Abstract
Organisms throughout biology need to maintain the integrity of their genome. From bacteria to vertebrates, life has established sophisticated mechanisms to detect and eliminate foreign genetic material or to restrict its function and replication. Tremendous progress has been made in the understanding of these mechanisms which keep foreign or unwanted nucleic acids from viruses or phages in check. Mechanisms reach from restriction-modification systems and CRISPR/Cas in bacteria and archaea to RNA interference and immune sensing of nucleic acids, altogether integral parts of a system which is now appreciated as nucleic acid immunity. With inherited receptors and acquired sequence information, nucleic acid immunity comprises innate and adaptive components. Effector functions include diverse nuclease systems, intrinsic activities to directly restrict the function of foreign nucleic acids (e.g., PKR, ADAR1, IFIT1), and extrinsic pathways to alert the immune system and to elicit cytotoxic immune responses. These effects act in concert to restrict viral replication and to eliminate virus-infected cells. The principles of nucleic acid immunity are highly relevant for human disease. Besides its essential contribution to antiviral defense and restriction of endogenous retroelements, dysregulation of nucleic acid immunity can also lead to erroneous detection and response to self nucleic acids then causing sterile inflammation and autoimmunity. Even mechanisms of nucleic acid immunity which are not established in vertebrates are relevant for human disease when they are present in pathogens such as bacteria, parasites, or helminths or in pathogen-transmitting organisms such as insects. This review aims to provide an overview of the diverse mechanisms of nucleic acid immunity which mostly have been looked at separately in the past and to integrate them under the framework nucleic acid immunity as a basic principle of life, the understanding of which has great potential to advance medicine.
Keywords: Innate immunity, RNAi, CRISPR/Cas, Antiviral immunity, Immune sensing of nucleic acids, cGAS, Sting, RIG-I, OAS, ADAR1, SAMHD1, TLR3, TLR7, TLR8, TLR9, Trex1, DNase, RNase H, RNase L, CpG-DNA, cGAMP, Oligonucleotide
1. Introduction
Immunology is typically categorized in innate and adaptive immunity. While the term innate is associated with conserved molecular patterns detected by germline-encoded receptors, adaptive immunity refers to T cells and B cells which use recombination and clonal selection to specifically adapt their immune receptors (T cell receptor, B cell receptors, and antibodies) in order to target foreign protein antigens. In this well-established concept, the innate immune system in the form of myeloid immune cells (macrophages, dendritic cells) provides information whether new protein antigens are associated with potential pathogens or damage. A limited number of germline-encoded innate immune receptors have been identified in the last two decades which are specialized to detect different classes of pathogen- or damage-associated molecules. Among them are several groups of immune receptors which are specialized on the detection of foreign or damage-associated nucleic acids. One of these groups of nucleic acid-sensing immune receptors are the Toll-like receptors (TLRs) TLR3, TLR7, TLR8, and TLR9 (TLR13 not existent in humans) which are preferentially located in the endolysosomal compartment of distinct immune cell subsets and certain somatic cells. Nucleic acid-detecting immune receptors located in the cytosol include the RIG-I family of helicases (RIG-I, MDA5, LGP2), cGAS, and AIM2. Although these nucleic acid-sensing immune receptors as part of the innate immune system participate in the regulation of protein antigen-directed adaptive immunity, they are now appreciated as part of a larger system of nucleic acid-directed immunity which functions to detect and eliminate foreign nucleic acids, whereas protein-directed adaptive immunity evolved to eliminate foreign proteins. Although there are crossregulatory functions of both systems, nucleic acid-directed immunity has a purpose on its own. This is underlined by the fact that protein-directed adaptive immunity developed more recently in evolution, whereas nucleic acid-directed immunity dates back to the earliest forms of life represented by bacteria and archaea. With the additions of RNAi and the CRISPR/Cas system, specific nucleases including the restriction-modification (R-M) systems, and antiviral effector proteins partially discovered only recently in the context of rare hereditary inflammatory diseases, the new concept of nucleic acid immunity evolves. With CRISPR/Cas and RNAi, biology has established two mechanisms which acquire new sequence information of pathogens and memorize this information for later defense against the same type of pathogen, characteristics which functionally correspond to adaptive immunity of T cells and B cells. The various mechanisms comprising nucleic acid immunity are highly relevant for the understanding of many inflammatory and infectious diseases. This review summarizes the currently known nucleic acid recognition-based antiviral response strategies. Antiviral response strategies span from ancient sequence or nucleic acid modification-dependent degradation systems (R-M, CRISPR, RNAi) to modern innate immunity in vertebrates, in which innate nucleic acid-sensing receptors induce a broad spectrum of antiviral alarm and effector mechanisms as well as subsequent adaptive immune responses.
2. Principles of Nucleic Acid Immunity in Different Species
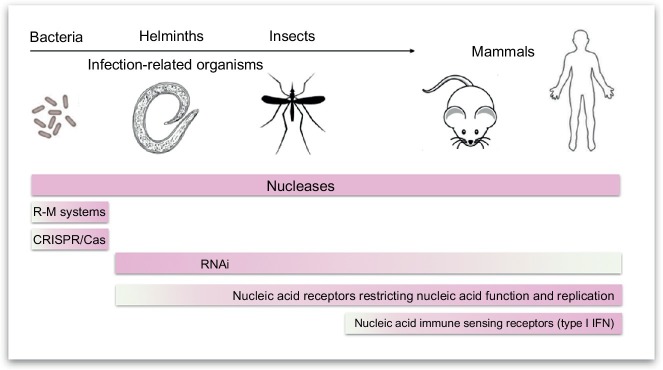
Foreign nucleic acids can be introduced by viruses or bacteriophages. However, species differ in their arsenal of defense mechanisms against such foreign nucleic acid invaders (Fig. 1 ). All species from bacteria to humans have established different types of nucleases which cleave nucleic acids that have identified themselves as foreign by their specific structure, by abundance and localization. One of the earliest forms of nucleases are the R-M systems in bacteria and archaea. In this system, modification enzymes and restriction endonucleases (REases) are directed to certain DNA sequence motifs in self DNA. Since modified DNA is not cleaved by the corresponding nucleases, DNA sequence motifs without modification are identified as foreign and are degraded (for further details, see later).
Fig. 1.
Mechanisms of nucleic acid immunity in species relevant for human disease. Biology has evolved a number of mechanisms to detect and eliminate foreign nucleic acids as introduced by viruses or bacteriophages. All species from bacteria to humans have established nucleases to directly degrade nucleic acids with structural characteristics or localizations which allow to distinguish them from regular cellular self nucleic acids. Other mechanisms are predominant in certain groups of species. Restriction-modification systems in bacteria and archaea apply sequence-specific modification of self nucleic acids which allows the specific detection and degradation of foreign nucleic acids (restriction endonucleases). Acquired sequence information is used by the CRISPR/Cas system in which new sequence information about pathogenic nucleic acids is integrated into the genome and thereby memorized in order to sequence-specifically degrade foreign nucleic acids. Sequence information is also used by RNA interference which serves antiviral nuclease functions (siRNA/DICER) as well as regulatory (microRNA) functions in higher multicellular organisms. In vertebrates, innate immune-sensing receptors including DICER-related helicases RIG-I and MDA5 dominate over RNAi as antiviral defense mechanism. While innate nucleic acid immune-sensing receptors elicit signaling pathways resulting in antiviral functions, a number of nucleic acid receptors (e.g., PKR, ADAR1, IFIT1) directly detect and restrict nucleic acid function and replication. Since the principles of nucleic acid immunity are either established in mammals themselves or in pathogens (bacteria, parasites, helminths) or pathogen-transmitting insects (e.g., mosquitoes), nucleic acid immunity as such is highly relevant for human health and disease.
Besides the R-M systems, a variety of RNases and DNases are established in evolution. DNA outside the nucleus is degraded by DNases I, II, and III. The RNA in DNA–RNA hybrids is degraded by RNase H. Long double-stranded RNA in the cytosol is subject to Dicer which cleaves RNA down to short double-stranded oligoribonucleotides which enter the RNAi pathway. Another way to acquire sequence information for antiviral defense is used by the CRISPR/Cas system. In this system, nucleic acid sequences derived from pathogens are integrated into the genome which allows the sequence-specific identification of the same type of pathogen during a subsequent challenge. In vertebrates, a number of highly specialized innate immune-sensing receptors such as TLR9 or RIG-I evolved to detect pathogen-associated nucleic acids and to induce appropriate immune responses. While innate nucleic acid immune-sensing receptors elicit antiviral signaling pathways, a number of nucleic acid-detecting effector proteins (viral restriction factors, e.g., PKR, ADAR1, IFIT1) directly detect and restrict nucleic acid function and replication. Various principles of nucleic acid immunity apply to different species, with only a subset applying to mammals. However, if it comes to infectious diseases, pathogens (bacteria, parasites, helminths) and pathogen-transmitting insects (e.g., mosquitoes) use additional mechanisms not present in mammals, which contribute to the interaction of the pathogen with transmitting organisms, and thus represent potential prophylactic or therapeutic targets.
3. Historic Overview of Different Fields Merging Into Nucleic Acid Immunity
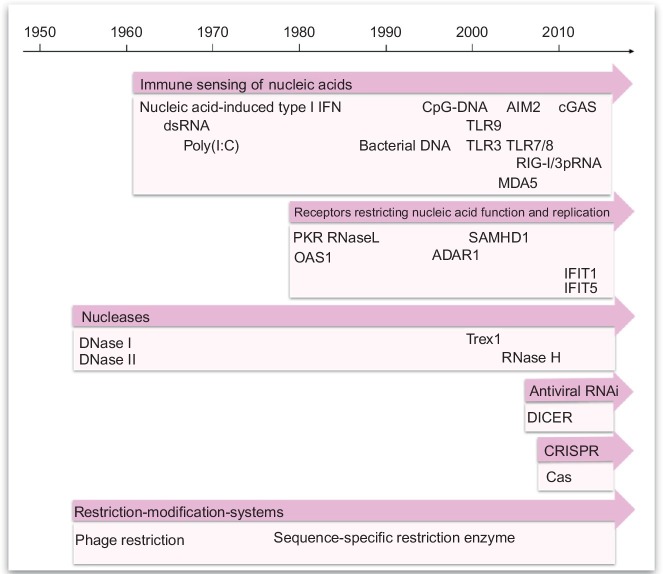
Several fields initially developed as independent lines of research and only recently were appreciated to closely cooperate in a defense system specialized in the detection and elimination of foreign genetic material. Fig. 2 provides a rough time line of key discoveries of principles, receptors, and ligands emerging from different areas of research all in the context of nucleic acid immunity. This brief overview cannot be comprehensive or provide the exact timing of each single discovery. The idea rather is to provide a picture how different fields evolved over the years. For more detailed information on the different receptors and pathways, the reader is referred to the respective specific paragraphs of this chapter below (Fig. 5, Fig. 6).
Fig. 2.
Overview of the time line of discoveries in nucleic acid immunity. This graph provides a noncomprehensive overview of the time lines when important principles, receptors, and ligands contributing to nucleic acid immunity have been described. Immune sensing of nucleic acids dates back to the early 1960s with the observation that nucleic acids such as long double-stranded RNA and specifically poly(I:C) can induce type I interferon. Later, it was appreciated that bacterial DNA is more active than vertebrate DNA. In 1995, the activity of bacterial DNA was attributed to a higher frequency of unmethylated CpG motifs in bacterial DNA. In 2000, TLR9 was identified as the immune receptor for the detection of unmethylated CpG motifs in DNA in the endosomal compartment. Sensing of cytoplasmic DNA remained unclear until in 2009 AIM2 and in 2012 cGAS were identified as the cytosolic receptors responsible for DNA-induced inflammasome activation and type I IFN induction, respectively. For immune sensing of RNA, the story of discoveries continued in 2001 with reports on TLR3-sensing long double-stranded RNA and was continued in 2004 with the appreciation of TLR7 and TLR8 as receptors sensing shorter forms of unmodified single and double-stranded RNA with great implications for the application of siRNA. Another milestone was reached with the immune sensing of cytoplasmic forms of RNA, specifically the detection of 5′-triphosphate short double-stranded forms of RNA by the cytosolic receptor RIG-I. The RIG-I-like receptor MDA5 added another cytosolic receptor which explained the induction of type I IFN by long double-stranded forms of RNA as observed early on in the 1960s. PKR identified in the late 1970s was the first of the receptors restricting nucleic acid function and replication without activating immunity and cytokines. SAMHD1 (depletion of dNTPs) and ADAR1 (A-to-I conversion in dsRNA) entered the field more recently in the context of genetic alterations in these genes identified in the context of inherited inflammatory syndromes (e.g., AGS). IFIT1 and IFIT5 are two other examples of more recently described receptors which inhibit the translation of mRNA. OAS1 was identified early on soon after PKR as a factor restricting viral replication by activating RNase L. Other nucleases contributing to nucleic acid immunity include RNase H structurally resolved in 2004, which degrades the RNA in DNA–RNA hybrids; furthermore, extracellular DNase I and endolysosomal DNase II are known since the mid-1950s. Knowledge around the function of the cytoplasmic DNase III which is also called Trex1 accumulated since 1999 and gained great impact on nucleic acid immunity like SAMHD1 and ADAR1 more recently in the context of inherited type I IFN-dependent inflammatory syndromes. Antiviral RNAi and the role of Dicer were first described in 2005, while the bacterial version of sequence-specific antiviral immunity, CRISPR/Cas, was identified in 2011. Restriction-modification systems are studied since the 1950s.
Fig. 5.
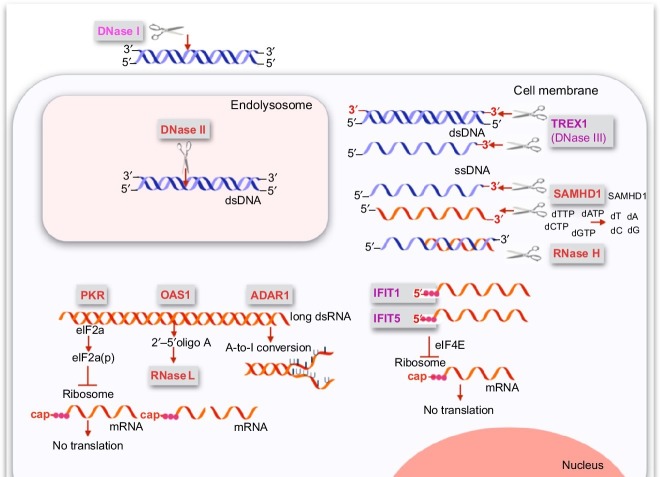
Receptors and nucleases restricting function and replication of foreign nucleic acids. This graph provides an overview of the proteins which target foreign nucleic acids without involving the classical immune functions such as the induction of cytokines or the activation of immune cells. Such proteins can directly act on the foreign nucleic acid, or they can elicit pathways indirectly acting on the foreign nucleic acid. The endonuclease DNase I is the most abundant DNase in the extracellular space which degrades DNA down to tetramers. DNase II is the predominant endonuclease in the endolysosomal compartment of cells. The cytoplasmic DNase III (Trex1) is a 3′-to 5′ exonuclease which degrades both double- and single-stranded DNA. The cytoplasmic RNase H recognizes DNA–RNA hybrids and cleaves the RNA in such hybrids. In contrast, RNase L is indirectly activated by oligoadenylates which are formed by OAS1 upon binding to long double-stranded RNA. Furthermore, ADAR1 modifies long double-stranded RNA by A-to-I conversions destabilizing the double strand resulting in changes in the coding sequence of proteins. SAMHD1 depletes the pool of dNTPs which is the prerequisite for DNA formation. SAMHD1 hydrolyzes the triphosphate in dNTPs resulting in deoxynucleosides. At the same time, SAMHD1 has been proposed to be a 3′-exonuclease for single-stranded DNA and RNA. PKR and IFIT1/5 inhibit mRNA translation by phosphorylation of the elF2a and by replacing elF4 in the ribosomal complex, respectively. While PKR is activated by long double-stranded RNA, IFIT1 and IFIT5 bind 5′-triphosphate ends of single-stranded RNA.
Fig. 6.
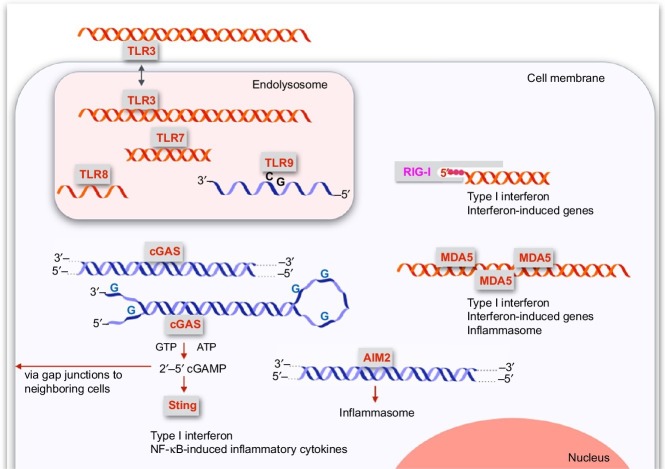
Immune-sensing receptors-detecting foreign nucleic acids and inducing indirect effector responses. This graph provides an overview of immune-sensing receptors of nucleic acids. TLR3 is the only one which besides its endosomal localization is also reported to be expressed on the cell membrane. TLR3 binds long double-stranded RNA which is not present in the cytosol of normal cells and is an indicator of foreign. TLR3 is expressed in myeloid immune cells and in a number of somatic cells including fibroblasts and endothelial cells. The other three TLRs expressed in the endolysosomal compartment of distinct immune cell subsets are TLR7, TLR8, and TLR9. TLR7 detects even short RNA, preferentially double-stranded and containing G and U. TLR8 detects single-stranded RNA. While TLR8 is expressed in human myeloid immune cells, TLR7 and TLR9 are predominantly expressed in human B cells and plasmacytoid dendritic cells. TLR9 detects single-stranded DNA containing unmethylated CpG dinucleotides. In the cytoplasm, RIG-I specifically detects RNA if it contains at least a short double strand with a blunt end and a 5′-triphosphate. The RIG-I-like receptor MDA5 detects long irregular forms of double-stranded RNA, but the exact definition of the ligand structure is unclear. Both RIG-I and MDA5 are widely expressed in immune cells and nonimmune cells, and induce a broad array of cell autonomous and extracellular antiviral responses including the production of type I interferon. MDA5 ligands also activate multiple other receptor pathways that depend on the detection of long double-stranded RNA, including PKR, ADAR1, and TLR3. The cytosolic receptor AIM2 detects long double-stranded DNA and activates the inflammasome. The other key receptor for the detection of DNA in the cytoplasm is cGAS. cGAS is activated by long double-stranded DNA and short forms of double-stranded DNA with single-stranded overhangs containing Gs, a structure which was termed Y-form DNA and which is presented during retroviral infection or by endogenous retroelements. Upon activation, cGAS catalyzes the formation of 2′–5′-cGAMP from GTP and ATP. 2′–5′-cGAMP acts as a second messenger which binds to the downstream signaling protein Sting which induces type I interferon via TBK1 and IRF3. 2′–5′-cGAMP can travel to and alarm neighboring cells via gap junctions. Sting also activates NF-κB activation and inflammatory cytokines.
Immune sensing of nucleic acids dates back to the early 1960s with the observation that nucleic acids such as long double-stranded RNA and specifically poly(I:C) can induce the antiviral factor type I interferon (Isaacs, Cox, & Rotem, 1963) which was first described in 1957 (Isaacs, 1957, Lindenmann et al., 1957). Three decades later, it was reported that bacterial DNA induces type I IFN much more vigorously than genomic DNA of vertebrates (Yamamoto et al., 1988, Yamamoto, Yamamoto, et al., 1992b). It was speculated that bacterial DNA in Sir William Coley's bacterial lysates was responsible for the antitumor activity seen using bacterial lysates for the treatment of tumor patients around 100 years earlier (Wiemann & Starnes, 1994). Efforts were undertaken to generate synthetic oligodeoxynucleotides which mimic the type I IFN-inducing activity of bacterial DNA. Palindromic DNA sequences were identified, and oligonucleotides containing such palindromes induced type I IFN in vitro (Yamamoto, Yamamoto, Kataoka, et al., 1992) but only showed weak activity in tumor models in vivo due to rapid degradation by DNases. In 1995, the immunological activity of bacterial DNA was attributed to a higher frequency of unmethylated CpG motifs in bacterial DNA (Krieg et al., 1995). Unmethylated CpG motifs were contained in the former palindromic sequences, but a palindromic sequence was not required for the type I IFN-inducing activity. The introduction of the phosphorothioate modification in DNA first described in 1977 (Vosberg & Eckstein, 1977) was used to stabilized these so-called CpG oligonucleotides which now could be successfully applied for treatment in experimental tumor models in vivo (Heckelsmiller et al., 2002). In 2000, TLR9 was identified as the innate immune receptor required for the detection of unmethylated CpG motifs in DNA (Hemmi et al., 2000). Notably, TLR9 was the first innate immune receptor reported to detect a specific type of nucleic acid and to induce an immune response. Despite intensive research, it took almost a decade to identify AIM2 as the next innate immune receptor-detecting DNA (Hornung & Latz, 2010). However, since AIM2 activates the inflammasome but not type I IFN, the field of DNA sensing struggled until end of 2012, when the cytosolic DNA-binding enzyme cyclic GMP-AMP synthase (cGAS) was discovered which generates cGAMP as second messenger for the downstream signaling molecule Sting, resolving a big question mark in the field. At that time, Sting was already known to be required for immune sensing of cytosolic DNA resulting in IFN induction (Barber, 2014), but Sting was unable to bind DNA directly.
Although RNA molecules were the first nucleic acids found to induce type IFN as described earlier, this line of research continued only in 2001 with the identification of double-stranded RNA as ligand for TLR3 (Alexopoulou, Holt, Medzhitov, & Flavell, 2001). Although the activation of TLR3 induces some type I IFN, it could not explain the massive amounts of type I IFN induced upon cytosolic delivery of the double-stranded RNA mimic poly(I:C). In parallel with the discovery of RNAi and siRNA, techniques of chemical synthesis of RNA rapidly progressed and highly pure synthetic RNA oligonucleotides at high quantities and reasonable costs became available. With easier access to highly pure synthetic RNA oligonucleotides, it was found that not only long double-stranded RNA but also single-stranded RNA stimulated type I IFN, specifically in immune cells, and the immune receptors involved were found to be TLR7 and TLR8 (TLR8 nonfunctional in mouse) (Diebold et al., 2004, Heil et al., 2004, Judge et al., 2005). Then it was noted that even siRNA induce type I IFN in TLR7 expressing immune cells, a surprising finding at that time since siRNA was thought to be short enough to not stimulate interferon responses (Hornung et al., 2005). This work also reported that the immune stimulatory activity of siRNA can be avoided by introducing chemical modifications such as 2′-O-methylation or by introducing pseudouridine which is used since then to avoid immunostimulation by siRNA applied in cells expressing TLR7 in vitro or in vivo (Hornung et al., 2005).
At that time, a cheap way to generate siRNA was the use of in vitro transcription. However, it was soon realized that siRNA made by in vitro transcription induces high amounts of type I IFN when transfected even in cells not expressing TLR7, including human myeloid immune cells. This stimulated research on the molecular mechanism responsible for type I IFN induction by in vitro-transcribed siRNA in myeloid immune cells. Finally, the cytosolic helicase RIG-I was identified to detect 5′-triphosphate ends in short blunt end double-stranded RNA (Hornung et al., 2006, Pichlmair et al., 2006, Schlee, Roth, et al., 2009). RIG-I was reported earlier as immune receptor involved in antiviral responses (Kato et al., 2005). Notably, in vitro transcription but not chemical synthesis of siRNA generates such 5′-triphosphate ends. The presence of unmodified 5′-triphsophate ends in the cytosol indicates the presence of RNA polymerase activity in the cytosol which only occurs in the course of viral replication. Another member of the RIG-I-like helicase family of receptors is MDA5 which was found to be responsible for the long sought after type I IFN-inducing activity of cytosolic long double-stranded RNA including poly(I:C) (Yoneyama et al., 2005). As of today, most of the type I IFN-inducing activities of nucleic acids can be assigned to specific immune receptors. Future may still keep some surprises for the field, for example, in the context of immune sensing in the nucleus or in the context of DNA damage repair.
While activation of the immune receptors described above results in the induction of immunologically active cytokines and immune responses, there is a group of nucleic acid receptors directly restricting nucleic acid function and replication largely without inducing an immune response. PKR was one of the first of these. Binding of long double-stranded RNA activates PKR to phosphorylate elF2a leading to the inhibition of ribosomal translation of mRNA to proteins (Clemens and Elia, 1997, Sen et al., 1978, Thomis et al., 1992, Thomis and Samuel, 1992). Soon after PKR, the 2′–5′-oligoadenylate synthetase (OAS) system was reported (Rebouillat and Hovanessian, 1999, Yang et al., 1981). In parallel to the OAS system, RNase L was identified (Baglioni et al., 1978, Brennan-Laun et al., 2014). Upon binding of OAS1 to long double-stranded RNA, OAS1 generates 2′-5′-linked oligoadenylates (2′5′-OA). 2′5′-OA activate RNAse L which then cleaves cellular RNAs thereby restricting viral propagation. SAMHD1 (sterile alpha motif and histidine–aspartate-domain-containing protein 1), originally described as IFN-inducible gene in 2000 (Li, Zhang, & Cao, 2000), has triphosphohydrolase activity that rapidly converts dNTPs to the corresponding deoxynucleoside and inorganic triphosphate, thereby depleting the supply of dNTP for reverse transcriptase activity of retroviruses. In 2009, it was learned that mutations in SAMHD1 cause inherited inflammatory syndromes with a type I IFN signature (e.g., AGS) (Rice et al., 2009), but the exact mechanism leading to type I IFN induction in this context is not well understood. Originally cloned in 1994 (Kim, Wang, Sanford, Zeng, & Nishikura, 1994), the RNA-editing enzyme ADAR1 binds to double-stranded RNA and converts A to I thereby contributing to self vs nonself recognition of RNA (Nishikura, 2016). In 2012, it was realized that mutations in ADAR1 cause inflammatory syndromes associated with a type I IFN signature (Rice et al., 2012). IFIT1 and IFIT5 are known for many years as type I IFN-inducible RNA-binding proteins which bind to single-stranded RNA lacking 2′-O-methylation at their 5′-end and inhibiting RNA translation (Hyde & Diamond, 2015). More recent work from 2011 (Pichlmair et al., 2011) added the information that IFIT1 and IFIT5 preferentially bind to viral RNA containing a 5′-triphosphate group, completing the picture how these proteins distinguish self from nonself single-stranded RNA.
Of the proteins which function primarily as nucleases, extracellular DNase I and endolysosomal DNase II are known since the mid-1950s. The cytoplasmic exonuclease Trex1 has been identified decades later in 1999 (Hoss et al., 1999). Only since 2006, we know that loss of function in Trex1 causes the type I IFN-associated inflammatory syndrome AGS (Crow et al., 2006), suggesting that Trex1 is critically involved in the clearance self DNA within the cytoplasm of cells. RNases H are widely expressed enzymes that hydrolyze RNA in RNA/DNA hybrids (Cerritelli & Crouch, 2009). While reports on RNase H activity date back to 1969 (Stein & Hausen, 1969), the heterotrimeric functional complex in eukaryotes was only described in 2004 (Jeong, Backlund, Chen, Karavanov, & Crouch, 2004) and human in 2009 (Chon et al., 2009). In 2007, it was reported that mutations in any of the three subunits of human RNase H2 cause Aicardi–Goutiéres syndrome (AGS) (Rice et al., 2007).
Antiviral RNAi and the role of Dicer were first described in 2006 (Wang et al., 2006, Zambon et al., 2006), while the role of RNA interference (RNAi) in mammalian innate immunity is still poorly understood. A bacterial counterpart of acquired sequence-specific antiviral immunity is the CRISPR/Cas system in prokaryotes first described in 2007 (Barrangou et al., 2007, Marraffini and Sontheimer, 2010). This prokaryotic immune system confers resistance to foreign genetic elements such as those present within plasmids and phages. Information around R-M systems accumulated since the early 1950s (Luria & Human, 1952) with REases first described in 1975 (Nathans & Smith, 1975).
4. Functional Components of Nucleic Acid Immunity
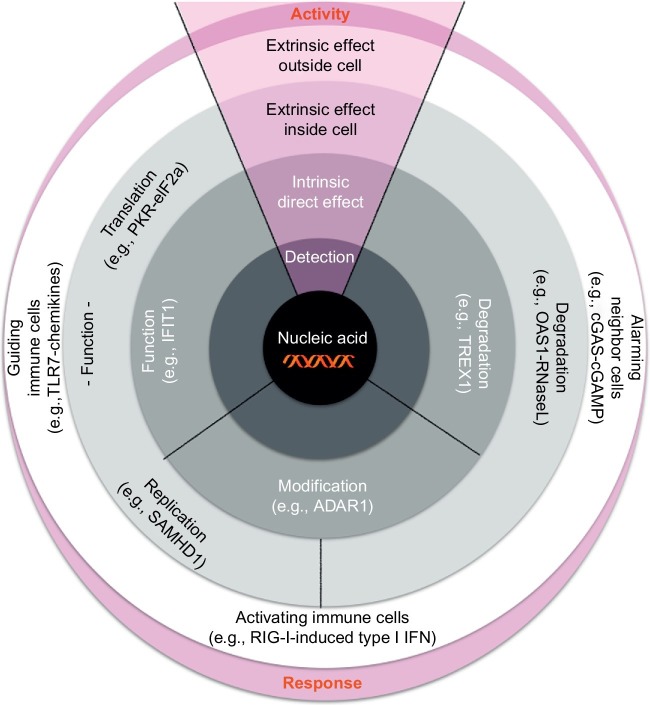
The concept of nucleic acid immunity integrates different functional components which have been studied and reviewed separately in the past. Only in the last few years and with the elegant genetic work on rare human inflammatory disorders associated with a type I IFN signature (of Crow and others (Crow & Rehwinkel, 2009)), it became evident that many of the antiviral restriction factors and various nuclease systems are tightly connected with innate immune sensing of nucleic acids inducing type I IFN. Altogether, biology has established a broad spectrum of effector functions which cover most of the molecular and mechanistic possibilities to restrict the propagation of foreign genetic material. Effector functions reach from direct actions on the detected nucleic acid to the elimination of cells containing foreign genetic material. Fig. 3 illustrates the functional components of nucleic acid immunity. Central to all components is the detection of foreign nucleic acids by the molecular interaction of a protein (immune-sensing receptor, restriction factor, or effector protein) or a specific nucleic acid (RNAi, CRISPR/Cas) with the targeted nucleic acid. The molecular challenge on this level is the specificity of the detection and the distinction of self vs foreign. A specific molecular signature of self nucleic acid (e.g., 2′-O-methylation at the N1 position in capped mRNA), compartmentalization of self nucleic acids, and the rapid clearance of surplus self nucleic acids are three examples which enable specific detection of foreign nucleic acid. Structural differences such as long double-stranded RNA not present under physiological circumstances allow the highest confidence level of detection. Upon detection, the system generates different types of responses. Detection of a foreign nucleic acid can trigger an intrinsic effect which acts directly on the detected nucleic acid. Examples are degradation (e.g., TREX1, CRISPR/Cas), structural modification (e.g., A-to-I conversion by ADAR1), or disabling the function (e.g., inhibition of translation of mRNA by IFIT1 or RNAi) (see Fig. 3, middle gray layer). Extrinsic effects upon detection of foreign nucleic acids require a signaling cascade finally resulting in an effect on the detected nucleic acid. Extrinsic effects can occur solely inside the same cell, or they can involve functions outside the cell. Extrinsic effects inside the same cell include degradation of the nucleic acid (e.g., RNase L activated by 2′–5′-OA generated by OAS1 upon binding of long double-stranded RNA). Extrinsic effect inside the same cell can also impact on the function of the detected nucleic acid. Examples for such functional effects are inhibition of translation (e.g., phosphorylation of elF2a by activated PKR) or inhibition of replication (e.g., restricting replication of retroviruses by depleting intracellular dNTP pools by IFN-inducible SAMHD1) (see Fig. 3, light gray layer). Extrinsic effects that involve functions outside the cell in which the nucleic acid is primarily detected appear as immune responses. They range from alarming neighboring cells (e.g., secretion of type I IFN or release of cGAMP via gap junctions) to the guidance of immune cells to the respective cell (e.g., TLR7-induced release of IP-10) and the activation of immune cells at the local site and systemically (e.g., activation of NK cells by RIG-I). Intrinsic and extrinsic effects act in concert to minimize the danger associated with foreign nucleic acids.
Fig. 3.
Overview of functional components in nucleic acid immunity. The primary detection of specific forms of nucleic acids by highly specialized proteins is the central part of nucleic acid immunity. Upon binding of nucleic acids, the participating specialized proteins can either exert intrinsic direct effects on the nucleic acid which they have bound, or they can have indirect extrinsic effects which require the participation of additional signaling. Extrinsic effects that restrict viral replication and function can be located inside or outside cells, or both. Intrinsic direct effects include degradation or structural modification of the bound nucleic acids, or direct inhibition of translation. Extrinsic indirect effects via signaling pathways include mechanisms that restrict translation or replication, or that lead to degradation of nucleic acids. Extrinsic effects with activities beyond the infected cell include alarming neighboring cells, activating immune effector cells, and guiding immune cells to the site of infection. Together, these intrinsic and extrinsic activities represent the repertoire of nucleic acid immunity to restrict viral infection.
5. Innate and Adaptive Components in Nucleic Acid Immunity
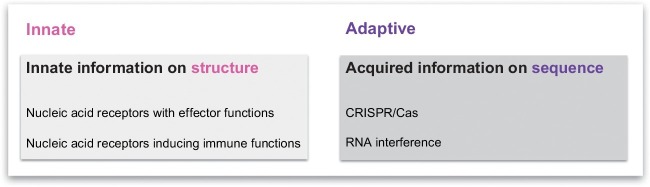
In classical immunology, we distinguish innate and adaptive immunity. While innate immunity relies on receptors encoded in the germline, adaptive immunity acquires information about pathogens during the life span and memorizes such information for later use. While adaptive immunity directed against proteins relies on the mechanism of genetic recombination to adapt to novel pathogen-derived proteins, in the adaptive part of nucleic acid immunity information on pathogen-derived nucleic acid sequences is acquired and memorized (CRISPR/Cas and RNAi) (see Fig. 4 ). Although the adaptive part of nucleic acid immunity requires the participation of germline-encoded proteins such as DICER, RISC, or CRISPR and Cas, the detection of foreign nucleic acids is mediated via acquired sequence information. In contrast, the innate components of nucleic acid immunity solely rely on germline-encoded receptors which via protein–nucleic acid interaction detect certain structures which are characteristic of foreign genetic materials. Innate immune-sensing receptors are identical throughout the life span. However, it is important to note that epigenetic regulation of gene expression still allows to adapt quantitatively to different environments (e.g., low or high burden of viral pathogens).
Fig. 4.
Innate and adaptive components in nucleic acid immunity. In classical immunology, we distinguish innate and adaptive immunity. While innate immunity relies on receptors encoded in the germline, adaptive immunity acquires information about pathogens during the life span and memorizes such information for later usage. While adaptive immunity directed against proteins relies on the mechanism of genetic recombination to adapt to novel pathogen-derived proteins, in the adaptive part of nucleic acid immunity information about pathogen-derived nucleic acid sequences is acquired and memorized (CRISPR/Cas and RNA interference). Unlike the adaptive components, the innate components of nucleic acid immunity rely on germline-encoded receptors which detect certain structures indicating viral pathogens. Therefore, these receptors are identical throughout the life span, but regulation of receptor expression (e.g., by means of epigenetics) still allows adaptation to different environments (e.g., low or high burden of viral pathogens).
6. Innate and Adaptive Nucleic Acid Immunity in Prokaryotes
The perpetual arms race between bacteria and phages has resulted in the evolution of efficient resistance systems that protect bacteria from phage infection. Such systems include R-M systems and CRISPR–Cas. The prokaryotic DNA R-M systems are based on the contrasting enzymatic activities of a sequence-specific REase and a matching sequence-specific host methyltransferase (MTase) (Vasu & Nagaraja, 2013). By transferring a methyl group to the C-5 carbon or the N4 amino group of cytosine or to the N6 amino group of adenine host-specific MTases protect potential cleavage sites of host DNA from REases, which on the other hand recognize and cleave foreign unmethylated or “inappropriately” methylated DNA from invading phages. This R-M systems can be considered as an innate defense system. On the other hand, the CRISPR–Cas system in prokaryotes represents a highly sophisticated adaptive immune system in which short fragments of invading DNA are integrated into the CRISPR loci. After transcription and processing of these loci, short CRISPR RNAs (CrRNAs) are generated which guide the nuclease activity of Cas proteins to complementary DNA or RNA resulting in target cleavage (Goldfarb et al., 2015, van der Oost et al., 2014).
7. Receptors and Nucleases Not Involving Classical Immune Functions
This chapter provides an overview of the well-established proteins targeting foreign nucleic acids without involving the classical immune functions such as the induction of cytokines or the activation of immune cells. Such proteins can directly act on the foreign nucleic acid, or they can elicit pathways indirectly acting on the foreign nucleic acid (see Fig. 5 ). For the family of APOBEC proteins which detect and modify viral nucleic acids, we refer to detailed reviews by others (Harris & Dudley, 2015).
7.1. ADAR1
Adenosine to inosine (A-to-I) RNA editing was originally discovered as enzymatic activity unwinding double-stranded RNA in Xenopus laevis oocytes and embryos (Bass & Weintraub, 1987). Soon after, it became clear that this activity is carried out by an adenosine deaminase acting on RNA (ADAR) (Bass and Weintraub, 1988, Wagner et al., 1989). Adenosine deaminases perform C6 deamination of adenosine in base-paired RNA structures resulting in A-to-I conversions, a process termed A-to-I RNA editing (Hogg, Paro, Keegan, & O'Connell, 2011). The type I IFN-inducible isoform of ADARs, ADAR1, first cloned in 1994 (Kim et al., 1994) contains three repeats of a double-stranded RNA-binding motif, and sequences conserved in the catalytic center of other deaminases. Transcription from separate promoters generates two isoforms of ADAR1, a full-length, interferon-inducible ADAR1p150 and a shorter and constitutively expressed ADAR1p110. Interestingly, both ADAR1p150 and ADAR1p110 isoforms shuttle between nucleus and cytoplasm (Nishikura, 2016). A-to-I editing frequently occurs in noncoding regions that contain inverted Alu repeats but can also occur in protein-coding regions of mRNAs resulting in the expression of altered proteins with sequences that are not encoded in the genome. Recent studies indicate that ADAR1 is also found in complex with Dicer to promote miRNA processing and RNAi efficacy (Ota et al., 2013), suggesting that both RNAi and ADAR are functionally related.
Viral dsRNA formed at different stages of replication of many viruses are substrates for RNA editing by ADAR. It is well established that ADAR enzymes interfer with the virus–host interaction with ADARs acting as pro- or antiviral factors. The biological consequences of A-to-I changes during viral infection is not completely understood (Tomaselli, Galeano, Locatelli, & Gallo, 2015). The current concept is that two effects oppose each other: on the one hand, ADAR1-mediated A-to-I editing of viral double-stranded RNA directly restricts the correct function of the edited RNA and thus directly inhibits viral replication. On the other hand, A-to-I editing of double-stranded RNA destabilizes long double-stranded RNA thereby reducing the recognition of long double-stranded RNA by double-stranded RNA receptors such as MDA5. As a consequence, depending on the type of virus, ADAR1 has the potential to negatively interfer or to support viral replication and thus can act as proviral or antiviral factor (George, John, & Samuel, 2014).
Independently of the presence of viral infection, the lack of ADAR1 in a cell results in the activation of MDA5 by endogenous RNA species. The most likely scenario is that A-to-I editing masks endogenous RNAs from detection by MDA5. The consequence is that if a virus actively inhibits the function of ADAR1, endogenous RNA ligands will form which activate MDA5 and thus induce a type I IFN response. Indeed, sequencing studies demonstrated that ADAR1-deficient cells display stretches of endogenous double-stranded RNA (Liddicoat et al., 2015). Thus, A-to-I editing of endogenous dsRNA is an essential function of ADAR1 preventing the activation of the cytosolic dsRNA response by endogenous transcripts.
7.2. SAMHD1
SAMHD1 is composed of a SAM and a HD domain. While the SAM domain of SAMHD1 mediates protein–protein interactions, the HD domain possesses the triphosphohydrolase activity through which SAMHD1 hydrolyzes dNTPs to deoxynucleosides (Goldstone et al., 2011, Powell et al., 2011). SAMHD1 expression has been demonstrated in monocytes, macrophages, myeloid dendritic cells, plasmacytoid dendritic cells (PDCs), and CD4 T cells (Baldauf et al., 2012, Gelais et al., 2012, Kim et al., 2012, Laguette et al., 2011, Pauls et al., 2014). The involvement of SAMHD1 in innate immunity was initially proposed based on its mouse ortholog Mg11 which is IFN-inducible in macrophages and dendritic cells (Li et al., 2000), hence the alternative name dendritic cell-derived IFN-γ-induced protein. Subsequent studies showed an increased SAMHD1 expression upon stimulation of macrophages with double-stranded DNA (Rice et al., 2009) and its upregulation in the context of viral infections (Hartman et al., 2007). Mutations in SAMHD1 have been shown to be responsible for 5% of patients with AGS which is characterized by inappropriate and aberrant type I IFN secretion causing symptoms reminiscent of a congenital infection (Rice et al., 2009). A loss of function of SAMHD1 results in spontaneous type I IFN production in AGS patients and SAMHD1 −/− mice (Behrendt et al., 2013, Rehwinkel et al., 2013). SAMHD1 was identified as a potent restriction factor for HIV (Hrecka et al., 2011, Laguette et al., 2011, Simon et al., 2015), other nonhuman retroviruses (Gramberg et al., 2013), and herpesviruses, including HSV-1 (Hollenbaugh et al., 2013, Kim et al., 2013). The current model is that SAMHD1 through its function as dNTP triphosphohydrolase decreases intracellular dNTP pools in nondividing cells below the threshold level required for efficient viral reverse transcriptase or viral DNA polymerase activity (Lahouassa et al., 2012, Wu, 2013). The observation that functional loss of SAMHD1 leads to a spontaneous type I IFN response suggests that uncontrolled activity of endogenous retroelements may be a source of the IFN-inducing nucleic acids, but the identity of such endogenous ligands is unknown to date. Besides dNTP triphosphohydrolase activity, a metal-dependent 3′- to 5′-exonuclease activity of SAMHD1 for ssDNA and ssRNA was demonstrated (Beloglazova et al., 2013). The RNAse activity was reported to directly degrade HIV-1 RNA (Ryoo et al., 2014), but further work will be necessary to confirm and exactly characterize the proposed nuclease activity of SAMHD1 (Rice et al., 2009).
7.3. PKR
The interferon-inducible, double-stranded RNA-activated protein kinase (protein kinase RNA-activated, PKR; also known as eukaryotic translation initiation factor 2-alpha kinase 2, EIF2AK2) was first cloned in 1992 and represents a key mediator of antiviral activities (Feng et al., 1992, Garcia et al., 2006, Thomis et al., 1992, Thomis and Samuel, 1992). PKR contains an N-terminal dsRNA-binding domain (dsRBD) which consists of two tandem copies of a conserved double-stranded RNA-binding motif, dsRBM1 and dsRBM2, and a C-terminal kinase domain. Binding of PKR to long double-stranded RNA (longer 30 bp) activates PKR by inducing dimerization and subsequent autophosphorylation. Activated PKR inhibits 5′-cap-dependent mRNA translation by phosphorylation of the eukaryotic translation initiation factor eIF2a thereby preventing viral protein synthesis (Farrell et al., 1978, Levin and London, 1978). Besides long double-stranded RNA, PKR has been shown to recognize RNA with limited secondary structures (RNA with a length of about 47 nt and weak structure; short stem-loops) containing uncapped 5′-triphosphates (Nallagatla et al., 2007). Antiviral functions of PKR beyond the mechanism of translation inhibition are still controversial. PKR affects diverse transcriptional factors such as interferon regulatory factor 1, STATs, p53, activating transcription factor 3, and NF-κB. In particular, how PKR triggers a cascade of events involving IKK phosphorylation of IkappaB and NF-κB nuclear translocation has been intensively studied. PKR was also reported to enhance but not being required for NF-κB-dependent type I IFN induction (Chu et al., 1999, Kumar et al., 1994, Maggi et al., 2000). Involvement of PKR in inflammasome activation is controversial (He et al., 2013, Lu et al., 2012). PKR-mediated 5′-cap-specific inhibition of translation is expected to perturb the proteome. Since PKR-activated elF2a is involved in the initiation of the translation from an AUG codon, the alternative non-AUG initiation takes place instead. An example of mRNAs using non-AUG initiation are mRNAs for the heat shock proteins. Another effect is the selected reduction of proteins with short half-life. Reduced translation of the NF-κB inhibitor protein IkappaB-alpha is one plausible explanation for the activation of the NF-κB pathway in response to PKR activation (McAllister, Taghavi, & Samuel, 2012). Other signaling pathways may be affected in the same way by the removal of an inhibitor with short half-life resulting in the activation of the pathway.
7.4. IFIT1 and IFIT5
Interferon-induced proteins with tetratricopeptide repeats (IFITs) are among the most abundantly expressed proteins of the group of interferon-stimulated genes (ISGs). They represent innate immune effector molecules that confer antiviral defense downstream of type I IFN through disruption of the host translation initiation machinery (Daffis et al., 2010, Pichlmair et al., 2011). They are evolutionarily conserved from fish to mammals. In humans, there are four well-characterized paralogues, IFIT1 (p56/ISG56), IFIT2 (p54/ISG54), IFIT3 (p60/ISG60), and IFIT5 (p58/ISG58). Like RIG-I, productive binding of both IFIT1 and IFIT5 was shown to depend on the presence of cytosolic 5′-triphosphate RNA and is nonsequence specific. Unlike RIG-I, IFIT1 and IFIT5 preferentially bind to single-stranded RNA or to double-stranded RNA with a minimum three (IFIT5) or five (IFIT1)-nucleotide overhangs containing an uncapped triphosphate group at the 5′-end of RNA (Abbas et al., 2013, Habjan et al., 2013, Kumar et al., 1994). Replacing the triphosphate with a 5′-cap, a 5′-monophosphate or 5′-OH diminishes the binding significantly (Abbas et al., 2013). IFIT1 competes with eIF4E, the endogenous 5′-cap binding and translation factor, in the 48S initiation complex formation. However, while in vitro competition experiments convincingly show that IFIT1 can compete with eIF4E for binding at completely unmethylated cap0 RNA, the out-competing of eIF4E from N7 methylated cap (cap0) structures is unclear. Binding of eIF4E to cap0 structures in lysates of IFN-primed cells is rather enhanced than reduced, suggesting additional mechanisms beyond eIF4E competition for 48S disruption (Habjan et al., 2013). A key role for IFIT1 in negative-strand RNA viruses (VSV, Influenza) and positive-strand RNA viruses (WNV, MHV) except picornaviruses was reported (Daffis et al., 2010, Habjan et al., 2013, Pichlmair et al., 2011).
7.5. OAS
The human 2′–5′-OAS family comprise four type I IFN-inducible genes: OAS1, OAS2, OAS3, and OASL (OAS-like protein) (Melchjorsen et al., 2009). Upon binding to long double-stranded RNA, OAS1, OAS2, and OAS3 catalyze the formation of 2′–5′-OA, whereas OASL has no enzymatic activity but still has potent antiviral activity due to its coactivating role in the RIG-I pathway (Schoggins et al., 2011, Zhu et al., 2014). The formation of 2′–5′-oligomers of adenosine (2′–5′-OA) upon exposure to dsRNA and subsequent inhibition of translation has been described early on (Clemens and Williams, 1978, Farrell et al., 1978, Hovanessian et al., 1977, Zilberstein et al., 1978). 2′–5′-OA function as second messenger of OAS binding to RNase L leading to dimer formation and subsequent degradation of cellular and viral RNA (Dong & Silverman, 1997). The structural mechanisms of RNase L activation by 2′–5′-OA and its dimer formation have recently been described (Han et al., 2014, Huang et al., 2014). All three human OAS isoforms are activated by dsRNA in vitro which is the presumed ligand in vivo as well. The full activation of the OAS system in virally infected cells leads to the inhibition of protein synthesis and the induction of apoptosis, thereby interfering with viral replication (Castelli et al., 1998). Activation of the OAS–RNase L system restricts replication of a variety of viruses, in particular positive-strand viruses (e.g., picornaviruses, flaviviruses, and alphaviruses) which produce high numbers of dsRNA during replication (Silverman, 2007). Virus-encoded inhibitors of the OAS–RNase L system such as the nonstructural protein 2 (NS2) of murine coronavirus or inhibitors expressed by picornaviruses support a key role of this system in the restriction of viruses. It is interesting to note that OAS and cGAS (see later) share similar structural features and enzymatic function. Both OAS and cGAS catalyze the uncommon 2′–5′ phosphodiester linkage upon binding to a nucleic acid ligand (Hornung, Hartmann, Ablasser, & Hopfner, 2014).
7.6. RNaseH
RNases H are a family of widely expressed nonsequence-specific endonucleases that hydrolyze solely the RNA of RNA/DNA hybrids resulting in 3′-hydroxyl and 5′-phosphate terminated products and an intact DNA strand (Cerritelli & Crouch, 2009). RNases H play crucial roles in the biochemical processes associated with DNA replication, gene expression, and DNA repair where RNA/DNA hybrids can occur. Furthermore, RNases H degrade RNA/DNA hybrids generated during viral replication. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes. Unlike in prokaryotes and in single-cell eukaryotes, in higher eukaryotes RNases H are essential for development.
The catalytic subunit of eukaryotic RNase H2, RNASEH2A, is well conserved and similar to the monomeric prokaryotic RNase HII. In contrast, the RNASEH2B and RNASEH2C subunits share very little homology between human and Saccharomyces cerevisiae or bacteria. RNASEH2B and RNASEH2C serve as a nucleation site for the addition of RNASEH2A to form an active RNase H2. Furthermore, they contain interaction sites with other proteins to support functions other than RNase H nuclease activity, but these functions are not well-defined yet.
RNase H2 deficiency can cause a number of pathogenetic principles including the occurrence of ribonucleoside monophosphates accumulating in genomic DNA and activating the DNA damage-response pathway. RNase H2 deficiency leads to abundance of cytosolic RNA–DNA hybrids and to an increase in retroelements which both represent potential ligands for the cGAS–STING signaling pathway (Mankan et al., 2014, Rigby et al., 2014). In fact, mutations in any of the three subunits RNASEH2A-, RNASEH2B-, or RNASEH2C of human RNase H2 cause AGS, a human neurological disorder with devastating consequences (Rice et al., 2007). Mutations that impair RNase H2 are also associated with systemic lupus erythematosus (SLE). Pathogenicity is supported by mouse models of AGS-associated mutations of RNase H which show a spontaneous cGAS/STING-dependent type I IFN-driven phenotype (Mackenzie et al., 2016, Pokatayev et al., 2016).
7.7. DNases
7.7.1. DNase I
Deoxyribonuclease I (DNase I) is an endonuclease which is secreted to cleave DNA in the extracellular space down to an average of tetranucleotides with 5′ monophosphate and 3′ hydroxyl DNA ends (Baranovskii, Buneva, & Nevinsky, 2004). Both single-stranded DNA and double-stranded DNA are degraded by DNase I. This nuclease appears to account for the major nucleolytic activity on DNA in serum and is responsible for the degradation of the majority of circulating DNA derived from apoptotic and necrotic cell death and from neutrophil extracellular traps. In addition to its role in the serum, it has been proposed as one of the deoxyribonucleases responsible for DNA fragmentation in the process of apoptosis (Samejima & Earnshaw, 2005). Notably, DNase1L3 complements the activity of DNase I. Although DNase1L3 harbors nuclear localization signals, its main function appears to be in the serum, where it can degrade protein-complexed DNA (Napirei, Ludwig, Mezrhab, Klockl, & Mannherz, 2009).
In the absence of DNase I, degradation of extracellular DNA is heavily reduced resulting in the activation of DNA-sensing immune receptors. Mice deficient in DNase I display a lupus-like phenotype with increased antinuclear antibody titers and glomerulonephritis (Napirei et al., 2000). Mutations in the human DNase I gene and factors inactivating DNase I have been associated with SLE (Hakkim et al., 2010, Yasutomo et al., 2001). In a subset of SLE patients, the presence of DNase I inhibitors or autoantibodies was associated with impaired DNA clearance and poor prognosis, suggesting that decreased DNase I activity can also be acquired (Hakkim et al., 2010). Notably, loss-of-function variants in DNase1L3 have also been associated with familiar SLE, supporting an important functional contribution of DNase1L3 in clearing DNA (Al-Mayouf et al., 2011).
7.7.2. DNase II
DNase II is a mammalian endonuclease with highest activity at low pH and in the absence of divalent cations. DNase II cleaves DNA between 5′-phosphate and 3′-hydroxyl resulting in the formation of nucleoside-3′-phosphates. DNase II is the predominant DNase located in lysosomes of cells in various tissues including macrophages (Evans and Aguilera, 2003, Yasuda et al., 1998). With its lysosomal localization and ubiquitous tissue distribution, this enzyme plays a pivotal role in the degradation of exogenous DNA encountered by endocytosis. It has been demonstrated that digestion of large DNA molecules and of CpG-A (Hartmann et al., 2003) by DNase II creates short DNA fragments which are sensed by TLR9 (Chan et al., 2015, Pawaria et al., 2015).
DNase II deficiency is another example of a cell-intrinsic nuclease defect driving autoimmunity. Loss of DNase II leads to a defect in the disposal of DNA within lysosomal compartments (Kawane et al., 2006, Yoshida et al., 2005). Accumulation of undigested DNA can result in the translocation of DNA into the cytoplasm which is then sensed by the cGAS–STING pathway (Ahn et al., 2012, Gao et al., 2015) as well as the AIM2 inflammasome (Baum et al., 2015, Jakobs et al., 2015). Mice lacking DNase II display an inflammatory response that depends on both cGAS and AIM2. Besides cell-extrinsic sources of DNA (e.g., nuclei expelled from erythroid precursor cells), recent work in DNase II-deficient mice suggests that damaged nuclear DNA is also subject to DNase II degradation and might stimulate cytosolic DNA immune-sensing receptors if not properly degraded (Lan, Londono, Bouley, Rooney, & Hacohen, 2014). In a model of cardiac-specific deletion of DNase II, severe myocarditis and dilated cardiomyopathy developed which was attenuated if immune sensing of accumulating mitochondrial DNA by TLR9 was inhibited (Oka et al., 2012).
7.7.3. DNase III/Trex1
The cytoplasmic DNase III (3′-repair exonuclease 1, Trex1) has been identified decades later than DNase I and II (Hoss et al., 1999). Trex1 is a 3′- to 5′- exonuclease which degrades both double- and single-stranded DNA. Most DNA reaching the cytosol is promptly removed by Trex1. Modifications have been reported which render DNA resistant to Trex1. For example, oxidation of guanine bases to 8-hydroxydeoxyguanine (8-OHdG) protects DNA from TREX1-dependent degradation leading to accumulation and cGAS-mediated recognition of oxidized DNA in the cytosol (Gehrke et al., 2013).
Only since 2006, it is known that loss of function in Trex1 causes the type I IFN-associated inflammatory syndrome AGS (Crow et al., 2006), suggesting that Trex1 is critically involved in the clearance self DNA within the cytoplasm of cells which otherwise is recognized by the immune sensor cGAS. Defects in Trex1 have been associated with SLE (Lee-Kirsch, Gong, et al., 2007) and with familial chilblain lupus (Lee-Kirsch, Chowdhury, et al., 2007); furthermore, genetic defects in Trex1 can cause retinal vasculopathy with cerebral leukodystrophy (Richards et al., 2007).
Trex1-deficient mice develop severe autoimmunity (Gall et al., 2012, Morita et al., 2004). The pathology is fully rescued by additional genetic defects in cGAS (Ablasser et al., 2014, Gao et al., 2015, Gray et al., 2015) or the type I IFN system demonstrating the critical pathogenic role of the IFN response triggered by endogenous DNA species. The source and identity of this DNA remain controversial but may derive from endogenous retroelements (Beck-Engeser et al., 2011, Stetson et al., 2008). Alternatively, DNA ligands originating during chromosomal replication have been proposed (Yang, Lindahl, & Barnes, 2007).
8. RNA Interference
RNAi is considered one of the major mechanism for sequence-specific detection and elimination of RNA genome-based viruses in plants and invertebrates (Szittya and Burgyan, 2013, Zhou and Rana, 2013). Besides its antiviral function, RNAi regulates gene expression in many organisms. By suppressing transcription or translation or by targeted degradation of mRNA, it controls many cellular developmental and physiological processes (Burger & Gullerova, 2015). RNAi is initiated by RNAse III family nucleases (nuclear Drosha and cytosolic Dicer) that cleave endogenous or exogenous double-stranded RNA to finally yield short 21–23 bp exogenous siRNA or endogenous miRNA (Bernstein et al., 2001, Elbashir et al., 2001, Lee et al., 2003). The miRNAs/siRNAs are then integrated in the RNA-induced silencing complex (RISC) which target complementary RNA for degradation or inhibition of translation (Iwakawa & Tomari, 2015). AGO family proteins in the RISC complex determine its effector function: perfectly matched mi/siRNAs mediate direct target cleavage by AGO2, while imperfectly matched mi/siRNAs inhibit translation of target mRNAs by AGO1, 3, or 4 and recruit additional effector proteins which in turn can degrade target RNA (Doench et al., 2003, Meister et al., 2004).
While an important role for RNAi for the antiviral responses in helminths, insects, and plants is well established, the contribution to antiviral immunity in vertebrates is under debate. Evidence accumulates for an antiviral role of RNAi in mammalian cells (Gantier, 2014), specifically if the otherwise dominating Dicer-related RIG-I-like helicases are inhibited. The major obstacle is that the contribution of a siRNA-mediated antiviral response cannot be studied by the knockout of Dicer which is lethal at early stages of mouse embryo development (Bernstein et al., 2003). It was reported that type I IFN-dominated innate immune responses suppress RNAi and vice versa (Seo et al., 2013). The absence of Dicer products from small RNA libraries of (+)ssRNA virus (YFV, HCV) infections strengthens this assumption (Pfeffer et al., 2005). However, more recent studies using the more sensitive next-generation sequencing indeed provide evidence for the generation of short virus-derived small RNAs (vsRNAs) in complex with AGO proteins and conforming to Dicer cleavage fragments of 24–31 bp (siRNA and piRNA) in vertebrate infection systems (Parameswaran et al., 2010). In a seminal work, suckling mice were infected by Nodamura virus (NoV) or a mutant virus lacking the NoV virus-encoded suppressor of RNAi, B2 (Li, Lu, Han, Fan, & Ding, 2013). NoV is a mosquito-transmissible (+)ssRNA virus, which is highly virulent to suckling mice and suckling hamsters. Loss of B2 leads to abundant occurrence of viral siRNAs and rendered mice completely resistant to NoV titers which are lethal in the presence of B2 (Li et al., 2013). Since B2 appears not to prevent recognition by RIG-I (Fan, Dong, Li, Ding, & Wang, 2015), an important innate immune sensor of (+)ssRNA-based viruses, the data suggest a strong role of virus RNA-specific RNAi during NoV infection. Still, an impact of B2 on endogenous miRNA networks or inhibition of other dsRNA-sensing receptors (MDA5, PKR, OAS) as a major cause of lethality is not completely excluded in this work and requires further analysis. Nevertheless, this study adds to the concept that virus-encoded suppressors of RNAi mask the actual role of RNAi in antiviral defense of vertebrates (VA1 noncoding RNA, Influenza NS1, vaccinia virus E3L, Ebola virus VP35, primate foamy virus Tas, HIV-1 Tat West Nile virus sfRNA) (Bennasser et al., 2005, Haasnoot et al., 2007, Lecellier et al., 2005, Li et al., 2004, Lu and Cullen, 2004, Schnettler et al., 2012, Svoboda, 2014). Importantly, murine or rat oocytes or embryonic stem cells in rat and mouse express a shortened form of Dicer (DicerO) with enhanced cleavage activity for long dsRNA which complicate the interpretation of results (Flemr et al., 2013). On the other hand, the finding that poly(I:C) stimulation or infection with DNA or RNA viruses leads to PARP13 induced poly-ADP-ribosylation of AGO2 which induces AGO2 degradation in all tested cells strongly indicates a competition rather than a cooperation between antiviral RNAi and nucleic acid immune-sensing pathways (Seo et al., 2013).
9. Immune-Sensing Receptors
Although some recent work propose a role of RNAi in the antiviral defense of vertebrates as described earlier, the current concept is that immune-sensing receptors represent the major antiviral defense strategy in vertebrates. Immune-sensing receptors recognize characteristic features of foreign and invading nucleic acids such as unusual localization, specific structural elements, and modifications. Stimulation of nucleic acid-sensing receptors results in the induction of cytokines (e.g., type I interferons) and chemokines to alarm neighboring cells, in the recruitment of immune cells, in the activation of cell autonomous mechanisms interfering with virus assembly and protein translation, or in the induction of several types of cell death including apoptosis, necroptosis, and pyroptosis. This chapter provides an overview of the most important immune-sensing receptors of nucleic acids for which robust evidence exists regarding the molecular mechanism of detection, the structural aspects of receptor ligand interaction, and the downstream signaling pathways (Fig. 6 ).
9.1. TLR3
TLR3 detects long double-stranded RNA (Alexopoulou et al., 2001), and unlike other nucleic acid-sensing TLRs, besides its endolysosomal localization, it is also expressed on the surface of certain cell types (Matsumoto et al., 2002, Pohar et al., 2013). TLR3 detects dsRNA longer than 40 bp. The ectodomains of two TLR3 molecules bind one dsRNA molecule in a way that the cytoplasmic C-terminal signaling domains are juxtaposed to each other resulting in downstream signaling (Liu et al., 2008). TLR3 interacts with the ribose-phosphate backbone of dsRNA and has no specific sequence requirements. Given the absence of long dsRNA under physiological conditions, TLR3 should be inactive in the absence of an infection. Still, a number of studies proposed recognition of endogenous dsRNA by TLR3 in situations of sterile tissue damage, but the specific ligand is not well defined.
9.2. TLR7 and TLR8
The identification of ligand specificities of TLR7 and TLR8 has been hampered by their mutually exclusive expression in different cell types and by considerable differences between mouse and human. TLR7 and TLR8 are examples where distinct function of nucleic acid-sensing TLRs is determined by their differential expression in immune cell subsets. While the expression of TLR7 in the human immune system is almost restricted to B cells and PDC, TLR8 is preferentially expressed in myeloid immune cells. Consequently, TLR7 ligands drive B cell activation and the production of large amounts of IFN-alpha in PDC, while TLR8 induces the secretion of high amounts of IL-12p70 in myeloid immune cells.
TLR7 and TLR8 are preferentially activated by polyU or by G- and U-rich sequences (Diebold et al., 2004, Heil et al., 2004, Hornung et al., 2005, Judge et al., 2005). However, confounding factors need to be considered while interpreting these results (Forsbach et al., 2008). Furthermore, it has been demonstrated that TLR8 selectively detects ssRNA, while TLR7 primarily detects short stretches of dsRNA but can also accommodate certain ssRNA oligonucleotides (Ablasser et al., 2009, Sioud, 2006). However, since neither polyU or G- and U-rich sequences nor ssRNA or short dsRNA structures are overrepresented in microbial or viral RNA, the distinction of self vs nonself by TLR7 and TLR8 requires additional information. In fact, endogenous RNAs are posttranscriptionally modified at their nucleobases and backbone. The current concept is that the addition of certain modifications to self RNA inside the nucleus provides a signature for self. One example is 2′-O-methylation which is a common nuclear modification of RNA performed by a specific MTase located in the nucleolus. The MTase adds a methyl group to the 2′-hydroxyl group of the ribose. This modification represents a marker of self in higher eukaryotic cells and prevents the recognition of endogenous RNA by TLR7 and TLR8 (Hornung et al., 2005, Judge et al., 2005, Kariko et al., 2005). Other modifications of RNA molecules which potently inhibit the detection of transfer RNA and ribosomal RNA by TLR7 and TLR8 are the incorporation of pseudouridine (Ψ), 5-methylcytidine (m5C), 2-thio-uridine (s2U), or N6-methyladenosine (m6A) (Kariko et al., 2005). The presence of such modifications in part explains the lack of immunostimulation of host-derived RNA vs microbial RNA (Gehrig et al., 2012, Jockel et al., 2012). However, endogenous RNA from apoptotic or dying cells still activates TLR7 and TLR8 once entering the endolysosomal compartment (Busconi et al., 2006, Ganguly et al., 2009, Vollmer et al., 2005). Thus, additional factors such as intracellular localization and degradation by nucleases likely support a faithful discrimination of self from nonself by TLR7 and TLR8. It is interesting to note that there is an obvious need to sense and eliminate certain endogenous RNAs as well. In this context, it has been reported that the loss of TLR7 function causes retroviral viremia (Yu et al., 2012) indicating that endogenous RNAs transcribed from RNA polymerase II promoters are not generally excluded from TLR-mediated recognition.
9.3. TLR9
TLR9 senses DNA in the endolysosomal compartment of certain immune cells (Hemmi et al., 2000). Like TLR7 and TLR8, TLR9 travels to the endolysosomal compartment via the chaperone protein UNC93B1 (protein unc-93 homolog B1) (Latz et al., 2004, Pelka et al., 2016). There, TLR9 is proteolytically processed at a defined protruding loop structure without disrupting the horseshoe shape of the protomer (Bauer, 2013, Onji et al., 2013, Peter et al., 2009). Cleavage is necessary for the activation of TLR signaling (Ewald et al., 2008, Park et al., 2008).
TLR9 preferentially detects DNA with unmethylated (no methylation at the C5 carbon of cytosine) CpG dinucleotides (CpG DNA) with a preference for certain sequence contexts (hexamer CpG motifs, in humans 5′-GTCGTT-3) which vary between species and which altogether are less frequent in eukaryotic self DNA (Hartmann and Krieg, 1999, Hartmann and Krieg, 2000, Hartmann et al., 2000, Hartmann et al., 1999, Hemmi et al., 2000, Krieg et al., 1995). Activation of TLR9 signaling is preceded by dimer formation where two CpG DNA molecules symmetrically bind two TLR9 molecules (Ohto et al., 2015). Both CpG DNA molecules bind to the C-terminal fragment of one protomer and the CpG-binding groove in the N-terminal fragment of the other. Inhibitory DNA oligonucleotides only bind to the N-terminal fragment. Methylated single-stranded CpG DNA and double-stranded DNA exhibit lower binding to TLR9 and are less potent to induce TLR9 dimer formation. Of note, digestion of DNA molecules by the lysosomal endonuclease DNase II creates short TLR9-stimulatory DNA fragments (Chan et al., 2015, Pawaria et al., 2015).
Notably, specificity for unmethylated CpG motifs is reduced if the CpG motif is within a phosphorothioate-modified DNA often used to stabilize oligodeoxynucleotides against DNases. Nevertheless, a high degree of specificity is well established for unmethylated CpG motif containing DNA within a natural phosphodiester backbone including microbial DNA (Coch et al., 2009, Hartmann and Krieg, 2000). It is specifically noteworthy that genomic microbial DNA displays a much stronger activity to stimulate TLR9 as compared to genomic DNA of vertebrates. Although eukaryotic DNA presents with a lower frequency of nonmethylated CpG motifs than microbial DNA, this difference in frequency of unmethylated CpG motifs does not allow a clear cut distinction of self from nonself on a structural basis. Endogenous DNA at high concentrations can activate TLR9 once delivered into the endolysosome (Marshak-Rothstein, 2006).
It is also of great importance to be aware of the differences of TLR9-mediated DNA recognition between species. In humans, TLR9 is almost exclusively expressed in B cells and PDC (Hornung et al., 2002), while in mice, TLR9 is expressed more widely including myeloid immune cells. In humans, TLR9 predominantly induces type IFN production in PDCs and polyclonal activation in B cells via MyD88/IRF7-dependent signaling. Species-specific expression patterns of TLR9 are responsible for the fundamental functional differences of TLR9 in mouse and man. Another important issue is that in preparations of human immune cell subsets, minute amounts of PDC indirectly activate other immune cell subsets such as monocytes, myeloid dendritic cells, or NK cells. This needs to be carefully considered when direct effects of TLR9 activation in immune cells other than B cells and PDC are claimed, such as direct TLR9 effects in human myeloid immune cells and NK cells. Three different classes of CpG oligonucleotides have been described, CpG-A, CpG-B, and CpG-C (Avalos et al., 2009, Hartmann et al., 2003, Kerkmann et al., 2003, Krug et al., 2003, Rothenfusser et al., 2004). Based on the palindromic structure, CpG-A spontaneously forms nanoparticle-like complexes (Kerkmann et al., 2005) that explain much higher type I IFN-inducing capacity in PDC as compared to CpG-B which are monomeric. Monomeric CpG-B potently activates B cells which do not internalize larger particles of DNA as with CpG-A complexes. CpG-C potently stimulates both B cells and PDCs. In cell culture, delayed TLR9 activation due to slower uptake of CpG-A nanoparticles allows a longer self-priming of PDC by minute amounts of spontaneously released type I IFN. Priming of PDC results in higher IFN-inducing activity of CpG-A seen in cell culture (Kim et al., 2014).
9.4. RIG-I
RIG-I belongs to the cytosolic DExD/H box RNA helicases and is one of three members of the so-called family of RIG-I-like helicases (others: MDA5 and LGP2). RIG-I is closely related to the Dicer family of helicases of the RNAi pathway. RIG-I contains a RNA helicase domain and a two N-terminal CARD domains which relay the signal to the downstream signaling adaptor MAVS (mitochondrial antiviral-signaling protein). RIG-I signaling via MAVS not only leads to the induction of type I IFN responses via TBK1 and IRF7/8, it also activates caspase-8-dependent apoptosis, preferentially in tumor cells (Besch et al., 2009, El Maadidi et al., 2014, Glas et al., 2013, Kumar et al., 2015). Furthermore, RIG-I was also found to mediate MAVS-independent inflammasome activation (Poeck et al., 2010), specifically in the context of viral infection (Poeck et al., 2010, Pothlichet et al., 2013).
RIG-I detects blunt ends of double-stranded RNA containing a 5′-triphosphate or a 5′-diphosphate (Goubau et al., 2014, Hornung et al., 2006, Marq, Hausmann, et al., 2010, Marq, Kolakofsky and Garcin, 2010, Pichlmair et al., 2006, Schlee, Roth, et al., 2009). Such RNA ligands are presented, for example, by negative-strand RNA viruses which form panhandle structures with their matching 5′- and 3′-ends of the single-stranded genomic RNA (Rehwinkel et al., 2010, Schlee, Roth, et al., 2009). While crystal structures confirmed the structural requirements as determined in functional studies (Civril et al., 2011, Jiang et al., 2011, Kowalinski et al., 2011, Luo et al., 2011, Wang et al., 2010), the minimum length of the double-strand required for RIG-I activation is still controversial. While approaches with synthetic or highly purified enzymatic double-stranded 5′-triphosphate RNA revealed a minimum length of 18–19 bp (Marq, Hausmann, et al., 2010, Schlee, Hartmann, et al., 2009), 10 bp were demonstrated to be sufficient for a hairpin forming oligonucleotide (Kohlway, Luo, Rawling, Ding, & Pyle, 2013). However, alternatively to the predicted hairpin, these oligonucleotides may form 20mer duplexes when entering the cell.
Although oligomerization of 2CARD modules of each RIG-I protein along the RIG-I filament bound to a longer double-stranded RNA molecule induces robust RIG-I signaling, the minimal signaling unit is sufficient for RIG-I to trigger signal transduction. In the latter case, a 2CARD tetramer is stabilized by ubiquitin chains (Wu & Hur, 2015). Furthermore, it is interesting to note that RIG-I mutants deficient in ATP hydrolysis of their helicase domain cannot detach from suboptimal endogenous RNA ligands leading to erroneous signaling which can cause autoimmunity (Lassig et al., 2015). Importantly, N1-methylation (2′-O-methylation at the first nucleotide of capped RNA) serves as a signature of self RNA and completely abrogates RIG-I sensing of RNA, while in the absence of N1 methylation, RIG-I binding is hardly impaired by the 5′ppp5′-linked m7G cap structure itself (Schuberth-Wagner et al., 2015).
RIG-I is ubiquitously expressed in all cell types including tumor cells. However, the type of RIG-I induced responses differs between cells. While normal healthy cells such as melanocytes and fibroblasts are quite resistant to RIG-I-induced apoptosis, tumor cells are highly susceptible to RIG-I-induced cell death (Besch et al., 2009, Kubler et al., 2010). Based on this tumor selective activity and a favorable toxicity profile, RIG-I-specific ligands are currently being developed for immunotherapy of cancer (Duewell et al., 2014, Duewell et al., 2015, Ellermeier et al., 2013, Schnurr and Duewell, 2013, Schnurr and Duewell, 2014). Part of the potent antitumor activity of RIG-I ligands is its ability to promote crosspresentation of antigens to CD8 T cells and to induce cytotoxic activity (Hochheiser et al., 2016). RIG-I ligands show strong therapeutic activity in viral infection models such as influenza (Weber-Gerlach & Weber, 2016). Notably, RIG-I has also been shown to be involved in the detection of intracellular bacteria (Abdullah et al., 2012). Rare genetic gain-of-function variants of RIG-I have been associated with an atypical form of Singleton Merten syndrome (Jang et al., 2015).
9.5. MDA5 and LGP2
Like RIG-I, melanoma differentiation associated gene 5 (MDA5) is a cytosolic DExD/H box RNA helicase which signals through MAVS and IRF3/IRF7 (Yoneyama et al., 2005). Despite its similar structure, MDA5 senses a different type of ligand which has been described as higher order RNA structures (Pichlmair et al., 2009). So far, a MDA5-specific ligand has not been described. Double-stranded RNA ligands activating MDA5 are typically promiscuous ligands, such as poly(I:C) which also activates TLR3 and antiviral effector proteins which inhibit translation upon binding to double-stranded RNA, such as PKR and OAS. Multiple effects of MDA5 ligands cause a high degree of toxicity in vivo strictly limiting the clinical application of MDA5 ligands.
Unlike RIG-I which primarily binds to the ends of RNA, MDA5 proteins bind double-stranded RNA internally, independently of its terminal structures. Additional MDA5 molecules then closely stack in a helical head-to-tail arrangement around dsRNA resulting in the formation of long MDA5 filaments which initiate signaling toward activation of MAVS (del Toro Duany, Wu, & Hur, 2015). LGP2 (laboratory of genetics and physiology 2) is a third cytosolic RIG-I-like helicase lacking CARD domains for signaling. LGP2 appears to contribute to the fine tuning of immune responses by inhibition of RIG-I and supporting MDA5 signaling (Rothenfusser et al., 2005, Venkataraman et al., 2007). While MDA5 contains the signaling CARD domains but has relatively weak binding to double-stranded RNA, LGP2 readily detects diverse double-stranded RNA species but lacks a signaling domain. The current concept is that LGP2 assists the interaction of MDA5 with double-stranded RNA and filament formation, thereby enhancing MDA5-mediated antiviral signaling (Bruns and Horvath, 2015, Bruns et al., 2014). Notably, genetic gain-of-function variants of MDA5 have been associated with autoimmune disorders (Junt and Barchet, 2015, Kato and Fujita, 2015).
9.6. AIM2
The HIN-200 (hematopoietic interferon-inducible nuclear proteins with a 200-amino acid repeat) family member AIM2 (absent in melanoma 2) binds and oligomerizes on cytoplasmic double-stranded DNA through its C-terminal HIN domain in a sequence-independent manner (Fernandes-Alnemri et al., 2009, Hornung et al., 2009, Roberts et al., 2009). DNA binding of the HIN domain relieves the autoinhibitory conformation of AIM2 and allows the N-terminal pyrin domain of multiple AIM2 proteins to form a helical structure which nucleates the helical assembly of ASC (apoptosis-associated speck-like protein containing a CARD) filaments (Lu, Kabaleeswaran, et al., 2014, Lu, Magupalli, et al., 2014, Lu et al., 2015) thereby forming an inflammasome that results in the release of IL-1beta. The formation of an AIM2 inflammasome requires a minimal length of double-stranded DNA of 50–80 bp (Jin et al., 2012).
9.7. cGAS/Sting
The cytosolic immune-sensing receptor cGAS (Cai et al., 2014, Wu et al., 2012) detects long double-stranded DNA (dsDNA) or short dsDNA with unpaired open ends containing guanosines (Y-form DNA) as, for example, presented in highly structured single-stranded DNA of retroviruses or certain endogenous retroelements (Herzner et al., 2015). It is important to note that Trex1 has a gate keeper function for cGAS. Usually, cytosolic DNA is efficiently degraded by TREX1. Only in the case of excess cytosolic DNA, or DNA modifications rendering DNA resistant to Trex1-mediated degradation, DNA gains access to cGAS resulting in downstream signaling. Along these lines, it has been reported that oxidized DNA (e.g., 8-hydroxyguanosine, 8-OHG) as occurring in the context of UV radiation or upon exposure to reactive oxygen species resists Trex1-mediated degradation (Gehrke et al., 2013). This results in an accumulation of DNA in the cytosol. Oxidized DNA has the same affinity to cGAS than nonoxidized DNA.
cGAS is monomeric in its unligated state. However, two cGAS molecules bind to two dsDNA molecules in a way that each cGAS protomer presents an additional interaction site with the DNA bound to the other cGAS protein. Upon activation by cytosolic DNA, cGAS catalyzes the formation of 2′–5′-cGAMP from GTP and ATP (Ablasser, Goldeck, et al., 2013, Gao, Ascano, et al., 2013a). 2′–5′-cGAMP acts as a second messenger which binds to the downstream signaling protein Sting (Gao, Ascano, Zillinger, et al., 2013) which induces type I interferon via TBK1 and IRF3. 2′–5′-cGAMP can travel to and alarm neighboring cells via gap junctions (Ablasser, Schmid-Burgk, et al., 2013). Sting activation is also associated with NF-κB activation and prominent induction of inflammatory cytokines such as IL-6 and TNF-a.
10. Conclusions
The general view is that most of the relevant immune receptors, effector proteins, and pathways participating in nucleic acid immunity have now been identified. These different players have in common that they all serve the function to detect and to disable foreign nucleic acids. Altogether they constitute the system of nucleic acid immunity. All of these pathways are relevant for human disease, either as part of the human antiviral defense system, or indirectly by being active in pathogens or pathogen-transmitting organisms. Examples are arboviruses (Zika, Dengue, Yellow-fever, West Nile) which are transmitted by arthropod vectors. Successful arboviruses need to escape both RNAi in insects and immunoreceptors such as RIG-I in humans. Only if they manage to inhibit both pathways, they can establish as pathogens. Therefore, it will be interesting to understand the molecular evolution of escape strategies in emerging viruses such as Zika, which may lead to the identification of the molecular step that allowed the virus to spread more efficiently (Rasmussen & Katze, 2016). Another example is the important role of RNAi in pathogens such as filariae and other human pathogenic helminths. There, RNAi may be useful as a therapeutic target. Another example is the presence of endosymbionts such as Wolbachia, a genus of bacteria, in filarial nematodes (Taylor, Bandi, & Hoerauf, 2005). The release of Wolbachia nucleic acids may contribute to the pathogenesis of filarial infection. Furthermore, CRISPR/Cas is involved in the evolution of pathogenic bacteria.
Nucleic acid sensing in vertebrates is required for antimicrobial immunity and is involved in the pathogenesis of many inflammatory diseases. Although much is known about the structure and the function of the single pathways, the functional interaction of the different pathways is far from being understood. Distinct expression patterns of the receptors in different cell types and cell-type-dependent differences in the expression of downstream signaling components and transcription factors contribute to the complexity of nucleic acid immunity. As a result, the same type of ligand can have different functional outcomes in different cell types. Furthermore, since different receptor pathways are activated by the same type of ligand, it is currently unclear whether competition of receptor binding or distinct molecular trafficking to the corresponding receptors or both impact on the functional outcome of ligand exposure. For example, long double-stranded DNA in principle binds to both AIM2 and cGAS, and functional cooperation or inhibition of the respective pathways are unclear.
Now since most of the individual molecular pathways of nucleic acid immunity are on the table, we see ourselves just at the dawn of an exciting new research field which is expected to advance medicine specifically in the areas of infection and inflammation and with broad implication for human diseases.
Acknowledgments
This work was supported by the DFG-funded excellence cluster ImmunoSensation, and the Helmholtz-funded German Center for Infection Research (Deutsches Zentrum für Infektionsforschung, DZIF).
References
- Abbas Y.M., Pichlmair A., Gorna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah Z., Schlee M., Roth S., Mraheil M.A., Barchet W., Bottcher J. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. The EMBO Journal. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K.A., Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature Immunology. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Rohl I. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A., Hemmerling I., Schmid-Burgk J.L., Behrendt R., Roers A., Hornung V. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. Journal of Immunology. 2014;192:5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- Ablasser A., Schmid-Burgk J.L., Hemmerling I., Horvath G.L., Schmidt T., Latz E. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J., Gutman D., Saijo S., Barber G.N. STING manifests self DNA-dependent inflammatory disease. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Al-Mayouf S.M., Sunker A., Abdwani R., Abrawi S.A., Almurshedi F., Alhashmi N. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nature Genetics. 2011;43:1186–1188. doi: 10.1038/ng.975. [DOI] [PubMed] [Google Scholar]
- Avalos A.M., Latz E., Mousseau B., Christensen S.R., Shlomchik M.J., Lund F. Differential cytokine production and bystander activation of autoreactive B cells in response to CpG-A and CpG-B oligonucleotides. Journal of Immunology. 2009;183:6262–6268. doi: 10.4049/jimmunol.0901941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglioni C., Minks M.A., Maroney P.A. Interferon action may be mediated by activation of a nuclease by pppA2'p5'A2'p5'A. Nature. 1978;273:684–687. doi: 10.1038/273684a0. [DOI] [PubMed] [Google Scholar]
- Baldauf H.M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nature Medicine. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskii A.G., Buneva V.N., Nevinsky G.A. Human deoxyribonucleases. Biochemistry. Biokhimiia. 2004;69:587–601. doi: 10.1023/b:biry.0000033731.50496.01. [DOI] [PubMed] [Google Scholar]
- Barber G.N. STING-dependent cytosolic DNA sensing pathways. Trends in Immunology. 2014;35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bass B.L., Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- Bass B.L., Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Bauer S. Toll-like receptor 9 processing: The key event in Toll-like receptor 9 activation? Immunology Letters. 2013;149:85–87. doi: 10.1016/j.imlet.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Baum R., Sharma S., Carpenter S., Li Q.Z., Busto P., Fitzgerald K.A. Cutting edge: AIM2 and endosomal TLRs differentially regulate arthritis and autoantibody production in DNase II-deficient mice. Journal of Immunology. 2015;194:873–877. doi: 10.4049/jimmunol.1402573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck-Engeser G.B., Eilat D., Wabl M. An autoimmune disease prevented by anti-retroviral drugs. Retrovirology. 2011;8:91. doi: 10.1186/1742-4690-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt R., Schumann T., Gerbaulet A., Nguyen L.A., Schubert N., Alexopoulou D. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Reports. 2013;4:689–696. doi: 10.1016/j.celrep.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova N., Flick R., Tchigvintsev A., Brown G., Popovic A., Nocek B. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. The Journal of Biological Chemistry. 2013;288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y., Le S.Y., Benkirane M., Jeang K.T. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z. Dicer is essential for mouse development. Nature Genetics. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Besch R., Poeck H., Hohenauer T., Senft D., Hacker G., Berking C. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. The Journal of Clinical Investigation. 2009;119:2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan-Laun S.E., Ezelle H.J., Li X.L., Hassel B.A. RNase-L control of cellular mRNAs: Roles in biologic functions and mechanisms of substrate targeting. Journal of Interferon & Cytokine Research: The Official Journal of the International Society for Interferon and Cytokine Research. 2014;34:275–288. doi: 10.1089/jir.2013.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns A.M., Horvath C.M. LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine. 2015;74:198–206. doi: 10.1016/j.cyto.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns A.M., Leser G.P., Lamb R.A., Horvath C.M. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Molecular Cell. 2014;55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger K., Gullerova M. Swiss army knives: Non-canonical functions of nuclear Drosha and Dicer. Nature Reviews. Molecular Cell Biology. 2015;16:417–430. doi: 10.1038/nrm3994. [DOI] [PubMed] [Google Scholar]
- Busconi L., Lau C.M., Tabor A.S., Uccellini M.B., Ruhe Z., Akira S. DNA and RNA autoantigens as autoadjuvants. Journal of Endotoxin Research. 2006;12:379–384. doi: 10.1179/096805106X118816. [DOI] [PubMed] [Google Scholar]
- Cai X., Chiu Y.H., Chen Z.J. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Molecular Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Castelli J.C., Hassel B.A., Maran A., Paranjape J., Hewitt J.A., Li X.L. The role of 2'-5' oligoadenylate-activated ribonuclease L in apoptosis. Cell Death and Differentiation. 1998;5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- Cerritelli S.M., Crouch R.J. Ribonuclease H: The enzymes in eukaryotes. The FEBS Journal. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M.P., Onji M., Fukui R., Kawane K., Shibata T., Saitoh S. DNase II-dependent DNA digestion is required for DNA sensing by TLR9. Nature Communications. 2015;6:5853. doi: 10.1038/ncomms6853. [DOI] [PubMed] [Google Scholar]
- Chon H., Vassilev A., DePamphilis M.L., Zhao Y., Zhang J., Burgers P.M. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Research. 2009;37:96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W.M., Ostertag D., Li Z.W., Chang L., Chen Y., Hu Y. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- Civril F., Bennett M., Moldt M., Deimling T., Witte G., Schiesser S. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Reports. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M.J., Elia A. The double-stranded RNA-dependent protein kinase PKR: Structure and function. Journal of Interferon & Cytokine Research: The Official Journal of the International Society for Interferon and Cytokine Research. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- Clemens M.J., Williams B.R. Inhibition of cell-free protein synthesis by pppA2'p5'A2'p5'A: A novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell. 1978;13:565–572. doi: 10.1016/0092-8674(78)90329-x. [DOI] [PubMed] [Google Scholar]
- Coch C., Busch N., Wimmenauer V., Hartmann E., Janke M., Abdel-Mottaleb M.M. Higher activation of TLR9 in plasmacytoid dendritic cells by microbial DNA compared with self-DNA based on CpG-specific recognition of phosphodiester DNA. Journal of Leukocyte Biology. 2009;86:663–670. doi: 10.1189/jlb.0509314. [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nature Genetics. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: Linking nucleic acid metabolism with autoimmunity. Human Molecular Genetics. 2009;18:R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Toro Duany Y., Wu B., Hur S. MDA5-filament, dynamics and disease. Current Opinion in Virology. 2015;12:20–25. doi: 10.1016/j.coviro.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes & Development. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Silverman R.H. A bipartite model of 2-5A-dependent RNase L. The Journal of Biological Chemistry. 1997;272:22236–22242. doi: 10.1074/jbc.272.35.22236. [DOI] [PubMed] [Google Scholar]
- Duewell P., Beller E., Kirchleitner S.V., Adunka T., Bourhis H., Siveke J. Targeted activation of melanoma differentiation-associated protein 5 (MDA5) for immunotherapy of pancreatic carcinoma. Oncoimmunology. 2015;4 doi: 10.1080/2162402X.2015.1029698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duewell P., Steger A., Lohr H., Bourhis H., Hoelz H., Kirchleitner S.V. RIG-I-like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8(+) T cells. Cell Death and Differentiation. 2014;21:1825–1837. doi: 10.1038/cdd.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Maadidi S., Faletti L., Berg B., Wenzl C., Wieland K., Chen Z.J. A novel mitochondrial MAVS/Caspase-8 platform links RNA virus-induced innate antiviral signaling to Bax/Bak-independent apoptosis. Journal of Immunology. 2014;192:1171–1183. doi: 10.4049/jimmunol.1300842. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Development. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier J., Wei J., Duewell P., Hoves S., Stieg M.R., Adunka T. Therapeutic efficacy of bifunctional siRNA combining TGF-beta1 silencing with RIG-I activation in pancreatic cancer. Cancer Research. 2013;73:1709–1720. doi: 10.1158/0008-5472.CAN-11-3850. [DOI] [PubMed] [Google Scholar]
- Evans C.J., Aguilera R.J. DNase II: Genes, enzymes and function. Gene. 2003;322:1–15. doi: 10.1016/j.gene.2003.08.022. [DOI] [PubMed] [Google Scholar]
- Ewald S.E., Lee B.L., Lau L., Wickliffe K.E., Shi G.P., Chapman H.A. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Dong S., Li Y., Ding S.W., Wang M. RIG-I-dependent antiviral immunity is effective against an RNA virus encoding a potent suppressor of RNAi. Biochemical and Biophysical Research Communications. 2015;460:1035–1040. doi: 10.1016/j.bbrc.2015.03.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell P.J., Sen G.C., Dubois M.F., Ratner L., Slattery E., Lengyel P. Interferon action: Two distinct pathways for inhibition of protein synthesis by double-stranded RNA. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:5893–5897. doi: 10.1073/pnas.75.12.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G.S., Chong K., Kumar A., Williams B.R. Identification of double-stranded RNA-binding domains in the interferon-induced double-stranded RNA-activated p68 kinase. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5447–5451. doi: 10.1073/pnas.89.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J.W., Datta P., Wu J., Alnemri E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemr M., Malik R., Franke V., Nejepinska J., Sedlacek R., Vlahovicek K. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Forsbach A., Nemorin J.G., Montino C., Muller C., Samulowitz U., Vicari A.P. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. Journal of Immunology. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- Gall A., Treuting P., Elkon K.B., Loo Y.M., Gale M., Jr., Barber G.N. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D., Chamilos G., Lande R., Gregorio J., Meller S., Facchinetti V. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. The Journal of Experimental Medicine. 2009;206:1983–1994. doi: 10.1084/jem.20090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantier M.P. Processing of double-stranded RNA in mammalian cells: A direct antiviral role? Journal of Interferon & Cytokine Research: The Official Journal of the International Society for Interferon and Cytokine Research. 2014;34:469–477. doi: 10.1089/jir.2014.0003. [DOI] [PubMed] [Google Scholar]
- Gao P., Ascano M., Wu Y., Barchet W., Gaffney B.L., Zillinger T. Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Ascano M., Zillinger T., Wang W., Dai P., Serganov A.A. Structure-function analysis of STING activation by c[G(2',5')pA(3',5')p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Li T., Li X.D., Chen X., Li Q.Z., Wight-Carter M. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C. Impact of protein kinase PKR in cell biology: From antiviral to antiproliferative action. Microbiology and Molecular Biology Reviews: MMBR. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig S., Eberle M.E., Botschen F., Rimbach K., Eberle F., Eigenbrod T. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. The Journal of Experimental Medicine. 2012;209:225–233. doi: 10.1084/jem.20111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke N., Mertens C., Zillinger T., Wenzel J., Bald T., Zahn S. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity. 2013;39:482–495. doi: 10.1016/j.immuni.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Gelais C., St., de Silva S., Amie S.M., Coleman C.M., Hoy H., Hollenbaugh J.A. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4 + T-lymphocytes cannot be upregulated by interferons. Retrovirology. 2012;9:105. doi: 10.1186/1742-4690-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George C.X., John L., Samuel C.E. An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1) Journal of Interferon & Cytokine Research: The Official Journal of the International Society for Interferon and Cytokine Research. 2014;34:437–446. doi: 10.1089/jir.2014.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glas M., Coch C., Trageser D., Dassler J., Simon M., Koch P. Targeting the cytosolic innate immune receptors RIG-I and MDA5 effectively counteracts cancer cell heterogeneity in glioblastoma. Stem Cells. 2013;31:1064–1074. doi: 10.1002/stem.1350. [DOI] [PubMed] [Google Scholar]
- Goldfarb T., Sberro H., Weinstock E., Cohen O., Doron S., Charpak-Amikam Y. BREX is a novel phage resistance system widespread in microbial genomes. The EMBO Journal. 2015;34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone D.C., Ennis-Adeniran V., Hedden J.J., Groom H.C., Rice G.I., Christodoulou E. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5'-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramberg T., Kahle T., Bloch N., Wittmann S., Mullers E., Daddacha W. Restriction of diverse retroviruses by SAMHD1. Retrovirology. 2013;10:26. doi: 10.1186/1742-4690-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray E.E., Treuting P.M., Woodward J.J., Stetson D.B. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of aicardi-goutieres syndrome. Journal of Immunology. 2015;195:1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J., de Vries W., Geutjes E.J., Prins M., de Haan P., Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathogens. 2007;3 doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M., Hubel P., Lacerda L., Benda C., Holze C., Eberl C.H. Sequestration by IFIT1 impairs translation of 2'O-unmethylated capped RNA. PLoS Pathogens. 2013;9 doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkim A., Furnrohr B.G., Amann K., Laube B., Abed U.A., Brinkmann V. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Donovan J., Rath S., Whitney G., Chitrakar A., Korennykh A. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science. 2014;343:1244–1248. doi: 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman Z.C., Kiang A., Everett R.S., Serra D., Yang X.Y., Clay T.M. Adenovirus infection triggers a rapid, MyD88-regulated transcriptome response critical to acute-phase and adaptive immune responses in vivo. Journal of Virology. 2007;81:1796–1812. doi: 10.1128/JVI.01936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann G., Battiany J., Poeck H., Wagner M., Kerkmann M., Lubenow N. Rational design of new CpG oligonucleotides that combine B cell activation with high IFN-alpha induction in plasmacytoid dendritic cells. European Journal of Immunology. 2003;33:1633–1641. doi: 10.1002/eji.200323813. [DOI] [PubMed] [Google Scholar]
- Hartmann G., Krieg A.M. CpG DNA and LPS induce distinct patterns of activation in human monocytes. Gene Therapy. 1999;6:893–903. doi: 10.1038/sj.gt.3300880. [DOI] [PubMed] [Google Scholar]
- Hartmann G., Krieg A.M. Mechanism and function of a newly identified CpG DNA motif in human primary B cells. Journal of Immunology. 2000;164:944–953. doi: 10.4049/jimmunol.164.2.944. [DOI] [PubMed] [Google Scholar]
- Hartmann G., Weeratna R.D., Ballas Z.K., Payette P., Blackwell S., Suparto I. Delineation of a CpG phosphorothioate oligodeoxynucleotide for activating primate immune responses in vitro and in vivo. Journal of Immunology. 2000;164:1617–1624. doi: 10.4049/jimmunol.164.3.1617. [DOI] [PubMed] [Google Scholar]
- Hartmann G., Weiner G.J., Krieg A.M. CpG DNA: A potent signal for growth, activation, and maturation of human dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9305–9310. doi: 10.1073/pnas.96.16.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Franchi L., Nunez G. The protein kinase PKR is critical for LPS-induced iNOS production but dispensable for inflammasome activation in macrophages. European Journal of Immunology. 2013;43:1147–1152. doi: 10.1002/eji.201243187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckelsmiller K., Rall K., Beck S., Schlamp A., Seiderer J., Jahrsdorfer B. Peritumoral CpG DNA elicits a coordinated response of CD8 T cells and innate effectors to cure established tumors in a murine colon carcinoma model. Journal of Immunology. 2002;169:3892–3899. doi: 10.4049/jimmunol.169.7.3892. [DOI] [PubMed] [Google Scholar]
- Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Herzner A.M., Hagmann C.A., Goldeck M., Wolter S., Kubler K., Wittmann S. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nature Immunology. 2015;16:1025–1033. doi: 10.1038/ni.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheiser K., Klein M., Gottschalk C., Hoss F., Scheu S., Coch C. Cutting edge: The RIG-I ligand 3pRNA potently improves CTL cross-priming and facilitates antiviral vaccination. Journal of Immunology. 2016;196:2439–2443. doi: 10.4049/jimmunol.1501958. [DOI] [PubMed] [Google Scholar]
- Hogg M., Paro S., Keegan L.P., O'Connell M.A. RNA editing by mammalian ADARs. Advances in Genetics. 2011;73:87–120. doi: 10.1016/B978-0-12-380860-8.00003-3. [DOI] [PubMed] [Google Scholar]
- Hollenbaugh J.A., Gee P., Baker J., Daly M.B., Amie S.M., Tate J. Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathogens. 2013;9 doi: 10.1371/journal.ppat.1003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature Medicine. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Hornung V., Hartmann R., Ablasser A., Hopfner K.P. OAS proteins and cGAS: Unifying concepts in sensing and responding to cytosolic nucleic acids. Nature Reviews. Immunology. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Latz E. Intracellular DNA recognition. Nature Reviews. Immunology. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- Hornung V., Rothenfusser S., Britsch S., Krug A., Jahrsdorfer B., Giese T. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. Journal of Immunology. 2002;168:4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- Hoss M., Robins P., Naven T.J., Pappin D.J., Sgouros J., Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. The EMBO Journal. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian A.G., Brown R.E., Kerr I.M. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature. 1977;268:537–540. doi: 10.1038/268537a0. [DOI] [PubMed] [Google Scholar]
- Hrecka K., Hao C., Gierszewska M., Swanson S.K., Kesik-Brodacka M., Srivastava S. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Zeqiraj E., Dong B., Jha B.K., Duffy N.M., Orlicky S. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Molecular Cell. 2014;53:221–234. doi: 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J.L., Diamond M.S. Innate immune restriction and antagonism of viral RNA lacking 2-O methylation. Virology. 2015;479–480:66–74. doi: 10.1016/j.virol.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A. Particle counts and infectivity titrations for animal viruses. Advances in Virus Research. 1957;4:111–158. doi: 10.1016/s0065-3527(08)60597-7. [DOI] [PubMed] [Google Scholar]
- Isaacs A., Cox R.A., Rotem Z. Foreign nucleic acids as the stimulus to make interferon. Lancet. 1963;2:113–116. doi: 10.1016/s0140-6736(63)92585-6. [DOI] [PubMed] [Google Scholar]
- Iwakawa H.O., Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends in Cell Biology. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- Jakobs C., Perner S., Hornung V. AIM2 drives joint inflammation in a self-DNA triggered model of chronic polyarthritis. PloS One. 2015;10 doi: 10.1371/journal.pone.0131702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M.A., Kim E.K., Now H., Nguyen N.T., Kim W.J., Yoo J.Y. Mutations in DDX58, which encodes RIG-I, cause atypical singleton-merten syndrome. American Journal of Human Genetics. 2015;96:266–274. doi: 10.1016/j.ajhg.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.S., Backlund P.S., Chen H.C., Karavanov A.A., Crouch R.J. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Research. 2004;32:407–414. doi: 10.1093/nar/gkh209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Ramanathan A., Miller M.T., Tang G.Q., Gale M., Jr., Patel S.S. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T., Perry A., Jiang J., Smith P., Curry J.A., Unterholzner L. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockel S., Nees G., Sommer R., Zhao Y., Cherkasov D., Hori H. The 2'-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. The Journal of Experimental Medicine. 2012;209:235–241. doi: 10.1084/jem.20111075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge A.D., Sood V., Shaw J.R., Fang D., McClintock K., MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature Biotechnology. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Junt T., Barchet W. Translating nucleic acid-sensing pathways into therapies. Nature Reviews. Immunology. 2015;15:529–544. doi: 10.1038/nri3875. [DOI] [PubMed] [Google Scholar]
- Kariko K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Kato H., Fujita T. RIG-I-like receptors and autoimmune diseases. Current Opinion in Immunology. 2015;37:40–45. doi: 10.1016/j.coi.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kawane K., Ohtani M., Miwa K., Kizawa T., Kanbara Y., Yoshioka Y. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- Kerkmann M., Costa L.T., Richter C., Rothenfusser S., Battiany J., Hornung V. Spontaneous formation of nucleic acid-based nanoparticles is responsible for high interferon-alpha induction by CpG-A in plasmacytoid dendritic cells. The Journal of Biological Chemistry. 2005;280:8086–8093. doi: 10.1074/jbc.M410868200. [DOI] [PubMed] [Google Scholar]
- Kerkmann M., Rothenfusser S., Hornung V., Towarowski A., Wagner M., Sarris A. Activation with CpG-A and CpG-B oligonucleotides reveals two distinct regulatory pathways of type I IFN synthesis in human plasmacytoid dendritic cells. Journal of Immunology. 2003;170:4465–4474. doi: 10.4049/jimmunol.170.9.4465. [DOI] [PubMed] [Google Scholar]
- Kim S., Kaiser V., Beier E., Bechheim M., Guenthner-Biller M., Ablasser A. Self-priming determines high type I IFN production by plasmacytoid dendritic cells. European Journal of Immunology. 2014;44:807–818. doi: 10.1002/eji.201343806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Nguyen L.A., Daddacha W., Hollenbaugh J.A. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. The Journal of Biological Chemistry. 2012;287:21570–21574. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U., Wang Y., Sanford T., Zeng Y., Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.T., White T.E., Brandariz-Nunez A., Diaz-Griffero F., Weitzman M.D. SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. Journal of Virology. 2013;87:12949–12956. doi: 10.1128/JVI.02291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlway A., Luo D., Rawling D.C., Ding S.C., Pyle A.M. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Reports. 2013;14:772–779. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Krieg A.M., Yi A.K., Matson S., Waldschmidt T.J., Bishop G.A., Teasdale R. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- Krug A., Rothenfusser S., Selinger S., Bock C., Kerkmann M., Battiany J. CpG-A oligonucleotides induce a monocyte-derived dendritic cell-like phenotype that preferentially activates CD8 T cells. Journal of Immunology. 2003;170:3468–3477. doi: 10.4049/jimmunol.170.7.3468. [DOI] [PubMed] [Google Scholar]
- Kubler K., Gehrke N., Riemann S., Bohnert V., Zillinger T., Hartmann E. Targeted activation of RNA helicase retinoic acid-inducible gene-I induces proimmunogenic apoptosis of human ovarian cancer cells. Cancer Research. 2010;70:5293–5304. doi: 10.1158/0008-5472.CAN-10-0825. [DOI] [PubMed] [Google Scholar]
- Kumar A., Haque J., Lacoste J., Hiscott J., Williams B.R. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Ingle H., Mishra S., Mahla R.S., Kumar A., Kawai T. IPS-1 differentially induces TRAIL, BCL2, BIRC3 and PRKCE in type I interferons-dependent and -independent anticancer activity. Cell Death & Disease. 2015;6 doi: 10.1038/cddis.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Segeral E. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E.C., Dragin L. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nature Immunology. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y.Y., Londono D., Bouley R., Rooney M.S., Hacohen N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Reports. 2014;9:180–192. doi: 10.1016/j.celrep.2014.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassig C., Matheisl S., Sparrer K.M., de Oliveira Mann C.C., Moldt M., Patel J.R. ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. eLife. 2015;4:e10859. doi: 10.7554/eLife.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E., Schoenemeyer A., Visintin A., Fitzgerald K.A., Monks B.G., Knetter C.F. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nature Immunology. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- Lecellier C.H., Dunoyer P., Arar K., Lehmann-Che J., Eyquem S., Himber C. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch M.A., Chowdhury D., Harvey S., Gong M., Senenko L., Engel K. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. Journal of Molecular Medicine. 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch M.A., Gong M., Chowdhury D., Senenko L., Engel K., Lee Y.A. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nature Genetics. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- Levin D., London I.M. Regulation of protein synthesis: Activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:1121–1125. doi: 10.1073/pnas.75.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.X., Li H., Lu R., Li F., Dus M., Atkinson P. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lu J., Han Y., Fan X., Ding S.W. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Zhang W., Cao X. Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunology Letters. 2000;74:221–224. doi: 10.1016/s0165-2478(00)00276-5. [DOI] [PubMed] [Google Scholar]
- Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmann J., Burke D.C., Isaacs A. Studies on the production, mode of action and properties of interferon. British Journal of Experimental Pathology. 1957;38:551–562. [PMC free article] [PubMed] [Google Scholar]
- Liu L., Botos I., Wang Y., Leonard J.N., Shiloach J., Segal D.M. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Cullen B.R. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. Journal of Virology. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A., Kabaleeswaran V., Fu T., Magupalli V.G., Wu H. Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. Journal of Molecular Biology. 2014;426:1420–1427. doi: 10.1016/j.jmb.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A., Li Y., Yin Q., Ruan J., Yu X., Egelman E. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discovery. 2015;1:15013. doi: 10.1038/celldisc.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A., Magupalli V.G., Ruan J., Yin Q., Atianand M.K., Vos M.R. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Nakamura T., Inouye K., Li J., Tang Y., Lundback P. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488:670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S.E., Human M.L. A nonhereditary, host-induced variation of bacterial viruses. Journal of Bacteriology. 1952;64:557–569. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K.J., Carroll P., Lettice L., Tarnauskaite Z., Reddy K., Dix F. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. The EMBO Journal. 2016;35:831–844. doi: 10.15252/embj.201593339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi L.B., Jr., Heitmeier M.R., Scheuner D., Kaufman R.J., Buller R.M., Corbett J.A. Potential role of PKR in double-stranded RNA-induced macrophage activation. The EMBO Journal. 2000;19:3630–3638. doi: 10.1093/emboj/19.14.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan A.K., Schmidt T., Chauhan D., Goldeck M., Honing K., Gaidt M. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. The EMBO Journal. 2014;33:2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marq J.B., Hausmann S., Veillard N., Kolakofsky D., Garcin D. Short double-stranded RNAs with an overhanging 5' ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. The Journal of Biological Chemistry. 2010;286:6108–6116. doi: 10.1074/jbc.M110.186262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marq J.B., Kolakofsky D., Garcin D. Unpaired 5' ppp-nucleotides, as found in arenavirus double-stranded RNA panhandles, are not recognized by RIG-I. The Journal of Biological Chemistry. 2010;285:18208–18216. doi: 10.1074/jbc.M109.089425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L.A., Sontheimer E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nature Reviews. Genetics. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshak-Rothstein A. Tolling for autoimmunity-prime time for 7. Immunity. 2006;25:397–399. doi: 10.1016/j.immuni.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Kikkawa S., Kohase M., Miyake K., Seya T. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochemical and Biophysical Research Communications. 2002;293:1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- McAllister C.S., Taghavi N., Samuel C.E. Protein kinase PKR amplification of interferon beta induction occurs through initiation factor eIF-2alpha-mediated translational control. The Journal of Biological Chemistry. 2012;287:36384–36392. doi: 10.1074/jbc.M112.390039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Molecular Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Melchjorsen J., Kristiansen H., Christiansen R., Rintahaka J., Matikainen S., Paludan S.R. Differential regulation of the OASL and OAS1 genes in response to viral infections. Journal of Interferon & Cytokine Research: The Official Journal of the International Society for Interferon and Cytokine Research. 2009;29:199–207. doi: 10.1089/jir.2008.0050. [DOI] [PubMed] [Google Scholar]
- Morita M., Stamp G., Robins P., Dulic A., Rosewell I., Hrivnak G. Gene-targeted mice lacking the Trex1 (DNase III) 3'- > 5' DNA exonuclease develop inflammatory myocarditis. Molecular and Cellular Biology. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla S.R., Hwang J., Toroney R., Zheng X., Cameron C.E., Bevilacqua P.C. 5'-Triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- Napirei M., Karsunky H., Zevnik B., Stephan H., Mannherz H.G., Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nature Genetics. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- Napirei M., Ludwig S., Mezrhab J., Klockl T., Mannherz H.G. Murine serum nucleases—Contrasting effects of plasmin and heparin on the activities of DNase1 and DNase1-like 3 (DNase1l3) The FEBS Journal. 2009;276:1059–1073. doi: 10.1111/j.1742-4658.2008.06849.x. [DOI] [PubMed] [Google Scholar]
- Nathans D., Smith H.O. Restriction endonucleases in the analysis and restructuring of DNA molecules. Annual Review of Biochemistry. 1975;44:273–293. doi: 10.1146/annurev.bi.44.070175.001421. [DOI] [PubMed] [Google Scholar]
- Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nature Reviews. Molecular Cell Biology. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U., Shibata T., Tanji H., Ishida H., Krayukhina E., Uchiyama S. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520:702–705. doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- Oka T., Hikoso S., Yamaguchi O., Taneike M., Takeda T., Tamai T. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onji M., Kanno A., Saitoh S., Fukui R., Motoi Y., Shibata T. An essential role for the N-terminal fragment of Toll-like receptor 9 in DNA sensing. Nature Communications. 2013;4:1949. doi: 10.1038/ncomms2949. [DOI] [PubMed] [Google Scholar]
- Ota H., Sakurai M., Gupta R., Valente L., Wulff B.E., Ariyoshi K. ADAR1 forms a complex with dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran P., Sklan E., Wilkins C., Burgon T., Samuel M.A., Lu R. Six RNA viruses and forty-one hosts: Viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathogens. 2010;6 doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B., Brinkmann M.M., Spooner E., Lee C.C., Kim Y.M., Ploegh H.L. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nature Immunology. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls E., Ruiz A., Riveira-Munoz E., Permanyer M., Badia R., Clotet B. p21 regulates the HIV-1 restriction factor SAMHD1. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1322–E1324. doi: 10.1073/pnas.1322059111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawaria S., Moody K., Busto P., Nundel K., Choi C.H., Ghayur T. Cutting Edge: DNase II deficiency prevents activation of autoreactive B cells by double-stranded DNA endogenous ligands. Journal of Immunology. 2015;194:1403–1407. doi: 10.4049/jimmunol.1402893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka K., Shibata T., Miyake K., Latz E. Nucleic acid-sensing TLRs and autoimmunity: Novel insights from structural and cell biology. Immunological Reviews. 2016;269:60–75. doi: 10.1111/imr.12375. [DOI] [PubMed] [Google Scholar]
- Peter M.E., Kubarenko A.V., Weber A.N., Dalpke A.H. Identification of an N-terminal recognition site in TLR9 that contributes to CpG-DNA-mediated receptor activation. Journal of Immunology. 2009;182:7690–7697. doi: 10.4049/jimmunol.0900819. [DOI] [PubMed] [Google Scholar]
- Pfeffer S., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grasser F.A. Identification of microRNAs of the herpesvirus family. Nature Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Pichlmair A., Lassnig C., Eberle C.A., Gorna M.W., Baumann C.L., Burkard T.R. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nature Immunology. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O. Activation of MDA5 requires higher-order RNA structures generated during virus infection. Journal of Virology. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H., Bscheider M., Gross O., Finger K., Roth S., Rebsamen M. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nature Immunology. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Pohar J., Pirher N., Bencina M., Mancek-Keber M., Jerala R. The role of UNC93B1 protein in surface localization of TLR3 receptor and in cell priming to nucleic acid agonists. The Journal of Biological Chemistry. 2013;288:442–454. doi: 10.1074/jbc.M112.413922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokatayev V., Hasin N., Chon H., Cerritelli S.M., Sakhuja K., Ward J.M. RNase H2 catalytic core Aicardi-Goutieres syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. The Journal of Experimental Medicine. 2016;213:329–336. doi: 10.1084/jem.20151464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothlichet J., Meunier I., Davis B.K., Ting J.P., Skamene E., von Messling V. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathogens. 2013;9 doi: 10.1371/journal.ppat.1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell R.D., Holland P.J., Hollis T., Perrino F.W. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. The Journal of Biological Chemistry. 2011;286:43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.L., Katze M.G. Genomic signatures of emerging viruses: A new era of systems epidemiology. Cell Host & Microbe. 2016;19:611–618. doi: 10.1016/j.chom.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouillat D., Hovanessian A.G. The human 2',5'-oligoadenylate synthetase family: Interferon-induced proteins with unique enzymatic properties. Journal of Interferon & Cytokine Research: The Official Journal of the International Society for Interferon and Cytokine Research. 1999;19:295–308. doi: 10.1089/107999099313992. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Maelfait J., Bridgeman A., Rigby R., Hayward B., Liberatore R.A. SAMHD1-dependent retroviral control and escape in mice. The EMBO Journal. 2013;32:2454–2462. doi: 10.1038/emboj.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Tan C.P., Goubau D., Schulz O., Pichlmair A., Bier K. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Rice G.I., Bond J., Asipu A., Brunette R.L., Manfield I.W., Carr I.M. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nature Genetics. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Kasher P.R., Forte G.M., Mannion N.M., Greenwood S.M., Szynkiewicz M. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nature Genetics. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G., Patrick T., Parmar R., Taylor C.F., Aeby A., Aicardi J. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. American Journal of Human Genetics. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A., van den Maagdenberg A.M., Jen J.C., Kavanagh D., Bertram P., Spitzer D. C-terminal truncations in human 3'-5' DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nature Genetics. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- Rigby R.E., Webb L.M., Mackenzie K.J., Li Y., Leitch A., Reijns M.A. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. The EMBO Journal. 2014;33:542–558. doi: 10.1002/embj.201386117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T.L., Idris A., Dunn J.A., Kelly G.M., Burnton C.M., Hodgson S. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B.G., Schoenemeyer A. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. Journal of Immunology. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- Rothenfusser S., Hornung V., Ayyoub M., Britsch S., Towarowski A., Krug A. CpG-A and CpG-B oligonucleotides differentially enhance human peptide-specific primary and memory CD8 + T-cell responses in vitro. Blood. 2004;103:2162–2169. doi: 10.1182/blood-2003-04-1091. [DOI] [PubMed] [Google Scholar]
- Ryoo J., Choi J., Oh C., Kim S., Seo M., Kim S.Y. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nature Medicine. 2014;20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima K., Earnshaw W.C. Trashing the genome: The role of nucleases during apoptosis. Nature Reviews. Molecular Cell Biology. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- Schlee M., Hartmann E., Coch C., Wimmenauer V., Janke M., Barchet W. Approaching the RNA ligand for RIG-I? Immunological Reviews. 2009;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W. Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnettler E., Sterken M.G., Leung J.Y., Metz S.W., Geertsema C., Goldbach R.W. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. Journal of Virology. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr M., Duewell P. Breaking tumor-induced immunosuppression with 5'-triphosphate siRNA silencing TGFbeta and activating RIG-I. Oncoimmunology. 2013;2 doi: 10.4161/onci.24170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr M., Duewell P. Induction of immunogenic cell death by targeting RIG-I-like helicases in pancreatic cancer. Oncoimmunology. 2014;3 doi: 10.4161/21624011.2014.955687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.M., Zillinger T., Goldeck M. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2'O-methylated self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.C., Taira H., Lengyel P. Interferon, double-stranded RNA, and protein phosphorylation. Characteristics of a double-stranded RNA-activated protein kinase system partially purified from interferon treated Ehrlich ascites tumor cells. The Journal of Biological Chemistry. 1978;253:5915–5921. [PubMed] [Google Scholar]
- Seo G.J., Kincaid R.P., Phanaksri T., Burke J.M., Pare J.M., Cox J.E. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host & Microbe. 2013;14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman R.H. Viral encounters with 2',5'-oligoadenylate synthetase and RNase L during the interferon antiviral response. Journal of Virology. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V., Bloch N., Landau N.R. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nature Immunology. 2015;16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: A central role for 2'-hydroxyl uridines in immune responses. European Journal of Immunology. 2006;36:1222–1230. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]
- Stein H., Hausen P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA hybrids: Effect on DNA-dependent RNA polymerase. Science. 1969;166:393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P. Renaissance of mammalian endogenous RNAi. FEBS Letters. 2014;588:2550–2556. doi: 10.1016/j.febslet.2014.05.030. [DOI] [PubMed] [Google Scholar]
- Szittya G., Burgyan J. RNA interference-mediated intrinsic antiviral immunity in plants. Current Topics in Microbiology and Immunology. 2013;371:153–181. doi: 10.1007/978-3-642-37765-5_6. [DOI] [PubMed] [Google Scholar]
- Taylor M.J., Bandi C., Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Advances in Parasitology. 2005;60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- Thomis D.C., Doohan J.P., Samuel C.E. Mechanism of interferon action: cDNA structure, expression, and regulation of the interferon-induced, RNA-dependent P1/eIF-2 alpha protein kinase from human cells. Virology. 1992;188:33–46. doi: 10.1016/0042-6822(92)90732-5. [DOI] [PubMed] [Google Scholar]
- Thomis D.C., Samuel C.E. Mechanism of interferon action: Autoregulation of RNA-dependent P1/eIF-2 alpha protein kinase (PKR) expression in transfected mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:10837–10841. doi: 10.1073/pnas.89.22.10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaselli S., Galeano F., Locatelli F., Gallo A. ADARs and the balance game between virus infection and innate immune cell response. Current Issues in Molecular Biology. 2015;17:37–51. [PubMed] [Google Scholar]
- van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nature Reviews. Microbiology. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu K., Nagaraja V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiology and Molecular Biology Reviews: MMBR. 2013;77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T., Valdes M., Elsby R., Kakuta S., Caceres G., Saijo S. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. Journal of Immunology. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- Vollmer J., Tluk S., Schmitz C., Hamm S., Jurk M., Forsbach A. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. The Journal of Experimental Medicine. 2005;202:1575–1585. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg H.P., Eckstein F. Incorporation of phosphorothioate groups into fd and phi X174 DNA. Biochemistry. 1977;16:3633–3640. doi: 10.1021/bi00635a020. [DOI] [PubMed] [Google Scholar]
- Wagner R.W., Smith J.E., Cooperman B.S., Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.H., Aliyari R., Li W.X., Li H.W., Kim K., Carthew R. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H. Structural and functional insights into 5'-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nature Structural & Molecular Biology. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Gerlach M., Weber F. Standing on three legs: Antiviral activities of RIG-I against influenza viruses. Current Opinion in Immunology. 2016;42:71–75. doi: 10.1016/j.coi.2016.05.016. [DOI] [PubMed] [Google Scholar]
- Wiemann B., Starnes C.O. Coley's toxins, tumor necrosis factor and cancer research: A historical perspective. Pharmacology & Therapeutics. 1994;64:529–564. doi: 10.1016/0163-7258(94)90023-x. [DOI] [PubMed] [Google Scholar]
- Wu L. Cellular and biochemical mechanisms of the retroviral restriction factor SAMHD1. ISRN Biochemistry. 2013;2013:728392. doi: 10.1155/2013/728392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Hur S. How RIG-I like receptors activate MAVS. Current Opinion in Virology. 2015;12:91–98. doi: 10.1016/j.coviro.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Sun L., Chen X., Du F., Shi H., Chen C. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2012;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Kuramoto E., Shimada S., Tokunaga T. In vitro augmentation of natural killer cell activity and production of interferon-alpha/beta and -gamma with deoxyribonucleic acid fraction from mycobacterium bovis BCG. Japanese Journal of Cancer Research: Gann. 1988;79:866–873. doi: 10.1111/j.1349-7006.1988.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Yamamoto T., Kataoka T., Kuramoto E., Yano O., Tokunaga T. Unique palindromic sequences in synthetic oligonucleotides are required to induce IFN [correction of INF] and augment IFN-mediated [correction of INF] natural killer activity. Journal of Immunology. 1992;148:4072–4076. [PubMed] [Google Scholar]
- Yamamoto S., Yamamoto T., Shimada S., Kuramoto E., Yano O., Kataoka T. DNA from bacteria, but not from vertebrates, induces interferons, activates natural killer cells and inhibits tumor growth. Microbiology and Immunology. 1992;36:983–997. doi: 10.1111/j.1348-0421.1992.tb02102.x. [DOI] [PubMed] [Google Scholar]
- Yang Y.G., Lindahl T., Barnes D.E. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Yang K., Samanta H., Dougherty J., Jayaram B., Broeze R., Lengyel P. Interferons, double-stranded RNA, and RNA degradation. Isolation and characterization of homogeneous human (2'-5')(a)n synthetase. The Journal of Biological Chemistry. 1981;256:9324–9328. [PubMed] [Google Scholar]
- Yasuda T., Takeshita H., Iida R., Nakajima T., Hosomi O., Nakashima Y. Molecular cloning of the cDNA encoding human deoxyribonuclease II. The Journal of Biological Chemistry. 1998;273:2610–2616. doi: 10.1074/jbc.273.5.2610. [DOI] [PubMed] [Google Scholar]
- Yasutomo K., Horiuchi T., Kagami S., Tsukamoto H., Hashimura C., Urushihara M. Mutation of DNASE1 in people with systemic lupus erythematosus. Nature Genetics. 2001;28:313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. Journal of Immunology. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Okabe Y., Kawane K., Fukuyama H., Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nature Immunology. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- Yu P., Lubben W., Slomka H., Gebler J., Konert M., Cai C. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 2012;37:867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Zambon R.A., Vakharia V.N., Wu L.P. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cellular Microbiology. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- Zhou R., Rana T.M. RNA-based mechanisms regulating host-virus interactions. Immunological Reviews. 2013;253:97–111. doi: 10.1111/imr.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhang Y., Ghosh A., Cuevas R.A., Forero A., Dhar J. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity. 2014;40:936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberstein A., Kimchi A., Schmidt A., Revel M. Isolation of two interferon-induced translational inhibitors: A protein kinase and an oligo-isoadenylate synthetase. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:4734–4738. doi: 10.1073/pnas.75.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]