Abstract
The circRNA is a newly defined noncoding RNA and characterized by its unique splicing reactions to form circles. However, the function of circRNAs during viral infection remains largely unknown. In this study, the circRNA expression profile during porcine endemic diarrhea virus (PEDV) infection in IPEC-J2 cell line was investigated using the next-generation sequencing technology. A total of 26670 circRNA candidates were identified. The functional annotation analysis revealed that the parent genes of differentially expressed circRNAs might be associated with host response to PEDV infection. Further analysis verified the existence of eight selected circRNAs and confirmed that PEDV infection alerted the expression patterns of circRNAs and their linear parent genes in IPEC-J2 cell line. The circRNA-miRNA interaction network was also constructed to elucidate their potential targets. Our results provided not only the first large-scale profile analysis of circRNAs associated with PEDV infection but also a novel direction to elucidate host-PEDV interactions.
Keywords: PEDV, circRNA, IPEC-J2, qRT-PCR, ceRNA network
1. Introduction
The circular RNA (circRNA) is novel type of endogenous noncoding RNAs expressed in all eukaryotic cells. In contrast to linear RNAs, the circRNA is characterized by back-splicing events that a downstream 5′ splice site joins with an upstream 3′ splice site (Jeck et al., 2013). Thus, the terminated 5′ caps and 3′ tails modifying the mature mRNAs are not found within circRNAs. With these characteristics, circRNAs were once thought to be the by-product of splicing errors when they were first detected in genomes of viriod plant pathogens and Hepatitis delta virus in 1976 (Halbreich et al., 1980, Sanger et al., 1976). Later, with the development and improvement of RNA-sequencing technologies and bioinformatics tools, circRNAs were discovered in nearly all the species (Gao et al., 2015). The circRNAs were even found to play vital roles in regulating gene expression.
Porcine epidemic diarrhea (PED) is one of the enteric infectious diseases in pigs, characterized by severe enteritis, vomiting, watery diarrhea, and up to 90% death rate in piglets younger than one week old. Since its re-emergence in 2010, this disease has caused huge economic loss to the swine industry in North America and East Asia (Davies, 2015, Lee and Lee, 2014, Temeeyasen et al., 2014, Vlasova et al., 2014, Wang et al., 2016). Porcine epidemic diarrhea virus (PEDV), the causative agent of PED, is a single stranded, positive-sense enveloped RNA virus, belonging to the Alphacoronavirus genus within the Coronaviridae family. Although the pathogenesis and immune mechanisms of other coronavirus such as SARS-CoV and MERS-CoV have been understood quite well (Channappanavar et al., 2014, de Wit et al., 2016, Qian et al., 2013, van den Brand et al., 2015), they remain largely unknown in PEDV infection, especially for the host–pathogen interactions. RNAs, a bridge between DNA and protein, have the potential to regulate the host response to the viral infection earlier and faster than the proteins. However, the detailed functions of RNAs, especially circRNAs, remain largely undefined during viral infection.
In this study, the circRNA expression profile in PEDV infected IPEC-J2 cells, a porcine intestinal epithelial cell line, was identified using the next-generation sequencing (NGS) technology. Some circRNA candidates that might potentially relate to the regulation of viral infection were selected and validated for their existence by RT-PCR. The dynamic expression levels of selected circRNAs and their primary linear genes were also verified using real-time RT-PCR assay. Finally, the circRNA-miRNA interaction analysis was performed to better understand the host-pathogen interactions. The circRNAs expression profile obtained from swine intestinal epithelial cells will greatly advance our knowledge about circRNAs and improve our understanding on host responses to PEDV infection.
2. Results
2.1. Samples at immune active stages were selected for circRNA sequencing
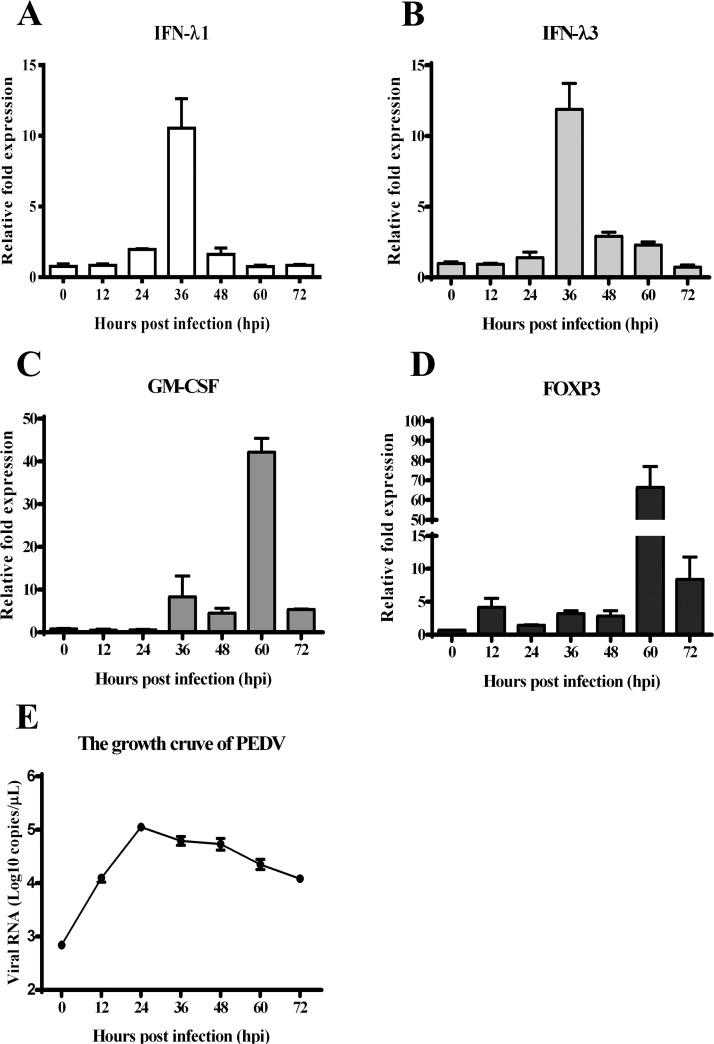
The expression of circRNAs has been widely detected in the life cycle among all eukaryotic cells. They are thought to be expressed in a spatio-temporal manner. Therefore, it is of great importance to select proper time points for profiling circRNAs expression during PEDV infection. To do so, IPEC-J2 cells were inoculated with PEDV at an MOI = 0.01 or mock-treated and harvested at 12, 24, 36, 48, 60 and 72 h post infection (hpi). The total RNA was extracted and subjected to the real-time RT-PCR analysis to evaluate the host immune responses induced by PEDV on IPEC-J2 cells. The type III interferon has been reported to play a vital role in maintaining the antiviral state of the mucosal epithelial cell surface in the gut and resist PEDV infection (Li et al., 2017, Shen et al., 2016, Zhang et al., 2018). Therefore, the IFN-λ1 and IFN-λ3 were selected to monitor antiviral responses and results showed that their expression levels were significantly upregulated and reached their peaks at 36 hpi and dropped to the normal level thereafter ( Fig. 1A, B). Foxp3 and granulocyte-macrophage are the major helper cells for IgA responses to gut microbiota (Cong et al., 2009, Gommerman et al., 2014, Nei et al., 2012, Russler-Germain et al., 2017). We next checked the expression of GM-CSF and Foxp3 to reflect the immune responses induced by PEDV infection. The results illustrated that they were both activated upon PEDV infection, with the highest expression levels appeared at 60 hpi then dropped down at 72 hpi (Fig. 1C, D). The growth curve of PEDV infection on IPEC-J2 demonstrated the number of viral RNA copies peaked at 24 hpi and slowly decreased thereafter (Fig. 1E). All these data indicated that the host cells responded to PEDV infection with a time depended manner. Therefore, the samples from 36 hpi, 60 hpi and 72 hpi were selected for library construction and circRNA sequencing.
Fig. 1.
PEDV infection induced the expression of cytokines and IFNs. IPEC-J2 cells plated in six-well plates were infected with PEDV LJX01/GS/2014 strain at an MOI = 0.01 or mock infected. Cell pellets were used for RNA extraction. The cDNA were prepared with hexamer random primers and subjected to real-time PCR analysis for the expressions of A) IFN-λ1, B) IFN-λ3, C) GM-CSF and D) FOXP3 at indicated time points. The relative expression levels were calculated by 2-∆∆Ct method. E) The copy numbers of PEDV genome were measured by real-time RT-qPCR analysis at each time point. All experiments were carried out triplicates.
2.2. The identification and characterization of circRNA candidates
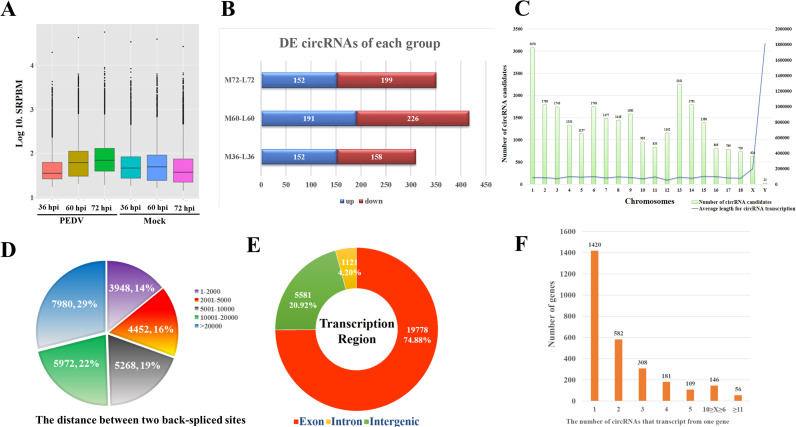
The rRNA-depleted RNA libraries were sequenced and computationally analyzed for screening back-splicing reads to identify circRNA candidates. The filtered reads were then mapped to the pig genome (GCA_000003025.4, Ensemble) using TopHat-Fusion. The genomic distance between two splice sites was less than 100 kb, and each circRNA was supported by at least 2 reads in each sample. Based on the mapping results, CIRI (v2.0) was used to detect circRNA candidates from transcriptome data by employing multiple filtration strategies (Gao et al., 2015). We identified 26670 novel circRNA candidates in all six samples. In each sample, the spliced reads per billion mapping (SRPBM) of all circRNA candidates was displayed by the box plot ( Fig. 2A). There were 1078 differently expressed (DE) circRNA candidates obtained from selected time points, among which 495 circRNAs were upregulated while 583 were downregulated (Fig. 2B).
Fig. 2.
Profiles of circRNA candidates. A total of 26670 circRNA candidates were obtained from PEDV infected and mock infected IPEC-J2 cells at 36, 60 and 72 hpi. A) The box plot indicates the expression levels of circRNA candidates from each sample. B) A total of 1078 differential circRNAs were obtained in these groups. Several circRNA candidates were differently expressed in all three groups. C) The histogram shows the number of circRNA generated from each chromosome. The line chart shows the average length for every circRNA transcription. D) The pie chart shows the length distribution of back-spliced sites. E) The sunburst chart shows the number and ratios of exon, intron and intergenic circRNAs. F) Different circRNAs can be transcribed from one gene. The histogram shows the number of genes that different circRNAs can be transcribed from.
We then analyzed the distribution of circRNAs transcription and found that all these circRNAs were transcribed from sus scrofa chromosomes (SSC). The number of circRNA candidate transcribed from SSC varied according to the length of chromosomes. Meanwhile, the average distance between splice sites for each circRNA transcription was nearly the same in all chromosomes except X and Y (Fig. 2C). Previous researches claimed that the length of most circRNAs were shorter than 2 kb (Fan et al., 2015, Lasda and Parker, 2014, Liang et al., 2017). However, our results showed that the distance between the two splice sites varied from 2 kb to 20 kb, even with 29% of circRNAs larger than 20 kb (Fig. 2D). It is also worth to mention that the distance between the splice sites does not represent the actual length of circRNAs. Nearly three quarters of circRNA candidates were transcribed from exons while only one quarter of them were transcribed from both intron and intergenic regions (Fig. 2E). Among all genes that circRNAs were transcribed from, there were 56 genes that could generate more than 10 circRNA candidates (Fig. 2F), indicating that some genes were more active for circRNA generation. These genes were mainly involved in ubiquitin signaling pathway, modification and transport of proteins, binding and transport of DNA and RNA. Some of these genes were reported to have the functional role in regulating viral infection (Park et al., 2013, Sorensen et al., 2005, Su et al., 2013, Wang et al., 2012). A detailed list of these genes was presented in Supplementary Table 1.
2.3. The functional analysis of the parent genes to circRNA candidates
As showed in Fig. 2E, most of identified circRNA candidates were derived from exonic regions and carried protein-coding sequence. The exon(s) incorporated into circRNAs might not be the part of mRNAs derived from the same primary transcript. Hence, the production of circRNAs that removes the translational codon from corresponding linear transcript may reduce the number of primary transcripts and related mature, linear transcripts (Ashwal-Fluss et al., 2014, Jeck and Sharpless, 2014, Lasda and Parker, 2014). However, there was no definite correlation between the expression of circRNAs and their corresponding mRNA. Functional analysis is usually employed to predict how the circRNAs affect their linear transcripts. In this study, there were a total of 83 parent genes changed their expression levels during viral infection, accounting for 56.5% of all 147 analyzed parent genes. Therefore, functional analysis of the parent linear genes to DE circRNA candidates was firstly performed to predict the effect of the production of circRNAs. Both Gene Ontology (GO) and KEGG databases were used in this process. GO annotation analysis revealed that the parent genes to DE circRNAs were mainly involved in catalytic and be part of macromolecular complex at all selected time points ( Fig. 3A, B, C). There were also several reports demonstrated that circRNAs were actively participated in the metabolism process (Granados-Riveron and Aquino-Jarquin, 2016, Ye et al., 2015, Zou et al., 2017). For functional analysis, the DE genes at all the time points were searched in KEGG pathway database. The ubiquitin mediated proteolysis, relevant to induction of IFNs and ISGs (Haas et al., 1987, Heaton et al., 2016, Hermant and Michiels, 2014, Kim et al., 2004), was found to be highly related to the DE genes at 36 hpi (Fig. 3D). Previous results in this study already showed the expression levels of IFN-λ1 and IFN-λ3 were significantly upregulated at 36 hpi (Fig. 1A, B), suggesting that the production of circRNAs potentially affect this biological process. At 60 hpi, KEGG analysis showed that some DE genes were involved in the T cell receptor signaling pathway (Fig. 3E). This was proved by our previous results, showing the FoxP3 and GM-CSF, two of key regulators of T cell maturation and division, were highly upregulated at this time point (Fig. 1C, D). Most PEDV-infected cells showed the sign of apoptosis at 72 hpi, resulting in many genes closely related to the degradation process (Fig. 3F). All these data above indicated that the parent genes of circRNA candidates involve in host responses to viral infection, implying that this process may be affected by circRNAs.
Fig. 3.
The functional analysis of the parent genes to identified circRNA candidates. The parent genes of differently expressed circRNA were mapped to the terms in the GO database for functional significance, and to the KEGG pathway database for pathway analysis. The enriched Go terms at A) 36 hpi, B) 60 hpi and C) 72 hpi shows the catalytic activity and macromolecular complex are the main process and cellular component of the related host genes. The KEGG pathway analysis shows that the parent genes of circRNAs were involved in many pathways related with host immune response at D) 36 hpi, E) 60 hpi and F) 72 hpi.
2.4. Verification of circRNAs candidates
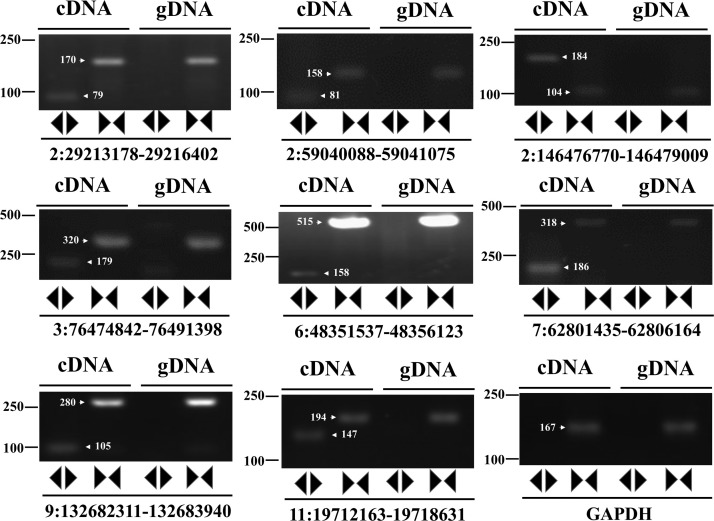
Based on the above functional analysis results, eight circRNA candidates were selected to verify their existence and expression using reverse transcription–polymerase chain reaction (RT-PCR) assay. The selection of all candidate circRNAs was based on the following three rules. 1). The expression levels of all candidates and their parent genes should be significantly changed (fold>2, p < 0.05); 2). The parent genes of all candidates should be related to immune response, antiviral effect, or facilitate viral infection; 3). All candidate cricRNAs should be identified or published by other researchers. (Liang et al., 2017). The validation assay was carried out in three independent infection experiments. The divergent and convergent PCR primer sets ( Table 1) were designed to amplify the selected circRNA candidates from the total RNA and genomic DNA. As expected, the expression and back-splicing junctions of the selected circRNAs, including circRNA2: 29213178-29216402, 59040088-59041075, 146476770-146479009, circRNA3: 76474842-76491398, circRNA6: 48351537-48356123, circRNA7: 62801435-62806164, circRNA 9: 132682311-132683940 and circRNA11: 19712163-19718631 were confirmed by RT-PCR in the cDNA but not the genomic DNA samples ( Fig. 4). Meanwhile, there was no any circRNAs amplicons obtained from both cDNA and genomic DNA (gDNA) when using the convergent primer sets to perform PCR amplification (Fig. 4). These results indicated that the circRNA candidates in the RNA-seq datasets originated from the CIRC explorer pipeline was reliable.
Table 1.
The divergent and convergent PCR primers for identification of circRNAs.
| circRNA | Primer type | Divergent primers | Convergent primers |
|---|---|---|---|
| 2:29213178-29216402 | F: | GCTGGATTCATAGGCTGTG | TTTGCGCCATAAGGTCCTGT |
| R: | ACTTGTAGGATTCAATGTTGGATT | TGACGGACCTCTGTCACTGT | |
| 2:59040088-59041075 | F: | AAGTGCCTTGATGTTGTC | TACGTTCCAACGTCTACCCG |
| R: | CTGAAGCTGGACCACTAC | GCTACAGAGGGCCTCACATT | |
| 2:146476770-146479009 | F: | ACAGGCAACATAAACTTCAAAT | ACTGTTGGAGCCTCTTGTCAC |
| R: | GACAAGAGGCTCCAACAG | TAACCTTGCCCCGTGTTTGT | |
| 3:76474842-76491398 | F: | TGAACAACAAGATATGGATT | TCATGCTCCAGGAAACCAACA |
| R: | ATGGCTATTGACAGGATT | AATCTCCATTTGCAGCGGGT | |
| 6:48351537-48356123 | F: | CCAAAGTTGCCAAAGTTAG | ACACGTCTCGTTCTGGTTGC |
| R: | GGTTGTCTCAGATGAATCA | CTCCTATCAGGCCTCTAGCC | |
| 7:62801435-62806164 | F: | ACTGCCAGATTACATCAT | GTCAAGTTCCTCATGGCCCG |
| R: | TACTGAACTGTCCACTTG | GCTGTGGCAAGGGTTCAATC | |
| 9:132682311-132683940 | F: | CTCTCAGCAGTTACATACAC | GGGACAAGCCCCCATCTTAAT |
| R: | CTATGGAGGTGGAGGAAT | TTGCAAACATGCCTTGCTCC | |
| 11:19712163-19718631 | F: | TTCTCTGCCAACATATCACAAT | CTGGGGTCCAGAGACTCACA |
| R: | CGCTCAGACTGTTATGAATACTAT | ACCACGGATTCATGCGACAA |
Fig. 4.
Validation of circRNAs by RT-PCR. Expression of eight selected circRNAs was validated by RT-PCR using both divergent primers covering the back-splice junction and convergent primers. The linear GADPH gene served as an internal control.
2.5. PEDV infection induced differential expression of circRNAs and their parent genes
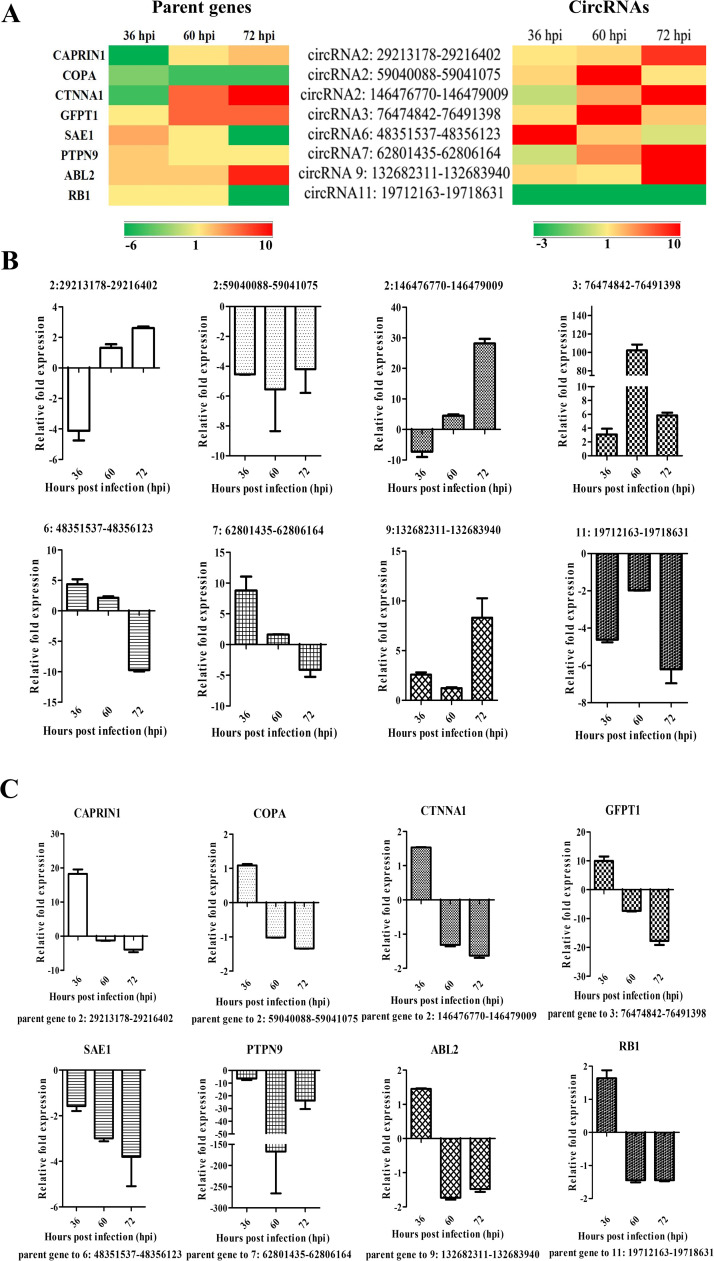
IPEC-J2 cells infected with PEDV and mock-infected were harvested at 36, 60 and 72 hpi. Total RNAs were extracted from each sample and equally divided into two parts. One part was treated with RNase R and subjected to real-time RT-PCR analysis while the other part was used for GAPDH detection, served as an internal control. The heat-map showed the differential expression patterns of selected circRNA candidates and their parent genes based on the RNA-seq results ( Fig. 5A). Our real-time RT-PCR results targeted to these selected circRNA candidates showed a coincident expression pattern with the RNA-seq results showed in the heat-map (Fig. 5A, B). Next, the expression levels of their parent linear genes were then detected using real-time RT-PCR assay. Some parent genes showed the same expression pattern with their circRNA transcripts but some other parent genes showed an opposite expression pattern with their circRNAs (Fig. 5C). These data suggested that all circRNAs and their parent genes were active during PEDV infection and might be potentially involved in host response against viral infection.
Fig. 5.
PEDV infection induces the differential expression of circRNAs and their parent genes A) The heat map shows both the parent genes and the selected circRNAs from infected samples compared with mock-treated samples. B) The expression of eight selected circRNAs (circRNA2: 29213178-29216402, circRNA2: 59040088-59041075, circRNA2: 146476770-146479009, circRNA3: 76474842-76491398, circRNA6: 48351537-48356123, circRNA7: 62801435-62806164, circRNA 9: 132682311-132683940 and circRNA11: 19712163-19718631) and C) their parent genes was validated with real-time PCR. The expression levels of circRNAs and their parent genes were normalized to GAPDH (internal control). The 2-ΔΔCt method was used to normalize the relative gene expression data. All experiments were done in triplicate.
2.6. Function associated analysis of the circRNA parent genes
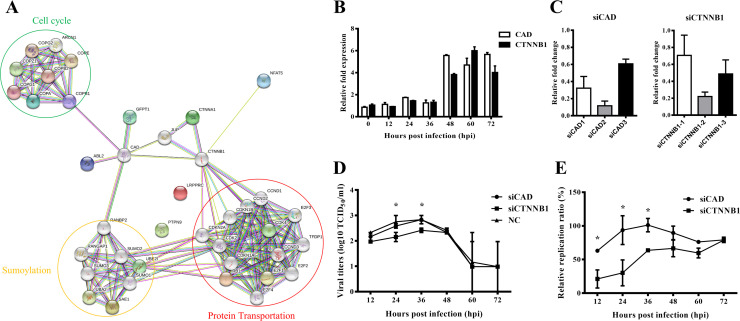
Based on the above results, the functional protein associated network analysis of these detected parent genes were then performed using STRING analysis (https://tring-db.org). The results demonstrated that the genes with their associated partners were mainly divided into three parts. Posttranslational modification of proteins by sumoylation, transportation of proteins between the endoplasmic reticulum and Golgi compartments, and the regulation of cell cycle were the main function among these proteins ( Fig. 6A). SAE1, RB1 and COPA with their interaction partners were involved in these biological processes. CAD and CTNNB1 served as the key nodes which connected these three parts.
Fig. 6.
The functional associated analysis of the parent genes for selected circRNAs A) The STRING analysis shows these parent genes of selected circRNAs are functional associated. These genes with their associated partners are mainly involved in the process of posttranslational modification of proteins by sumoylation, transport of proteins between the endoplasmic reticulum and Golgi compartments, and the regulation of cell cycle. CAD and CTNNB1 served as the key nodes in the interaction network. B) The expression levels of CAD and CTNNB1 at different time points post PEDV infection were measured real-time PCR analysis. C) The siRNA candidates were transfected into IPEC-J2 cells to knockdown the expressions of CAD2 and CTNNB1–2. D) The CAD and CTNNB1 in IPEC-J2 cells were knocked-down by specific siRNAs. The kinetics of PEDV replication in these cells was measured by TCID50 assay. Scramble siRNA was served as negative control. E) The ratios of PEDV genome copies in siCAD and siCTNNB1 cells were normalized by that in NC cells.
We then detected the expression of CAD and CTNNB1 during PEDV infection using the same RNA from RNA-seq analysis. Real-time PCR analysis showed that expression levels of CAD and CTNNB1 remained at baseline before 36 hpi and significantly increased thereafter (Fig. 6B). To investigate the relationship between PEDV replication and CAD and CTNNB1, the siRNAs targeting CAD and CTNNB1 were then synthesized and transfected into IPED-J2 cells. The results demonstrated that siCAD-2 and siCTNNB1–2 among their siRNA candidates exhibited the best inhibition efficacy (Fig. 6C). The siCAD-2 and siCTNNB1–2-transfected IPEC-J2 cells were inouculated with PEDV and harvested at different time points. The viral titters were determined by TCID50 assay. The results showed that knocking-down of CTNNB1 but not CAD significantly decreased the viral loads of PEDV at early stage (Fig. 6D, E), suggesting that CTNNB1 might be involved in PEDV infection and replication.
2.7. Interaction network of circRNA-miRNA
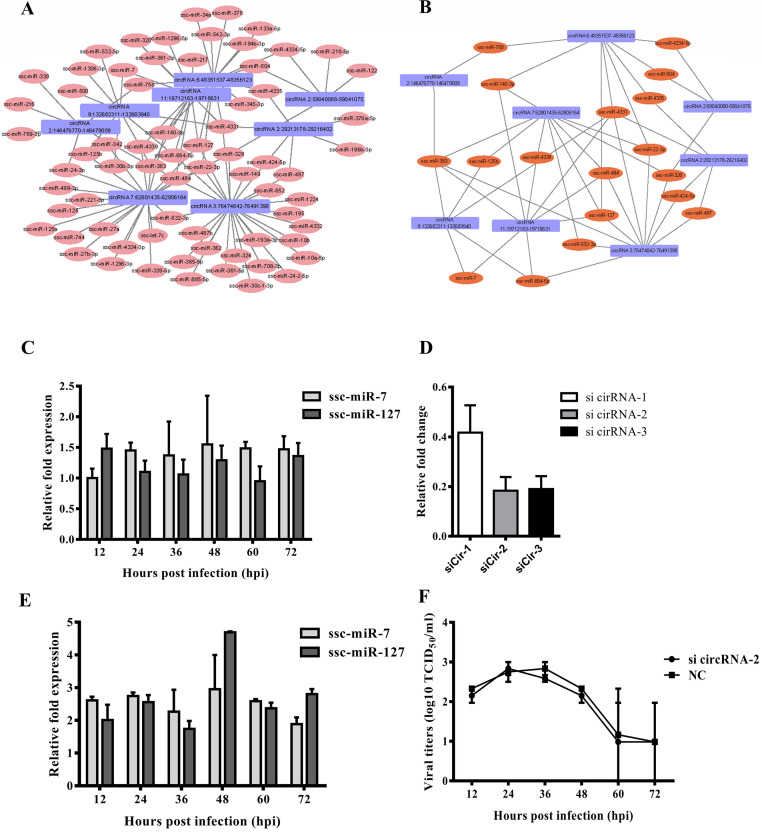
The functions of circRNA were recently identified as regulators of transcription and splicing of their parent genes, miRNA sponges, adaptors for protein–protein interaction and protein-coding, modulators of rRNA and tRNA biogenesis. Apart from the main function in regulating transcription and splicing of their parent genes, circRNAs can also indirectly regulate gene expression by competitively binding miRNAs with genes (Hansen et al., 2013a, Kulcheski et al., 2016). As most miRNAs have not been studied, an integrated analysis of interaction between the selected circRNAs and their target miRNAs was performed to gain insights into their functional connections. The binding site analysis of miRNAs in circRNAs showed that a total of 102 miRNAs (see Supplementary Table 2 for details) had potential interactions with selected circRNAs ( Fig. 7A). Then the connection nodes among different circRNAs were extracted for a detailed ceRNA network analysis. As showed in Fig. 7B, 18 miRNAs interact with these 8 selected circRNAs. Among the 18 miRNAs, the expression levels of ssc-miR-127and ssc-miR-7 elevated > 1.5 folds at several time points (Fig. 7C). The analysis of circRNA-miRNA interaction network revealed that circRNA11: 19712163-19718631 may act as a miRNA sponge for ssc-miR-127 and ssc-miR-7. To investigate this hypothesis, a set of siRNAs against the back-splicing site of circRNA11: 19712163-19718631 were then synthesized and transfected into IPED-J2 cells. The results demonstrated that sicirRNA-2 targeting circRNA11: 19712163-19718631 exhibited the best inhibition efficacy (Fig. 7D). Once the circRNA11: 19712163-19718631 was knocked-down by sicirRNA-2, the expression of ssc-miR-127 and ssc-miR-7 increased 2–5 folds (Fig. 7E), implying that the circRNA11: 19712163-19718631 is a miRNA sponge for ssc-miR-127 and ssc-miR-7. However, down-regulation of circRNA11: 19712163-19718631 didn’t affect PEDV replication in IPEC-J2 cells (Fig. 7F and Supplementary Fig. 1). Although some of the predicated target genes to these miRNAs might be involved in viral infection and antiviral effect, the role of these miRNAs still remained largely unknown.
Fig. 7.
The circRNA–miRNA interaction network A) The circRNA-miRNA regulatory circuit mediated by cellular miRNAs upon PEDV infection in IPEC-J2 cells. B) miRNAs that sponged at least two circRNAs may play important roles in the regulation. The rectangle nodes represent circRNAs while circle nodes represent miRNAs. C) The expression levels of ssc-miR-7 and ssc-miR-127 at different time points post PEDV infection were measured real-time PCR analysis. D) The siRNA candidates were transfected into IPEC-J2 cells to knockdown the expressions of circRNA11: 19712163-19718631. E) Following down-regulation of circRNA11: 19712163-19718631 by siRNA, the expression levels of ssc-miR-7 and ssc-miR-127 were measured by real-time RT-PCR. F) The circRNA11: 19712163-19718631 in IPEC-J2 cells were knocked-down by specific siRNAs. The kinetics of PEDV replication in these cells was measured by TCID50 assay. Scramble siRNA was served as negative control.
3. Discussion
Recently, accumulating researches have shown that the circRNA is a transcriptional production in various tissues and cell types of eukaryotes (Danan et al., 2012, Guo et al., 2014, Lu et al., 2015, Veno et al., 2015). The functions of circRNAs have been primarily illustrated in the disease progression and cancer (Hsiao et al., 2017, Li et al., 2015a, Lu, 2017, Xu et al., 2017, Yang et al., 2016). However, the expression profiles and functions of circRNAs during viral infection remain largely unknown. To date, there have only been quite a few reports regarding the identification and characterization of circular RNAs during the infection of ebola virus, hepatitis B virus, avian leukosis virus subgroup-J and DNA tumor virus SV40 (Cui et al., 2018, Shi et al., 2017, Wang et al., 2017, Zhang et al., 2017). There is still a long way to elucidate the detailed functions and mechanism of circRNAs in viral infection.
In this study, we investigated the dynamic circRNA expression profile of IPEC-J2 cells infected with PEDV. In order to investigate the relationship between circRNAs and PEDV-induced immune responses in the IPEC-J2 cells, we collected samples at different time points post PEDV infection based on the expression of IFN-λ, Foxp3, and GM-CSF, for circRNAs sequencing and identified 26670 circRNA candidates. With GO and KEGG analysis, we demonstrated that the parent genes of DE circRNAs were closely related to the metabolism process, the ubiquitin mediated proteolysis, and the T cell receptor signaling pathway at the early stage of PEDV infection, and the degradation process at late stage, suggesting some circRNAs might be involved in these processes. Eight circRNAs were further selected and validated their existence using RT-PCR assay. The expression levels of their linear parent genes were also detected. The ceRNA network was constructed to define the interaction between selected circRNAs and their target miRNAs. All these data suggested that circRNAs could potentially restrict or facilitate PEDV infection through regulating gene expression.
The cell tropism of PEDV infection is mainly targeted to the small intestines, which is the largest mucosal tissue across whole body. There are lots of microorganisms residing in the intestinal tract, which makes it more complicated for the investigations of host-pathogen interactions and immune responses in the gut. The Vero-E6 cell line is widely used for the isolation, culture and innate immunity studies of PEDV. However, the results from Vero-E6 may mislead the understanding of the mechanism immune responses and pathogenicity of PEDV infection because it is an African green monkey kidney epithelial cells lack of genes encoding type I interferons (IFN), instead of porcine intestinal epithelial cells. In this study, we investigated the expression profile of circRNAs in PEDV infected IPEC-J2 cells, a porcine ileum epithelium cell line. We believe that this cell line will help us get more reliable information about PEDV-host interactions at RNA level.
The major question in the field of circRNAs research is to precisely figure out their biological functions. The circRNAs regulate gene expression through several different ways. The generation of circRNAs derived from exonic regions may potentially affect the protein expression from the parent genes. The exon-intron circRNAs localized to the promoter of their parental genes and associated with RNA polymerase II enhance the transcription efficiency through the interactions between U1 snRNA/U1A/U1C complex and the 50-splice site remained in the intron (Li et al., 2015b). Another most important way is called “ceRNA hypothesis” (Taulli et al., 2013), in which circRNAs can sequestrate miRNA and impair its activity, thereby up-regulating or down-regulating miRNA target gene expression (Thomson and Dinger, 2016). CiRS-7 is the first circRNA identified as miRNA sponge (Hansen et al., 2013b). It has more than 70 conserved binding sites of miRNA-7 and is densely bound by Argonaute (AGO) proteins (Hansen et al., 2013a). In addition to parent gene regulation and miRNA sponges, circRNAs can interact with RNA binding proteins, such as circ-Foxo3 (Du et al., 2016), circ-MBL (muscleblind) (Memczak et al., 2013) or coding proteins (Legnini et al., 2017; Yang et al., 2017; Yang et al., 2018). But the study of the detailed regulation mechanism is still in its preliminary phase. Most circRNAs have been proved to play important roles via the former two mechanisms. Therefore, we performed the functional analysis to predict the roles of identified circRNAs candidates according to their parent genes and their target miRNAs.
In this study, functional analysis revealed that the ubiquitin mediated proteolysis and the T cell receptor signal pathway were closely related to parent genes of DE circRNAs at 36 hpi and 60 hpi. At 72 hpi, most expressed genes were involved in the process of cell degradation. These gene expression patterns were highly coincident with the outputs of IPEC-J2 cells post PEDV infection, showing upregulated type III IFNs expression at 36 hpi, cytokine secretion at 60 hpi, and cell apoptosis at 72 hpi. The parent gene of circRNA6: 48351537-48356123 was SAE1, a SUMO activating enzyme subunit, which could play a key role in the regulation of IFNs. The circRNA6: 48351537-48356123 was upregulated at 36 hpi but downregulated thereafter, which correlated with the dynamic expression levels of Type III IFNs. The parent gene of circRNA2: 59040088-59041075 and circRNA3: 76474842-76491398 was involved in the vesicles transportation and secretion. These processes were vital to the function of cytokines. The expressions of all the circRNA7: 62801435-62806164, circRNA 9: 132682311-132683940 and circRNA11: 19712163-19718631 were drastically affected at 72 hpi. Their parent genes were components of the P body or involved in autophagy and cell cycle regulation. These processes were also related with cell death. All these data above demonstrated that the circRNAs we identified in this study have the potential to play important role in host response to PEDV infection.
In additional, we used the software miRanda-3.3a to predict the microRNA targeting sites for circRNAs. A gene co-expression network between selected circRNAs and miRNAs was then constructed for the further functional analysis. Nine key node miRNAs among all predicated miRNAs were detected in our studies and presented as interesting molecules for further study. No target mRNAs of these miRNAs were identified and their roles in viral infection or the life cycle of cells had not been reported. The detailed functions of these miRNAs should be elaborated in the future study.
In summary, this study identified and characterized 26670 circRNA candidates related to PEDV infection on IPEC-J2 cells. Eight circRNAs were selected and identified their existence. To our knowledge, this is the first piece of data showing circRNA expression profile induced by PEDV infection on swine ileum epithelial cells. We believe that the results from this study will provide a solid basis for understanding the detailed functions of circRNA between virus-host interactions.
4. Materials and methods
4.1. Cells culture and virus infection
IPEC-J2 cells were cultured in DMEM/F12 (Gibco) medium supplemented with 10% fetal bovine serum (Gemini, USA) free of bovine viral diarrhea virus (BVDV) and anti-BVDV antibodies, 100 units/ml penicillin, 5 μg/ml insulin/selenium/transferrin (Life, USA) and 5 ng/ml epidermal growth factors (EGF) (Life, USA) in 5% CO2, incubator at 37 °C. The PEDV LJX01/GS/2014 strain was isolated from a pig farm and propagated in our laboratory into a cell-adapted virus. To analyze host responses to PEDV infection, 80% confluent IPEC-J2 cells were inoculated with PEDV at an MOI = 0.01 or mock infected. Following incubation at 37 °C for 1 h, the IPEC-J2 cells were then washed with PBS three times and supplied with culture medium. At different time points post infection, both the supernatant and the cell pellets were collected for further analysis.
4.2. RNA processing and library preparation
Total RNA was extracted from both PEDV infected and mock infected cells at different time points (36, 60 and 72 hpi) by TRIzol reagent (Invitrogen, USA). Total RNA of each sample was quantified and qualified by the Agilent 2100 Bioanalyzer (Agilent Technologies, USA), and 1 μg total RNA with RIN value above 7 was used for following library preparation. Pair-End index libraries were constructed according to the manufacturer's protocol (NEBNext® Ultra™RNA Library Prep Kit for Illumina®). Then libraries with different indexes were multiplexed and loaded on an Illumina HiSeq instrument according to manufacturer's instructions (Illumina, USA). Sequencing was carried out using a 2 × 125 paired-end (PE) or 2 × 150 paired-end (PE) configuration; image analysis and base calling were conducted by the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina, USA) on the HiSeq instrument. The sequences were processed and analyzed by GENEWIZ Biotechnology Co.
4.3. CircRNA prediction and mapping
After sequencing, the QC-checked paired end (PE) reads of each samples were mapped to reference genome (Sscrofa10.2, Ensemble) using Bwa (v0.7.5a-r405) (Langmead and Salzberg, 2012). The duplicate reads due to PCR amplification during library preparation were removed by samtools. CIRI (V2.0) was then used to search for a set of predicted circRNA loci from the data obtained (Gao et al., 2015).
4.4. Differential expression analysis of circRNA candidates
In our study, three samples of each group were collected and pooled for sequencing. The DESeq Bioconductor package (v1.18.0), a model based on the negative binomial distribution, was employed for differential expression analysis (Anders and Huber, 2010). The read count data were generated from the negative binomial distribution. For each gene, the count Yij was given by Yij ~ NB (μij, σij2). Here, σi is the per-gene dispersion calculated by the CR-APL method, and the expected value μi is a function of the library size. After being adjusted by Benjamini and Hochberg's approach for controlling the false discovery rate, P-value of detected genes were set at < 0.05 for differential expression.
4.5. Experimental validation of circRNAs
In order to validate the existence of selected circRNAs, both convergent and divergent primers were designed according to the predicated sequences of circRNA candidates. The sequences of primer sets are listed in Table 1. The PCR conditions were as follows: an initial 90 s step at 94 °C followed by 35 cycles of 30 s denaturing at 94 °C, 30 s annealing at 56 °C and 30 s extension at 72 °C. Genomic DNA was extracted using a Cell-Tissue DNA Midi Preparation Kit (Bio Teke).
4.6. RNA Preparation and qRT-PCR analysis
For the removing of linear RNA, 1 µg of total RNA from each sample was mixed with 1 unit of Ribonuclease R (RNase R) (Epicentre) and 2 µl reaction buffer. The reaction mixtures were incubated at 37 °C for 10 min. For controls, the RNA was treated without RNase R. After digestion, the RNAs were then purification by RNeasy MinElute Cleanup Kit (Qiagen, Germany). To quantify the amount of circRNA and mRNA, the prepared RNA of each sample was reverse transcribed into cDNA with Revertaid first strand cDNA synthesis kit (Thermo, USA). The real-time PCR was performed in ABI 7500 system with Fast Start Universal SYBR Green Master (Roche, Switzerland), and primer sets listed in Table 2. To quantify the expression levels of CAD and CTNNB1, the primer sets (CAD-F: ACAAGGAGGACTTTGCCTCG, CAD-R: GCTCCGAGGAACAAGGTGTA; CTNNB1-F: AGACATGCCATCATGCGTTC, CTNNB1-R: CAGTACAGCGAGCCGTTTC; and GAPDH-F: ATCCCGCCAACATCAAATGG, GAPDH-R: ACTTCTCATGGTTCACGCCC) were designed and synthesized from TsingKe Biotech company (Beijing, China). For ssc-miR-7 and ssc-miR-127 quantification, the miRcute miRNA cDNA synthesis kit and primer sets were purchased from Tiangen Biotech Company (Beijing, China). Briefly, the reactions were incubated at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 30 s. All reactions were run in triplicate. The 2-∆∆Ct method was used to measure expression level of target circRNAs and mRNAs.
Table 2.
The qPCR primers of the parent genes to selected circRNAs.
| circRNA | Parent gene | Primer sets |
|---|---|---|
| 2:29213178-29216402 | CAPRIN1 | F: AGGCTTGATGAATGGATA |
| R: TTGGAGTGTTAGAGAATGA | ||
| 2:59040088-59041075 | COPA | F: TCTTCACATTGCTTGGACAT |
| R: ACACTCGGATGGTCTGAT | ||
| 2:146476770-146479009 | CTNNA1 | F: CTTTGCTCTCTGCTGTTA |
| R: GATACCATCTTCTACGACTT | ||
| 3:76474842-76491398 | GFPT1 | F: AACACAATGAGAGGAAGA |
| R: CGCAAGCAATAAGGATTA | ||
| 6:48351537-48356123 | SAE1 | F: CTACCACGGATACACATT |
| R: CTTCTACTCCTTGACTAACT | ||
| 7:62801435-62806164 | PTPN9 | F: CACACTCAGACAGATTAC |
| R: ACGATAGGTATTCTCCAA | ||
| 9:132682311-132683940 | ABL2 | F: CGCTGAAGGAGGATACTA |
| R: CCAGATTAGGATGCTTGATT | ||
| 11:19712163-19718631 | RB1 | F: CGAAGAAGAACGATTATCCATT |
| R: AGCCATTACAACCTCAAGA |
4.7. RNA interference
To knockdown CAD, CTNNB1 and circRNA11: 19712163-19718631, small interfering RNA (siRNA) were designed and synthesized by RiboBio company. The detailed sequences of these siRNAs were listed in Table 3. Following the manufacturer's instructions, 200 nM siRNAs were transfected into IPEC-J2 cells using X-tremeGENE siRNA Transfection reagent (Roche, Switzerland). Cells were harvested at different time points for further analysis.
Table 3.
The sequences of siRNAs.
| Target genes | siRNA name | siRNA sequence (5′-3′) |
|---|---|---|
| CAD | siCAD-1 | CAAGACATCACTGCCAAGA |
| siCAD-2 | GGAGAACATGGCCATCATT | |
| siCAD-3 | AGATGTCTCACCTGTTCAA | |
| CTNNB1 | siCTNNB1-1 | TGGACAGTATGCAATGACT |
| siCTNNB1–2 | CTCAAGCTTTAGTGAATAT | |
| siCTNNB1–3 | ATACCATTCCATTGTTTGT | |
| circRNA11: 19712163-19718631 | sicirRNA-1 | ACATTCAGAATCCACTTCT |
| sicirRNA-2 | TCAGAATCCACTTCTGATT | |
| sicirRNA-3 | GAATCCACTTCTGATTTCA |
4.8. GO annotation and KEGG pathway analysis
Gene Ontology (GO) functional significance and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to determine the roles of the parent genes of circRNAs in GO terms (biological processes, cellular components, and molecular functions) and biological pathways. The q value yields the rich factor indicating the significance of pathway correlations.
4.9. The construction of circRNA–miRNA co-expression network
The miRNA targeting sites for the circRNAs was predicated by miRanda-3.3a. The circRNA–miRNA co-expression network was then constructed based on the prediction of miRNA binding sites and the correlations between circRNA and miRNA. The circRNA–miRNAs with the lowest p value were selected to generate a network map with Cytoscape software version 3.2.1 (Shannon et al., 2003).
4.10. Statistical analysis
The differences between matched groups were examined for statistical significance using Student's t-test. An unadjusted P value of less than 0.05 was considered to be significant.
4.11. Availability of data and materials
The raw data for this article were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under BioProject number PRJNA 505270.
Acknowledgements
The authors thank Ms. Manita Aryal for critical proofreading of this manuscript. This work was supported by National Key R&D Program of China (2016YFD0500103), National Natural Science Foundation of China (31572498 and 31702209), the Elite Youth Program of CAAS, and partly by China Central Public-interest Scientific Institution Basal Research Fund.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2018.11.014.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- van den Brand J.M., Smits S.L., Haagmans B.L. Pathogenesis of middle east respiratory syndrome coronavirus. J. Pathol. 2015;235:175–184. doi: 10.1002/path.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014;59:118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Feng T., Fujihashi K., Schoeb T.R., Elson C.O. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc. Natl. Acad. Sci. USA. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S., Qian Z., Chen Y., Li L., Li P., Ding H. Screening of up- and downregulation of circRNAs in HBV-related hepatocellular carcinoma by microarray. Oncol. Lett. 2018;15:423–432. doi: 10.3892/ol.2017.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danan M., Schwartz S., Edelheit S., Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies P.R. The dilemma of rare events: porcine epidemic diarrhea virus in North America. Prev. Vet. Med. 2015;122:235–241. doi: 10.1016/j.prevetmed.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Zhang X., Wu X., Guo H., Hu Y., Tang F., Huang Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015;16:148. doi: 10.1186/s13059-015-0706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wang J., Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommerman J.L., Rojas O.L., Fritz J.H. Re-thinking the functions of IgA(+) plasma cells. Gut Microbes. 2014;5:652–662. doi: 10.4161/19490976.2014.969977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Riveron J.T., Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim. Biophys. Acta. 2016;1859:1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A.L., Ahrens P., Bright P.M., Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- Halbreich A., Pajot P., Foucher M., Grandchamp C., Slonimski P. A pathway of cytochrome b mRNA processing in yeast mitochondria: specific splicing steps and an intron-derived circular DNA. Cell. 1980;19:321–329. doi: 10.1016/0092-8674(80)90506-1. [DOI] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- Heaton S.M., Borg N.A., Dixit V.M. Ubiquitin in the activation and attenuation of innate antiviral immunity. J. Exp. Med. 2016;213:1–13. doi: 10.1084/jem.20151531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermant P., Michiels T. Interferon-lambda in the context of viral infections: production, response and therapeutic implications. J. Innate Immun. 2014;6:563–574. doi: 10.1159/000360084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K.Y., Lin Y.C., Gupta S.K., Chang N., Yen L., Sun H.S., Tsai S.J. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.I., Giannakopoulos N.V., Virgin H.W., Zhang D.E. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulcheski F.R., Christoff A.P., Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg. Infect. Dis. 2014;20:1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., Laneve P., Rajewsky N., Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Molecular cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. (e29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang J., Zhou P., Le Y., Zhou C., Wang S., Xu D., Lin H.K., Gong Z. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am. J. Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- Li L., Fu F., Xue M., Chen W., Liu J., Shi H., Chen J., Bu Z., Feng L., Liu P. IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antivir. Res. 2017;140:76–82. doi: 10.1016/j.antiviral.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., Zhu P., Chang Z., Wu Q., Zhao Y., Jia Y., Xu P., Liu H., Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- Liang G., Yang Y., Niu G., Tang Z., Li K. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res.: Int. J. Rapid Publ. Rep. Genes Genomes. 2017;24:523–535. doi: 10.1093/dnares/dsx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Cui L., Zhou Y., Zhu C., Fan D., Gong H., Zhao Q., Zhou C., Zhao Y., Lu D., Luo J., Wang Y., Tian Q., Feng Q., Huang T., Han B. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.Y. Roles of the circular RNA circ-Foxo3 in breast cancer progression. Cell Cycle. 2017;16:589–590. doi: 10.1080/15384101.2017.1278935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Nei T., Urano S., Motoi N., Takizawa J., Kaneko C., Kanazawa H., Tazawa R., Nakagaki K., Akagawa K.S., Akasaka K., Ichiwata T., Azuma A., Nakata K. IgM-type GM-CSF autoantibody is etiologically a bystander but associated with IgG-type autoantibody production in autoimmune pulmonary alveolar proteinosis. American journal of physiology. Lung Cell. Mol. Physiol. 2012;302:L959–L964. doi: 10.1152/ajplung.00378.2011. [DOI] [PubMed] [Google Scholar]
- Park S.W., Han M.G., Park C., Ju Y.R., Ahn B.Y., Ryou J. Hantaan virus nucleocapsid protein stimulates MDM2-dependent p53 degradation. J. Gen. Virol. 2013;94:2424–2428. doi: 10.1099/vir.0.054312-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Travanty E.A., Oko L., Edeen K., Berglund A., Wang J., Ito Y., Holmes K.V., Mason R.J. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am. J. Respir. Cell Mol. Biol. 2013;48:742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russler-Germain E.V., Rengarajan S., Hsieh C.S. Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol. 2017;10:1375–1386. doi: 10.1038/mi.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Zhang C., Guo P., Liu Z., Sun M., Sun J., Li L., Dong J., Zhang J. Short communication: antiviral activity of porcine IFN-lambda3 against porcine epidemic diarrhea virus in vitro. Virus Genes. 2016;52:877–882. doi: 10.1007/s11262-016-1374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Hu N., Li J., Zeng Z., Mo L., Sun J., Wu M., Hu Y. Unique expression signatures of circular RNAs in response to DNA tumor virus SV40 infection. Oncotarget. 2017;8:98609–98622. doi: 10.18632/oncotarget.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen K.D., Sorensen A.B., Quintanilla-Martinez L., Kunder S., Schmidt J., Pedersen F.S. Distinct roles of enhancer nuclear factor 1 (NF1) sites in plasmacytoma and osteopetrosis induction by Akv1-99 murine leukemia virus. Virology. 2005;334:234–244. doi: 10.1016/j.virol.2005.01.039. [DOI] [PubMed] [Google Scholar]
- Su W.C., Chen Y.C., Tseng C.H., Hsu P.W., Tung K.F., Jeng K.S., Lai M.M. Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc. Natl. Acad. Sci. USA. 2013;110:17516–17521. doi: 10.1073/pnas.1312374110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulli R., Loretelli C., Pandolfi P.P. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat. Struct. Mol. Biol. 2013;20:541–543. doi: 10.1038/nsmb.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temeeyasen G., Srijangwad A., Tripipat T., Tipsombatboon P., Piriyapongsa J., Phoolcharoen W., Chuanasa T., Tantituvanont A., Nilubol D. Genetic diversity of ORF3 and spike genes of porcine epidemic diarrhea virus in Thailand. Infect., Genet. Evol.: J. Mol. Epidemiol. Evolut. Genet. Infect. Dis. 2014;21:205–213. doi: 10.1016/j.meegid.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- Veno M.T., Hansen T.B., Veno S.T., Clausen B.H., Grebing M., Finsen B., Holm I.E., Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Deng X., Yan W., Zhu Z., Shen Y., Qiu Y., Shi Z., Shao D., Wei J., Xia X., Ma Z. Stabilization of p53 in influenza A virus-infected cells is associated with compromised MDM2-mediated ubiquitination of p53. J. Biol. Chem. 2012;287:18366–18375. doi: 10.1074/jbc.M111.335422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Guo Z.D., Li J.M., Zhao Z.Z., Fu Y.Y., Zhang C.M., Zhang Y., Liu L.N., Qian J., Liu L.N. Genome-wide search for competing endogenous RNAs responsible for the effects induced by Ebola Virus replication and transcription using a trVLP system. Front. Cell. Infect. Microbiol. 2017;7:479. doi: 10.3389/fcimb.2017.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhang M., Zheng X., Yi P., Lan C., Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Du W.W., Li X., Yee A.J., Yang B.B. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y., Wong C.C., Xiao X., Wang Z. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H., Huang S., Xie B., Zhang N. Novel role of FBXW7 circular RNA in repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C.Y., Chen L., Liu C., Zhu Q.H., Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III Interferon Restriction by Porcine Epidemic Diarrhea Virus and the Role of Viral Protein nsp1 in IRF1 Signaling. J. Virol. 2018;92(4):e01677–17. doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yan Y., Lei X., Li A., Zhang H., Dai Z., Li X., Chen W., Lin W., Chen F., Ma J., Xie Q. Circular RNA alterations are involved in resistance to avian leukosis virus subgroup-J-induced tumor formation in chickens. Oncotarget. 2017;8:34961–34970. doi: 10.18632/oncotarget.16442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Ding Z., Jiang J., Lu F., Xia X., Ma X. Confirmation and preliminary analysis of circRNAs potentially involved in human intervertebral disc degeneration. Mol. Med. Rep. 2017;16:9173–9180. doi: 10.3892/mmr.2017.7718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
The raw data for this article were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under BioProject number PRJNA 505270.