Abstract
RNA interference (RNAi) is a sequence-specific, post-transcriptional process of mRNA degradation. Here, we described specific silencing of five white spot syndrome virus (WSSV) genes in Litopenaeus vannamei in vivo with sequence-specific siRNAs. These genes included DNA polymerase (dnapol), ribonucleotide reductase small subunit (rr2), thymidine kinase and thymidylate kinase (tk-tmk), vp24 and vp28. At 6 days post-challenged, the relative survival rates of shrimp injected with siDNApol, siRR2, siTK-TMK, siVP24 and siVP28 (siRNAs for dnapol, rr2, tk-tmk, vp24 and vp28 genes) reached 50%, 50%, 66%, 33% and 33%, respectively. Specific siRNAs of the five WSSV genes could result in suppression of the target genes and a significant reduction in the viral proliferation. In negative controls, sequence-independent siRNA (mutant siRNA) could not inhibit expression of these five genes or viral replication. Consequently, injection of sequence-dependent siRNA could induce anti-WSSV activity in shrimp. These results suggest that siRNA can suppress WSSV efficiently in shrimp, and it may provide a potential approach to the therapy of aquaculture viral disease.
Keywords: WSSV, RNA interference, siRNA, Semi-quantitative RT-PCR
1. Introduction
White spot syndrome virus (WSSV), one of the most virulent pathogens in cultured penaeid shrimp, has been reported worldwide since the early 1990s and has caused significant economic losses in shrimp culture worldwide (Takahashi et al., 1994, Chou et al., 1995, Huang et al., 1995, Wang et al., 1995, Wongteerasupaya et al., 1995). The major characteristic of the WSSV infection is that the carapace and the body develop obvious white spots. In China, the outbreaks and prevalence of WSSV were greatly reduced with some control measures, such as eliminating infection source, cutting off the infection pathways of transmission, strengthening the control of water quality, and enhancement of diseases resistance in shrimp (He et al., 2000, He et al., 2003a). The production of cultured penaeid shrimp accounted for more than 600 thousand tons in 2004 in China. Up to now, the prevalence of WSSV remains and there is no effective antiviral treatment to control the outbreaks and prevalence of WSSV.
Based on the Eighth Report of the International Committee on Taxonomy of Viruses (ICTV), WSSV is a species of the genus Whispovirus, the new family Nimaviridae. It contained a single molecule of circular dsDNA with an approximate size of 300 kb (Fauquet et al., 2005). The complete genomic sequence of WSSV has been determined during the past several years (van Hulten et al., 2001a, Yang et al., 2001) and it contains approximately 180 putative open reading frames (ORFs), of which only a few could be assigned a putative function (Fauquet et al., 2005).
RNAi is a common mechanism for post-transcriptional gene silencing. It is a phenomenon recently observed in a variety of eukaryotic organisms. RNAi is initiated by the double-stranded RNA (dsRNA)-specific endonuclease or Dicer, which promotes cleavage of long dsRNA into 21–23-mer short interfering RNA (siRNA). Then siRNA molecules induce sequence-specific degradation of homologous single-stranded RNA (Elbashir et al., 2001, Sharp, 2001, Zamore, 2002). RNAi has been employed to manipulate gene expression, elucidate signal pathways, and knock down specific genes to evaluate their physiological roles or perhaps control pathogen infection (Fire et al., 1998, Kennerdell and Carthew, 1998, Ngo et al., 1998, Wianny and Zernicka-Goetz, 2000). In addition, RNAi has been widely proven as an effective mechanism to suppress the viral infection or replication of many viruses, including the several important human pathogens such as poliovirus (Gitlin et al., 2002), HIV-1 (Jacque et al., 2002), hepatitis B virus (McCaffrey et al., 2003, Shlomai and Shaul, 2003), hepatitis C virus (Kapadia et al., 2003), foot and mouth disease virus (FMDV) (Kahana et al., 2004), influenza virus A (Ge et al., 2003), Dengue virus (Adelman et al., 2002), and SARS coronavirus (SCV) (He et al., 2003b, Li et al., 2005). The results of these studies suggest the possibility of using RNAi as an antiviral tool in disease prevention in shrimp aquaculture.
Recently, a few groups have reported that viruses from the aquatic animals could be inhibited by exogenously synthetic long dsRNAs or siRNAs (Robalino et al., 2004, Robalino et al., 2005, Tirasophon et al., 2005, Westenberg et al., 2005, Xie et al., 2005). They showed that long dsRNA molecules could induce a sequence-independent antiviral immunity in shrimp (Robalino et al., 2004, Robalino et al., 2005), and viral replication could be effectively inhibited by long dsRNA molecules and siRNA molecules in shrimp or fish cells (Tirasophon et al., 2005, Xie et al., 2005). siRNA cannot act on non-specific sequence. However, it has been shown that non-specific siRNAs also reduce mortality in shrimp (Westenberg et al., 2005), and neither sequence-dependent nor sequence-independent siRNAs could induce the antiviral action (Robalino et al., 2005). In this paper, we have synthesized five siRNAs targeting the five WSSV genes and studied their effectiveness in inhibiting WSSV replication in vivo. The five genes include DNA polymerase gene (dnapol), ribonucleotide reductase small subunit gene (rr2), thymidine kinase and thymidylate kinase gene (tk-tmk), two structural protein genes vp24 and vp28. We showed that siRNAs targeting the mRNA of these genes effectively inhibited WSSV replication in shrimp, but the mutant siRNA could not inhibit expression of target genes of WSSV and viral replication. Our results suggest that siRNA technology might represent a potential therapeutic or preventive approach against shrimp viral diseases.
2. Materials and methods
2.1. Animals
Litopenaeus vannamei shrimp were obtained from a shrimp culture pond in Donghai Island, Zhanjiang, Guangdong Province, PR China. The average body length was 7 to 8 cm, and the average weight was 6 to 7 g. Before WSSV infection, 10 shrimp were randomly chosen for WSSV detection by one-step PCR to confirm the absence of WSSV in those shrimp. They were kept in 1000-L aquaria for 6 days, fed with approximately 0.25 g (4% of shrimp weight) commercial feed daily. The water temperature was kept at 28 ± 1 °C, with an 80% daily water exchange and air supply. The shrimp were divided into 3 trial groups: the treated group, positive control group and negative control groups (Table 1 ). There were 15 shrimp for each group, and 6 of them were for the observation of survival rate. The time of the observation lasted for 10 days.
Table 1.
The groups in the experiment and the numbers of shrimp in each group
| Groups | Treatment | Number of shrimp | |
|---|---|---|---|
| Trial groups | siDNApol | Injecting with siDNApol and WSSV simultaneously | 15 |
| siRR2 | Injecting with siRR2 and WSSV simultaneously | 15 | |
| siTK-TMK | Injecting with siTK-TMK and WSSV simultaneously | 15 | |
| siVP24 | Injecting with sivp24 and WSSV simultaneously | 15 | |
| siVP28 | Injecting with siVP28 and WSSV simultaneously | 15 | |
| siMUT | Injecting with siMUT and WSSV simultaneously | 15 | |
| Negative control groups | siDNApol | Injecting with siDNApol only | 15 |
| siRR2 | Injecting with siRR2 only | 15 | |
| siTK-TMK | Injecting with siTK-TMK only | 15 | |
| siVP24 | Injecting with siVP24 only | 15 | |
| siVP28 | Injecting with siVP28 only | 15 | |
| siMUT | Injecting with siMUT only | 15 | |
| PBS | Injecting with PBS only | 15 | |
| Positive control group | Injecting with WSSV only | 15 | |
2.2. siRNA
siRNAs were generated using in vitro Transcription T7 kit (TaKaRa, Japan) as previously described by Xie et al. (2005). Briefly, the interference site of each gene was designed from the conserved domain and the target sequences of RNAi were selected via an on-line software (http://www.ambion.com/techlib/misc/siRNA_finder.html). The designed sequences were showed in Table 2 . Mutant siRNA target sequence was derived from siVP28 target sequence with 6 nt changed. Appropriate GC content sequences of the type AA (N21) [N, any nucleotide (nt)] were searched within the conservative domain of selected genes, in order to obtain a 21-nt sense and 21-nt antisense strand with 2-nt 3′ overhangs. The siRNAs were prepared by hybridizing two complementary ssRNAs that were in vitro transcribed from separate dsDNA templates containing a T7 promoter. The siRNAs were purified by ethanol precipitation and quantified by measuring the absorbance at 260 nm (A260) with a spectrophotometer.
Table 2.
The siRNA sequence (sense strand) in this study
| Name | Target gene | siRNA target sequence | Positiona |
|---|---|---|---|
| siDNApol | DNA polymerase gene (dnapol) | 5′-ATTGACGAGTGCAATCAGATT-3′ | 2920–2940 |
| siRR2 | Ribonucleotide reductase small subunit gene (rr2) | 5′-CGTCCGAATTGAACAAGAATT-3′ | 849–869 |
| siTK-TMK | Thymidine kinase and thymidylate kinase gene (tk-tmk) | 5′-TACTTTTGGTGTCGAAAGATT-3′ | 1020–1040 |
| siVP24 | vp24 | 5′-AATGCTACAGGAAGGGAATTT-3′ | 433–453 |
| siVP28 | vp28 | 5′-GATTAACCCATCAAAGGCCTT-3′ | 426–446 |
| siMUTb | Mutant | 5′-GATTACAAACGCAAAGGCCTT-3′ |
Positions of siDNApol, siRR2, siTK-TMK, siVP24 and siVP28 target sequences were according to the WSSV genome (GenBank accession no. AF332093).
Mutated siRNA (siMUT) has six mismatch nucleotides siVP28.
2.3. siRNA delivery and virus infection in vivo
WSSV-containing extract used to challenge shrimp was prepared as described previously (Jian et al., 2005). Briefly, moribund L. vannamei shrimp with white spots on the cuticle and positive for the presence of WSSV by one-step PCR were used to prepare inoculums for the challenge tests. Muscle (approximately 100 mg) was homogenized in 10 volumes of PBS buffer (pH 7.4) on ice, and then centrifuged at 4250 ×g for 15 min at 4 °C. The supernatant was passed through a 0.45 μm filter unit and filtrates were diluted to appropriate concentrates in which filtrates could induce 100% mortality in health shrimp over a large number of experiments. 50 μl (1 × 104 copies/μl) viral solution and 10 μl (10 μg siRNA) siRNA per shrimp were injected into L. vannamei shrimp intramuscularly between the third and fourth abdominal segment.
2.4. Quantitative PCR assay
Real-time quantitative PCR method was used to measure copies of virus in shrimp that were injected with siRNA and virus extract in this study. DNA was extracted from 30-mg muscle of the fourth abdominal segment using Tissue DNA kit (Omiga, USA). Primers and probe were 5′-CCA CCA ATT CTA CTC ATG TAC CAA A-3′ (forward), 5′-TCC TTG CAA TGG GCA AAA TC-3′ (reverse), 5′-CTG GGT TAC GAG TCT AA-mgb-3′ (probe) designed according to a Hind III fragment of WSSV DNA sequence from Penaeus monodon (Deng et al., 2000). This PCR fragment was cloned into PUC18 vector, and the recombinant plasmid DNA containing the target fragment was used as standard to quantify the template to make the standard curve. The results (Ct value) of viral quantity would be converted into copy number of virus based on the standard curve. PCR mix consisted of ABI Taqman Universal PCR Master Mix containing AmpliTaq Gold DNA polymerase, AmpErase uracil-N-glycosylase (UNG), dNTPs with dUTP and optimized buffer components (Applied Biosystems, Inc.). A sample of 10 to 50 ng of DNA from shrimp was added to a PCR mixture containing 0.45 μM of each primer and 0.125 μM of TaqMan probe in a final volume of 25 μl. Amplification was performed with the following program: 2-min reaction for AmpErase UNG at 50 °C, and activation of the AmpliTaq for 10 min at 95 °C, 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The WSSV quantity of each sample was determined by ABI Prism 7000 Sequence Detection System Software (SD1.0). Each sample was tested in triplicate.
2.5. Analysis of viral gene expression in shrimp
Specific silencing of target genes was confirmed by semi-quantitative RT-PCR. Total muscle RNA of the fourth abdominal segment was isolated with Trizol reagent (Invitrogen, USA) according to the manufacturer's protocol. The samples were extracted with acidic hydroxybenzene after treatment with Trizol reagent to eliminate traces of DNA. To detect the corresponding mRNA expression of WSSV genes in shrimp, 2 μg of RNA extract was used as template for RT-PCR amplification with the one-step RT-PCR system (Invitrogen, USA). Semi-quantitative PCR were performed using dnapol gene primers (FW: 5′-TTA AGG AAC GCA ACA AG TTA-3′; RV: 5′-AAG CAG AGT CCA TAC ACA GC-3′), rr2 gene primers (FW: 5′-AAC GAT TTG GCG GAA CTA A-3′; RV: 5′-CAG GCA GGG AAA CTG TGA G-3′), tk-tmk gene primers (FW: 5′-CGA CAG GAG CAG CCA TAC G-3′; RV: 5′-AGC AGA GCA CCA CTC AAC G-3′), vp24 gene primers (FW: 5′-GCT ATA CTG GCG GGT TTG-3′; RV: 5′-GTC CAC TGT TAT ATC CCT CTT TG-3′), vp28 gene primers (FW: 5′-CAC AAC ACT GTG ACC AAG-3′; RV: 5′-TTT ACT CGG TCT CAG TGC CAG-3′) and β-actin gene primers (FW: 5′-ATG TGT GAC GAC GAA GTA GC-3′; RV: 5′-GGT GGT CGT GAA GGT GTA AC-3′), respectively. The PCR products were analyzed by agarose gel electrophoresis described elsewhere previously (Xie et al., 2005). Briefly, the gels were photographed and images were analyzed by the AlphaEase FC imaging system (Alpha Innotech Corporation, USA) after electrophoresis. The pixel value (intensity) and area of each band in every image and the background of certain image were counted. The average value (AVG) was calculated as follows: AVG = [Σ(each pixel value − background)] / area. The results can eliminate the error of different exposure time from image to image, and the error of different band area generated by various gel running conditions. The datum of AVG of band of our selected genes fragments was normalized to the RT-PCR fragment of β-actin gene to get normalized AVG (nAVG). nAVG of each siRNA treated samples at 3 time-intervals indicated expression condition of each gene. All RT-PCR amplifications were performed in the linear range.
3. Results
3.1. WSSV inhibited by specific siRNAs in shrimp
To investigate whether the target genes are related to virulence, siRNAs and virus extract were co-injected intramuscularly into the shrimp. The genes with important roles in WSSV replication, nucleotide metabolism and virus–host interactions were chosen for the experiment. For example, DNA polymerase is an important enzyme for DNA replication of WSSV (Chen et al., 2002). Both of ribonucleotide reductase (RR) and thymidine kinase & thymidylate kinase (TK-TMK) are the key enzymes of nucleotide metabolism (Tsai et al., 2000, Tzeng et al., 2002). VP28 is the major WSSV envelope protein related to systemic virus infection (van Hulten et al., 2001b), and VP24 is the basic WSSV nucleocapsid protein (van Hulten et al., 2000). The mutant siRNA was used as non-specific control for interference action of siRNAs. Delivery of the selected sequence-specific siRNAs resulted in increase of survival rate of shrimp infected by WSSV, as compared to the virus injected controls (Fig. 1 ). At 6 days post-infection (p.i.), the survival rates of shrimp injected with siRNAs for dnapol, rr2, tk-tmk, vp24 and vp28 genes were 50%, 50%, 66%, 33% and 33%, respectively, while the survival rate of the control shrimp was 0% (Fig. 1). At 10 days p.i. after siRNA and virus injection, the survival rates of shrimp of siDNApol, siRR2, siTK-TMK and siVP28 groups were 33%, and that of siVP24 group was 17%. Thus, the protection provided by specific siRNAs of WSSV genes was significant. However, the protection rate could not reach 100% by one injection. At the same time, the shrimp of control groups only injected with sequence-specific siRNA or PBS were all alive. These results indicated that these sequence-specific siRNAs were safe for shrimp.
Fig. 1.
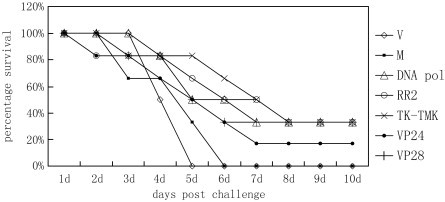
Treatments of targeted genes-siRNAs protected shrimp from WSSV challenge. Shrimp were treated with virus only (V, filled rhombuses), siMUT + virus (M, filled squares), siDNApol + virus (filled triangles), siRR2 + virus (open circles), siTK-TMK + virus (decussation), siVP24 + virus (filled circles), and siVP28 + virus (crisscross), respectively. Survival rates of specific siRNA groups were higher than that of positive controls and siMUT groups (n = 6).
3.2. Suppression of selected WSSV gene expression by specific siRNAs
To examine if treatment of specific and mutant siRNAs resulted in a reduced expression level of the selected WSSV genes in shrimp in vivo, semi-quantitative and highly sensitive RT-PCR analyses were used to compare the relative silencing effects. In the specific siRNA injected groups, samples at 3 time-intervals (24, 36, 48 h) from the virus positive and mutant siRNA controls were analyzed (Fig. 2 ). β-actin of L. vannamei was used as an internal control, and the relative quantity of each target gene expression was analyzed using the AlphaEase FC imaging system (Fig. 2C). Delivery of specific siRNAs resulted in a significant suppression of the corresponding WSSV mRNA (Fig. 2A). At 24 h p.i., a reduction in each WSSV mRNA was observed. There was no degradation of the targeted gene transcripts from positive controls (Fig. 2B). Our results showed that the synthesis of targeted mRNAs was inhibited by sequence-specific siRNAs in vivo. Moreover, the suppression effect of WSSV gene expression was low at 48 h p.i. (Fig. 2). Thus siRNA silencing was specific and transient in this system.
Fig. 2.
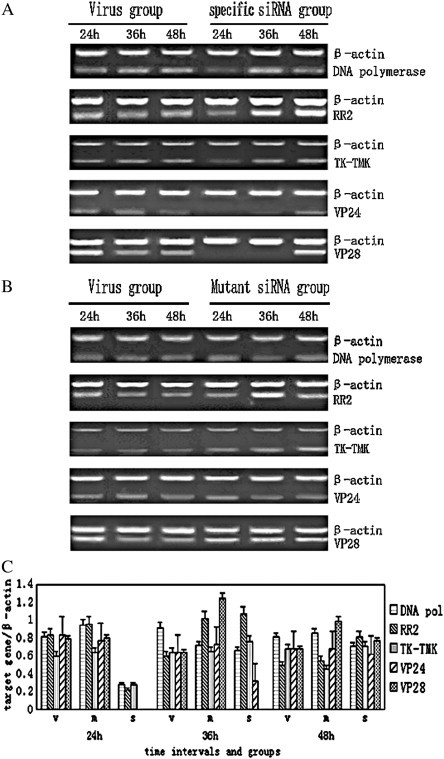
(A) Expression of target genes of each specific siRNA group was examined following siRNA injection and virus challenge. Virus group was infected WSSV without siRNA injection. Specific siRNA group was injected specific siRNAs of selected WSSV genes and WSSV at the same time. β-actin of L. vannamei was selected for inner-control. (B) Expression of each selected gene of mutant siRNA group was examined after siRNA injection and virus challenge. Virus group was infected WSSV without siRNA injection. Mutant siRNA (siMUT) group was injected siMUTs and WSSV at the same time. (C) The relative quantity of each target gene expression was analyzed by the AlphaEase FC imaging system. X-axis showed different groups at 24 h, 36 h and 48 h test and there were 3 shrimp in each time. v was positive group, m was siMUT group, and s was specific siRNA group. Y-axis showed the ratio of electrophoresis band intensity of each target gene to that of β-actin. Each group included five ratios, they were dnapol/β-actin (rectangles with transverse lines), rr2/β-actin (diagonal rectangles), tk-tmk/β-actin (rectangles with vertical stripes), vp24/β-actin (filled rectangles) and vp28/β-actin (rectangles with gridding), respectively.
3.3. Inhibition of WSSV replication in shrimp
To ensure that the specific siRNAs inhibit replication of virus, the quantity of WSSV in shrimp from each group was analyzed at 24, 36 and 48 h p.i. with quantitative real-time PCR. The viral copies were converted into logarithm to construct histogram. The results showed that sequence-specific siRNAs significantly inhibited replication of WSSV relative to that of positive controls and the mutant siRNA injected group at 24 h during experiment (Fig. 3 ). However, at both 36 and 48 h p.i., there were no significant differences in the amount of virus from shrimp of specific siRNA groups and positive control. Apparently, the inhibition of target genes expression of WSSV via sequence-dependent siRNAs resulted in reduction of WSSV in shrimp.
Fig. 3.
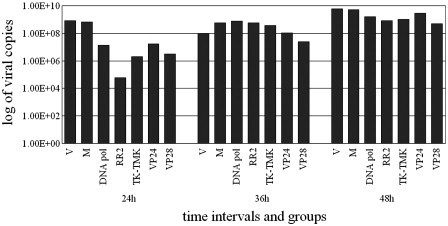
Quantitative PCR of WSSV from shrimp of each group at 24, 36 and 48 h after siRNA and virus injection. The amount of virus detected was extrapolated from a standard curve, and the results were expressed as a log of copies of virus.
3.4. Sequence-independent siRNA failed to induce antiviral effect in shrimp
We designed a mutant siRNA (siMUT) as a sequence-independent control by changing 6 nt of siVP28 in this study. The effect of siMUT to suppress the expression of each selected WSSV gene was determined by using semi-quantitative RT-PCR and real-time quantitative PCR, the effect to reduce the viral replication was detected. As predicted, siMUT could neither inhibit the expression of vp28 nor the other four selected WSSV genes (Fig. 2B), and the viral replication was not reduced by siMUT (Fig. 3). These results suggested that the inhibition of target genes and the reduction of viral replication were specific. Sequence-dependent siRNAs delayed death and enhanced survival rate of shrimp infected with WSSV, but sequence-independent siRNA did not.
4. Discussion
Recently, RNAi-based gene therapies, especially in viral diseases, have become more and more interesting and promising. As a powerful tool, RNAi technology plays an important role in protecting host from viral infection (Ge et al., 2003), suppressing the transcription of viral genome (Yoshinouchi et al., 2003) and blocking viral replication (Hamasaki et al., 2003). The present work indicates that RNAi directed against WSSV genome could effectively block expression of target genes and viral replication. Thus, it is possible to use RNAi to silence viral gene expression and to inhibit viral replication in shrimp. RNAi also provides a useful tool for further study of gene functions in shrimp and for prevention and treatment of viral diseases in aquatic animals.
In this study, siRNAs were synthesized to target several major genes of WSSV, including structural proteins and enzymes involved in virus replication, transcription, and virus–host interaction (Chen et al., 2002, Tsai et al., 2000, van Hulten et al., 2000). Among these five genes, dnapol, rr2 and tk-tmk were early expression genes and the other two (vp24 and vp28) were late expression genes (Liu et al., 2005, Marks et al., 2003). In mammalian cells, ribonucleotide reductase (RR) catalyzes the reaction in which 2′-deoxyribonucleotides (dADP, dGDP, dUDP, and dCDP) are synthesized from the corresponding ribonucleoside 5′-diphosphates (ADP, GDP, UDP, and CDP) (Engström et al., 1985). In addition, the level and activity of ribonucleotide reductase are highly regulated by the cell cycle (Yao et al., 2003). Consequently, some large DNA viruses such as herpesviruses and poxviruses can synthesis their own ribonucleotide reductases to replicate in non-dividing cells in which the cellular ribonucleotide reductase expression level is low (Lin et al., 2002). Besides ribonucleotide reductase, thymidine kinase (TK) and thymidylate kinase (TK-TMK) are also important enzymes of nucleotide biosynthesis in WSSV replication. TK and TMK are related to virulence influence (Stanberry et al., 1985, Tzeng et al., 2002). VP28 is identified as a major envelope protein of WSSV and is involved in viral infection (van Hulten et al., 2001b). In contrast, VP24 is a nucleocapsid protein of WSSV and its function is still unclear (van Hulten et al., 2000). We designed one specific siRNA (21 nt) corresponding the conservative region of each selected genes, all of which proved to be effective (Fig. 2B). The results showed that delivery of sequence-specific siRNAs could result in significant reduction of the corresponding gene expression (Fig. 2A), and decrease the number of WSSV virions (Fig. 3). It was more important that the death of shrimp infected by WSSV was delayed after injecting sequence-dependent siRNAs into the shrimp (Fig. 1). Therefore we concluded that the anti-WSSV activity is probably the result of siRNA-mediated WSSV RNA degradation in shrimp. The silencing of the non-structural viral protein genes (dnapol, rr2 and tk-tmk) increased the shrimp survival rate more than the silencing of the structural protein genes (vp24 and vp28). But the reduction of vp24 and vp28 gene expression was greater than the reduction of the other 3 genes. siRNA silencing experiments were repeated three times, and the results were consistent. It is worth further investigation to see whether the metabolism related genes expressed in early replication stage would play a more critical role in WSSV replication.
Additionally, we designed a mutant siRNA (siMUT) as sequence-independent siRNA control by changing 6 nt of siVP28. The results showed that siMUT could not inhibit the expression of vp28 and the other four selected genes of WSSV (Fig. 2B). Moreover, the viral replication did not decrease (Fig. 3) and the death of shrimp infected by WSSV did not delay by the control siMUT (Fig. 1). Thus, the observation of this study suggested that siRNAs mediated significant reduction specifically, and not a global down-regulation resulting from activation of the dsRNA-activated RNA-dependent protein kinase (PKR), which could lead to an innate immunoreaction in a non-sequence-specific manner (Hannon, 2002; Robalino et al., 2004). Meanwhile, our results showed that interference was sequence-specific using siRNAs. However, in a recent RNAi research in shrimp in vivo (Westenberg et al., 2005), GFP-siRNA (non-specific siRNA control) induced reduction of shrimp mortality. In other species such as mice (Flynn et al., 2004, Tompkins et al., 2004), GFP-siRNA as controls did not inhibit the expression of target gene and viral replication in vivo. This phenomenon was interesting and worth further investigation.
Our data strongly indicated that sequence-specific siRNAs significantly inhibited expression of target genes and viral replication to protect shrimp from WSSV. Moreover, many studies demonstrated that sequence-specific siRNAs successfully suppressed expression of corresponding genes in animals in vivo such as mice (McCaffrey et al., 2002, Saito et al., 2005, Song et al., 2003), zebrafish (Dodd et al., 2004) and shrimp (Westenberg et al., 2005). However, the investigation of the effect of WSSV VP19-siRNA molecules on the response of shrimp to WSSV infection concluded that siRNAs delivered into shrimp had poor biological activity, not only in gene silencing but also as inducers of sequence-specific antiviral responses (Robalino et al., 2005). The possible explanation for this discrepancy could be that different selection of target sequences for RNAi would result in completely dissimilar interference impacts (Li et al., 2005, Zheng et al., 2004).
So far, RNAi has become an essential experimental tool in many research fields of life sciences. Although WSSV infection is a major problem in shrimp aquaculture worldwide, there is no effective antiviral treatment. siRNA technology may provide a possible therapeutic strategy against WSSV infection. Recently, several researches about RNAi in shrimp and the results of our study suggested that effective time of RNAi in vivo is limited, and the delivery efficiency of dsRNA or siRNA is not difficult to control and detect (Robalino et al., 2004, Westenberg et al., 2005). The half-life of synthetic RNA duplexes is very short. In some circumstances, these siRNAs will not stay long enough for complete elimination of virus replication in an infected cell. Further studies are necessary to construct high expression vectors and better transfection approaches of dsRNA/siRNA in shrimp in vivo. Therefore, it is necessary to study siRNA injection continuously into shrimp instead of single injection so that the siRNA may perform more effectively. Moreover, a permanent shrimp cell line would bring further advantages.
In conclusion, we have developed a specific siRNA approach that can effectively inhibit replication of WSSV. Although there are a number of important issues to be resolved, these siRNAs may be a potential candidate for therapeutic measures against the current disease caused by WSSV.
Acknowledgments
This study was supported by the National Natural Science Foundation of China under grant Nos. 30325035 and U0631008; the National Basic Research Program of China under grant No. 2006CB1011802; the National High Technology Research and Development Program of China (863 Program) under grant Nos. 2006AA09Z445, 2006AA100312, and 2006AA100309; Natural Science Foundation of Guangdong Province of China under grant No. 20023002; and Science and Technology Bureau of Guangdong Province. We thank Dr. Xiao-Qiang Yu, Division of Cell Biology and Biophysics, School of Biological Sciences University of Missouri-Kansas City, for thoroughly revising and commenting on this manuscript.
References
- Adelman Z.N., Sanchez-Vargas I., Travanty E.A., Carlson J.O., Beaty B.J., Blair C.D., Olson K.E. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 2002;76:12925–12933. doi: 10.1128/JVI.76.24.12925-12933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., Wang H.C., Huang C.J., Peng S.E., Chen Y.G., Lin S.J., Chen W.Y., Dai C.F., Yu H.T., Wang C.H., Lo C.F., Kou G.H. Transcriptional analysis of the DNA polymerase gene of white spot syndrome virus. Virology. 2002;301:136–147. doi: 10.1006/viro.2002.1536. [DOI] [PubMed] [Google Scholar]
- Chou H.Y., Huang C.Y., Wang C.H., Chiang H.C., Lo C.F. Pathogenicity of a baculovirus infection causing white spot syndrome in cultured penaeid shrimp in Taiwan. Dis. Aquat. Org. 1995;23:165–173. [Google Scholar]
- Deng M., He J.G., Lü L., Zuo T., Long Q.X., Chan S.-M. Partial cloning of the genome library of white spot syndrome virus from Penaeus monodon and DNA probe detection methods [abstract in English] J. Fish China. 2000;24:161–166. [Google Scholar]
- Dodd A., Chambers S.P., Love D.R. Short interfering RNA-mediated gene targeting in the zebrafish. FEBS Lett. 2004;561:89–93. doi: 10.1016/S0014-5793(04)00129-2. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström Y., Eriksson S., Jildevik I., Skog S., Thelander L., Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. J. Biol. Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A. Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press; London: 2005. Nimaviridae; pp. 187–192. [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Flynn M.A., Casey D.G., Todryk S.M., Mahon B.P. Efficient delivery of small interfering RNA for inhibition of IL-12p40 expression in vivo. J. Inflam. (Lond.) 2004;1:4. doi: 10.1186/1476-9255-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q., McManus M.T., Guyen T.N., Shen C.H., Sharp P.A., Eisen H.N., Chen J. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L., Karelsky S., Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- Hamasaki K., Nakao K., Matsumoto K., Ichikawa T., Ishikawa H., Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003;543:51–54. doi: 10.1016/s0014-5793(03)00400-9. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- He J.G., Deng M., Long Q.X., Zhou H.M., Li G.S., Yao P., Lü L., Chen Y.G., Weng S.P., Miao S.Y., Jiang S.G., Wu Y.Y., Huang X., Zuo T., Yang X.M., Yu N., Xie S.T., Zhong W., Li G.F., Ye Q.Z., Jiang J.B., Chan S.-M., Mo F. vol. 39. 2000. Theory and strategies for controlling White Spot Syndrome (WSS) of cultured Penaeus monodon in South China; pp. 147–153. (Acta Scientarium Naturalium Universitatis Sunyasteni). (supplement) [Google Scholar]
- He J.G., Chen Y.G., Deng M., Yao P., Zhou H.M., Weng S.P., Jiang S.G., Long Q.X., Chan S.-M. Natural host range and pathogenicity of white spot syndrome virus infection in shrimp and crab species in China. In: Phillips B., Megrcy B.A., Zhou Y., editors. Proceedings of the Third World Fisheries Congress: Feeding the World with Fish in the Next Millennium—the Balance between Production and Environment. vol. 38. Bethesda; Maryland: 2003. pp. 195–203. (American Fisheries Society, Symposium). [Google Scholar]
- He M.L., Zheng B., Peng Y., Peiris J.S., Poon L.L., Yuen K.Y., Lin M.C., Kung H.F., Guan Y. Inhibition of SARS-associated coronavirus infection and replication by RNA interference. JAMA. 2003;290:2665–2666. doi: 10.1001/jama.290.20.2665. [DOI] [PubMed] [Google Scholar]
- Huang J., Song X.L., Yu J., Yang C. Baculoviral hypodermal and hematopoietic necrosis: study on the pathogen and pathology of the explosive epidemic disease of shrimp [in Chinese] Mar. Fish. Res. 1995;16:1–10. [Google Scholar]
- Jacque J.M., Triques K., Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X.F., Lu L., Chen Y.G., Chan S.M., He J.G. Comparison of a novel in situ polymerase chain reaction (ISPCR) method to other methods for white spot syndrome virus (WSSV) detection in Penaeus vannamei. Dis. Aquat. Org. 2005;67:171–176. doi: 10.3354/dao067171. [DOI] [PubMed] [Google Scholar]
- Kahana R., Kuznetzova L., Rogel A., Shemesh M., Hai D., Yadin H., Stram Y. Inhibition of foot-and-mouth disease virus replication by small interfering RNA. J. Gen. Virol. 2004;85:3213–3217. doi: 10.1099/vir.0.80133-0. [DOI] [PubMed] [Google Scholar]
- Kapadia S.B., Brideau-Andersen A., Chisari F.V. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell J.R., Carthew R.W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Li B.J., Tang Q., Cheng D., Qin C., Xie F.Y., Wei Q., Xu J., Liu Y., Zheng B.J., Woodle M.C., Zhong N., Lu P.Y. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.T., Chang Y.S., Wang H.C., Tzeng H.F., Chang Z.F., Lin J.Y., Wang C.H., Lo C.F., Kou G.H. Ribonucleotide reductase of shrimp white spot syndrome virus (WSSV): expression and enzymatic activity in a baculovirus/insect cell system and WSSV-infected shrimp. Virology. 2002;304:282–290. doi: 10.1006/viro.2002.1696. [DOI] [PubMed] [Google Scholar]
- Liu W.J., Chang Y.S., Wang C.H., Kou G.H., Lo C.F. Microarray and RT-PCR screening for white spot syndrome virus immediate-early genes in cycloheximide-treated shrimp. Virology. 2005;334:327–341. doi: 10.1016/j.virol.2005.01.047. [DOI] [PubMed] [Google Scholar]
- Marks H., Mennens M., Vlak J.M., van Hulten M.C.M. Transcriptional analysis of the white spot syndrome virus major virion protein genes. J. Gen. Virol. 2003;84:1517–1523. doi: 10.1099/vir.0.19018-0. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Meuse L., Pham T.T., Conklin D.S., Hannon G.J., Kay M.A. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Nakai H., Pandey K., Huang Z., Salazar F.H., Xu H., Wieland S.F., Marion P.L., Kay M.A. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Ngo H., Tschudi C., Gull K., Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robalino J., Browdy C.L., Prior S., Metz A., Parnell P., Gross P., Warr G. Induction of antiviral immunity by double-stranded RNA in a marine invertebrate. J. Virol. 2004;78:10442–10448. doi: 10.1128/JVI.78.19.10442-10448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robalino J., Bartlett T., Shepard E., Prior S., Jaramillo G., Scura E., Chapman R.W., Gross P.S., Browdy C.L., Warr G.W. Double-stranded RNA induces sequence-specific antiviral silencing in addition to nonspecific immunity in a marine shrimp: convergence of RNA interference and innate immunity in the invertebrate antiviral response. J. Virol. 2005;79:13561–13571. doi: 10.1128/JVI.79.21.13561-13571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Yokota T., Mitani T., Ito K., Anzai M., Miyagishi M., Taira K., Mizusawa H. Transgenic small interfering RNA halts amyotrophic lateral sclerosis in a mouse model. J. Biol. Chem. 2005;280:42826–42830. doi: 10.1074/jbc.M507685200. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. RNA interference—2001. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- Shlomai A., Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- Song E., Lee S.K., Wang J., Ince N., Ouyang N., Min J., Chen J., Shankar P., Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Stanberry L.R., Kit S., Myers M.G. Thymidine kinase-deficient herpes simplex virus type 2 genital infection in guinea pigs. J. Virol. 1985;55:322–328. doi: 10.1128/jvi.55.2.322-328.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Itami T., Kondo M., Maeda M., Fujii R., Tomonaga S., Supamattaya K., Boonyaratpalin S. Electron microscopy evidence of bacilliform virus infection in kuruma shrimp (Penaeus japonicus) Fish Pathol. 1994;29:121–125. [Google Scholar]
- Tirasophon W., Roshorm Y., Panyim S. Silencing of yellow head virus replication in penaeid shrimp cells by dsRNA. Biochem. Biophys. Res. Commun. 2005;334:102–107. doi: 10.1016/j.bbrc.2005.06.063. [DOI] [PubMed] [Google Scholar]
- Tompkins S.M., Lo C.Y., Tumpey T.M., Epstein S.L. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.F., Yu H.T., Tzeng H.F., Leu J.H., Chou C.M., Huang C.J., Wang C.H., Lin J.Y., Kou G.H., Lo C.F. Identification and characterization of a shrimp white spot syndrome virus (WSSV) gene that encodes a novel chimeric polypeptide of cellular-type thymidine kinase and thymidylate kinase. Virology. 2000;277:100–110. doi: 10.1006/viro.2000.0597. [DOI] [PubMed] [Google Scholar]
- Tzeng H.F., Chang Z.F., Peng S.E., Wang C.H., Lin J.Y., Kou G.H., Lo C.F. Chimeric polypeptide of thymidine kinase and thymidylate kinase of shrimp white spot syndrome virus: thymidine kinase activity of the recombinant protein expressed in a baculovirus/insect cell system. Virology. 2002;299:248–255. doi: 10.1006/viro.2002.1480. [DOI] [PubMed] [Google Scholar]
- van Hulten M.C.W., Goldbach R.W., Vlak J.M. Three functionally diverged major structural proteins of white spot syndrome virus evolved by gene duplication. J. Gen. Virol. 2000;81:2525–2529. doi: 10.1099/0022-1317-81-10-2525. [DOI] [PubMed] [Google Scholar]
- van Hulten M.C.W., Witteveldt J., Peters S., Kloosterboer N., Tarchini R., Fiers M., Sandbrink H., Lankhorst R.K., Vlak J.M. The white spot syndrome virus DNA genome sequence. Virology. 2001;286:7–22. doi: 10.1006/viro.2001.1002. [DOI] [PubMed] [Google Scholar]
- van Hulten M.C.W., Witteveldt J., Snippe M., Vlak J.M. White spot syndrome virus envelope protein VP28 is involved in the systemic infection of shrimp. Virology. 2001;285:228–233. doi: 10.1006/viro.2001.0928. [DOI] [PubMed] [Google Scholar]
- Wang C.H., Lo C.F., Leu J.H., Chou C.M., Yeh P.Y., Chou H.Y., Tung M.C., Chang C.F., Su M.C., Kou G.H. Purification and genomic analysis of baculovirus associated with white spot syndrome (WSBV) of Penaeus monodon. Dis. Aquat. Org. 1995;23:239–242. [Google Scholar]
- Westenberg M., Heinhuis B., Zuidema D., Vlak J.M. siRNA injection induces sequence-independent protection in Penaeus monodon against white spot syndrome virus. Virus Res. 2005;114:133–139. doi: 10.1016/j.virusres.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Wianny F., Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- Wongteerasupaya C., Vickers J.E., Sriurairatana S., Nash G.L., Akarajamorn A., Boonsaeng V., Panyim S., Tassanakajon A., Withyachumnarnkul B., Flegel T.W. A non-occluded, systemic baculovirus that occurs in cells of ectodermal and esodermal origin and causes high mortality in the black tiger prawn Penaeus monodon. Dis. Aquat. Org. 1995;21:69–77. [Google Scholar]
- Xie J., Lü L., Deng M., Weng S., Zhu J., Wu Y., Gan L., Chan S.-M., He J. Inhibition of reporter gene and iridovirus-tiger frog virus in fish cell by RNA interference. Virology. 2005;338:43–52. doi: 10.1016/j.virol.2005.04.040. [DOI] [PubMed] [Google Scholar]
- Yang F., He J., Lin X.H., Li Q., Pan D., Zhang X.B., Xu X. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 2001;75:11811–11820. doi: 10.1128/JVI.75.23.11811-11820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R.J., Zhang Z., An X.X., Bucci B., Perlstein D.L., Stubbe J.A., Huang M.X. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinouchi M., Yamada T., Kizaki M., Fen J., Koseki T., Ikeda Y., Nishihara T., Yamato K. In vitro and in vivo growth suppression of human papillomavirus 6-positive cervical cancer cells by E6 siRNA. Molec. Ther. 2003;8:762–768. doi: 10.1016/j.ymthe.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Zamore P.D. Ancient pathways programmed by small RNAs. Science. 2002;296:1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- Zheng B.J., Guan Y., Tang Q., Du C., Xie F.Y., He M.L., Chan K.W., Wong K.L., Lader E., Woodle M.C., Lu P.Y., Li B., Zhong N. Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus. Antivir. Ther. 2004;9:365–374. [PubMed] [Google Scholar]


