Abstract
Porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV) and porcine deltacoronavirus (PDCoV) share tropism for swine intestinal epithelial cells. Whether mixing of viral components during co-infection alters pathogenic outcomes or viral replication is not known. In this study, we investigated how different coronavirus nucleocapsid (CoV N) proteins interact and affect PEDV replication. We found that PDCoV N and TGEV N can competitively interact with PEDV N. However, the presence of PDCoV or TGEV N led to very different outcomes on PEDV replication. While PDCoV N significantly suppresses PEDV replication, overexpression of TGEV N, like that of PEDV N, increases production of PEDV RNA and virions. Despite partial interchangeability in nucleocapsid oligomerization and viral RNA synthesis, endogenous PEDV N cannot be replaced in the production of infectious PEDV particles. Results from this study give insights into functional compatibilities and evolutionary relationship between CoV viral proteins during viral co-infection and co-evolution.
Keywords: Porcine epidemic diarrhea virus, Swine coronavirus, Nucleocapsid protein
Highlights
-
•
PDCoV N and TGEV N interact with PEDV N in a competitive, RNA-dependent manner.
-
•
PEDV replication in cell culture is enhanced by overexpression of TGEV or PEDV N but strongly suppressed by that of PDCoV N.
-
•
Both TGEV and PDCoV N can partially rescue viral RNA and protein synthesis functions of PEDV N, albeit to different degrees.
-
•
Neither TGEV nor PDCoV N can completely replace PEDV N in the production of PEDV infectious virions.
1. Introduction
Porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV) and porcine deltacoronavirus (PDCoV) belong to the family Coronaviridae (Enjuanes, 2000). PEDV and TGEV have been classified into the alphacoronavirus genus, whereas PDCoV belongs to the deltacoronavirus genus (Jung et al., 2016a; Jung and Saif, 2015a). They share similar genome architectures, with a 25–30 kb positive-sense, single-stranded RNA genome. The 5′ two-thirds of the viral genome encodes non-structural proteins from open reading frames (ORF) 1a and 1b necessary for viral genome replication. The rest of the genome encodes a number of unique accessory proteins such as PEDV ORF3, TGEV 3a/3b/7, PDCoV NS6/NS7, and four common structural proteins, namely the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins (Kocherhans et al., 2001; Lee and Lee, 2014; Penzes et al., 2001). These enteric swine coronaviruses (CoVs) infect epithelial cells lining the small intestine and cause villous atrophy, resulting in malabsorption and severe diarrhea (Jung et al., 2016a). An outbreak of these viruses, especially PEDV, can lead to up-to-100% mortality in neonatal piglets, prompting huge economic losses in the swine production industry worldwide. Unless they are examined by laboratory-level diagnosis, these CoVs produce almost indistinguishable pathogenesis.
Co-infection of enteric pathogens are common. TGEV and PDCoV have been found to co-circulate with PEDV in the field (Song et al., 2015; Wang et al., 2016). In PDCoV-positive samples, the rate of PEDV co-infection as detected by RT-PCR varies from 33% to 50% (Jung et al., 2016a; Jung and Saif, 2015a). Although TGEV infection has become rarer nowadays, it has still been detectable in samples in China, and often together with PEDV and/or PDCoV (Dong et al., 2015; Wang et al., 2013). Despite substantial epidemiological evidence of co-infection, the effects of these events on disease outcomes have not yet been formally described. Since these enteric swine CoVs share cell tropism, co-infection of these viruses can theoretically cause mixing of viral components in the same cellular compartments, possibly leading to direct or indirect effects on viral replication kinetics or pathogenic outcomes. To the best of our knowledge, there are currently no reports on studies at molecular or cellular levels on how viral components from different CoV species interact with or affect other viruses. Investigation of possible molecular interactions between components of PEDV, PDCoV and TGEV and their influence on replication of each virus would provide a crucial insight into comprehensive understanding of these CoVs.
Of all viral proteins, we have chosen to start with the N protein, as it is among the most abundant and ubiquitous structural proteins in infected cells. The CoV N protein is functionally conserved across the family Coronaviridae (Chang et al., 2009; Cong et al., 2017), with its primary function being to form a scaffold for packaging viral genomic RNA (gRNA) into the internal core of virions (de Haan and Rottier, 2005). Besides scaffolding, other functions of the CoV N protein (primarily based on studies of common representatives of the family like severe acute respiratory syndrome-CoV (SARS-CoV) or mouse hepatitis virus (MHV)) include acting as RNA chaperones (Zuniga et al., 2007, 2010), promoting viral genome transcription or replication (Hurst et al., 2010, 2013; Masters et al., 1994; Zuniga et al., 2007, 2010), facilitating viral assembly (de Haan and Rottier, 2005; Kuo et al., 2016), suppressing antiviral RNA-interference activity from their hosts (Cui et al., 2015), and suppressing host immunity (Ding et al., 2014, 2017; Xu et al., 2013; Zhang et al., 2018).
Based on sequence alignment and limited structural data from some representative CoVs, all CoV N proteins are predicted to consist of three structural domains: the N-terminal domain (NTD), linker region (LKR) and C-terminal domain (CTD) (Chang et al., 2014; McBride et al., 2014). NTD binds RNA through electrostatic interaction with its charged amino acids as well as interaction between conserved aromatic residues in the proteins and nucleotide bases in the RNA (Chang et al., 2014; Huang et al., 2004; Tan et al., 2006). LKR is a disordered domain between NTD and CTD. Studies of SARS-CoV N indicate roles for LKR in RNA binding, virion assembly and self-association (Chang et al., 2009, 2013; He et al., 2004a). Although it is also reported to have RNA binding capacity, CTD is a more hydrophobic domain mainly responsible for self-association to form stable dimers and subsequent oligomers of CoV N (Chen et al., 2007; Yu et al., 2006). Studies of N proteins from SARS-CoV and MHV revealed that N dimerization followed by multimerization is a common process in the formation of viral ribonucleoprotein that initiates viral genome packaging in the virion assembly process among CoVs (Cong et al., 2017; Fan et al., 2005; Ma et al., 2010; Yu et al., 2006).
Examples from other viruses suggest that cross-association between viral nucleocapsid proteins could determine different outcomes during co-infection. For instance, mixed infection between two types of plant tospoviruses results in interspecies interaction between their N proteins and more severe symptoms compared to single infections, suggesting synergy between these viruses (Bag et al., 2012; Tripathi et al., 2015). On the other hand, intertypic interference between types A and B influenza viruses could be partially explained by the inhibitory effect of type B influenza virus nucleoprotein exerted on its type A counterpart (Aoki et al., 1984; Jaru-ampornpan et al., 2014). Given structural similarities, we expect that CoV N proteins would likely cross-interact with each other during a co-infection event. Whether this cross-interaction has beneficial or detrimental effects on viral replication has never been explored. Investigation of the interaction between CoV N proteins could lead to insights into virus evolution, anti-CoV drug development and vaccine design (Chang et al., 2016; Lo et al., 2013).
2. Materials and methods
2.1. Biological materials
Human embryonic kidney (HEK) 293T cells, wild-type VeroE6 cells and VeroE6-based cell lines stably expressing CoV N were maintained in OptiMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics at 37 °C with 5% CO2. The virus PEDV-AVCT12-mCherry (PEDV-mCherry), its infectious clone (pSMART-BAC- mCherry-PEDVAVCT12 [pPEDV-mCherry]) and pCAGGS-PEDV N-Myc, a plasmid for C-terminally Myc-tagged PEDV N expression, have been described previously (Jaru-Ampornpan et al., 2017; Jengarn et al., 2015).
2.2. Plasmid construction
TGEV and PDCoV N encoding genes were codon-optimized for high expression in mammalian cells and to avoid expression of PDCoV NS7 embedded in the PDCoV N gene (Fang et al., 2017) (GenBank: AAG30228.1, AFD29191.1, respectively). PCR products of the corresponding N genes with indicated tags at the C-termini were ligated into pCAGGS vectors to give plasmids for expression of tagged CoV N (pCAGGS-TGEV N-FLAG and pCAGGS-PDCoV N-HA). pCAGGS-HA-PEDV N for N-terminally HA-tagged PEDV N expression was similarly constructed based on the same PEDV N DNA sequence in pCAGGS-PEDV N-Myc. pGEX-4T3-PEDV N was constructed by in-frame insertion of the PEDV N gene sequence, codon-optimized for bacterial expression based on the sequence from AVCT12 strain (accession number LC053455), into the pGEX-4T3 vector (Amersham) following the coding sequence of N-terminal GST. All plasmids were verified by DNA sequencing.
2.3. Western blot analysis and antibodies
Cells were lysed with lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 5% glycerol, supplemented with a protease inhibitor cocktail (Halt™ Protease inhibitor cocktail, Thermo Scientific)). Proteins in the cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto nitrocellulose membranes (Bio-Rad Laboratories). The membrane was blocked in 5% skim milk prior to incubation with indicated primary antibodies. Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Biolegend) or (HRP)-conjugated donkey anti-rabbit IgG (BioLegend) was used as secondary antibodies. Primary antibodies used in this study include mouse-anti-Myc (Thermo Scientific), rabbit-anti-FLAG (Cell Science Technology), rabbit-anti-HA (Cell Science Technology), mouse-anti-S1 (a kind gift from Dr. Qigai He, Huazhong Agricultural University), and mouse-anti-PEDV N (SD 6–29, Medgene Labs).
2.4. Co-immunoprecipitation (Co-IP)
HEK293T cells were co-transfected with pCAGGS-PEDV N-Myc (or the empty pCAGGS vector for negative controls) and either pCAGGS-HA-PEDV N, pCAGGS-TGEV N-FLAG or pCAGGS-PDCoV N-HA using FuGENE HD (Promega) according to the manufacturer's instructions. At 48 h post-transfection (hpt), cell lysates were prepared as described previously and pre-adsorbed to control agarose beads (Thermo Scientific) for 1 h at 4 °C to reduce non-specific binding. Then, the treated lysates were incubated with anti-Myc-conjugated agarose beads (Thermo Scientific) at 4 °C overnight. The mixtures were washed three times with lysis buffer supplemented with 250 mM NaCl. IP complexes were eluted by boiling the bead mixtures in 2× non-reducing sample buffer (Thermo Scientific) for 5 min and supplemented with 2 mM DTT before SDS-PAGE and western blot analysis.
2.5. GST-pulldown assay
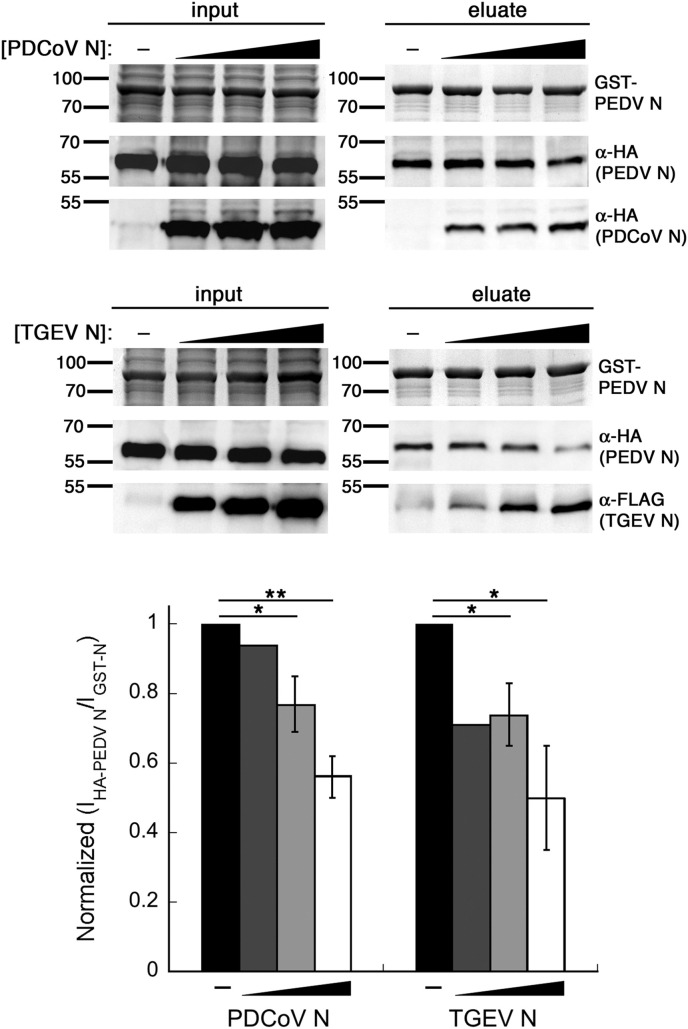
GST and GST-PEDV N were expressed in BL21 (DE3*) cells. After 3-h protein induction by 0.5 mM isopropyl ß-d-1-thiogalactopyranoside, bacteria were lysed by sonication in 1× phosphate-buffered saline (PBS). The lysates were clarified and stored in 100-μl aliquots at –80 °C until use. HEK293T cells were transfected with plasmids as indicated and were lysed at 48 hpt with lysis buffer. The bacterial lysate containing the bait protein (GST/GST-PEDV N) and the HEK293T cell lysate containing the prey protein (HA-PEDV N, TGEV N-FLAG, PDCoV N-HA, or combinations thereof) were incubated with pre-equilibrated GSH FF resin (Amersham) overnight and washed twice in high-salt GST buffer and four times in low-salt GST buffer (20 mM Tris-HCl pH 8.0, 20% glycerol, 1 mM EDTA, 0.1% NP-40 and 1 M/100 mM NaCl) (Nguyen and Goodrich, 2006). The bound proteins were released by boiling the resin in 2× SDS-PAGE loading buffer and analyzed by Coomassie staining (for bait) and western blotting with indicated antibodies (for prey). Figures were representatives of three independent experiments. For Fig. 2 , quantification of eluted protein bands in each GST pulldown reaction was performed using the BioRad Image Lab software 6.0.1 and expressed as the ratios of pulled-down PEDV N (western blot with anti-HA) to eluted GST-PEDV N (Coomassie stain). Because of variability between independent experiments, these ratios were normalized to the conditions without extra CoV N protein performed side-by-side in each experiment before statistical comparison. For the RNaseA experiment, the combined lysates were treated with 10 μg of RNaseA (Thermo Scientific) or PBS buffer for 30 min at 4 °C to avoid protein precipitation at high temperature. To verify the absence of RNA after the treatment, total RNAs were purified from parts of treated lysates by RNA purification kit (Thermo Scientific) and were used at equal volumes as templates for RT-PCR. The RT-PCR products were visualized by Ethidium bromide staining.
Fig. 2.
Other CoV N proteins bind competitively to PEDV N. GST pulldown by GST-PEDV N was performed with HEK293T cell lysates containing HA-PEDV N and varying amounts (0–2 μg transfected plasmids) of PDCoV N-HA or TGEV N-FLAG. The input and eluates were separated by SDS-PAGE. Coomassie staining was used to detect GST-PEDV N, and western blotting was used to detect HA- and FLAG-tagged proteins. The figure is representative of three independent experiments. Quantification was performed using the BioRad Image Lab software 6.0.1 as described in Materials and Methods. The pulldown ratios from the conditions without PDCoV or TGEV N proteins in each independent experiment were set to one. *p < 0.05, **p < 0.01 compared to the conditions without PDCoV or TGEV N proteins.
2.6. Generation of VeroE6 cell lines stably expressing CoV N
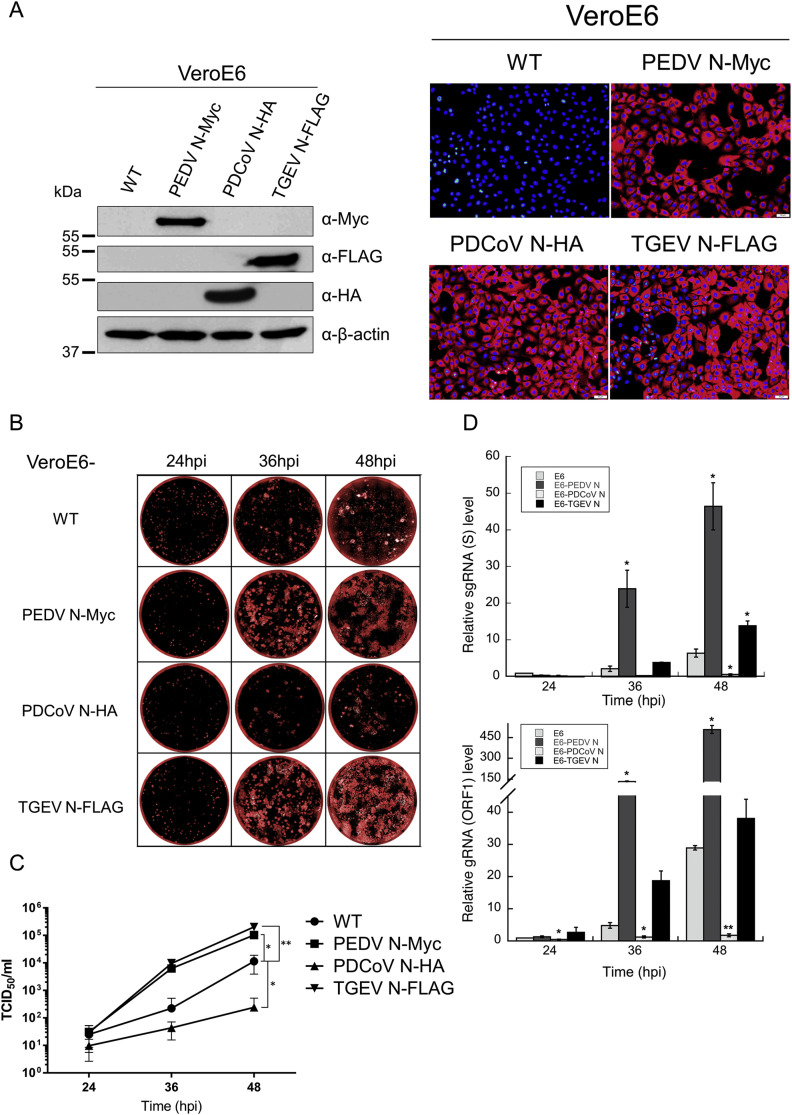
VeroE6 cell lines expressing PEDV N-Myc, TGEV N-FLAG and PDCoV N-HA were constructed using the same protocol as VeroE6-PEDV N cells as described previously (Liwnaree et al., 2019). Briefly, lentiviruses carrying each CoV N gene were generated by co-transfection of pSIN-CSGW-UbEM carrying the inserted CoV N gene with a packaging plasmid encoding Gag, Pol, Rev and Tat (pCMV-ΔR8.91) and a plasmid expressing lentiviral VSV envelope glycoprotein (pMD2.G) into HEK293T cells. At 48 hpt, supernatants containing the lentiviruses were harvested and filtered through a 0.45-μm filter. The filtered supernatants were then used to transduce VeroE6 cells. A single clone of the transduced cells expressing the corresponding N protein was selected based on similar levels of the transgene protein expression.
2.7. Immunofluorescence analysis
Uniform expression of exogenous N proteins in the engineered VeroE6 cell lines was verified by immunofluorescence analysis. Briefly, cells were grown in an eight-well slide. At 24 h, cells were fixed with 80% cold acetone for 10 min. After washing twice with PBS, cells were blocked with 10% FBS, 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature. Cells were then incubated with mouse anti-PEDV N antibodies (SD 6–29) or anti-PDCoV N (SD55-197, Medgene Labs) or anti-TGEV N (211.56, Median Diagnostics, Republic of Korea). After washing twice with PBS, cells were incubated with goat anti-mouse IgG alkaline phosphatase antibodies (Abcam). Fluorescence was developed by adding alkaline phosphatase substrate (ImmPACT™ Vector® Red). Fluorescence images were taken under a fluorescence microscope (Olympus).
2.8. Construction of the N-deficient PEDV infectious clone (pPEDV-mCherry-ΔN)
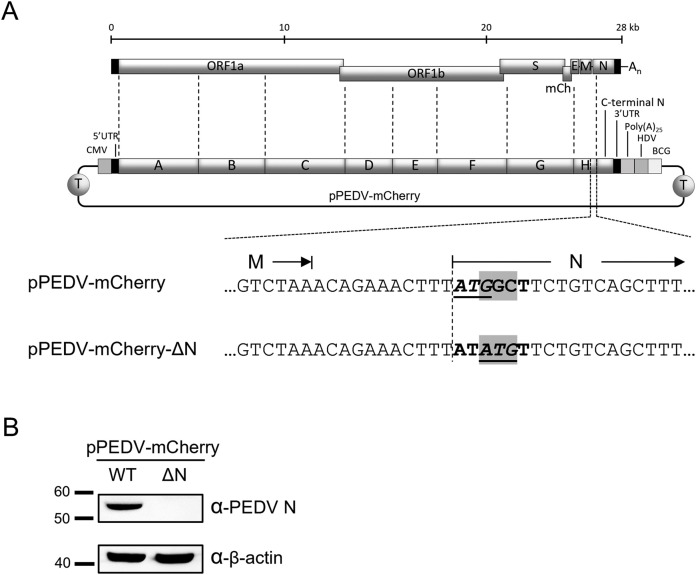
The N-deficient PEDV infectious clone (pPEDV-mCherry-ΔN) was constructed using a strategy described previously (Jengarn et al., 2015; Wanitchang et al., 2019). A frameshift mutation was used to silence N expression by mutating the original start codon in the N ORF, ATGGCT, to ATATGT in an intermediate plasmid, pTZ-GH, containing a PEDV genome fragment (designated ‘GH’; Fig. 6A) with the 5′ end of the N gene as described previously (Jengarn et al., 2015). The site-directed mutagenesis was performed in the pTZ-GH cloning vector. The mutated fragment was amplified by primers that facilitated subsequent In-Fusion ligation (In-Fusion HD, Clontech) into a pre-digested pSMART-BAC plasmid containing the rest of the PEDV genome (Jengarn et al., 2015) to yield pSMART-BAC-PEDV-mCherry-ΔN (designated pPEDV-mCherry-ΔN).
Fig. 6.
Construction of the N-deficient PEDV infectious clone, pPEDV-mCherry-ΔN. (A) Schematic representation of a frameshift mutation strategy to silence N expression from the pPEDV-mCherry-derived infectious clone to yield pPEDV-mCherry-ΔN. CMV, cytomegalovirus immediate-early promoter; HDV, hepatitis delta virus ribozyme self-cleavage site; BGH, bovine growth hormone termination; T, transcription terminator. (B) Western blot analysis of lysates prepared from HEK293T cells transfected with pPEDV-mCherry or pPEDV-mCherry-ΔN (2 μg each) at 72 hpt.
2.9. Rescue of reverse genetics-derived PEDV
HEK293T cells were transfected with 2 μg of the indicated infectious clone (or co-transfected with the indicated amount of a CoV N expressing plasmid) using FuGENE HD (Promega) according to the manufacturer's instructions. At 72 hpt, cell lysates were prepared for western blot analysis as described previously, and supernatants were transferred for adsorption on VeroE6 cells for 1 h at 37 °C. For viral rescue of PEDV-mCherry-ΔN, VeroE6 cells stably expressing PEDV N was further transfected with 2 μg of pCAGGS-PEDV N for 24 h before virus adsorption to ensure sufficient expression of PEDV N in the infected cells. The inocula were then removed, and the cells were washed once with PBS and supplied with 2 ml fresh OptiMEM with 0.1% TrypLE (Thermo Scientific). At indicated time points, infected cells were imaged under a fluorescence microscope (Olympus), and the supernatants were collected for subsequent experiments or viral titer analysis by TCID50 assay.
2.10. Virus infection
VeroE6-based cell lines (5×105 cells/ml) were plated in a six-well plate. At 24 h, the cells were washed once with PBS and treated with 1 ml of virus at the indicated multiplicity of infection (MOI). At 1 h post-adsorption, the inoculum was removed, and the cells were washed once with PBS and supplied with 2 ml of fresh OptiMEM containing 0.1% TrypLE. Extent of PEDV infection was monitored by mCherry fluorescence under a fluorescence microscope. Cell lysates or supernatants were harvested at the indicated time points for further analysis. For viral infection in the transient CoV N expression experiment, VeroE6 cells were transfected with 1 or 2 μg of pCAGGS-PEDV N-Myc, pCAGGS-PDCoV N-HA, pCAGGS-TGEV N-FLAG, or the empty pCAGGS vector and were incubated for 24 h to allow for protein expression. Cells were then infected with PEDV-mCherry (MOI = 0.0001) as described above.
2.11. TCID50 assay
Monolayers of VeroE6 cells in 96-well plates were washed once with PBS. One hundred microliters of 10-fold serially diluted virus in OptiMEM with 0.1% TrypLE was added to the cells (8 wells per each dilution). At 72 h post-infection (hpi), the infected cells were scored by mCherry expression under a fluorescence microscope. TCID50 titers were calculated using the Reed-Muench method (Reed and Muench, 1938).
2.12. RT-qPCR
For analysis of viral RNA synthesis, total RNA was extracted from VeroE6 cell lines expressing various N infected with PEDV-mCherry (MOI = 0.0001) at indicated time points using the RNA extraction kit (Thermo Scientific). DNaseI (Fermentas) was used to treat the RNA (15 min at 37 °C) before inactivation with EDTA (10 min at 65 °C). One-step RT-qPCR was performed with the Luna Universal One-Step RT-qPCR mix (New England Biolabs) as described previously (Liwnaree et al., 2019). Relative quantities of RNA accumulation were evaluated using the 2– ΔΔ Ct method normalized against viral RNA levels from infected VeroE6 cells at 24 hpi. Values are reported as averages ± SEM from three independent experiments.
2.13. Statistical analysis
GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA, USA) was used for figure preparation and statistical analyses of virus titers in Fig. 4, Fig. 5 . KaleidaGraph 4.1 (Synergy) was used for figure preparation and statistical analyses of GST pulldown in Fig. 2 and RT-qPCR results in Fig. 5.
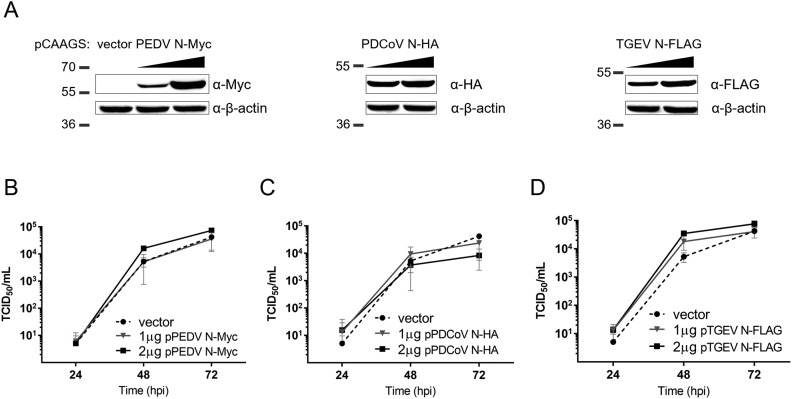
Fig. 4.
Effect of CoV N transient expression on PEDV replication in VeroE6 cells. VeroE6 cells were transfected with 1 or 2 μg of pCAGGS plasmid expressing N from PEDV, PDCoV or TGEV. At 24 hpt, cells were then infected with PEDV-mCherry (MOI = 0.0001). (A) Western blot analysis of protein expression from the transfections. (B–D) Virus titers from cell supernatants collected at 24, 48 and 72 hpi from VeroE6 cells transiently expressing tagged N proteins from PEDV (B), PDCoV (C) and TGEV (D) were determined by TCID50 assay. Values are averages ± SEM of three independent experiments.
Fig. 5.
PEDV replication is promoted by TGEV N and suppressed by PDCoV N. (A) VeroE6 clones stably expressing CoV N from either PEDV, PDCoV or TGEV were generated by lentiviral transduction and selected based on relatively equal expression as determined by western blot and immunofluorescence analyses. The selected clones of VeroE6 cells were infected with PEDV-mCherry (MOI 0.0001) and monitored at the indicated time points for PEDV spread by (B) fluorescence microscopy and (C) production of infectious particle by TCID50 assay. Values shown are averages ± SEM of three independent experiments. (D) Total RNA was extracted from infected cells in (B) to monitor subgenomic mRNA (top panel) and genomic RNA (bottom panel) synthesis by RT-qPCR with primer pairs specific to sgRNA of S or to ORF1a, respectively. Values are expressed relative to those from VeroE6 cells at 24 hpi (set to one) and are averages ±SEM of three independent experiments. *p < 0.05, **p < 0.01 compared to VeroE6 cells at the same time point.
3. Results
3.1. Cross-species interactions occur between swine enteric coronavirus nucleocapsid proteins
During co-infection, viruses can influence the course of infection of other viruses via direct physical interactions between viral components. To explore the possibility that swine CoV N proteins might affect replication of other co-infecting CoVs, we first investigated cross-species interaction between these N proteins. Specifically, we asked if N proteins from TGEV or PDCoV can interact with PEDV N and possibly affect PEDV replication. We first assessed interspecies protein-protein interaction by co-immunoprecipitation. HEK293T cells were co-transfected with pCAGGS-PEDV N-Myc and pCAGGS expressing HA-PEDV N, PDCoV N-HA or TGEV N-FLAG. At 48 hpt, lysates were prepared and incubated with agarose beads coupled to anti-Myc antibodies prior to elution and analysis by western blotting. As expected, HA-tagged PEDV N could be precipitated with Myc-tagged PEDV N but not with empty beads, indicating specificity of the homo-oligomeric interaction (Fig. 1A). Interestingly, both N proteins from related CoVs also displayed specific interaction with PEDV N, both co-eluting with Myc-tagged PEDV N (Fig. 1A). Nevertheless, we observed slight non-specific binding in the case of PDCoV N-HA despite stringent washing conditions. Therefore, we utilized a GST pulldown assay as an alternative and independent verification of these results.
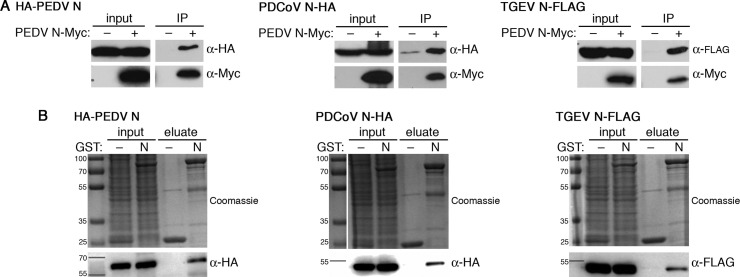
Fig. 1.
Interspecies association between CoV N proteins. (A) HEK293T cell lysates prepared from co-transfection of 500 ng of the empty pCAGGS vector (–) or pCAGGS-PEDV N-Myc with each of the pCAGGS expressing the indicated CoV N protein were used for co-immunoprecipitation with anti-Myc antibody followed by western blot analysis with indicated antibodies. Input, 1:20 dilution of cell lysates; IP, eluted immunoprecipitate. (B) GST- or GST-PEDV N-containing bacterial lysates and HEK293T cell lysates prepared from transfection with each of the pCAGGS expressing the indicated CoV N protein were used for GST pulldown with GSH FF resin. The input and eluates were analyzed with SDS-PAGE and western blotting.
For GST pulldown, the bait proteins, GST and GST-PEDV N, were expressed in bacterial BL21 (DE3*) cells, and the prey proteins, HA-PEDV N, PDCoV N-HA and TGEV N-FLAG, were expressed in HEK293T cells. Bacterial and HEK293T cell lysates were incubated with GSH FF resin overnight to allow for bait-prey interaction. After removing non-specific binding by multiple rounds of washing, the bound proteins were released and analyzed by SDS-PAGE. Bait and prey proteins were detected by Coomassie staining and western blotting with indicated antibodies, respectively. HA-PEDV N could only be observed when pulled down with GST-PEDV N, while no HA-PEDV N was observed when pulled down with GST, demonstrating the specificity of the assay (Fig. 1B). Next, we performed the GST pulldown experiment with PDCoV N and TGEV N as prey proteins. Similarly, we found that PDCoV N-HA and TGEV N-FLAG only co-eluted with GST-PEDV N, indicating that N proteins from both TGEV and PDCoV bound to PEDV N specifically (Fig. 1B). Both co-immunoprecipitation and GST pulldown results indicated that interspecies hetero-oligomers of CoV N proteins are possible, suggesting conservation of the oligomerization motifs and function of nucleocapsid proteins among swine CoVs. These results encouraged us to further probe the factors affecting the nature of these interactions.
The cross-association capability of PDCoV N or TGEV N could possibly interrupt homo-oligomerization between PEDV N and lead to competitive binding between PEDV N and other CoV N proteins. Using the GST pulldown assay, we further probed the interplay between these CoV N proteins to see how PEDV N homo-oligomers would be affected in the presence of other CoV N proteins. HEK293T cells were co-transfected with pCAGGS-HA-PEDV N and varying amounts of pCAGGS-PDCoV N-HA or pCAGGS-TGEV N-FLAG. At 48 hpt, lysates were prepared and incubated with GST-PEDV N-containing bacterial lysate and GSH FF resin overnight. GST pulldown revealed that, upon increasing amounts of co-eluting PDCoV or TGEV N, co-eluting HA-tagged PEDV N decreased reciprocally (Fig. 2). This indicates, to a certain level, a competition between homo-oligomerization of PEDV N and hetero-oligomerization with nucleocapsid proteins from other CoVs, and suggests that these CoV N proteins utilize the same motifs or binding interfaces in oligomeric complex formation or harness common factors such as RNA that can mediate oligomeric interactions.
One of the primary functions of CoV N proteins is to bind and organize viral RNA genomes for viral assembly (McBride et al., 2014). However, there have been conflicting reports about the necessity of viral RNA in promoting or assisting CoV N self-oligomerization. Some reports observed RNaseA susceptibility of the oligomeric complex (Narayanan et al., 2003; Verheije et al., 2010), while others detected intact higher CoV N oligomers despite the absence of RNA (Cong et al., 2017; Jayaram et al., 2006; Ma et al., 2010). To investigate whether the presence of RNA influences interspecies association between these swine enteric CoV N proteins, GST pulldown experiments were performed with or without RNaseA treatment. First, however, we tested whether or not RNA affected PEDV N homo-oligomerization in this assay setup. HEK293T cell lysate containing HA-tagged PEDV N was mixed with GST-PEDV N bacterial lysate and split into two equal parts. The lysate mixtures were either treated with RNaseA or PBS buffer for 30 min at 4 °C. The absence of RNA after the treatment was assured by performing RT-PCR reactions with primers specific to the PEDV N gene (Fig. 3). The lysate mixtures were then incubated with GSH FF resin overnight to compare the amount of HA-PEDV N pulled down by GST-PEDV N in the presence or absence of RNA. Our results across three independent experiments consistently showed a slight but noticeable increase in GST-PEDV N in the elution fraction after RNaseA treatment (Fig. 3). Moreover, the amount of HA-PEDV N pulled down in the absence of RNA seemed to be greater than that observed in the presence of RNA, suggesting that RNA interferes with the ability of PEDV N to bind to each other. We also consistently observed a slight decrease in the level of tagged PEDV N in the input fraction after the lysates were treated with RNaseA, implying decreased overall stability of PEDV N in the absence of RNA. Similar decreases in protein levels were not observed with TGEV or PDCoV N proteins (see below), suggesting specificity of the phenomenon.
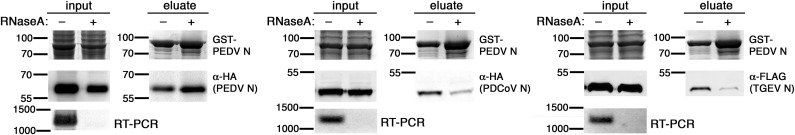
Fig. 3.
Effect of RNA on homo- and hetero-oligomerization of PEDV N. GST pulldown by GST-PEDV N was performed with HEK293T cell lysates containing HA-PEDV N, PDCoV N-HA or TGEV N-FLAG. The lysate mixtures were treated with 10 μg RNaseA (+) or buffer (–) for 30 min at 4 °C prior to incubation with GSH FF resin. The input and eluates were analyzed with SDS-PAGE (Coomassie staining for GST-PEDV N) and western blotting for the other N proteins. RNA was extracted from a portion of the lysate mixtures, treated with DNaseI, and subjected to RT-PCR with primers specific to the nucleocapsid genes studied in each experiment. Expected RT-PCR products were about 1200 bpand were analyzed by agarose gel electrophoresis.
Interestingly, when the experiments were performed on lysates containing PDCoV N-HA or TGEV N-FLAG, a completely opposite trend was observed for hetero-oligomerization. While increased GST-PEDV N was still similarly observed in the eluted fraction in the absence of RNA, both PDCoV N and TGEV N were pulled down by GST-PEDV N much less efficiently in the absence of RNA (Fig. 3). It should be noted that the total amount of the tagged CoV N proteins did not decrease, arguing against the possibility that TGEV or PDCoV N proteins were more prone to degradation without the bound RNA (Fig. 3, ‘input’ lanes). These results suggest that the presence of RNA immensely helps strengthen hetero-oligomerization between PEDV N and other CoV N proteins but renders homo-oligomerization between PEDV N weakened. Although not an absolute requirement for interspecies complex formation, RNA could act as a bridge for N proteins from different species to form the complex, implying the heterologous protein-protein interactions are significantly weaker or less specific than the homologous interaction. Even though the levels of RNA purified from RNaseA-treated lysates were below the detection limit of spectrophotometer (data not shown) and could not produce visible RT-PCR products, we could not rule out the possibility of leftover small RNA fragments mediating heterologous CoV N protein binding, giving rise to much fainter TGEV N or PDCoV N bands that co-eluted with GST-PEDV N. In summary, the results in this section demonstrate conserved association capability among different CoV N proteins but suggest some differentiating features, especially the influence of RNA on each complex, between homo- and hetero-oligomers involving PEDV N.
3.2. PEDV replication is promoted by TGEV N but suppressed by PDCoV N
Hetero-oligomer formation as demonstrated in the previous section formed a basis for our investigation into N-mediated virus-virus interaction. As it has been shown for other viruses, it is not unreasonable to assume that the specific and competitive binding between N proteins of PEDV and other CoVs could interrupt regular functions of PEDV N during viral growth. To investigate how this protein-protein interaction might affect PEDV replication, we transiently transfected VeroE6 cells with varying amounts of the pCAGGS plasmid expressing N proteins from either PDCoV or TGEV for 24 h before infection with PEDV-mCherry (MOI = 0.0001) and followed the course of viral replication for each condition. The levels of CoV N expression were verified by western blotting against tags attached to each N protein (Fig. 4A). Cell culture supernatants were collected at 24, 48 and 72 hpi to determine virus titers by TCID50 assay (Fig. 4B–D). The results showed that VeroE6 cells transiently transfected with either PEDV N or TGEV N showed a dose-dependent increase, up to an order of magnitude, in PEDV-mCherry titers compared to VeroE6 cells transfected with a blank vector (Fig. 4B and D). Interestingly, VeroE6 cells transiently transfected with PDCoV N showed suppression on PEDV-mCherry growth, especially at high levels of PDCoV N expression (Fig. 4C).
Due to relatively low transfection efficiencies in VeroE6 cells and the uncontrollable proportion of exogenous N expression and PEDV infection occurring in the same cells, the actual effects of these CoV N proteins on PEDV growth might be even more pronounced than observed from the transfection experiments. To test this, we constructed VeroE6-based cell lines stably expressing N from PEDV, PDCoV and TGEV by lentivirus transduction. VeroE6 clones expressing relatively equal amounts of N proteins from either PEDV, PDCoV and TGEV were selected based on western blot analysis (Fig. 5A). Uniform expression of exogenous N proteins in these engineered VeroE6 cells was also assessed by immunofluorescence to eliminate the concern from the transient transfection experiment (Fig. 5A). Equal numbers of cells were plated to confluence in 24-well plates and infected with PEDV-mCherry (MOI = 0.0001). Spread of PEDV infection and syncytium formation was monitored daily by fluorescence microscopy. Supernatants were collected at 24, 36 and 48 hpi to monitor PEDV growth by TCID50 assay. VeroE6 cells expressing N from PEDV and TGEV clearly accelerated PEDV syncytia formation and spread. By 36 hpi, most cells displayed signs of profuse infection; by 48 hpi, infected cells were dead and detached compared to infection in wild-type VeroE6 cells (Fig. 5B). Interestingly, PDCoV N stably expressed in VeroE6 cells significantly suppressed PEDV spread; even at 48 hpi, no extensive syncytia had formed (Fig. 5B). Quantification of supernatant PEDV-mCherry titers by TCID50 assay also reflected the extent of syncytium formation. At 36 hpi, VeroE6 cells expressing TGEV N or PEDV N yielded about a hundred times more infectious PEDV virus particles than VeroE6 cells, while those expressing PDCoV N resulted in an infectious titer about ten times lower. By 48 hpi, PEDV replication in wild-type VeroE6 cells had started to catch up and closed the gap to about one order of magnitude. However, VeroE6-PDCoV N cells still showed significantly slower PEDV replication kinetics (Fig. 5C). As expected, we noted that the suppressive effect from VeroE6-PDCoV N cells was stronger than that observed in transiently transfected VeroE6 cells. This could be because the stable cell line homogeneously expresses PDCoV N which can directly exert an effect on the infecting virus, while only a fraction of infected cells was transfected (resulting in the effect being diluted out by cells that were infected but not transfected).
Previously, we observed a significant enhancement of PEDV RNA synthesis and increased viral titers in VeroE6-PEDV N cells (Liwnaree et al., 2019). To explore if similar mechanisms were employed by TGEV N and PDCoV N on PEDV replication, we then probed viral RNA production inside these VeroE6-based cell lines using RT-qPCR. Total RNA was extracted from cells infected with PEDV-mCherry at 24, 36 and 48 hpi. Specific primers were used to quantify viral genomic RNA (ORF1a gene; gRNA) and subgenomic RNA (5′UTR- S gene; sgRNA) production. The levels of each RNA species were normalized to the levels of GAPDH mRNA from the same conditions and were expressed as relative to RNA levels at 24 hpi from infected VeroE6 cells. As previously observed, VeroE6-PEDV N cells drastically increased sgRNA and gRNA production at 36 and 48 hpi (Fig. 5D). On the other hand, VeroE6-PDCoV N cells displayed stunted PEDV RNA transcription and replication; even at 48 hpi, the levels of sgRNA and gRNA were still lower than those observed in VeroE6 cells at 24 hpi (Fig. 5D). VeroE6-TGEV N cells moderately enhanced PEDV RNA synthesis compared to wild-type VeroE6 cells but did not reach the same level as VeroE6-PEDV N cells (Fig. 5B). According to the results, the presence of extraneous N proteins posed greater effects on gRNA replication than sgRNA transcription. These data corroborated with visual observation and quantitative measurement of infectious viral particle production. Together, they strengthened the findings in the transient expression experiments and suggest opposing directions of influence on PEDV replication mediated by N proteins derived from related CoVs.
3.3. Other CoV N proteins can partially replace some functions of PEDV N
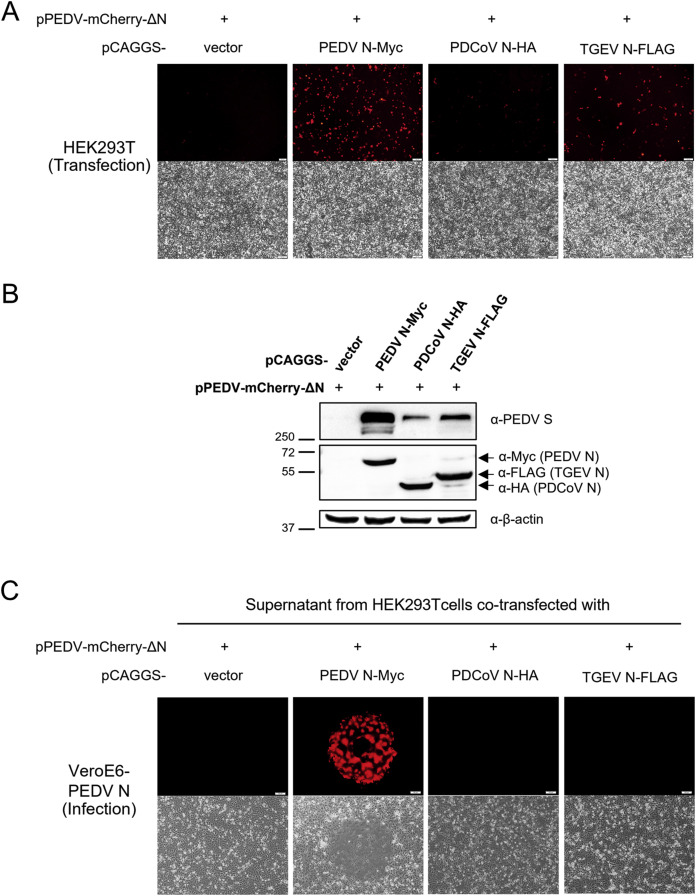
Based on their ability to cross-associate and affect PEDV RNA synthesis and replication, we speculated that N proteins from other CoVs, especially TGEV, may have interchangeable functions with PEDV N in PEDV replication. In an attempt to answer which roles and functions of PEDV N could be replaced by other CoV N proteins, we constructed an N-deficient PEDV infectious clone, pPEDV-mCherry-ΔN, and determined whether supplying CoV N proteins in trans could replace the missing PEDV N protein from the PEDV's viral genome. Since the nucleotide fragment of the N gene in the viral genome may play other important roles in viral genome packaging or transcription, we employed a frameshift mutation strategy to silence N expression from the infectious clone while minimally disturbing the nucleotide sequence of the N gene. The original start codon in the N ORF, ATGGCT, was mutated to ATATGT, thereby shifting the start codon by two nucleotides, resulting in a +2 frameshift during translation (Fig. 6A). The mutated sequence theoretically produced a short unrelated peptide of 21 amino acids with early termination. We intentionally kept the mCherry gene in the construct as a visual indicator for sub-genomic mRNA transcription and protein expression in the virus rescue process. To verify the lack of N expression during PEDV reverse genetics rescue, we transfected the N-deficient infectious clone into HEK293T cells. Western blot analysis confirmed that N expression was completely diminished in cells transfected with the N-deficient infectious clone compared to the wild-type pPEDV-mCherry (Fig. 6 B).
With the N-deficient infectious clone in hand, we next investigated PEDV reverse genetics rescue in the presence of other CoV N proteins. HEK293T cells were co-transfected with pPEDV-mCherry-ΔN and a pCAGGS plasmid expressing CoV N protein. Expression of mCherry from the infectious clone was used as a visual proxy for production of viral RNA and proteins in HEK293T cells during the reverse genetics step. No detectable fluorescence was observed from co-transfection with the empty pCAGGS vector, confirming the role of PEDV N protein during at least the first round of viral RNA transcription and viral protein translation (Fig. 7 A). Co-transfection with pCAGGS-PEDV N-Myc yielded a substantial fraction of red cells, while slightly lower amount of mCherry protein expression was observed from cells co-transfected with pCAGGS-TGEV N-FLAG. Co-transfection with pCAGGS-PDCoV N-HA resulted in very low, yet detectable, numbers of mCherry-positive cells (Fig. 7A).
Fig. 7.
Other CoV N proteins can partially function to replace PEDV N. For PEDV rescue experiments, HEK293T cells were co-transfected with pPEDV-mCherry-ΔN and 1 μg of pCAGGS plasmid expressing N from PEDV, PDCoV or TGEV. At 72 hpt, (A) cells were imaged under fluorescence microscopy to monitor mCherry expression from pPEDV-mCherry-ΔN, and (B) cell lysates were prepared for western blot analysis with an anti-S antibody. Supernatants from (A) were adsorbed onto VeroE6-PEDV N cells, which were additionally transfected with 2 μg pCAGGS-PEDV N to ensure sufficient N expression. At 72 hpi, (C) cells were imaged under fluorescence microscopy to monitor mCherry expression and syncytium formation as a sign for successful PEDV infection into VeroE6-PEDV N cells (Scale bar, 100 μm).
Cell lysates were prepared at 72 hpt and probed for S expression using anti-S antibody to corroborate visual inspection. As a negative control, cells transfected with pPEDV-mCherry-ΔN and pCAGGS vector displayed no detectable trace of S (Fig. 7B). Cells co-transfected with pCAGGS expressing CoV N proteins showed varying levels of S expression, with PEDV N-Myc giving the highest S expression and PDCoV N-HA giving the lowest (Fig. 7B). TGEV N-FLAG expression resulted in noticeably lower S expression compared to PEDV N-Myc but still higher than that of PDCoV N-HA (Fig. 7B). The levels of spike protein expression from the N-deficient clone in each condition correlated with the level of mCherry expression as observed by fluorescence microscopy (Fig. 7A and B). These results suggested that CoV N proteins from alphacoronaviruses can aid viral RNA and protein production from infectious PEDV clones more efficiently than that from deltacoronavirus.
To determine if PEDV rescue successfully produced viable infectious particles with help from various CoV N proteins in trans, HEK293T cell supernatants were collected at 72 hpt to inoculate VeroE6-PEDV N cells pre-transfected with pCAGGS-PEDV N. We found that additional transfection could ensure sufficient PEDV N expression in infected cells and facilitated PEDV replication better than untreated VeroE6-PEDV N cells (data not shown). Limited syncytium formation was observed only in VeroE6-PEDV N cells inoculated with the supernatant derived from HEK293T cells co-transfected with pCAGGS-PEDV N-Myc (Fig. 7C). However, the infecting viruses could not continuously propagate to produce extensive syncytia as routinely observed with the rescue of pPEDV-mCherry. This is probably due to insufficient PEDV N protein levels as supplied by the stable cell line compared to virally encoded PEDV N. Remarkably, supernatants from HEK293T cells co-transfected with a plasmid encoding TGEV N or PDCoV N did not contain infectious progeny from virus rescue, displaying no second-round PEDV-mCherry replication in VeroE6-PEDV N cells as determined by mCherry expression or syncytia formation (Fig. 7C). These results suggest that, despite some conserved function of N proteins among porcine enteric CoVs in self-oligomerization and assistance during viral RNA synthesis or protein expression, N is not fully interchangeable among CoVs for generating PEDV infectious virus progeny. These results imply that the suppressive or enhancing effects on PEDV replication in cells expressing PDCoV N or TGEV N may reflect their effects on viral RNA replication and/or viral protein expression, rather than their abilities to supply complementary N proteins during RNP core formation or virion assembly.
4. Discussion
Although the swine industry has focused on PEDV alone as one of its largest threats, few have attempted to understand the implications of swine enteric CoV co-infection despite multiple epidemiological studies. As the true impact of co-infection on disease outcomes still awaits further investigation, these viruses could predictably exert influence on each other, considerably shaping the course of their infection or evolution. Molecular interactions underlying these possible viral interferences also remain to be identified and characterized. In this work, we tried to understand how different enteric swine CoV N proteins, one of the most abundant structural proteins, interact and affect functions of PEDV N and replication kinetics of PEDV.
First, we provided evidence of cross-interaction between CoV N proteins despite low sequence similarity among them. Amino acid sequence analyses on PEDV, TGEV and PDCoV N proteins revealed that the proteins can be divided roughly into two structural domains (NTD and CTD) with relatively high conservation and three intrinsically disordered regions (IDRs) with relatively low conservation located in the middle and at the two termini (Fig. 8 ). This domain analysis is consistent with previous crystallographic studies on SARS-CoV and MHV N, which also suggested conserved general structural organization of CoV N (Chang et al., 2009; Cong et al., 2017; Ma et al., 2010; Yu et al., 2006). Moreover, immunological data documenting cross-reactivity of antibodies against N proteins from TGEV or PDCoV with PEDV N further implied shared topological similarity between these CoV N proteins (Gimenez-Lirola et al., 2017; Lin et al., 2015; Ma et al., 2016). Nevertheless, precise molecular mechanisms mediating interspecies oligomerization of CoV N proteins remain to be determined. Structural and biochemical studies of SARS-CoV N strongly suggest a role of CTD as a major mediator for CoV N dimerization. Moreover, CTDs of N proteins from SARS-CoV and porcine reproductive and respiratory syndrome virus (PRRSV), despite diverse sequences, could adopt well-conserved structures, implying similar approaches to oligomerization, at least at a gross structure level (Yu et al., 2006). However, since CoV N CTDs formed intricate molecular interaction networks during dimerization (Chang et al., 2014), it is unlikely that CoV N proteins from different species could replace all these specific molecular interactions. Consistent with our results that hetero-oligomer formation is highly sensitive to the presence of RNA, a more likely possibility is that RNA-protein interactions play a more dominant role in interspecies CoV N interactions than protein-protein interactions.
Fig. 8.
Analysis of CoV N sequences reveals low sequence similarity but conserved structural organization. Multiple amino acid sequence alignment was performed with T-coffee (Notredame et al., 2000). The color scheme denotes consistency between different alignment methods ranging from bad (low agreement; blue and green) to good (high agreement; pink). Bold and normal lettering denote ordered and disordered regions as predicted by the PrDOS program (Ishida and Kinoshita, 2007). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Next, we asked if N proteins from different CoVs can replace PEDV N function during viral RNA and protein synthesis. During the rescue of the N-deficient PEDV infectious clone, we showed that supplying N protein from any CoV in trans can initiate first-round sgRNA transcription and protein expression. Most likely, this function stems from the shared intrinsic ability of CoV N to bind viral RNA and act as an RNA chaperone that initiates RNA replication and/or transcription processes (Zuniga et al., 2007, 2010). These results support previous notions that CoV N proteins play important roles during CoV rescue. For instance, rescue of TGEV infectious RNA required at least co-transfection of mRNA encoding the N protein (Yount et al., 2000). Schelle et al. demonstrated that an N-deficient HCoV229E RNA vector replicon carrying the GFP gene can produce sgRNA and express GFP from the vector when co-transfected with N from PEDV, a closely related virus, but not from MHV, a more distantly-related one (Schelle et al., 2005). We noted a similar preference in our experiments. Between TGEV N and PDCoV N, the former is more effective in replacing the indigenous PEDV N than the latter. Since both PEDV and TGEV belong to the alphacoronavirus genus, while PDCoV is classified as a deltacoronavirus, TGEV N would be expected to be functionally and structurally closer to PEDV N, and hence able to form replication/transcription complexes and participate in PEDV replication with higher compatibility than PDCoV N.
Although both TGEV N and PDCoV N can oligomerize and perform some functions during g/sgRNA transcription, they alone cannot totally replace all PEDV N's functions. Without native PEDV N, infectious PEDV virions could not be detected in the presence of TGEV or PDCoV N, implying that other functions are not interchangeable. These critical functions might involve viral core assembly as CoV N participates in many steps during viral core or virion assembly (McBride et al., 2014). For instance, several reports showed that CoV N is involved, directly or indirectly, with recognition of specific packaging signals for genome packaging (Hsieh et al., 2005; Hsin et al., 2018; Kuo et al., 2014). Since RNA packaging signals among CoVs are distinct in terms of length and sequence, even closely related CoV N might not be able to recognize PEDV packaging signals. Similar compatibility issues have been raised in the case of viral interference between different types of influenza viruses (Baker et al., 2014). Another critical step that could be disrupted by heterologous CoV N is viral assembly through interactions between N and M proteins. It is widely accepted that CoV N and M proteins interact to form the viral core, but different CoVs utilize different regions of N in binding to their own M proteins. For instance, while MHV utilizes the very C-terminal end of N to bind M, SARS-CoV N uses the middle disordered linker for the interaction (He et al., 2004b; Narayanan et al., 2000). In addition, although both TGEV and MHV N proteins used their C-termini to bind M, the N binding sites on TGEV M and MHV M are very distinct (Hurst et al., 2005; Kuo and Masters, 2002), pointing to high specificity for each virus. It could be argued that our use of C-terminally tagged CoV N proteins might have contributed to failure in rescuing infectious PEDV from pPEDV-mCherry-ΔN, possibly due to interference with N-M protein interaction. Nevertheless, our results showed that addition of a Myc tag at the C-terminus of PEDV N did not completely ruin protein-protein interaction required for viral assembly as PEDV N-Myc remained capable of producing infectious virions. Moreover, using untagged PEDV N and TGEV N during reverse genetics rescue of pPEDV-mCherry-ΔN gave similar results to those presented in Fig. 7 (data not shown), suggesting that the inability of TGEV N or PDCoV N to rescue infectious PEDV lacking endogenous nucleocapsid protein stemmed largely from incompatibility of interspecies protein interaction rather than the interference from epitope tags. In all, it is conceivable that not all of N functions could be replaced by the protein originating from other viral species, even though N proteins from PEDV, TGEV and PDCoV can cross-assemble, form high-order oligomers and aid in RNA synthesis processes.
Notably, we observed completely opposite effects of TGEV and PDCoV N proteins on PEDV replication. We noticed that, in the presence of RNA, homologous binding between PEDV N itself is weakened and cross binding between PEDV N and the other N proteins is strengthened. During co-infection, where viral gRNA is ubiquitous, the RNA could act as a tethering bridge to stabilize the formation of heterologous oligomers, which may not be a productive core structure for PEDV virion assembly and may interrupt the optimal processes in viral replication that prefer the homo-oligomer of N. Such interruption could explain the suppressive effect of PDCoV N. Nevertheless, the existence of interspecies CoV N oligomers might not be the only factor dictating the effect of extraneous CoV N proteins on PEDV replication. TGEV N also forms RNA-dependent interspecies oligomers with PEDV N and could have resulted in similar suppression. Instead, we observed a small enhancement effect on PEDV replication in the presence of TGEV N. This could be the net outcome of other oligomerization-independent roles of TGEV N that indirectly enhance PEDV replication. A study by Zhao et al. showed that, when compared to single-virus infection, co-infection of TGEV and PEDV could synergistically enhance rupture to tight and adherens junctions of the intestinal epithelial cells IPEC-J2 by suppressing expression of proteins that help form the junction (Zhao et al., 2014). Such altered barrier integrity is possibly caused by CoV N and facilitate viral replication. Another possible role of PEDV or TGEV N that could lead to enhancement of PEDV replication is to act as a viral suppressor of RNA silencing (Cui et al., 2015). Using MHV N as a model, they found that CoV N can bind and suppress host antiviral RNAi, leading to increased viral protein expression and subsequent viral replication. More remarkably, they found that N proteins of alphacoronaviruses, including TGEV and PEDV, also have this ability. Indeed, among different CoV N proteins, they found that TGEV N was among the most active and MERS-CoV N was the least active (Cui et al., 2015). Therefore, N from TGEV may also employ this mechanism in promoting PEDV replication, while some other CoV N proteins might not be capable of this function. These are interesting hypotheses currently under investigation, which could reveal more complexity of co-infection between these viruses.
In summary, this work shows how other swine enteric CoV N proteins can substitute for endogenous PEDV N in some functions but not in others, demonstrating both conservation and specificity among related viruses in the Coronaviridae family. Furthermore, co-expression experiments revealed interesting phenomena in which different CoV N proteins exert vastly different effects on PEDV replication. These results raise even more questions. For example, in the context of co-infection, in which all other viral components are present, what would be the net outcomes on replication kinetics of each virus? Are there effects from other viral components? Considering the opposing effects on PEDV replication of N proteins from TGEV, an older virus emerging prior to PEDV, and PDCoV, a newly emerged one, is it possible that nucleocapsid proteins could act as one of the factors in permitting or limiting emergence of enteric swine CoVs? If so, what is the effect of PEDV N on newer CoVs such as swine acute diarrhea syndrome coronavirus (SADS-CoV), which has been detected predominantly in farms encountering PEDV epidemics (Zhou et al., 2018), and vice versa? Further investigation into these questions would give valuable insights for us to better understand co-infection events and their implications for the emergence of novel enteric swine viruses.
Declaration of competing interest
All authors of this manuscript have no conflict of interest to declare.
Acknowledgements
The authors would like to thank Dr. Qigai He for anti-PEDV S antibody, and Benjamas Liwnaree for technical assistance. We thank the members of Virology and Cell Technology Research Team, especially Dr. Samaporn Teeravechyan, for critical comments and language help on the manuscript. This work was supported by BIOTEC’s Fellow Grant [grant number P-15-51261].
References
- Aoki H., Nishiyama Y., Tsurumi T., Shibata M., Ito Y., Seo H., Yoshii S., Maeno K. Mechanism of interference between influenza A/WSN and B/Kanagawa viruses. J. Gen. Virol. 1984;65(Pt 8):1385–1393. doi: 10.1099/0022-1317-65-8-1385. [DOI] [PubMed] [Google Scholar]
- Bag S., Mitter N., Eid S., Pappu H.R. Complementation between two tospoviruses facilitates the systemic movement of a plant virus silencing suppressor in an otherwise restrictive host. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.F., Nogales A., Finch C., Tuffy K.M., Domm W., Perez D.R., Topham D.J., Martinez-Sobrido L. Influenza A and B virus intertypic reassortment through compatible viral packaging signals. J. Virol. 2014;88:10778–10791. doi: 10.1128/JVI.01440-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Chen C.M., Chiang M.H., Hsu Y.L., Huang T.H. Transient oligomerization of the SARS-CoV N protein--implication for virus ribonucleoprotein packaging. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein--forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Lo S.C., Wang Y.S., Hou M.H. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov. Today. 2016;21:562–572. doi: 10.1016/j.drudis.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Kriegenburg F., de Haan C.A.M., Reggiori F. Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers. Sci. Rep. 2017;7:5740. doi: 10.1038/s41598-017-06062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Wang H., Ji Y., Yang J., Xu S., Huang X., Wang Z., Qin L., Tien P., Zhou X., Guo D., Chen Y. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. Virol. 2015;89:9029–9043. doi: 10.1128/JVI.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Rottier P.J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Yuan S., Zhao L., Wang X., Long S., Wang M., Wang D., Foda M.F., Xiao S. The nucleocapsid proteins of mouse hepatitis virus and severe acute respiratory syndrome coronavirus share the same IFN-beta antagonizing mechanism: attenuation of PACT-mediated RIG-I/MDA5 activation. Oncotarget. 2017;8:49655–49670. doi: 10.18632/oncotarget.17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Spaan W., Snijder E., Cavanagh D. Nidovirales. In: van Regenmortel C.M.F.M.H.V., Bishop D.H.L., Carsten E.B., Estes M.K., Lemon S.M., Mayo M.A., McGeoch D.J., Pringle C.R., Wickner R.B., editors. Virus Taxonomy. Academic Press; New York, N.Y: 2000. pp. 827–834. [Google Scholar]
- Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J. vol. 13. Structure; London, England: 2005. pp. 1859–1868. (The Nucleocapsid Protein of Coronavirus Infectious Bronchitis Virus: Crystal Structure of its N-Terminal Domain and Multimerization Properties). 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Hong Y., Liu X., Dong N., Ma P., Bi J., Wang D., Xiao S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 2017;98:173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Lirola L.G., Zhang J., Carrillo-Avila J.A., Chen Q., Magtoto R., Poonsuk K., Baum D.H., Pineyro P., Zimmerman J. Reactivity of porcine epidemic diarrhea virus structural proteins to antibodies against porcine enteric coronaviruses: Diagnostic implications. J. Clin. Microbiol. 2017;55:1426–1436. doi: 10.1128/JCM.02507-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N., Cutts T., Andonov A., Cao J., Booth T.F., Plummer F.A., Tyler S., Baker L., Li X. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N., Feldmann H., Strocher U., Theriault S., Cutts T., Cao J., Booth T.F., Plummer F.A., Tyler S., Li X. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P.-K., Chang S.C., Huang C.-C., Lee T.-T., Hsiao C.-W., Kou Y.-H., Chen I.Y., Chang C.-K., Huang T.-H., Chang M.-F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin W.C., Chang C.H., Chang C.Y., Peng W.H., Chien C.L., Chang M.F., Chang S.C. Nucleocapsid protein-dependent assembly of the RNA packaging signal of Middle East respiratory syndrome coronavirus. J. Biomed. Sci. 2018;25:47. doi: 10.1186/s12929-018-0449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Kuo L., Koetzner C.A., Ye R., Hsue B., Masters P.S. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 2005;79:13285–13297. doi: 10.1128/JVI.79.21.13285-13297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Ye R., Goebel S.J., Jayaraman P., Masters P.S. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 2010;84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaru-Ampornpan P., Jengarn J., Wanitchang A., Jongkaewwattana A. Porcine epidemic diarrhea virus 3C-like protease-mediated nucleocapsid processing: possible link to viral cell culture adaptability. J. Virol. 2017;91 doi: 10.1128/JVI.01660-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaru-ampornpan P., Narkpuk J., Wanitchang A., Jongkaewwattana A. Nucleoprotein of influenza B virus binds to its type A counterpart and disrupts influenza A viral polymerase complex formation. Biochem. Biophys. Res. Commun. 2014;443:296–300. doi: 10.1016/j.bbrc.2013.11.100. [DOI] [PubMed] [Google Scholar]
- Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., Lescar J., Prasad B.V. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jengarn J., Wongthida P., Wanasen N., Frantz P.N., Wanitchang A., Jongkaewwattana A. Genetic manipulation of porcine epidemic diarrhoea virus recovered from a full-length infectious cDNA clone. J. Gen. Virol. 2015;96:2206–2218. doi: 10.1099/vir.0.000184. [DOI] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23:137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Hurst-Hess K.R., Koetzner C.A., Masters P.S. Analyses of coronavirus assembly interactions with interspecies membrane and nucleocapsid protein chimeras. J. Virol. 2016;90:4357–4368. doi: 10.1128/JVI.03212-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Koetzner C.A., Hurst K.R., Masters P.S. Recognition of the murine coronavirus genomic RNA packaging signal depends on the second RNA-binding domain of the nucleocapsid protein. J. Virol. 2014;88:4451–4465. doi: 10.1128/JVI.03866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76:4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. 2014. Complete Genome Characterization of Korean Porcine Deltacoronavirus Strain KOR/KNU14-04/2014. Genome Announcements 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Gao X., Oka T., Vlasova A.N., Esseili M.A., Wang Q., Saif L.J. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J. Virol. 2015;89:3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liwnaree B., Narkpuk J., Sungsuwan S., Jongkaewwattana A., Jaru-Ampornpan P. Growth enhancement of porcine epidemic diarrhea virus (PEDV) in Vero E6 cells expressing PEDV nucleocapsid protein. PLoS One. 2019;14 doi: 10.1371/journal.pone.0212632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.S., Lin S.Y., Wang S.M., Wang C.T., Chiu Y.L., Huang T.H., Hou M.H. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013;587:120–127. doi: 10.1016/j.febslet.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Tong X., Xu X., Li X., Lou Z., Rao Z. Structures of the N- and C-terminal domains of MHV-A59 nucleocapsid protein corroborate a conserved RNA-protein binding mechanism in coronavirus. Protein & cell. 2010;1:688–697. doi: 10.1007/s13238-010-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Oglesbee M., Krakowka S., Niehaus A., Wang G., Jia A., Song H., Li J. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine deltacoronavirus. Vet. Microbiol. 2016;186:90–96. doi: 10.1016/j.vetmic.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Koetzner C.A., Kerr C.A., Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Kim K.H., Makino S. Characterization of N protein self-association in coronavirus ribonucleoprotein complexes. Virus Res. 2003;98:131–140. doi: 10.1016/j.virusres.2003.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.N., Goodrich J.A. Protein-protein interaction assays: eliminating false positive interactions. Nat. Methods. 2006;3:135–139. doi: 10.1038/nmeth0206-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Penzes Z., Gonzalez J.M., Calvo E., Izeta A., Smerdou C., Mendez A., Sanchez C.M., Sola I., Almazan F., Enjuanes L. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the purdue virus cluster. Virus Genes. 2001;23:105–118. doi: 10.1023/A:1011147832586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method OF estimating fifty per cent ENDPOINTS12. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: Identification, prevalence and full-length genome sequence analysis. Transboundary and emerging diseases. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi D., Raikhy G., Pappu H.R. Movement and nucleocapsid proteins coded by two tospovirus species interact through multiple binding regions in mixed infections. Virology. 2015;478:137–147. doi: 10.1016/j.virol.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Verheije M.H., Hagemeijer M.C., Ulasli M., Reggiori F., Rottier P.J., Masters P.S., de Haan C.A. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84:11575–11579. doi: 10.1128/JVI.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhao P., Guo L., Liu Y., Du Y., Ren S., Li J., Zhang Y., Fan Y., Huang B., Liu S., Wu J. Porcine epidemic diarrhea virus variants with high pathogenicity, China. Emerg. Infect. Dis. 2013;19:2048–2049. doi: 10.3201/eid1912.121088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanitchang A., Saenboonrueng J., Kaewborisuth C., Srisutthisamphan K., Jongkaewwattana A. A single V672F substitution in the spike protein of field-Isolated PEDV promotes Cell(-)Cell fusion and replication in VeroE6 cells. Viruses. 2019;11 doi: 10.3390/v11030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y., Liu H.J., Tong D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp 1 in IRF1 signaling. J. Virol. 2018;92 doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Gao J., Zhu L., Yang Q. Transmissible gastroenteritis virus and porcine epidemic diarrhoea virus infection induces dramatic changes in the tight junctions and microfilaments of polarized IPEC-J2 cells. Virus Res. 2014;192:34–45. doi: 10.1016/j.virusres.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Cruz J.L., Sola I., Mateos-Gomez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Sola I., Moreno J.L., Sabella P., Plana-Duran J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]