Abstract
Human enterovirus 71 (EV71) belongs to the genus Enterovirus in the family Picornaviridae and has been recognized as one of the most important pathogens that cause emerging infectious disease. Despite of the importance of EV71, the nonstructural protein 3AB from this virus is little understood for its function during EV71 replication. Here we expressed EV71 3AB protein as recombinant protein in a eukaryotic expression system and uncovered that this protein possesses a nucleic acid helix-destabilizing and strand annealing acceleration activity in a dose-dependent manner, indicating that EV71 3AB is a nucleic acid chaperone protein. Moreover, we characterized the RNA chaperone activity of EV71 3AB, and revealed that divalent metal ions, such as Mg2+ and Zn2+, were able to inhibit the RNA helix-destabilizing activity of 3AB to different extents. Moreover, we determined that 3B plus the last 7 amino acids at the C-terminal of 3A (termed 3B+7) possess the RNA chaperone activity, and five amino acids, i.e. Lys-80, Phe-82, Phe-85, Tyr-89, and Arg-103, are critical and probably the active sites of 3AB for its RNA chaperone activity. This report reveals that EV71 3AB displays an RNA chaperone activity, adds a new member to the growing list of virus-encoded RNA chaperones, and provides novel knowledge about the virology of EV71.
Keywords: Enterovirus 71, RNA chaperone, RNA duplex unwinding, Replication, Antivirals strategies
Introduction
Enterovirus 71 (EV71), a member of the genus Enterovirus in the family Picornaviridae, is the major causative pathogen for hand-foot-and-mouth disease (HFMD) in children (Hagiwara et al., 1978), and can also cause neurological syndromes, such as aseptic meningitis, encephalitis, poliomyelitis-like paralysis, and even death (Chang et al., 1999, Ooi et al., 2010). EV71 has been recognized as one of the most important pathogens that cause emerging infectious disease; moreover, increased occurrence of EV71 epidemic activities has been reported throughout the Asia-Pacific region in recent years (Huang et al., 2008, Yang et al., 2009, Zhang et al., 2010). Like other enteroviruses, EV71 contains a single-stranded, positive-sense RNA genome, which is approximately 7.4 kb in length and consisting of a single open reading frame (ORF) flanked by 5′ and 3′ untranslated regions (UTRs). Enteroviral ORF encodes a long polyprotein that is subsequently cleaved at specific sites to produce separate structural and nonstructural proteins (Brown and Pallansch, 1995). Previous studies from enteroviruses, mainly poliovirus, have shown that some of enteroviral nonstructural proteins, including 3DPol, the RNA-dependent RNA polymerase (RdRp), 3CD, 3AB and 3B, as well as certain host factors are involved in viral RNA replication (Hey et al., 1987).
For RNA viruses including enteroviruses, their viral RNAs (vRNAs) require proper secondary and tertiary structures to form diverse cis-acting elements within their 5′-UTR, 3′-UTR or ORF region, which play essential or critical roles in vRNA replication, translation, and encapsidation (Musier-Forsyth, 2010, Rajkowitsch et al., 2007, Zuniga et al., 2009). Moreover, the formation and deformation of RNA tertiary structures should be a dynamically regulated process for vRNAs to function properly in various events. However, correct folding of RNA molecules is quite challenging, because RNAs would be easily trapped in intermediate structures that are thermodynamically stable (i.e. kinetic trap) (Musier-Forsyth, 2010, Woodson, 2010). In response to this challenge, hosts or viruses encode a variety of RNA “helper” or remodeling proteins, generally including ATP-dependent RNA helicases and ATP-independent RNA chaperones, which help kinetically trapped RNAs overcome thermodynamic barriers and refold to form proper RNA structures (Singleton et al., 2007). RNA helicases contain ATPase activity, and utilize the energy of ATP binding and hydrolysis to unwind RNA duplexes. These proteins are thought to participate in most ATP-dependent remodeling of structured RNAs (Singleton et al., 2007). On the other hand, RNA chaperones are a heterogeneous group of proteins that can destabilize RNA duplexes and accelerate correct RNA folding by helping misfolded RNAs escape kinetic traps (Herschlag, 1995). For enteroviruses, although nonstructural protein 2CATPase has long been predicted as a putative helicase based on its AAA+ ATPase activity and conserved helicase motifs, the putative helicase activity associated with 2CATPase has not been formally determined (Sweeney et al., 2010). In contrast, previous studies of poliovirus have shown that poliovirus nonstructural protein 3AB possesses an RNA chaperone activity in vitro, which probably involves in polioviral RNA synthesis by 3DPol (DeStefano and Titilope, 2006).
Enteroviral 3AB is a ~12 kDa and 108 amino acids (a.a.) protein, containing N-terminal 3A (~10 kDa) and C-terminal 3B/VPg (~2 kDa) domains. Protein 3AB is derived from the ~84 kDa P3 precursor of enteroviral polyprotein, which also contains 3DPol and 3C protease (3CPro). Finally, 3AB will be proteolytically cleaved into proteins 3A and 3B/VPg (Pallai et al., 1989). During enteroviral RNA replication, proteins 3A, 3B/VPg as well as 3AB play critical roles in vRNA synthesis (Plotch and Palant, 1995). Among them, protein 3A is 87 a.a. in length and has an effect on protein trafficking (Belov et al., 2005). For all picornaviruses, the 3B/VPg is a small peptide (21–24 a.a.) that functions as a primer to initiate vRNA synthesis by 3DPol, which is covalently linked to the 5′-end of all newly synthesized vRNAs (Pathak et al., 2007). Moreover, proteins 3A and 3AB have been reported to play key roles in the formation of enteroviral replication complex.
In human enteroviruses, poliovirus is the type specie and has long been recognized as the most important enterovirus since it causes poliomyelitis (Nathanson and Langmuir, 1963). Following the mostly complete eradication of poliovirus and poliomyelitis in most regions around the world (Friedrich, 2000), EV71 has emerged as the most important human neuron-tropic enterovirus (Ishimaru et al., 1980). So far, no effective vaccine or antiviral therapy is available for EV71 (Chong et al., 2012). Previous study has shown that mutants resistant to Enviroxime, an antiviral drug blocking polioviral RNA replication, map to the coding sequence of 3A, indicating that 3A (or 3AB) plays a critical role in vRNA replication and is an ideal target for antiviral therapy against other enteroviruses (Sadeghipour et al., 2012). Thus, a potential anti-EV71 strategy is to develop novel antivirals targeting 3A or 3AB (Kuo and Shih, 2013, Tan et al., 2014). However, although poliovirus 3AB has been reported to display an in vitro RNA chaperone activity, the function of EV71 3AB has been poorly studied. Due to the importance of 3AB for enteroviruses, a thorough understanding to its roles in EV71 virology would be desired for combating this emerging virus, and may serve as a basis for future studies.
In this study, we expressed the 3AB domain of EV71 polyprotein in a eukaryotic expression system and determined that EV71 3AB displays nonspecific nucleic acid helix-destabilizing and annealing acceleration activities. Further characterization of EV71 3AB revealed that divalent metal ions, such as Mg2+, Zn2+, inhibit the RNA helix-destabilizing of 3AB to different extents. Moreover, we demonstrated that 3B plus the last 7 amino acids at the C-terminal of 3A (termed 3B+7) still possesses RNA chaperone activity. Using a point mutagenesis approach, we uncovered that two of the point mutations were able to abolish the RNA helix-destabilizing activity of 3B+7, while the remaining mutants inhibited the activity to different extents. The results indicated that it is probable that the five amino acids (Lys-80, Phe-82, Phe-85, Tyr-89, and Arg-103) are the active sites for the RNA chaperone activity of 3AB, which may serve as the target for anti-EV71 therapy.
Results
EV71 3AB protein can destabilize both RNA and DNA helices
RNA chaperones are usually able to destabilize or unwind short RNA helices (Rajkowitsch and Schroeder, 2007). To examine whether EV71 3AB possesses RNA helix-destabilizing activity, we expressed and purified EV71 3AB as MBP fusion protein (MBP-3AB) using baculovirus expression system ( Fig. 1B). To assess helix-destabilizing activity, a short HEX-labeled RNA (RNA1) and a long non-labeled RNA (RNA2) were annealed to generate a standard RNA substrate with both 5′ (22 bases) and 3′ (23 bases) single-stranded overhangs ( Fig. 2A, left panel and Table 2). The helix-destabilizing reaction was conducted by incubating RNA helix substrate with MBP-3AB, followed by gel electrophoresis. Our data show that the HEX-labeled RNA strand was released from the standard RNA helix substrate in the presence of MBP-3AB, whereas the negative control MBP had no effect on the same substrate (Fig. 2A, right panel), indicating that EV71 3AB has the activity to destabilize RNA helix. In this and subsequent experiments, boiled duplex substrates were used as positive controls.
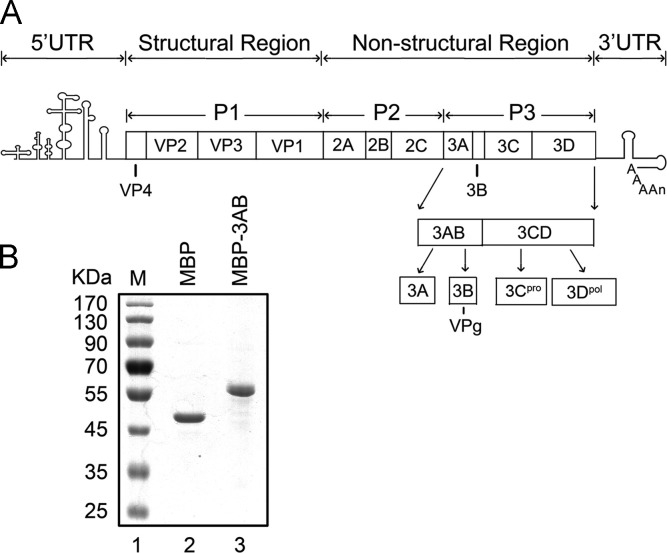
Fig. 1.
Organization of an EV71 genome and polyprotein processing. (A) The 5′ and 3′UTR contain RNA sequences forming RNA structural elements. The coding region is divided into three sections, including the P1, P2 and P3 precursor. Protein 3AB is derived from the P3 precursor of enteroviral polyprotein, which also contains 3DPol and 3C protease (3CPro). 3AB will be proteolytically cleaved into proteins 3A and 3B/VPg. (B) Electrophoresis analysis of purified MBP-3AB and MBP alone. Proteins were loaded onto a 10% SDS-PAGE gel and then visualized via Coomassie blue staining. Lane 1, protein molecular mass markers; lane 2, purified MBP; lane 3, purified MBP-3AB.
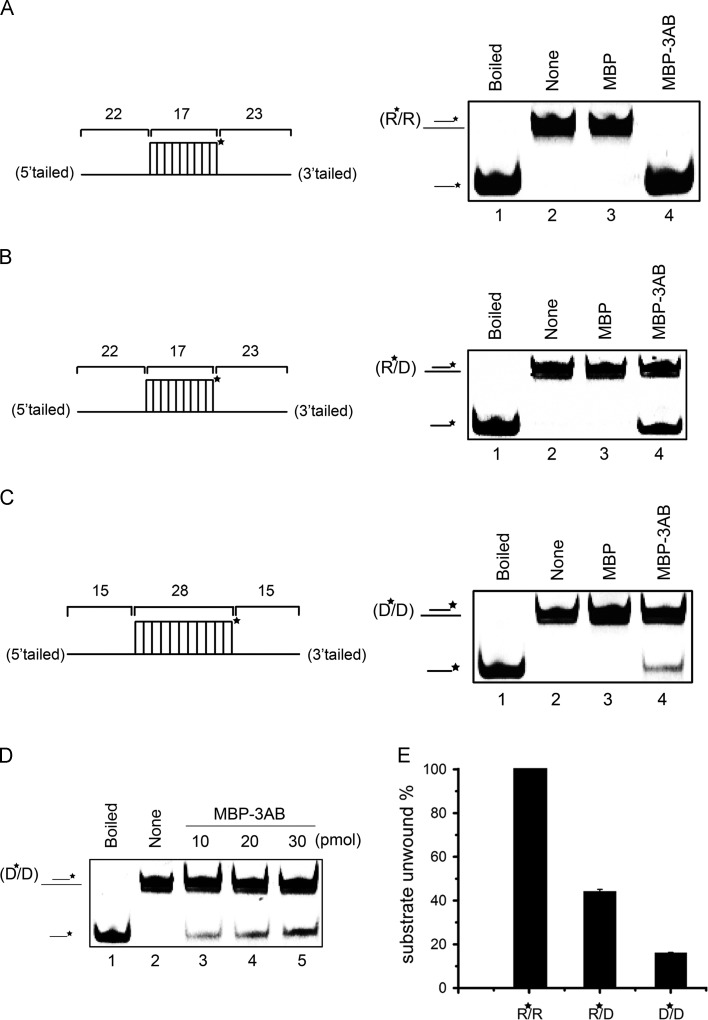
Fig. 2.
EV71 3AB can destabilize both RNA and DNA helices. (A–C) Purified MBP-3AB was incubated with standard RNA duplex (R*/R substrate), RNA/DNA hybrid duplex (R*/D substrate) and DNA duplex (D*/D substrate) as illustrated in left panels. Asterisk (*) indicates HEX-labeled strand. The preparations of destabilizing substrates are indicated in Materials and Methods. The 0.1 pmol substrate was incubated in standard reaction mixtures in the presence or absence of 10 pmol MBP-3AB as indicated, and the destabilizing activity was assessed via gel electrophoresis and scanning on a Typhoon 9200. (A) R*/R substrate (left panel). Lane 1, boiled reaction mixture without enzyme addition; Lane 2, reaction mixture without enzyme addition; Lane 3, complete reaction mixture with negative control MBP alone; Lane 4, complete reaction mixture with MBP-3AB. (B) R*/D substrate (left panel). Lanes 1 and 2: boiled (lane 1) or native (lane 2) reaction mixtures without enzyme addition; Lane 3, complete reaction mixture with negative control MBP alone; Lane 4, complete reaction mixture with MBP-3AB. (C) D*/D substrate (left panel). Lanes 1 and 2: boiled (lane 1) or native (lane 2) reaction mixtures without enzyme addition; Lane 3, complete reaction mixture with negative control MBP alone; Lane 4, complete reaction mixture with MBP-3AB. (D) Increasing amount of MBP-3AB was incubated with 0.1 pmol DNA duplex (D*/D) as indicated, and the destabilizing activity was assessed via gel electrophoresis and scanning on a Typhoon 9200. Asterisk (*) indicates HEX-labeled strand. (E) The helix destabilization was plotted as the percentage of the released RNA or DNA from the total helix substrates (Y-axis) at indicated helix substrates as X-axis.
Table 2.
The list of Oligonucleotides.
| Oligonucleotides | Sequence (5′-3′) |
|---|---|
| RNA1 | GUGUAGUAAUCGUCCAU |
| RNA2 | AGUUUAAAACGCACGAGACACAAUGGACGAUUACUACACCAACAGAUCAAUUGAGCAAUCCG |
| RNA3 | AUGGACGAUUACUACACCAACAGAUCAAUUGAGCAAUCCG |
| RNA4 | AGUUUAAAACGCACGAGACACAAUGGACGAUUACUACAC |
| RNA5 | AUGGACGAUUACUACAC |
| RNA6 | CAUUAUCGGAUAGUGGAACCUAGCUUCGACUAUCGGAUAAUC |
| RNA7 | UUUUUUUUUUUUUUUUUUUUGAUUAUCCGAUAGUCGAAGCUAGGUUCCACUAUCCGAUAA UGUUUUUUUUUUUUUUUUUUUU |
| RNA8 | GUAACUAUAUUCGUCAUUAUCUCAUUUCCCAAAUUAAUACCAUAAAUUC |
| RNA9 | CUCGAAUGUCAGCGACAGAAUUUAUAAAUACAAUAGAUGGAAUUGAAAUACCUAAUUCACAU AUAAUAUACUUUGGAAUAGAUACCCCUUUUUGUGAAAUAACGAGAAAAGCACCACGAAUUUA UGGUAUUAAUUUGGGAAAUGAGAUAAUGACGAAUAUAGUUACACCGCUAGCUUUACCAGACA AUGAGUUUUCGCCUUUACGUAUUAUGAAAAAUAUUAAAGAUGCGGAAACCAAACCAUAUAUC AGUCAGUCUGGUUGGGUCACUACUGUGUUAUCAGCUG |
| DNA1 | AGTTTAAAACGCACGAGACACAATGGACGATTACTACACCAACAGATCAATTGAGCAATCCG |
| DNA2 | CACCACAACCACCACCACCACACCATGG |
| DNA3 | TTTTTTTTTTTTTTTCCATGGTGTGGTGGTGGTGGTTGTGGTGTTTTTTTTTTTTTTT |
The synthesized single strands with florescence labeling are indicated with boldfaced characters.
Previous studies have shown that a variety of RNA chaperones are also able to destabilize/unwind DNA-DNA or DNA-RNA duplexes (Cheng et al., 2013), thus being named nucleic acid chaperones (DeStefano and Titilope, 2006). To further examine whether the helix-destabilizing activity of EV71 3AB is specific for RNA helix, we constructed two different nucleic acid helix substrates, R⁎/D (Fig. 2B, left panel) via annealing a short HEX-labeled RNA (RNA1) and a long non-labeled DNA (DNA2) and D⁎/D (Fig. 2C, left panel) via annealing a short HEX-labeled DNA (DNA1) and a long non-labeled DNA (DNA3) (Table 2). Each helix substrate was reacted with MBP-3AB under the same conditions as in Fig. 2A. Our results revealed that EV71 3AB was able to destabilize both RNA-DNA and DNA-DNA helix substrates (Fig. 2B and C). Moreover, we reacted DNA-DNA helix substrate with different concentrations of MBP-3AB, and the data show that 3AB can destabilize DNA-DNA helix in a dose-dependent manner (Fig. 2D). The destabilizing activities of EV71 3AB to different helix substrates were also quantified (Fig. 2E), suggesting that EV71 3AB unwinds RNA-RNA helix more efficiently. Altogether, our data show that EV71 3AB has a nucleic acid helix-destabilizing activity for both RNA and DNA helices, and this feature is similar with the nonstructural protein 2C from Ectropis obliqua picorna-like virus (EoV), which is also able to destabilize both RNA and DNA helices (Cheng et al., 2013).
The helix destabilization by EV71 3AB exhibits no directionality
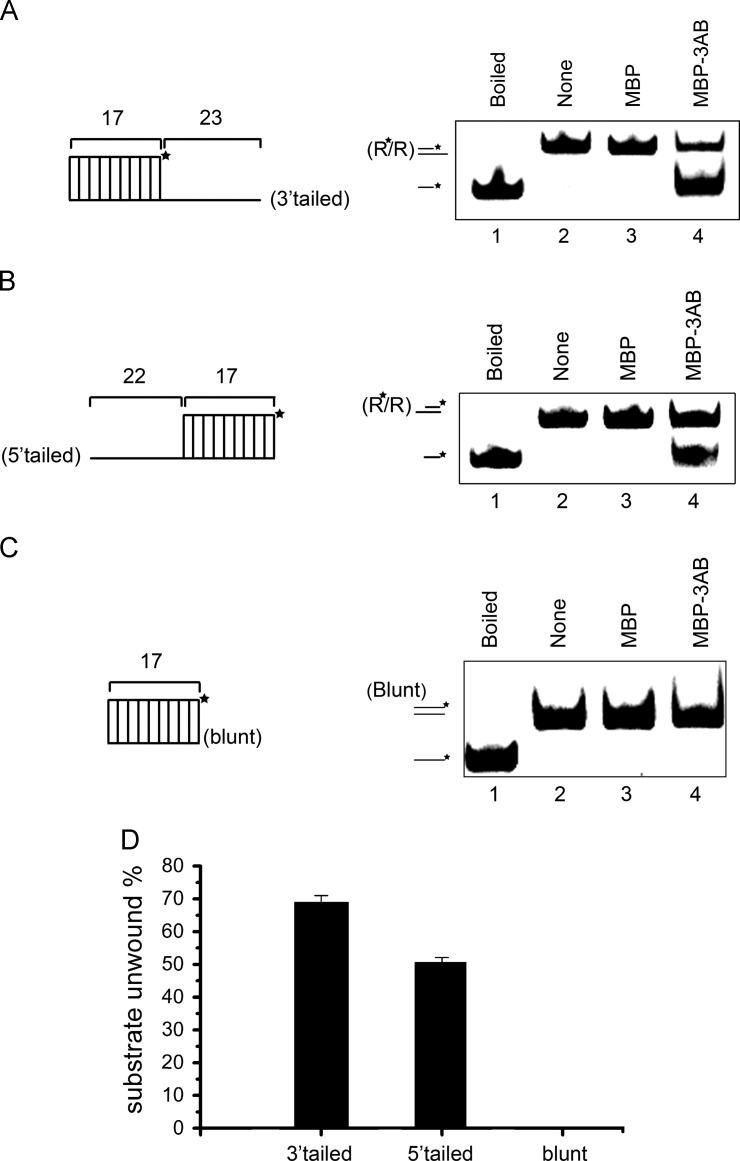
A major difference between RNA helicases and RNA chaperones is that the helix unwinding by helicases has directionality (also called polarity), while that of RNA chaperones is usually bidirectional (Singleton et al., 2007). After determining that EV71 3AB has nucleic acid helix-destabilizing activity, we sought to examine if the helix destabilization by 3AB has any directionality. Notably, although EV71 3AB can destabilize both DNA and RNA helices, we only used RNA helices in the subsequent experiments of the current work, since EV71 is an RNA virus and no DNA is involved in its life cycle. To this end, three different RNA helix substrates, containing a 3′ 23-nt single-stranded overhang, a 5′ 22-nt overhang, and blunt ends (as illustrated in Fig. 3A–C, left panels) were generated. These substrates were then reacted with MBP-3AB under our standard destabilizing condition. Our results showed that MBP-3AB was able to destabilize both 3′-tailed and 5′-tailed RNA helices, but not blunt-ended one (Fig. 3A–C). These experiments have been independently repeated several times. Altogether, our data demonstrate that the helix-destabilizing activity of EV71 3AB has no directionality or polarity.
Fig. 3.
EV71 3AB destabilizes RNA helices in a bidirectional manner. (A) Negative control MBP alone (lane 3) and MBP-3AB (10 pmol) were reacted with 3′-tailed (lane 4) RNA helix substrate (0.1 pmol) as illustrated in upper panel, respectively, under standard reaction conditions. Asterisk (*) indicates HEX-labeled strand. (B) Negative control MBP alone (lane 3) and MBP-3AB (10 pmol) were reacted with 5′-tailed (lane 4) RNA helix substrate (0.1 pmol) as illustrated in upper panel, respectively, under standard reaction conditions. For panels A and B, Boiled 3′-tailed (A) or 5′-tailed (B) substrate (lane 1) or substrates alone (lanes 2) were used as controls. (C)MBP-3AB (10 pmol) was reacted with blunt-ended RNA helix (0.1 pmol) as illustrated in upper panel. (D) The unwinding activity was plotted as the percentage of the released RNA from the total RNA helix substrates (Y-axis) at indicated helix substrates as X-axis. Error bars represent standard deviation values from three separate experiments.
Kinetics study of the helix-destabilizing reaction of EV71 3AB with respect to rate constants
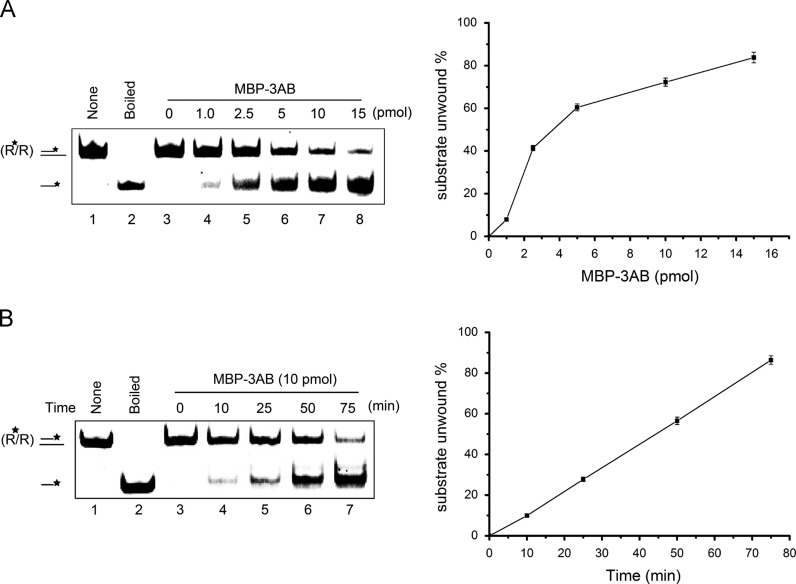
In order to calculate the rate constants of EV71 3AB to evaluate its nucleic acid chaperone activity, we examined its helix-destabilizing activity under different concentrations of MBP-3AB and different reaction times. We found that the RNA helix-destabilizing activity of MBP-3AB was dose dependent, as increasing the dose of 3AB gradually enhanced helix destabilization ( Fig. 4A). In addition, the RNA helix-destabilizing activity increases with the increase of reaction time (Fig. 4B). The quantitated data obtained using different 3AB doses and reaction times can be used to calculate the rate constant. The helix-destabilizing reaction of EV71 3AB belongs to class of zero-order reactions for the RNA helix substrates, and belongs to one half-order reactions for protein 3AB. Using Origin curve fitting method, we calculated that the rate constant of the helix-destabilizing reaction of EV71 3AB is 6.29×10−12 (mol/L)1/2 s−1.
Fig. 4.
Kinetics study of the helix-destabilizing reaction of EV71 3AB with respect to rate constants. (A) Standard RNA helix substrate (3′ tailed) (0.1 pmol) was reacted with increasing amounts of MBP-3AB (0 to 15 pmol) for 1h. (B) Standard RNA helix substrate (3′ tailed) (0.1 pmol) was reacted with MBP-3AB (10 pmol) at different reaction times (0–75 min). For (A and B, right panels), the destabilizing activities were plotted as the percentage of the released RNA from the total RNA helix substrates (Y-axis) at indicated amounts of MBP-3AB (A) or reaction time intervals (B) as X-axis. Error bars represent standard deviation values from three separate experiments.
Biochemical conditions for the RNA helix-destabilizing activity of EV71 3AB
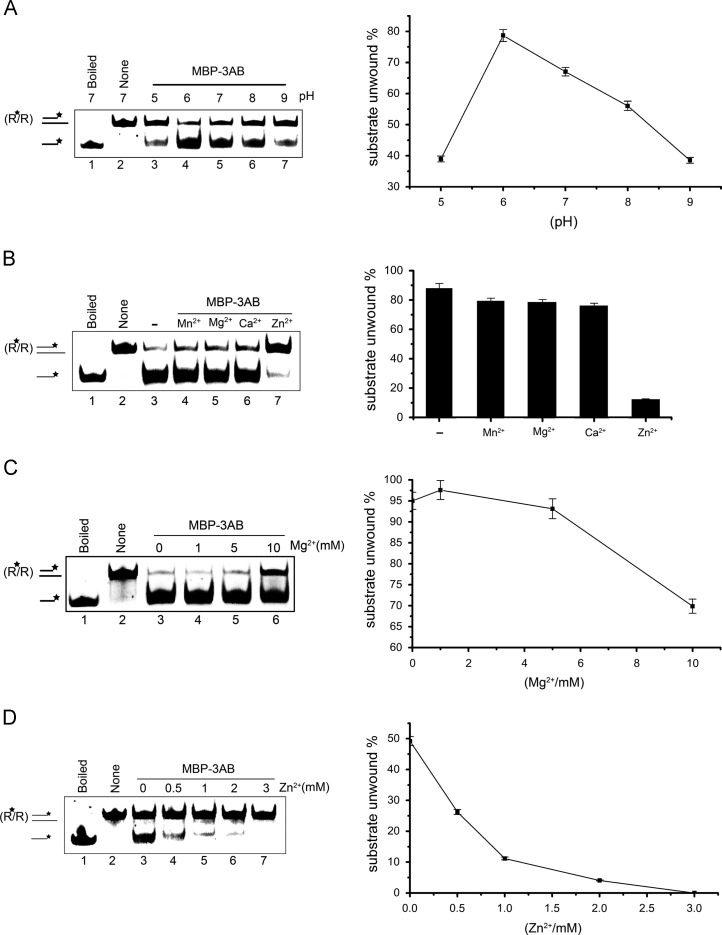
To further characterize the biochemical reaction conditions for the RNA helix-destabilizing activity of MBP-3AB, we examined its activity under various conditions, such as pH values, divalent metal ions and ion concentrations. Of note, for this and subsequent experiments, a 3′-tailed RNA helix substrate was used.
We also examined the effects of different pH on the helix-destabilizing activity of MBP-3AB. Our results showed that MBP-3AB prefers mildly acidic pH, as MBP-3AB exhibited the highest helix-destabilizing activity at pH 6.0 relative to others ( Fig. 5A). Furthermore, we examined the requirement for Mg2+ as well as three other divalent metal ions (Mn2+, Ca2+, and Zn2+) for the helix-destabilizing activity of EV71 3AB. For helicases, Mg2+ is usually required for their helix unwinding activities (Warrener and Collett, 1995, Yang et al., 2006). Here we found that MBP-3AB did not require the presence of 2 mM Mg2+ or other divalent metal ions for its helix-destabilizing activity (Fig. 5B, lanes 3–7), and the presence of 2 mM Zn2+ even blocked RNA helix destabilization (Fig. 5B, lane 7). We further assessed the impacts of Mg2+ and Zn2+ at various concentrations on the helix-destabilizing activity of MBP-3AB. As shown in Fig. 5C, the helix-destabilizing activity of EV71 3AB was insensitive to Mg2+ when its concentration is lower than 5 mM, while high Mg2+ concentration (10 mM) is even moderate inhibitory. These results indicate that Mg2+ is not required for the helix-destabilizing activity of EV71 3AB. Our results also showed that when the concentration of Zn2+ reached 1 mM, the helix-destabilizing activity of MBP-3AB was dramatically inhibited (Fig. 5D, lanes 5–7), while at lower (0.5 Mm) Zn2+ concentrations, the helix-destabilizing activity of MBP-3AB was moderately inhibited (Fig. 5D, lane 4).
Fig. 5.
Biochemical conditions for the RNA helix-destabilizing activity of EV71 3AB. (A) Helix-destabilizing activity was determined at the indicated pH in the absence of divalent metal ions. (B) Standard RNA helix substrate (3′ tailed) (0.1 pmol) was reacted with MBP-3AB in the presence of the indicated divalent metal ions at 2 mM. (C and D) 3′-tailed RNA helix substrate (0.1 pmol) was reacted with MBP-3AB in the presence of indicated concentrations of MgCl2 (C) or ZnCl2 (D). Asterisks (*) indicate the HEX-labeled strand. For panels A–D, boiled and native substrates without MBP-3AB addition were used as controls (lane 1 and lane 2). For (A-D, right panels), the unwinding activity was plotted as the percentage of the released RNA from the total RNA helix substrates (Y-axis) at indicated pH values (A), divalent metal ions (B), Mg2+ concentrations (C), or Zn2+ concentration (D) as X-axis.
EV71 3AB destabilizes structured RNA strands and stimulates RNA strand annealing
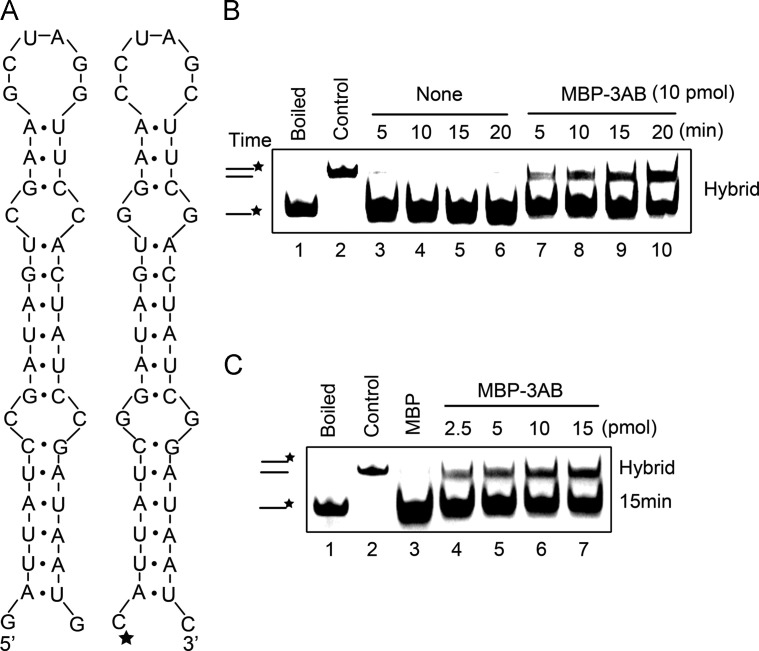
To further characterize and verify the RNA chaperone activity of EV71 3AB, a classic assay for measuring helix destabilization and annealing stimulation of RNA chaperones (Cheng et al., 2013, DeStefano and Titilope, 2006, Gangaramani et al., 2010, Heath et al., 2003, Yang et al., 2014) was adapted for EV71 3AB. As illustrated in Fig. 6A, two 42-nt complementary RNA strands that form defined stem-loop structures were used, one of which was HEX-labeled at its 5′ end (Fig. 6A, right). 0.1 pmol of HEX-labeled strands and non-labeled strands were mixed in the presence and absence of 10 pmol MBP-3AB, followed by gel shift assay to measure the hybridization of the two complementary RNA strands. Our results show that in the absence of 3AB, hybridization was not observed as incubation time increased (Fig. 6B, lanes 3–6), while the presence of 3AB dramatically enhanced the hybridization (Fig. 6B, lanes 7–10). In addition, we observed that the annealing stimulation by EV71 3AB was in a dose-dependent manner, as increasing the dose of 3AB resulted in an increase in hybridization (Fig. 6C), indicating that EV71 3AB is able to destabilize RNA secondary structures and enhance the formation of more stable RNA hybrids.
Fig. 6.
EV71 3AB destabilizes structured RNA strands. (A) Schematic illustrations of the predicted stem-loop structures of the two 42-nt RNA substrates. The two RNA strands are complementary. One strand has HEX-labeled 5′ end as indicated by asterisk (right), while the other strand was not labeled (left). (B) The two complementary strands were mixed (0.1 pmol each), and reacted in the absence (lanes 3–6) or presence (lanes 7–10) of MBP-3AB (10 pmol) for indicated time (5–20 min). (C) The two complementary strands were mixed (0.1 pmol each), and reacted with increasing amounts of MBP-3AB (2.5–15 pmol) for 15 min. Complete reaction mixture with negative control MBP alone (10 pmol) (lane 3). For (B-C), the mix of the two strands was boiled (to prevent spontaneous annealing) (lane 1) or treated with a thermal cycler (lane 2) as negative or positive control, respectively. Samples were subjected to gel electrophoresis (see Materials and methods).
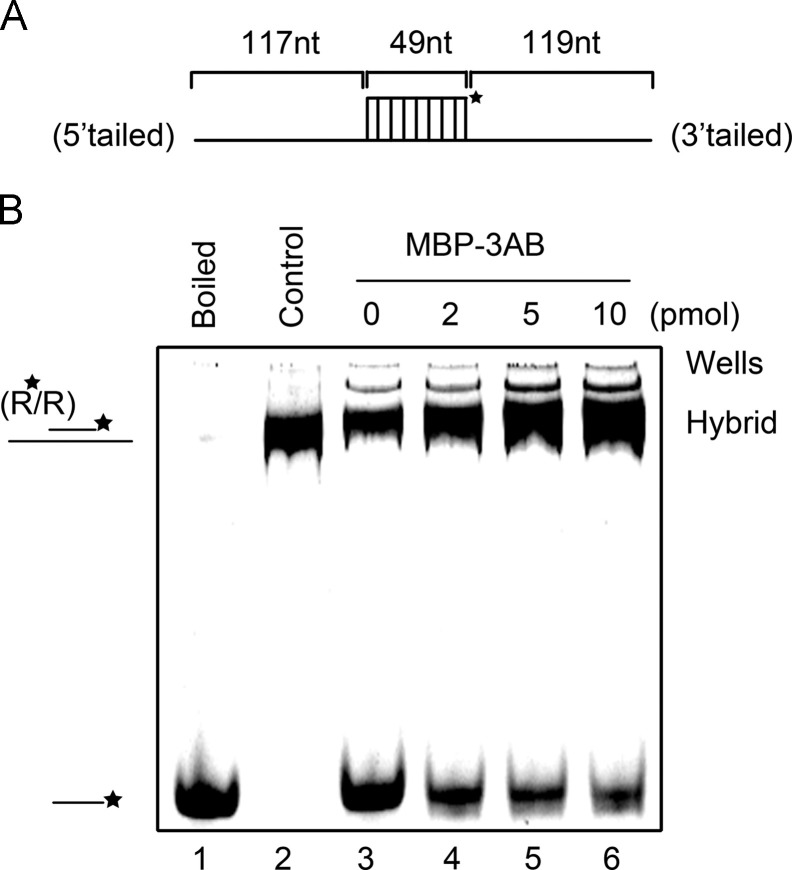
For RNA chaperones, an important characteristic is to stimulate annealing of complementary RNA strands. To determine whether EV71 3AB has this activity, a short HEX-labeled RNA strand (49 nt) was incubated with a long non-labeled RNA strand (285 nt) ( Fig. 7A) in the absence or presence of MBP-3AB. Moreover, RNA chaperone normally functions well when its amount is largely excessive over that of its substrate (Levin et al., 2005). To examine whether this notion also applies to EV71 3AB, the two RNA strands (0.1 pmol each strand) were mixed, and then reacted with increasing amounts of MBP-3AB (0–10 pmol) for 10 min. After that, gel shift assay was performed to measure the annealing of the two partially complementary strands. Our data show that increasing the amount of MBP-3AB resulted in stronger stimulating effects on strand annealing in a dose-dependent manner (Fig. 7B, lanes 4–6). Taken together, we conclude that EV71 3AB possesses an RNA chaperone activity that is able to destabilize RNA helix and stimulate RNA strand annealing.
Fig. 7.
EV71 3AB stimulates the annealing of RNA strands. (A) Schematic illustrations of the 49-nt RNA strand (upper) that is complementary to the 285-nt RNA strand. The 49-nt strand is HEX labeled as indicated by asterisk. (B) The two strands were mixed (0.1 pmol for each strand), and reacted with increasing amounts of MBP-3AB (0–10 pmol) for 10 min. The mix of the two strands was boiled (to prevent spontaneous annealing) (lane 1) or treated with a thermal cycler (lane 2) as negative or positive control, respectively. Samples were subjected to gel electrophoresis (see Materials and methods).
Mutational analysis of the 3B+7 region of 3AB
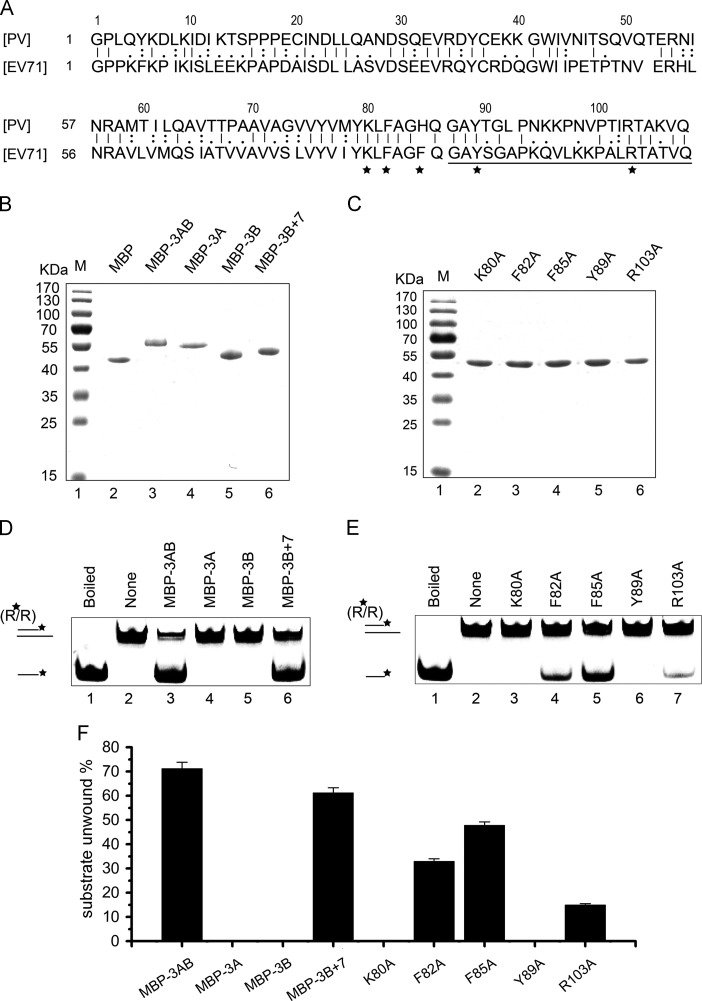
Previous study of poliovirus 3AB has reported that the 3B region plus the C-terminal last 7 a.a. of 3A is the core region for the RNA chaperone activity of 3AB (Gangaramani et al., 2010). To determine which region of EV71 3AB is required for its RNA chaperone activity, we expressed EV71 3A (1–86 a.a.), 3B (87–108 a.a.) and 3B+7 (80–108 a.a., termed 3B+7) ( Fig. 8A) as MBP fusion proteins (MBP-3A, MBP-3B, MBP-3B+7) in a eukaryotic (baculovirus) expression system, and subsequently purified the proteins, followed by SDS-PAGE (Fig. 8B). After that, 10 pmol of each protein was incubated with RNA helix substrates. Our results showed that neither MBP-3A nor MBP-3B was able to destabilize RNA helix (Fig. 8D, lanes 4–5), while MBP-3B+7 effectively destabilized RNA helix. Moreover, the efficiency of MBP-3B+7 to destabilize helix is only slightly less than that of MBP-3AB (Fig. 8D, lane 3 and lane 6).
Fig. 8.
Mutational analysis of the 3B+7 region of 3AB. (A) The amino acid sequence alignment analysis of 3AB proteins between EV71 and poliovirus. Regions of EV71 3AB: 3A, 1–86; 3B (VPg), 87–108(underlined); 3B+7, 82–108. Amino acids with an asterisk (*) were mutagenesis sites. (B and C) Electrophoresis analysis of expressed and purified eukaryotic proteins as illustrated in upper panel. Proteins were subjected to 10% SDS-PAGE, and protein bands were stained with Coomassie blue. (D and E) 3′-tailed RNA helix substrate (0.1 pmol) was reacted with proteins (10 pmol) as illustrated in upper panel, respectively, under standard reaction conditions. (F) The destabilizing activity was plotted as the percentage of the released RNA from the total RNA helix substrates (Y-axis) at indicated proteins as X-axis. Error bars represent standard deviation values from three separate experiments.
Since 3B+7 contains most RNA chaperone activity of 3AB but either 3A or 3B does not, it is reasonable to speculate that the residues within the 3B+7 region and nearby the junction between 3A and 3B are critical for the RNA chaperone activity. Based on the amino acid sequence alignment analysis of 3AB proteins between EV71 and poliovirus, five residues K80, F82, F85, Y89 and R103, whose corresponding residues have been reported to be critical for the RNA chaperone activity of poliovirus 3AB (Gangaramani et al., 2010), were selected; and then, each residue was point mutated to Alanine (A) in 3B+7 (as indicated in Fig. 8A). The 3B+7 mutants were eukaryotically expressed as MBP-fusion proteins and then purified (Fig.8C). After that, helix-destabilizing assays were conducted. As shown in Fig. 8E, the K80A or Y89A mutation abolished the helix-destabilizing activity of 3B+7 (lanes 3 and 6), while R103A, F82A and F85A mutations resulted in different extents of inhibition on the helix-destabilizing activity of 3B+7 (lanes 4, 5 and 7), indicating that these five residues are the critical sites for the RNA chaperone activity of EV71 3AB.
Discussion
In this report, we show that the nonstructural protein 3AB from EV71 has an RNA chaperone activity. For RNA viruses, their vRNA molecules may adopt diverse structures, some of which are misfolded inactive intermediates. The role of RNA chaperones is usually to facilitate the refolding of misfolded RNA intermediates, and assists in the formation of more globally stable and active RNAs. Thus, RNA chaperone activity is generally believed to be required for efficient replication and translation of vRNAs (Ivanyi-Nagy et al., 2005, Mir and Panganiban, 2006a, Zúñiga et al., 2009). So far, the list of virus-encoded RNA chaperones keeps growing and includes retroviruses nucleocapsid (NC) protein, HIV-1 Vif and Tat, coronavirus nucleocapsid (N) protein, hantavirus nucleocapsid (N) protein, flavivirus core protein, hepatitis D virus small delta antigen (SdAg), cypovirus capsid protein VP5, EoV nonstructural protein 2C, and poliovirus 3AB (Baric et al., 1988, Batisse et al., 2012, Cheng et al., 2013, DeStefano and Titilope, 2006, Huang et al., 2003, Ivanyi-Nagy et al., 2008, Kuciak et al., 2008, Mir and Panganiban, 2006b, Rein et al., 1998, Yang et al., 2014). Our current study adds EV71 3AB as a new member of this list.
Previous study has reported that poliovirus 3AB possesses an RNA chaperone activity; moreover, our current study determined the RNA chaperone activity of EV71 3AB. Despite of the numerous commonalities between poliovirus and EV71, the 3AB proteins from these two enteroviruses still exhibited some difference. For example, poliovirus 3AB has been reported to be much more potent than 3B+7 for RNA chaperone activities (Gangaramani et al., 2010); however, we observed that for EV71, the RNA chaperone activity of wild-type 3AB is only moderately stronger than that of 3B+7 (Fig. 8D). Moreover, poliovirus 3AB has been observed to shift single-stranded (ss) RNA in a gel mobility shift assay (DeStefano and Titilope, 2006); however, EV71 3AB failed to cause gel shift on ssRNA (data not shown). Interestingly, EoV 2C, a well-defined RNA chaperone, also failed to shift ssRNA (Cheng et al., 2013). Since either EV71 3AB or EoV 2C displays RNA chaperone activity to RNA duplex or structured RNAs, it is clear that they must have direct interaction with RNA molecules. A reasonable explanation for the difficulty to observe ssRNA shift is that the binding of chaperones to RNA is too weak or transient, thus preventing the interaction to be detected by a gel shift assay.
RNA chaperones are a heterogeneous group of proteins that share no common consensus sequences or motifs, and the mechanism(s) by which RNA chaperones destabilize RNA helix and stimulate strand annealing is still poorly understood. Currently, two models have been proposed to explain the activities of RNA chaperones. One is the “entropy transfer” model in which intrinsic disordered regions of RNA chaperones can transfer their entropy or disorder to misfolded RNAs, thereby facilitating these RNAs to escape thermodynamic barriers (Rajkowitsch et al., 2007, Zuniga et al., 2009). The other model is that some RNA chaperones remodel RNA molecules through transient ionic interaction with negatively charged phosphodiester backbone of RNAs, thereby disrupting the stability of RNAs (Woodson, 2010). For 3AB proteins from both poliovirus and EV71, our disorder prediction using DisProt Predictor VL3, the algorithm used to predict intrinsic disorders for certain viral RNA chaperones (Bracken et al., 2004), indicates that these two enteroviral 3AB proteins do not contain intrinsic disorder (data not shown), similarly with EoV 2C (Cheng et al., 2013), implying that enteroviral 3AB does not use the entropy transfer mechanism to remodel RNAs. On the other hand, the RNA helix-destabilizing activity of EV71 3AB was tested under various conditions, and our results showed that Mg2+ resulted in moderate inhibition of the RNA helix-destabilizing activity of EV71 3AB at high concentrations (≥10 mM) (Fig. 5C). This observation suggests that EV71 3AB is likely to remodel RNA molecules through transient ionic interaction with negatively charged RNA backbones, since divalent metallic ions like Mg2+ may compete with RNA chaperone to stabilize RNAs.
Interestingly, comparing to Mg2+, Zn2+ efficiently inhibited the helix destabilization by EV71 3AB even at low concentrations (≤0.5 mM), while higher concentrations of Zn2+ (≥1 mM) almost abolished the helix destabilization (Fig. 5D). This blocking effect of Zn2+ suggests that Zn2+ inhibits the RNA destabilizing activity of EV71 3AB through a different mechanism to Mg2+. It is plausible that Zn2+ directly binds EV71 3AB and regulates its RNA destabilizing activity by inducing conformational change of the protein (Pfister et al., 2000). Moreover, EV71 3AB exhibited the highest activity at pH 6. EV71 3AB is a basic protein (pI=9.13, tested by Compute pI/Mw tool), and it is proposed that this protein needs a mildly acidic environment as its optimal reaction condition.
During the replication of EV71, 3AB will be proteolytically processed into 3A and 3B (VPg) (Fig. 1A), neither of which has RNA chaperone activity, while the putative C-terminal cytoplasmic domain of 3AB (termed 3B+7) possesses RNA chaperone activity. Five point mutations of the 3B+7 demonstrated weakened or abolished chaperone activity. Particularly, the K80A or Y89A mutation abolished the helix-destabilizing activity of 3B+7, while R103A, F82A and F85A mutations inhibited the helix-destabilizing activity of 3B+7 by different extents (Fig. 8). Furthermore, we also examined the annealing stimulation activity by the five point mutations (data not shown), and got similar results to the RNA helix-destabilizing activity. The results indicated that the five amino acids (Lys-80, Phe-82, Phe-85, Tyr-89, and Arg-103) are critical and probably active sites of 3AB for its RNA chaperone activity. These sites may also serve as the potential target for the development of anti-EV71 compound or therapy.
During the life cycle of EV71, the RNA chaperone activity of 3AB may participate in diverse processes, such as coating and protecting viral genomic RNA, facilitating the formation and/or deformation of vRNA cis-acting elements, vRNA synthesis by RdRp, and translation (Gangaramani et al., 2010). However, due to technical limitations, it is difficult to reveal the exact role of an RNA chaperone in a particular process. Based on the studies of poliovirus and EV71, our current work suggests that the RNA chaperone activity is a general function for nonstructural protein 3AB in the genus Enterovirus, the family Picornaviridae, and even the order Picornavirales, and provide novel knowledge about the replication of EV71.
Materials and methods
Plasimd constructions
Standard procedures were used for extraction of viral genome RNA. A cDNA fragment of EV71 3AB (amino acids 1441 to 1548 of polyprotein ORF) was inserted into vector pFastBac™HTA-MBP. The vector pFastBac™HTA-MBP was originated from the vector pFastBac™HTA (Invitrogen, Carlsbad, CA) (Li et al., 2012), in which the Maltose-Binding Protein (MBP) was N-terminally fused modified by our lab as previously described (Han et al., 2013, Wang et al., 2012). Mutations were generated in the same manner. Point mutations were introduced into the corresponding templates with appropriate primers containing the desired nucleotide changes. The primers used in this study are shown in Table 1. The resulting plasmids were subjected to Bac-to-Bac Baculovirus Expression System to express the fusion proteins with an MBP-tag at the N-terminus of EV71 3AB.
Table 1.
The list of Primers.
| Primers | Sequence (5′-3′) |
|---|---|
| Primers | |
| 3AB-F | GAATTCGGTCCACCTAAGTTCA (EcoR I) |
| 3AB-R | CTCGAGTTACTGTACTGTTGCTGTGCGAA (Xho I) |
| 3A F | GAATTCGGTCCACCTAAGTTCA (EcoR I) |
| 3A R | CTCGAGTTACTGAAACCCTGCAAAGA (Xho I) |
| 3B F | GAATTCGGTGCGTATTCTGGT (EcoR I) |
| 3B R | CTCGAGTTACTGTACTGTTGCTGT (Xho I) |
| 3B+7-F | GAATTCAAGCTCTTTGCAG (EcoR I) |
| 3B+7-R | CTCGAGTTACTGTACTGTTGCTGT (Xho I) |
| K80A-F | GAATTCgcaCTCTTTGCAGGGTT (EcoR I) |
| K80A-R | CTCGAGTTACTGTACTGTTGCTGT (Xho I) |
| F82A-F | GAATTCAAGCTCgcaGCAGGGTTTCA (EcoR I) |
| F82A-R | CTCGAGTTACTGTACTGTTGCTGT (Xho I) |
| F85A-F | GAATTCAAGCTCTTTGCAGGGgcaCAGGGTGCGTAT (EcoR I) |
| F85A-R | CTCGAGTTACTGTACTGTTGCTGT (Xho I) |
| Y89A-F | GAATTCAAGCTCTTTGCAGGGTTTCAGGGTGCGgcaTCTGGTGCTC (EcoR I) |
| Y89A-R | CTCGAGTTACTGTACTGTTGCTGT (Xho I) |
| R103A-F | GAATTCAAGCTCTTTGCAG (EcoR I) |
| R103A-R | CTCGAGTTACTGTACTGTTGCTGTtgcAAGAGCAGGTTTC (Xho I) |
Underlined characters indicate restriction endonuclease sites; types are shown in parentheses. Substituted nucleotides for mutagenesis are shown with lowercase characters.
Expression and purification of recombinant proteins
The expression and purification of MBP alone, MBP-3AB and its derivates were performed as previously described. Sf9 cells at a density of 2–106 cells/ml were infected with the recombinant baculoviruses and the recombinant proteins were harvested at 72 h after infection. Cell pellets were resuspended in a binding buffer [20 mM Tris–HCl (pH 7.4), 200 mM NaCl, 1 mM EDTA, 10 mM 2-Mercaptoethanol] supplemented with protease inhibitors cocktail (Sigma, St. Louis, MO, USA). After being resuspended, cells were lysed via sonication and then debris was removed by centrifugation for 30 min at 11,000 g. The protein in the supernatant was purified using amylose affinity chromatography (New England BioLabs, Ipswich, MA), and then concentrated using Amicon Ultra-15 filters (Millipore, Schwalbach, Germany). The buffer was replaced by 20 mM HEPES-KOH (pH 7.0). All proteins were quantified using the Bradford method and stored at −70 °C in aliquots. Proteins were confirmed by 10% SDS-PAGE as described previously (Wang et al., 2013, Zhou et al., 2013).
Preparation of oligonucleotide duplexes
RNA helices/duplexes consist of two complementary oligonucleotide strands were annealed and gel purified. Of the two strands, the longer one was unlabeled and referred to the template strand, while the smaller strand, referred to the release strand, was labeled at 5′ end hexachlorofluorescein (HEX) (Takara, Dalian) (Warrener and Collett, 1995). All unlabeled DNA strands were purchased from Invitrogen, and HEX-labeled DNA and RNA strands were purchased from Takara. Unlabeled RNA strands were synthesized from in vitro transcription using T7 RNA polymerase (Promega, Madison, WI). The posttranscriptional RNAs were electrophoresed in a 5% polyacrylamide 8 M urea gel and further purified by the use of a Poly-Gel RNA extraction kit (Omega Bio-Tek) according to the manufacturer′s instructions.
To generate oligonucleotide duplexes, the labeled release strands and unlabeled template strands were incubated at a proper ratio in a 10 μl reaction mixture containing 25 mM HEPES-KOH (pH, 7.0) and 50 mM NaCl. The mixture was heated to 95 °C for 5 min and then cooled gradually to room temperature. Extracted oligonucleotides were ethanol precipitated and dissolved in nuclease-free water. The standard RNA helix substrate with both 5′ and 3′ tails was annealed with RNA1 and RNA2, the R⁎/D substrate was annealed with RNA1 and DNA1, the D⁎/D substrate was annealed with DNA2 and DNA3, the 3′-tailed RNA helix substrate was annealed with RNA1 and RNA3, the 5′-tailed RNA helix substrate was annealed with RNA1 and RNA4, and the blunt ended substrate was annealed with RNA1 and RNA5. The R⁎/R substrates with 42 matched base pairs were annealed with RNA6 and RNA7. The R⁎/R substrates with 49 matched base pairs were annealed with RNA8 and RNA9. All DNA and RNA oligonucleotides are listed in Table 2.
Nucleic acid helix-destabilizing assays
The total reaction volume is 10 μl, incubating 10 pmol of recombinant protein to 0.1 pmol of HEX-labeled helix substrates in a buffer containing 50 mM HEPES-KOH (pH 7.0), 2.5 mM MgCl2, 2 mM DTT, 0.01% BSA and 1.5 U/μl RNasin. After incubation at 37 °C for 1 h, the reaction mixture was treated with proteinase K (final concentration at 1 μg/μl) at 37 °C for 15 min. The digestion reaction was terminated by the addition of 5×loading buffer (100 mM Tris–HCl, 20 mM EDTA, 1% SDS, 0.04% Triton X-100, 50% glycerol and bromophenol blue [pH7.0]). Meanwhile, substrates were heat denatured at 95 °C for 5 min as the positive control. Mixtures were electrophoresed on 12% native-PAGE gels. Gels were scanned with a Typhoon 9200 (GE Healthcare, Piscataway, NJ). Image-Quant software (GE Healthcare) was used to quantify the ratio of released single strands versus the total of substrates.
RNA strand hybridization assays
An RNA strand hybridization assay was carried out at a 10 ul reaction volume, containing indicated amount of recombinant protein incubated with 0.1 pmol of HEX-labeled and unlabeled RNA strands in 37 °C in a buffer containing 50 mM HEPES-KOH (pH 7.0), 2.5 mM MgCl2, 2 mM DTT, 0.01% BSA and 1.5 U/μl RNasin. The reaction was terminated and analyzed as described above. The mixtures were electrophoresed using 12% native-PAGE gels. Gels were scanned with a Typhoon 9200 (GE Healthcare, Piscataway, NJ).
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program, 2014CB542603), the National Natural Science Foundation of China Grant nos. 31270190 (to X.Z.), No. 81201292 (to X.Z.) and No. 31270189 (to Y.H.); Fundamental Research Funds for the Central Universities Grant 204202004 (to F.T.) and the Chinese 111 Project Grant no. B06018.
Contributor Information
Yuanyang Hu, Email: yyhu@whu.edu.cn.
Xi Zhou, Email: zhouxi@whu.edu.cn.
References
- Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N., Stohlman S.A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batisse J., Guerrero S., Bernacchi S., Sleiman D., Gabus C., Darlix J.L., Marquet R., Tisne C., Paillart J.C. The role of Vif oligomerization and RNA chaperone activity in HIV-1 replication. Virus Res. 2012;169:361–376. doi: 10.1016/j.virusres.2012.06.018. [DOI] [PubMed] [Google Scholar]
- Belov G.A., Fogg M.H., Ehrenfeld E. Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor. J. Virol. 2005;79:7207–7216. doi: 10.1128/JVI.79.11.7207-7216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C., Iakoucheva L.M., Romero P.R., Dunker A.K. Combining prediction, computation and experiment for the characterization of protein disorder. Curr. Opin. Struct. Biol. 2004;14:570–576. doi: 10.1016/j.sbi.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Brown B.A., Pallansch M.A. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995;39:195–205. doi: 10.1016/0168-1702(95)00087-9. [DOI] [PubMed] [Google Scholar]
- Chang L.Y., Lin T.Y., Hsu K.H., Huang Y.C., Lin K.L., Hsueh C., Shih S.R., Ning H.C., Hwang M.S., Wang H.S., Lee C.Y. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet. 1999;354:1682–1686. doi: 10.1016/S0140-6736(99)04434-7. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Yang J., Xia H., Qiu Y., Wang Z., Han Y., Xia X., Qin C.F., Hu Y., Zhou X. The nonstructural protein 2C of a Picorna-like virus displays nucleic acid helix destabilizing activity that can be functionally separated from its ATPase activity. J. Virol. 2013;87:5205–5218. doi: 10.1128/JVI.00245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong P., Hsieh S.Y., Liu C.C., Chou A.H., Chang J.Y., Wu S.C., Liu S.J., Chow Y.H., Su I.J., Klein M. Production of EV71 vaccine candidates. Human Vaccin. Immunother. 2012;8:1775–1783. doi: 10.4161/hv.21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano J.J., Titilope O. Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities. J. Virol. 2006;80:1662–1671. doi: 10.1128/JVI.80.4.1662-1671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich F. Molecular evolution of oral poliovirus vaccine strains during multiplication in humans and possible implications for global eradication of poliovirus. Acta Virol. 2000;44:109–117. [PubMed] [Google Scholar]
- Gangaramani D.R., Eden E.L., Shah M., Destefano J.J. The twenty-nine amino acid C-terminal cytoplasmic domain of poliovirus 3AB is critical for nucleic acid chaperone activity. RNA Biol. 2010;7:820–829. doi: 10.4161/rna.7.6.13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A., Tagaya I., Yoneyama T. Epidemic of hand, foot and mouth disease associated with enterovirus 71 infection. Intervirology. 1978;9:60–63. doi: 10.1159/000148922. [DOI] [PubMed] [Google Scholar]
- Han Y., Wang Q., Qiu Y., Wu W., He H., Zhang J., Hu Y., Zhou X. Periplaneta fuliginosa densovirus nonstructural protein NS1 contains an endonuclease activity that is regulated by its phosphorylation. Virology. 2013;437:1–11. doi: 10.1016/j.virol.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Heath M.J., Derebail S.S., Gorelick R.J., DeStefano J.J. Differing roles of the N- and C-terminal zinc fingers in human immunodeficiency virus nucleocapsid protein-enhanced nucleic acid annealing. J.Biol. Chem. 2003;278:30755–30763. doi: 10.1074/jbc.M303819200. [DOI] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Hey T.D., Richards O.C., Ehrenfeld E. Host factor-induced template modification during synthesis of poliovirus RNA in vitro. J. Virol. 1987;61:802–811. doi: 10.1128/jvi.61.3.802-811.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.C., Hsu Y.W., Wang H.C., Huang S.W., Kiang D., Tsai H.P., Wang S.M., Liu C.C., Lin K.H., Su I.J., Wang J.R. Appearance of intratypic recombination of enterovirus 71 in Taiwan from 2002 to 2005. Virus Res. 2008;131:250–259. doi: 10.1016/j.virusres.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Huang Z.S., Su W.H., Wang J.L., Wu H.N. Selective strand annealing and selective strand exchange promoted by the N-terminal domain of hepatitis delta antigen. J. Biol. Chem. 2003;278:5685–5693. doi: 10.1074/jbc.M207938200. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y., Nakano S., Yamaoka K., Takami S. Outbreaks of hand, foot, and mouth disease by enterovirus 71. High incidence of complication disorders of central nervous system. Arch Dis. Child. 1980;55:583–588. doi: 10.1136/adc.55.8.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Davidovic L., Khandjian E.W., Darlix J.L. Disordered RNA chaperone proteins: from functions to disease. Cell. Mol. Life Sci.: CMLS. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Lavergne J.P., Gabus C., Ficheux D., Darlix J.L. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucl. Acids Res. 2008;36:712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuciak M., Gabus C., Ivanyi-Nagy R., Semrad K., Storchak R., Chaloin O., Muller S., Mely Y., Darlix J.L. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucl. Acids Res. 2008;36:3389–3400. doi: 10.1093/nar/gkn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo R.L., Shih S.R. Strategies to develop antivirals against enterovirus 71. Virol. J. 2013;10:28. doi: 10.1186/1743-422X-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucl. Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Li S.F., Wang H.L., Hu Z.H., Deng F. Genetic modification of baculovirus expression vectors. Virol. Sin. 2012;27:71–82. doi: 10.1007/s12250-012-3236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. RNA (New York, N.Y.) 2006;12:272–282. doi: 10.1261/rna.2101906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. Characterization of the RNA chaperone activity of hantavirus nucleocapsid protein. J. Virol. 2006;80:6276–6285. doi: 10.1128/JVI.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musier-Forsyth K. RNA remodeling by chaperones and helicases. RNA Biol. 2010;7:632–633. doi: 10.4161/rna.7.6.14467. [DOI] [PubMed] [Google Scholar]
- Nathanson N., Langmuir A.D. The cutter incident poliomyelitis following formaldehyde-inactivated poliovirus vaccination in the united states during the spring of 1955: ii Relationship of poliomyelitis to cutter vaccine. Am. J. Epidemiol. 1963;78:29–60. doi: 10.1093/oxfordjournals.aje.a120328. [DOI] [PubMed] [Google Scholar]
- Ooi M.H., Wong S.C., Lewthwaite P., Cardosa M.J., Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- Pallai P.V., Burkhardt F., Skoog M., Schreiner K., Bax P., Cohen K.A., Hansen G., Palladino D.E., Harris K.S., Nicklin M.J. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J Biol. Chem. 1989;264:9738–9741. [PubMed] [Google Scholar]
- Pathak H.B., Arnold J.J., Wiegand P.N., Hargittai M.R., Cameron C.E. Picornavirus genome replication: assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex. J. Biol. Chem. 2007;282:16202–16213. doi: 10.1074/jbc.M610608200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister T., Jones K.W., Wimmer E. A cysteine-rich motif in poliovirus protein 2C(ATPase) is involved in RNA replication and binds zinc in vitro. J. Virol. 2000;74:334–343. doi: 10.1128/jvi.74.1.334-343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch S.J., Palant O. Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J. Virol. 1995;69:7169–7179. doi: 10.1128/jvi.69.11.7169-7179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L., Chen D., Stampfl S., Semrad K., Waldsich C., Mayer O., Jantsch M.F., Konrat R., Blasi U., Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- Rajkowitsch L., Schroeder R. Dissecting RNA chaperone activity. RNA (New York, N.Y.) 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A., Henderson L.E., Levin J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- Sadeghipour S., Bek E.J., McMinn P.C. Selection and characterisation of guanidine-resistant mutants of human enterovirus 71. Virus Res. 2012;169:72–79. doi: 10.1016/j.virusres.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Sweeney T.R., Cisnetto V., Bose D., Bailey M., Wilson J.R., Zhang X., Belsham G.J., Curry S. Foot-and-mouth disease virus 2C is a hexameric AAA+ protein with a coordinated ATP hydrolysis mechanism. J. Biol. Chem. 2010;285:24347–24359. doi: 10.1074/jbc.M110.129940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Lai J.K., Sam I.C., Chan Y.F. Recent developments in antiviral agents against enterovirus 71 infection. J. Biomed. Sci. 2014;21:14. doi: 10.1186/1423-0127-21-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Han Y., Qiu Y., Zhang S., Tang F., Wang Y., Zhang J., Hu Y., Zhou X. Identification and characterization of RNA duplex unwinding and ATPase activities of an alphatetravirus superfamily 1 helicase. Virology. 2012;433:440–448. doi: 10.1016/j.virol.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Qiu Y., Liu Y., Qi N., Si J., Xia X., Wu D., Hu Y., Zhou X. Characterization of a nodavirus replicase revealed a de novo initiation mechanism of RNA synthesis and terminal nucleotidyltransferase activity. J. Biol. Chem. 2013;288:30785–30801. doi: 10.1074/jbc.M113.492728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrener P., Collett M.S. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J. Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson S.A. Taming free energy landscapes with RNA chaperones. RNA Biol. 2010;7:677–686. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Zhang J., Cai D., Li D., Chen W., Jiang H., Hu Y. Biochemical characterization of Periplaneta fuliginosa densovirus non-structural protein NS1. Biochem. Biophys. Res. Commun. 2006;342:1188–1196. doi: 10.1016/j.bbrc.2006.02.053. [DOI] [PubMed] [Google Scholar]
- Yang F., Ren L., Xiong Z., Li J., Xiao Y., Zhao R., He Y., Bu G., Zhou S., Wang J., Qi J. Enterovirus 71 outbreak in the People′s Republic of China in 2008. J. Clin. Microbiol. 2009;47:2351–2352. doi: 10.1128/JCM.00563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Cheng Z., Zhang S., Xiong W., Xia H., Qiu Y., Wang Z., Wu F., Qin C.F., Yin L., Hu Y., Zhou X. A cypovirus VP5 displays the RNA chaperone-like activity that destabilizes RNA helices and accelerates stranding annealing. Nucl. Acids Res. 2014;42:2538–2554. doi: 10.1093/nar/gkt1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Sola I., Cruz J.L.G., Enjuanes L. Role of RNA chaperones in virus replication. Virus Res. 2009;139:253–266. doi: 10.1016/j.virusres.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu Z., Yang W., Ren J., Tan X., Wang Y., Mao N., Xu S., Zhu S., Cui A., Zhang Y., Yan D., Li Q., Dong X., Zhang J., Zhao Y., Wan J., Feng Z., Sun J., Wang S., Li D., Xu W. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang city of China. Virol. J. 2010;7:94. doi: 10.1186/1743-422X-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Han Z., Wang L.F., Shi Z. Identification of immunogenic determinants of the spike protein of SARS-like coronavirus. Virol. Sin. 2013;28:92–96. doi: 10.1007/s12250-013-3292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Sola I., Cruz J.L., Enjuanes L. Role of RNA chaperones in virus replication. Virus Res. 2009;139:253–266. doi: 10.1016/j.virusres.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]