Abstract
Coronavirus membrane (M) protein is the most abundant structural protein playing a critical role in virion assembly. Previous studies show that the N-terminal ectodomain of M protein is modified by glycosylation, but its precise functions are yet to be thoroughly investigated. In this study, we confirm that N-linked glycosylation occurs at two predicted sites in the M protein ectodomain of infectious bronchitis coronavirus (IBV). Dual mutations at the two sites (N3D/N6D) did not affect particle assembly, virus-like particle formation and viral replication in culture cells. However, activation of the ER stress response was significantly reduced in cells infected with rN3D/N6D, correlated with a lower level of apoptosis and reduced production of pro-inflammatory cytokines. Taken together, this study demonstrates that although not essential for replication, glycosylation in the IBV M protein ectodomain plays important roles in activating ER stress, apoptosis and proinflammatory response, and may contribute to the pathogenesis of IBV.
Keywords: Coronavirus, Glycosylation, Particle assembly, ER stress, Apoptosis, Pro-inflammatory response, Viral pathogenesis
1. Introduction
Coronaviruses are a group of enveloped viruses with non-segmented, single-stranded, and positive sense RNA genome of 27–30 kb in length. Apart from infecting a variety of economically important vertebrates, several coronaviruses have been shown to infect human hosts, among which severe acute respiratory syndrome coronavirus (SARS-CoV) and middle east respiratory syndrome coronavirus (MERS-CoV) are zoonotic and highly pathogenic coronaviruses causing regional and global outbreaks (Lim et al., 2016).
In cells infected with coronaviruses, a 3′-coterminal nested set of six to nine mRNAs species, including the genome-length mRNA (mRNA1) and five to eight subgenomic mRNA species (mRNA2-9), is expressed. The genome-length mRNA1 of coronavirus infectious bronchitis virus (IBV) encodes two overlapping replicase proteins in the form of polyproteins 1a and 1ab, which are processed by viral proteinases into 15 nonstructural proteins (Nsp2-Nsp16) (Lim et al., 2016, Masters, 2006). The subgenomic mRNAs encode four structural proteins: the highly glycosylated spike protein (S), the small membrane-associated envelope protein (E), the integral membrane protein (M) and the phosphorylated nucleocapsid protein (N), are encoded by different subgenomic mRNAs (Masters, 2006). In addition, several accessory proteins, such as 3a, 3b, 5a, and 5b are also encoded by subgenomic mRNAs (Liu and Inglis, 1991, Liu et al., 1991).
Upon replication and transcription of viral RNA and translation of viral proteins, coronaviruses assemble and bud at the endoplasmic reticulum (ER) and Golgi intermediate compartment (ERGIC) (Klumperman et al., 1994, Stertz et al., 2007). The particles are then transported in smooth-wall vesicles, trafficked via the secretory pathway, and finally release from cells by exocytosis. Coronavirus M protein plays a pivotal role during the assembly and budding processes (Masters, 2006, Bárcena et al., 2009, Neuman et al., 2011). It is a multispanning membrane protein, consisting of a short amino terminus exposed on the exterior of the virion (the ectodomain), three hydrophobic transmembrane domains and a large carboxy-terminal region situated in the interior of the virion (Masters, 2006). Current assembly model supports that this integral membrane protein may adopt a region of the intracellular membrane for virion assembly (Lim and Liu, 2001). By interacting with other structural components, M protein would also be able to draw other structural proteins, such as S and E and ribonucleoprotein (RNP) into virions (Lim and Liu, 2001, Ye et al., 2004, Luo et al., 2006). Meanwhile, a network of M-M interactions would result in the exclusion of host cell membrane proteins from the viral envelope (De Haan et al., 2000, Neuman et al., 2008). The ectodomain, which is the least conserved region of M protein, is also glycosylated. Whereas M protein of some lineage A Betacoronaviruses, such as mouse hepatitis virus (MHV), bovine coronavirus and human coronavirus-OC43 (HCoV-OC43), is modified by O-linked glycosylation, M protein of other coronaviruses have been shown to be exclusively N-linked glycosylated (Holmes et al., 1981, Lapps et al., 1987, Mounir and Talbot, 1992, Hogue and Nayak, 1990, Utiger et al., 1995, Stern and Sefton, 1982, Dea et al., 1990). Different functions have been assigned to the oligosaccharide side chains, including protein folding, structure stability, intracellular sorting, and the induction of immune response (Drickamer and Taylor, 1998, Helenius and Aebi, 2001, Fung and Liu, 2018).
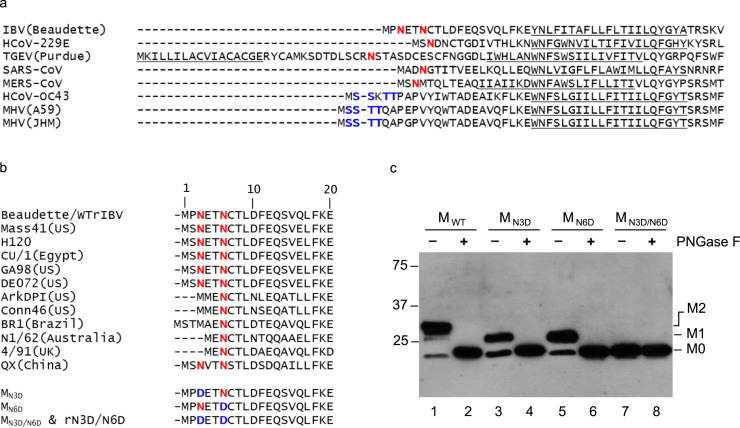
IBV is a prototype coronavirus and the etiological agent of infectious bronchitis, which impairs the respiratory and urogenital tracts of chickens (Cavanagh, 2007). IBV M protein contains 225 amino acids with three transmembrane domains (Binns et al., 1986, Rottier et al., 1986). The N-terminal luminal domain consists of 20 amino acids (Cavanagh et al., 1986), with two consensus sites for N-linked glycosylation near its N terminus at amino acid position 3 and 6, respectively ( Fig. 1a). Early studies confirmed that IBV M protein is modified by N-linked oligosaccharides, as the glycosylated forms were no longer detectable in cells treated with tunicamycin, a specific inhibitor of N-linked glycosylation (Stern and Sefton, 1982). Interestingly, treatment of tunicamycin did not inhibit virion assembly, although these particles were essentially non-infectious (Stern and Sefton, 1982).
Fig. 1.
Alignment of the N-terminal sequences of coronavirus M proteins and the effects of glycosylation of IBV M protein on the protein expression. a. Alignment of the N-terminal sequences of coronavirus M proteins. The proposed transmembrane domains are underlined. Putative N-linked and O-linked glycosylation sites are indicated in red and blue, respectively. b. Alignment of the N-terminal sequences of M proteins in different IBV strains. The proposed transmembrane domains are underlined. Putative N-linked glycosylation sites are indicated in red. Generation of three mutants, MN3D, MN6D, and MN3D/N6D. Asn to Asp substitutions were introduced into the M protein to change the either one or both of the two predicted N-linked glycosylation sites at the amino acid positions 3 and 6, respectively. c. Expression and glycosylation of wild type and mutant M proteins. Total cell lysates prepared from HeLa cells expressing IBV MWT, MN3D, MN6D, and MN3D/N6D were either treated with or without PNGase F. Polypeptides were separated by SDS-PAGE and analyzed by Western blot with anti-IBV M antiserum. Numbers on the left indicate molecular masses in kilo Daltons. The unglycosylated (M0), glycosylated at one position (M1) and glycosylated at two positions (M2) forms of the M protein are indicated.
In the present study, the effect of mutations at the N-linked glycosylation sites in the N-terminal ectodomain of IBV M protein on virus assembly and virus-host interaction was investigated by biochemical assays and reverse genetics. The result confirm that mutations in the IBV M glycosylation sites (N3D/N6D) did not affect particle assembly, virus-like particle (VLP) formation and IBV replication in cell culture. However, compared with the wild type control, activation of ER stress response, apoptosis, and induction of pro-inflammatory cytokines were all significantly reduced in cells infected with rN3D/N6D, although the in vivo virulence in terms of ELD50 was only marginally affected. Taken together, the data demonstrate that although glycosylation in the M ectodomain is not essential for IBV replication, it contributes to virus-host interactions and viral pathogenesis.
2. Materials and methods
2.1. Viruses and cells
The egg-adapted Beaudette strain of IBV (ATCC VR-22) was obtained from the American Type Culture Collection (ATCC) and was adapted to Vero cells as previously described (Liu et al., 1998). To prepare the virus stock, monolayers of Vero cells were infected at a multiplicity of infection (MOI) of approximately 0.1 and cultured in plain Dulbecco Modified Eagle Medium (DMEM, Gibco) at 37 °C for 24 h. After three freeze/thaw cycles, cell lysate was clarified by centrifugation at 1500g at 4 °C for 30 min. The supernatant was aliquot and stored at − 80 °C as virus stock. The titer of the virus stock was determined by plaque assays. Mock lysate was prepared by same treatment of uninfected Vero cells.
Vero cells were cultured in DMEM (Gibco) supplemented with 5% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin (Gibco), and grown in a 37 °C incubator supplied with 5% CO2.
2.2. Construction and recovery of recombinant IBV
To obtain a full-length IBV cDNA clone, five plasmids which contain five fragments (A to E) spanning the entire IBV genome were constructed as previously described (Fang et al., 2007). Briefly, the fragments were amplified by RT-PCR from total RNA of IBV-infected Vero cells. To facilitate the assembly of the full-length cDNA in vitro, restriction sites for either BsmBI or BsaI were introduced into both the 5′ and 3′ ends of the fragments. The PCR products were purified and cloned into pKT0, pCR-XL-TOPO (Invitrogen) or pGEM-T Easy (Promega) vectors. In fragment A, T7 promoter sequence was inserted immediately upstream of the 5′ end of the IBV genome to facilitate in vitro transcription by T7 polymerase. Plasmids were digested with either BsmBI (fragment A) or BsaI (fragments B, C, D, and E), and resolved using 0.8% agarose gels pre-stained with crystal violet. Bands corresponding to each of the fragments were excised and purified with QIAquick gel extraction kit (QIAGEN Inc.). Fragments A, B, C, D, and E were ligated with T4 DNA ligase at 16 °C overnight. The ligation products were extracted with phenol/chloroform, precipitated with ethanol, and detected by electrophoresis on 0.4% agarose gels.
Full-length transcripts were generated in vitro using the mMessage mMachine T7 kit (Ambion) according to the manufacturer's instructions with certain modifications. Briefly, 30 µl of transcription reaction with a 1:1 ratio of GTP to cap analog was sequentially incubated at 40.5 °C for 25 min, 37.5 °C for 50 min, 40.5 °C 25 min and 37.5 °C for 20 min. The N transcripts were generated by using a linearized pKT0-IBV-N plasmid (containing IBV N gene and the 3′-UTR region) as a template. A 1:2 ratio of GTP to cap analog was used for the transcription of IBV N gene.
The in vitro synthesized full-length RNA and N transcripts were treated with DNaseI and purified with phenol/chloroform. Vero cells were grown to 90% confluency, trypsinized, washed twice with ice cold PBS, and resuspended in PBS. RNA transcripts were added to 400 µl of Vero cell suspension in a 4 mm electroporation cuvette (Bio-Rad) and electroporated with one pulse at 450 V, 50 µF with a Bio-Rad Gene Pulser II electroporator. The electroporated Vero cells were cultured overnight in 1% FBS containing MEM in a 60 mm dish or a six-well plate and further cultured in MEM without FBS.
2.3. Plaque assay
Confluent Vero cells in 6 well plates were infected with 100 µl of 100-fold diluted virus stock. After 1 h of incubation, cells were washed twice with PBS and cultured in 2 ml DMEM containing 1% carboxymethylcellulose (Sigma) and 1% FBS. After 4 days of incubation, the cells were fixed with 4% paraformaldehyde for 15 min and visualized by staining the intact cells with 0.1% toluidine blue (Sigma) for 30 min. The yield of virus was calculated in terms of plaque-forming unit (PFU) per ml.
2.4. Virus titration by TCID50 method
Confluent monolayers of Vero cells on 6-well plates were infected with wild-type and recombinant IBV and harvested at different time points post-infection. Viral stocks were prepared by three freeze/thaw cycles. Vero cells on 96-well plates were infected with 10-fold serial dilutions of the viral stock. Three days post-inoculation, the number of infected wells was counted, and the 50% tissue culture infection dose (TCID50) of each sample was determined by using the Reed and Muench method.
2.5. Western blot analysis
Total protein was harvest by lysing cells with 1× RIPA buffer at the indicated time points. After mixing with 5× Laemmli (containing 0.1 M DTT), the cell lysate was boiled at 95 °C for 5 min and subjected to SDS–PAGE. Proteins were transferred to PVDF membrane (Bio-Rad) and blocked overnight at 4 °C in blocking buffer containing 5% skim milk powder in PBST buffer (80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl, 0.1% Tween 20, pH 7.5). The membrane was incubated with primary antibodies for 1 h. The antibodies against β-actin (#4967), were purchased from Cell Signaling Technology. The antisera against IBV M protein and N protein were isolated from rabbits immunized with bacterial expressed fusion proteins as previously described (Liu and Inglis, 1991, Li et al., 2005). After washing three times with PBST, the membrane was incubated with anti-mouse or anti-rabbit IgG antibodies conjugated with horseradish peroxidase (DAKO) for 1 h. After washing three times with PBST, the polypeptides were detected with a chemiluminescence detection kit (ECL, Amersham Pharmacia Biotech) according to the instructions of the manufacturer.
2.6. Indirect immunofluorescence
To detect subcellular localization of IBV M protein, Vero cells were grown on coverslips, fixed at 8 h post-infection with 4% paraformaldehyde for 10 min. Cells were then permeated by incubation with 0.2% Triton X-100 in PBS at room temperature for 10 min, followed by rinsing in PBS three times. Cells were then incubated with anti-IBV M antiserum for at least 1 h, washed with PBS three times, and incubated with anti-rabbit IgG conjugated to FITC (DAKO) for 2 h. After three washes with PBS, cells were mounted on a glass slide using a fluorescence mounting medium containing DAPI (DAKO). Images were collected with a META 510 confocal laser-scanning microscope (Zeiss).
2.7. Purification of IBV particles by sucrose gradient ultracentrifugation
Vero cells in T75 Flask were washed once with PBS and infected with IBV (MOI~2). After incubation for 90 min, the inoculum was replaced with 12 ml of DMEM. At 18 h post-infection the supernatant was collected and clarified by centrifugation at 450 × g for 15 min. IBV particles were partially purified, as previously described (Liu and Inglis, 1991), by ultracentrifugation on a 20% sucrose cushion in a Beckman SW28 rotor at 75,000×g for 3 h. The pellet was resuspended in 1 ml TNE buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM EDTA), applied on a discontinuous sucrose gradient (20–60% sucrose), and centrifuged at 28,000 rpm for 18 h. Subsequently, 10 fractions were collected. Each fraction was subjected to Western blot analysis with anti-IBV M antiserum.
2.8. Preparation and analysis of VLP
VLP formation was prepared essentially as previously described (Lim and Liu, 2001). Briefly, COS-7 cells were seeded one day prior to achieving 90% confluency for infection at an MOI of 10 with vTF7-3 (Fuerst et al., 1986). Cells were transfected with pKT-IBV E, in combination with pKT-IBV-MWT or pKT-IBV-MN3D/N6D using Lipofectamine 2000 (Invitrogen). At 15 h post-infection, the culture supernatant was harvested and clarified at 1500×g for 10 min at 4 °C. VLPs were collected by pelleting the clarified media through a 20% sucrose gradient cushion for 2 h in a Beckman SW41Ti rotor at 38,000 rpm at 4 °C. Pellets were resuspended directly in 1× SDS loading buffer. VLP samples and the corresponding whole cell lysates were analyzed by Western Blot using anti-IBV M and anti-IBV E antisera.
2.9. Quantitative real-time PCR
Total RNA from cultured cells was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The extracted total RNA was reverse transcribed using the FastKing gDNA Dispelling RT SuperMix (TIANGEN) according to the manufacturer's instructions. cDNAs were then subjected to qPCR using appropriate primers. Primers used for IBV gRNA were 5′-GTTCTCGCATAAGGTCGGCTA-3′(forward) and 5′-GCTCACTAAACACCACCAGAAC-3′(reverse). Primers used for IBV sgRNA2 were 5′-GCCTTGCGCTAGATTTTTAACTG-3′(forward) and 5′-AGTGCACACAAAAGAGTCACTA-3′(reverse). Primers used for monkey GRP78 were 5′-GAAAGAAGGTTACCCATGCAGT-3′(forward) and 5′-CAGGCCATAAGCAATAGCAGC-3′(reverse). Primers used for monkey CHOP were 5′-GAACGGCTCAAGCAGGAAATC-3′(forward) and 5′-TTCACCATTCGGTCGATCAGAG-3′(reverse). Primers used for monkey XBP1-Total were 5′-CCCTCCAGAACATCTCCCCAT-3′(forward) and 5′-ACATGACTGGGTCCAAGTTGT-3′(reverse). Primers used for monkey XBP1-Spliced were 5′-TGCTGAGTCCGCAGCAGGTG-3′(forward) and 5′-ACATGACTGGGTCCAAGTTGT-3′(reverse). Primers used for monkey IL-6 were 5′-GTGCAAATGAGTACAAAAGTCCTGA-3′(forward) and 5′-GTTCTGCGCCTGCAGCTTC-3′(reverse). Primers used for monkey IL-8 were 5′-AAGACGTACTCCAAACCTATCCAC-3′(forward) and 5′-TCTGTATTGACGCAGTGTGGTC-3′(reverse). Primers used for monkey GAPDH were 5′-CTGGGCTACACTGAGCACC-3′(forward) and 5′-AAGTGGTCGTTGAGGGCAATG-3′(reverse). Quantitative real-time RT-PCR (qPCR) was performed using the Talent qPCR PreMix (SYBR Green) (TIANGEN) according to the manufacturer's instructions. Briefly, 1 µl cDNA product was mixed with 0.3 µl forward primer (10 µM), 0.3 µl reverse primer (10 µM), 7.5 µl 2X SYBR select master mixture and 5.9 µl RNase-Free ddH2O to make up a typical 15 µl qPCR reaction. The mixture was then subjected to thermo cycling in a 7500 real-time PCR system (Applied Biosystems). The standard protocol includes enzyme activation at 50 °C for 3 min, initial denaturation at 95 °C for 3 min, followed by 40 cycles of denaturing (95 °C, 5 s) and annealing/extension (60 °C, 30 s) with fluorescent acquisition at the end of each cycle. The results obtained were in the form of threshold cycles (CT values). The relative abundance of the mRNA was calculated using GAPDH as an internal control and normalized to 0 h post infection samples (in time course experiments).
2.10. 50% Embryo Lethal Dose (ELD50) measurement
The virus stock of WTrIBV or rN3D/N6D were 10-fold serially diluted and 0.2 ml diluted virus was injected into the allantoic cavity of 10-day old embryonated specific pathogen free (SPF) eggs and incubated at 37 °C for 5 days. The number of live or dead embryos was counted and the 50% Embryo Lethal Dose (ELD50) was calculated using the Reed and Muench method (Fung and Liu, 2017).
2.11. Construction of plasmids
Wild type IBV M gene was amplified from cDNA of IBV-infected Vero cells using the forward primer 5′-TGGAAGATCTCCACCATGCCCAACGAGAC AAATTG-3′ and reverse primer 5′-CCGGAATTCTTATGTGTAAAGACTACTTC-3′. Similarly, IBV M mutant N3D was amplified with forward primer 5′-TGGAAGATCTCCACCATGCCCGACGAGACAAATTGTACT-3′ and reverse primer 5′-CCGGAATTCTTATGTGTAAAGACTACTTC-3′; IBV M mutant N6D was amplified with forward primer 5′-TGGAAGATCTCCACCATGCCCAACGA GACAGATTGTACTCTTG-3′ and reverse primer 5′-CCGGAATTCTTATGTGTAA AGACTACTTC-3′; IBV M mutant N3D/N6D was amplified with forward primer 5′-TGGAAGATCTCCACCATGCCCGACGAGACAGATTGTACTCTTG-3′ and reverse primer 5′-CCGGAATTCTTATGTGTAAAGACTACTTC-3′. All PCR products were cloned to vector pKT0 using BglII/EcoRI restriction sites.
3. Results
3.1. Confirmation of the N-linked glycosylation sites in IBV M protein
In the 20-amino-acid N-terminal ectodomain domain of the M protein of IBV Beaudette strain, two consensus N-linked glycosylation sites at amino acid positions 3 (N3) and 6 (N6), respectively, were predicted (Fig. 1a). Analysis by multiple alignments of IBV variants from different geographical locations and isolation times showed that the N6 site is strictly conserved in all IBV variants, but the N3 site is less conserved (Fig. 1b). As shown in Fig. 1b, the N3 site is missing in some strains (Fig. 1b).
To identify the N-linked glycosylation sites and to study the functional role of N-linked glycosylation in IBV M protein, Asn to Asp substitutions were introduced into the M protein to change the two predicted N-linked glycosylation sites at the amino acid positions 3 and 6, respectively. Three mutant M proteins, MN3D, MN6D, and MN3D/N6D were generated and expressed (Fig. 1b). Western blot analysis of HeLa cells expressing wild type M protein (MWT) detected three bands (Fig. 1c, lane 1), representing the nonglycosylated form (M0), glycosylated M at one position (M1) and at both positions (M2), respectively. In cells expressing the MN3D and MN6D, both M0 and M1 bands were detected, but the M2 band was absent (Fig. 1c, lane 3 and 5). For the double mutant MN3D/N6D, only the lowest band representing the unglycosylated form of M protein was detected (Fig. 1c, lane 7).
To further analyze the N-linked glycosylation status of M protein, cells expressing wild type and mutant M were treated with PNGase F and analyzed by Western blot with anti-M polyclonal antibodies. As shown in Fig. 1c, PNGase F treatment removed the two upper bands in wild type M protein and the upper band in each single mutant M protein (Fig. 1c, lane 2, 4 and 6). No difference was observed for the MN3D/N6D with or without PNGase F treatment (Fig. 1c lane 7 and 8). These results confirm that IBV M protein is indeed N-linked glycosylated at both N3 and N6 positions.
3.2. Effect of N3D/N6D mutations on the growth properties of the mutant IBV
To characterize the functional effect of N-linked glycosylation on M protein, the mutant M construct containing Asn-Asp substitution at both N3 and N6 positions was introduced into a full-length infectious cDNA clone (Fang et al., 2007). The full-length transcript together with the N transcript was electroporated into Vero cells. Typical CPEs were observed in cells transfected with either WTrIBV or rN3D/N6D full length genomic transcripts at 4 days post-electroporation and recombinant viruses were recovered. Nucleotide sequencing of the RT-PCR fragments covering region containing the amino acid mutations confirmed that the mutations were introduced into and maintained in the viral genome of the recovered IBV.
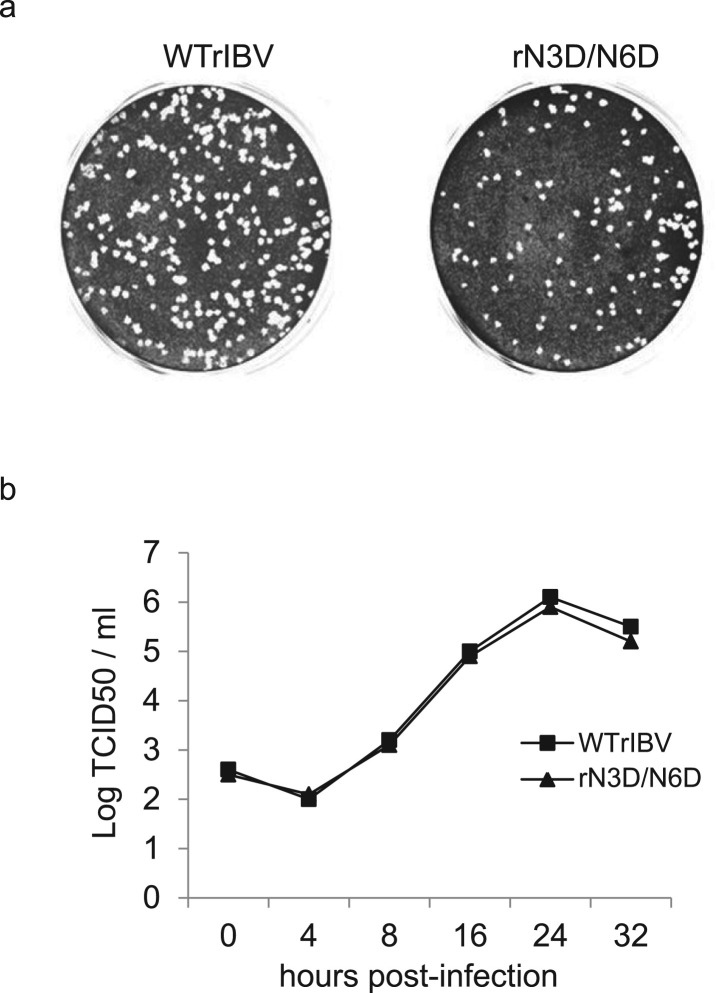
The growth properties of rN3D/N6D on Vero cells were then tested by analysis of plaque sizes and growth curves. Compared to cells infected with WTrIBV, similar plaques in terms of size and shape were observed in Vero cells infected with rN3D/N6D ( Fig. 2a). It was noted that the number of plaques for rN3D/N6D was fewer than that of WTrIBV (Fig. 2a). This was due to the fact that the original titer of the rN3D/N6D stock used in this experiment was lower.
Fig. 2.
Plaque morphologies and growth kinetics of WTrIBV and the rN3D/N6D mutant. a. Plaque sizes and morphologies of WTrIBV and rN3D/N6D. Monolayers of Vero cells on a 6-well plate were infected with 100 µl of 1000-fold diluted virus stocks and cultured in the presence of 0.5% carboxymethyl cellulose at 37 °C for 3 days. Cells were then fixed and stained with 0.1% toluidine. b. The growth curves of WTrIBV and rN3D/N6D. Vero cells were infected with the viruses and harvested at 0, 4, 8, 16, 24 and 32 h post-inoculation, respectively. Viral stocks were prepared by freezing/thawing the cells three times, and Vero cells on 96-well plates were infected with 10-fold serial dilution of the viral stock. TCID50 was determined by using the Reed and Muench method.
The mutant virus also displayed very similar growth kinetics as wild type virus (Fig. 2b). These results indicate that glycosylation of M protein at both N3 and N6 positions does not significantly influence the growth properties of IBV in cell culture. The genetic stability of rN3D/N6D was tested by propagation on Vero cells for 20 passages. Sequencing of the M protein of rN3D/N6D showed that the mutations were stable.
3.3. Effects of N3D/N6D mutations on the subcellular localization of the protein and sedimentation properties of the mutant IBV
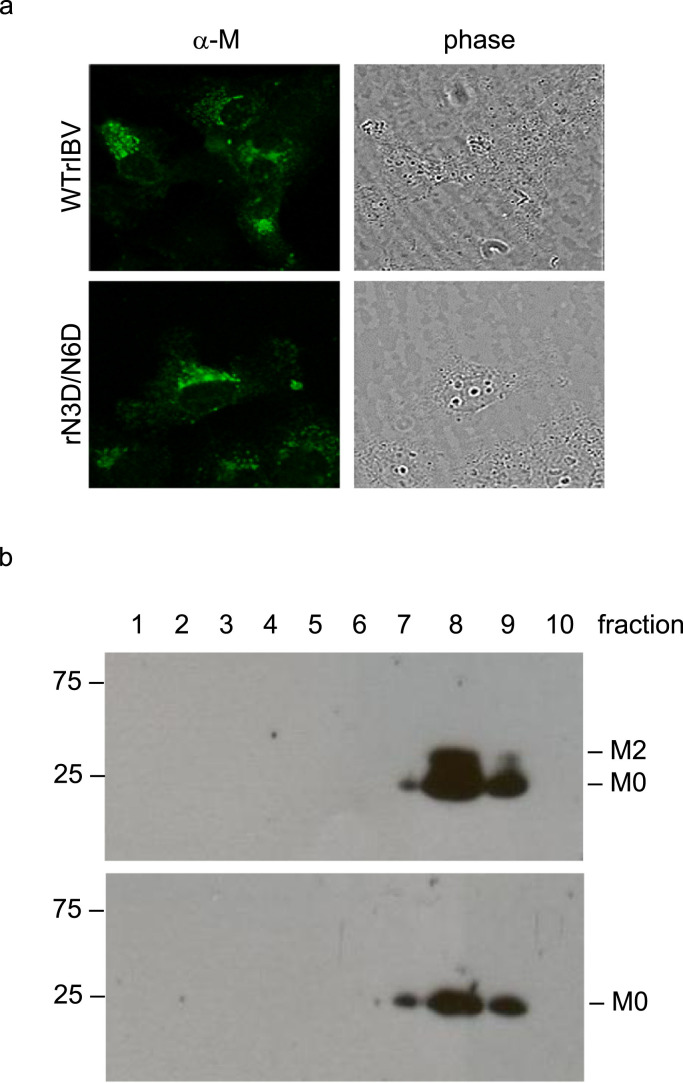
Addition of a carbohydrate chain is important for folding, structural stability and intracellular sorting of the modified protein. It is well established that IBV M localizes to the Golgi apparatus in the infected cells (Swift and Machamer, 1991). Subcellular localization of M protein in Vero cells infected with WTrIBV or rN3D/N6D was determined by indirect immunofluorescence using anti-IBV M antiserum. In both infected cells, a perinuclear, dot-like staining pattern indicative of Golgi localization was observed ( Fig. 3a). This suggests that removal of the N-glycans from M protein does not affect the normal transport and localization pattern of the protein.
Fig. 3.
The subcellular localization of M protein and the sedimentation properties of WTrIBV and the rN3D/N6D mutant. a. Intracellular distribution of M protein in cells infected with WTrIBV or rN3D/N6D. Vero cells were infected with WTrIBV or rN3D/N6D before being fixed, permeabilized and incubated with anti-IBV M antiserum. The bound primary antibodies were detected using FITC-labelled anti-rabbit secondary antibodies. b. Sedimental analysis of WTrIBV and the rN3D/N6D mutant. Vero cells were infected with WTrIBV or rN3D/N6D. Viral particles in the culture media were pelleted through 30% sucrose cushion and then sedimented into a discontinuous sucrose gradient consisting of 20%, 30%, 40%, 50% and 60% sucrose 2 ml each in TNE buffer. Ten fractions, 1 ml each, were collected from the top to the bottom for analysis by Western blot with anti-IBV M antiserum.
IBV particles are heterogeneous in a sucrose gradient and sediment in a broad band of density (Collins et al., 1976, Macnaughton and Davies, 1980). Purified virions of rN3D/N6D and WTrIBV were compared by ultracentrifugation in 20–60% discontinuous sucrose gradients. Fractions were collected and analyzed by Western blot with anti-M antiserum. The sedimentation profiles were similar, with most of rN3D/N6D and WTrIBV particles detected in fractions 8 and 9 (Fig. 3b). This result suggests that removal of the N-glycans from M protein does not significantly affect the sedimentation properties of the particles.
3.4. Effect of N3D/N6D mutations on VLP assembly
Subsequently, the question of whether M protein-mediated VLP formation would be sensitive to N3D/N6D mutations was addressed by co-expression of the mutant M protein with wild type E protein using the recombinant vaccinia virus expressing T7 RNA polymerase (vTF7-3) (Fuerst et al., 1986). At 18 h post-transfection, the culture media were removed from cells and clarified by centrifugation. VLPs were isolated from the clarified media by centrifugation through a sucrose cushion. Cell lysates and extracellular VLPs were analyzed by SDS-PAGE and Western blotting, showing that only the unglycosylated M was incorporated into VLPs ( Fig. 4), consistent with our previous report (Lim and Liu, 2001). The mutant M protein could form VLPs in a similar efficiency as wild type M protein, as shown by the presence of a similar amount of extracellular M (Fig. 4), indicating that mutation of the N-linked glycosylation sites in the N-terminal ectodomain of M protein does not affect VLP formation.
Fig. 4.
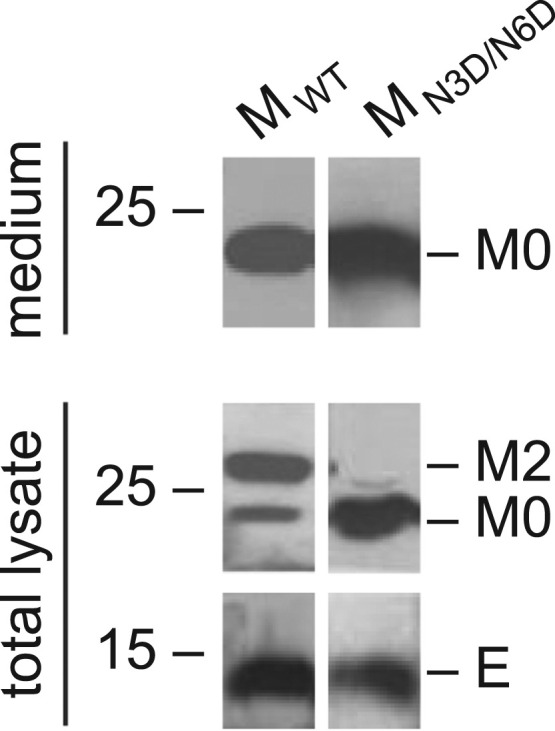
The effect of N3D/N6D mutation of IBV M protein on VLP assembly. COS-7 cells were infected with the recombinant vaccinia/T7 virus (vTF7-3) and transfected with a plasmid encoding wild type IBV E gene, together with a plasmid encoding MWT or MN3D/N6D, respectively. Intracellular cell lysates and pelleted extracellular VLPs were analyzed by Western blot with antisera against IBV M and E proteins.
3.5. Effect of N3D/N6D mutations on IBV-induced ER stress response and induction of proinflammatory cytokines
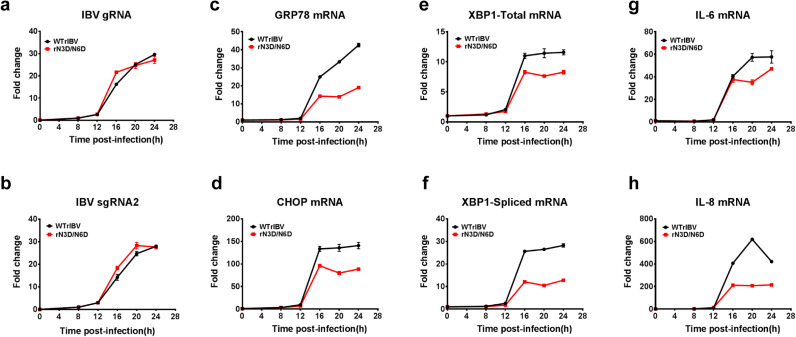
Although IBV M protein is presumably post-translationally inserted into the ER membrane, N-linked glycosylation of its ectodomain is nonetheless dependent on modification enzymes inside the ER lumen (Fung and Liu, 2018). We have previously shown that IBV infection activates the PERK-eIF2α and IRE1-XBP1 branches of the UPR signaling pathway. To determine whether N-linked glycosylation on the M protein may contribute to the IBV-induced ER stress, Vero cells were infected with WTrIBV or rN3D/N6D at MOI~2, respectively, and total RNA was harvested in a time-course experiment. RT-qPCR analysis revealed that the accumulation of IBV genomic RNA (gRNA) and subgenomic RNA2 (sgRNA2) was comparable in cells infected with WTrIBV and rN3D/N6D at the same time points, suggesting that mutations of the N-glycosylation sites did not affect the replication and transcription of IBV RNAs ( Fig. 5a and b). In cells infected with WTrIBV, the mRNA levels of several ER stress-related genes, such as GRP78, CHOP, total XBP1 and spliced XBP1, were significantly upregulated at late stages (16 hpi onwards) of IBV infection. In contrast, induction of these genes was considerably lower in cells infected with rN3D/N6D (Fig. 5c–f). A similar pattern was also observed in the expression profiles of two proinflammatory cytokines IL-6 and IL-8. Compared with the WTrIBV control, the induction of IL-6 and IL-8 at late stages of IBV infection was significantly dampened in cells infected with rN3D/N6D (Fig. 5g and h).
Fig. 5.
Induction ER stress response and pro-inflammatory cytokines in cells infected with WTrIBV or the rN3D/N6D mutant. Vero cells were infected with WTrIBV or rN3D/N6D at MOI~2 and cellular RNA was harvested in a time course experiment. The amounts of IBV genomic RNA (gRNA), IBV subgenomic mRNA2 (sgRNA2), ER stress-related genes and two proinflammatory cytokines were determined by RT-qPCR, using GAPDH as an internal control. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
3.6. Effect of N3D/N6D mutations on IBV-induced apoptosis
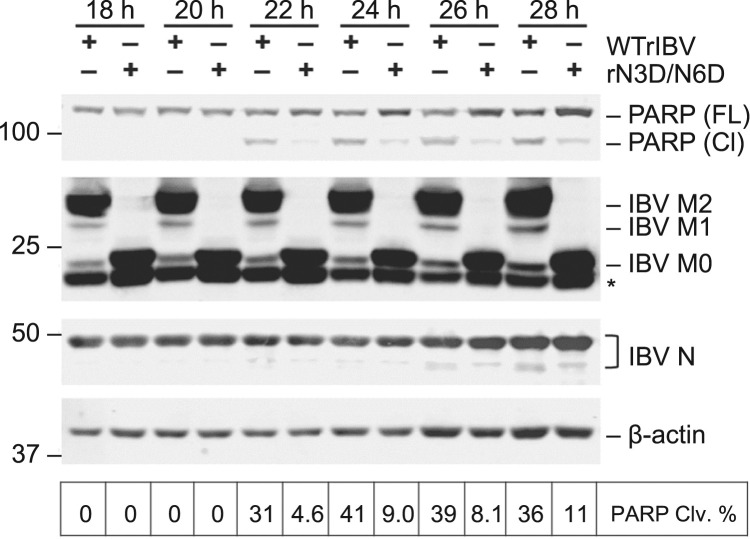
IBV infection was previously shown to induce caspase-dependent and p53-independent apoptosis in the late stage infected cells (Liu et al., 2001), and that ER stress effector proteins, such as CHOP and XBP1, modulate IBV-induced apoptosis (Liao et al., 2013, Fung et al., 2014a). To determine whether glycosylation on the M protein affects IBV-induced apoptosis, Vero cells were infected with WTrIBV or rN3D/N6D at MOI~2, respectively, and harvested at 18 hpi onwards in 2 h intervals. As shown in Fig. 6, most of the M protein is fully or partially glycosylated in cells infected with WTrIBV, whereas strictly no M glycosylation was observed in cells infected with rN3D/N6D. Similar amounts of IBV N protein were detected at the same time points for WTrIBV and rN3D/N6D, further confirming that mutations in the N-linked glycosylation sites did not significantly affect IBV replication in Vero cells. To determine the induction of apoptosis, cleavage of poly (ADP-ribose) polymerase (PARP) was used as an apoptotic marker. PARP is a substrate of caspase 3, and the percentage of its cleavage from the full-length protein (116 kDa) to the cleaved form (89 kDa) has been widely used as a benchmark of apoptosis induction (Fung and Liu, 2017). In cells infected with WTrIBV, significant PARP cleavage could be detected starting from 22 hpi, which remained at a considerable level until the end of the time course (Fig. 6). In sharp contrast, only marginally detectable PARP cleavage was observed in cells infected with rN3D/N6D. This result suggests that N-linked glycosylation in the M protein may contribute to the induction of apoptosis during IBV infection.
Fig. 6.
Induction of apoptosis in cells infected with WTrIBV or the rN3D/N6D mutant. Vero cells were infected with WTrIBV or rN3D/N6D at MOI~2. Protein lysates were harvested at 18 hpi onwards in 2 h intervals and were subjected to Western blot analysis using the indicated antisera or antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kilo Dalton were indicated on the left. Degree of cell apoptosis was calculated as the band intensity of cleaved PARP protein divided by the band intensity of the total protein. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
3.7. Effect of N3D/N6D mutations on the pathogenesis of IBV in embryonated eggs
The result above suggested that although N-linked glycosylation of the IBV M protein did not significantly affect viral replication in cell culture, it could modulate important aspects of virus-host interactions. N-linked glycosylation of the IBV M ectodomain may therefore contribute to the virulence and pathogenesis in vivo. To address this possibility, 0.2 ml of serially diluted WTrIBV or rN3D/N6D was injected into the allantoic cavity of 10-day old embryonated SPF chicken eggs respectively, and incubated at 37 °C for 5 days. The number of live or dead embryos was counted and the 50% Embryo Lethal Dose (ELD50) calculated using the Reed and Muench method (Reed and Muench, 1938). It was found that ELD50 of WTrIBV was slightly higher than to that of rN3D/N6D ( Fig. 7), indicating that N-linked glycosylation of M protein might render a certain enhancement effect on the replication and pathogenesis of IBV in vivo.
Fig. 7.
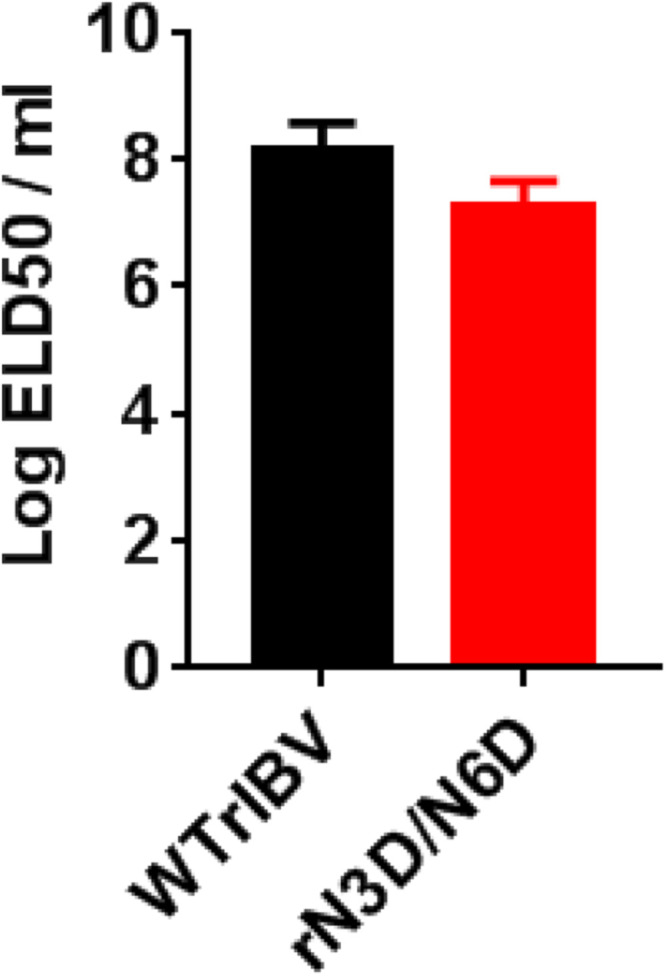
ELD50 of WTrIBV and the rN3D/N6D mutant. The virus stocks of WTrIBV and rN3D/N6D of the same titers were 10-fold serially diluted and 0.2 ml diluted virus was injected into the allantoic cavity of 10-day old embryonated SPF eggs and incubated at 37 °C for 5 days. The numbers of live or dead embryos were counted and the 50% embryo lethal dose (ELD50) was calculated using the Reed and Muench method. The bar chart shows the results from two independent experiments with standard deviations.
4. Discussion
Coronavirus M protein plays a central role in particle assembly by interacting with other viral structural components, leading to the capture and incorporation of the structural components into virions at the budding sites. In this study, the two predicted N-linked glycosylation sites at the N-terminal ectodomain of IBV M protein were confirmed to be glycocylated and the functional involvement of the N-linked glycosylation of M protein in IBV virion assembly, IBV-induced ER stress response, apoptosis, proinflammatory response and pathogenesis was investigated, by combining reverse genetics, biochemical and cell biology approaches.
Coronavirus M proteins are invariably glycosylated in the luminal ectodomain. While the M protein of some lineage A Betacoronaviruses is O-linked glycosylated, those of other coronaviruses are exclusively N-linked (Fung and Liu, 2018). Only a single N-glycosylation site has been identified in the SARS-CoV M protein at N4 (Hu et al., 2003, Voss et al., 2006). When expressed alone with a C-terminal FLAG-tag, SARS-CoV M protein acquired high-mannose N-glycans in the ER, which were modified into complex glycans in the Golgi (Nal et al., 2005). However, SARS-CoV M protein in the infected cells and purified virions was later found to retain high-mannose N-glycans sensitive to endoglycosidase H, indicating that the maturation of N-linked glycans might be inhibited in cells infected with SARS-CoV (Voss et al., 2006). In this study, we confirmed that M protein of the Vero-adapted Beaudette strain of IBV was glycosylated at N3 and N6. In both transiently transfected cells expressing M protein and IBV-infected cells, IBV M protein was predominantly present in a fully glycosylated form (glycosylated on both N3 and N6), while the partially (N3 or N6) glycosylated and the non-glycosylated form were the minor forms, suggesting that both N3 and N6 in the IBV M ectodomain are efficiently glycosylated in cell culture.
Previous studies have shown that neither N-linked nor O-linked glycosylation is essential for virion assembly. For example, a mutant TGEV with disruption of the N-linked glycosylation site of M protein was isolated, and no obvious consequences on viral replication and infectivity were observed (Laude et al., 1992). Compared with the wild type control, a glycosylation-deficient recombinant SARS-CoV (N4Q) exhibited normal virion morphology and similar growth kinetics in cultured cells (Voss et al., 2009). Similarly, O-linked glycosylation-deficient recombinant MHV replicated normally in cell culture (de Haan et al., 1998, de Haan et al., 2003). Data presented in this study showed that recombinant IBV harboring mutations in both glycosylation sites of the M ectodomain (N3D/N6D) exhibited normal plaque morphology and replicated similarly as the wild type control. Furthermore, no obvious differences in the subcellular localization of M protein and the sedimental properties of the purified virus were not detected. Taken together, these results suggest that similar to other coronaviruses, glycosylation of the M ectodomain is not essential for the assembly and morphogenesis of IBV in cell culture.
Interestingly, most of the IBV M protein incorporated into the VLPs was the unglycosylated form. However, M protein in the IBV virions is glycosylated as shown in the sedimentation analysis. The difference in the M protein glycosylation status between VLPs and IBV virions indicated that the mechanisms underlying the VLP assembly and viral assembly in the study systems used might not be exactly the same. The discrepancy between the VLP assembly and viral assembly was also found in the MHV M protein C-terminal mutant (de Haan et al., 1998). Further characterization of this difference would be required to deepen our understanding of the assembly process of coronavirus particles and to clarify the usefulness of VLP formation as a tool for studying this important process in coronavirus life cycles.
During coronavirus replication, a massive amount of viral structural proteins is synthesized in the ER, which saturates its folding capacity and lead to ER stress (Fung et al., 2014b). Previous studies have shown that coronavirus infection or overexpression of the S glycoprotein may trigger ER stress, resulting in the activation of the UPR signaling pathways that modulated apoptosis and pro-inflammatory response (Liao et al., 2013, Fung et al., 2014a, Versteeg et al., 2007, Fung and Liu, 2014). Although not as highly glycosylated as the S protein, M protein is the most abundant structural protein in the coronavirus particle as well as in the infected cells, and its glycosylation also depends on host modification enzymes in the ER lumen. In this study, we found that induction of GRP78, a well-characterized ER stress marker gene at the mRNA level was significantly impaired in cells infected with rN3D/N6D compared with the wild type control. Consistently, activation of the PERK and IRE branch of UPR was also impaired as indicated by the lower mRNA levels of the downstream effector gene CHOP and XBP1-spliced, compared to the cells infected with wild type virus. Notably, the reduced ER stress level also correlated with a decreased level of apoptosis and reduced induction of proinflammatory cytokines IL-6 and IL-8. These results suggest that N-linked glycosylation of the M ectodomain also contributes to the induction of ER stress response during IBV infection, resulting in a reduced level of IBV-induced apoptosis and a proinflammatory response.
The genomic regions encoding all structural genes were sequenced in WTrIBV and rN3D/N6D, and it was found that the two viruses share identical sequences in the S and N genes. As the coding sequence of the last nine amino acids in E gene overlaps with the N-terminal region of the M protein, the N3D/N6D mutations inevitably result in the simultaneous Q102R/K105R substitutions in the E protein. Our preliminary data suggest overexpression of wild type and mutant IBV E protein did not induce ER stress, apoptosis and the production of pro-inflammatory cytokines. However, we cannot totally rule out at this stage the possibility that these two substitutions in the IBV E protein may have a certain effect on IBV-induced ER stress, apoptosis and the production of pro-inflammatory cytokines in the infected cells.
Although the M protein is the most abundant structural proteins produced inside a coronavirus-infected cell, overexpression of MHV M protein per se was not sufficient to induce ER stress and upregulate IL-8 (CXCL2) transcription in cultured cells, even when the recombinant vaccinia virus (vTF7-3) was used to strongly promote transgene expression (Versteeg et al., 2007). We have also overexpressed IBV M protein in Vero cells with or without vTF7-3 infection, but no significant induction of ER stress or IL-8 production was observed compared with the vector control (unpublished data). There are two possible explanations. First, the induction of ER stress and IL-8 production may require additional viral factors (such as S glycoprotein) produced during coronavirus infection. Secondly, the level of overexpressed M protein, even with the help of vTF7-3 infection, may not be as high as that in IBV-infected cells. Therefore, the involvement of M glycosylation in ER stress/IL-8 induction can only be determined during actual infection.
Although dispensable for virion assembly and in vitro infectivity, glycosylation of the M ectodomain was previously shown to play a role in coronavirus pathogenesis and/or replication in vivo. For example, the induction of type I interferon was significantly reduced in cells infected with glycosylation-deficient mutant of TGEV or MHV, compared with the wild type control (Laude et al., 1992, de Haan et al., 2003). Also, although in vivo interferogenic capability was not affected, O-linked glycosylation-deficient mutant of MHV did not replicate as well in the liver of infected mice (de Haan et al., 2003). In this study, we have observed a minor difference in the ELD50 between WTrIBV and rN3D/N6D. It is uncertain that the differential induction of apoptosis and pro-inflammatory response observed in cell culture experiments may contribute to the slight difference observed in the in vivo virulence between wild type and the mutant viruses. More specific and sensitive approaches would be required to uncover the effect of M glycosylation on IBV replication and pathogenesis in vivo.
Acknowledgment
This work was partially supported by Guangdong Natural Science Foundation Grant 2018A030313472, and Guangdong Province Key Laboratory of Microbial Signals and Disease Control Grants MSDC-2017-05 and MSDC-2017-06, Guangdong, People's Republic of China.
Acknowledgments
Conflict of interest
The authors declare no conflict of interest.
References
- Bárcena M., Oostergetel G.T., Bartelink W., Faas F.G.A., Verkleij A., Rottier P.J.M., Koster A.J., Bosch B.J. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. USA. 2009;106:582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns M.M., Boursnell M.E., Tomley F.M., Brown T.D. Nucleotide sequence encoding the membrane protein of the IBV strain 6/82. Nucleic Acids Res. 1986;14:5558. [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Pappin D.J. Coronavirus IBV glycopolypeptides: locational studies using proteases and saponin, a membrane permeabilizer. Virus Res. 1986;4:145–156. doi: 10.1016/0168-1702(86)90038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M.S., Alexander D.J., Harkness J.W. Heterogeneity of infectious bronchitis virus grown in eggs. Arch. Virol. 1976;50:55–72. doi: 10.1007/BF01318001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dea S., Verbeek A.J., Tijssen P. Antigenic and genomic relationships among turkey and bovine enteric coronaviruses. J. Virol. 1990;64:3112–3118. doi: 10.1128/jvi.64.6.3112-3118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drickamer K., Taylor M.E. Evolving views of protein glycosylation. Trends Biochem. Sci. 1998;23:321–324. doi: 10.1016/s0968-0004(98)01246-8. [DOI] [PubMed] [Google Scholar]
- Fang S., Chen B., Tay F.P.L., Ng B.S., Liu D.X. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology. 2007;358:136–147. doi: 10.1016/j.virol.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T.R., Niles E.G., Studier F.W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014:5. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Activation of the c-Jun NH2-terminal kinase pathway by coronavirus infectious bronchitis virus promotes apoptosis independently of c-Jun. Cell Death Dis. 2017;8:3215. doi: 10.1038/s41419-017-0053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liao Y., Liu D.X. The ER stress sensor IRE1α protects cells from apoptosis induced by coronavirus infectious bronchitis virus. J. Virol. 2014;88:12752–12764. doi: 10.1128/JVI.02138-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Huang M., Liu D.X. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus–host interactions. Virus Res. 2014;194:110–123. doi: 10.1016/j.virusres.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., de Wit M., Kuo L., Montalto-Morrison C., Haagmans B.L., Weiss S.R., Masters P.S., Rottier P.J.M. The glycosylation status of the murine hepatitis coronavirus M protein affects the interferogenic capacity of the virus in vitro and its ability to replicate in the liver but not the brain. Virology. 2003;312:395–406. doi: 10.1016/S0042-6822(03)00235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A., Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Hogue B.G., Nayak D.P. Expression of the porcine transmissible gastroenteritis coronavirus M protein. Adv. Exp. Med. Biol. 1990;276:121–126. doi: 10.1007/978-1-4684-5823-7_18. [DOI] [PubMed] [Google Scholar]
- Holmes K.V., Doller E.W., Sturman L.S. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology. 1981;115:334–344. doi: 10.1016/0042-6822(81)90115-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Wen J., Tang L., Zhang H., Zhang X., Li Y., Wang J., Han Y., Li G., Shi J., Tian X., Jiang F., Zhao X., Wang J., Liu S., Zeng C., Wang J., Yang H. The M protein of SARS-CoV: basic structural and immunological properties. Genom. Proteom. Bioinforma. 2003;1:118–130. doi: 10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapps W., Hogue B.G., Brian D.A. Deduced amino acid sequence and potential O-glycosylation sites for the bovine coronavirus matrix protein. Adv. Exp. Med. Biol. 1987;218:123–129. doi: 10.1007/978-1-4684-1280-2_14. [DOI] [PubMed] [Google Scholar]
- Laude H., Gelfi J., Lavenant L., Charley B. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J. Virol. 1992;66:743–749. doi: 10.1128/jvi.66.2.743-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.Q., Xiao H., Tam J.P., Liu D.X. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579:2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Fung T.S., Huang M., Fang S.G., Zhong Y., Liu D.X. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 2013;87:8124–8134. doi: 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P., Liu D.X. The missing link in coronavirus assembly retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001;276:17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim X.Y., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Xu H.Y., Liu D.X. Induction of caspase-dependent apoptosis in cultured cells by the avian coronavirus infectious bronchitis virus. J. Virol. 2001;75:6402–6409. doi: 10.1128/JVI.75.14.6402-6409.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Cavanagh D., Green P., Inglis S.C. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991;184:531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Shen S., Xu H.Y., Wang S.F. Proteolytic mapping of the coronavirus infectious bronchitis virus 1b polyprotein: evidence for the presence of four cleavage sites of the 3C-like proteinase and identification of two novel cleavage products. Virology. 1998;246:288–297. doi: 10.1006/viro.1998.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Wu D., Shen C., Chen K., Shen X., Jiang H. Severe acute respiratory syndrome coronavirus membrane protein interacts with nucleocapsid protein mostly through their carboxyl termini by electrostatic attraction. Int. J. Biochem Cell Biol. 2006;38:589–599. doi: 10.1016/j.biocel.2005.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnaughton M.R., Davies H.A. Two particle types of avian infectious bronchitis virus. J. Gen. Virol. 1980;47:365–372. doi: 10.1099/0022-1317-47-2-365. [DOI] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounir S., Talbot P.J. Sequence analysis of the membrane protein gene of human coronavirus OC43 and evidence for O-glycosylation. J. Gen. Virol. 1992;73(Pt 10):2731–2736. doi: 10.1099/0022-1317-73-10-2731. [DOI] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.-Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., Wüthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Rottier P.J., Welling G.W., Welling-Wester S., Niesters H.G., Lenstra J.A., Van, der Zeijst B.A. Predicted membrane topology of the coronavirus protein E1. Biochemistry. 1986;25:1335–1339. doi: 10.1021/bi00354a022. [DOI] [PubMed] [Google Scholar]
- Stern D.F., Sefton B.M. Coronavirus proteins: structure and function of the oligosaccharides of the avian infectious bronchitis virus glycoproteins. J. Virol. 1982;44:804–812. doi: 10.1128/jvi.44.3.804-812.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift A.M., Machamer C.E. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 1991;115:19–30. doi: 10.1083/jcb.115.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utiger A., Tobler K., Bridgen A., Ackermann M. Identification of the membrane protein of porcine epidemic diarrhea virus. Virus Genes. 1995;10:137–148. doi: 10.1007/BF01702594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., Van De Nes P.S., Bredenbeek P.J., Spaan W.J.M. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J. Virol. 2007;81:10981–10990. doi: 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss D., Kern A., Traggiai E., Eickmann M., Stadler K., Lanzavecchia A., Becker S. Characterization of severe acute respiratory syndrome coronavirus membrane protein. FEBS Lett. 2006;580:968–973. doi: 10.1016/j.febslet.2006.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss D., Pfefferle S., Drosten C., Stevermann L., Traggiai E., Lanzavecchia A., Becker S. Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein. Virol. J. 2009;6:79. doi: 10.1186/1743-422X-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R., Montalto-Morrison C., Masters P.S. Genetic analysis of determinants for spike glycoprotein assembly into murine coronavirus virions: distinct roles for charge-rich and cysteine-rich regions of the endodomain. J. Virol. 2004;78:9904–9917. doi: 10.1128/JVI.78.18.9904-9917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]