Abstract
This study seeks to understand the general distribution of virome abundance and diversity in tropical freshwater ecosystems in Singapore and the geospatial distribution of the virome under different landuse patterns. Correlations between diversity, environmental parameters and land use patterns were analyzed and significant correlations were highlighted. Overall, the majority (65.5%) of the annotated virome belonged to bacteriophages. The percentage of Caudovirales was higher in reservoirs whereas the percentages of Dicistroviridae, Microviridae and Circoviridae were higher in tributaries. Reservoirs showed a higher Shannon-index virome diversity compared to upstream tributaries. Land use (urbanized, agriculture and parkland areas) influenced the characteristics of the virome distribution pattern. Dicistroviridae and Microviridae were enriched in urbanized tributaries while Mimiviridae, Phycodnaviridae, Siphoviridae and Podoviridae were enriched in parkland reservoirs. Several sequences closely related to the emerging zoonotic virus, cyclovirus, and the human-related virus (human picobirnavirus), were also detected. In addition, the relative abundance of PMMoV (pepper mild mottle virus) sequences was significantly correlated with RT-qPCR measurements (0.588 < r < 0.879, p < 0.05). This study shows that spatial factors (e.g., reservoirs/tributaries, land use) are the main drivers of the viral community structure in tropical freshwater ecosystems.
Keywords: Virome, Land use, Geospatial distribution, Diversity, Human-related viruses, Freshwater ecosystems
Graphical abstract
Highlights
-
•
Land use is the main driver of the viral community structure in surface waters.
-
•
66 viral families were detected and 66% of the annotated virome was bacteriophages.
-
•
Reservoirs showed a higher Shannon diversity (10.4) than upstream tributaries (8.4).
-
•
Sequences closely related to human picobirnavirus and cyclovirus were detected.
-
•
The relative abundance of PMMoV sequences was correlated with RT-qPCR measurements.
1. Introduction
Viruses are the most abundant living entities on Earth and all living entities are associated with at least one virus which can control microbial communities (Ackermann, 2003). Viral metagenomics have been reported widely in water cycles, especially marine waters, to evaluate the role of viruses in microbial diversity and biogeochemical cycling (Angly et al., 2006; Breitbart et al., 2002; Cassman et al., 2012; Culley et al., 2006).
Determining the factors in constructing the viral community is important for both understanding and manipulating ecosystems (Dinsdale et al., 2008). Factors shaping the viral community in freshwater ecosystems can include temporal factors, geospatial factors, natural disturbances (e.g., typhoon) and human activities (Djikeng et al., 2009; Emerson et al., 2012; Fancello et al., 2013; Ge et al., 2013; Hwang et al., 2017; López-Bueno et al., 2009; Skvortsov et al., 2016; Tseng et al., 2013).
Among all the factors, land use activities is a major factor in shaping waterborne viromes. Land use changes are the primary drivers of the viral community and a range of associated infectious waterborne disease outbreaks (Patz et al. 2004, 2008). In the water cycle, agriculture brings excess nutrients and agricultural chemicals to surface waters, causing oxygen depletion and increasing algal blooms (Foley et al., 2005). Urbanization degrades water quality through surface runoff, and human pathogenic viruses have been detected more frequently in watersheds with dominant urban and agricultural land cover (Corsi et al., 2014; Lenaker et al., 2017). Furthermore, human expansion into wildlife habitats or construction of zoos and animal parks provide opportunities for human-wildlife interactions, thus, increasing the risk for the possible transmission of zoonotic viruses to human populations (Patz et al., 2008). These different anthropogenic activities harbor diverse and distinct viral hosts, including bacteria, plants, wild animals and humans. It is hypothesized that the environment surrounding these reservoirs may favor distinct viral predators and further change the viral community characterization. However, up till now, studies of land use impacts on the virome community in freshwater ecosystems are still limited as they mainly rely on traditional methodology (culture-based method or qPCR/RT-qPCR), which focuses on limited human virus targets without considering the whole picture of the viral community in the water environment (Corsi et al., 2014; Lenaker et al., 2017). To date, there is no systematic report focusing on the geospatial distribution and diversity of viromes in natural surface waters and how they may be impacted by human activities and the effect of different land use. In addition, emerging viral pathogens of zoonotic origin could also be discovered through viral metagenomics, for which information is very limited.
Singapore is a highly urbanized island located in the tropics with an area of 719.1 km2, and a population of 5.61 million in 2016 (Department of Statistics Singapore, 2016). Water is a scarce resource in this country and a total of 17 reservoirs are used to collect rainwater and surface waters for potable water supplies. Increasingly, selected reservoirs are being used as focal points for sporting events (kayaking and dragon boating) and recreational activities so that the public can enjoy and appreciate water resources. As such, good water quality is needed to protect recreational users of these water bodies. Previous studies have detected waterborne viral pathogens (e.g., norovirus GI/GII, adenovirus, rotavirus and astrovirus) in Singapore water bodies using qPCR/RT-qPCR (Aw and Gin, 2011; Aw et al., 2009; Rezaeinejad et al., 2014). Thus, continued surveillance of these viral targets as well as a broader range of viral targets expanded to the virome community coupled with land use information, could provide important data and new dimensions for managing the safety of water resources.
For these reasons, understanding the geospatial distribution of viromes is needed. In this study, seven tropical reservoirs with diverse upstream land use functions (i.e., urbanized, agricultural and parkland areas) were examined. The viral community structure and virome populations specific to each of these environments were systematically investigated together with the characteristics of the watershed. By conducting a complete virome analysis of these freshwater ecosystems (especially viral pathogens and fecal viral indicators), a comprehensive picture of the links between the virome community structure and specific land use activities could be elucidated and thus, used to conduct risk assessment of associated waterborne disease. This information would be important for environmental management at a macroscopic level to protect public health.
Thus, the objectives of this study were to: 1) investigate the overall virome distribution and diversity in diverse freshwater ecosystems (reservoirs/tributaries) in a tropical environment, 2) compare the virome community based on the different land use patterns, 3) assess the extent of human-related pathogenic viruses in surface waters, especially emerging zoonotic and human-related viruses, which may have been undetected before.
2. Materials and methods
2.1. Site description and sample collection
A total of seven reservoirs and three catchments were sampled in Singapore during January (Northeast Monsoon) and April (Inter-Monsoon period), 2015 (Table S1). In total, 19 sampling points were surveyed, comprising of 10 locations in the reservoirs and 9 locations in the tributaries. Only reservoirs 1, 2 and 4 had corresponding tributary sampling points. The study sites were divided into 3 categories based on their geospatial characteristics: urbanized, agricultural and parkland areas (Table 1 ). Apart from storing water and preventing flood control, some of the reservoirs (i.e., R1-3, 6–7) also catered for recreational activities such as kayaking, dragon boating and water skiing. All the upstream tributary sites were designated as non-recreational areas.
Table 1.
Landuse percentage and population density in different reservoirs and catchments.
| Sampling location | Landuse percentage |
Population density (people/km2) | ||||
|---|---|---|---|---|---|---|
| Residential | Urban | Green | Agricultural | |||
| Reservoirs (10) |
Site 1 | 38.1 | 25.9 | 35.9 | 0 | 7147.8 |
| Site 2 | 43.5 | 42.7 | 13.8 | 0 | 12280.0 | |
| Site 3 | 41.7 | 43.3 | 15 | 0 | 4758.5 | |
| Site 4 | 16.8 | 9.8 | 62.8 | 10.6 | 5.1 | |
| Site 5a | 0 | 8.68 | 91.32 | 0 | 0.3 | |
| Site 5b | 0 | 3.14 | 96.86 | 0 | 0.3 | |
| Site 5c | 0 | 3.64 | 96.13 | 0 | 0.3 | |
| Site 6a | 39.15 | 33.16 | 27.69 | 0 | 13332.1 | |
| Site 6b | 47.8 | 38.8 | 13.4 | 0 | 13332.1 | |
| Site 7 | 0 | 0 | 100 | 0 | 0.3 | |
| Tributaries (9) |
Site 1a | 59.33 | 29.39 | 11.28 | 0 | 457.7 |
| Site 1b | 46.65 | 43.87 | 9.48 | 0 | 6133.0 | |
| Site 1c | 33.06 | 42.89 | 24.05 | 0 | 12128.8 | |
| Site 2a | 61.97 | 27.79 | 10.24 | 0 | 19492.9 | |
| Site 2b | 45.88 | 34.24 | 19.88 | 0 | 19492.9 | |
| Site 2c | 32.44 | 50.16 | 17.4 | 0 | 19492.9 | |
| Site 4a | 0 | 0 | 47.58 | 52.42 | 5.1 | |
| Site 4b | 0 | 1.29 | 84.05 | 14.66 | 5.1 | |
| Site 4c | 0 | 0 | 32.58 | 67.42 | 5.1 | |
2.2. Measurements of environmental parameters
Physical-chemical parameters, including temperature, pH, turbidity, conductivity, salinity, total dissolved solids (TDS) and dissolved oxygen (DO) were measured on site using a Hanna Meter probe (HI9828 Multiparameter Meter; Hanna Instruments). 24-hour rainfall data were obtained from the Singapore Historical Daily Records ( http://www.weather.gov.sg/climate-historical-daily/ ).
2.3. Viral concentration and nucleic acids extraction
2.3.1. Primary concentration, secondary concentration and viral nucleic acids extraction
30-L water samples were collected from each sampling location in three 10-L carboys. The raw-water sample was immediately transported to the lab and concentrated through a hollow fiber ultrafiltration unit with blocking and elution buffer (Hemoflow Fresenius HF80S, Germany) to a final volume of 600 mL. The hollow fiber ultrafilter was purged with nanopure water for 5 min and pre-treated with 500 mL of blocking solution (0.1 g of NaPP in 1 L of nanopure water) for 15 min. After that, the water sample was recirculated until a final volume of approximately 200–250 mL was reached. An elution step was carried out by recirculating around 300 mL of elution buffer (0.1 g of NaPP, 5 mL of Tween 80, and 10 μl of Antiform in 1 L of nanopure water) for 5 min. Both retentate and eluent were combined to a final volume of 600 mL during primary concentration. 200 mL of primary concentrate was further processed to enrich viral particles in a secondary concentration step through polyethylene glycol (PEG) precipitation (10% PEG 8000 (w/v) and 0.3M NaCl) after pH adjustment (pH = 7.2) (Jaykus et al., 1996). After incubation of the mixture at 4 °C for 18 h followed by 14,000 g for 45 min, the pellet was dissolved in 10 mL of phosphate buffer saline (PBS, pH7.2) with an equal volume of chloroform. After centrifugation at 3000 g for 45 min, the supernatant was filtered through a 0.22 μm sterile syringe and further concentrated to a final volume of 1 mL using a 30 kDa ultracentrifugal filter device (Merck Millipore, Ireland). After primary and secondary concentration, 140 μl of viral nucleic acids (DNA and RNA) was extracted using the QIAamp Viral RNA Mini kit (QIAGEN, Hilden, Germany) and then stored at −80 °C (Saeidi et al., 2017).
2.3.2. Reverse transcription, qPCR/RT-qPCR analysis
In order to quantify the human pathogens in reservoirs and their tributaries using qPCR/RT-qPCR, 14 viral targets were performed to include: (1) Four genotypes of male-specific coliphages (FRNA GI-FRNA GIV); (2) Ten human viral pathogens and (3) One plant viral pathogen/microbial indicator (i.e., PMMoV) (Table S3). The majority of targets belonged to Group IV ssRNA (+) except for adenovirus (group I dsDNA) and rotavirus (group III dsRNA). The details of qPCR/RT-qPCR primers and probes are listed in Table S4. The extracted viral nucleic acids were reverse transcribed using ImProm-II Reverse Transcription System for detecting RNA viruses following manufacturer's instructions (Promega, USA). RT products were stored at −20 °C for later analysis. qPCR/RT-qPCR was carried out in a StepOnePlus Real-Time PCR system (Applied Biosystems, USA) using FastStart Universal Probe Master (Rox) (Roche, Germany) following MIQE guidelines (Bustin et al., 2009).
2.4. Random amplification and sequencing of viromes
The extracted nucleic acids were reverse transcribed and amplified to obtain a sufficient quantity of DNA and cDNA (Wang et al., 2002). To ensure there was no microbial contamination, the negative control was run in 1% agarose gel and ascertained that no DNA gel band was present in the negative control lane. After purification of viral DNA and cDNA, samples were sent to SCELSE (Singapore Centre on Environmental Life Sciences Engineering). In total, 38 libraries and one negative control library using Illumina TruSeq Nano DNA Library Kit were constructed. The sequencing libraries with a corresponding insert size and adapters were prepared accordingly as previously described (Ng et al., 2017). The Illumina's PhiX control library was used as standard. The libraries were then pooled and sequenced in one lane with equimolar concentrations on an Illumina HiSeq2500 sequencer in rapid mode at a final concentration of 10 pM and a read-length of 250 bp paired-end (V2 sequencing reagents). The overview of the virome datasets is detailed in Table S6. The effect of the negative control is ignored, as explained in the supplementary information.
2.5. 16s rRNA gene amplicon sequencing and processing
Meanwhile, 16s rRNA gene amplicon sequencing was applied to the same batch of samples at Sites 1–7 to characterize the bacteria community. A total volume of 10 mL water samples from primary concentration was centrifuged at 10,000 g for 20 min. After removing the supernatant, the pellet was transferred to a PowerSoil® Bead Tube (Mo Bio Laboratories, Inc., Carlsbad, CA) for DNA extraction with a final volume of 120 μl. Genomic DNA (gDNA) was amplified for the marker gene targeting the 16S rRNA variable regions V6 to V8 using the universal pyrotaq primer set (926wF and 1392R) (Rinke et al., 2014). Samples were sequenced in the University of Illinois, Research Resources Center - DNA Services Facility and sequence preprocessing as described previously (Te et al., 2017). The biological data (subsampled OTUs) were square-root transformed to reduce the dominance of major OTUs before putting into PRIMERv7 for PCoA and correlation analyses.
2.6. Processing of viromes
The sequencing data were trimmed to remove adaptors, low quality reads, “Primer B” sequences which were used for random amplification and Phix reads following the BBtools instruction (version 35.43). Reads were then de novo assembled into contigs by CLC Genomics (version 8.0.3) through cross-assembly strategy. The assembly setting in CLC Genomics is: minimum contig length of 1000 bp and similarity fraction 0.95. Contigs were then uploaded into the Metavir2 and Virsorter pipeline. Briefly, Metavir2 pipeline will first extract open reading frames (ORFs) from contigs through the MetaGeneAnnotator. All predicted ORFs were then compared to the National Centre for Biotechnology Information (NCBI) RefseqVirus protein database (2015.01.05) using a reference-dependent taxonomic analysis, with the threshold of 50 bit score and an e-value cut-off of 10−3. After getting the best BLAST hit affiliation of each predicted gene, annotation of each contig was made based on the lowest common ancestor (Roux et al., 2014). In addition, Metavir2 pipeline will compute the genes affiliated to the PFam database for function analysis. Virsorter identified category 1 viral signal (pretty sure) and category 2 viral signal (quite sure) from the assembled sequence and the viral signal included bacterial and archaeal viruses (Roux et al., 2015).
In particular, human-related viruses were extracted from Metavir2 results and further examined to search against NCBI non-redundant nucleotide database (downloaded in June 2016) through BLASTn search for nucleotide similarity and query coverage (E-value <1E-5).
2.6.1. Quantification of virome
In order to quantify the relative abundance of each contig within different libraries, reads were remapped to contigs using Novoalign (Hercus, 2012). This absolute matrix was used for quantifying the relative abundance of taxonomy assignment. Since every contig has its weight in quantifying virus concentration, to standardize and quantify every contig's relative abundance between each library, RPKM (Reads per kilobase of “transcript” per million mapped reads) was used after remapping trimmed reads to each contig across different libraries (Mortazavi et al., 2008; Reyes et al., 2015).
RPKM was calculated as followed:
This matrix was equivalent to an OTU table for PCoA analysis.
2.6.2. Viral α-diversity
The non-normalized matrix of reads mapping to each contig per sample (absolute reads matrix) was used and rarefied to 245,176 reads per sample (the smallest sample size in absolute reads matrix of taxonomy affiliated viruses was 245,176) to calculate α-diversity. “Observed species” and “Shannon diversity index” were calculated for each sample using MacQIIME (version 1.9.1) with the iteration set at 10.
2.6.3. PCoA analysis
Multivariate analysis of viral community composition under different treatments (e.g., landuse, reservoir/tributaries, Monsoon/Inter-Monsoon season) was conducted using ANOSIM in PRIMERv7 and PERMANOVA+. The similarity of each pair of samples in terms of biotic characteristics was calculated using Bray-Curtis similarity. RPKM of the total contigs and of the virsorter category 1 and 2 phages were log plus one transformed before inputting into PRIMERv7 for PCoA and correlation analyses.
2.6.4. Mapping reads to reference genome
All the reference genomes of qPCR/RT-qPCR viral targets were downloaded from NCBI and accession numbers are detailed in Table S5. Mapping was performed using the CLC Genomics Workbench 8.0.3 based on the default setting (mismatch gap = 2, with the linear gap cost, length fraction: 0.5, similarity fraction is based on the specific targets, at medium (50%) medium-high (70%) and high (90%, 95% or 99%) three levels) (Table S5). The threshold of three levels could represent the nucleotide variation within groups of these viral targets (de Graaf et al., 2016; Doceul et al., 2016). The proportion of mapping reads equals the number of mapping reads in each library divided by the total reads in the library.
2.7. Geospatial analyses
All geospatial analyses were performed using ArcGIS v.10 (ESRI, Aylesbury, UK) and the percentage of different land use in different reservoirs and their tributaries are detailed in Table 1. Land-use shape files were obtained from the Singapore land authority and PUB (Singapore's National Water Agency). Four different layers of map were used for analysis: (i) Catchment land-use shape files, (ii) River shape files, (iii) Sub-catchment shape files and (iv) Drain line maps. WGS_1984_Web_Mercator_Auxiliary_Sphere was used as the projection coordinate system while GCS_WGS_1984/SVY21 Singapore was used as the geographic coordinate system.
Population data was downloaded from the Singapore Statistics website (http://www.singstat.gov.sg/statistics/browse-by-theme/geographic-distribution, Basic demographic characteristic 2015) and the population density was calculated based on the total population within the planning area divided by the planning area which covered the drainage area for that sampling point. The shape file of master plan of 2014 planning boundary no sea was used as the reference map for the planning area classification (https://data.gov.sg/dataset/master-plan-2014-planning-area-boundary-no-sea, Department of Statistics Singapore).
2.8. Statistical tests and plots
All the statistical tests including one-way ANOVA and Spearman rank correlations were performed using IBM SPSS 23 (IBM, Portsmouth, UK). Environmental parameters were log plus one transformed before correlating with α-diversity index as the raw data were right skewed. Graphs were plotted using Microsoft Excel (2013) and OriginPro 2017 (OriginLab, Northampton, MA).
2.9. Data deposition
Virome datasets were deposited in NCBI Sequence Read Archive (SRA) under accession No. SRR5995660-5995697. 16s rRNA sequences were deposited in NCBI SRA under accession No. SRR5902299-5902336.
3. Results
3.1. General water quality and land use classification
Environmental parameters of the study sites (i.e., reservoirs and tributaries points) are illustrated in Table S2. The temperature remained relatively stable for all the sampling points, ranging from 27 to 29.4 °C with a mean value 28.6 °C. pH ranged from 6.5 to 8.5 with a mean value 7.6. Other environmental parameters varied more, including conductivity (139.5–545.5 μS/cm, mean: 316.85 μS/cm), TDS (69.8–273 ppm, mean: 165 ppm), DO (1.69–10.3 ppm, mean: 6.48 ppm) and turbidity (3.5–22.7 FNU, mean: 13.8 FNU). The 24-hour rainfall and 5-day rainfall ranged from 0 to 23.9 mm (mean: 5.6 mm) and 0–13.3 mm (mean: 4.8 mm), respectively. The 24-hour rainfall and 5-day rainfall had higher mean values (i.e., 9.69 mm and 5.19 mm, respectively) during the Northeast Monsoon (Jan) and lower mean values (i.e., 1.42 mm and 4.45 mm, respectively) during the Inter-Monsoon (Apr).
Table 1 summarizes the land use percentage and the population density for the 7 reservoir catchments. To better evaluate land use impacts on the viral community, the sampling sites were classified into three categories: urbanized areas, agriculture areas and parkland areas. Sites 1, 2 and 3 included approximately 65–90% residential and urban land use, and as such can be regarded as urbanized areas. Site 4 had less than 30% of residential and urban areas, with about 33–84% green and 11–67% agricultural areas. Site 4 was the only sampling location with a reasonable percentage of agriculture area; therefore, Site 4 was considered as agricultural. Site 5 and Site 7 had similar land use percentage with more than 95% of the land covered with green areas; thus, it was considered as parkland. Site 6 included 80% of residential and urban categories. However, unlike Sites 1–4, there was no drain directly connected to the reservoir at this site and hence, the chance for possible viral contamination was relatively small. Therefore, Site 6 was classified as parkland areas as well as Site 5 and Site 7. The land use category clustering of the seven different sites illustrated the reasonability of our land use category classification (Fig. S1).
The population density was obtained from the Singapore Department of Statistics. Among the sample locations, Sites 1, 2 and 6 had the highest population densities (>5000 people/km2) while Sites 4, 5 and 7 had the lowest (<10 people/km2).
3.2. Overall virome description
A total of 1.156 hundred million high-quality Hiseq sequencing reads of 38 samples were obtained from different reservoirs and their tributaries in Singapore during the Monsoon and Inter-Monsoon periods (Table S6). These were assembled across different libraries and 156,706 contigs were generated with an average length of 2035bp (N50 = 2020bp). On average, 58.67% ± 12.7% reads mapped back to the assembled contigs per library (min: 34.20%, max 85.67%).
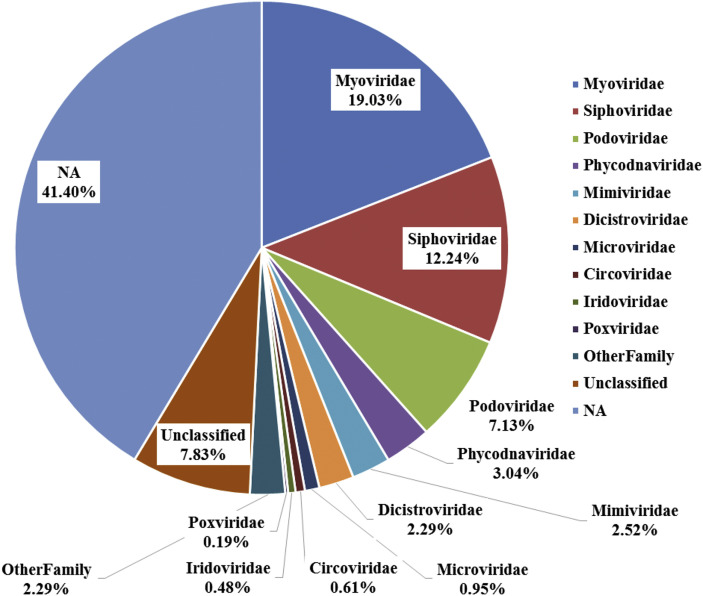
Of the 156,706 contigs, 65,598 contigs were annotated using Metavir2 pipeline. Based on the Absolute Reads matrix (the number of reads mapped to each family in each sample) and the Metavir2 annotation result, on average, 58.6% was annotated and a total of 66 families were identified (Fig. 1 ). The majority (38.4%) of these reads belonged to the Caudovirales (Myoviridae, Siphoviridae and Podoviridae). This was followed by reads for Phycodnaviridae and Mimiviridae (whose hosts belong to algae and amoeba) which were retrieved at percentages of 3.0% and 2.5%, respectively. Apart from these major viral families, other families such as Dicistrovidae, Microviridae, Circoviridae, Iridoviridae and Poxviridae were also present in our study. 7.8% were unclassified viral sequences and more than 40% were unassigned (NA) sequences.
Fig. 1.
Relative abundance of taxonomic assignment of virome based on family level using Absolute Reads matrix from Metavir2 pipeline. (Abbreviations: NA, not assigned sequences).
Function analysis of the 156,706 viral contigs identified 483,461 genes and 185,519 (38.4%) of the genes encoded hypothetical proteins or conserved hypothetical proteins. The 30 most retrieved PFAM annotations showed that phage terminase enzyme, phage portal and phage integrase were responsible for the phage function (Table S7).
3.3. Taxonomy assignment of virome in both reservoirs and their tributaries
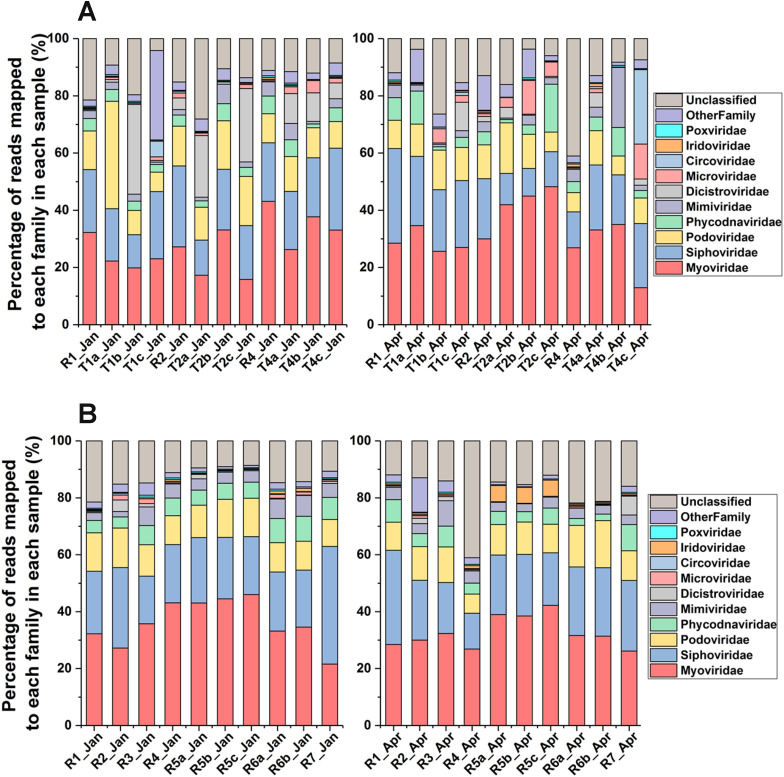
Fig. 2 A shows the taxonomic percentage of virome in 3 reservoirs and their tributaries (Sites 1, 2 and 4). Fluctuation could be observed within tributaries at the same sampling site. In contrast, virome composition in reservoirs was similar across several sampling points in Singapore and shows a more stable pattern than the tributaries (Fig. 2B). On average, Caudovirales accounts for 59.8% among all the annotated viruses in all the tributaries (Sites 1, 2 and 4) with a proportion of 58.1% and 61.7% in Jan and Apr, respectively. For the reservoirs, Caudovirales comprises an average of 70.65% among all the annotated viruses with a proportion of 72.9% and 67.8% in Jan and Apr, respectively. Note that the percentage of Caudovirales was higher in all the reservoirs than in all the tributaries. In contrast, the percentage of Dicistroviridae was higher in tributaries (11.8%) than in reservoirs (1.1%) in Jan (Tables S8 and S9). The percentage of Microviridae and Circoviridae in tributaries was approximately 4.6% (reservoirs: 0.34%) and 3.2% (0.21%), respectively. The shift in composition was observed as the percentage of Caudovirales decreased while the percentage of Dicistroviridae, Microviridae and Circoviridae subsequently increased, as locations changed from reservoirs to tributaries.
Fig. 2.
Taxonomic percentage of annotated sequences in Metavir2 at (A) Sites 1, 2 and 4 in both reservoirs and their tributaries (B) Sites 1–7 in reservoirs in Jan, Apr 2015 based on absolute reads matrix. "R" and "T" stand for Reservoir and Tributary, respectively.
Viral communities in different reservoirs in terms of land use impact were compared and the majority of viral communities were largely conserved and stable at the family level with Myoviridae, Siphoviridae and Podoviridae as the main family level, and small differences observed in Dicistroviridae and other families (Fig. 2B).
3.4. α-diversity and its correlation with environmental parameters and land use
Both the richness and diversity (observed species and Shannon diversity) was calculated on the taxonomically affiliated viruses from the subsampled absolute reads matrix. The species richness of viral samples from reservoirs with tributaries comprising mostly agriculture and urbanized areas had a similar amount of species while those from parkland areas showed lower viral richness, which was reasonable as parkland areas would generally receive less viral contamination. It was also noted that the viral species richness from tributaries was also lower than their corresponding reservoirs. The average Shannon diversity of reservoirs and tributaries was computed as 10.4 and 8.4 respectively and a one-way ANOVA test showed that the Shannon-diversity of tributaries was significantly smaller than those from reservoirs (P < 0.05). This indicated that tributaries were more likely to be dominated by selected species compared to the reservoirs (Fig. S2) and this may be related to the viral host's presence (e.g., bacteria, archaea, algae, insects, plants, animals). Future studies could explore this further.
A Spearman rank correlation was carried out between richness and diversity (observed species and Shannon index calculated by MacQIIME), environmental parameters and the land use pattern (Table 2 ). The correlation coefficient showed that the richness index of observed species was significantly positively correlated with rainfall and agriculture while the Shannon index was negatively correlated with pH.
Table 2.
Spearman rank correlations between α-diversity index, environmental parameters and landuse.
| Environmental parameters and landuse | Correlation coefficient a |
||
|---|---|---|---|
| pH | One-day Rainfall | Agriculture | |
| Observed species | .329b | .342b | |
| Shannon index | -.399b | ||
Only significant correlations are shown.
Correlation is significant (p < 0.05, 2-tailed t test).
3.5. β-diversity and its correlation with land use impacts
When looking at the β-diversity of the microbial community (<0.22 μm) structure, the distribution of RPKM (<0.22 μm microbial community) varied significantly between land use (PERMANOVA, p = 0.0001, Pseudo-F = 3.29) and between reservoirs and tributaries (PERMANOVA, p = 0.0002, Pseudo-F = 3.35). However, no significant difference was observed between Monsoon and Inter-Monsoon periods (PERMANOVA, p = 0.0923, Pseudo-F = 1.47) and there was no significant cross linking between the three factors (Table S10). To further evaluate the influence of land use category (urban, agriculture and parkland areas) in shaping the viral community (ANOSIM global test, R = 0.45, significance level = 0.1%), pairwise tests in the ANOSIM test showed that the major differences were between urban and parkland (R = 0.765, significance level = 0.1%) and between agriculture and parkland (R = 0.811, significance level = 2%). However, the microbial community (<0.22 μm) from the urbanized category appeared to have a similar structure to that of the agriculture category (R = 0.051, significant level = 35.7%).
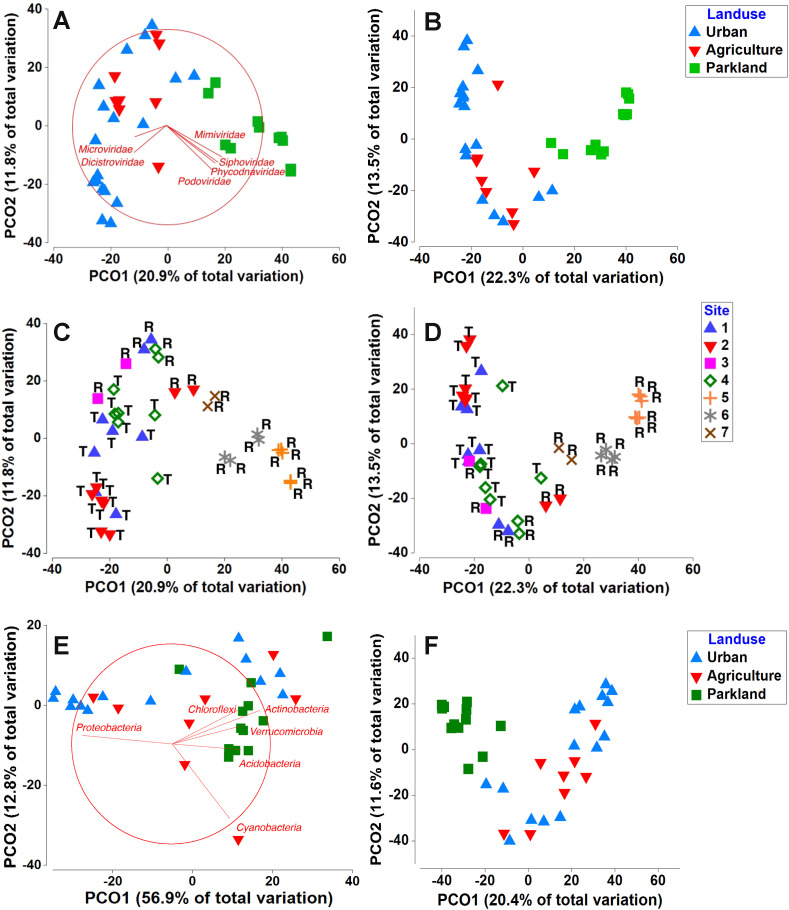
PCoA analysis suggested that land use had an impact on both microbial community (<0.22 μm) composition and the virsorter category 1 and 2 phage community. Both shared similar patterns where parkland samples grouped together while samples from urbanized and agricultural areas were dispersed in the left corner (Fig. 3 A and B). This pattern was consistent with the bacterial community geospatial distribution (Fig. 3E and F). Mimiviridae, Phycodnaviridae, Podoviridae and Siphoviridae were enriched in parkland areas (Spearman R = 0.36–0.52 to PCO1) whereas Dicistroviridae and Microviridae were enriched in urbanized areas (Spearman R = −0.37 to PCO1) (Fig. 3A). Acidobacteria, Actinobacteria and Verrucomicrobia were enriched in parkland areas (Spearman R = −0.70 to 0.90 to PCO1), Cyanobacteria were enriched in agricultural areas (R = −0.75 to PCO2) whereas Proteobacteria were enriched in urbanized and agricultural areas (Spearman R = −0.90 to PCO1) (Fig. 3E), comparable to a recent study conducted in Singapore showing that Proteobacteria were enriched in horticultural and residential samples (Nshimyimana et al., 2017).
Fig. 3.
PCoA analysis of (A) Metagenome (<0.22μm), (B) Virsorter Cat 1, Cat 2 phages in samples from different land use, (C) Metagenome (<0.22μm), (D) Virsorter Cat 1, Cat 2 phages in samples in all sites (labels in plot means R: Reservoir, T: Tributary), (E) 16s rRNA gene amplicon sequencing distinguished by Phylum level and (F) 16s rRNA gene amplicon sequencing distinguished by OTU level based on Bray-Curtis similarity index. In (A), individual viral family (only dominant viral families (>0.1% of all the virome) are selected) contributing to variation were determined by Spearman rank correlation (R > 0.4) to the first two PCO axis and are represented by vectors and in (E), individual bacteria phylum contributing to variation were determined by Spearman rank correlation (R > 0.7) to the first two PCO axis and are represented by vectors.
The microbial community (<0.22 μm) and virsorter category 1 and 2 phage community in the seven reservoirs both showed distinctly different distributions within reservoirs/tributaries and within different sites (Fig. 3C and D). Different sampling points of reservoirs at Sites 5 and 6 did not show any difference. For Sites 1, 3 and 4, the microbial community shared a similar contig profile even though they did not share the same land use pattern (i.e., Sites 1 and 3: urbanized area; Site 4: agriculture area). Noticeably, urbanized area Site 2 showed a slightly different pattern when compared with the other three reservoirs, perhaps due to potential different viral host communities (Site 2 is near the construction sites). The microbial community in the tributaries for Sites 1, 2 and 4 were much more variable spatially and temporally, compared to the reservoir microbial community.
3.6. Human and plant related viruses
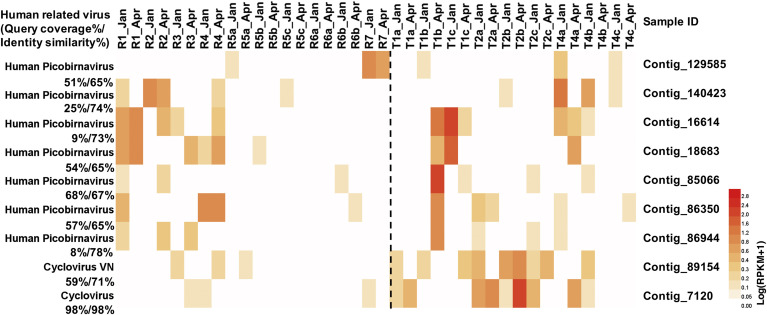
In our study, 7 contigs were affiliated to human picobirnaviruses and 2 contigs were affiliated to cycloviruses. They had a higher relative abundance in the urbanized and agricultural reservoirs and all the tributaries (Fig. 4 ). Possible viral contamination could be from urban drains including those from housing estates, sewer leakage and construction sites.
Fig. 4.
Heatmap of human-related virus across all the sampling points (log(RPKM+1)).
Note: Samples from reservoirs and tributaries are on the left and right part of the dashed line, respectively.
Although previous studies have detected enteric viruses in urban catchments in Singapore (Aw and Gin, 2011; Aw et al., 2009; Liang et al., 2015; Rezaeinejad et al., 2014), our metagenomics sequencing was unable to find contigs assigned to the enteric viruses commonly detected in Singapore surface waters (e.g., norovirus GI, norovirus GII, adenovirus, rotavirus). However, several fecal indicators including PMMoV, FRNAGI (MS2) and FRNAGIII (QBeta), were detected in our samples. Interestingly, in our study, the 6 PMMoV-affiliated contigs had a higher nucleotide similarity (97.25%–99.82%) with the average query coverage of 98.5% (Table 3 ), suggesting that PMMoV is a highly conserved virus with a lower evolution rate in the surface water environment. This result is similar with a recent study, where PMMoV detected in 85% of surface water samples shared 99–100% nucleotide identity with PMMoV reference genome (Rosiles-González et al., 2017).
Table 3.
Contigs affiliated as PMMoV.
| Contig_ID | Metavir2 taxonomy assignment | GeneBank Accession | Contig Length | BLASTn Identity (%)/E-value |
Query Coverage (%) | |
|---|---|---|---|---|---|---|
| PMMoV_Contig1 | 300 | Virgaviridae; Tobamovirus; Pepper mild mottle virus | LC082099.1 | 1707 | 99%/0 | 100 |
| PMMoV_Contig2 | 301 | Virgaviridae; Tobamovirus; Pepper mild mottle virus | KX399389.1 | 1708 | 99%/0 | 100 |
| PMMoV_Contig3 | 327 | Virgaviridae; Tobamovirus; Pepper mild mottle virus | KX883742.1 | 2277 | 99%/0 | 97 |
| PMMoV_Contig4 | 328 | Virgaviridae; Tobamovirus; Pepper mild mottle virus | AB000709.2 | 2292 | 99%/0 | 97 |
| PMMoV_Contig5 | 1112 | Virgaviridae; Tobamovirus; Pepper mild mottle virus | LC082099.1 | 1800 | 99%/0 | 100 |
| PMMoV_Contig6 | 1113 | Virgaviridae; Tobamovirus; Pepper mild mottle virus | KU646837.1 | 1777 | 100%/0 | 97 |
3.7. Correlation of metagenomics data with molecular assays for selected viral targets
qPCR/RT-qPCR was conducted to detect and quantify the concentration of the common enteric and zoonotic viruses in the surveyed communities. 14 viral targets (9 viral pathogens, 4 F+ male-specific coliphages and 1 plant virus) were measured and the detection frequency was between 0 and 65.79% across all the samples (Table S5). The 9 viral pathogens detected in this study using qPCR/RT-qPCR included enteric viruses (adenovirus, astrovirus, rotavirus, norovirus GI, norovirus GII, enterovirus and aichi virus) and other zoonotic viruses (e.g. hepatitis E virus, saporo virus).
Reads mapping to human-infective viruses were rare and were not observed to assemble into contigs. High-quality reads from different libraries were aligned to the viral pathogen database (ViPR/IRD) and mapped to viral reference genomes in NCBI (Table S5). Few reads (1–9 reads per library) were assigned to the human-infective viruses norovirus and saporo virus using both VIP pipeline and standalone pipeline (Results not shown) (Li et al., 2016), and 0–23 reads per library were mapped to human viral pathogens reference genome at the 90–99% nucleotide similarity level. No significant correlations were observed between qPCR measurements and read mapping for these viruses. These results suggest that metagenomics is less sensitive at detecting low-abundance viral pathogens against a background of a large proportion of bacteriophages.
A total of 3 targets were found in both the metagenomics (contigs) and qPCR data. Of these, PMMoV showed a good spearman rank correlation (p < 0.05) (Table 4 ). Spearman rank correlation was carried out between the PMMoV molecular results and the corresponding metagenomics data in terms of contigs level and reads level (Table 5 ). In terms of contigs level, all 6 PMMoV-affiliated contigs showed a significant good correlation between molecular results and metagenomics data (0.588 < r < 0.795, p < 0.05). In terms of reads level, out of the 38 libraries, 23 libraries (60.5%) had reads mapping to the PMMoV reference genome (NC_003630.1) with 99% identity to the reference genome. Based on all the 38 samples, the Spearman rank correlation coefficient was 0.879 (p < 0.05) (Table 5).
Table 4.
Summary of Spearman rank correlation between metagenome and RT-qPCR on indicators PMMoV, MS2 and Qbeta (N = 38).
| Targets | Number of contigs | Number of contigs having significant correlation (p < 0.05) | Number of contigs having significant and good correlation (r > 0.6, p < 0.05) | Correlation coefficient range (p < 0.05) |
|---|---|---|---|---|
| PMMoV | 6 | 6 | 5 | 0.588–0.795 |
| FRNAGI (MS2a) | 12 | 1 | 0 | 0.358 |
| FRNAGIII (QBetaa) | 18 | 10 | 2 | 0.355–0.658 |
MS2 belongs to the group of FRNA GI (animal-specific indicator) and QBeta belongs to the group of FRNA GIII (human-specific indicator).
Table 5.
Spearman rank correlation between qPCR (GC/100 ml) and metagenomics data (RPKM) on PMMoV (N = 38).
| Correlation coefficient |
|||||||
|---|---|---|---|---|---|---|---|
| PMMoV_contig1 | PMMoV_contig2 | PMMoV_contig3 | PMMoV_contig4 | PMMoV_contig5 | PMMoV_contig6 | PMMoV reads mapped to contigs per library (%) | |
| PMMoV-qPCR | .795a | .588a | .653a | .721a | .785a | .639a | .879a |
Correlation is significant (p < 0.01, 2-tailed t-test).
4. Discussion
This study is the first to correlate land use impact with freshwater water viromes in an urban tropical environment using Illumina Hiseq sequencing. It is noteworthy that metagenomics of DNA viruses and RNA viruses have been often considered separately and that studies of DNA viruses are far more than RNA virome analysis. This disparity is largely a result of technical challenges presented by RNA metagenomics as RNA is fragile and needs to be reverse transcribed into DNA. Thus up till now, the majority of the virome metagenomics papers studied either DNA or RNA (but not both). Moreover, the amplification protocol reported in these earlier studies may have influenced the final outcome (López-Bueno et al., 2009; Rodriguez-Brito et al., 2010). For DNA viruses, the GenomiPhi Kit (GE Healthcare) has been used for amplification through DNA polymerase phi29 but it was discovered that it has a bias towards single-stranded circular DNA viruses. For RNA virome analysis, random priming mediated sequence independent single primer amplification (RP-SISPA) was used in Lake Needwood, Maryland (Djikeng et al., 2009). Wang et al. (2002) developed a protocol capable of amplifying DNA and RNA viruses simultaneously. This was subsequently tested in reclaimed water, plasma samples, wastewater and ballast water (Bibby and Peccia, 2013; Kim et al., 2015; Rosario et al., 2009; Wylie et al., 2012). RNA viruses account for more than 70% of the viral pathogens due to their adaptive abilities (mutations, recombinations or reassortments) to infect novel hosts easily (Temmam et al., 2014). In our study, 92.3% of viruses are DNA viruses whereas 7.6% are RNA viruses among annotated viral families. Among all the viral pathogens detected by metagenomics, 10 contigs affiliated to human picobirnaviruses are DNA viruses whereas 2 contigs affiliated to cyclovirus are RNA viruses. The simultaneous application of combining both DNA and RNA metagenomics enabled the identification of a much larger numbers of viral sequences, especially for human-related viruses. This was the approach taken in our study here.
4.1. Geospatial distribution of virome
In this geospatial metagenomics study, we identified virome abundance and diversity in different reservoirs and tributaries. Due to insufficient viral genome sequencing information in the NCBI, 41.4% of the virome genome had no significant hits and bacteriophages were the most abundant organisms in the matrix, which is in agreement with previous studies of surface water viromes (Mohiuddin and Schellhorn, 2015; Skvortsov et al., 2016; Tseng et al., 2013). Phages, considered to be the most abundant and diverse biological entities on Earth, play an important role in shaping biological and geochemical processes at the global scale (Díaz-Muñoz and Koskella, 2014). They may also be responsible for human health in spite of the fact that they do not infect humans. This is because some phages can convey new properties of coding for toxin production to the host bacteria, thus converting harmless bacteria into pathogens (Grabow, 2001), for example, the shiga toxin-converting phages have been known to change the pathogenicity of E. coli O157:H7 (Muniesa and Jofre, 2000).
Tributaries were found to have a higher proportion of Microviridae and Dicistroviridae. The hosts of Microviridae belonged to Enterobacteriaceae and obligate parasitic bacteria (Roux et al., 2012). Most of the Enterobacteriaceae taxa strains are pathogenic, for example, virulent strains of E. coli and Klebsiella pneumonia. Enterobacteriaceae occurred more in tributaries, which may contribute to the distribution and abundance of Microviridae in tributaries. The hosts of Dicistroviridae are soil-inhabiting invertebrates (e.g., aphids and ants, which are common in tropical Singapore). Thus, the fact that tributaries harbor the viral hosts, indicate that the viruses may be released from catchments and be washed into the tributary periodically.
The smaller spatial variation pattern of larger reservoirs compared with tributaries was not surprising, as reservoirs tend to be more resilient to urban and storm water runoff than their tributaries. In previous studies, freshwater microbial communities have been found to be resilient to natural disturbances (Tseng et al., 2013).
4.2. Influence of environmental parameters, land use and human activities on α-diversity and community structure
When correlating α-diversity with environmental parameters and land use factors, it was found that pH had a significant correlation among all the environmental parameters (r = −0.399, p < 0.05). pH is a key factor in determining virus infectivity where low pH (pH < 4) has been found to significantly reduce phage survivability (70–100%) (Jurczak-Kurek et al., 2016), Even though the pH range in this study is relatively small (6.185–8.78), it is likely that these differences could make a difference in viral α-diversity in our study.
A moderate correlation with rainfall (r = 0.329, p < 0.05) was observed which could be due to heavy rainfall flushing terrestrial bacteria, viruses and nutrients to the reservoir (Tseng et al., 2013). Precipitation was found to be the major factor affecting the microbial community in a subtropical reservoir in Taiwan (Tseng et al., 2013). Singapore is characterized by two main monsoon seasons: the Northeast Monsoon (December–March) and Southwest Monsoon season (June–September). 24-hour rainfall during the sampling times during the Monsoon and Inter-Monsoon season showed a significant difference between January 2015 (9.69 mm) and April 2015 (1.42 mm) (one-way ANOVA, F = 3.018, p-value = 0.091), which could have contributed to the characterization of the virome composition.
Based on land use category, observed species were also found to be significantly correlated with agriculture land use (r = 0.342, p < 0.05). This could be expected as intensive agriculture has been reported to bring nutrients and agricultural chemicals to the water cycle (Foley et al., 2005), which in turn, disturbs the microbial community indirectly (Tseng et al., 2013). Agricultural intensification has been associated with pathogen emergence transmission between wildlife and domestic animal populations and human populations, which in turn, could increase the species of zoonotic viruses (Pulliam et al., 2012). Jones et al. (2008) concluded that agricultural practices are one of the socioeconomic drivers in the spatial distribution of emerging infectious diseases. However, the non-significant correlation between diversity and other land use types does not necessarily suggest weak connections between the viral community and land use factor, since α-diversity indices (Shannon diversity and Observed species) are just one aspect of characterizing the viral community and does not necessarily represent a complete view. β-diversity analysis indicated that Mimiviridae, Phycodnaviridae, Siphoviridae and Podoviridae were enriched in reservoirs of parkland areas while Dicistroviridae and Microviridae were enriched in tributaries of urban areas (Fig. 3A). Siphoviridae, Podoviridae and Microviridae belong to bacteriophage, which may be associated with parkland and urban enriched bacteria (i.e., Actinobacteria, Verrucomicrobia, Chloroflexi, Acidobacteria and Proteobacteria) in our study (Fig. 3E).
Overall, viral diversity in the surveyed reservoirs (8 < H < 12) was higher than that reported in a subtropical reservoir in Taiwan (5.1 < H < 7.0) (Tseng et al., 2013). These differences could be due to the geographical (latitudinal gradient) differences in microbial diversity, as low-latitude tropical ecosystems tend to lead to higher biological diversity (Chown and Convey, 2007; Fuhrman et al., 2008). Kim et al. (2016) reported that viruses had higher richness near the equator and lower richness at higher latitude, similar to human pathogen species (Guernier et al., 2004). However, these comparisons between the diversity in freshwater virome studies need further confirmation due to different indices used (chao1, Simpson index, Shannon index, and observed species) and diverse calculation methods applied (e.g., PHACCs, QIIME, Catchall). By using PHACCs based on the contig spectra generated by Circonspect, Tseng et al. (2013) derived a Shannon diversity (H) range from 5.1 to 7 and from 7.8k to 21.8k viral genotypes in one subtropical freshwater reservoir, which was lower than the ocean's virome Shannon diversity in British Columbia, the Gulf of Mexico and the Sargasso Sea (H of 10.8, 8.21 and 7.74, respectively) (Angly et al., 2009). The difference in methods used in published papers makes it difficult to draw comparable conclusions. A standard pipeline in deriving the viral diversity in ecology is required in order to make comparisons of viral diversity across diverse aquatic ecosystems from different studies.
The graphs of PCoA analysis in both reservoirs and tributaries indicated that the land use pattern around the surveyed areas had an important impact on characterizing the virome community. Even though the viral communities at the family level were conserved across different reservoirs, the PCoA plots suggested a dynamic shift in differences between contig levels of the viral community in terms of land use. The geospatial distribution patterns in the surface water environment could have resulted from both direct and indirect factors. On the one hand, the different land use patterns could also introduce foreign viral contamination into water bodies directly through urban/agriculture runoff, precipitation, leaking sewers, etc., while the viral community itself can be indirectly changed and characterized by the relationship to their hosts in specific environments. The runoff from the surrounding areas can also bring specific bacteria, or other vectors and nutrients into the reservoirs and the tributaries (Tseng et al., 2013). Even though previous studies have shown the correlation between land use and water-borne viral pathogens (Corsi et al., 2014; Lenaker et al., 2017), ours is the first study correlating land use cover with the whole viral community, thus overcoming the limitations of investigating specific viruses and providing comprehensive information on the community structure with the relationship of land use cover.
4.3. Human/plant related viruses and correlations between qPCR/RT-qPCR and metagenomics
Viral pathogens could be introduced into reservoirs and tributaries through urban and storm water runoff. Although enteric viruses (e.g., adenovirus, norovirus, rotavirus, enterovirus, etc.) could not be detected with viral metagenomics in our study, past studies have shown that they are prevalent in tributaries using qPCR and thus, need to be carefully monitored for quantifying risk assessment and providing guidelines for water recreational activities (Aw and Gin, 2011; Aw et al., 2009). Vergara et al. (2016) quantified illness risks of norovirus in an urban catchment in Singapore by using quantitative microbial risk assessment. The finding reported mean probability of illness associated with norovirus were 0.0061 and 0.0089 in the scenario of primary contact recreation of adults and children, which is below the USEPA guideline value of 0.036 (USEPA, 2012). In addition, other potential emerging viral pathogens (e.g., hepatitis E virus, coronavirus, cyclovirus, bird flu virus), which may originate from wildlife and indigenous animals in tropical forests or animal parks can also spread to neighboring tributaries, potentially causing disease to humans. According to Jones et al. (2008), the emerging infectious diseases are dominated by zoonoses (60.3%) and the majority of these are from wildlife (71.8%), with an increasing trend and more hotspots concentrated in lower-latitude developing countries.
In this study, we observed sequences related to the human-infective virus, human picobirnavirus and the emerging zoonotic virus, cyclovirus. Human picobirnavirus, a bi-segmented double-stranded RNA virus, has been detected in both healthy and unhealthy human beings. It has also been found to be prevalent (100% detection frequency) in raw sewage samples collected in the United States (Symonds et al., 2009). The pathogenicity of human picobirnaviruses has not been established and it has been suggested as an opportunistic pathogen which might cause diarrhea (Giordano et al., 1999; Grohmann et al., 1993). As metagenomics can only provide relative abundance, further studies are needed using qPCR in order to determine absolute concentrations and to better evaluate health risks.
Viruses belonging to the Circoviridae (approximately 0.6% of the virome) may be involved in disease in vertebrate animals and plants. A large proportion was found to belong to swan circoviruses (59%) and Circoviridae 2 LDMD-2013 (30%). Two contigs found in the present study were assigned to the suggested cyclovirus-VN which originated from human samples in Vietnam ( Garigliany et al., 2014). Indeed, cyclovirus VN was initially reported to be restricted to central and southern Vietnam but was subsequently detected in both farm animals and human clinical samples from Africa, indicating their geographic transmission capacity (Garigliany et al., 2014; van Doorn et al., 2013). In Singapore, human cyclovirus VS5700009 (CyCV-VS5700009) was previously found in Singapore harbor water by using a metagenomics approach (Kim et al., 2016). The contig 89154 shared a 97.79% nucleotide similarity with human cyclovirus VN (KF031466) with a query coverage of 98.15%. The discovery of cyclovirus VN in freshwater tributaries in the densely populated area of Site 2 in our study indicates a possible transmission of the emerging human cyclovirus in the Singapore urban water cycle. Further risk assessment of host populations should be conducted for these water environments (van Doorn et al., 2013).
Samples from Sites 5, 6 and 7 had lower occurrence rate (both RPKM and absolute reads matrix) and concentration of human picobirnavirus and cyclovirus affiliated viruses, consistent with lower human population density (<10 people/km2) except for reservoir 6. The exception of reservoir 6 (explained in Results 3.1) suggested that drainage points could also be significant drivers in shaping the viral communities as well as land use cover and population density. Higher population density could result in high occurrence of human-related viruses (Lenaker et al., 2017). Here, the population density to some extent could reflect the hotspots of human-related viruses. Limitation of using population density to quantify human activities and human-related viral hosts in our study exists as the population density referred to is the resident population. However, the mobile population such as those associated with modern transportation, tourism, business travel and immigration could also contribute to dissemination of these high-impact pathogens (Arguin et al., 2009). In addition, public holidays could introduce variation in population numbers to commercial and recreational areas based on lifestyle choices. Thereafter, more detailed data will be needed in order to track and investigate the human-related virus transmission patterns through virus-host interaction.
In our sequencing data, the absence of the majority of enteric viruses in surface waters is reasonable. A potentially relatively low abundance of human-related viruses in our surface water system and insufficient sequencing coverage could have resulted in rare sequences not being assembled into contigs for further downstream analysis. In contrast with our freshwater ecosystem, a previous study of sewage sludge samples revealed a large number of human viral pathogens, unveiling 43 types of human viruses (Bibby and Peccia, 2013). This result is expected as the sludge matrix harbors large amounts of human-related viruses and the concentrations of viral pathogens are much higher.
In our study, a good Spearman rank correlation (0.588 < r < 0.795, p < 0.05) for the indicator virus PMMoV was discovered as the qPCR/RT-qPCR concentrations of this target are relatively higher (PMMoV geomean: 958.45GC/L). This suggests that viral metagenomics, to some extent, is a conservative estimate of the true viral abundance, based on validation of RT-qPCR data with RPKM in relative abundance of contigs, especially for contigs with a higher qPCR concentration. Similar results were also obtained in a previous study where a significant correlation (p < 0.0001) between qPCR and RPKM across viral taxa in clinical samples was obtained (Graf et al., 2016). Overall, viral metagenomics has its advantage in the simultaneous discovery of the entire set of targets in the community in spite of the intrinsic limitation of downstream bioinformatics (i.e., assembly efficiency and database bias). It can pave the way in finding emerging zoonotic viruses and alternative plausible fecal indicators in predicting viral pathogens in the future. After zooming into the specific and desired targets using high throughput sequencing, gene-specific PCR or qPCR (e.g., cyclovirus-VN) could be further investigated to confirm the presence of these potential viral pathogens and zoonotic viruses to reveal the epidemiology or transmission patterns of these viral pathogens (Kim et al., 2015).
5. Conclusion
This study has shed light on the diversity of the viral communities in tropical reservoirs and their tributaries with different land use. Correlations between the diversity index, physical-chemical parameters and land use patterns showed that environmental parameters (i.e., pH and precipitation) and spatial factors (e.g., reservoirs/tributaries, land use) are the main drivers of the viral community structure. Although enteric viruses were not detected by viral metagenomics, human-related viruses, including emerging zoonotic viruses, were detected in our samples indicating the importance of continued monitoring of these environments where specific hosts could be harbored. In addition, the links between qPCR/RT-qPCR and metagenomics were shown using both contigs and reads, confirming that our metagenomics quantification path is reliable and scientific. Even though metagenomics sequencing technology cannot replace qPCR/RT-qPCR due to its relatively low sensitivity in detecting gene-specific viral pathogens, its wide coverage of viral targets could add valuable information, such as detecting new, emerging zoonotic viruses and finding alternative fecal indicators or markers of contamination.
Acknowledgements
This research grant is supported by the Singapore National Research Foundation under its Environment and Water Research Programme and administered by PUB, Singapore's national water agency (Ref: 1301-IRIS-37 [IDD 90301/1/65]). We would like to thank National University of Singapore and Center for Environmental Sensing and Modeling (CENSAM) for supporting this research.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.watres.2018.03.017.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ackermann H.-W. Bacteriophage observations and evolution. Res. Microbiol. 2003;154(4):245–251. doi: 10.1016/S0923-2508(03)00067-6. [DOI] [PubMed] [Google Scholar]
- Angly F.E., Felts B., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H. The marine viromes of four oceanic regions. PLoS Biol. 2006;4(11) doi: 10.1371/journal.pbio.0040368. e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly F.E., Willner D., Prieto-Davó A., Edwards R.A., Schmieder R., Vega-Thurber R., Antonopoulos D.A., Barott K., Cottrell M.T., Desnues C. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput. Biol. 2009;5(12) doi: 10.1371/journal.pcbi.1000593. e1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguin P.M., Marano N., Freedman D.O. Globally mobile populations and the spread of emerging pathogens. Emerg. Infect. Dis. 2009;15(11):1713–1714. doi: 10.3201/eid1511.091426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw T., Gin K.H. Prevalence and genetic diversity of waterborne pathogenic viruses in surface waters of tropical urban catchments. J. Appl. Microbiol. 2011;110(4):903–914. doi: 10.1111/j.1365-2672.2011.04947.x. [DOI] [PubMed] [Google Scholar]
- Aw T.G., Gin K.Y.-H., Oon L.L.E., Chen E.X., Woo C.H. Prevalence and genotypes of human noroviruses in tropical urban surface waters and clinical samples in Singapore. Appl. Environ. Microbiol. 2009;75(15):4984–4992. doi: 10.1128/AEM.00489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47(4):1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M., Salamon P., Andresen B., Mahaffy J.M., Segall A.M., Mead D., Azam F., Rohwer F. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99(22):14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cassman N., Prieto-Davó A., Walsh K., Silva G.G., Angly F., Akhter S., Barott K., Busch J., McDole T., Haggerty J.M. Oxygen minimum zones harbour novel viral communities with low diversity. Environ. Microbiol. 2012;14(11):3043–3065. doi: 10.1111/j.1462-2920.2012.02891.x. [DOI] [PubMed] [Google Scholar]
- Chown S.L., Convey P. Spatial and temporal variability across life's hierarchies in the terrestrial Antarctic. Philosophical Transactions of the Royal Society B: Biological Sciences. 2007;362(1488):2307–2331. doi: 10.1098/rstb.2006.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi S.R., Borchardt M., Spencer S., Hughes P.E., Baldwin A.K. Human and bovine viruses in the Milwaukee River watershed: hydrologically relevant representation and relations with environmental variables. Sci. Total Environ. 2014;490:849–860. doi: 10.1016/j.scitotenv.2014.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A.I., Lang A.S., Suttle C.A. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312(5781):1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- de Graaf M., van Beek J., Koopmans M.P. Human norovirus transmission and evolution in a changing world. Nat. Rev. Microbiol. 2016;14(7):421–433. doi: 10.1038/nrmicro.2016.48. [DOI] [PubMed] [Google Scholar]
- Department of Statistics Singapore . 2016. Population in Brief 2016. [Google Scholar]
- Díaz-Muñoz S.L., Koskella B. Bacteria-phage interactions in natural environments. Adv. Appl. Microbiol. 2014;89(135) doi: 10.1016/B978-0-12-800259-9.00004-4. 10.1016. [DOI] [PubMed] [Google Scholar]
- Dinsdale E.A., Edwards R.A., Hall D., Angly F., Breitbart M., Brulc J.M., Furlan M., Desnues C., Haynes M., Li L. Functional metagenomic profiling of nine biomes. Nature. 2008;452(7187):629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Djikeng A., Kuzmickas R., Anderson N.G., Spiro D.J. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One. 2009;4(9):e7264. doi: 10.1371/journal.pone.0007264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doceul V., Bagdassarian E., Demange A., Pavio N. Zoonotic hepatitis E virus: classification, animal reservoirs and transmission routes. Viruses. 2016;8(10):270. doi: 10.3390/v8100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J.B., Thomas B.C., Andrade K., Allen E.E., Heidelberg K.B., Banfield J.F. Dynamic viral populations in hypersaline systems as revealed by metagenomic assembly. Appl. Environ. Microbiol. 2012;78(17):6309–6320. doi: 10.1128/AEM.01212-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancello L., Trape S., Robert C., Boyer M., Popgeorgiev N., Raoult D., Desnues C. Viruses in the desert: a metagenomic survey of viral communities in four perennial ponds of the Mauritanian Sahara. ISME J. 2013;7(2):359–369. doi: 10.1038/ismej.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J.A., DeFries R., Asner G.P., Barford C., Bonan G., Carpenter S.R., Chapin F.S., Coe M.T., Daily G.C., Gibbs H.K. Global consequences of land use. Science. 2005;309(5734):570–574. doi: 10.1126/science.1111772. [DOI] [PubMed] [Google Scholar]
- Fuhrman J.A., Steele J.A., Hewson I., Schwalbach M.S., Brown M.V., Green J.L., Brown J.H. A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl. Acad. Sci. Unit. States Am. 2008;105(22):7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigliany M.-M., Hagen R.M., Frickmann H., May J., Schwarz N.G., Perse A., Jöst H., Börstler J., Shahhosseini N., Desmecht D. Cyclovirus CyCV-VN species distribution is not limited to Vietnam and extends to Africa. Sci. Rep. 2014;4:7552. doi: 10.1038/srep07552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Wu Y., Wang M., Wang J., Wu L., Yang X., Zhang Y., Shi Z. Viral metagenomics analysis of planktonic viruses in East Lake, Wuhan, China. Virol. Sin. 2013;28(5):280–290. doi: 10.1007/s12250-013-3365-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M.O., Martinez L.C., Rinaldi D., Espul C., Martinez N., Isa M.B., Depetris A.R., Medeot S.I., Nates S.V. Diarrhea and enteric emerging viruses in HIV-infected patients. AIDS Res. Hum. Retrovir. 1999;15(16):1427–1432. doi: 10.1089/088922299309937. [DOI] [PubMed] [Google Scholar]
- Grabow W. Bacteriophages: update on application as models for viruses in water. Water Sa. 2001;27(2):251–268. [Google Scholar]
- Graf E.H., Simmon K.E., Tardif K.D., Hymas W., Flygare S., Eilbeck K., Yandell M., Schlaberg R. Unbiased detection of respiratory viruses by use of RNA sequencing-based metagenomics: a systematic comparison to a commercial PCR panel. J. Clin. Microbiol. 2016;54(4):1000–1007. doi: 10.1128/JCM.03060-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann G.S., Glass R.I., Pereira H.G., Monroe S.S., Hightower A.W., Weber R., Bryan R.T. Enteric viruses and diarrhea in HIV-infected patients. N. Engl. J. Med. 1993;329(1):14–20. doi: 10.1056/NEJM199307013290103. [DOI] [PubMed] [Google Scholar]
- Guernier V., Hochberg M.E., Guégan J.-F. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004;2(6) doi: 10.1371/journal.pbio.0020141. e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercus C. Novocraft Technologies; Selangor: 2012. Novoalign. [Google Scholar]
- Hwang J., Park S.Y., Park M., Lee S., Lee T.-K. Seasonal dynamics and metagenomic characterization of marine viruses in Goseong Bay, Korea. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0169841. e0169841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaykus L.-A., De Leon R., Sobsey M.D. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl. Environ. Microbiol. 1996;62(6):2074–2080. doi: 10.1128/aem.62.6.2074-2080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczak-Kurek A., Gąsior T., Nejman-Faleńczyk B., Bloch S., Dydecka A., Topka G., Necel A., Jakubowska-Deredas M., Narajczyk M., Richert M. Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Scientific Reports. 2016;6 doi: 10.1038/srep34338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Aw T.G., Rose J.B. Transporting ocean viromes: invasion of the aquatic biosphere. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0152671. e0152671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Aw T.G., Teal T.K., Rose J.B. Metagenomic investigation of viral communities in ballast water. Environ. Sci. Technol. 2015;49(14):8396–8407. doi: 10.1021/acs.est.5b01633. [DOI] [PubMed] [Google Scholar]
- Lenaker P.L., Corsi S., Borchardt M.A., Spencer S.K., Baldwin A.K., Lutz M.A. Hydrologic, land cover, and seasonal patterns of waterborne pathogens in Great Lakes tributaries. Water Res. 2017;113:11–21. doi: 10.1016/j.watres.2017.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang H., Nie K., Zhang C., Zhang Y., Wang J., Niu P., Ma X. VIP: an integrated pipeline for metagenomics of virus identification and discovery. Sci. Rep. 2016;6 doi: 10.1038/srep23774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Goh S., Vergara G., Fang H., Rezaeinejad S., Chang S., Bayen S., Lee W., Sobsey M.D., Rose J. Alternative fecal indicators and their empirical relationships with enteric viruses, Salmonella enterica, and Pseudomonas aeruginosa in surface waters of a tropical urban catchment. Appl. Environ. Microbiol. 2015;81(3):850–860. doi: 10.1128/AEM.02670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bueno A., Tamames J., Velázquez D., Moya A., Quesada A., Alcamí A. High diversity of the viral community from an Antarctic lake. Science. 2009;326(5954):858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- Mohiuddin M., Schellhorn H. Spatial and temporal dynamics of virus occurrence in two freshwater lakes captured through metagenomic analysis. Front. Microbiol. 2015;6:960. doi: 10.3389/fmicb.2015.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Muniesa M., Jofre J. Occurrence of phages infecting Escherichia coli O157: H7 carrying the Stx 2 gene in sewage from different countries. FEMS Microbiol. Lett. 2000;183(1):197–200. doi: 10.1111/j.1574-6968.2000.tb08957.x. [DOI] [PubMed] [Google Scholar]
- Ng C., Tay M., Tan B., Le T.-H., Haller L., Chen H., Koh T.H., Barkham T.M.S., Gin K.Y.-H. Characterization of metagenomes in urban aquatic compartments reveals high prevalence of clinically relevant antibiotic resistance genes in wastewaters. Front. Microbiol. 2017;8:2200. doi: 10.3389/fmicb.2017.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nshimyimana J.P., Freedman A.J.E., Shanahan P., Chua L.C., Thompson J.R. Variation of bacterial communities with water quality in an urban tropical catchment. Environ. Sci. Technol. 2017;51:5591–5601. doi: 10.1021/acs.est.6b04737. [DOI] [PubMed] [Google Scholar]
- Patz J.A., Daszak P., Tabor G.M., Aguirre A.A., Pearl M., Epstein J., Wolfe N.D., Kilpatrick A.M., Foufopoulos J., Molyneux D. Unhealthy landscapes: policy recommendations on land use change and infectious disease emergence. Environ. Health Perspect. 2004:1092–1098. doi: 10.1289/ehp.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz J.A., Olson S.H., Uejio C.K., Gibbs H.K. Disease emergence from global climate and land use change. Medical Clinics of North America. 2008;92(6):1473–1491. doi: 10.1016/j.mcna.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Pulliam J.R., Epstein J.H., Dushoff J., Rahman S.A., Bunning M., Jamaluddin A.A., Hyatt A.D., Field H.E., Dobson A.P., Daszak P. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. Journal of the Royal Society Interface. 2012;9:89–101. doi: 10.1098/rsif.2011.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Blanton L.V., Cao S., Zhao G., Manary M., Trehan I., Smith M.I., Wang D., Virgin H.W., Rohwer F. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112(38):11941–11946. doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaeinejad S., Vergara G., Woo C., Lim T., Sobsey M.D., Gin K. Surveillance of enteric viruses and coliphages in a tropical urban catchment. Water Res. 2014;58:122–131. doi: 10.1016/j.watres.2014.03.051. [DOI] [PubMed] [Google Scholar]
- Rinke C., Lee J., Nath N., Goudeau D., Thompson B., Poulton N., Dmitrieff E., Malmstrom R., Stepanauskas R., Woyke T. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat. Protoc. 2014;9(5):1038. doi: 10.1038/nprot.2014.067. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B., Li L., Wegley L., Furlan M., Angly F., Breitbart M., Buchanan J., Desnues C., Dinsdale E., Edwards R., Felts B., Haynes M., Liu H., Lipson D., Mahaffy J., Martin-Cuadrado A.B., Mira A., Nulton J., Pasic L., Rayhawk S., Rodriguez-Mueller J., Rodriguez-Valera F., Salamon P., Srinagesh S., Thingstad T.F., Tran T., Thurber R.V., Willner D., Youle M., Rohwer F. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4(6):739–751. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- Rosario K., Nilsson C., Lim Y.W., Ruan Y., Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 2009;11(11):2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- Rosiles-González G., Ávila-Torres G., Moreno-Valenzuela O.A., Acosta-González G., Leal-Bautista R.M., Grimaldo-Hernández C.D., Brown J.K., Chaidez-Quiroz C., Betancourt W.Q., Gerba C.P. Food and Environmental Virology; Mexico: 2017. Occurrence of Pepper Mild Mottle Virus (PMMoV) in Groundwater from a Karst Aquifer System in the Yucatan Peninsula; pp. 1–11. [DOI] [PubMed] [Google Scholar]
- Roux S., Enault F., Hurwitz B.L., Sullivan M.B. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3 doi: 10.7717/peerj.985. e985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S., Krupovic M., Poulet A., Debroas D., Enault F. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0040418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S., Tournayre J., Mahul A., Debroas D., Enault F. Metavir 2: new tools for viral metagenome comparison and assembled virome analysis. BMC Bioinformatics. 2014;15:76. doi: 10.1186/1471-2105-15-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeidi N., Gu X., Goh S.G., Xin C.L.Y., Gin K.Y.-H. Evaluating the efficacy of commercial kits for viral DNA/RNA extraction. Water Pract. Technol. 2017;12(1):80–86. [Google Scholar]
- Skvortsov T., de Leeuwe C., Quinn J.P., McGrath J.W., Allen C.C., McElarney Y., Watson C., Arkhipova K., Lavigne R., Kulakov L.A. Metagenomic characterisation of the viral community of lough neagh, the largest freshwater lake in Ireland. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0150361. e0150361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Griffin D.W., Breitbart M. Eukaryotic viruses in wastewater samples from the United States. Appl. Environ. Microbiol. 2009;75(5):1402–1409. doi: 10.1128/AEM.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te S.H., Tan B.F., Thompson J.R., Gin K.Y.-H. Relationship of microbiota and cyanobacterial secondary metabolites in planktothricoides-dominated bloom. Environ. Sci. Technol. 2017;51(8):4199–4209. doi: 10.1021/acs.est.6b05767. [DOI] [PubMed] [Google Scholar]
- Temmam S., Davoust B., Berenger J.-M., Raoult D., Desnues C. Viral metagenomics on animals as a tool for the detection of zoonoses prior to human infection? Int. J. Mol. Sci. 2014;15(6):10377–10397. doi: 10.3390/ijms150610377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.-H., Chiang P.-W., Shiah F.-K., Chen Y.-L., Liou J.-R., Hsu T.-C., Maheswararajah S., Saeed I., Halgamuge S., Tang S.-L. Microbial and viral metagenomes of a subtropical freshwater reservoir subject to climatic disturbances. ISME J. 2013;7(12):2374–2386. doi: 10.1038/ismej.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA . 2012. Recretional Water Quality Criteria. 820-F-12—058. [Google Scholar]
- van Doorn H.R., Nghia H.D.T., Chau T.T.H., de Vries M., Canuti M., Deijs M., Jebbink M.F., Baker S., Bryant J.E., Tham N.T. Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections. MBio. 2013;4(3) doi: 10.1128/mBio.00231-13. e00231–00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara G., Rose J., Gin K. Risk assessment of noroviruses and human adenoviruses in recreational surface waters. Water Res. 2016;103:276–282. doi: 10.1016/j.watres.2016.07.048. [DOI] [PubMed] [Google Scholar]
- Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99(24):15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie K.M., Mihindukulasuriya K.A., Sodergren E., Weinstock G.M., Storch G.A. Sequence analysis of the human virome in febrile and afebrile children. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0027735. e27735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.