Abstract
Coronavirus infection induces the generation of autophagosomes, and certain host proteins regulating cellular autophagy are hijacked by some coronaviruses to facilitate the formation of double membrane vesicles. However, mechanisms underlying coronavirus-induced autophagy remain largely unknown. In this study, we demonstrate that autophagosome formation and apparent autophagic flux are induced in cells infected with infectious bronchitis virus (IBV) – a gammacoronavirus. Notably, IBV-induced autophagy was dependent on autophagy related 5 (ATG5) but not beclin1 (BECN1), although both are essential proteins in the canonical autophagy pathway. Moreover, the ER stress sensor inositol requiring enzyme 1 (IRE1), but not its substrate X-box protein 1 (XBP1), was also essential for the induction of autophagy during IBV infection. Finally, the anti-apoptotic extracellular signal-regulated kinase 1/2 (ERK1/2) also contributed to IBV-induced autophagy. Our findings add new knowledge to the regulatory mechanisms governing coronavirus-induced autophagy, highlighting an extensive cross-talk among cellular signaling pathways during coronavirus infection.
Keywords: Coronavirus, Autophagy, ER stress, Unfolded protein response, Mitogen-activated protein kinase
1. Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved “self-eating” process where part of the cytoplasm and/or organelles are sequestered within a double membrane vesicle (named autophagosome), which ultimately fuses with the lysosome for bulk degradation (Yang and Klionsky, 2010). Under basal conditions, autophagy allows cells to break down long-lived proteins and damaged organelles. When cells are under starvation or growth factor deprivation, autophagy is also induced to recycle amino acids and fatty acids, thus maintaining the cellular metabolism and ensuring the cell survival. Autophagy is also activated under a variety of cellular stress conditions, such as hypoxia, reactive oxygen species, DNA damage, protein aggregation, or infection by intracellular pathogens (Kroemer et al., 2010). In most scenarios, autophagy facilitates stress adaptation and cell survival. However, in other settings, autophagy constitutes an alternative pathway of cell death called autophagic cell death (Maiuri et al., 2007).
The complete autophagy pathway can be divided into four steps: initiation, nucleation, elongation and lysosomal fusion. Each step is tightly regulated by numerous highly conserved autophagy-related genes (ATGs) (Mizushima et al., 2008). In the canonical signaling pathway of starvation-induced autophagy, the coiled-coil, myosin-like BCL2 interacting protein (BECN1, also known as ATG6 in yeast) is an essential component of the class III phosphatidylinositol-3-OH kinase (PI3K) complex, which is essential for the isolation membrane nucleation and phagophore assembly (Liang et al., 1999; Furuya et al., 2005). On the other hand, autophagy related 5 (ATG5), together with ATG12 and ATG16, constitutes a ubiquitination-like system that is essential for the elongation of isolation membranes and autophagosome formation (Mizushima et al., 1998; Suzuki et al., 2001). A critical event catalyzed by the ATG12-ATG5-ATG16L1 complex is the conversion of microtubule-associated proteins 1A/1B light chain 3 (MAP1LC3, hereafter referred to as LC3) from a soluble form (LC3-I) to a lipidated form (LC3-II). LC3-II is stably associated with both the inner and outer membrane of autophagosomes, and its biochemical and microscopic detection has been commonly used to monitor autophagy (Klionsky et al., 2016).
Coronaviruses are a group of enveloped viruses with large (27–32 kb), single-stranded, positive-sense RNA genomes. Inside the infected cells coronavirus infection induces the formation of double membrane vesicles (DMVs), to which viral replication/transcription complexes (RTCs) are associated (Knoops et al., 2008a; Lim et al., 2016; Fung and Liu, 2018). The morphological similarity between autophagosomes and coronavirus-induced DMVs led to some early speculations that cellular autophagy machineries may be hijacked to benefit coronavirus replication. However, follow-up studies have failed to detect the co-localization of autophagosomal markers and the viral replicase proteins (Snijder et al., 2006; de Haan and Reggiori, 2008). Remarkably, Reggiori et al. have recently shown that mouse hepatitis virus (MHV) hijacks short-living, ER-derived and LC3-coated vesicles (called EDEMosomes) for the formation of DMVs, but the conversion of LC3-I to LC3-II is not required for this process (Reggiori et al., 2010). This is consistent with the findings that depletion of ATG5 or treatment with autophagy inhibitor does not affect the coronavirus replication (Zhao et al., 2007; Cottam et al., 2011).
Although not essential for coronavirus replication, autophagy is undoubtedly activated during the infection with different coronaviruses including TGEV, MHV, IBV and other viruses belonging to the order Nidovirales (Cong et al., 2017). Notably, overexpression of the non-structural protein 6 (nsp6) of IBV, MHV, and SARS-CoV was shown to induce the formation of autophagosomes in the transfected cells (Cottam et al., 2011). Moreover, the autophagosomes induced by coronavirus nsp6 have smaller diameters compared with those induced by starvation, suggesting that nsp6 might also restrict the expansion of autophagosomes (Cottam et al., 2014). However, nsp6 overexpression does not activate the mTOR pathway or the ER stress response, so how it induces autophagy remains elusive. Although coronavirus-induced autophagy has been implicated in critical aspects of virus-host interaction, such as innate immune response (Chen et al., 2014) and apoptosis (Zhu et al., 2016), the detailed mechanisms are still largely unknown.
In this study, using the gammacoronavirus IBV as a model, we show that IBV infection induces autophagosome formation and apparent autophagic flux in the infected cells. By adopting the RNA interference approach, it was found that while ATG5 was required for IBV-induced autophagy, BECN1 - another upstream protein of the canonical starvation-induced pathway is dispensable. Among the three sensors of the unfolded protein response (UPR), only inositol requiring enzyme 1 (IRE1) was required for the induction of autophagy during IBV infection. Interestingly, neither of the two known downstream effectors of IRE1 – X-box protein 1 (XBP1) and c-Jun N-terminal kinase (JNK), is required for IBV-induced autophagy. On the other hand, the mitogen-activated protein (MAP) kinase extracellular signal-regulated kinase 1/2 (ERK1/2) was found to be involved in autophagy induction, which may protect the infected cells from apoptosis during IBV infection. Taken together, this study provides new insights into the cellular factors modulating coronavirus-induced autophagy. Moreover, the current data suggest that intricate cross-talks between multiple cellular signaling pathways contribute to the host response during coronavirus infection.
2. Materials and methods
2.1. Virus and cell lines
The egg-adapted Beaudette strain of IBV (ATCC VR-22) was obtained from American Type Culture Collection (ATCC) and adapted to Vero cells as described (Ng and Liu, 1998). To prepare the virus stock, monolayers of Vero cells were infected and cultured in plain Dulbecco modified Eagle medium (DMEM, Gibco) at 37 °C for 24 h. After three freeze/thaw cycles, cell lysate was clarified by centrifugation at 1500×g at 4 °C for 30 min. The supernatant was aliquot and stored at −80 °C as virus stock. The titer of the virus stock was determined by plaque assays. The mock lysate was prepared by the same treatment of uninfected Vero cells.
H1299 cells were cultured in RPMI1640 supplemented with 5% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin (Gibco). All cells were grown in a 37 °C incubator supplied with 5% CO2. In all the experiments, a monolayer of cells was washed twice with the serum-free medium before infected with IBV at MOI∼2 or incubated with an equal volume of UV-inactivated IBV or mock lysate in serum-free medium. After 2 h of adsorption, cells were washed twice and incubated at 37 °C before harvested at the indicated time points.
2.2. Antibodies, chemicals, and reagents
The anti-serum against IBV S and N protein were from rabbits immunized with bacterial expressed fusion proteins as previously described (Liu and Inglis, 1991; Li et al., 2005). The antibodies against LC3 (#3868), β-actin (#4967), ATG5 (#2630), ERK1/2 (#9102), GFP(#2555), IRE1(#3294), JNK (#9258), PERK(#3192), phosphor-ERK (#9101), phosphor-JNK (#4668) and CHOP(#2895) were purchased from Cell Signaling Technology and used for Western blot according to the manufacturer's instructions. The antibody against BECN1 (#11427) was from Santa Cruz Biotechnology.
2.3. Plasmid constructions and stable transfection
The cDNA of human LC3 was amplified from H1299 cells by reverse transcriptase-polymerase chain reaction (RT-PCR) using the forward primer: 5′-CCGGAATTCCATGCCGTCGG AGAAGAC-3′ and reverse primer: 5′-CGGGGTACCAACAATTCTAGAAGAGCTGCA-3′). The PCR product was cloned to the C-terminus of the green fluorescent protein (GFP) between EcoRI and KpnI in pEGFP-C1 (Clontech), and the resulting plasmid was named pGFP-LC3. pmRFP-GFP-LC3 was obtained from Addgene as previously described (Kimura et al., 2007).
Transfection of plasmids DNA to H1299 cells was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. To select for stably transfected cells, H1299 cells ∼90% confluent in 35 mm dishes were transfected with 2 μg plasmid DNA or mock transfected. At 16 h post transfection, cells were trypsinized, diluted 100 times and plated on 100 mm dishes with RPMI 1640 containing 0.5 mg/ml G418. The G418 containing medium was replaced every 4 days and cells were selected for up to 3 weeks until all the cells in the mock-transfected control were dead. Stable cell colonies were transferred to 24-well plates and expanded.
2.4. RNA interference
ATF6 siRNA (+): 5′-GCAACCAAUUAUCAGUUUA dTdT-3′, BECN1 siRNA (+): 5′- GAUUGAAGACACAGGAGGC dTdT-3′, IRE1 siRNA (+): 5′-GGACGUGAGCGACAGAAUA dTdT-3′, JNK siRNA (+): 5′- AAAGAAUGUCCUACCUUCUdTdT-3′, XBP1 siRNA (+): 5′- ACAGCAAGUGGUAGAUUUA dTdT-3′ and control EGFP siRNA (+): 5′-GCUGACCCUGAAGUUCAUC dTdT-3′ were purchased from Sigma. ERK1/2 siRNA was from Cell Signaling Technology. The PERK, CHOP and non-targeting control siRNA (siNC) were purchased from Ambion. Transfection of siRNA to H1299 cells was performed using DhamaFECT transfection reagent according to the manufacturer's instructions. At 48 h post-transfection, cells were infected with IBV at MOI∼2 or mock infected, and incubated for indicated time before harvested.
2.5. RNA extraction and RT-PCR analysis
Total RNA from cultured cells was extracted with TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. Briefly, cells were lysed with 1 ml TRIzol per 10 cm2 effective growth area and the lysates were mixed with a one-fifth volume of chloroform. After centrifugation at 12,000×g at 4 °C for 15 min, the aqueous phase was mixed with an equal volume of isopropanol. RNA was pelleted by centrifugation at 12,000×g at 4 °C for 15 min, washed with 70% ethanol twice and dissolved in RNase-free H2O. The concentration of the total RNA was measured using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific).
The cDNA was reverse transcribed from total RNA with oligo(dT) with ImProm-II™ Reverse Transcription System (Promega) according to the manufacturer's instructions. The following primers were used for PCR: XBP1 forward 5′-CAGCGCTTGGGGATGGATGC-3′ and XBP1 reverse 5′- CCATGGGGAGATGTTCTGGA-3'; ATF6 forward 5′- CATCCGCAGAAGGGGAGACACA-3′ and ATF6 reverse 5′-CTATTGTAATGACTCAGGGA -3’; GAPDH forward 5′-GGGCTCATCTGAAGGGTGGTGCTA-3′ and GAPDH reverse 5′-GTGGACGCTGGGATGATGTTCTGG-3';
2.6. SDS-PAGE and Western blot analysis
Cells were infected with IBV and harvested at indicated times points using cell scrapers (Corning). After centrifugation at 16,000×g for 1 min, the supernatant was discarded and the pellets were lysed in 1 × RIPA buffer. After clarifying by centrifugation and determination of protein concentration by spectrophotometer, the cell lysates were mixed with Laemmli sample buffer containing 100 mM dithiothreitol. The protein samples were boiled at 90 °C for 5 min and centrifuged at 16,000×g for 5 min. An equal amount of protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to 0.2 μm nitrocellulose membranes (Bio-Rad). After the nonspecific antibody binding sites were blocked with 5% skim milk in Tris-buffered saline (20 mM Tris-HCl pH 7.4, 150 mM NaCl) containing 0.1% Tween 20, the membranes were incubated with 1 μg/ml primary antibodies at 4 °C overnight. After washing with Tris-buffered saline, the membranes were incubated with 1:2000 diluted anti-mouse or anti-rabbit IgG antibodies conjugated with horseradish peroxidase (DAKO) at room temperature for 2 h. The membranes were washed and the proteins detected with a chemiluminescence detection kit (Amersham Biosciences) and medical X-ray films (Fujifilm) according to the manufacturer's instructions. The films were scanned as grayscale 8-bit images and the density of bands was determined by the NIH software ImageJ. All experiments were repeated for at least three times with similar result, and one of the representative results was shown.
2.7. Immunofluorescence assay
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 30 min and permeabilized with ice-cold absolute methanol at −20 °C for 10 min. After washing and blocking with blocking buffer (5% goat serum and 0.3% Triton-X100 in PBS) at room temperature for 1 h, cells were incubated with anti-IBV N anti-serum (1:300) at 4 °C overnight (Liang et al., 2019). After washing, cells were further incubated in goat anti-rabbit secondary antibody conjugated with Alexa Fluor 594 (1:1000, Molecular Probes) at 4 °C for 1 h. The cell nuclei were then stained with 0.5 μg/ml 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI) from Sigma. The cells were then washed, mounted with coverslip using an antifade mounting medium (DAKO) and sealed with clear nail polish. Fluorescence images were captured using the confocal microscope LSM510 (Zeiss).
3. Results
3.1. IBV infection induces the formation of autophagosomes
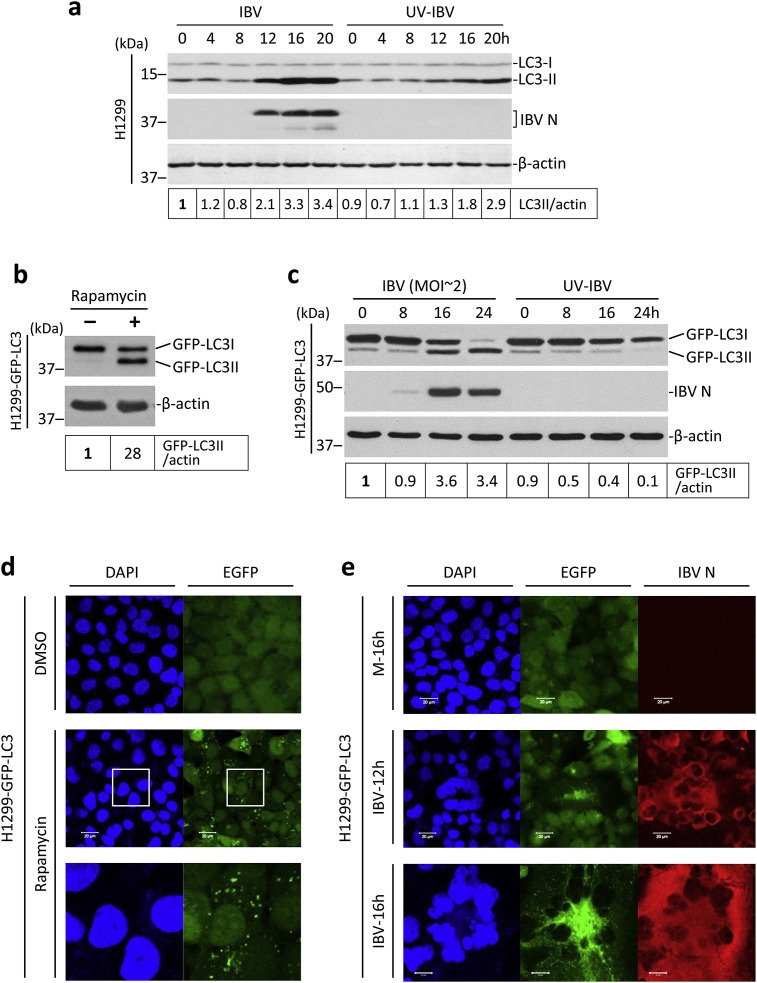
The induction of autophagy has been observed in cells infected with several coronaviruses (Zhao et al., 2007; Cottam et al., 2011). To validate that IBV also induces autophagy, and to obtain a temporal profile of IBV-induced autophagy, a time course experiment was performed. As shown in Fig. 1 a, H1299 cells were infected with IBV at MOI∼2 or incubated with UV-IBV for the indicated time. The conversion of the LC3 protein from the non-lipidated LC3-I form into the lipidated LC3-II form was widely used as an indicator of autophagy induction. According to a well-established guideline for autophagy study, the ratio between LC3-II and the loading control (such as β-actin or β-tubulin) should be used as a measurement of autophagy induction (Klionsky et al., 2016). The LC3-II/actin ratio remained stable from 0 to 8 hpi in both IBV-infected cells and the UV-IBV-treated control (Fig. 1a). In cells infected with IBV, the LC3-II/actin ratio significantly increased at 12 hpi, peaked at 16 hpi, and remained stable till the end of the time course. In cells incubated with UV-IBV, the LC3-II/actin ratio also gradually increased. Serum withdrawal has been known to induce autophagy (Klionsky et al., 2016). Since cells were incubated in serum-free medium in both the IBV-infected group (to prevent the inhibitory effect of serum on IBV replication) and the UV-IBV-treated control group, the high LC3-II/actin ratio observed at later time points may be partially contributed by the effect of serum withdrawal. Nonetheless, the LC3-II/actin ratio was always higher in the IBV-infected cells compared with control of the same time point after 12 hpi, and almost 2 times higher at the peak of 16 hpi (Fig. 1a).
Fig. 1.
Autophagy is induced in cells infected with IBV
(a) Lipidation of endogenous LC3 during IBV infection. H1299 cells were infected with IBV at MOI∼2 or incubated with UV-IBV. Protein lysates were harvested at the indicated time points and subjected to Western blot analysis using antibodies against LC3 and IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. The ratio of LC3-II to the corresponding β-actin band was determined and normalized to the IBV-infected 0 h sample. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(b) H1299-GFP-LC3 cells were treated with 0.5 μM rapamycin or the same volume of DMSO for 5 h before harvested for Western blot with EGFP antibody. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. The ratio of GFP-LC3-II to the corresponding β-actin band was determined and normalized to the solvent control (−). The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(c) H1299-GFP-LC3 cells were infected with IBV as in (a) and subjected to Western blot using antibodies against EGFP and IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated. GFP-LC3II to actin ratio was determined as in (b) and normalized to the IBV-infected 0 h sample. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(d) H1299-GFP-LC3 cells were treated as in (b) and fixed. Cell nuclei were stained with DAPI. Fluorescent images were captured with a confocal microscope. The square region was enlarged in the bottom panel to highlight the GFP-LC3II puncta. The experiment was repeated three times with similar results, and the result of one representative experiment is shown. (e) H1299-GFP-LC3 cells were infected as in (c). Cells were harvested at the indicated time points, fixed and stained with antibody against IBV N (red). Nuclei were stained with DAPI. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
The high basal level of endogenous LC3-II rendered it difficult to study the specific effect of IBV infection on autophagy induction. Therefore, we shifted to use another well-characterized autophagy reporter system in which the coding sequence of LC3 was fused with an N-terminal EGFP tag. We reasoned that, since the GFP-LC3 reporter is overexpressed at a much higher level than the endogenous LC3 protein, more pronounced GFP-LC3-I to GFP-LC3-II conversion would be expected in IBV-infected cells compared with that of the UV-IBV control. To this end, H1299 cells were stably transfected with the pEGFP-LC3 plasmid DNA to generate the H1299-GFP-LC3 cell line. As shown in Fig. 1b, H1299-GFP-LC3 cells were treated with the commonly used autophagy inducer rapamycin. A considerable conversion of GFP-LC3-I into GFP-LC3-II was observed in the rapamycin-treated cells, whereas only a very low basal level of GFP-LC3-II could be detected in the solvent control, suggesting that the GFP-LC3-II to actin ratio could be used as an indicator of autophagy induction. The stable cells were then infected with IBV at MOI∼2 or incubated with UV-IBV. As shown in Fig. 1c, the protein level of GFP-LC3-I in UV-IBV-treated cells was significantly higher than that of GFP-LC3-II throughout the time course, with the GFP-LC3-II/actin ratio steadily reduced. In sharp contrast, in the IBV-infected H1299-GFP-LC3 cells, significant conversion of GFP-LC3-I to GFP-LC3-II could be observed starting from 16 hpi. Indeed, the GFP-LC3-II to actin ratio significantly increased at 16 and 24 hpi in the IBV-infected cells (Fig. 1c). Band intensities of total GFP-fusion proteins were gradually decreased in both the IBV-infected cells and the UV-IBV control, possibly due to the degradation of the reporter protein. As expected, the GFP-LC3 reporter system was less sensitive to autophagy induced by serum withdrawal (as observed in the UV-IBV control), thus autophagy specifically induced by IBV infection could be determined with a low background.
Another advantage provided by the GFP-LC3 reporter is that autophagy induction could be determined by the formation of GFP-LC3-II puncta using fluorescent microscopy (Klionsky et al., 2016). As shown in Fig. 1d, the GFP-LC3-I in DMSO-treated cells was distributed diffusely in the cytoplasm. Rapamycin treatment caused the lipidation of GFP-LC3, thus the cytosolic GFP-LC3-II puncta were clearly visible. Next, H1299-GFP-LC3 cells were infected with IBV or mock infected before subjecting to immunofluorescence analysis using an antiserum against IBV N. As shown in Fig. 1e, no specific signal for IBV N was detected in the mock-infected cells and the GFP-LC3 protein had a diffuse pattern, suggesting an absence of autophagy induction. In cells infected with IBV for 12 h, most cells were positively stained with IBV N protein, featuring a cytoplasmic localization as previously reported (Masters, 2006). Moreover, clusters of cells expressing a high level of IBV N protein fused with each other to form multinucleated syncytia. Notably, GFP-LC3-II puncta excluded from the nuclei could be readily detected inside these syncytia. On the other hand, in the surrounding mononucleated cells expressing a low level of IBV N, the GFP-LC3 protein is still diffusely located in the cytoplasm (Fig. 1e). At a later time point (16 hpi), infected cells underwent massive fusion and generated large syncytia that was stained strongly with the IBV N antiserum. Moreover, extensive GFP-LC3-II puncta were observed scattered in the cytoplasm of the syncytia but were excluded from the nuclei (Fig. 1e). The result consolidates the Western blot data and further suggests that autophagy induction is dependent on the stage of IBV infection. Significant LC3 lipidation and puncta formation can be observed only at a relatively late stage of infection when extensive syncytia are formed. In the following experiments where the GFP-LC3 reporter was used, cells were harvested for analysis at late time points.
3.2. IBV infection induces apparent autophagic flux
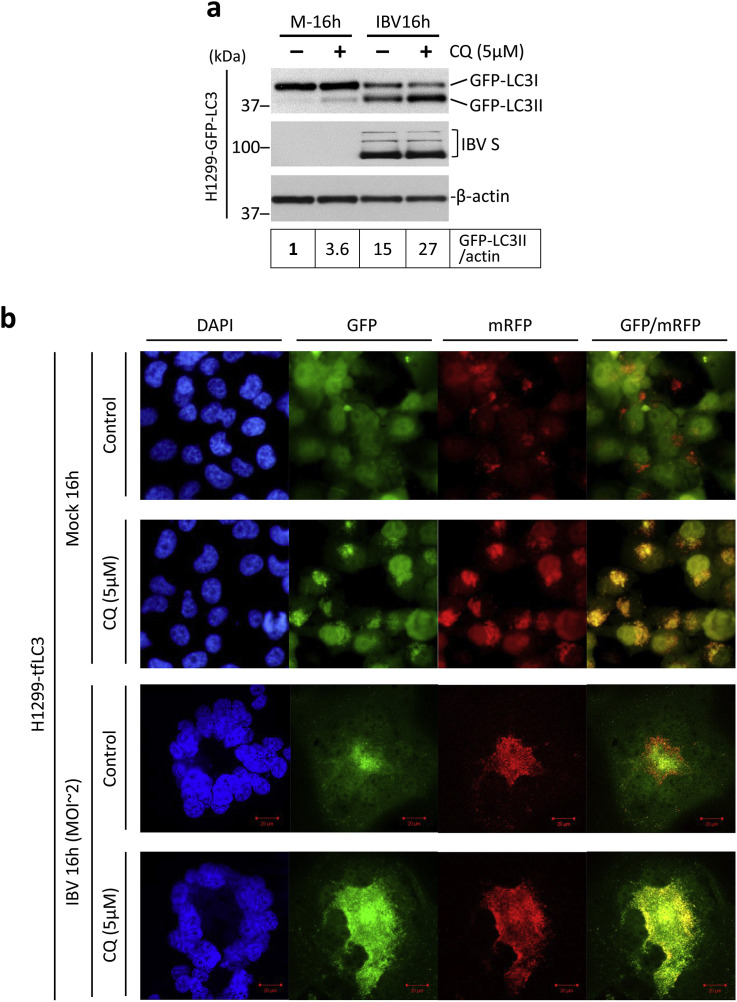
The elevated level of LC3 lipidation/punctation only suggests an increased formation of autophagosomes, which may result from either the increase of de novo autophagy activation or the inhibition of autophagosome degradation. To differentiate these two potential mechanisms, cells were treated with chloroquine (CQ), a chemical that inhibits the lysosomal degradation of autophagosomes. As shown in Fig. 2 , H1299-GFP-LC3 cells were infected with IBV at MOI∼2 or mock infected, and cells were also simultaneously treated with either 5 μM CQ or the same volume of PBS. Treatment with CQ slightly increased GFP-LC3-II/actin ratio in the mock-infected cells, suggesting that the basal level of autophagosome degradation was inhibited by CQ. Treatment of CQ at 5 μM did not significantly affect IBV replication, as indicated by similar levels of IBV S protein in CQ-treated cells compared with the PBS control. As expected, IBV infection significantly increased the GFP-LC3-II/actin ratio compared with the mock-infected cells. Notably, treatment of CQ in the IBV-infected cells further increased the GFP-LC3-II/actin ratio (Fig. 2a). The result suggested that IBV infection led to further autophagosome accumulation even when lysosomal degradation was blocked. Therefore, the increased level of LC3 lipidation/punctation observed in IBV-infected cells was at least partially contributed by a de novo increase of autophagy induction.
Fig. 2.
IBV infection induces apparent autophagic flux.
(a) H1299-GFP-LC3 cells were infected with IBV at MOI∼2 or mock infected. After 2 h of adsorption, cells were washed, treated with 5 μM chloroquine or an equal volume of PBS, and incubated before harvested. Cell lysates were subjected to Western blot as in Fig. 1c except for that antibody of IBV S was used in the place of IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated. GFP-LC3II to actin ratio was determined as in Fig. 1b and normalized to the mock infected PBS treated control. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(b) H1299-tfLC3 cells were infected with IBV or mock-infected as in (a). After 2 h of adsorption, cells were treated with CQ or an equal volume of PBS as in (a). Cells were harvest and fixed, and cell nuclei were stained as in Fig. 1d. The confocal images from GFP and mRFP channels were merged to show the co-localization of GFP and mRFP. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
On the other hand, to determine whether IBV infection might affect autophagy flux by inhibiting autophagosome degradation, we adopted another well-characterized reporter system featuring the tandem fluorescence-labeled LC3 (tfLC3) (Kimura et al., 2007). In this system, LC3 is fused with monomeric red fluorescence protein (mRFP) and GFP in tandem at the N-terminal. Inside the pH neutral autophagosomes, both GFP and mRFP could be detected. However, as the autophagosomes fuse with the lysosomes, GFP is quenched by the acidic lysosomal environment whereas mRFP retained its fluorescence. Therefore, only red but not green fluorescence could be detected for the autolysosomes. As a result, in a GFP/mRFP channel merged image, the autophagosomes will appear as yellow puncta (colocalized GFP and mRFP), while the autolysosomes will appear as red puncta (mRFP-only). H1299 stably expressing the tfLC3 reporter was generated (referred to H1299-tfLC3). As shown in Fig. 2b (first row), in the mock-infected and PBS-treated cells, GFP signal was diffusely distributed in the cytoplasm (similar to GFP-LC3 localization in mock-infected cells), while mRFP exhibited both a diffusely cytoplasmic and a punctated localization. Consequently, no yellow puncta could be detected in the GFP/mRFP merged image. This result suggested that autophagy flux was highly efficient in the mock-infected untreated cells, as the autophagosomes were rapidly converted to the autolysosomes. On the other hand, in the mock-infected cells treated with 5 μM CQ, both GFP and mRFP formed a large number of puncta (Fig. 2b, second row). The GFP/mRFP merged image indicated almost complete colocalization of GFP and mRFP, with bearly any mRFP-only puncta. Therefore, the CQ treatment almost completely inhibited the conversion of autophagosomes into autolysosomes, resulting in the blockage of autophagy flux. Notably, both GFP and mRFP signals were significantly higher in CQ-treated cells compared with untreated cells. This likely resulted from the accumulation of tfLC3 reporter protein due to the inhibition of its lysosomal degradation.
When H1299-tfLC3 was infected with IBV in the absence of 5 μM CQ, a considerable level of GFP puncta was detected (similar to GFP-LC3 localization in IBV-infected cells), while the mRFP puncta were even more pronounced (Fig. 2b, third row). In the GFP/mRFP merged image, both yellow autophagosomes and red autolysosomes could be observed. This result suggested that IBV infection saturated the autophagy flux, thus some autophagosomes could no longer be immediately converted to autolysosomes and accumulated in the infected cells. However, autophagy flux was not effectively blocked in IBV-infected cells, as a considerable amount of the mRFP-only puncta could still be detected, indicating that a large proportion of autophagosomes could still be converted into autolysosomes in IBV-infected cells. In contrast, when H1299-tfLC3 cells were treated with 5 μM CQ and infected with IBV, a complete colocalization of GFP and mRFP puncta was observed, suggesting a complete blockage of autophagy flux (Fig. 2b, fourth row). Taken together, studies using CQ and tfLC3 reporter suggested that IBV infection induced apparent autophagic flux in the infected cells, which proceeded to the stage of lysosomal fusion.
3.3. ATG5 but not Beclin1 is required for IBV-induced autophagy
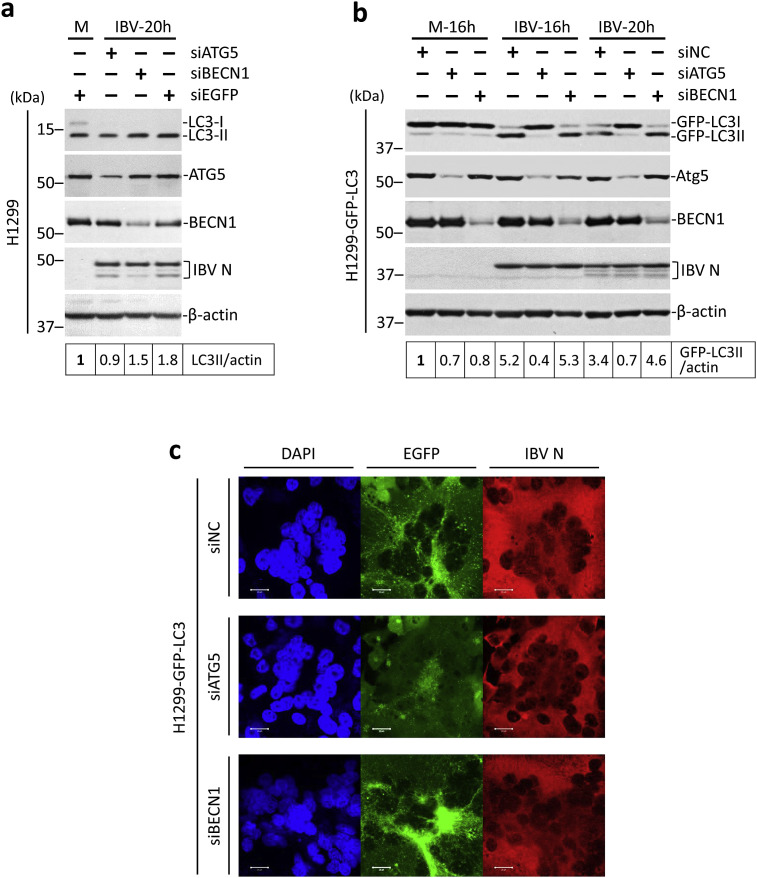
Autophagy is regulated by a number of proteins encoded by the autophagy-related genes. In the canonical starvation-induced autophagy pathway, BECN1 is essential for the early step of nucleation of the isolation membrane, while ATG5 is required for the later step of membrane expansion and autophagosome formation. To see whether these two canonical proteins were required for IBV-induced autophagy, the siRNA-mediated knockdown approach was adopted. H1299 cells were transfected with siATG5, siBECN1, or siEGFP before infected with IBV at MOI∼2. As shown in Fig. 3 a, the knockdown of ATG5 and BECN1 could be determined by the reduced protein levels in the Western blot. Notably, a significant level of LC3 lipidation was detected even in the siEGFP-transfected mock-infected cells. The high basal level of autophagy induction most likely resulted from transfection with cationic liposomes, which has been reported by others previously (Man et al., 2010). Nevertheless, knockdown of either gene did not significantly affect IBV replication, as suggested by the similar levels of IBV N protein compared with the siEGFP control. As expected, IBV infection induced a ∼2-fold increase of the LC3-II/actin ratio in the siEGFP control, as compared with the mock-infected cells (Fig. 3a). Notably, no significant increase of the LC3-II/actin ratio could be detected in the ATG5-knockdown cells infected with IBV compared with the mock-infected control. In cells transfected with siBECN1 and infected with IBV, LC3-II/actin ratio was considerably higher than the mock-infected cells, although slightly lower than the IBV-infected siEGFP control (Fig. 3a).
Fig. 3.
IBV-induced autophagy is ATG5-dependent but BECN1-independent.
(a) H1299 cells were transfected with siATG5, siBECN1 or siEGFP before infected with IBV at MOI∼2 or mock infected. Protein lysates were harvested at the indicated time points and subjected to Western blot analysis using antibodies against ATG5, BECN1, LC3, and IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. The LC3-II/actin ratio was determined as in Fig. 1a and normalized to the mock-infected siEGFP control. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(b) H1299-GFP-LC3 cells were transfected and infected as in (a) except that siNC was used to replace siEGFP. Western blot analysis was performed as in (a). Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. GFP-LC3II to actin ratio was determined as in Fig. 1c and normalized to the mock-infected siNC control. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(c) H1299-GFP-LC3 cells were transfected and infected as in (b). Cells were harvested at the indicated time, fixed and stained with antiserum against IBV N (red). Nuclei were stained with DAPI. Fluorescent images were captured as in Fig. 1e. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
The high basal level of autophagy in the mock-infected cells prompted us to shift to the H1299-GFP-LC3 cells. As shown in Fig. 3b, a similar knockdown experiment was performed in H1299-GFP-LC3 cells and the knockdown efficiency was determined as described above. A non-targeting siRNA (siNC) was used in place of siEGFP, because siEGFP would suppress the expression of the GFP-LC3 reporter. A similar low level of GFP-LC3 lipidation was observed in the mock-infected cells, providing a relatively low background to examine autophagy specifically induced by the IBV infection. Compared with the mock-infected cells, ∼5-fold and ∼3-fold increase in the GFP-LC3-II/actin ratio could be observed in cells transfected with siNC at 16 hpi and 20 hpi respectively, indicating a potent induction of autophagy (Fig. 3b). Similarly, ∼5-fold increase in the GFP-LC3-II/actin ratio could be observed in the IBV-infected BECN1-knockdown cells compared with mock-infected control. In sharp contrast, in ATG5-knockdown cells infected with IBV, the GFP-LC3-II/actin ratio remained at a similar level as in the mock-infected cells, indicating that IBV-induced autophagy was significantly inhibited by the depletion of ATG5. The result was further confirmed by the GFP-LC3 puncta analysis. As shown in Fig. 3c, significant levels of GFP-LC3-II punta formation induced by IBV infection could be observed in the siNC or siBECN1-transfected cells. In contrast, the GFP-LC3 puncta formation was detected at a much lower level in the IBV-infected ATG5-knockdown cells compared with the siNC control. In summary, our result shows that IBV-induced autophagy is only dependent on ATG5 but not BECN1.
3.4. The ER stress sensor IRE1 is involved in IBV-induced autophagy
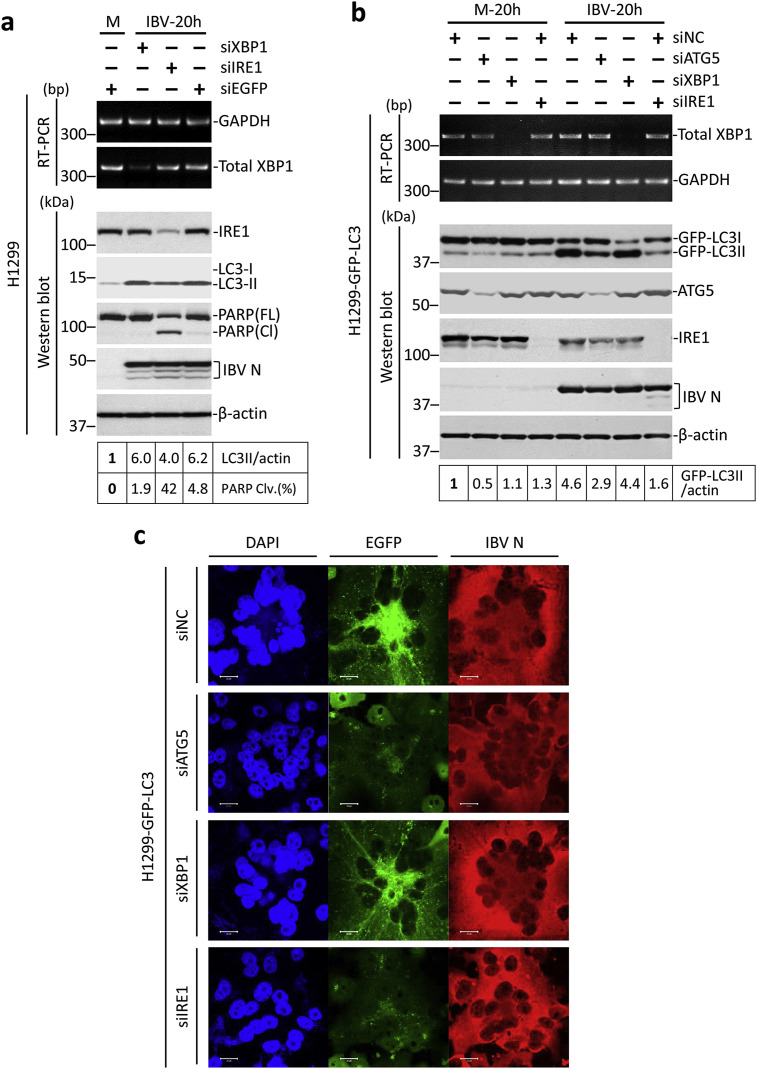
Since IBV-induced autophagy seems to require ATG5 but not BECN1, it is possible that early steps (initiation and nucleation) of IBV-induced autophagy might utilize separate pathway(s) distinct from the BECN1-dependent canonical pathway. Previously, we have shown that IBV infection induces ER stress and activates the inositol-requiring protein 1 (IRE1) branch of unfolded protein response (Fung et al., 2014). Notably, ER stress has been shown to induce cytoprotective autophagy through the IRE1 pathway (Ogata et al., 2006). Therefore, we proceeded to investigate the involvement of IRE1 and its downstream effector XBP1 in IBV-induced autophagy. To this end, H1299 cells were transfected with siIRE1, siXBP1, or siEGFP before infected with IBV at MOI∼2. As shown in Fig. 4 a, the knockdown of IRE1 could be determined by Western blot. No specific protein band could be detected for XBP1, thus its knockdown was determined by RT-PCR. The knockdown of IRE1 and XBP1 did not significantly affect IBV replication, as indicated by similar levels of IBV N protein compared with the siEGFP control. Compared to the mock-infected control, both siXBP1- and siEGFP-transfected cells had a significant increase in the LC3-II/actin ratio after infected with IBV for 20 h. In contrast, knockdown of IRE1 slightly inhibited IBV-induced autophagy, as indicated by the reduced LC3-II/actin ratio compared with the IBV-infected siEGFP control (Fig. 4a). Consistent with our previous publication, more prominent PARP cleavage was observed in the IBV-infected IRE-knockdown cells compared to the control, suggesting a pro-survival role of IRE1 during IBV infection.
Fig. 4.
IBV-induced autophagy requires the ER stress sensor IRE1.
(a) H1299 cells in duplicate were transfected with siXBP1, siIRE1 or siEGFP before infected with IBV at MOI∼2 or mock infected. Protein lysates were harvested at the indicated time points and subjected to Western blot analysis using antibodies against IRE1, LC3 and IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. The LC3-II/actin ratio was determined as in Fig. 3a. The percentage of PARP cleavage [PARP Clv. (%)] was calculated as the intensity of cleaved PARP [PARP(Cl)] divided by the total intensities of full-length PARP [PARP(FL)] and PARP(Cl). In the second set of cells, total RNA was extracted and subjected to RT-PCR using primers specific for XBP1 and GAPDH. Sizes of DNA ladders in bp were indicated on the left. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(b) H1299-GFP-LC3 cells induplicate were transfected as in (a) except that siATG5 was also included and siNC was used to replace siEGFP. Cells were infected with IBV or mock infected and Western blot was performed using antibodies against ATG5, IRE1, GFP, and IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. GFP-LC3II to actin ratio was determined as in Fig. 3b. In the second set of cells, total RNA was extracted and subjected to RT-PCR using primers specific for XBP1 and GAPDH. Sizes of DNA ladders in bp were indicated on the left. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(c) H1299-GFP-LC3 cells were infected with IBV or mock infected. Cells were harvested at the indicated time, fixed and stained using antiserum against IBV N (red). Nuclei were stained with DAPI. Fluorescent images were captured as in Fig. 1e. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
To validate this result, H1299-GFP-LC3 cells were similarly transfected, with siATG5 also included as a positive control. As shown in Fig. 4b, the knockdown of ATG5, IRE1, and XBP1 was determined by either Western blot or RT-PCR. The levels of IBV N protein were similar, suggesting that the gene knockdown did not significantly affect IBV replication. Consistent with the data presented above, IBV infection induced autophagy in cells transfected with siNC, indicated by the increased GFP-LC3/actin ratio compared to the mock-infected control. A similar degree of autophagy induction was detected in the XBP1-knockdown cells infected with IBV. In contrast, IBV-induced lipidation of GFP-LC3 was significantly reduced in cells transfected with siATG5 or siIRE1 (Fig. 4b). The result was then further confirmed by the GFP-LC3 puncta analysis. While a large amount of GFP-LC3-II puncta could be readily observed in the IBV-infected control and the XBP1-knockdown cells, they were only detected at a very low level in either ATG5-or IRE1-knockdown cells infected with IBV (Fig. 4c). Therefore, the ER stress sensor IRE1 was indeed required for IBV-induced autophagy.
3.5. The ER stress sensors ATF6 and PERK are not required for IBV-induced autophagy
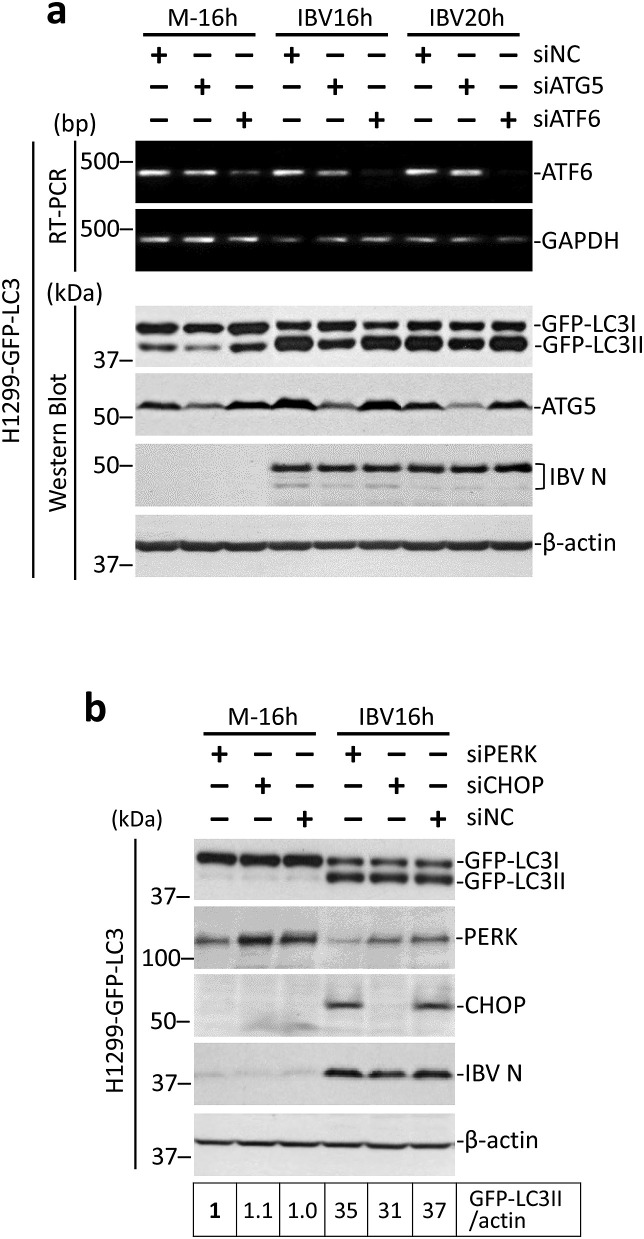
Apart from the IRE1 branch, the unfolded protein response consists of two additional pathways, regulated by the sensor proteins ATF6 and PERK respectively. To see whether these two sensors were also involved in IBV-induced autophagy, similar knockdown experiments were performed in H1299-GFP-LC3 cells. The transcription factor C/EBP homologous protein (CHOP) downstream of the PERK pathway is also investigated, as it has been shown to be induced during IBV infection and promotes IBV-induced apoptosis (Liao et al., 2013). As shown in Fig. 5 a and b, efficient knockdown of ATG5, PERK or CHOP could be determined by Western blot analysis. No specific protein band could be detected for ATF6, thus its knockdown was determined by RT-PCR. The replication of IBV was not significantly affected by the silencing of ATG5, ATF6, PERK or CHOP, as indicated by the similar levels of IBV N protein compared with the siNC control of the same time point. Consistent with the above results, the knockdown of ATG5 reduced the lipidation of GFP-LC3 during IBV infection. In contrast, in the ATF6-, PERK-, or CHOP-knockdown cells infected with IBV, the GFP-LC3-II/actin ratios were all similar to that of the siNC control of the same time points (Fig. 5a and b). Taken together, it appeared that ATF6, PERK, and CHOP were not required for autophagy induction during IBV infection.
Fig. 5.
ATF6 and PERK is not involved in IBV-induced autophagy.
(a) H1299-GFP-LC3 cells in duplicate were transfected with siATG5, siATF6 or siNC and infected with IBV or mock infected. Western blot was performed using antibodies against ATG5, GFP and IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. GFP-LC3II to actin ratio was determined as in Fig. 3b. In the second set of cells, total RNA was extracted and subjected to RT-PCR using primers specific for ATF6 and GAPDH. Sizes of DNA ladders in bp were indicated on the left. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(b) H1299-GFP-LC3 cells were transfected with siPERK, siCHOP or siNC and infected with IBV or mock infected. Protein lysates were harvested at the indicated time points and subjected to Western blot analysis using antibodies against PERK, CHOP and IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. The LC3-II/actin ratio was determined as in Fig. 1a and normalized to the mock-infected siEGFP control. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
3.6. MAP kinase ERK1/2, but not JNK, is required for IBV-induced autophagy
MAP kinases are important regulators of major cellular processes during coronavirus infection (Fung et al., 2016). Previously, we have shown that JNK phosphorylation is negatively regulated by IRE1 and serves a pro-apoptotic role during IBV infection (Fung et al., 2014; Fung and Liu, 2017). Importantly, previous studies have shown that signaling through the IRE1-JNK pathway is crucial for the induction of pro-survival autophagy in cells under ER stress (Ogata et al., 2006). Therefore, we went on to look at the potential involvement of JNK.
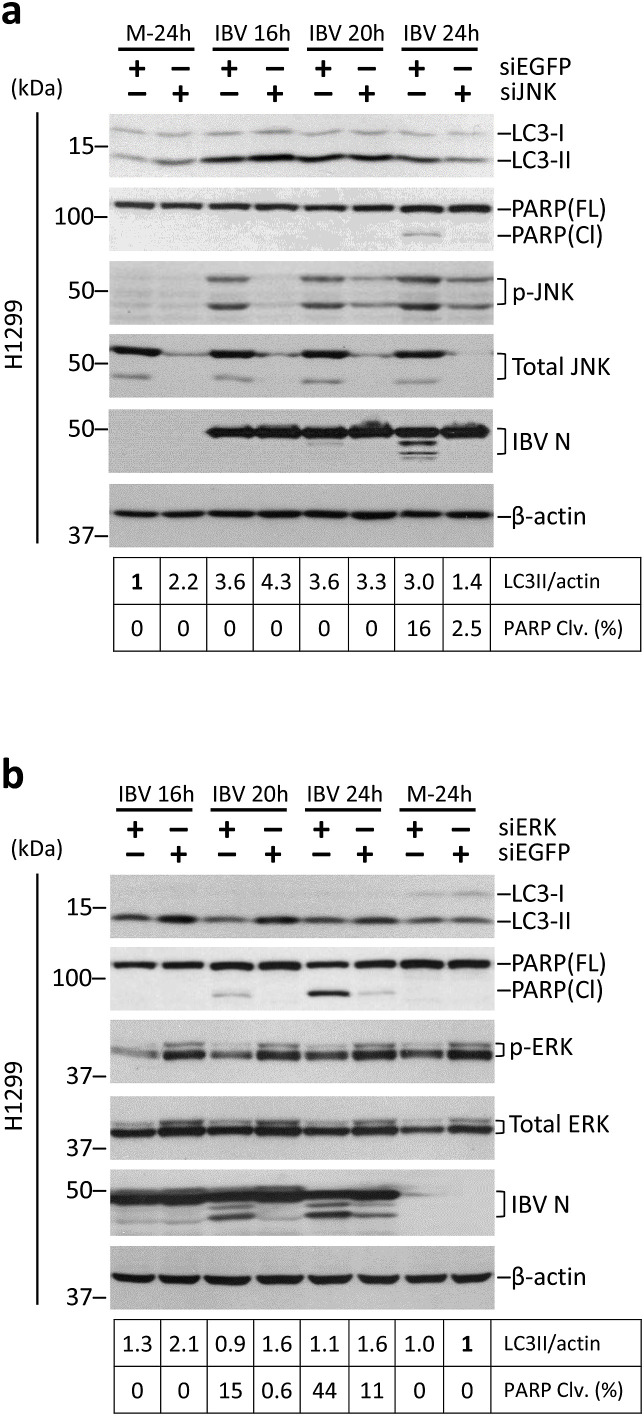
H1299 cells were transfected with siJNK or siEGFP (as negative control) before infected with IBV at MOI∼2. As shown in Fig. 6 a, the protein expression of total JNK was almost completely abolished in cells transfected with siJNK compared with the control. JNK phosphorylation could be detected at a high level in the control cells infected with IBV, but was markedly reduced in the JNK-knockdown cells of the same time point. Replication of IBV was not significantly affected by the knockdown of JNK, as indicated by the similar levels of IBV N protein. Consistent with the result above, IBV infection led to a significant increase of LC3II/actin ratio in the control cells. Notably, a similar level of LC3 lipidation was observed in JNK-knockdown cells compared with the control at 16 and 20 h post-infection (Fig. 6a), although a lower LC3II/actin ratio was detected at 24 hpi. This result suggests that the activation of JNK is not essential for autophagy induction during IBV infection. Consistent with our previous finding, a lower level of PARP cleavage was detected in JNK-knockdown cells at 24 hpi, suggesting that JNK was pro-apoptotic at the late stage of IBV infection.
Fig. 6.
IBV-induced autophagy requires the MAP kinase ERK but not JNK.
(a) H1299 cells were transfected with siJNK or siEGFP and infected with IBV or mock infected. Protein lysates were harvested at the indicated time points and subjected to Western blot analysis using antibodies against LC3, PARP, phosphor-JNK, total JNK, and IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. The LC3-II/actin ratio and percentage of PARP cleavage was determined as in Fig. 4a and normalized to the mock-infected siEGFP control. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
(b) H1299 cells were transfected with siERK or siEGFP and infected with IBV or mock infected. Protein lysates were harvested at the indicated time points and subjected to Western blot analysis using antibodies against LC3, PARP, phosphor-ERK, total ERK, and IBV N. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. The LC3-II/actin ratio and percentage of PARP cleavage was determined as in (a) and normalized to the mock-infected siEGFP control. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
Besides JNK, another MAP kinase – ERK1/2 is also activated during coronavirus infection. Previously, we have shown that ERK1/2 phosphorylation is negatively regulated by the PERK-eIF2α-CHOP branch of UPR and promotes the survival of IBV-infected cells (Liao et al., 2013). A similar knockdown experiment was performed as shown in Fig. 6b. The knockdown of ERK was determined by Western blot, and reduced protein levels of both total ERK and phosphorylated ERK could be detected in the siERK-transfected cells compared with the control cells. The knockdown of ERK did not affect IBV replication, as indicated by the similar levels of IBV N protein compared with the control cells of the same time point. Notably, while IBV infection induced significant LC3II lipidation in the siEGFP control cells, the LC3II/actin ratio remained low in the ERK-knockdown cells (Fig. 6b). Also consistent with our published data, a significantly higher level of PARP cleavage was detected in ERK-knockdown cells infected with IBV, compared with the control cells of the same time point. Taken together, ERK1/2 was required for autophagy induction in IBV-infected cells, and served a pro-survival role at the late stage of IBV infection.
4. Discussion
Autophagy is a highly conserved cellular process contributing to the homeostasis and stress adaptation in cells under various stimulations. In this study, we found that autophagy was induced in cells infected with the gammacoronavirus IBV. Using the inhibitor CQ and the tfCL3 reporter system, we showed that IBV infection induced apparent autophagic flux that proceeded to the stage of lysosomal fusion. Moreover, using RNA interference, we demonstrated that IBV-induced autophagy was dependent on ATG5, but was independent on BECN1, although both proteins are essential for the canonical pathway in starvation-induced autophagy. In an attempt to identify potential regulatory host factors, we showed that the ER stress sensor IRE1 and the MAP kinase ERK were both involved in autophagy triggered by IBV infection.
Previous studies on coronavirus-induced autophagy mainly relied on the biochemical detection of LC3 lipidation (Zhao et al., 2007) or the fluorescent detection of endogenous LC3 redistribution (Cottam et al., 2011). There are several limitations with either of these methods. For instance, commercial LC3 antibodies are known to display higher specificity against LC3-II than LC3-I, therefore densitometry analysis comparing LC3-I and LC3-II bands is usually misleading, and thus autophagy induction should be gauged by the ratio of LC3-II normalized to internal control such as β-actin (Klionsky et al., 2016). On the other hand, because LC3 is known to be hijacked by coronaviruses for the formation DMVs (Reggiori et al., 2010), the LC3-II puncta detected by fluorescent microscopy might represent coronavirus-induced, LC3-I-coated vesicles but not genuine LC3-II-coated autophagosomes. Moreover, an elevated level of LC3 lipidation/punctation only indicates the accumulation of autophagosomes, which could be resulted from increased de novo autophagy induction or a blockage of autophagy maturation.
In this study, we validated the induction of autophagy during coronavirus infection, using both the endogenous LC3 lipidation assay and the GFP-LC3 reporter. Compared with the endogenous LC3, the GFP-LC3 protein is expressed at a much higher level and thus more sensitive to autophagy activation. Also, because Western blot is performed using an antibody against GFP, comparison of the intensities of GFP-LC3-I and GFP-LC3-II bands is meaningful and represents their true stoichiometric ratio. Importantly, compared to endogenous LC3, the GFP-LC3 reporter is also far less sensitive to the “background” autophagy induced by the siRNA/plasmid DNA transfection and/or the serum withdrawal during IBV infection, making it possible to determine the autophagy specifically induced by IBV infection. Furthermore, we also examine the apparent autophagy flux using the lysosomal inhibitor chloroquine and the tfLC3 reporter system. Because the lipidation of GFP-LC3 was further increased by the chloroquine treatment and mRFP-only autolysosomes could be observed in H1299-tfLC3 cells infected with IBV, it was concluded that IBV infection did not significantly inhibit the lysosomal fusion of autophagosomes. Therefore, our result suggested that apparent autophagic flux was induced in cells infected with IBV.
In a previous study by Cottam et al., autophagy was activated in cells overexpressing nsp6 of IBV, MHV or SARS-CoV, or nsp5-7 of the related arterivirus porcine reproductive and respiratory syndrome virus (PRRSV) (Cottam et al., 2011). Coronavirus nsp6 is an ER-localized, highly hydrophobic protein that contains multiple transmembrane domains (Oostra et al., 2008). Notably, the co-expression of SARS-CoV nsp3, nsp4 and nsp6 is sufficient to induce DMV formation in the transfected cells (Angelini et al., 2013). However, it is worth noting that in coronavirus-infected cells, non-structural proteins are usually produced at levels significantly lower than those in the transfected cells where their overexpression was driven by a strong promoter. Therefore, the phenotypes observed in the transfection studies might not always be physiologically relevant, and further characterization is required using suitable recombinant viruses in a genuine time course infection experiment. Unfortunately, despite our best efforts, FLAG-tagged IBV nsp6 could not be expressed at a detectable level in either H1299 or H1299-GFP-LC3 cells (data not shown). However, our result suggested that neither the BECN1-dependent signaling of the canonical autophagy pathway nor the PERK-CHOP branch of UPR is required for IBV-induced autophagy, which is consistent with the previously published result that nsp6 overexpression did not activate the starvation-induced mammalian target of rapamycin (mTOR)-mediated signaling or induce the expression of CHOP (Cottam et al., 2011).
In starvation-induced autophagy, BECN1 is required for the production of PI3P and the nucleation of autophagosomes (Hosokawa et al., 2009; Backer, 2008), whereas ATG5 constitutes one of the ubiquitin-like conjugation systems required for the expansion of isolation membranes (Ohsumi, 2001; Geng and Klionsky, 2008). Previously, it has been shown that the inhibition of PI3K does not affect autophagy induction in MHV-infected cells, while the knockdown or knockout of ATG5 significantly reduces autophagy induction in cells infected with MHV or IBV (Zhao et al., 2007; Cottam et al., 2011). Consistently, we found that the GFP-LC3 lipidation/punctation during IBV infection was not affected in BECN1-knockdown cells, but almost abolished in ATG5-knockdown cells. Therefore, autophagy signaling in IBV-infected cells may utilize alternative upstream pathways independent of BECN1, eventually converging with the canonical autophagic signaling at the ATG5-dependent stage of isolation membrane expansion.
It is not uncommon for RNA viruses to utilize only a subset of the components of the canonical autophagic pathway. A recent study showed that three RNA viruses (poliovirus, dengue virus, and Zika virus) utilize ATG9 for membrane sculpting and recruit LC3 directly to the membranes, bypassing the requirement of ATG5-mediated LC3 lipidation (Abernathy et al., 2019). Additionally, BECN1 is not required for the replication of poliovirus or dengue virus, while the upstream Unc-like autophagy-activating kinase may or may not be required for poliovirus-induced autophagy (Abernathy et al., 2019; Corona Velazquez et al., 2018). Similarly, it has been previously shown that LC3 is hijacked by MHV-infected cells to facilitate the formation of DMVs, although this process appears to be independent of LC3 lipidation (Reggiori et al., 2010). In this study, we found that ATG5 was required for LC3 lipidation/punctation in IBV-infected cells, but its knockdown did not affect IBV replication. Thus, the proteins co-opted to support coronavirus replication (such as LC3-I) may constitute only a subset of the components in the autophagic pathway, while other proteins (such as ATG5) may still be required for autophagy induction in the coronavirus-infected cells.
Coronavirus replication is structurally (Knoops et al., 2008b; Maier et al., 2013) and functionally (Reggiori et al., 2010) associated with the ER, and has been shown to induce ER stress and activate all three branches of the UPR (Fung et al., 2014; Liao et al., 2013; Fung and Liu, 2014). In this study, we found that among the three ER stress sensors, only IRE1, but not ATF6 or PERK, was required for autophagy induction during IBV infection. As for the two proteins downstream of IRE1 signaling pathway, neither XBP1 nor JNK was required for the induction of autophagy in IBV-infected cells. This is quite different from cells treated with tunicamycin or thapsigargin, where ER stress engages the IRE1-JNK pathway to induce pro-survival autophagy (Ogata et al., 2006). Presumably, during IBV infection, some unknown protein(s) may be responsible to mediate the signaling from activated IRE1 to ATG5 (or other ATGs in the complex), thereby bridging the signaling pathways of ER stress and autophagy.
Similar to IRE1, ERK also serves an anti-apoptotic role during IBV infection (Liao et al., 2013). While a direct signaling pathway between IRE1 and ERK has not been reported, ERK-dependent autophagy has been described in several systems, such as human colon cancer HT29 cells depleted of amino acid (Ogier-Denis et al., 2000), human breast cancer MCF-7 cells treated with TNF-α (Sivaprasad and Basu, 2008), and cells infected with coxsackievirus A16 (Shi et al., 2015) or porcine circovirus type 2 (Zhu et al., 2012). ERK-dependent autophagy is sometimes characterized by extensive cell rounding and the formation of cytoplasmic macrovacuoles associated with autophagic cell death (Cagnol and Chambard, 2010; Cagnol et al., 2006). On the other hand, signaling through the IRE1-ASK1-JNK pathway is required for ER stress-dependent cell death (Urano et al., 2000). Therefore, proteins such as IRE1 and ERK may be at the center of an integrated signaling network, modulating autophagy, cell death, and other important pathways in coronavirus-infected cells.
A growing number of viruses have been demonstrated to induce autophagy in infected cells (Wong et al., 2008; Ke and Chen, 2011; Heaton and Randall, 2011). For certain viruses such as poliovirus, coxsackievirus B3 and hepatitis C virus, autophagy is induced by the viruses, which in turn promotes viral replication. Whereas for other viruses such as Sindbis virus, vesicular stomatitis virus and herpes simplex virus-1, autophagy acts as a host defense mechanism to either eliminate the virus or to activate the innate immune response (Kim et al., 2010). For coronaviruses, the membrane-associated papain-like protease 2 (PLP2) has been shown to interact with BECN1 and induce incomplete autophagy, which in turn modulates antiviral innate immunity (Chen et al., 2014). On the other hand, selective autophagy of mitochondria was observed in cells infected with TGEV, which counteracts oxidative stress and apoptosis induced by TGEV infection (Zhu et al., 2016). Therefore, although not essential for coronavirus replication, autophagy constitutes a very crucial aspect of virus-host interaction. Further investigation of coronavirus-induced autophagy will lead to a better understanding of the biology and pathogenesis of this important virus family.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgment
This work was partially supported by Natural Science Foundation of Guangdong Province Grant 2018A030313472, and Guangdong Province Key Laboratory of Microbial Signals and Disease Control Grant MSDC-2017-05 and MSDC-2017-06, Guangdong, People's Republic of China.
References
- Abernathy E., Mateo R., Majzoub K., van Buuren N., Bird S.W., Carette J.E. Differential and convergent utilization of autophagy components by positive-strand RNA viruses. PLoS Biol. 2019 Jan 4;17(1) doi: 10.1371/journal.pbio.2006926. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6334974/ [cited 2019 Apr 28];17(1). Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4(4) doi: 10.1128/mBio.00524-13. e00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J.M. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 2008 Feb 15;410(1):1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Cagnol S., Chambard J.-C. ERK and cell death: mechanisms of ERK-induced cell death – apoptosis, autophagy and senescence. FEBS J. 2010 Jan 1;277(1):2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- Cagnol S., Van Obberghen-Schilling E., Chambard J.-C. Prolonged activation of ERK1,2 induces FADD-independent caspase 8 activation and cell death. Apoptosis Int. J. Prog. Cell Death. 2006 Mar;11(3):337–346. doi: 10.1007/s10495-006-4065-y. [DOI] [PubMed] [Google Scholar]
- Chen X., Wang K., Xing Y., Tu J., Yang X., Zhao Q. Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell. 2014 Dec;5(12):912–927. doi: 10.1007/s13238-014-0104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Verlhac P., Reggiori F. The interaction between Nidovirales and autophagy components. Viruses. 2017 Jul 11;9(7) doi: 10.3390/v9070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona Velazquez A., Corona A.K., Klein K.A., Jackson W.T. Poliovirus induces autophagic signaling independent of the ULK1 complex. Autophagy. 2018;14(7):1201–1213. doi: 10.1080/15548627.2018.1458805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011 Nov;7(11):1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014 Aug;10(8):1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Reggiori F. Are nidoviruses hijacking the autophagy machinery? Autophagy. 2008 Apr;4(3):276–279. doi: 10.4161/auto.5241. [DOI] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014 Jun 17;5:296. doi: 10.3389/fmicb.2014.00296. http://journal.frontiersin.org/article/10.3389/fmicb.2014.00296/abstract [cited 2018 Mar 20];5. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Activation of the c-Jun NH2-terminal kinase pathway by coronavirus infectious bronchitis virus promotes apoptosis independently of c-Jun. Cell Death Dis. 2017 Dec 13;8(12):3215. doi: 10.1038/s41419-017-0053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13(6):405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liao Y., Liu D.X. The endoplasmic reticulum stress sensor IRE1α protects cells from apoptosis induced by the coronavirus infectious bronchitis virus. J. Virol. 2014 Nov 1;88(21):12752–12764. doi: 10.1128/JVI.02138-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liao Y., Liu D.X. Regulation of stress responses and translational control by coronavirus. Viruses. 2016 Jul 4;8(12):184. doi: 10.3390/v8070184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya N., Yu J., Byfield M., Pattingre S., Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005 Apr;1(1):46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- Geng J., Klionsky D.J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’’ review series. EMBO Rep. 2008 Sep;9(9):859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton N.S., Randall G. Dengue virus and autophagy. Viruses. 2011 Aug 4;3(8):1332–1341. doi: 10.3390/v3081332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke P.-Y., Chen S.S.-L. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J. Clin. Investig. 2011 Jan;121(1):37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Lee S., Jung J.U. When autophagy meets viruses: a double-edged sword with functions in defense and offense. Semin. Immunopathol. 2010 Dec;32(4):323–341. doi: 10.1007/s00281-010-0226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007 Sep;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Van Den Worm S.H.E., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008 Sep 16;6(9):e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Van Den Worm S.H.E., Zevenhoven-Dobbe J.C., Van Der Meer Y., Koster A.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Mariño G., Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010 Oct 22;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.Q., Xiao H., Tam J.P., Liu D.X. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005 Apr 25;579(11):2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999 Dec 9;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liang J.Q., Fang S., Yuan Q., Huang M., Chen R.A., Fung T.S. N-Linked glycosylation of the membrane protein ectodomain regulates infectious bronchitis virus-induced ER stress response, apoptosis and pathogenesis. Virology. 2019 May;531:48–56. doi: 10.1016/j.virol.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Fung T.S., Huang M., Fang S.G., Zhong Y., Liu D.X. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 2013 Jul 15;87(14):8124–8134. doi: 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y., Ng Y., Tam J., Liu D.X. Human coronaviruses: a review of virus–host interactions. Diseases. 2016 Jul 25;4(4):26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991 Dec;185(2):911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Hawes P.C., Cottam E.M., Mantell J., Verkade P., Monaghan P. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio. 2013;4(5):e00801–e00813. doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri M.C., Zalckvar E., Kimchi A., Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007 Sep;8(9):741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Man N., Chen Y., Zheng F., Zhou W., Wen L.-P. Induction of genuine autophagy by cationic lipids in mammalian cells. Autophagy. 2010 May;6(4):449–454. doi: 10.4161/auto.6.4.11612. [DOI] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D. A protein conjugation system essential for autophagy. Nature. 1998 Sep 24;395(6700):395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008 Feb 28;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.F., Liu D.X. Identification of a 24-kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology. 1998 Apr 10;243(2):388–395. doi: 10.1006/viro.1998.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell Biol. 2006 Dec;26(24):9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier-Denis E., Pattingre S., El Benna J., Codogno P. Erk1/2-dependent phosphorylation of Galpha-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J. Biol. Chem. 2000 Dec 15;275(50):39090–39095. doi: 10.1074/jbc.M006198200. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001 Mar;2(3):211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P., te Lintelo E.G., Rottier P.J. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 2008;82(24):12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010 Jun 25;7(6):500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., He X., Zhu G., Tu H., Liu Z., Li W. Coxsackievirus A16 elicits incomplete autophagy involving the mTOR and ERK pathways. PLoS One. 2015 Apr 8;10(4) doi: 10.1371/journal.pone.0122109. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4390341/ cited 2017 Jul 12];10(4). Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprasad U., Basu A. Inhibition of ERK attenuates autophagy and potentiates tumour necrosis factor-alpha-induced cell death in MCF-7 cells. J. Cell Mol. Med. 2008 Aug;12(4):1265–1271. doi: 10.1111/j.1582-4934.2008.00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J.M., van der Meulen J., Koerten H.K. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006 Jun;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001 Nov 1;20(21):5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H.P. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000 Jan 28;287(5453):664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Wong J., Zhang J., Si X., Gao G., Mao I., McManus B.M. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 2008 Sep;82(18):9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010 Sep;12(9):814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007 Dec;3(6):581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- Zhu B., Zhou Y., Xu F., Shuai J., Li X., Fang W. Porcine circovirus type 2 induces autophagy via the AMPK/ERK/TSC2/mTOR signaling pathway in PK-15 cells. J. Virol. 2012 Nov;86(22):12003–12012. doi: 10.1128/JVI.01434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Mou C., Yang X., Lin J., Yang Q. Mitophagy in TGEV infection counteracts oxidative stress and apoptosis. Oncotarget. 2016 May 10;7(19):27122–27141. doi: 10.18632/oncotarget.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]