Abstract
Although rodents are important reservoirs for RNA viruses, to date only one species of rodent coronavirus (CoV) has been identified. Herein, we describe a new CoV, denoted Lucheng Rn rat coronavirus (LRNV), and novel variants of two Betacoronavirus species termed Longquan Aa mouse coronavirus (LAMV) and Longquan Rl rat coronavirus (LRLV), that were identified in a survey of 1465 rodents sampled in China during 2011–2013. Phylogenetic analysis revealed that LAMV and LRLV fell into lineage A of the genus Betacoronavirus, which included CoVs discovered in humans and domestic and wild animals. In contrast, LRNV harbored by Rattus norvegicus formed a distinct lineage within the genus Alphacoronavirus in the 3CLpro, RdRp, and Hel gene trees, but formed a more divergent lineage in the N and S gene trees, indicative of a recombinant origin. Additional recombination events were identified in LRLV. Together, these data suggest that rodents may carry additional unrecognized CoVs.
Keywords: Coronavirus, Evolution, Phylogeny, Rodents, Recombination
Highlights
-
•
A novel coronavirus (CoV) was identified in rodents from China.
-
•
Three species of CoVs are co-circulating in rodents in Zhejiang province, China.
-
•
Recombination events were observed in the newly identified rodent CoVs.
Introduction
Coronaviruses (CoVs; family Coronaviridae) are the etiological agent(s) of respiratory, enteric, hepatic, and neurological diseases in animals and humans. The first coronavirus (infectious bronchitis virus) was isolated in chicken embryos in 1937 (Beaudette and Hudson, 1937), with subsequent viral isolations in rodents, domestic animals, and humans. However, until the emergence of severe acute respiratory syndrome (SARS) in China in 2002/3 (Drosten et al., 2003, Woo et al., 2009), coronaviruses had been of greater concern to agriculture than public health. Since the discovery of SARS-CoV intense scientific efforts have been directed toward characterizing additional coronaviruses in humans and other animals (Drexler et al., 2010, Guan et al., 2003, Lau et al., 2005, Li et al., 2005, Quan et al., 2010, van der Hoek et al., 2004, Woo et al., 2012). As a consequence, the number of coronaviruses identified has increased rapidly (Woo et al., 2009, Woo et al., 2012). Of particular importance was the recent discovery of a new severe respiratory illness with renal failure (Middle East Respiratory Syndrome, MERS) caused by a novel coronavirus (MERS-CoV) (Bermingham et al., 2012, van Boheemen et al., 2012), and which is also a zoonosis (Annan et al., 2013, Azhar et al., 2014, Reusken et al., 2013). It is highly likely that there are additional unrecognized coronaviruses circulating in animals.
Rodentia (rodents) is the largest order of mammals with approximately 2277 species worldwide, representing some 42% of all mammalian species (Wilson and Reeder, 2005). Rodents are a major zoonotic source of human infectious diseases (Meerburg et al., 2009, Luis et al., 2013), particularly as they often live at high densities and hence may harbor high levels of microbial diversity (Moya et al., 2004). In addition, some rodent species live in close proximity to humans, such that they represent an important zoonotic risk. To date, however, only one species of coronavirus – Murine coronavirus – has been associated with rodents (de Groot et al., 2011). The prototype virus, which was named mouse hepatitis virus (MHV), was first isolated in mice in 1949 (Cheever et al., 1949), with a variant then identified in rats in 1970 (where it was termed rat sialodacryoadenitis coronavirus) (Parker et al., 1970). No other rodent-associated CoVs have been discovered since this time.
Although RNA viruses are often characterized by their high rates of mutation, recombination may also be of evolutionary importance, and has been associated with such characteristics as the ability to infect new hosts and alter virulence (Holmes, 2013). Recombination appears to be commonplace in coronaviruses (Graham and Baric, 2010, Jackwood et al., 2012, Keck et al., 1987, Woo et al., 2006), and which may facilitate their emergence. For example, two types of feline CoVs (FCoV) – FCoV type I and II – have arisen by double recombination events between FCoV types I and canine Coronavirus (CCoV) (Herrewegh et al., 1998). Similarly, recombination generated the three genotypes (A, B and, C) of human coronavirus HKU1 (Woo et al., 2006), and homologous recombination has occurred in the evolutionary history of SARS-CoV (Graham and Baric, 2010).
To explore the diversity and evolution of CoVs in rodent populations we screened rodents collected from rural regions of Zhejiang province, China. This revealed a remarkable diversity of CoVs circulating in rodents, along with evidence for cross-species transmission and recombination.
Results
Collection of rodents, and the identification of coronaviruses
A total of 1465 rodents representing 10 different species were captured from three locations in Zhejiang province, China during 2011–2013 ( Table 1 and Fig. 1). RT-PCR targeting a conserved sequence of the viral RdRp (RNA-dependent RNA polymerase) gene was performed to detect coronaviruses. PCR products of the expected size were recovered from 10 Apodemus agrarius, 4 Rattus norvegicus, 14 R. lossea, 1 R. tanezumi, and 1 Niviventer confucianus, such that approximately 2% of rodents were positive for CoV (Table 1). The classification of these viruses as CoVs (Family Coronaviridae, Genus Alpha- and Beta-) was confirmed by genetic analyses (see below).
Table 1.
Prevalence of coronaviruses in rodents in Zhejiang Province, China.
| Species | Longquan |
Wencheng |
Lucheng | Total | ||
|---|---|---|---|---|---|---|
| Residential | field | Residential | field | Residential | ||
| Apodemus agrarius | – | 10/427 | – | 0/17 | – | 10/444 |
| Mus musculus | 0/3 | – | 0/4 | – | – | 0/7 |
| Microtus fortis | 0/44 | 0/261 | – | – | – | 0/305 |
| Micromys minutus | – | 0/2 | – | – | – | 0/2 |
| Niviventer confucianus | – | 1/58 | – | 0/27 | – | 1/85 |
| Rattus norvegicus | 3/214 | – | 0/31 | – | 1/17 | 4/262 |
| R. lossea | – | 14/300 | – | 0/1 | – | 14/301 |
| R. tanezumi | 0/25 | 1/7 | 0/18 | – | 0/3 | 1/53 |
| R. fulvescens | 0/1 | 0/3 | – | – | – | 0/4 |
| R. edwardsi | – | – | – | 0/2 | – | 0/2 |
| Total | 3/287 | 26/1058 | 0/53 | 0/47 | 1/20 | 30/1465 |
Note: CoV RNA positive specimens/total specimens; “–” no animals were captured.
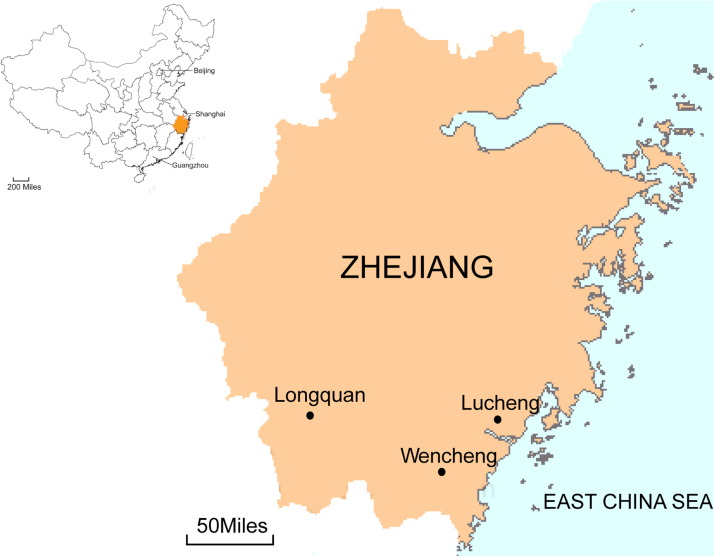
Fig. 1.
A map of Zhejiang province, China showing the location of trap sites in which rodents were captured and surveyed for coronaviruses.
Genetic characterization of viral sequences
To better characterize the rodent CoVs discovered here, complete viral RdRp gene sequences were recovered from 21 (70%) of the RNA positive rodent samples described above. Additionally, 1 complete and 4 near complete (>98%) viral genome sequences were successfully recovered from five positive CoV samples (Table S1). Genetic analysis indicated that two CoVs sampled from R. norvegicus in Lucheng and Longquan shared 50.6%–71.7% nucleotide sequence similarity with alpha coronaviruses; 15 CoVs from 13 R. lossea, 1 R. tanezumi, and 1 N. confucianus from Longquan had 78.4%–89.5% nucleotide sequence similarity with murine coronavirus and 76.4%–85.7% nucleotide sequence similarity with human coronavirus HKU1; and 13 CoVs from 10 A. agrarius, 2 R. norvegicus and 1 R. lossea from Longquan had 60.3%–85.9% nucleotide sequence similarity with rabbit coronavirus HKU14, isolated from domestic rabbits in Guangzhou, China (Lau et al., 2012) (see phylogenetic results below). Overall, we designated these newly described viruses as Longquan Aa mouse coronavirus (LAMV), Longquan Rl rat coronavirus (LRLV), and Lucheng Rn rat coronavirus (LRNV), reflecting their host species and the geographic location of sampling.
Further comparison of the CoV replicase domains [i.e. ADP-ribose 1″-phosphatase (ADRP), chymotrypsin-like protease (3CLpro), RdRp, helicase (Hel), 3′-to-5′ exonuclease (ExoN), nidoviral endoribonuclease specific for uridylate (NendoU) and ribose-2′-O-methyltransferase (O-MT)] revealed that LRNV Lucheng-19 was <90% similar in amino acid sequence to known members of the genus Alphacoronavirus (Table S2). Hence, these data suggest that LRNV is sufficiently divergent that it represents a novel species of coronaviruses according to the criteria for species demarcation in the subfamily Coronavirinae defined by the International Committee on Taxomony of Viruses (ICTV) (de Groot et al., 2011). With respect to the other two viruses, LRLV Longquan-189 was <90% similar to known members of the genus Betacoronavirus in the ADRP and NendoU regions, suggesting that it represents a new variant of murine coronavirus (see phylogenetic analysis). Although LAMV Longquan-343 was nearly 90% similar in the conserved replicase domains to betacoronavirus 1 (i.e. Human coronavirus OC43, Bovine coronavirus, Porcine hemagglutinating encephalomyelitis virus, and Rabbit coronavirus HKU14) and <90% to other members of the genus Betacoronavirus (Table S2), it does not exhibit sufficient sequence divergence to represent a new virus species. Hence, we classify it as a new variant of Betacoronavirus 1.
The viral genome sequences obtained in this study were compared with Rhinolophus bat coronavirus HKU2 virus, a member of the genus Alphacoronavirus (Lau et al., 2007), and MHV (Cheever et al., 1949), human coronavirus HKU1 (Woo et al., 2006), rabbit coronavirus HKU14 (Lau et al., 2012), which are all members of the genus Betacoronavirus, ( Fig. 2). A previous description of the genome organization of CoVs (de Groot et al., 2011) was used as a reference. LRNV (Lucheng-19) had a genome of 28,763 nucleotides, with a G+C content of 40.2%. Its genome organization was similar to those of members of the genus Alphacoronavirus, with the characteristic 5′-replicase ORF1ab-S-envelope(E)- membrane(M)-N-3′ gene order (Fig. 2, Table 2, Appendix A). The replicase ORF1ab (20,249 nucleotides in length) includes 16 predicted nonstructural proteins (nsp) (Table S3). In a manner similar to Rhinolophus bat coronavirus HKU2 and human coronavirus NL63, LRNV possessed the core part of the putative transcription regulatory sequence (TRS) 5′-AACUAA-3′ upstream of the 5′ end of each ORF with the exception NS4, with variable nucleotides matching the leader core sequences (Table 2). Additionally, LRNV contained NS7a and NS7b genes between the M and N genes (Fig. 2), and which are observed in no other members of the genus Alphacoronavirus. Perhaps the most striking feature of the LRNV genome was that NS2 encodes a putative nonstructural protein of 275 amino acids located between the replicase ORF1ab and the S gene (Fig. 2, Table 2). A BLAST search revealed that this NS2 had no amino acid sequence similarity with alpha-CoVs, but possessed approximately 42% amino acid identity with the NS2a of lineage A of beta-CoVs. Hence, this is suggestive of a homologous recombination event between alpha-CoVs and beta-CoVs (and which was confirmed in the phylogenetic analysis below).
Fig. 2.
Genome organization of coronaviruses. The three CoVs discovered in this study are shown in bold. The star signifies the presence of a betacoronavirus-like NS2a gene in LRNV.
Table 2.
Coding of potential and putative transcription regulatory sequences of the LRNV (Lucheng-19) genome sequence.
| ORF | Location (nt) | Length (nt) | Length (aa) | TRS location | No. of matching base pairs compared to leader TRS (body/leader) | TRS sequence (s) (distance in bases to AUG) |
|---|---|---|---|---|---|---|
| 1ab | 332–20,580 (shift at 12,538) | 20,249 | 6749 | 65 | CAACUCAACUAAACGA(251)AUG | |
| NS2 | 20,577–21,404 | 828 | 275 | 20,565 | 11/12 | CAACUUAACUAAAUG |
| S | 21,412–24,816 | 3405 | 1134 | 21,398 | 11/14 | UGACUAAACUAAACAUG |
| NS4 | 24813–25457 | 645 | 214 | |||
| E | 25,457–25,693 | 237 | 78 | 25,446 | 10/12 | CCACUUAACUAAUG |
| M | 25,703–26,449 | 747 | 248 | 25,690 | 9/13 | UUGAUCAACUAAAAUG |
| NS7a | 26,461–26,958 | 498 | 165 | 26,443 | 11/16 | GGUCUAAACUAAACCA(2)AUG |
| NS7b | 26,555–26,926 | 372 | 123 | 26,511 | 9/16 | CAUUAAAACUAAUUGU(28)AUG |
| N | 26,974–28,149 | 1176 | 391 | 26,960 | 8/14 | AGUUUCAACUAACAAUG |
| NS8a | 27,607–28,149 | 543 | 180 | 27,555 | 9/16 | UGAUAGAACUAAAGAA(36)AUG |
| NS9 | 28,151–28,465 | 315 | 104 | 28,134 | 9/16 | UGAUGAAACUAAUUGA(1)AUG |
Numbers in parentheses represent the number of nucleotides to the putative start codon. Start codons are underlined. The conserved TRS core sequence, AACUAA, is highlighted in bold.
In contrast, the genome organization of LRLV and LAMV were similar to those of members of the lineage A CoVs of the genus Betacoronavirus, with the characteristic 5′-ORF1ab-hemagglutinin-esterase (HE)-S-E-M-N-3′ gene order (Fig. 2). In both LAMV and LRLV the NS5a and NS5b nonstructural proteins were located between the S and E genes. Interestingly, however, NS5a was not observed in the strain Longquan-370 (LRLV). The TRS of LAMV and LRLV appeared in two forms – the CUAAAC and CCAAAC type. The S protein of LRLV were 1350–1366 amino acids in length, with 65%–89% amino acid identity to the S proteins of other lineage A CoVs of the genus Betacoronavirus, while the S protein of LAMV were 1358 amino acids in length with 62%–68% amino acid identity to other lineage A CoVs of the genus Betacoronavirus. Finally, the S proteins of both LRLV and LAMV contain a potential signal peptide, receptor binding domain, a potential S1 and S2 cleavage site, two heptad repeats and one transmembrane domain ( Table 3).
Table 3.
Characteristics of the spike protein in LRLV and LAMV.
| Spike protein | Strain | Signal peptide | Receptor binding domain | Cleavage site | Heptad repeat | Transmembrane domain |
|---|---|---|---|---|---|---|
| LRLV | Longquan-708 | 1–14 | 325–533; 586–669 | 754–755 | 1009–1122; 1257–1288 | 1307–1329 |
| Longquan-370 | 1–13 | 319–525 | 762–763 | 996–1109; 1244–1275 | 1294–1316 | |
| Longquan-189 | 1–13 | 319–525 | 762–763 | 996–1109; 1244–1275 | 1294–1316 | |
| LAMV | Longquan-343 | 1–15 | 327–497 | 776–777 | 1004–1117; 1252–1290 | 1302–1324 |
Phylogenetic relationships among the CoVs
To determine the evolutionary relationships among the novel CoVs discovered here and those found previously, we inferred phylogenetic trees based on the amino acid sequences of the 3CLpro, RdRp, Hel, S and N proteins ( Fig. 3, Fig. 4). Consistent with previous work (de Groot et al., 2011), all CoVs fell into two well supported groups, corresponding to the Alphacoronavirus and Betacoronavirus genera respectively. With respect to the viruses identified here, LRNV clustered within the genus Alphacoronavirus, while LAMV and LRLV fell into the genus Betacoronavirus.
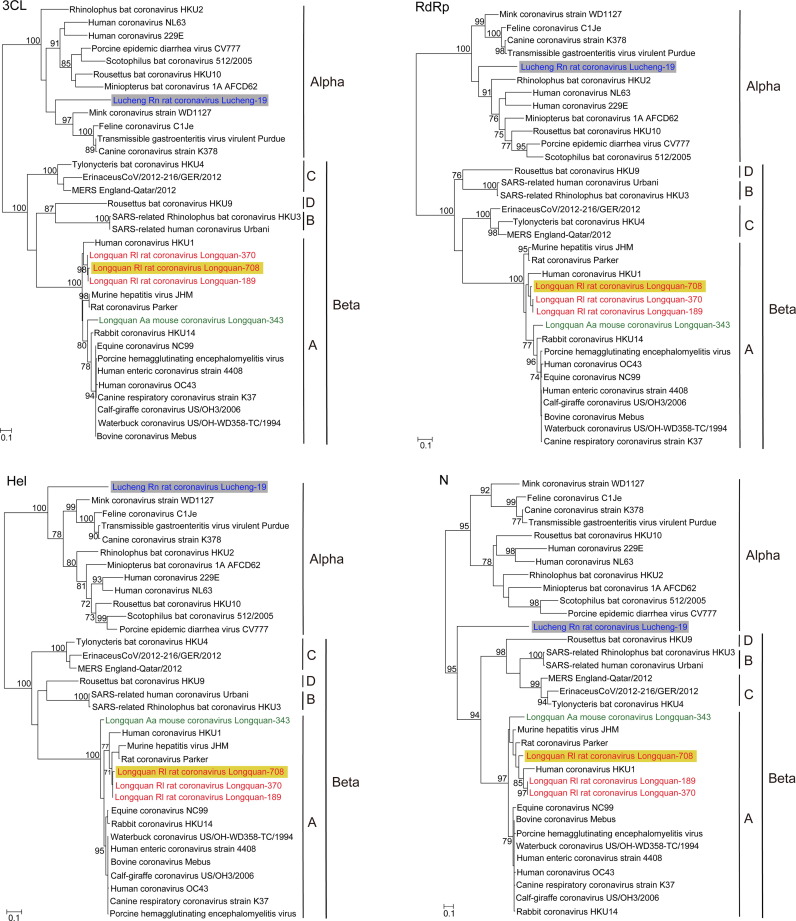
Fig. 3.
Phylogenetic analyses of the amino acid sequences of the 3CLpro, RdRp, Hel, and N genes of Lucheng Rn rat coronavirus (LRNV) Lucheng-19, Longquan Aa mouse coronavirus (LAMV) Longquan-343, and Longquan Rl rat coronaviruses (LRLV) Longquan-708, Longquan-189, and Longquan-370. Numbers (>70) above or below branches indicate percentage bootstrap values. The trees were mid-point rooted for clarity only. The scale bar represents the number of amino acid substitutions per site. The GenBank accession numbers of the viruses used in this analysis are shown in Table S1.
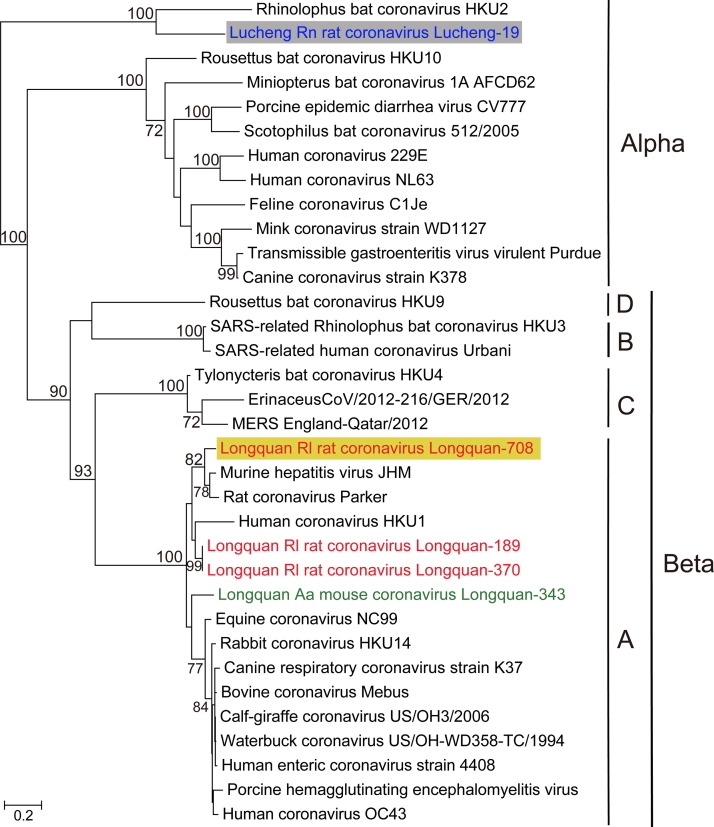
Fig. 4.
Phylogenetic analyses of the amino acid sequences of the S genes of LRNV, LAMV and LRLV. Numbers (>70) above or below branches indicate percentage bootstrap values. The trees were mid-point rooted for clarity only. The scale bar represents the number of amino acid substitutions per site. The GenBank accession numbers of other CoVs are given in Table S1.
One of the most striking observations from the phylogenetic analysis was that the sequences from LRNV were located at five different positions in the phylogenetic trees, strongly suggestive of recombination (Fig. 3, Fig. 4). Specifically, in the 3CLpro and RdRp gene trees, LRNV clustered as a member of the genus Alphacoronavirus. LRNV also fell with alphacoronaviruses in the Hel genes tree, but as a basal lineage. Strikingly, however, LRNV clustered with the betacoronaviruses in the N gene tree, and formed a divergent lineage in S gene tree with Rhinolophus bat coronavirus HKU2, which has previously been shown to be a recombinant (Lau et al., 2007).
In contrast, LAMV and LRLV consistently clustered within the lineage A of the genus Betacoronavirus, which also contained human CoV HKU1, MHV, rabbit coronavirus HKU14, and human CoV OC43 viruses. Notably, however, in the 3CLpro, RdRp, and N gene trees (Fig. 3), two strains (Longquan-189 and Longquan-370) formed a monophyletic group with human coronavirus HKU1 virus, which was isolated from a patient with pneumonia in Hong Kong in 2005 (Woo et al., 2005). Interestingly, Longquan-708 was closely related to Longquan-370 and Longquan-189 in the 3CLpro, RdRp, and Hel genes, yet clustered with MHV and rat coronavirus in the S gene tree (Fig. 4), and represented a distinct lineage in the N gene tree (Fig. 3). Such variable grouping suggests that Longquan-708 may be a recombinant between two rodent CoV lineages. Finally, in the 3CLpro, RdRp, and S gene trees, LAMV (Longquan-343) occupied the most divergent phylogenetic position in a group of viruses that contained human CoV OC43 (St-Jean et al., 2004) as well as viruses sampled from domestic and wild animals (Hasoksuz et al., 2007, Lau et al., 2012, Lim et al., 2013, Vijgen et al., 2006). However, LAMV comprised a distinct lineage in the Hel and N gene trees.
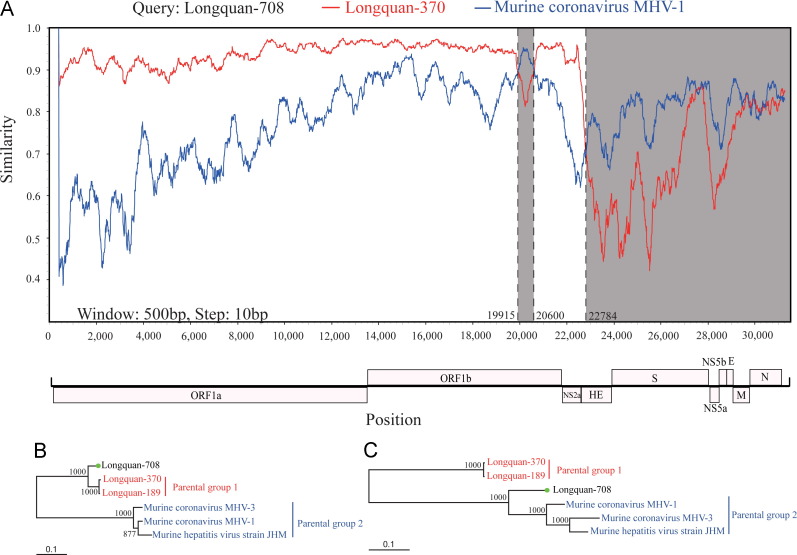
To investigate these putative recombination events in more detail, we undertook additional sequence analyses ( Fig. 5 and Fig. S1). Multiple methods within the RDP program supported statistically significant recombination events in LRLV strain Longquan-708 (p<1.022−146 to p<5.908−17). Similarity plots suggested the presence of three recombination breakpoints at nucleotide positions 19,294, 19,979, and 22,112, which separated the genome into four regions (Fig. 5A). In turn, these could be grouped into two putative ‘parental regions’; region A (nt 1 to 19,915 and 20,600 to 22,784) and region B (19,915 to 20,600 and 22,784 to the end of the sequence). In parental region A, Longquan-708 was most closely related to Longquan Rl Rat coronaviruses, while in parental region B it was more closely related to MHV. This recombination event was confirmed by phylogenetic analyses, in which the alternative grouping of Longquan-708 was supported with high bootstrap values (Fig. 5B and C).
Fig. 5.
Recombination within the genome of LRLV Longquan-708. A sequence similarity plot (A) reveals three recombination break-points shown by black dashed lines, with their locations indicated at the bottom. The plot shows genome scale similarity comparisons of the Longquan-708 (query) against Longquan Rl Rat Coronavirus (parental group 1, red) and Murine hepatitis virus (parental group 2, blue). The background color of parental region A is white, while that of parental region B is gray. Phylogenies of parental region A (B) and parental region B (C) are shown below the similarity plot. Numbers (>70) above or below branches indicate percentage bootstrap values. The GenBank accession numbers of the viruses used in this analysis are shown in Table S1.
In contrast, although readily apparent in the amino acid phylogenies, the recombination event involving LRNV did not receive significant statistical support in the RDP analysis, likely because the latter utilizes nucleotide sequences and these are highly divergent (for example, the S protein of LRNV differs from those of alphacoronaviruses by >78% at the nucleotide sequence analysis). Similar suggestions have previously been made with respect to recombination in Rhinolophus bat coronavirus HKU2 (Lau et al., 2007).
Discussion
We screened for coronaviruses in 1465 rodents representing 10 different species sampled in three locations in Zhejiang province, southeastern China. This survey identified a novel and phylogenetically distinct coronavirus in R. norvegicus – Lucheng Rn rat coronavirus (LRNV) – which belonged to the genus Alphacoronavirus. According to the criteria defined by ICTV (de Groot et al., 2011), LRNV was sufficiently genetically distinct that it should be recognized as a distinct species within the family Coronaviridae. However, the other two viruses identified – Longquan Aa mouse coronavirus (LAMV) and Longquan Rl rat coronavirus (LRLV) – belong to the established species betacoronavirus 1 and murine coronavirus, respectively. More generally, the presence of all three viruses indicates that genetically diverse CoVs co-circulate in rodents in Zhejiang province.
It is notable that rodent-associated CoVs comprise a major proportion of the known genetic diversity in lineage A CoVs of the genus Betacoronavirus. This lineage contains viruses that cause enteric and respiratory diseases in humans (human coronavirus HKU1 and OC43) as well as in domestic animals (e.g. hemagglutinating encephalomyelitis in pigs) (St-Jean et al., 2004, Vijgen et al., 2006, Woo et al., 2005, Zhang et al., 1994). Clearly, the role by rodents in the evolution of lineage A CoVs of the genus Betacoronavirus merits further investigation.
This study also provides the first evidence of CoVs of the genus Alphacoronavirus in Rattus rats (R. norvegicus), in the form of LRNV. However, our phylogenetic analysis suggested that this virus had a recombinant origin, with its N gene sequence more closely related to those of the genus Betacoronavirus (Fig. 4). Recombination appears to be commonplace in coronaviruses (Woo et al., 2009), particularly within closely related viruses such as MHV variants (Smits et al., 2005). Nevertheless, only a few examples of inter-genotype recombination, involving CoVs from bats (Hon et al., 2008) and felines (Herrewegh et al., 1998), have been documented to date. Hence, the observation that LRNV has a recombinant origin is significant because it means that recombination can occur between viruses assigned to different genera.
Alpha- and beta-CoVs are largely associated with mammals, whereas gamma- and delta-CoVs are largely harbored by avian species (Woo et al., 2012). Because much of the genetic diversity of alpha- and beta-CoVs is associated with infections in bats, it has been suggested that bats are the main reservoir hosts for both alpha- and beta- CoVs (Woo et al., 2009). Herein, we discovered three phylogenetically distinct lineages of rodent-associated CoVs within a limited geographic area in China, all of which are distinct from those viruses associated in bats. Consequently, it is clear that large-scale surveillance is needed to fully understand the role played by rodents in the evolution and emergence of coronaviruses.
Material and methods
Ethics statement
This study was reviewed and approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention of the Chinese CDC. All animals were treated strictly according to the guidelines for the Laboratory Animal Use and Care from the Chinese CDC and the Rules for the Implementation of Laboratory Animal Medicine (1998) from the Ministry of Health, China, under the protocols approved by the National Institute for Communicable Disease Control and Prevention. All surgery was performed under ether anesthesia, and all efforts were made to minimize suffering.
Specimen collection
Rodents were trapped in cages using cooked food as bait during 2011–2013 in Zhejiang province, China (Fig. 1) (Mills et al., 1995). All animals were initially classified to a specific rodent species by morphological examination, and were further confirmed by sequence analysis of the mt-cyt b gene (Guo et al., 2013). All animals were anesthetized with ether before they were sacrificed, and every effort was made to minimize suffering. Tissue samples of liver, spleen, lung, kidney, and rectum were collected from animals for the detection of CoVs.
CoV detection and full genome sequencing
Total RNA was extracted from fecal or tissue samples using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer׳s instructions. The RNA was eluted with RNase-free water and was used as the template for reverse transcription-PCR (RT-PCR) and deep sequencing. CoV RNA was detected by nested RT-PCR which amplified the RNA-dependent RNA polymerase gene (RdRp) of CoVs using conserved primers (sequence available on request). Reverse transcription was undertaken using AMV reverse transcriptase (Promega, Beijing) according to the manufacturer׳s protocol. The cDNA was amplified with the following PCR protocol: 35 cycles of demodulation at 94 °C for 40 s, annealing at 44 °C for 40 s and extending at 72 °C for 40 s, with ddH2O as a negative control. For CoV positive RNA extractions, pair-end (90 bp) sequencing was performed on the HiSeq 2000 (Illumina) platform. The library preparation and sequencing steps were performed by the BGI Tech Corporation (Shenzhen, China) following a standard protocol provided by Illumina. The resulting sequencing reads were then assembled de novo by the Trinity program (Grabherr et al., 2011) into 152,684 contigs (>200 bp). BLASTx was performed to retrieve the CoV full genome sequences from the assembled contigs. These sequences were further verified using Sanger sequencing methods with primers designed based on the deep-sequencing results. To amplify the terminal ends, 3′ and 5′ RACE kits (TaKaRa, Dalian, China) were used.
Nucleotide sequence accession numbers
The sequences generated in this study have been deposited in GenBank and assigned accession numbers KF294379-KF294380, KF294358-KF294372, KF294345-KF294357 for those representing coronaviruses, and KF294387-KF294416 for the host mt-cyt b genes (Table S1).
Evolutionary analyses
Because of extensive sequence divergence between the nucleotide sequences of different coronavirus genera, all phylogenetic analyses were based on amino acid sequences. Accordingly, amino acid sequence alignments were performed using the MAFFT algorithm (Katoh and Standley, 2013). After alignment, gaps and ambiguously aligned regions were removed with Gblocks (v0.91b) (Talavera and Castresana, 2007). Phylogenetic analyses were then performed using the sequences of five CoV proteins: (i) 3CLpro, (ii) RdRp, (iii) Hel, (iv) spike protein (S), and (v) the nucleocapsid protein (N). Phylogenetic trees were estimated using the maximum likelihood (ML) method implemented in PhyML v3.0 (Guindon et al., 2010) with bootstrap support values calculated from 1000 replicate trees. The best-fit amino acid substitution models (LG+Γ for 3CLpro, LG+Γ+I for Hel, RdRp, S and N) were determined using MEGA version 5 (Tamura et al., 2011). The following data set sizes were used in the final analysis: 3CLpro=290 amino acids (aa), RdRp=869 aa, Hel=581 aa, S=429 aa, N=138 aa.
The TMHMM program (version 2.0; www.cbs.dtu.dk/services/TMHMM/) was used to predict the transmembrane domains, while the Signal P program (version 4.0; http://www.cbs.dtu.dk/services/ SignalP/) was to determine signal sequences. Protein family analysis was performed using PFAM and InterProScan (Apweiler et al., 2001, Bateman et al., 2002).
Following visual inspection of the amino acid phylogenies, potential recombination events were identified in complete genome (nucleotide) sequences using the Recombination Detection Program v4 (RDP4), employing the RDP, GENECONV, bootscan, maximum chi square, Chimera, SISCAN, and 3SEQ methods (Martin et al., 2010) (with default parameters). All analyses were performed with a Bonferroni corrected P-value cutoff of 0.01. When putative recombination events were observed by two or more methods and with significant phylogenetic (topological) incongruence, the viral sequences were considered as potentially recombinant. To further characterize these recombination events, particularly the location of breakpoints, we inferred similarity plots using Simplot version 3.5.1 (Lole et al., 1999). For each of the putative recombinant regions, phylogenies were estimated using the ML method performed with PhyML v3.0 (Guindon et al., 2010) under the best-fit substitution model determined by jModelTest (Posada, 2008).
Acknowledgments
This study was supported by the 12th Five-Year Major National Science and Technology Projects of China (2014ZX10004001-005), National Natural Science Foundation of China (Grants 81290343, 81273014). ECH is funded by an NHMRC Australia Fellowship (AF30). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2014.10.017.
Appendix A. Supplementary materials
Supplementary data Fig. S1 further illustrate recombination events among the novel CoVs described here, phylogenetic analyses were performed based on the nucleotide sequences of the RdRp, S and N genes of LRNV, LAMV and LRLV. Numbers (>70) above or below branches indicate percentage bootstrap values. The trees were mid-point rooted for clarity only. The scale bar represents the number of nucleotide substitutions per site. Putative recombinant lineages are shaded with gray and yellow, respectively. Schematic genetic exchanges of parental genomes and of the recombinant offspring in different colors are shown below the trees. The GenBank accession numbers of other CoVs are given in Table S1.
Supplementary data
Supplementary data
Supplementary data
References
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K., Lina P.H., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human Betacoronavirus 2c EMC/2012-related viruses in bats. Ghana and Eur. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., Durbin R., Falquet L., Fleischmann W., Gouzy J., Hermjakob H., Hulo N., Jonassen I., Kahn D., Kanapin A., Karavidopoulou Y., Lopez R., Marx B., Mulder N.J., Oinn T.M., Pagni M., Servant F., Sigrist C.J., Zdobnov E.M. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucl. Acids. Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L. The Pfam protein families database. Nucl. Acids. Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudette F.R., Hudson C.B. Cultivation of the virus of infectious bronchitis. J. Am. Vet. Med. Assoc. 1937;90:51–58. [Google Scholar]
- Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R., Pebody R.G., Green H.K., Boddington N.L., Gopal R., Price N., Newsholme W., Drosten C., Fouchier R.A., Zambon M. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro. Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- Cheever F.S., Daniels J.B., Pappenheimer A.M., Bailey O.T. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin. J. Exp. Med. 1949;90:181–194. doi: 10.1084/jem.90.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. Family Coronaviridae. In: King A.M.Q., Lefkowitz E., Adams M.J., Carstens E.B., editors. Virus Taxonomy: 9th report of the International Committee on Taxonomy of Viruses. Elsevier, CA; San Diego: 2011. pp. 806–828. [Google Scholar]
- Drexler J.F., Gloza-Rausch F., Glende J., Corman V.M., Muth D., Goettsche M., Seebens A., Niedrig M., Pfefferle S., Yordanov S., Zhelyazkov L., Hermanns U., Vallo P., Lukashev A., Muller M.A., Deng H., Herrler G., Drosten C. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J. Virol. 2010;84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guo W.P., Lin X.D., Wang W., Tian J.H., Cong M.L., Zhang H.L., Wang M.R., Zhou R.H., Wang J.B., Li M.H., Xu J., Holmes E.C., Zhang Y.Z. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013;9:e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasoksuz M., Alekseev K., Vlasova A., Zhang X., Spiro D., Halpin R., Wang S., Ghedin E., Saif L.J. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J. Virol. 2007;81:4981–4990. doi: 10.1128/JVI.02361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A., Smeenk I., Horzinek M.C., Rottier P.J., de Groot R.J. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C. Virus evolution. In: Fields B.N., Knipe D.M., Howley P.M., editors. Fields Virology. 6th ed. Wolters Kluwer/Lippincott Williams & Wilkins Health; Philadelphia, PA: 2013. pp. 286–313. (pp.) [Google Scholar]
- Hon C.C., Lam T.Y., Shi Z.L., Drummond A.J., Yip C.W., Zeng F., Lam P.Y., Leung F.C. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hall D., Handel A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012;12:1305–1311. doi: 10.1016/j.meegid.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck J.G., Stohlman S.A., Soe L.H., Makino S., Lai M.M. Multiple recombination sites at the 5′-end of murine coronavirus RNA. Virology. 1987;156:331–341. doi: 10.1016/0042-6822(87)90413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Wang M., Lam C.S., Xu H., Guo R., Chan K.H., Zheng B.J., Yuen K.Y. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Yip C.C., Fan R.Y., Huang Y., Wang M., Guo R., Lam C.S., Tsang A.K., Lai K.K., Chan K.H., Che X.Y., Zheng B.J., Yuen K.Y. Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J. Virol. 2012;86:5481–5496. doi: 10.1128/JVI.06927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lim S.I., Choi S., Lim J.A., Jeoung H.Y., Song J.Y., Dela Pena R.C., An D.J. Complete genome analysis of canine respiratory coronavirus. Genome Announc. 2013;1:e00093–e000112. doi: 10.1128/genomeA.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis A.D., Hayman D.T., O’Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R., Mills J.N., Timonin M.E., Willis C.K., Cunningham A.A., Fooks A.R., Rupprecht C.E., Wood J.L., Webb C.T. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. Biol. Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerburg B.G., Singleton G.R., Kijlstra A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009;35:221–270. doi: 10.1080/10408410902989837. [DOI] [PubMed] [Google Scholar]
- Mills J.N., Childs J.E., Ksiazek T.G., Peters C.J., Velleca W.M. Methods For Trapping And Sampling Small Mammals For Virologic Testing. Centers for Disease Control and Prevention; Atlanta: 1995. pp. 15–18. [Google Scholar]
- Moya A., Holmes E.C., Gonzalez-Candelas F. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2004;2:279–288. doi: 10.1038/nrmicro863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.C., Cross S.S., Rowe W.P. Rat coronavirus (RCV): a prevalent, naturally occurring pneumotropic virus of rats. Arch. Gesamte Virusforsch. 1970;31:293–302. doi: 10.1007/BF01253764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Quan P.L., Firth C., Street C., Henriquez J.A., Petrosov A., Tashmukhamedova A., Hutchison S.K., Egholm M., Osinubi M.O., Niezgoda M., Ogunkoya A.B., Briese T., Rupprecht C.E., Lipkin W.I. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. MBio. 2010;1:e00208–e00210. doi: 10.1128/mBio.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Müller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Smits-De Vries L., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortázar-Schmidt C., Drosten C., Koopmans M.P. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Gerwig G.J., van Vliet A.L., Lissenberg A., Briza P., Kamerling J.P., Vlasak R., de Groot R.J. Nidovirus sialate-O-acetylesterases: evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J. Biol. Chem. 2005;208:6933–6941. doi: 10.1074/jbc.M409683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean J.R., Jacomy H., Desforges M., Vabret A., Freymuth F., Talbot P.J. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473–e00512. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.E., Reeder D.M. Mammal Species of the World. A Taxonomic and Geographic Reference. 3rd ed. Johns Hopkins University Press; Baltimore, Maryland: 2005. [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Yip C.C., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.M., Herbst W., Kousoulas K.G., Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 1994;44:152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data Fig. S1 further illustrate recombination events among the novel CoVs described here, phylogenetic analyses were performed based on the nucleotide sequences of the RdRp, S and N genes of LRNV, LAMV and LRLV. Numbers (>70) above or below branches indicate percentage bootstrap values. The trees were mid-point rooted for clarity only. The scale bar represents the number of nucleotide substitutions per site. Putative recombinant lineages are shaded with gray and yellow, respectively. Schematic genetic exchanges of parental genomes and of the recombinant offspring in different colors are shown below the trees. The GenBank accession numbers of other CoVs are given in Table S1.
Supplementary data
Supplementary data
Supplementary data