase targets in many viral proteins and the stability of their processed products suggest that caspase cleavage of viral polypeptides can constitute an evolutionary advantage that benefits virus replication. In this study we have provided compelling evidence that caspases activated during ARV-induced apoptosis catalyze the cleavage of ARV nonstructural muNS protein to produce the 17-kDa N-terminal peptide muNSN and the 55-kDa C-terminal protein muNSC. In the literature to date, there are several examples of other non-structural viral proteins that are caspase targets, like the NS1 protein of Aleutian mink disease parvovirus, the NS5A protein of hepatitis C virus, the NS1′ protein of Japanese encephalitis virus, and the EIA protein of adenovirus 12 (Best et al., 2003, Grand et al., 2002, Satoh et al., 2000, Sun et al., 2012). We also observed that a substantial amount of full-length muNS remains uncleaved both in infected cells and after 6 h of incubation with recombinant caspase 3, implying that caspase-catalyzed muNS cleavage is a regulated process, as has been reported for several cellular and viral proteins (Best et al., 2003, Chaudhry et al., 2012, Cheng et al., 2010, Mashima et al., 1999, Sun et al., 2012). Incomplete processing of a precursor viral protein could be a strategy to reduce the activity of the intact protein or it could be used to increase the protein repertoire encoded by size-limited viral genomes, thus generating cleaved viral products with novel properties/activities.
That avian and mammalian reoviruses use different mechanisms to express their muNSC isoforms (Busch et al., 2011), and that both the muNS precursor and its truncated forms coexist in avian/mammalian reovirus-infected cells suggest that there should be distinct functional roles for both the precursor and its truncated proteins. It should be noted however that avian and mammalian reovirus muNSC proteins display different properties. Thus, while mammalian reovirus muNSC still maintains its precursor׳s ability to associate with the major core protein lambda1 (Miller et al., 2010), this ability is not shared by its ARV counterpart (Fig. 6B), because sequences of ARV muNS required for binding to lambdaA are lacking in muNSC. It has been reported that the release of astrovirus from infected cells depends on both the processing of the capsid precursor polypeptide VP90 and the activities of executioner caspases (Banos-Lara and Mendez, 2010). Accordingly, the possibility exists that muNS processing is required for enhanced release and dissemination of ARV, although it is unlikely, since apoptosis has also been reported to promote the release of mammalian reovirus from infected cells without enhancing the production of its muNSC protein (Marcato et al., 2007). In the absence of an established reverse genetics system for avian reoviruses, the consequences of muNS cleavage on viral replication and spread remain unclear.
Despite the facts that ARV muNSC is able to form inclusions when expressed in isolation and that it collects into inclusions when co-expressed with muNS (Brandariz-Nuñez et al., 2010b), muNSC associates with factories of infected cells more weakly than full-length muNS, sigmaNS or lambdaA, as evidenced by extraction with TX buffer (Fig. 6C and D). This observation suggests that ARV factories are complex structures formed by a diverse array of protein–protein and protein–RNA interactions and that these entities are much more complex than muNS-derived inclusions. This is supported by our observation that the inclusions formed by expressing muNS versions in baculovirus-infected insect cells can be easily purified (Brandariz-Nuñez et al., 2010a), whereas we are unable to purify the factories formed in ARV-infected cells.
The results shown in Fig. 5 demonstrate that muNS is a substrate for mammalian caspase 3, yet the absence of both commercial recombinant caspases of avian origin and caspase 3-deficient avian cells does not allow us to identify the avian caspase that catalyzes ARV muNS cleavage in infected avian cells. Nevertheless, this cleavage is probably catalyzed by an avian caspase 3-like protease, since caspase sequences and targets are highly conserved in different species (Sakamaki and Satou, 2009), and since we found both that caspase 3/7 is active in ARV-infected avian cells and that muNS-to-muNSC processing is promoted by the caspase 3 activator staurosporine (Chae et al., 2000). The presence of a caspase 3/7 consensus sequence (151DSPD↓A155) within the muNS cleavage region allowed us to map the muNS cleavage site between residues Asp-154 and Ala-155, by using site-directed mutagenesis. This finding suggests that this region of muNS is placed in a flexible and exposed loop accessible to caspases. Interestingly, the caspase 3/7 consensus sequence found in ARV S1133 muNS is not fully conserved among avian reoviruses isolated from chickens, since it is only present in 8 isolates, and in one quail isolate, but not in other 9 chicken isolates. Thus, in the muNS protein from 4 chicken isolates Asp-151 is replaced by His, while in the protein from another 5 isolates Asp-151 and Ser-152 are replaced by His and Glu, respectively ( Fig. 8). If cleavage of these “nonconsensus” ARV muNS proteins still occurs between Asp-154 and Ala-155, it would imply that accessibility is more important than the presence of specific amino acids at positions 3 and 4 of the caspase consensus sequence, which is in agreement with previous reports indicating that caspases do not have a strict requirement for positions 2–4 of the consensus cleavage site (Timmer and Salvesen, 2007, Thornberry et al., 1997). Curiously, the muNS protein encoded by both duck and goose reoviruses does not contain a caspase 3/7 consensus sequence spanning residues 151–155, because Asp-154 is replaced by a Gly residue. However, these muNS proteins contain two putative caspase targets, one spanning residues 142–146 (142GTMDA146), and the other comprising residues 156–160 (156SVPDV160). Surprisingly, the former caspase target is also present in the muNS protein from four chicken isolates (Fig. 8). It should be of interest to check whether duck/goose reoviruses induce apoptosis, whether the muNS protein of these viruses is cleaved in infected cells and whether its cleavage takes place at one or both of these caspase consensus sequences. Nevertheless, the presence of specific residues in the muNS 140–177 region could serve to assign a poultry reovirus as an avian/quail or a goose/duck reovirus. Thus, the muNS from reoviruses of avian/quail origin has leucine in position 144, alanine or valine in 150, aspartic acid in 154, cysteine in 156 and valine in 159, whereas the muNS from reoviruses of duck/goose origin has methionine, glycine, glycine and aspartic acid in those positions.
Fig. 8.
Alignment of the deduced amino acid sequences of the muNS 140–177 region from different poultry reoviruses. Sources of the host birds, virus strains, deduced amino acid sequences of the muNS region 140–177 and protein accession numbers are indicated. Putative consensus caspase cleavage sequences are boxed.
Methods
Cells, viruses, antibodies and reagents
Primary cultures of CEFs were prepared from 9- to 10-day-old chicken embryos and grown in monolayers in medium 199 supplemented with 10% tryptose phosphate broth and 5% calf serum. Avian DF-1, human HeLa and MCF-7, and monkey Vero cell lines, all were grown in monolayers in Dulbecco׳s modified Eagle׳s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS). MCF-7 cells were a kind gift from María Dolores Blanco (Universidad Complutense de Madrid). Sf9 cells were grown in suspension culture in serum-free Sf-900 II media at 27 °C. Strains S1133, 1733 and 2408 of avian reovirus were grown in semiconfluent monolayers of primary CEFs. Conditions for growing and titrating these viruses have been described previously (Grande and Benavente, 2000). The recombinant baculovirus AcNPV-S1133-muNS was grown in Sf9 cells as previously described (Brandariz-Nuñez et al., 2010b).
Rabbit polyclonal sera against ARV S1133 reovirions and cores, as well as ARV S1133 proteins muNS and sigmaNS were raised in our laboratory (Touris-Otero et al., 2004b). Monoclonal anti-gamma histone H2A.X (phospho S139) antibody was purchased from Abcam plc. Rabbit anti-actin polyclonal antibody was purchased from Santa Cruz Biotechnology. Peroxidase-conjugated goat anti-rabbit antibody was purchased from Sigma. Pancaspase inhibitors Z-VAD-FMK and Q-VD-OPh were from Calbiochem. Caspases 3 and 7 were from Enzo Life Science (Cat. # C-3: ALX-201-059; C-7: ALX-201-061-U025). All other reagents were purchased from Sigma.
Viral infections and protein analysis
Semiconfluent monolayers of cells were infected with 10 PFU/cell of ARVs for the times indicated for each experiment. In all experiments, except for some of Fig. 1 and those of Fig. 5A, CEF cell monolayers were infected with the ARV S1133. For metabolic radiolabeling, the cultured medium was removed and the cells were incubated for the indicated times in methionine/cysteine-free medium containing 100 μCi/ml of [35S]methionine–cysteine. For immunoprecipitation and Western blot analyses the cells were lysed in RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP40, 0.5% DOC, 0.1% SDS, supplemented with “Complete protease inhibitor cocktail” from Roche Diagnostics), and muNS-derived proteins were detected with a muNS-specific antiserum (Touris-Otero et al., 2004b).
To isolate TX-soluble and -insoluble fractions, 2×106 cells were lysed in 200 μl of TX buffer (10 mM Pipes pH 6.8, 3 mM MgCl2, 100 mM KCl, 300 mM sucrose, 1% Triton X-100, supplemented with “Complete protease inhibitor cocktail” from Roche Diagnostics) and the soluble fraction was removed. The plates were washed again with TX buffer and the insoluble fraction that remained attached to the plates was directly solubilized in 200 μl of RIPA buffer (Touris-Otero et al., 2004a). For pulse-chase analysis, mock-infected and reovirus-infected cells were incubated for 2 h at 12 h p.i. in medium lacking methionine and cysteine, and then incubated for 10 min in the same medium supplemented with 500 μCi/ml [35S]amino acids. The cells were chased for the indicated times in non-radioactive medium supplemented with an excess of nonradiolabelled methionine and cysteine. Infection of Sf9 insect cells with the recombinant baculovirus AcNPV-S1133-muNS and purification of recombinant muNS have already been described (Brandariz-Nuñez et al., 2010b).
Plasmid construction and cell transfection
The construction of recombinant plasmids expressing full-length ARV S1133 muNS, GFP-muNS, and the aminoterminal truncations comprising muNS residues 84-635, 127–635 and 140–635 has already been described (Brandariz-Nuñez et al., 2010b). Plasmid pCINeo-muNS was used as template to generate constructs expressing the following proteins. The D154A muNS point mutant was generated with the QuikChange site-directed mutagenesis kit (Stratagene), according to the manufacturer׳s specifications. The forward primer was: 5′–CCACCGCTGATTCCCCCGCTGCCTGCGTCCCAGTCACC–3′; and the reverse primer was: 5′–GGTGACTGGGACGCAGGCAGCGGGGGAATCAGCGGTGG–3′. To generate the construct expressing muNS(1–154), the forward primer was: 5′–GCGGAATTCATCATGGCGTCAACCAAGTGG–3′; and the reverse primer was: 5′–GCGTCTAGATTAATCGGGGGAATCAGCGGTGG–3′. To generate the construct expressing muNS (155–635), the forward primer was: 5′–GCGGAATTCATCATGGCCTGCGTCCCAGTC–3′; and the reverse primer was: 5′–GCGTCTAGATCACAGATCATCCACCAATTCTTC–3′. The correctness of the recombinant plasmids was confirmed by nucleotide sequencing.
Transfection of preconfluent cell monolayers was done using Lipofectamine Plus reagent (Invitrogen) following the manufacturer׳s instructions, with 1 μg of DNA per well of a 12-well dish. Transfected cells were incubated at 37 °C for 24 h, unless otherwise stated.
Indirect immunofluorescence microscopy
Cell monolayers grown on coverslips were infected or transfected as indicated in the figure legends. At the indicated times, the monolayers were washed twice with PBS and fixed in paraformaldehyde for 15 min at 4 °C. Fixed cells were washed twice with PBS, incubated for 10 min in permeabilizing buffer (0.1% Triton X-100 in PBS), incubated 20 min with blocking buffer (2% BSA in PBS), and then incubated for 1 h at room temperature with primary antibodies diluted in blocking buffer. The cells were washed three more times with PBS and then incubated with secondary antibodies and DAPI (4,6-diamidino-2-phenylindole) for 1 h at room temperature. Coverslips were then washed six times with PBS and mounted on glass slides. Images were obtained with an Olympus DP-71 digital camera mounted on an Olympus BX51 fluorescence microscope. Images were processed with Adobe Photoshop (Adobe Systems).
in vitro transcription and translation
The recombinant plasmids used as templates were linearized with NotI, purified by extraction with phenol/chloroform, precipitated with ethanol and resuspended in sterile water at a final concentration of 1 mg/ml. in vitro transcription from the T7 promoter was performed by using a RiboMAX Large Scale RNA Production System (Promega). in vitro translation was carried out by using a Rabbit Reticulocyte Lysate System (nuclease-treated; Promega) following the manufacturer׳s instructions, for 90 min at 30 °C in the presence of 50 mg/ml RNA and 0.4 mCi/ml [35S]methionine (Hartmann analytic KSM-01).
Intracellular caspase activity and in vitro protease cleavage assays
The intracellular activity of caspases 3/7 was determined with the Caspase-Glo® 3/7 Assay kit from Promega, following the manufacturer׳s instructions. in vitro caspase cleavage assays were performed by mixing 1 μg of muNS purified from baculovirus-infected insect cells, as described previously (Brandariz-Nuñez et al., 2010a), with 1 unit of recombinant caspase in 10 μl of caspase buffer (20 mM Hepes pH 7.4; 100 mM NaCl; 0.05% NP-40). In some samples 10 μM of the pancaspase inhibitor Q-VD-OPh was also included. Mixtures were incubated for 4 h a 37 °C, mixed with 5 μl of 3X Laemmli electrophoresis sample buffer and boiled for 5 min at 100 °C. Cleavage of muNS was analyzed by Western blotting.
To evaluate the capacity of extracts of ARV-infected cells to promote muNS cleavage, extracts from mock-infected and infected cells were prepared as follows. Samples of 2×107 CEF cells, mock-infected or infected with 0.1 PFU/ml of ARV S1133, in the presence or absence of 10 μM Q-VD-OPh, were washed twice with PBS at 24 hpi and overlayed with 1 ml of caspase buffer. The cells were then collected by scrapping and lysed by one cycle of freezing and thawing. A typical protease cleavage assay was performed by mixing 5 μl of cell extract with 5 μl of reticulocyte lysate containing 35S-radiolabeled muNS or its D154A mutant. Samples were incubated for 2 h at 37 °C, then supplemented with 5 μl of 3X Laemmli sample buffer and boiled for 5 min at 100 °C. Processing of muNS was monitored by SDS-PAGE and autoradiography.
Acknowledgments
We would like to thank Rebeca Menaya for technical assistance and other members of our laboratory for helpful discussions on the manuscript. We also thank Laboratorios MSD España (Salamanca, Spain) for providing pathogen-free embryonated eggs. This work was funded by grants from the Ministerio de Economia Competitividad (BFU2010-22228) and from the Xunta de Galicia (CN 2012/018).
References
- Bacon L.D., Hunt H.D., Cheng H.H. A review of the development of chicken lines to resolve genes determining resistance to diseases. Poult. Sci. 2000;79:1082–1093. doi: 10.1093/ps/79.8.1082. [DOI] [PubMed] [Google Scholar]
- Banos-Lara M.R., Mendez E. Role of individual caspases induced by astrovirus on the processing of its structural protein and its release from the cell by a non-lytic mechanism. Virology. 2010;401:322–332. doi: 10.1016/j.virol.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Benavente J., Martinez-Costas J. Early steps in avian reovirus morphogenesis. Curr. Top. Microbiol. Immunol. 2006;309:67–85. doi: 10.1007/3-540-30773-7_3. [DOI] [PubMed] [Google Scholar]
- Benavente J., Martinez-Costas J. Avian reovirus: structure and biology. Virus Res. 2007;123:105–119. doi: 10.1016/j.virusres.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Best S.M. Viral subversion of apoptotic enzymes: escape from death row. Annu. Rev. Microbiol. 2008;62:171–192. doi: 10.1146/annurev.micro.62.081307.163009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S.M., Shelton J.F., Pompey J.M., Wolfinbarger J.B., Bloom M.E. Caspase cleavage of the non-structural protein NS1 mediates replication of Aleutian mink disease parvovirus. J. Virol. 2003;77:5305–5312. doi: 10.1128/JVI.77.9.5305-5312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelon G., Labrada L., Martinez-Costas J., Benavente J. The avian reovirus genome segment S1 is a functionally tricistronic gene that expresses one structural and two nonstructural proteins in infected cells. Virology. 2001;290:181–191. doi: 10.1006/viro.2001.1159. [DOI] [PubMed] [Google Scholar]
- Brandariz-Nuñez A., Menaya-Vargas R., Benavente J., Martinez-Costas J. A versatile molecular tagging method for targeting proteins to avian reovirus muNS inclusions. Use in protein immobilization and purification. PLoS ONE. 2010;5:e13961. doi: 10.1371/journal.pone.0013961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandariz-Nuñez A., Menaya-Vargas R., Benavente J., Martinez-Costas J. Avian reovirus muNS protein forms homo-oligomeric inclusions in a microtubule-independent fashion, which involves specific regions of its C terminal domain. J. Virol. 2010;84:4289–4301. doi: 10.1128/JVI.02534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch L.K., Rodriguez-Grille J., Ignacio Casal J., Martinez-Costas J., Benavente J. Avian and mammalian reoviruses use different molecular mechanisms to synthesize their μNS isoforms. J. Gen. Virol. 2011;92:2566–2574. doi: 10.1099/vir.0.036459-0. [DOI] [PubMed] [Google Scholar]
- Caserta T.M., Smith A.N., Gultice A.D., Reedy M.A., Brown T.L. Q-VD-OPh, a broad spectrum caspase inhibitor with potent antiapoptotic properties. Apoptosis. 2003;8:345–352. doi: 10.1023/a:1024116916932. [DOI] [PubMed] [Google Scholar]
- Chae H-J., Kang J-S., Byun J-O., Han K-S., Kim D-U., Oh S-E., Kim H-M., Chae S-W., Kim H-R. Molecular mechanism of staurosporine-induced apoptosis in osteoblasts. Pharmacol. Res. 2000;42:373–381. doi: 10.1006/phrs.2000.0700. [DOI] [PubMed] [Google Scholar]
- Chaudhry P., Singh M., Parent S., Asselin E. Prostate apoptosis response 4 (Par-4), a novel substrate of caspase 3 during apoptosis activation. Mol. Cell Biol. 2012;32:826–839. doi: 10.1128/MCB.06321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Chen A.Y., Best S.M., Bloom M.E., Pintel D., Qiu J. The capsid proteins of Aleutian mink disease virus activate caspases and are specifically cleaved during infection. J. Virol. 2010;84:2687–2696. doi: 10.1128/JVI.01917-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem R.J. Baculoviruses and apoptosis: a diversity of genes and responses. Curr. Drug Targets. 2007;8:1069–1074. doi: 10.2174/138945007782151405. [DOI] [PubMed] [Google Scholar]
- Clem R.J., Fechheimer M., Miller L.K. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- Cook P.J., Ju B.G., Telese F., Wang X., Glass C.K., Rosenfeld M.G. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Kepp O., Morselli E., Vitale I., Senovilla L., Pinti M., Zitvogel L., Kroemer G. Viral strategies for the evasion of immunogenic cell death. J. Intern. Med. 2010;267:526–542. doi: 10.1111/j.1365-2796.2010.02223.x. [DOI] [PubMed] [Google Scholar]
- Grand R.J.A., Schmeiser K., Gordon E.M., Zhang X., Gallimore P.H., Turnell A.S. Caspase-mediated cleavage of adenovirus early region 1A proteins. Virology. 2002;301:255–271. doi: 10.1006/viro.2002.1586. [DOI] [PubMed] [Google Scholar]
- Grande A., Benavente J. Optimal conditions for the growth, purification and storage of the avian reovirus S1133. J. Virol. Methods. 2000;85:43–54. doi: 10.1016/s0166-0934(99)00155-x. [DOI] [PubMed] [Google Scholar]
- Jänicke R.U. MCF-7 breast carcinoma cells do not express caspase-3. Breast Cancer Res. Treat. 2009;117:219–221. doi: 10.1007/s10549-008-0217-9. [DOI] [PubMed] [Google Scholar]
- Ji W.T., Lin F.L., Wang Y.C., Shih W.L., Lee L.H., Liu H.J. Intracellular cleavage of σA protein of avian reovirus. Virus Res. 2010;149:71–77. doi: 10.1016/j.virusres.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Jones R.C. Avian reovirus infections. Rev. Sci. Tech. 2000;19:614–625. doi: 10.20506/rst.19.2.1237. [DOI] [PubMed] [Google Scholar]
- Kong B.W., Lee J.Y., Bottje W.G., Lassiter K., Lee J., Foster D.N. Genome-wide differential gene expression in immortalized DF-1 chicken embryo fibroblast cell line. BMC Genomics. 2011;12:571. doi: 10.1186/1471-2164-12-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuželová K., Grebeňová D., Brodská B. Dose-dependenteffects of the caspase inhibitor Q-VD-OPh on different apoptosis-related processes. J. Cell Biochem. 2011;112:3334–3342. doi: 10.1002/jcb.23263. [DOI] [PubMed] [Google Scholar]
- Labrada L., Bodelon J., Viñuela J., Benavente J. Avian reoviruses cause apoptosis in cultured cells: viral uncoating, but not viral gene expression, is required for apoptosis induction. J. Virol. 2002;76:7932–7941. doi: 10.1128/JVI.76.16.7932-7941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcato P., Shmulevitz M., Pan D., Stoltz D., Lee P.W. Ras transformation mediates reovirus oncolysis by enhancing virus uncoating, particle infectivity, and apoptosis-dependent release. Mol. Ther. 2007;15:1522–1530. doi: 10.1038/sj.mt.6300179. [DOI] [PubMed] [Google Scholar]
- Mashima T., Naito M., Tsuruo t. Caspase-mediated cleavage of cytoskeletal actine plays a positive role in the process of morphological apoptosis. Oncogene. 1999;18:2423–2430. doi: 10.1038/sj.onc.1202558. [DOI] [PubMed] [Google Scholar]
- Miller C.L., Arnold M.M., Broering T.J., Hastings C.E., Nibert M.L. Localization of mammalian orthoreovirus proteins to cytoplasmic factory-like structures via nonoverlapping regions of μNS. J. Virol. 2010;84:867–882. doi: 10.1128/JVI.01571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M., Partin K., Bhatia M., Partin J., Carter C. Association of avian reovirus proteins with the structural matrix of infected cells. Virology. 1987;159:265–277. doi: 10.1016/0042-6822(87)90464-8. [DOI] [PubMed] [Google Scholar]
- Richard A., Tulasne D. Caspase cleavage of viral proteins, another way for viruses to make the best of apoptosis. Cell Death Dis. 2012;3:e277. doi: 10.1038/cddis.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamaki K., Satou Y. Caspases: evolutionary aspects of their functions in vertebrates. J. Fish Biol. 2009;74:727–753. doi: 10.1111/j.1095-8649.2009.02184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh S., Hirota M., Noguchi T., Hijikata M., Handa H., Shimotonot k. Cleavage of hepatitis C virus nonstructural protein 5A by a caspase-like protease(s) in mammalian cells. Virology. 2000;270:476–487. doi: 10.1006/viro.2000.0287. [DOI] [PubMed] [Google Scholar]
- Schägger H. Tricine-SDS-PAGE. Nat. Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- Sun J., Yu Y., Deubel V. Japanese encephalitis virus NS1′ protein depends on pseudoknot secondary structure and is cleaved by caspase during virus infection and cell apoptosis. Microbes Infect. 2012;14:930–940. doi: 10.1016/j.micinf.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Teodoro J.G., Branton P.E. Regulation of apoptosis by viral gene products. J. Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry N.A., Rano T.A., Peterson E.P., Rasper D.M., Timkey T., Garcia Calvo M., Houtzager V.M., Nordstrom P.A., Roy S., Vaillancourt J.P., Chapman K.T., Nicholson D.W. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- Timmer J.C., Salvesen G.S. Caspase substrates. Cell Death Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- Touris-Otero F., Cortez-San Martin M., Martinez-Costas J., Benavente J. Avian reovirus morphogenesis occurs within viral factories and begins with the selective recruitment of σNS and λA to μNS inclusions. J. Mol. Biol. 2004;341:361–374. doi: 10.1016/j.jmb.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Touris-Otero F., Martinez-Costas J., Vakharia V.N., Benavente J. Avian reovirus nonstructural protein μNS forms viroplasm-like inclusions and recruits protein σNS to these structures. Virology. 2004;319:94–106. doi: 10.1016/j.virol.2003.10.034. [DOI] [PubMed] [Google Scholar]
- van der Heide L. The history of avian reovirus. Avian Dis. 2000;44:638–664. [PubMed] [Google Scholar]
- Varela. R., Martinez-Costas J., Mallo M., Benavente J. Intracellular posttranslational modifications of S1133 avian reovirus proteins. J. Virol. 1996;70:2974–2981. doi: 10.1128/jvi.70.5.2974-2981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Adamski R., Chen J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 2010;584:3717–3724. doi: 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]