Abstract
Fecal indicator bacteria, such as Escherichia coli (E.coli) and Enterococcus, have been widely used to indicate the presence of pathogens. However, the suitability of fecal indicator bacteria to represent health risks is still being challenged, particularly in tropical aquatic environments. The objective of this study is to understand the occurrence and prevalence of indicators and pathogens in areas with contrasting land use, as well as to identify the major correlations between indicators, pathogens and environmental parameters. The spatial and temporal variation of indicators and pathogens was studied to examine the distribution patterns for areas with different land use, and the impact of seasonal changes on microbial populations. A total of 234 water samples were sampled for two years from reservoirs and their tributaries, and tested for fecal indicator bacteria, coliphages, human specific markers, pathogenic bacteria and viruses. The prevalence of indicators and pathogens in reservoirs were generally low, while relatively high concentrations were observed in tributaries to varying degrees. Of the enteric viruses, norovirus GII was among the most prevalent and had the highest concentration. Although strong correlations were found between indicators, only relatively weak correlations were found between indicators and pathogens. The results in this study showed that none of the bacteria/phage indicators were universal predictors for pathogens. Inclusion of the alternative indicators, Methanobrevibacter smithii, Bacteroides and human polyomaviruses (HPyVs) to monitoring programs could help to determine whether the fecal source was human. The microbial distribution patterns allow the classification of sampling sites to different clusters and thus, help to identify sites which have poor water quality. This approach will be useful for water quality management to pinpoint factors that influence water quality and help to prioritize sites for restoration of water quality.
Keywords: Bacteria indicators, Pathogens, Enteric viruses, Coliphages, Occurrence, Correlations
Graphical abstract
Highlights
-
•
Norovirus GII was the most prevalent enteric viruses.
-
•
Land use classification was correlated to the distribution of pathogens/indicators.
-
•
Alternative indicators helped to improve the prediction of enteric viruses.
-
•
Cluster analysis identified sites with poor water quality.
1. Introduction
Increasing urbanization, industrialization and agricultural activity have greatly impaired surface water quality globally. Waterborne pathogens include viruses and bacteria which can be introduced into the urban water cycle via leaking sewers, urban runoff, agricultural runoff and wastewater discharges (Arnone and Walling, 2007). Enteric viruses are widely recognized as the commonest and most hazardous waterborne pathogens, causing both sporadic and outbreak-related illnesses (La Rosa et al., 2012). Among enteric viruses, noroviruses, astroviruses, enteroviruses, adenoviruses and hepatitis A virus are the most extensively studied (Aw et al., 2009; Ganesh and Lin, 2013). Of these, adenovirus, caliciviruses (includes norovirus), enterovirus and hepatitis A virus are listed in the USEPA CCL3, which are currently not subjected to any proposed or promulgated national primary drinking water regulations, and which may require regulation under the Safe Drinking Water Act (SDWA) (USEPA, 2009). Little is known about the epidemiology of enteric viruses as well as their occurrence and behavior in tropical aquatic environments, since most studies have been conducted in temperate and cold regions.
In addition to enteric viruses, other potential emerging viral pathogens (e.g. hepatitis E virus, sapovirus, nipah virus, coronavirus, bird flu virus) may originate from wild animals in tropical catchment forests. Hepatitis E virus (HEV), a causative agent of human hepatitis E, is a single-stranded positive-sense RNA virus from the genus Hepevirus (Nan et al., 2017). HEV is transmitted primarily in humans by the fecal-oral route through contaminated drinking water or food. Outbreaks normally occur in developing or undeveloped countries (Ahmad et al., 2010; Howard et al., 2010; Khan et al., 2011). However, recent evidence has shown that various animal species have serum antibodies to HEV, suggesting hepatitis E is a zoonotic disease (Meng, 2010; Nan et al., 2017). Li et al. (2005) reported that Genotype 3 HEV RNA was detected in both patient serum and wild boar meat (Li et al., 2005). This raises concern that HEV may be discharged into tributaries through animals which exist in forested areas. Sapovirus is another emerging enteric virus. It belongs to Caliciviruses, and causes acute viral gastroenteritis (Haramoto et al., 2008; Sano et al., 2011). As it has been identified in bats, dogs, pigs and mink (Oka et al., 2015), it may also exist in tropical catchment waters. Other potential zoonotic viruses include coronavirus (such as SARS) (Casanova et al., 2009; de Wit et al., 2016) and bird flu virus (influenza A virus subtypes H5N1 and H7N9) (Peiris et al., 2007; Shi et al., 2013; Wang et al., 2013).
Apart from viruses, bacteria that cause illness from recreational waters include Salmonella, Pseudomonas aeruginosa, Shigella, Campylobacter and Enteropathogenic Escherichia coli (Hlavsa et al., 2015; Sanborn and Takaro, 2013). P. aeruginosa is a ubiquitous opportunistic pathogen and occurs naturally in aquatic environments. However, the infections caused by P. aeruginosa in recreational waters should not be neglected, particularly for young children, elder people and immune-compromised population. Moreover, P. aeruginosa is currently one of the major bacteria that is capable of multiple-drug resistance (Aloush et al., 2006; Hirsch and Tam, 2010). Another bacterium that can cause recreational water-related illness is Salmonella. Although Salmonellae are usually known as foodborne pathogens, water could be a potential mode of transmission. Salmonella can be found in the feces of a large variety of vertebrates, including humans, reptiles, birds, farm and wild animals (Matias et al., 2016; Silva et al., 2014).
To date, water quality monitoring is still based on the detection of fecal indicator bacteria (i.e. E. coli and Enterococcus). Ideally, fecal indicator bacteria should originate from the same host as the pathogen and have similar survival pattern as the pathogen outside the host. However, substantial evidence shows the limitations of using only these indicators to represent the water quality from a fecal point of view (Byappanahalli et al., 2006; Field and Samadpour, 2007; Wu et al., 2011). Studies have shown the persistence of fecal indicator bacteria in tropical regions (Byappanahalli and Fujioka, 1998; Carrillo et al., 1985). This could be due to the higher humidity, warmer conditions, heavier rainfall with highly erosive rains that encourage the splash effect and transportation of microorganisms in tropical regions (Rochelle-Newall et al., 2015). Although there is less seasonal variation in tropical regions as compared to temperate regions, the monsoon season (wet season) and inter-monsoon season (dry season) showed significant difference in the occurrence of microorganisms (Chuah and Ziegler, 2018; Nguyen et al., 2016; Rochelle-Newall et al., 2016). Consequently, alternative indicators such as coliphages and human specific markers have been suggested to address the limitation of fecal indicator bacteria (Field and Samadpour, 2007; Harwood et al. 2013, 2014). The relationship between the presence of pathogens including viruses and alternative indicators has been studied. However, disparate results have been obtained. Harwood et al. (2014) and Jofre et al. (2016) conducted a comprehensive review of the correlations between alternative indicators and coliphages with epidemiological studies and pathogens. Wu et al. (2011) suggested several factors that could contribute to the varied results in different studies. Sample size and the number of positive samples for pathogens are among the main factors that determine the correlations between indicators and pathogens.
In recent years, it was reported that wild boar numbers in Singapore's forests are rapidly increasing (National Parks Board of Singapore, 2013), possibly resulting in potential contamination of HEV in tributaries although this has yet to be established. The close proximity of forests and wetlands, and urban populations in Singapore could lead to exposure of zoonotic pathogens through discharge into tributaries and reservoirs, potentially resulting in health risks to humans. To date, the emerging viral pathogens have not received sufficient attention, and there have been no studies on these viruses in Singapore's surface waters.
In this study, surveillance of indicators and pathogens was carried out to determine the prevalence of pathogens in catchments with different land use, providing a baseline of the surface water quality in tropical urban environments with a range of land use. As there are contrasting results in previous correlation studies between indicators (including alternative indicators) and pathogens, this study aims to identify the major correlations and examine if these indicators are associated with the occurrence of bacterial and viral pathogens for the tropical catchments studied. The spatial and temporal distributions of indicators and pathogens were also studied in order to identify the diversity patterns of these microorganisms in reservoirs and tributaries with different land use, as well as the impact of seasonal changes on microbial populations.
2. Material and methods
2.1. Sampling sites and land use studies
This study was conducted in Singapore (coordinates of Singapore “1.352083, 103.8198395”) with a land area of 719.1 km2 and population of 5.61 million people as of 2016 (Department of Statistics Singapore, 2016). Sampling sites and land use categories were mapped with ArcGIS version 10.3.1. Software (ESRI, Redlands, CA, USA). Land use shape files were obtained from the Singapore Land Authority (SLA) and PUB (Singapore's National Water Agency) where the land use subcategories were defined by the Urban Redevelopment Authority (URA). However, to reduce the number of parameters, subcategories were merged into 5 main categories: (i) Residential, (ii) Urban, (iii) Green, (iv) Agricultural and (v) Water body. The characteristics for each sampling location are described in Table 1 . In order to reduce the number of parameters, the weightage of land use (which is the combination of different land use categories) was calculated. For the calculation of weightage, numbers 4, 3, 2 and 1 were assigned to Residential, Urban, Agriculture and Green, respectively. A final weightage number was calculated according to the contribution of each land use category to each sampling site.
Table 1.
Study sites and catchment characteristics.
| Sampling area | A | B | C | D | E | F | G |
|---|---|---|---|---|---|---|---|
| Catchment size (m2) | 51,306,895 | 68,852,025 | 120,301,291 | 45,713,961 | 20,808,361 | 26,980,810 | 67,777,648 |
| Land use (%) | |||||||
|
31.28 | 16.02 | 35.00 | 17.31 | 34.95 | 34.04 | 0.67 |
|
38.95 | 19.49 | 37.20 | 27.25 | 36.69 | 43.85 | 1.55 |
|
24.68 | 48.99 | 20.90 | 47.09 | 20.39 | 18.62 | 79.20 |
|
0 | 7.00 | 0 | 0 | 0 | 0 | 0 |
|
5.08 |
8.50 |
6.90 |
8.35 |
8.01 |
3.49 |
18.58 |
|
Sample name | |||||||
| Sample collected from: | |||||||
|
A_reservoir | B_reservoir | C_reservoir | D_reservoir1 | E_reservoir1 | F_reservoir | G_reservoir |
| D_reservoir2 | E_reservoir2 | ||||||
| D_reservoir3 | |||||||
|
A_tributary1 | B_tributary1 | C_tributary1 | ||||
| A_tributary2 | B_tributary2 | C_tributary2 | |||||
| A_tributary3 | B_tributary3 | C_tributary3 | |||||
Note: Sampling area A is located in a high density residential area with several construction activities nearby; sampling area B is situated in an agricultural area with fish farms and farming activities; sampling area C receives runoff from highly urbanized commercial areas (designated as business district); sampling area D is surrounded by green areas and is near to wild life parks; sampling area E is located in a residential area, although there is no direct drain channel to the water bodies; sampling area F is located near an industrial park; and sampling area G is within a protected parkland area and is designated for secondary contact recreational water activities.
2.2. Water sampling and processing
Water samples (234 samples) were collected throughout the two-year study (November 2014 to February 2017) from 7 sites (10 reservoirs and 9 tributaries) in Singapore located in catchments with different land use (Table 1). Water samples from each location were collected approximately 10 cm below the water surface by grab sampling. Water samples were kept in a 5-L sterile carboy and 3 × 10-L sterile carboys, shipped back to the laboratory within 2 h for downstream sample processing and analyzing.
Physical-chemical parameters including temperature, pH, conductivity, salinity, total dissolved solids, dissolved oxygen and turbidity were measured on site with a portable probe (HI9828 Multiparameter Meter; Hanna Instruments). Rainfall data was obtained from the Meteorological Service Singapore (http://www.weather.gov.sg/climate-historical-daily/). Antecedent dry period was calculated from the number of dry days before sampling time.
2.3. Microbiological analyses
2.3.1. Culture-based detection
Colilert™, Enterolert™ and Pseudalert™ (IDEXX Laboratories, Inc., Westbrook, Maine) were used to quantify the cell numbers of E. coli, Enterococcus and P. aeruginosa respectively according to manufacturer's instructions. Male-specific and somatic coliphages were quantified using single agar layer (SAL) plaque assay method according to the standard protocols of US EPA Method 1602 (USEPA, 2001). (Detailed protocol in supplementary information).
2.3.2. Primary concentration
30-L of water sample was concentrated through hollow fiber ultrafiltration cartridge (Hemoflow Fresenius, HF 80S) to a final volume of 600 mL. The ultrafiltration cartridge was purged with nanopure water and pre-treated with blocking solution prior to the concentration step. The microbes were finally recovered through an elution step. (Detailed protocol in supplementary information).
2.3.3. Molecular-based detection
For human specific markers and bacterial pathogens detection, 10-mL of primary concentrated sample (equivalent to 500 mL of raw water sample) was centrifuged at 10,000 g for 20 min. The pellet was subjected to DNA extraction with PowerSoil® DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA). The primers, probes and thermal cycles used for each target are listed in Table S1.
For virus detection, 200-mL of primary concentrated sample (equivalent to 10 L of raw water sample) was used for the secondary concentration. The pH of primary concentrated sample was adjusted to 7.2 and polyethylene glycol (PEG) precipitation was then applied for the secondary concentration. The virus particles were separated from the secondary concentrated sample using chloroform and subsequently, filtered through a 0.22 μm syringe filter and ultra centrifugal filter (30 kDa, Amicon®, Merck, Germany). Viral DNA and RNA were co-extracted with QIAamp Viral RNA Mini Kit (Qiagen). For viral RNA, complementary DNA (cDNA) was synthesized using ImProm-II™ Reverse Transcription System (Promega, USA). The quantitative polymerase chain reaction (qPCR) assay was performed in a 20-μL reaction mixture containing 2.5 μL of DNA (for adenovirus) or cDNA (the rest of the target viruses), 1× of mastermix (as per Table S1), primers and probe (final concentration as per Table S1). The thermal conditions for each target are summarized in Table S1. (Detailed protocol in supplementary information).
2.4. Statistical analyses
Preliminary analysis to examine the distribution of data was carried out before the selection of statistical analysis method. The non-parametric statistical method, Kendall Tau-b was selected for the correlation analysis. The Kruskal-Wallis test was used to examine if there was a significant difference in microorganism population and environmental parameters for different sites and sampling duration. Cluster analysis was used to classify the sampling sites according to the microbial population. The dissimilarity of the microbial community between samples was analyzed with Principal coordinates analysis (PCO) plots. Cluster analysis and PCO plot were carried out with PRIMER software (version 7, PRIMER-E Ltd), while the rest of the statistical analyses were done with SPSS software (version 22.0, SPSS Inc.).
3. Results
3.1. Environmental parameters
The mean, standard deviation and minimum readings of environmental parameters are summarized in Table 2 . The water temperature and pH in local surface waters were relatively constant throughout the two-year study with a standard deviation of 1.36 °C and 0.97, respectively. A wider range of readings was found in other environmental parameters, such as salinity, conductivity, dissolved oxygen (DO) and turbidity (Table 2). The environmental parameters for each sampling area are presented in Fig. S2.
Table 2.
Summary of the microorganism concentrations and environmental parameters in water samples collected in this study.
| Variable | NumObs | % NDs | Meana | Std. deviation | Min | Kaplan-Meier Method |
||
|---|---|---|---|---|---|---|---|---|
| 90%ile | 95%ile | |||||||
| Fecal indicator bacteria |
E. coli (MPN/100 mL) | 231 | 7.3 | 111.7 | 31.7 | 0.5 | 9650 | 24707 |
|
Enterococcus (MPN/100 mL) |
234 |
8.6 |
40.7 |
22.4 |
0.5 |
3177 |
7760 |
|
| Coliphages |
Somatic coliphage (PFU/100 mL) | 227 | 23.2 | 14.8 | 16.2 | 0.5 | 500 | 1067 |
| F+ coliphage (PFU/100 mL) | 226 | 35.5 | 5.6 | 10.3 | 0.5 | 145.5 | 273.2 | |
| Total coliphage (PFU/100 mL) |
227 |
21.8 |
16.2 |
17.3 |
0.5 |
535.3 |
1781 |
|
| FRNA Genotypes |
FRNA G1 (GC/L) | 227 | 82.3 | 90.8 | 2.1 | 70 | 173.3 | 387.3 |
| FRNA G2 (GC/L) | 227 | 47.7 | 457.0 | 11.7 | 70 | 17411 | 34548 | |
| FRNA G3 (GC/L) | 227 | 95.0 | 81.6 | 2.2 | 70 | 70 | 72.98 | |
| FRNA G4 (GC/L) |
227 |
98.2 |
73.2 |
1.5 |
70 |
70 |
70 |
|
| Human specific markers |
B. theta (GC/100 mL) | 226 | 65.9 | 66.3 | 5.0 | 30 | 754.9 | 2346 |
| M. smithii (GC/100 mL) | 227 | 96.4 | 32.3 | 1.6 | 30 | 30 | 30 | |
| HPyVs (GC/100 mL) |
227 |
90.5 |
16.0 |
2.7 |
12 |
12 |
214.2 |
|
| Bacterial pathogens |
Salmonella (GC/100 mL) | 226 | 81.8 | 16.5 | 2.2 | 12 | 56.8 | 86.05 |
| P.aeruginosa (GC/100 mL) | 226 | 33.6 | 126.2 | 10.4 | 12 | 4205 | 8462 | |
| Pseudalert (MPN/100 mL) |
231 |
8.2 |
96.8 |
26.0 |
0.5 |
6268 |
18378 |
|
| Enteric viruses |
Adenovirus (GC/L) | 227 | 87.3 | 29.0 | 3.1 | 20 | 82.45 | 470.8 |
| Norovirus GI (GC/L) | 227 | 60.0 | 173.5 | 3.5 | 80 | 905.6 | 2628 | |
| Norovirus GII (GC/L) | 227 | 48.2 | 921.7 | 13.1 | 80 | 25213 | 36264 | |
| Rotavirus (GC/L) | 227 | 98.2 | 83.4 | 1.4 | 80 | 80 | 80 | |
| Astrovirus (GC/L) | 227 | 35.5 | 384.6 | 4.3 | 80 | 3105 | 4367 | |
| Enterovirus (GC/L) | 227 | 84.6 | 98.8 | 2.0 | 80 | 148.3 | 296.6 | |
| Hepatitis A virus (GC/L) | 227 | 60.9 | 33.4 | 2.2 | 20 | 96.7 | 141.2 | |
| Hepatitis E virus (GC/L) | 227 | 86.4 | 104.0 | 2.3 | 80 | 185.7 | 644.8 | |
| Aichi virus (GC/L) | 227 | 75.9 | 142.3 | 3.8 | 80 | 1141 | 4123 | |
| Sapovirus (GC/L) | 227 | 95.9 | 88.5 | 2.0 | 80 | 80 | 80 | |
| Influenza A virus (GC/L) |
227 |
34.6 |
352.4 |
3.8 |
80 |
1910 |
2512 |
|
| Plant virus |
PMMoV (GC/L) |
227 |
29.6 |
2027.1 |
19.5 |
80 |
125837 |
336514 |
| Environmental parameters | Temperature (°C) | 177 | – | 29.17 | 1.36 | 23.87 | ||
| pH | 230 | – | 7.56 | 0.97 | 3.85 | |||
| EC (μS/cm) | 221 | – | 295.07 | 155.12 | 3 | |||
| Salinity (psu) | 223 | – | 0.14 | 0.08 | 0 | |||
| DO (ppm) | 209 | – | 6.04 | 2.00 | 1.04 | |||
| Turbidity (FNU) | 220 | – | 16.24 | 27.42 | 0 | |||
| Daily rainfall (mm) | 224 | – | 7.53 | 13.58 | 0 | |||
| Antecedent dry period (d) | 224 | – | 1.66 | 2.84 | 0 | |||
Abbreviation: NumObs – Total number of observed samples; % NDs – Percentage of nondetects; Min – Minimum value; MPN – Most probable number; GC – Gene copy; PFU – Plaque forming unit; EC – Electrical conductivity; DO – Dissolved oxygen.
Geometric mean was calculated for microorganisms while arithmetic mean was calculated for environmental parameters.
3.2. Occurrence and abundance of microbial indicators and pathogens in surface waters
29 targets were tested, including fecal indicators, coliphages, human specific markers, bacterial pathogens, plant virus and enteric viruses (Table 2). Prior to the quantification of these microorganisms, a recovery test for pre-concentration was conducted using the bacteriophage MS2. The recovery of pre-concentration achieved in this study was 70–80% (Saeidi et al., 2017). All the samples were subjected to an inhibitory test for qPCR. Samples with inhibition level greater than 3 Ct units were diluted in order to achieve an inhibition level less than 3 Ct units.
The total number of observed samples (NumObs) and the percentage of nondetects (% NDs) are reported in Table 2. Fecal indicators, E. coli and Enterococcus, were detected in most of the water samples, with the percentage of nondetects of 7.3% and 8.6%, respectively. The percentage of nondetects in coliphages was between 21.8% and 35.5%. The genotypes of FRNA showed that FRNA G2 was more prevalent (52.3%) in environmental waters compared to other genotypes such as FRNA G1 (17.1%), G3 (5%) and G4 (1.8%). Bacteroides thetaiotaomicron (B. theta) was detected most frequently (34.1%) among the human specific markers, followed by HPyVs (9.5%) and M. smithii (3.6%). The opportunistic bacterial pathogen, P. aeruginosa, was detected more frequently using the IDEXX kit with Pseudalert™ (91.8%) compared to the qPCR method (66.4%). Nevertheless, the Wilcoxon signed-rank test showed that there was no statistically significant difference between the two methods (p = 0.536). Norovirus GII, astrovirus and influenza A virus were the three most prevalent enteric viruses detected in Singapore's surface waters (Table 2).
The cell concentration for each microorganism was reported as the geometric mean and respective standard deviation by substituting the nondetects with half the lowest detection limit. In addition, the cell concentration was also reported in higher percentile (i.e. 90%ile and 95%ile) with the Kaplan-Meirer method, as recommended by Helsel (2005) for data with more than 80% nondetects (Helsel, 2005). E. coli and Enterococcus were frequently detected in water samples collected with a geometric mean of 111.7 MPN/100 mL and 40.7 MPN/100 mL, respectively. In general, the coliphage counts were lower than the fecal indicator bacteria. FRNA G2 had higher counts (geometric mean = 457 GC/L) among the other genotypes. Pepper mild mottle virus (PMMoV) was generally more prevalent (70.4%) and at much higher concentration (geometric mean = 2027.1 GC/L) than the other human specific markers. Of the enteric viruses tested in this study, norovirus GII had the highest geometric mean (921.7 GC/L), followed by astrovirus (384.6 GC/L), influenza A virus (352.4 GC/L), norovirus GI (173.5 GC/L) and Aichi virus (142.3 GC/L).
3.3. Spatial variation of microbial indicators and pathogens
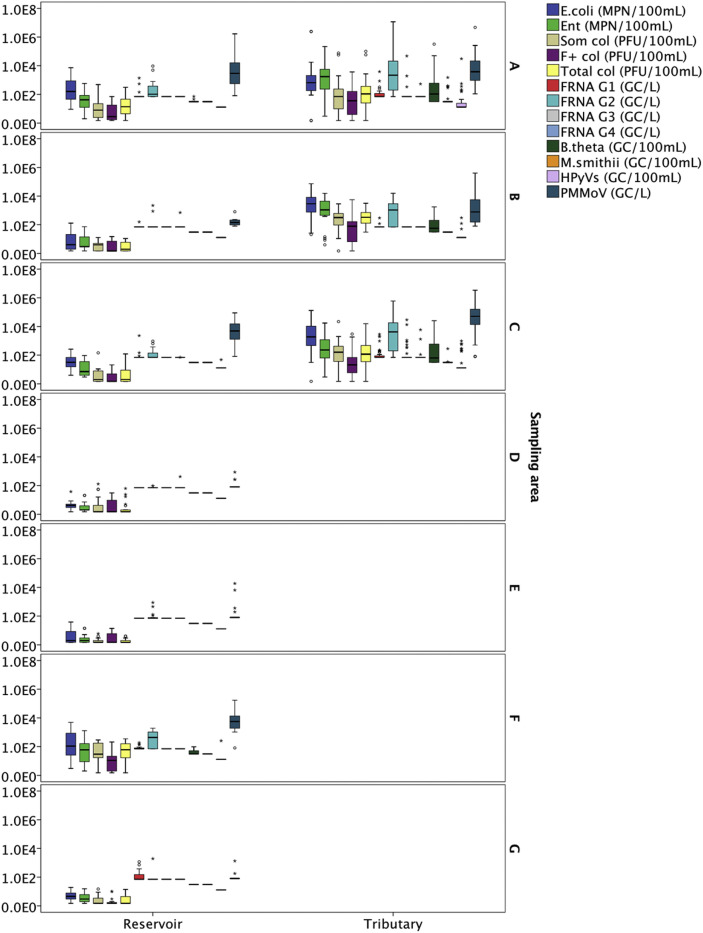
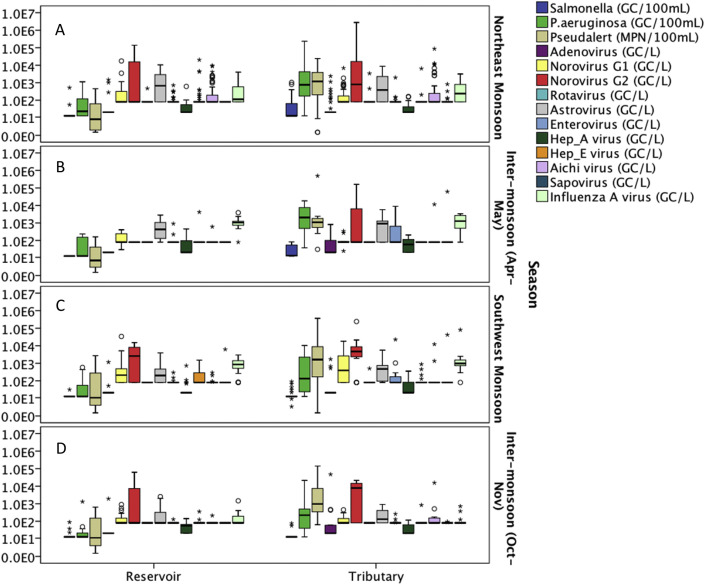
The samples were divided into 7 subgroups based on the sampling locations (Table 1). Water samples from both reservoir and tributaries were collected from sampling area A, B and C, while only reservoir samples were collected from sampling area D to G. Fig. 1 shows the boxplot of the occurrence of microbial indicators in water samples collected from these 7 sampling areas throughout the two-years sampling duration, where the data from reservoirs and tributaries are plotted separately (Fig. 1). In general, the cell concentrations of all microbial indicators were higher in water samples collected from tributaries than reservoirs. In particular, water samples collected from the tributaries at sampling area A showed significantly high concentrations of FRNA G2, B. theta and HPyVs. Similarly, sampling sites B, C and F had high concentrations of human specific markers (i.e. FRNA G2, B. theta and PMMoV). It is interesting to note that a relatively high concentration of FRNA G1was also observed in sampling area G (parkland area).
Fig. 1.
Box plots on the occurrence of microbial indicators in water samples collected from (A) Sampling area A – Highly residential and construction area; (B) Sampling area B – Agricultural area; (C) Sampling area C – Urbanized area; (D) Sampling area D – Green area near the zoo; (E) Sampling area E − Parks near residential area; (F) Sampling area F – Near industry area; and (G) Sampling area G – Parkland area, throughout the two-years sampling duration. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
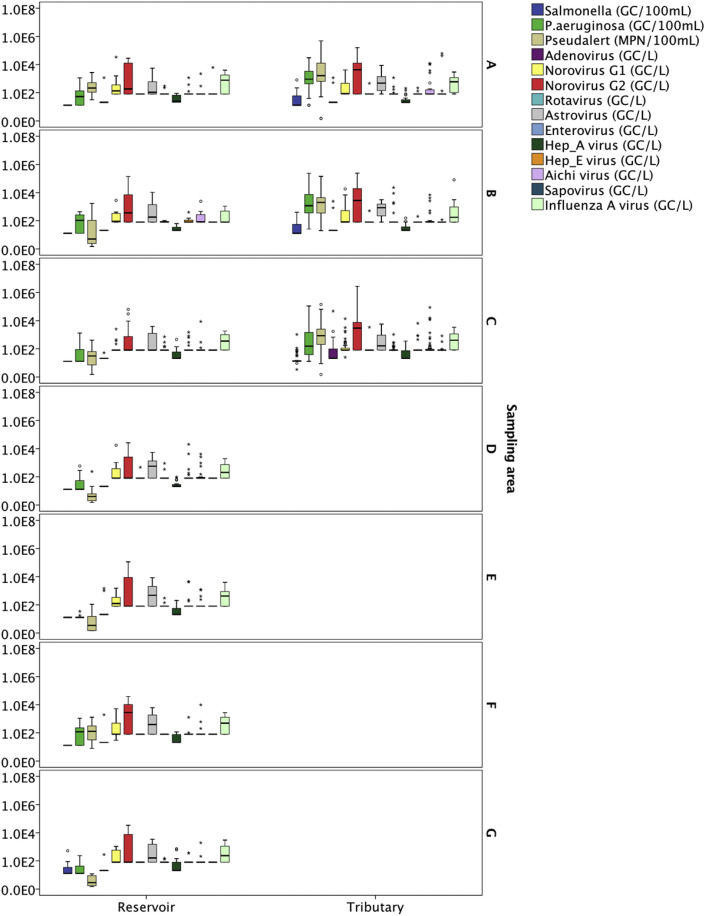
The occurrence of pathogens in term of spatial variation is presented in Fig. 2 (A – G). Salmonella spp. was commonly found in tributaries (detection frequency ranged from 25.5% to 39.13%) compared to reservoirs (detection frequency = 0%), except for reservoir samples at sampling area G. The concentration of Salmonella spp. at sampling area G ranged from 12 GC/100 mL to 523 GC/100 mL with a detection frequency of 37.5%. The concentration of P. aeruginosa in tributaries (geometric mean with qPCR = 558.10 GC/100 mL; geometric mean with Pseudalert™ = 1088.76 MPN/100 mL) were generally higher than reservoirs (geometric mean with qPCR = 34.96 GC/100 mL; geometric mean with Pseudalert™ = 13.23 MPN/100 mL). Despite the low occurrence of adenovirus in all the samples tested (overall detection frequency = 12.7%), the results showed that adenovirus was mostly detected in tributaries collected from sampling area C – urbanized commercial area (detection frequency = 33.33%; geometric mean = 48.03 GC/L). The concentration of norovirus GII was generally high in both tributaries and reservoirs, except for the reservoir samples at sampling area C. Sampling area A (residential area with construction sites) had the highest geometric mean of norovirus GII (1325.87 GC/L), followed by sampling area B (agriculture area) (1188.41 GC/L) and sampling area F (industrial area) (1100.30 GC/L), while sampling area D (green area near wild life parks) had the lowest geometric mean of norovirus GII (349.22 GC/L). Other highly prevalent enteric viruses found in local surface waters were astrovirus and influenza A virus. The geometric mean of astrovirus and influenza A virus at various sampling areas ranged from 274.45 GC/L to 543.62 GC/L and from 260.21 GC/L to 492.90 GC/L, respectively.
Fig. 2.
Box plots on the occurrence of pathogens in water samples collected from (A) Sampling area A – Highly residential and construction area; (B) Sampling area B – Agricultural area; (C) Sampling area C – Urbanized area; (D) Sampling area D – Green area near the zoo; (E) Sampling area E − Parks near residential area; (F) Sampling area F – Near industry area; and (G) Sampling area G – Parkland area, throughout the two-years sampling duration. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The non-parametric statistical analysis, Kruskal-Wallis test was carried out to test the significant difference in spatial distribution for microbial indicators and pathogens. All microbial indicators were significantly different between sampling sites, except for FRNA G1 and FRNA G4 (Table S2). These two indicators were infrequently detected in the water samples (% NDs = 82.3% and 98.2%, respectively). Meanwhile for pathogens, there were only two pathogens (i.e. P. aeruginosa and influenza A virus) which showed a significant difference in concentration between sampling sites.
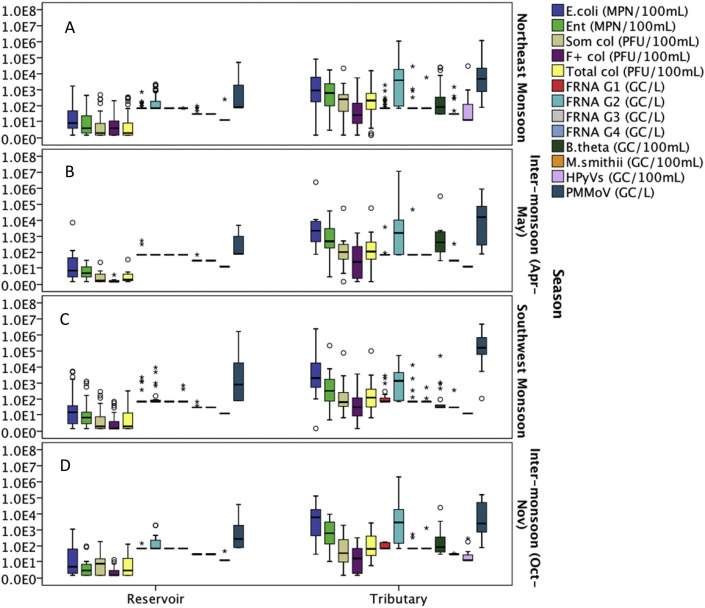
3.4. Temporal variation of microbial indicators and pathogens
The occurrence data were categorized into four seasons, (i) Northeast Monsoon (December–March), (ii) Inter-monsoon (April–May), (iii) Southwest Monsoon (June–September) and (iv) Inter-monsoon (October–November) based on the time when the samples were collected (Fig. 3, Fig. 4 ). Based on the Kruskal-Wallis test, none of the microbial indicators showed statistically significant difference at the p = 0.05 level between the seasons for the reservoir samples (Table S3). However, B. theta, HPyVs and PMMoV showed significant differences between seasons for the water samples collected from tributaries (P < 0.05). B. theta had a relatively lower concentration during the Southwest Monsoon compared to other seasons, while PMMoV had the opposite trend where the concentration during the Southwest Monsoon was higher than other seasons. HPyVs were mainly detected in December 2014 and January 2015, however, this human specific marker was not detected in the subsequent Northeast Monsoon.
Fig. 3.
Box plots on the occurrence of indicators in water samples collected from reservoirs and tributaries during the (A) Northeast Monsoon; (B) Inter-monsoon (Apr–May); (C) Southwest Monsoon; and (D) Inter-monsoon (Oct–Nov).
Fig. 4.
Box plots on the occurrence of pathogens in water samples collected from reservoirs and tributaries during the (A) Northeast Monsoon; (B) Inter-monsoon (Apr–May); (C) Southwest Monsoon; and (D) Inter-monsoon (Oct–Nov).
Among the pathogens in reservoir samples, norovirus GII, astrovirus, Aichi virus and influenza A virus showed a significant difference between seasons (Table S3). The occurrence and concentration of norovirus GII was low during the inter-monsoon (Apr–May) and pairwise comparisons showed that their counts were significantly different between the inter-monsoon (Apr–May) and Northeast Monsoon, as well as inter-monsoon (Apr–May) and Southwest Monsoon periods (Fig. S3). In general, relatively high concentrations of astrovirus, Aichi virus and influenza A virus were found during the Northeast Monsoon in reservoir samples. For water samples collected from tributaries, P. aeruginosa, norovirus GI, enterovirus and influenza A virus were the pathogens that showed significant differences in concentration between seasons (Table S3). The results revealed that norovirus GI and influenza A had the highest geometric mean during the Southwest Monsoon, while P. aeruginosa and enterovirus achieved the highest geometric means during the inter-monsoon (Apr–May) period. Overall, no universal trend can be drawn from the seasonal variation for microbes tested in this study. Rainfall intensity, rainfall duration, antecedent dry period and dilution factor could be some of the factors that affected the microbial concentration in different seasons. Kruskal-Wallis test revealed that rainfall, DO, temperature, salinity and electrical conductivity are significantly different during monsoon and inter-monsoon seasons (Table S3 and Fig. S1). Table S4 further described the rainfall pattern for different seasons in Singapore based on the data collected during the sampling events in this two-year study.
3.5. Correlations between microbial indicators, pathogens and environmental parameters
Correlation analysis was conducted with Kendal's Tau-b method. The correlations were examined (i) between indicators (Table 3 ); (ii) between indicators and pathogens (Table 4 ); (iii) among pathogens (Table 5 ); and (iv) between environmental parameters and indicators/pathogens (Table 6 ). This study found significant correlations among the fecal indicator bacteria (r = 0.670) and coliphages (r = 0.511), as well as between fecal indicator bacteria and coliphages (r = 0.432–0.801). Among the pathogens, P. aeruginosa was the only pathogen that had moderate to good correlations with fecal indicator bacteria/coliphages (r = 0.388–0.692) (Table 4), while weak correlations were found between fecal indicator bacteria/coliphages and Salmonella (r = 0.199–0.265), and also Aichi virus (r = 0.124–0.293). Alternative indicators which are suggested to be highly associated with human waste contamination were also examined in this study. Correlations analyses showed that FRNA G2 (r = 0.422–0.502), B. theta (r = 0.340–0.497) and PMMoV (r = 0.264–0.419) were correlated with fecal indicator bacteria/coliphages (Table 3). FRNA G2, B. theta and PMMoV which are more specific to human fecal pollution, were significantly correlated with most of the indicators except FRNA G4. Unfortunately, most of the enteric viruses did not have significant correlations greater than 0.3 with the alternative indicators, except for Aichi virus and sapovirus. Improved correlations were found between Aichi virus and HPyVs (r = 0.471) and also between Aichi virus and M. smithii (r = 0.315), while sapovirus was correlated with FRNA G3 (r = 0.382) and M. smithii (r = 0.332). Although alternative indicators did not improve the correlations with P. aeruginosa as compared with fecal indicator bacteria/coliphages, however, moderate correlations were found with B. theta, HPyVs and PMMoV (r = 0.325–0.480). Correlation with Salmonella also improved with B. theta (r = 0.321). The results from this study revealed very weak correlations among the pathogens (r < 0.3), except for Salmonella and P. aeruginosa detected with qPCR (r = 0.313). Note that there were two methods, qPCR and IDEXX method, used in this study for P. aeruginosa quantification and both methods were significantly correlated at r = 0.440.
Table 3.
Kendal's Tau-b correlations between microbial indicators.
| E.coli | Ent | Som col | F col | Total col | FRNA G1 | FRNA G2 | FRNA G3 | FRNA G4 | B.theta | M.smithii | HPyVs | PMMoV | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fecal indicator bacteria | |||||||||||||
| E.coli | 1 | .670** | .573** | .432** | .591** | .213** | .474** | .228** | .451** | .172** | .213** | .419** | |
| Ent | 670** | 1 | .549** | .495** | .592** | .215** | .425** | .209** | .497** | .221** | .258** | .363** | |
| Coliphages | |||||||||||||
| Som col | .573** | .549** | 1 | .511** | .801** | .144** | .457** | .199** | .415** | .205** | .233** | .374** | |
| F col | .432** | .495** | .511** | 1 | .554** | .165** | .422** | .226** | .340** | .183** | .234** | .264** | |
| Total col | .591** | .592** | .801** | .554** | 1 | .169** | .502** | .220** | .428** | .219** | .270** | .382** | |
| FRNA genotypes | |||||||||||||
| FRNA G1 | .213** | .215** | .144** | .165** | .169** | 1 | .202** | .178** | .144* | .262** | .147** | ||
| FRNA G2 | .474** | .425** | .457** | .422** | .502** | .202** | 1 | .309** | .374** | .261** | .324** | .448** | |
| FRNA G3 | .228** | .209** | .199** | .226** | .220** | .178** | .309** | 1 | .313** | .268** | .296** | .139* | .235** |
| FRNA G4 | .313** | 1 | |||||||||||
| Human specific markers | |||||||||||||
| B.theta | .451** | .497** | .415** | .340** | .428** | .144* | .374** | .268** | 1 | .333** | .351** | .296** | |
| M.smithii | .172** | .221** | .205** | .183** | .219** | .261** | .296** | .333** | 1 | .355** | .174** | ||
| HPyVs | .213** | .258** | .233** | .234** | .270** | .262** | .324** | .139* | .351** | .355** | 1 | .131* | |
| Plant virus | |||||||||||||
| PMMoV | .419** | .363** | .374** | .264** | .382** | .147** | .448** | .235** | .296** | .174** | .131* | 1 | |
Abbreviation:Ent – Enterococcus; Som col – Somatic coliphage; F col – F+ coliphage; Total col – Total coliphage.
**Correlation is significant at the 0.01 level (2-tailed).
*Correlation is significant at the 0.05 level (2-tailed).
Table 4.
Kendal's Tau-b correlations between microbial indicators and pathogens.
| AdeV | NoV_G1 | NoV_G2 | RotV | AstV | EntV | Hep_A | Hep_E | AichiV | SapoV | Inf_A_V | Salmonella | P.aeruginosa | Pseudalert | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fecal indicator bacteria | ||||||||||||||
| E.coli | .201** | .131** | .190** | .099* | .199** | .408** | .658** | |||||||
| Ent | .128* | -.107* | .171** | .176** | .138** | .258** | .500** | .692** | ||||||
| Coliphages | ||||||||||||||
| Som col | .137* | .118* | -.119* | .124* | .254** | .432** | .539** | |||||||
| F col | .126* | .293** | .155** | .203** | .388** | .440** | ||||||||
| Total col | .118* | .120* | .146** | .134* | .265** | .484** | .558** | |||||||
| FRNA genotypes | ||||||||||||||
| FRNA G1 | .126* | .179** | .183** | |||||||||||
| FRNA G2 | .191** | .168** | .201** | .221** | .201** | .339** | .427** | |||||||
| FRNA G3 | .215** | .240** | .178** | .382** | .129* | .157** | .241** | |||||||
| FRNA G4 | .117* | |||||||||||||
| Human specific markers | ||||||||||||||
| B.theta | .233** | .117* | -.160** | .182** | .213** | .321** | .480** | .452** | ||||||
| M.smithii | .156* | .171** | .315** | .332** | .211** | .216** | .221** | |||||||
| HPyVs | .127* | .471** | .161* | .243** | .325** | .218** | ||||||||
| Plant virus | ||||||||||||||
| PMMoV | .234** | .166** | .140** | .188** | .137** | .236** | .383** | |||||||
Abbreviation: Ent – Enterococcus; Som col – Somatic coliphage; F col – F+ coliphage; Total col – Total coliphage; AdeV – Adenovirus; NoV_G1 – Norovirus GI; NoV_G2 – Norovirus GII; RotV – Rotavirus; AstV – Astrovirus; EntV – Enterovirus; Hep_A – Hepatitis A virus; Hep_E – Hepatitis E virus; AichiV – Aichi virus; Sapo_V – Sapovirus; Inf_A_V – Influenza A virus.
**Correlation is significant at the 0.01 level (2-tailed).
*Correlation is significant at the 0.05 level (2-tailed).
Table 5.
Kendal's Tau-b correlations between pathogens.
| AdeV | NoV_G1 | NoV_G2 | RotV | AstV | EntV | Hep_A | Hep_E | AichiV | SapoV | Inf_A_V | Salmonella | P.aeruginosa | Pseudalert | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enteric viruses | ||||||||||||||
| AdeV | 1 | .133* | .191** | .169∗∗ | ||||||||||
| NoV_G1 | 1 | .170** | -.146** | |||||||||||
| NoV_G2 | .170** | 1 | .196** | .131* | -.133* | -.138** | ||||||||
| RotV | 1 | |||||||||||||
| AstV | .196** | 1 | -.149** | -.128* | ||||||||||
| EntV | .133* | 1 | .122* | .277** | .126* | |||||||||
| Hep_A | -.146** | -.149** | 1 | |||||||||||
| Hep_E | .131* | 1 | -.183** | -.106∗ | ||||||||||
| AichiV | .133* | -.128* | .122* | 1 | .223** | .200** | .192** | .132* | ||||||
| SapoV | .191** | .277** | .223** | 1 | .142* | .194** | ||||||||
| Inf_A_V | -.138** | .126* | 1 | .115* | ||||||||||
| Bacterial pathogens | ||||||||||||||
| Salmonella | .200** | .142* | 1 | .313** | .236** | |||||||||
| P.aeruginosa | -.183** | .192** | .313** | 1 | .440∗∗ | |||||||||
| Pseudalert | .169** | -.106* | .132* | .194** | .115* | .236** | .440** | 1 | ||||||
Abbreviation: AdeV – Adenovirus; NoV_G1 – Norovirus GI; NoV_G2 – Norovirus GII; RotV – Rotavirus; AstV – Astrovirus; EntV – Enterovirus; Hep_A – Hepatitis A virus; Hep_E – Hepatitis E virus; AichiV – Aichi virus; Sapo_V – Sapovirus; Inf_A_V – Influenza A virus.
**Correlation is significant at the 0.01 level (2-tailed).
*Correlation is significant at the 0.05 level (2-tailed).
Table 6.
Kendal's Tau-b correlations between environmental parameters and indicators/pathogens.
| Temp | pH | EC | Salinity | DO | Turbidity | Daily rainfall | Antecedent dry period | Weightage of land use | |
|---|---|---|---|---|---|---|---|---|---|
| Fecal indicator bacteria | |||||||||
| E.coli | -.234** | .257** | .254** | -.238** | .360** | .173** | -.190** | .322** | |
| Ent | -.279** | .091* | .301** | .309** | -.200** | .427** | .179** | -.146** | .287** |
| Coliphages | |||||||||
| Som col | -.302** | .182** | .196** | -.203** | .350** | .165** | -.147** | .260** | |
| F col | -.380** | .136** | .163** | .328** | .209** | -.148** | .233** | ||
| Total col | -.279** | .230** | .247** | -.214** | .365** | .163** | -.121* | .281** | |
| FRNA genotypes | |||||||||
| FRNA G1 | -.119* | .160** | .173** | .134* | |||||
| FRNA G2 | -.307** | .169** | .195** | -.166** | .226** | .133** | .393** | ||
| FRNA G3 | .150** | .265** | |||||||
| FRNA G4 | |||||||||
| Human specific markers | |||||||||
| B.theta | -.163** | .157** | .225** | .232** | -.160** | .228** | .305** | ||
| M.smithii | .116* | .119* | .153** | .139* | .153** | ||||
| HPyVs | -.270** | .137* | .155** | .189** | .140* | .135* | |||
| Plant virus | |||||||||
| PMMoV | -.165** | .205** | .203** | -.270** | .258** | .451** | |||
| Enteric viruses | |||||||||
| AdeV | .157** | ||||||||
| NoV_G1 | .117* | ||||||||
| NoV_G2 | -.141** | .118* | |||||||
| RotV | |||||||||
| AstV | .112* | ||||||||
| EntV | |||||||||
| Hep_A | |||||||||
| Hep_E | |||||||||
| AichiV | -.292** | .124* | .168** | -.114* | |||||
| SapoV | .148** | ||||||||
| Inf_A_V | .113* | ||||||||
| Bacterial pathogens | |||||||||
| Salmonella | -.156** | .151** | .160** | .122* | -.115* | ||||
| P.aeruginosa | -.236** | .279** | .274** | .281** | .136** | .202** | |||
| Pseudalert | -.200** | .113* | .269** | .282** | -.177** | .379** | .152** | -.143** | .305** |
Abbreviation: Ent – Enterococcus; Som col – Somatic coliphage; F col – F+ coliphage; Total col – Total coliphage; AdeV – Adenovirus; NoV_G1 – Norovirus GI; NoV_G2 – Norovirus GII; RotV – Rotavirus; AstV – Astrovirus; EntV – Enterovirus; Hep_A – Hepatitis A virus; Hep_E – Hepatitis E virus; AichiV – Aichi virus; Sapo_V – Sapovirus; Inf_A_V – Influenza A virus; Temp – Temperature; EC – Electrical conductivity; DO – Dissolved oxygen.
∗∗ Correlation is significant at the 0.01 level (2-tailed).
∗ Correlation is significant at the 0.05 level (2-tailed).
In terms of environmental factors, an increase in temperature significantly reduced the concentrations of fecal indicator bacteria, coliphages, FRNA G2, B. theta, HPyVs and bacterial pathogens, while EC and salinity showed the opposite trend. As expected, the lower the DO, the higher the concentrations of bacteria and coliphages. Turbidity was generally correlated with the concentrations of bacteria and coliphage, but not with the concentration of enteric viruses. The correlation analysis also showed that the impact of daily rainfall was positively, although weakly correlated with the occurrence of microbial indicators, while weak negative correlations were observed between antecedent dry period and microbial indicators. In addition, there was a greater influence of land use on the concentrations of bacteria and coliphages compared to enteric viruses. Among the indicators and pathogens, PMMoV was significantly correlated with land use (r = 0.451).
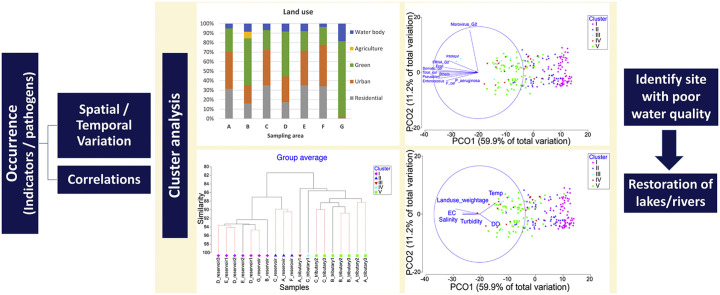
3.6. Spatial clustering within the microbial community and relationships with environmental parameters
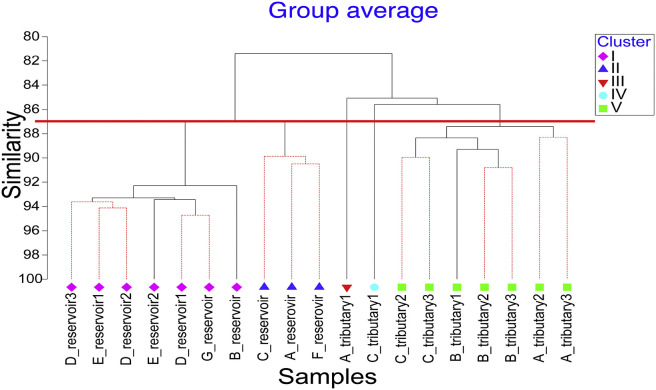
An assessment of spatial variations in surface water quality is important for identifying sites where water quality has been impaired, and subsequently, to allow management to make better decisions to control and restore the water quality. Cluster analysis was used to group the sampling sites based on the occurrence of microorganisms. Data was log transformed and resemblance was conducted with Bray-Curtis similarity. Hierarchical clustering was applied for the cluster analysis and the clustering result is illustrated by a dendrogram (Fig. 5 ). At a similarity of 87%, the sampling sites can generally be classified into five main clusters (i.e. I to V). The reservoir samples were clustered into two main clusters, I and II where Cluster I comprised 7 reservoir samples collected from sampling areas B, D, E and G. Sampling areas B, D and G are reservoirs located in areas where there are extensive forests and/or green areas with minimal impact from anthropogenic activities. Although sampling area E is a reservoir in a residential area, there is no direct surface runoff drain channeled into the reservoir. Hence, the water quality is less impacted by nearby residential land use. In comparison, reservoir samples from sampling areas A, C and F were grouped into Cluster II, where industrial, commercial and residential (including several construction sites) zones are predominant. The water samples collected from tributaries at sampling area A (sampling site: A_tributary1) and sampling area C (sampling site: C_tributary1) were classified into two independent clusters, Cluster III and Cluster IV, respectively. The rest of the tributaries had high similarity in term of the distribution of microorganisms and were classified into the same cluster, Cluster V.
Fig. 5.
Dendrogram showing clustering of sampling sites according to the occurrence of microorganisms.
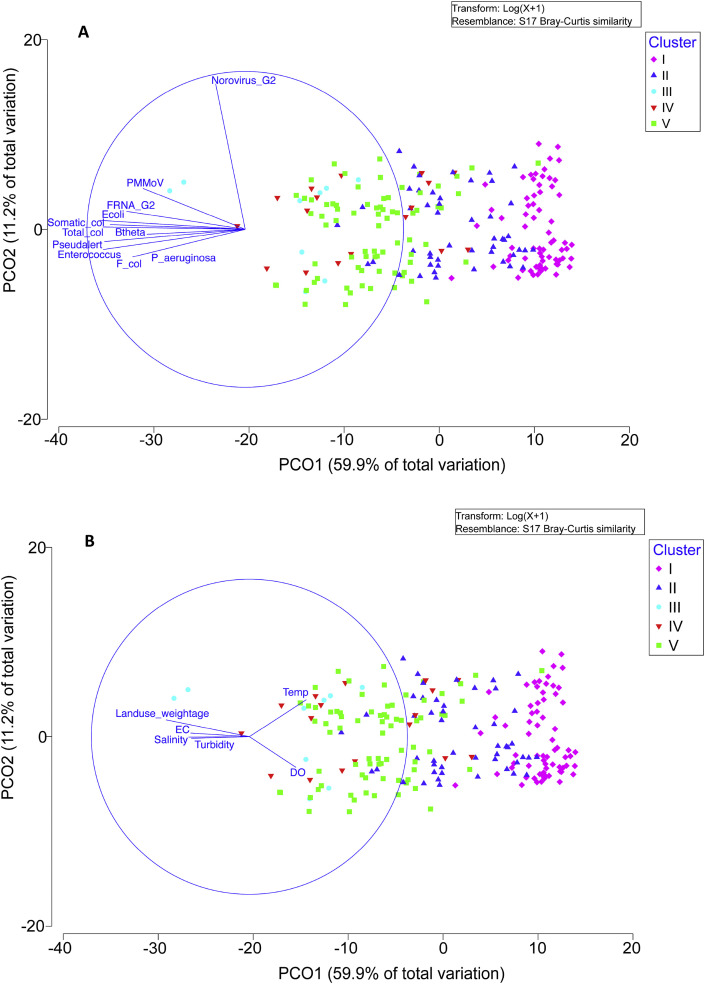
Due to the large complexity between samples, PCO analysis was conducted to understand the similarity or dissimilarity of the microorganism community between samples. The PCO plot revealed that 71.1% of the variation could be explained through PCO 1 and PCO2 (Fig. 6 ). By limiting the r value to greater than 0.5, the main variables that contributed to PCO1 were E. coli (r = -0.92), Enterococcus (r = −0.90), somatic coliphage (r = −0.85), F+ coliphage (r = −0.71), total coliphage (r = −0.99), FRNA G2 (r = −0.75), B. theta (r = −0.62), P. aeruginosa (r = −0.71), Pseudalert (r = −0.89) and PMMoV (r = −0.65). Norovirus GII was the only variable that contributed to PCO2 with r = 0.92.
Fig. 6.
PCO plot on (A) the dissimilarity of microorganism community between samples; and (B) impact of environmental variables on the distributions of microorganisms.
The impact of environmental variables on the distributions of microorganisms was also analyzed with the PCO plot (Fig. 6B). By projecting the environmental vectors on the same axes and setting the r value to greater than 0.3, temperature (r = 0.36), EC (r = −0.37), salinity (r = −0.39), turbidity (r = −0.37), DO (r = 0.3) and land use weightage (r = −0.53) were the main environmental variables that influenced the distribution of microorganisms. Further discussion can be found in the supplementary information.
4. Discussion
Our study examined the occurrence and the abundance of microbial indicators and pathogens in the surface waters over a period of two years. In this study, pre-concentration using hollow fiber ultrafiltration was used to simultaneously concentrate a wide range of microbes from the environment. This method allowed the parallel detection of viruses, bacteria and protozoa (data not shown). To our knowledge, this is the first comprehensive and systematic surveillance study done in tropical surface waters.
4.1. Microbial indicators
Studies have shown strong positive correlations between the membrane filter technique and IDEXX kits (Budnick et al., 1996; Kinzelman et al., 2005). As such, the readings from IDEXX kits were compared directly with USEPA recreational water quality guideline values. In this study, the geometric mean for E. coli (111.7 MPN/100 mL) was below the USEPA's recreational water quality guideline value of 126 CFU/100 mL for primary contact recreational activities (USEPA, 2012). Meanwhile, the geometric mean for Enterococcus (40.7 MPN/100 mL) was slightly above the USEPA's guideline value (35 CFU/100 mL). According to Singapore's recreational water quality guidelines, it is recommended that recreational waters are suitable for primary contact recreational activities if Enterococcus counts are below 200 counts/100 mL 95% of the time (source: www.nea.gov.sg). Based on reservoir data, Enterococcus counts were always low in reservoirs compared to tributaries and generally fulfilled the requirement for primary contact recreational activities, except for sampling area F (geometric mean = 41.5 MPN/100 mL; 80% of the time the Enterococcus counts were below 200 counts/100 mL). Nevertheless, all reservoirs in Singapore are currently not open for primary contact activities.
The coliphage analyses showed that somatic coliphages were generally more prevalent and had higher concentrations compared to F+ coliphages, which is in agreement with other studies (Aw and Gin, 2010; Benjamin-Chung et al., 2017; Lodder and de Roda Husman, 2005). The recent introduction of a new coliphage host (E. coli CB390) which can simultaneously detect both somatic coliphages and F+ coliphages was also used in this study (Guzmán et al., 2008). As reported in earlier studies, this study also found no statistical difference between the sum of concentrations detected using E. coli CN13 (somatic coliphage) and E. coli Famp (F+ coliphage) with the concentrations detected using E. coli CB390 (total coliphages) for the environmental water samples tested (Mann-Whitney test, p = 0.221) (Bailey et al., 2017).
Most of the serious threats to humans is believed to originate from human waste contamination rather than from animal waste (Field and Samadpour, 2007). Unfortunately, the source of contamination is difficult to track based on fecal indicator bacteria as these indicators could be released into environment by both humans and animals fecal sources (Souza et al., 1999). As a result, the human specific markers such as B. theta, HPyVs and M. smithii have been suggested as potential markers for human waste contamination (Carson et al., 2005; Harwood et al., 2014; Johnston et al., 2010; McQuaig et al. 2009, 2012; Yampara-Iquise et al., 2008). In this study, a high prevalence of B. theta was observed compared to HPyVs and M. smithii. An earlier study showed that B. theta was more sensitive towards the detection of human waste contamination, although HPyVs gave better specificity in terms of human source tracking (unpublished data). Carson et al. (2005) found a cross-reactivity with dogs (16% of 31 samples) in their B. theta assay (Carson et al., 2005). However, Griffith et al. (2009) showed that the detection of B. theta was among the most reliable methods to detect samples containing human fecal material with no false positive results (Griffith et al., 2009).
Studies have shown that the genotypes of FRNA are associated with different sources of contamination (Hsu et al., 1995; Stewart-Pullaro et al., 2006), i.e. the presence of FRNA G2 and G3 are typically associated with human waste contamination while FRNA G1 and G4 are typically associated with animal wastes. In this study, FRNA G2 was the most prevalent genotype among the four genotype groups. This may explain the high probability of human waste contamination in the water samples tested. However, studies also revealed the possible detection of FRNA G2 in poultry, pig and cattle feces (Schaper et al., 2002b), and the presence of FRNA G4 in human feces (Hsu et al., 1995). In this study, a higher prevalence of FRNA G2 (detection frequency = 52.3%) was found compared to FRNA G3 (detection frequency = 5%). This finding contradicts an earlier study which reported that FRNA G2 seems to be more prominent in cold regions, while FRNA G3 dominated in warm regions (Furuse et al., 1978). One of the possible reasons could be the higher persistence of FRNA G2 compared to FRNA G3 in natural environments (Schaper et al., 2002a). It is also interesting to note that a relatively high concentration of FRNA G1 was observed at sampling area G, a protected catchment with rich biodiversity. This is consistent with the fact that FRNA G1 is associated with fecal wastes from animals (Cole et al., 2003).
PMMoV, a virus that can infect pepper plants, has been suggested as one of the potential indicators for human fecal contamination due to its high abundance in human feces and high prevalence in the human population (Zhang et al., 2005). Although PMMoV showed high prevalence in local surface waters, the application of this indicator in human waste source tracking needs to be evaluated carefully, particularly in the context of wastewater reuse. Studies have shown the difficulty in removing PMMoV during wastewater treatment due to its high abundance and persistence (Kitajima et al., 2014).
In summary, the detection of these different human specific markers suggested a high probability of human waste contamination in the tributaries at sampling sites A, B, C and F. Although the tributaries where these markers were detected are not used for recreational water activities, the tracking of human source contamination would be useful to identify the upstream source of pollution and hence, provide better mitigation strategies to remedy downstream contamination problems.
4.2. Pathogens
Norovirus GII (51.8%), astrovirus (64.5%) and influenza A virus (65.4%) were the three most prevalent enteric viruses detected in Singapore's surface waters. Norovirus GII showed a similar range of detection rate (48%–69%) with earlier studies in Singapore (Aw and Gin, 2011; Liang et al., 2015; Rezaeinejad et al., 2014). A relatively high prevalence of norovirus GII was also reported in environmental surface waters at different locations (Calgua et al., 2013; Kishida et al., 2012; Kiulia et al., 2014; Lee and Kim, 2008). In this study, a higher prevalence of norovirus GII (51.8%) was found compared to norovirus GI (40%). A virus decay study conducted in groundwater revealed that norovirus GII was more persistent than norovirus GI (Charles et al., 2009). As currently there is no available cell line for the norovirus infectivity test, the detection of norovirus relies on molecular methods. Teunis et al. (2008) demonstrated the low infectious dose for norovirus, where the 50% infectious dose, LD50 was equivalent to 1015 genomes copies or 18 viruses (Teunis et al., 2008). Ngazoa et al. (2008) reported approximately 30% of norovirus genomes were detected in river water after 30 days incubation at 25 °C. This explains the high prevalence of norovirus in surface waters found in this study. Another study on viral persistence revealed that the reduction rate of nucleic acid of norovirus surrogates was significantly lower than the reduction rate of infectivity (Bae and Schwab, 2008). Nevertheless, extra precautions are needed when interpreting health risks brought by norovirus due to its low infectious dose.
In the case of astrovirus, the detection frequency (58%) was comparable to the results reported by Aw and Gin's study (2011) (50%). Although there is no clear explanation for the high prevalence of astrovirus in local surface waters compared to other places (Pusch et al., 2005; Steyer et al., 2011; Taylor et al., 2001), one of the reasons could be due to the persistence of certain serogroups of astrovirus in tropical conditions. Another possible explanation is the close genetic relationship between human astrovirus and animal astrovirus, as well the cross-infectivity of human and animal astrovirus (De Benedictis et al., 2011; Delwart, 2012).
Influenza A viruses are pathogens that can infect large numbers of warm-blooded animals, such as birds, pigs, horses and humans. It is believed that aquatic birds serve as a natural reservoir for all known subtypes of influenza A virus and are probably the ultimate source of human pandemic influenza strains (Taubenberger and Morens, 2010; Webster et al., 1992). Studies have shown that a high persistence of influenza A viruses in environmental waters which can remain infectious over a long period, depending on the strain types and environmental factors (e.g. salinity, temperature, pH) (Dublineau et al., 2011; Keeler et al. 2013, 2014; Stallknecht et al., 1990; Webster et al., 1992). In this study, a high detection rate of influenza A virus was found (64.5%), which is higher than those previously reported in France (44.4%)(Deboosere et al., 2011) and Netherlands (40%) (Heijnen and Medema, 2011). Heijnen and Medema et al.’s study suggested the discharge of treated sewage was not the primary source of influenza A virus detected in river water but rather, was mainly from aquatic birds due to the very low prevalence of influenza A virus in sewage samples (1 out of 10 samples). In our study, the highest mean concentration of influenza A virus was found at sampling area B (mean = 3063.85 GC/L), which was an area dominated by chicken farming.
Although other studies showed the high prevalence of adenovirus in natural environments (Jiang, 2006; Rigotto et al., 2010), the detection frequency of adenovirus observed in this study was only 12.7%, with a geometric mean concentration of 29 GC/L. A low prevalence of adenovirus was also reported in Choi and Jiang's study (2005), where 16% out of 114 river samples from California had positive detections of adenovirus. In the same study, negative results were reported for infection of adenovirus using two human tissue culture cell lines, HEK293A and A5949 (Choi and Jiang, 2005). Similarly, this study also found negative results for infectious adenovirus using the integrated cell culture-PCR (ICC-PCR) with Human colon carcinoma (CaCo-2) cell line in surface waters. In contrast, positive infectivity results were observed in samples collected from the influent of a wastewater treatment plant (data not shown). The absence of infectious adenovirus observed in surface waters could be due to the loss of viral infectivity after exposure to sunlight or other harsh conditions in the natural environment, in addition to the low concentrations detected. Comparing the occurrence data obtained in this study with earlier studies conducted in Singapore, a clear trend of reduction in enteric viruses (i.e. adenovirus, rotavirus and enterovirus) was found (Aw and Gin, 2011; Liang et al., 2015; Rezaeinejad et al., 2014). This showed the effectiveness of the public sewers rehabilitation programme to restore the structural integrity of the sewerage system and hence, minimize pollution to surface waters.
High prevalence of sapovirus has been reported in many environmental waters such as in Japan (20%) (Kitajima et al., 2010) and South Africa (14–92%) (Murray et al., 2013). However, a low detection frequency was found in this study (4.1%). As reported in Kitajima et al.'s study (2010), sapovirus was mostly detected during winter time (November to March). This may explain the low prevalence of sapovirus in Singapore, which has warm tropical temperature throughout the year.
Hepatitis E virus was detected sporadically at different locations with the highest geometric mean of 999.38 GC/L at sampling area D, an area surrounded by green areas (forests), as well as wild life parks. The seasonal distribution patterns revealed a higher prevalence and concentration of hepatitis E virus during the Northeast Monsoon and Southwest Monsoon (Fig. 4), consistent with heavy rain which could flush animals feces from green areas or parks into the tributaries and reservoirs.
Although P. aeruginosa is an opportunistic pathogen that is ubiquitously present in natural environments, the ability of P. aeruginosa to resist multiple antibiotics makes this opportunistic pathogen a clinically important pathogen (Lister et al., 2009). In this study, a high detection frequency of P. aeruginosa was observed using the IDEXX kit (i.e. Pseudalert™) compared to the qPCR method. This could be due to the lower detection limit that can be achieved with the IDEXX kit. In addition, the qPCR method only quantifies the toxin A synthesis regulating gene (regA gene) while the IDEXX kit quantifies P. aeruginosa strains. Although the correlation between the readings from Pseudalert™ and qPCR is not strong (r = 0.440, p < 0.01), the Wilcoxon sign-rank test showed that the detection of P. aeruginosa did not elicit a statistically significant difference with both methods (p = 0.536).
In this study, Salmonella spp. was not detected in reservoirs, except for sampling area G with concentrations up to 523 GC/100 mL and detection frequency of 37.5%. Reptiles, amphibians and birds are some animals that frequently carry Salmonella spp. (Silva et al., 2014). Sampling area G is a relatively protected forested area (low urban development), but the presence of these animals in the natural environment may have contributed to the higher counts and prevalence of Salmonella spp. in reservoirs here compared to other sites.
4.3. Correlations between indicators, pathogens and environmental variables
Several epidemiological studies have been conducted to relate the presence of indicator bacteria with gastroenteritis illness, however, some studies have shown limited correlations between fecal indicator bacteria and pathogens (Wu et al., 2011). Alternative indicators such as human specific markers (i.e. Bacteroides, M. smithii and HPyVs), coliphages (i.e. somatic and F+ coliphages), genotypes of FRNA phages and plant virus (i.e. PMMoV) have recently been proposed to allow for better prediction of human pathogens (Hughes et al., 2017; McQuaig et al. 2009, 2012; Vergara et al., 2015; Yampara-Iquise et al., 2008). As a result, a number of recent studies have focused on correlations between the alternative indicators and pathogens (Harwood et al., 2014; Jofre et al., 2016). Nevertheless, disparate results were reported. To our knowledge, none of the studies have related these indicators to a wide range of human viruses, particularly enteric viruses in waters.
As shown in the earlier studies, this study found significant correlations among fecal indicator bacteria and coliphages (Liang et al., 2015; Lodder et al., 2010; Rezaeinejad et al., 2014). This is expected as coliphages are the phages for E. coli hosts and all these indicators should have originated from the same sources. However, fecal indicator bacteria/coliphages did not correlate well with most of the pathogens, except for P. aeruginosa, similar to many other studies. Nevertheless, there are some studies which demonstrated limited correlations between indicator bacteria/coliphages and pathogens. For instance, significant weak correlation was found between F+ coliphage and norovirus GI (r = 0.233) and norovirus GII (r = 0.312) (Rezaeinejad et al., 2014), even though Lodder et al. (2010) and Liang et al. (2015) failed to show the correlation between coliphages and norovirus. The inclusion of alternative indicators in this study improved the correlations with pathogens, especially for Salmonella spp., sapovirus and Aichi virus. Liang et al.’s study (2015) showed an improvement in predictive stepwise linear regreasion models for Salmonella spp. and norovirus GII by incorporating the alternative indicators (i.e. HPyVs and M. smithii).
There are several factors that may contribute to the lack of correlation between indicators and pathogens. These factors include the survival and transport of indicators/pathogens in the natural environment, shedding patterns among host populations, detection method, percentage of positive detection, land uses, as well as geographic variation (Wu et al., 2011). The major drawback for fecal indicator bacteria and coliphages is that these bacteria/phages cannot indicate the source of pollution as they are present in both human and animal wastes (Havelaar et al., 1986). As such, it is not surprising that very weak or even no correlation was found between indicators/coliphages and enteric viruses, which are mainly excreted from infected humans. Apart from the different shedding patterns for human pathogens, fecal indicator bacteria are also able to persist for extended periods, and may even grow in the natural environment (Byappanahalli et al., 2006). In an earlier controlled laboratory study, the survival of indicators and human specific indicators was examined together with adenovirus under the effects of sunlight and salinity (Liang et al., 2017). The study showed that B. theta had slightly lower decay rates compared to adenovirus in both fresh and sea water, while M. smithii had higher decay rates than adenovirus in both fresh and seawaters. These differences may explain the slightly better correlations between B. theta and adenovirus compared to other indicators in this study (r = 0.233).
The weak correlations between fecal indicators/coliphages and pathogens could also be due to the different detection methods. These differences could have implications on the final results due to variations in sensitivity, recovery rate and specificity. Thus, similar detection methods will usually yield better correlations. In this study, P. aeruginosa quantified with IDEXX kits correlated better with indicators (examined with culture-based method) than P. aeruginosa quantified by the molecular method. The low shedding rate and low percentage of positive detection in pathogens also contributed to the lack of correlation with indicators. Among the pathogens, P. aeruginosa (quantified by IDEXX kit) is the only pathogen that achieved high detection frequency (91.8%) and hence, stronger correlations with indicators were observed. Apart from the factors discussed above, land use also played a major role in the correlation analysis. Compared with the earlier studies conducted in Singapore, relatively weak correlations were found in this study, particularly between the indicators and pathogens. The reason being this study extended the sampling sites to other major reservoirs in Singapore which covered various types of land use, whereas the previous study focused on a single urbanized reservoir and its tributaries. When separate correlation analysis was conducted based on specific sites, the correlations improved significantly.
While this study showed a higher concentration of microbes during the wet season compared to dry season (Rochelle-Newall et al., 2016), the study failed to draw a general seasonal variation pattern for microbes (except for human specific markers). Similarly, a study in Vietnam also showed no significant seasonal difference in E. coli (Nguyen et al., 2016). As the sampling events were conducted on a monthly basis, limited storm water samples were collected to represent a complete hydrology profile during the monsoon (wet) season. Nevertheless, correlation analysis showed consistent relationships, albeit weak, between the concentration of indicator microbes and rainfall (r = 0.163 to 0.209), as well as antecedent dry period (r = −0.121 to −0.190).
5. Conclusions
The present study reports the occurrence and distribution of microbial indicators, pathogenic bacteria and enteric viruses in tropical surface waters collected from 7 sampling areas (19 sampling sites) with varied land use over a period of two years. The application of hollow fiber ultrafiltration coupled with molecular detection methods allowed rapid and simultaneous detection of a wide range of microorganisms, including the presence of pathogenic viruses.
Despite the strong correlations among the indicators and coliphages, relative weak or no correlations were found between indicators and viral pathogens. The present study also failed to identify a universal indicator that was associated with all the pathogens tested. In addition, the correlations between indicators and pathogens could be highly variable depending on environmental factors, such as temperature, land use, rainfall, geographical location, etc. As such, future prediction of pathogens may require a suite of indicators rather than a single indicator. Machine learning models which enable learning from large data sets could be a better approach for future pathogen predictive models.
Statistical analyses revealed that the occurrence of indicators was significantly different between sampling sites compared to pathogens. Cluster analysis and PCO plots successfully identified potential sites which had degraded or poorer water quality. This will aid in management strategies to prioritize sites for restoration of water quality.
As current detection of enteric viruses was based on molecular methods, it was not possible to give strong evidence on the risks of these viruses, which rely mainly on cell culture infectivity tests. Nevertheless, preliminary studies on virus infectivity showed that a relatively low fraction of infectious adenovirus and astrovirus was present when measured with ICC-PCR compared to qPCR, which is in agreement with other studies (Choi and Jiang, 2005; Le Cann et al., 2004). As such, the interpretation of health risks brought by these enteric viruses need to be evaluated carefully.
Declaration of interest statement
None.
Acknowledgements
This research grant is supported by the Singapore National Research Foundation under its Environmental and Water Research Programme and administered by PUB (Ref: 1301-IRIS-37 [IDD 90301/1/65]). We would like to thank National University of Singapore for supporting this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.watres.2018.11.058.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmad T., Waheed Y., Tahir S., Safi S., Fatima K., Afzal M., Farooqi Z., Qadri I. Frequency of HEV contamination in sewerage waters in Pakistan. J. Infect. Develop. Ctries. 2010;4(12):842–845. doi: 10.3855/jidc.612. [DOI] [PubMed] [Google Scholar]
- Aloush V., Navon-Venezia S., Seigman-Igra Y., Cabili S., Carmeli Y. Multidrug-Resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob. Agents Chemother. 2006;50(1):43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnone R.D., Walling J.P. Waterborne pathogens in urban watersheds. J. Water Health. 2007;5(1):149. doi: 10.2166/wh.2006.001. [DOI] [PubMed] [Google Scholar]
- Aw T.G., Gin K.Y.-H., Ean Oon L.L., Chen E.X., Woo C.H. Prevalence and genotypes of human noroviruses in tropical urban surface waters and clinical samples in Singapore. Appl. Environ. Microbiol. 2009;75(15):4984–4992. doi: 10.1128/AEM.00489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw T.G., Gin K.Y.H. Environmental surveillance and molecular characterization of human enteric viruses in tropical urban wastewaters. J. Appl. Microbiol. 2010;109(2):716–730. doi: 10.1111/j.1365-2672.2010.04701.x. [DOI] [PubMed] [Google Scholar]
- Aw T.G., Gin K.Y.H. Prevalence and genetic diversity of waterborne pathogenic viruses in surface waters of tropical urban catchments. J. Appl. Microbiol. 2011;110(4):903–914. doi: 10.1111/j.1365-2672.2011.04947.x. [DOI] [PubMed] [Google Scholar]
- Bae J., Schwab K.J. Evaluation of murine norovirus, Feline Calicivirus, Poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl. Environ. Microbiol. 2008;74(2):477–484. doi: 10.1128/AEM.02095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey E.S., Price M., Casanova L.M., Sobsey M.D. E. coli CB390: an alternative E. coli host for simultaneous detection of somatic and F+ coliphage viruses in reclaimed and other waters. J. Virol Methods. 2017;250(Suppl. C):25–28. doi: 10.1016/j.jviromet.2017.09.016. [DOI] [PubMed] [Google Scholar]
- Benjamin-Chung J., Arnold B.F., Wade T.J., Schiff K., Griffith J.F., Dufour A.P., Weisberg S.B., Colford J.M. Coliphages and gastrointestinal illness in recreational waters: pooled analysis of six coastal beach cohorts. Epidemiology. 2017;28(5):644–652. doi: 10.1097/EDE.0000000000000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnick G.E., Howard R.T., Mayo D.R. Evaluation of Enterolert for enumeration of enterococci in recreational waters. Appl. Environ. Microbiol. 1996;62(10):3881–3884. doi: 10.1128/aem.62.10.3881-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli M.N., Fujioka R.S. Evidence that tropical soil environment can support the growth of Escherichia coli. Water Sci. Technol. 1998;38(12):171–174. [Google Scholar]
- Byappanahalli M.N., Whitman R.L., Shively D.A., Ting E.W.T., Tseng C.C., Nevers M.B. Seasonal persistence and population characteristics of Escherichia coli and enterococci in deep backshore sand of two freshwater beaches. J. Water Health. 2006;04(3):313–320. doi: 10.2166/wh.2006.518. [DOI] [PubMed] [Google Scholar]
- Calgua B., Fumian T., Rusiñol M., Rodriguez-Manzano J., Mbayed V.A., Bofill-Mas S., Miagostovich M., Girones R. Detection and quantification of classic and emerging viruses by skimmed-milk flocculation and PCR in river water from two geographical areas. Water Res. 2013;47(8):2797–2810. doi: 10.1016/j.watres.2013.02.043. [DOI] [PubMed] [Google Scholar]
- Carrillo M., Estrada E., Hazen T.C. Survival and enumeration of the fecal indicators Bifidobacterium adolescentis and Escherichia coli in a tropical rain forest watershed. Appl. Environ. Microbiol. 1985;50(2):468–476. doi: 10.1128/aem.50.2.468-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson C.A., Christiansen J.M., Yampara-Iquise H., Benson V.W., Baffaut C., Davis J.V., Broz R.R., Kurtz W.B., Rogers W.M., Fales W.H. Specificity of a Bacteroides thetaiotaomicron marker for human feces. Appl. Environ. Microbiol. 2005;71(8):4945–4949. doi: 10.1128/AEM.71.8.4945-4949.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles K.J., Shore J., Sellwood J., Laverick M., Hart A., Pedley S. Assessment of the stability of human viruses and coliphage in groundwater by PCR and infectivity methods. J. Appl. Microbiol. 2009;106(6):1827–1837. doi: 10.1111/j.1365-2672.2009.04150.x. [DOI] [PubMed] [Google Scholar]
- Choi S., Jiang S.C. Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Appl. Environ. Microbiol. 2005;71(11):7426–7433. doi: 10.1128/AEM.71.11.7426-7433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuah C.J., Ziegler A.D. Temporal variability of faecal contamination from on-site sanitation systems in the groundwater of Northern Thailand. Environ. Manag. 2018;61(6):939–953. doi: 10.1007/s00267-018-1016-7. [DOI] [PubMed] [Google Scholar]
- Cole D., Long S.C., Sobsey M.D. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 2003;69(11):6507–6514. doi: 10.1128/AEM.69.11.6507-6514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedictis P., Schultz-Cherry S., Burnham A., Cattoli G. Astrovirus infections in humans and animals – molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011;11(7):1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboosere N., Horm S.V., Pinon A., Gachet J., Coldefy C., Buchy P., Vialette M. Development and validation of a concentration method for the detection of influenza a viruses from large volumes of surface water. Appl. Environ. Microbiol. 2011;77(11):3802–3808. doi: 10.1128/AEM.02484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart E. Animal virus discovery: improving animal health, understanding zoonoses, and opportunities for vaccine development. Current Opinion in Virology. 2012;2(3):344–352. doi: 10.1016/j.coviro.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Statistics Singapore . 2016. Population in Brief 2016. [Google Scholar]
- Dublineau A., Batéjat C., Pinon A., Burguière A.M., Leclercq I., Manuguerra J.-C. Persistence of the 2009 pandemic influenza a (H1N1) virus in water and on non-porous surface. PloS One. 2011;6(11) doi: 10.1371/journal.pone.0028043. e28043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field K.G., Samadpour M. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res. 2007;41(16):3517–3538. doi: 10.1016/j.watres.2007.06.056. [DOI] [PubMed] [Google Scholar]
- Furuse K., Sakurai T., Hirashima A., Katsuki M., Ando A., Watanabe I. Distribution of ribonucleic acid coliphages in south and east Asia. Appl. Environ. Microbiol. 1978;35(6):995–1002. doi: 10.1128/aem.35.6.995-1002.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh A., Lin J. Waterborne human pathogenic viruses of public health concern. Int. J. Environ. Health Res. 2013;23(6):544–564. doi: 10.1080/09603123.2013.769205. [DOI] [PubMed] [Google Scholar]
- Griffith J.F., Cao Y., McGee C.D., Weisberg S.B. Evaluation of rapid methods and novel indicators for assessing microbiological beach water quality. Water Res. 2009;43(19):4900–4907. doi: 10.1016/j.watres.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Guzmán C., Mocé-Llivina L., Lucena F., Jofre J. Evaluation of Escherichia coli host strain CB390 for simultaneous detection of somatic and F-specific coliphages. Appl. Environ. Microbiol. 2008;74(2):531–534. doi: 10.1128/AEM.01710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Katayama H., Phanuwan C., Ohgaki S. Quantitative detection of sapoviruses in wastewater and river water in Japan. Lett. Appl. Microbiol. 2008;46(3):408–413. doi: 10.1111/j.1472-765X.2008.02330.x. [DOI] [PubMed] [Google Scholar]
- Harwood V.J., Boehm A.B., Sassoubre L.M., Vijayavel K., Stewart J.R., Fong T.-T., Caprais M.-P., Converse R.R., Diston D., Ebdon J., Fuhrman J.A., Gourmelon M., Gentry-Shields J., Griffith J.F., Kashian D.R., Noble R.T., Taylor H., Wicki M. Performance of viruses and bacteriophages for fecal source determination in a multi-laboratory, comparative study. Water Res. 2013;47(18):6929–6943. doi: 10.1016/j.watres.2013.04.064. [DOI] [PubMed] [Google Scholar]
- Harwood V.J., Staley C., Badgley B.D., Borges K., Korajkic A. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2014;38(1):1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- Havelaar A.H., Furuse K., Hogeboom W.M. Bacteriophages and indicator bacteria in human and animal faeces. J. Appl. Bacteriol. 1986;60(3):255–262. doi: 10.1111/j.1365-2672.1986.tb01081.x. [DOI] [PubMed] [Google Scholar]
- Heijnen L., Medema G. Surveillance of Influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in The Netherlands. J. Water Health. 2011;9(3):434–442. doi: 10.2166/wh.2011.019. [DOI] [PubMed] [Google Scholar]
- Helsel D.R. Wiley-Interscience; 2005. Nondetects and Data Analysis: Statistics for Censored Environmental Data. [Google Scholar]
- Hirsch E.B., Tam V.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 2010;10(4):441–451. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavsa M.C., Roberts V.A., Kahler A.M., Hilborn E.D., Mecher T.R., Beach M.J., Wade T.J., Yoder J.S. Centers for Disease Control and Prevention; 2015. Outbreaks of Illness Associated with Recreational Water - United States, 2011-2012; pp. 668–672. [PMC free article] [PubMed] [Google Scholar]
- Howard C.M., Handzel T., Hill V.R., Grytdal S.P., Blanton C., Kamili S., Drobeniuc J., Hu D., Teshale E. Novel risk factors associated with hepatitis E virus infection in a large outbreak in Northern Uganda: results from a case-control study and environmental analysis. Am. J. Trop. Med. Hyg. 2010;83(5):1170–1173. doi: 10.4269/ajtmh.2010.10-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F.C., Shieh Y.S., van Duin J., Beekwilder M.J., Sobsey M.D. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl. Environ. Microbiol. 1995;61(11):3960–3966. doi: 10.1128/aem.61.11.3960-3966.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes B., Beale D.J., Dennis P.G., Cook S., Ahmed W. Cross-comparison of human wastewater-associated molecular markers in relation to fecal indicator bacteria and enteric viruses in recreational beach waters. Appl. Environ. Microbiol. 2017;83(8) doi: 10.1128/AEM.00028-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S.C. Human adenoviruses in Water: occurrence and health Implications: a critical review. Environ. Sci. Technol. 2006;40(23):7132–7140. doi: 10.1021/es060892o. [DOI] [PubMed] [Google Scholar]
- Jofre J., Lucena F., Blanch A.R., Muniesa M. Coliphages as model organisms in the characterization and management of water resources. Water. 2016;8(5):199. [Google Scholar]
- Johnston C., Ufnar J.A., Griffith J.F., Gooch J.A., Stewart J.R. A real-time qPCR assay for the detection of the nifH gene of Methanobrevibacter smithii, a potential indicator of sewage pollution. J. Appl. Microbiol. 2010;109(6):1946–1956. doi: 10.1111/j.1365-2672.2010.04824.x. [DOI] [PubMed] [Google Scholar]
- Keeler S.P., Dalton M.S., Cressler A.M., Berghaus R.D., Stallknecht D.E. Abiotic factors affecting the persistence of avian influenza virus in surface waters of waterfowl habitats. Appl. Environ. Microbiol. 2014;80(9):2910–2917. doi: 10.1128/AEM.03790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeler S.P., Lebarbenchon C., Stallknecht D.E. Strain-related variation in the persistence of influenza A virus in three types of water: distilled water, filtered surface water, and intact surface water. Virol. J. 2013;10 doi: 10.1186/1743-422X-10-13. 13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Tanaka Y., Kurbanov F., Elkady A., Abbas Z., Azam Z., Subhan A., Razza S., Hamid S., Jafri W., Shih J., Xia N., Takahashi K., Mishiro S., Mizokami M. Investigating an outbreak of acute viral hepatitis caused by hepatitis E virus variants in Karachi, South Pakistan. J. Med. Virol. 2011;83(4):622–629. doi: 10.1002/jmv.22036. [DOI] [PubMed] [Google Scholar]
- Kinzelman J.L., Singh A., Ng C., Pond K.R., Bagley R.C., Gradus S. Use of IDEXX colilert-18® and quanti-tray/2000 as a rapid and simple enumeration method for the implementation of recreational water monitoring and notification programs. Lake Reservoir Manag. 2005;21(1):73–77. [Google Scholar]
- Kishida N., Morita H., Haramoto E., Asami M., Akiba M. One-year weekly survey of noroviruses and enteric adenoviruses in the Tone River water in Tokyo metropolitan area, Japan. Water Res. 2012;46(9):2905–2910. doi: 10.1016/j.watres.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes — identification of potential viral indicators. Sci. Total Environ. 2014;488–489(Suppl. C):290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Oka T., Haramoto E., Katayama H., Takeda N., Katayama K., Ohgaki S. Detection and genetic analysis of human sapoviruses in river water in Japan. Appl. Environ. Microbiol. 2010;76(8):2461–2467. doi: 10.1128/AEM.02739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiulia N.M., Mans J., Mwenda J.M., Taylor M.B. Norovirus GII.17 predominates in selected surface water sources in Kenya. Food and Environmental Virology. 2014;6(4):221–231. doi: 10.1007/s12560-014-9160-6. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Fratini M., Libera S., Iaconelli M., Muscillo M. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super Sanita. 2012;48(4):397–406. doi: 10.4415/ANN_12_04_07. [DOI] [PubMed] [Google Scholar]
- Le Cann P., Ranarijaona S., Monpoeho S., Le Guyader F., Ferré V. Quantification of human astroviruses in sewage using real-time RT-PCR. Res. Microbiol. 2004;155(1):11–15. doi: 10.1016/j.resmic.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Lee C., Kim S.-J. The genetic diversity of human noroviruses detected in river water in Korea. Water Res. 2008;42(17):4477–4484. doi: 10.1016/j.watres.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Li T.-C., Chijiwa K., Sera N., Ishibashi T., Etoh Y., Shinohara Y., Kurata Y., Ishida M., Sakamoto S., Takeda N., Miyamura T. Hepatitis E virus transmission from wild boar meat. Emerging Infectious Disease journal. 2005;11(12):1958. doi: 10.3201/eid1112.051041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Goh S.G., Gin K.Y.H. Decay kinetics of microbial source tracking (MST) markers and human adenovirus under the effects of sunlight and salinity. Sci. Total Environ. 2017;574(Suppl. C):165–175. doi: 10.1016/j.scitotenv.2016.09.031. [DOI] [PubMed] [Google Scholar]
- Liang L., Goh S.G., Vergara G.G.R.V., Fang H.M., Rezaeinejad S., Chang S.Y., Bayen S., Lee W.A., Sobsey M.D., Rose J.B., Gin K.Y.H. Alternative fecal indicators and their empirical relationships with enteric viruses, Salmonella enterica, and Pseudomonas aeruginosa in surface waters of a tropical urban catchment. Appl. Environ. Microbiol. 2015;81(3):850–860. doi: 10.1128/AEM.02670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister P.D., Wolter D.J., Hanson N.D. Antibacterial-Resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009;22(4):582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., de Roda Husman A.M. Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Appl. Environ. Microbiol. 2005;71(3):1453–1461. doi: 10.1128/AEM.71.3.1453-1461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., van den Berg H.H.J.L., Rutjes S.A., de Roda Husman A.M. Presence of enteric viruses in source waters for drinking water production in The Netherlands. Appl. Environ. Microbiol. 2010;76(17):5965–5971. doi: 10.1128/AEM.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias C.A.R., Pereira I.A., Araujo M.d.S.d., Santos A.F.M., Lopes R.P., Christakis S., Rodrigues D.d.P., Siciliano S. Characteristics of Salmonella spp. isolated from wild birds confiscated in illegal trade markets, Rio de Janeiro, Brazil. BioMed Res. Int. 2016:1–7. doi: 10.1155/2016/3416864. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaig S., Griffith J., Harwood V.J. Association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution. Appl. Environ. Microbiol. 2012;78(18):6423–6432. doi: 10.1128/AEM.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaig S.M., Scott T.M., Lukasik J.O., Paul J.H., Harwood V.J. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 2009;75(11):3379–3388. doi: 10.1128/AEM.02302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.J. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 2010;140(3–4):256. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray T.Y., Mans J., Taylor M.B. First detection of human sapoviruses in river water in South Africa. Water Sci. Technol. 2013;67(12):2776. doi: 10.2166/wst.2013.203. [DOI] [PubMed] [Google Scholar]
- Nan Y., Wu C., Zhao Q., Zhou E.-M. Zoonotic hepatitis E virus: an ignored risk for public health. Front. Microbiol. 2017;8(2396) doi: 10.3389/fmicb.2017.02396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngazoa E.S., Fliss I., Jean J. Quantitative study of persistence of human norovirus genome in water using TaqMan real-time RT-PCR. J. Appl. Microbiol. 2008;104(3):707–715. doi: 10.1111/j.1365-2672.2007.03597.x. [DOI] [PubMed] [Google Scholar]
- Nguyen H.T.M., Le Q.T.P., Garnier J., Janeau J.L., Rochelle-Newall E. Seasonal variability of faecal indicator bacteria numbers and die-off rates in the Red River basin, North Viet Nam. Sci. Rep. 2016;6:21644. doi: 10.1038/srep21644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Wang Q., Katayama K., Saif L.J. Comprehensive review of human sapoviruses. Clin. Microbiol. Rev. 2015;28(1):32–53. doi: 10.1128/CMR.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., de Jong M.D., Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin. Microbiol. Rev. 2007;20(2):243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch D., Oh D.-Y., Wolf S., Dumke R., Schröter-Bobsin U., Höhne M., Röske I., Schreier E. Detection of enteric viruses and bacterial indicators in German environmental waters. Arch. Virol. 2005;150(5):929–947. doi: 10.1007/s00705-004-0467-8. [DOI] [PubMed] [Google Scholar]
- Rezaeinejad S., Vergara G.G.R.V., Woo C.H., Lim T.T., Sobsey M.D., Gin K.Y.H. Surveillance of enteric viruses and coliphages in a tropical urban catchment. Water Res. 2014;58(Suppl. C):122–131. doi: 10.1016/j.watres.2014.03.051. [DOI] [PubMed] [Google Scholar]
- Rigotto C., Victoria M., Moresco V., Kolesnikovas C.K., Corrêa A.A., Souza D.S.M., Miagostovich M.P., Simões C.M.O., Barardi C.R.M. Assessment of adenovirus, hepatitis A virus and rotavirus presence in environmental samples in Florianopolis, South Brazil. J. Appl. Microbiol. 2010;109(6):1979–1987. doi: 10.1111/j.1365-2672.2010.04827.x. [DOI] [PubMed] [Google Scholar]
- Rochelle-Newall E., Nguyen T.M.H., Le T.P.Q., Sengtaheuanghoung O., Ribolzi O. A short review of fecal indicator bacteria in tropical aquatic ecosystems: knowledge gaps and future directions. Front. Microbiol. 2015;6(308) doi: 10.3389/fmicb.2015.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochelle-Newall E.J., Ribolzi O., Viguier M., Thammahacksa C., Silvera N., Latsachack K., Dinh R.P., Naporn P., Sy H.T., Soulileuth B., Hmaimum N., Sisouvanh P., Robain H., Janeau J.-L., Valentin C., Boithias L., Pierret A. Effect of land use and hydrological processes on Escherichia coli concentrations in streams of tropical, humid headwater catchments. Sci. Rep. 2016;6:32974. doi: 10.1038/srep32974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeidi N., Gu X., Goh S.G., Xin C.L.Y., Gin K.Y.-H. Evaluating the efficacy of commercial kits for viral DNA/RNA extraction. Water Pract. Technol. 2017;12(1):80. [Google Scholar]
- Sanborn M., Takaro T. Recreational water–related illness: office management and prevention. Can. Fam. Physician. 2013;59(5):491–495. [PMC free article] [PubMed] [Google Scholar]
- Sano D., Pérez-Sautu U., Guix S., Pintó R.M., Miura T., Okabe S., Bosch A. Quantification and genotyping of human sapoviruses in the Llobregat river catchment, Spain. Appl. Environ. Microbiol. 2011;77(3):1111–1114. doi: 10.1128/AEM.01721-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper M., Durán A.E., Jofre J. Comparative resistance of phage isolates of four genotypes of F-specific RNA bacteriophages to various inactivation processes. Appl. Environ. Microbiol. 2002;68(8):3702–3707. doi: 10.1128/AEM.68.8.3702-3707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper M., Jofre J., Uys M., Grabow W.O.K. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of faecal pollution in South Africa and Spain. J. Appl. Microbiol. 2002;92(4):657–667. doi: 10.1046/j.1365-2672.2002.01600.x. [DOI] [PubMed] [Google Scholar]
- Shi J., Deng G., Liu P., Zhou J., Guan L., Li W., Li X., Guo J., Wang G., Fan J., Wang J., Li Y., Jiang Y., Liu L., Tian G., Li C., Chen H. Isolation and characterization of H7N9 viruses from live poultry markets — implication of the source of current H7N9 infection in humans. Chin. Sci. Bull. 2013;58(16):1857–1863. [Google Scholar]
- Silva C., Calva E., Maloy S. In: One Health: People, Animals, and the Environment. Atlas R.M., Maloy S., editors. American Society for Microbiology; Washington, DC: 2014. pp. 137–148. [Google Scholar]
- Souza V., Rocha M., Valera A., Eguiarte L.E. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 1999;65(8):3373–3385. doi: 10.1128/aem.65.8.3373-3385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallknecht D.E., Kearney M.T., Shane S.M., Zwank P.J. Effects of pH, temperature, and salinity on persistence of avian influenza viruses in water. Avian Dis. 1990;34(2):412–418. [PubMed] [Google Scholar]
- Stewart-Pullaro J., Daugomah J.W., Chestnut D.E., Graves D.A., Sobsey M.D., Scott G.I. F+RNA coliphage typing for microbial source tracking in surface waters. J. Appl. Microbiol. 2006;101(5):1015–1026. doi: 10.1111/j.1365-2672.2006.03011.x. [DOI] [PubMed] [Google Scholar]
- Steyer A., Torkar K.G., Gutiérrez-Aguirre I., Poljšak-Prijatelj M. High prevalence of enteric viruses in untreated individual drinking water sources and surface water in Slovenia. Int. J. Hyg Environ. Health. 2011;214(5):392–398. doi: 10.1016/j.ijheh.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Taubenberger J.K., Morens D.M. Influenza: the once and future pandemic. Publ. Health Rep. 2010;125(Suppl. 3):16–26. [PMC free article] [PubMed] [Google Scholar]
- Taylor M.B., Cox N., Vrey M.A., Grabow W.O.K. The occurrence of hepatitis A and astroviruses in selected river and dam waters in South Africa. Water Res. 2001;35(11):2653–2660. doi: 10.1016/s0043-1354(00)00551-0. [DOI] [PubMed] [Google Scholar]
- Teunis P.F.M., Moe C.L., Liu P.E., Miller S., Lindesmith L., Baric R.S., Le Pendu J., Calderon R.L. Norwalk virus: how infectious is it? J. Med. Virol. 2008;80(8):1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- USEPA . USEPA; Washington, DC: 2009. Fact Sheet: Final Third Drinking Water Contaminant Candidate List (CCL 3) [Google Scholar]
- USEPA . USEPA; Washington D.C.: 2001. Method 1602: Male-Specific (F+) and Somatic Coliphages in Water by Single Agar Layer (SAL) Procedure. [Google Scholar]
- USEPA . 2012. 2012 Recreational Water Quality Criteria. [Google Scholar]
- Vergara G.G.R.V., Goh S.G., Rezaeinejad S., Chang S.Y., Sobsey M.D., Gin K.Y.H. Evaluation of FRNA coliphages as indicators of human enteric viruses in a tropical urban freshwater catchment. Water Res. 2015;79(Suppl. C):39–47. doi: 10.1016/j.watres.2015.04.022. [DOI] [PubMed] [Google Scholar]
- Wang B., Zhang H., Chen Q., Chen Z. Full genome sequence of an avian influenza H5N1 virus isolated from the environment in Human province, China. Genome Announc. 2013;1(3) doi: 10.1128/genomeA.00088-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Long S.C., Das D., Dorner S.M. Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J. Water Health. 2011;9(2):265. doi: 10.2166/wh.2011.117. [DOI] [PubMed] [Google Scholar]
- Yampara-Iquise H., Zheng G., Jones J.E., Carson C.A. Use of a Bacteroides thetaiotaomicron-specific α-1-6, mannanase quantitative PCR to detect human faecal pollution in water. J. Appl. Microbiol. 2008;105(5):1686–1693. doi: 10.1111/j.1365-2672.2008.03895.x. [DOI] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W.H., Run J.-Q., Wei C.L., Soh S.W.L., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2005;4(1):e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.