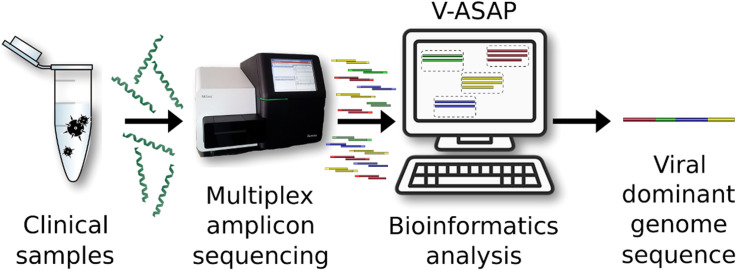
Abstract
Genome sequencing of virus has become a useful tool for better understanding of virus pathogenicity and epidemiological surveillance. Obtaining virus genome sequence directly from clinical samples is still a challenging task due to the low load of virus genetic material compared to the host DNA, and to the difficulty to get an accurate genome assembly. Here we introduce a complete sequencing and analyzing protocol called V-ASAP for Virus Amplicon Sequencing Assembly Pipeline. Our protocol is able to generate the viral dominant genome sequence starting from clinical samples. It is based on a multiplex PCR amplicon sequencing coupled with a reference-free analytical pipeline. This protocol was applied to 11 clinical samples infected with coronavirus OC43 (HcoV-OC43), and led to seven complete and two nearly complete genome assemblies. The protocol introduced here is shown to be robust, to produce a reliable sequence, and could be applied to other virus.
Keywords: Coronavirus, High-throughput sequencing, Complete genome, Bioinformatics
Graphical abstract
1. Introduction
When an outbreak occurred in a health-care setting, identification of the causative pathogen and epidemiological investigations need to be fast and effective to allow a targeted infection control response. During viral outbreaks, molecular diagnosis methods as real-time RT/PCR allow identifying the pathogen and eventually the index case of the outbreak. However, the outbreak investigation must also determine clusters of patients and the pathway of the dissemination of the causative agent to stop the viral dissemination. Until now, Sanger sequencing method is used to obtain partial sequence of viral genome necessary to identify clusters of cases. However, many technical limitations, especially the small amount of viral genome in the biological samples, make this approach not very effective for the management of the outbreak. High-throughput sequencing (HTS) technologies provide the possibility to rapidly obtain the full sequence of pathogen genomes. Notably whole-genome sequencing (WGS) of viruses is a powerful tool for the development of novel treatments and vaccines, for studying virus evolution and genetic association to diseases or for tracking outbreaks. Recently, HTS has been used to investigate viral outbreaks in health-care settings (Garvey et al., 2017; Houlihan et al., 2018; Vaughan et al., 2014). The depth of the sequencing data and the quality of the obtained sequences make this tool particularly efficient in this context. However, despite the relative small size of virus genomes, their sequencing often remains difficult. The small amount of virus genetic material compare to the host nucleic acid decreases viral sequencing output. In addition, one have to deal with the difficulty that several viral variants coexist in a single sample, presenting more or less variable sequences depending on the intrinsic mutation rate of the virus. All these points burden the sequencing and the assembly of viral genome. Today, numerous hospital laboratories have immediate access to HTS methods but the bioinformatics analysis remains difficult without bioinformatics skills. Consequently developing effective and easy-to-use protocols is a new challenge for the spreading of HTS methods as tools to investigate viral outbreaks in hospital.
Three main methods based on HTS are currently used for viral whole-genome sequencing: metagenomic sequencing, target enrichment sequencing and PCR amplicon sequencing, each showing benefits and drawbacks (Houldcroft et al., 2017). In metagenomic sequencing, total DNA (and/or RNA) from a sample including host but also bacteria, viruses and fungi is extracted and sequenced. It is a simple and cost-effective approach, and it is the only approach not requiring reference sequences. Instead, the other two HTS approaches, target enrichment and amplicon sequencing, both depend on reference information to design baits or primers. The limitation of metagenomic sequencing is that it requires a very high sequencing depth to obtain enough viral genome material. The target enrichment sequencing uses virus-specific capture oligonucleotides to enrich the viral genome preparation before sequencing. This method is more specific than metagenomics sequencing but implies higher costs and a more advanced technical expertise for sample preparation. Finally, the PCR amplicon sequencing is a well-established method consisting in specific viral genome amplification by PCR before sequencing. It is easily applicable on large number of samples in a routine use and so very adequate for clinical samples. The PCR amplification method, compared to the others, is particularly relevant for samples containing very low viral genetic material, it presents several disadvantages, though. The sequence of the virus of interest has to be known and not too variable to be correctly amplified by the set of designed primers. A second pitfall is due to the fact that the PCR cycles can introduce some amplification errors along the sequence which make the assembly step more prone to mistakes. Finally, this method can only be used for small genomes because of the number of PCR reactions which has to be limited.
A new protocol based on multiplex PCR method was recently introduced for virus sequencing (Quick et al., 2017) to obtain consensus genome sequences directly from samples. They showed that the numbers of reads with metagenomic approaches is often very low for Zika virus and applied the multiplex PCR method successfully. This innovative approach allows to rapidly (1–2 days) obtain a consensus sequence directly from clinical samples loaded with as few as 50 genome copies per reaction. This method is of great interest during outbreak and can also be used as an inexpensive and convenient method in the lab. However, it was not tested for viral genomes larger than 12 kb. Moreover, the authors also introduced in this study an analytical pipeline to obtain the consensus sequence from the alignment of reads against a reference sequence in addition to the complete and optimized sequencing protocol.
The bioinformatics analysis of virus sequencing data is often based on alignment, or mapping, of reads against a reference sequence followed by the consensus extraction by majority voting. However, the alignment step is known to introduce some biases (Archer et al., 2010, Posada-Cespedes et al., 2017). For example, if the studied virus sequence is divergent from the chosen reference sequence, the reads covering the regions of divergence could not be aligned correctly which will bias the resulting consensus. Moreover, the mapping step of reads in divergent, repetitive or low complexity regions is a difficult task which have to be carefully examined (Caboche et al., 2014). Finally, the choice of the reference sequence itself is a critical step from which the resulting consensus sequence will strongly depend.
Here we present an innovative complete protocol to rapidly obtain viral dominant sequence for a routine use in clinical context. This protocol is based on multiplex PCR amplicon sequencing. A set of reference sequences representative of the studied virus is required in order to identify conserved regions and design a set of primers. We also introduce a new analysis pipeline called V-ASAP for Virus Amplicon Sequencing Assembly Pipeline, which is able to build the dominant genome sequence from amplicon sequencing data without requiring any alignment step and so without having to choose a reference sequence. V-ASAP was first evaluated with data from sequencing of Zika virus (Quick et al., 2017) and then applied to sequence viral RNA extracted from 11 samples obtained from patients infected with OC43 coronaviruses (HCoV-OC43) and isolated at the University Hospital of Lille (France) from nasal swab or broncho-alveolar lavage. Belonging to the family of Coronaviridae and to the genus betacoronavirus, HCoV-OC43 are among the known viruses that cause the common cold, but can also cause severe lower respiratory tract infections, including pneumonia in infants, in elderly and in immunocompromised individuals. Coronaviruses are enveloped viruses possessing a positive single-stranded RNA genome with a length between 26.2 and 31.7 kb (ICTV taxonomy https://talk.ictvonline.org/p/coronavirus-genomes). Despite of the low load of HCoV-OC43 in samples and the relative high size of the genome sequence (30 kb), our protocol allowed to obtain fully finished genomes for 7 out of the 11 clinical samples. For 2 other genomes, 2 and 8 contigs were obtained covering respectively more than 99% and 94% of the entire genome.
2. Results and discussion
2.1. Protocol description
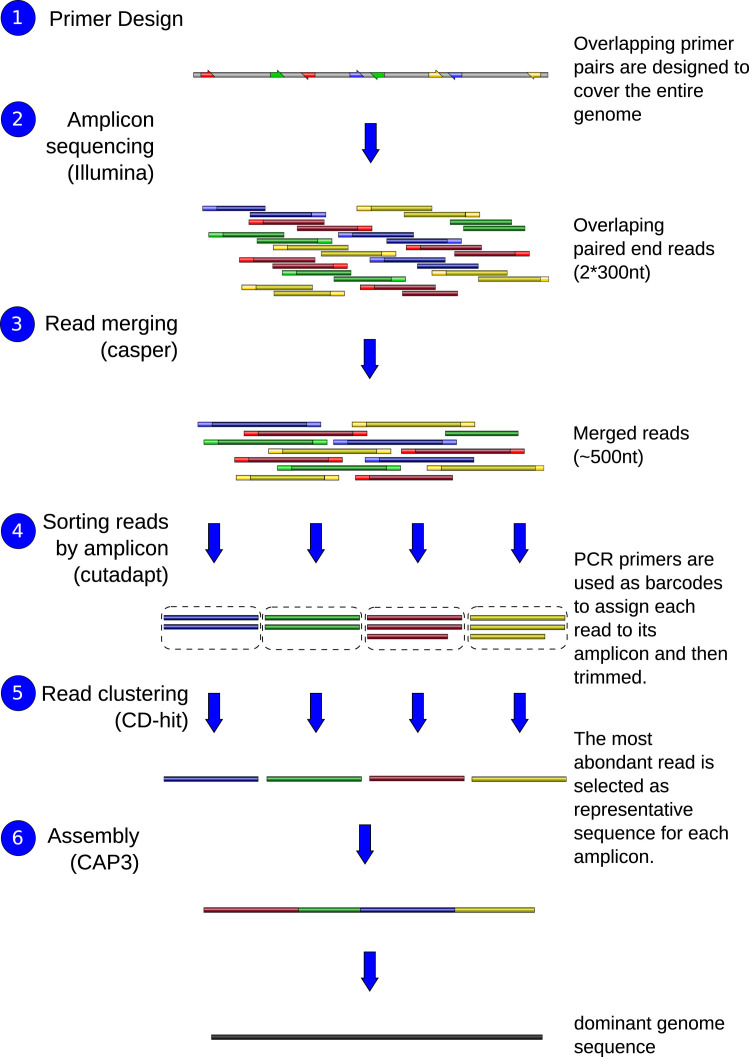
The complete protocol is schematically represented in Fig. 1. The first step consists in designing of overlapping primer pairs to cover the entire genome (Fig. 1, step 1). The next step (Fig. 1, step 2) is the amplification and sequencing of fragments which could be performed with Illumina paired-end library protocol allowing to obtain reads of 300 bp.
Fig. 1.
Schematic representation of the complete protocol for viral sequencing directly from clinical samples.
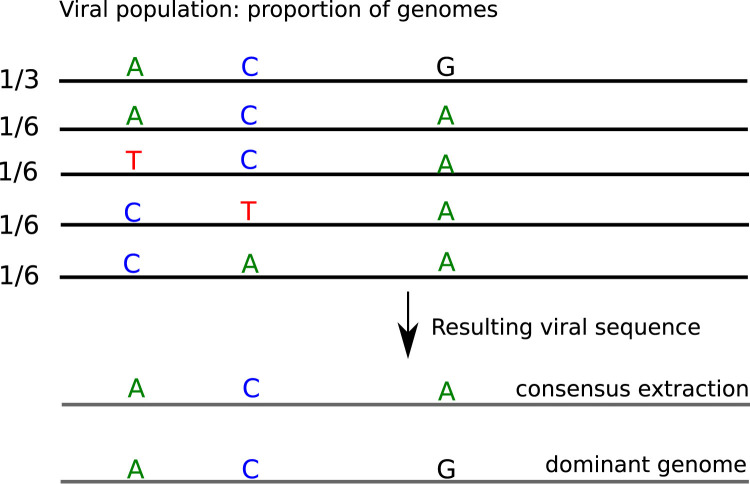
The analysis begins with the paired-end read merging step (Fig. 1, step 3) performed with CASPER (Kwon et al., 2014) and leading to reads of 500–550 bp. Each merged read is then assigned to its amplicon of origin. For this assignment, the PCR primers are used as barcodes in a demultiplexing strategy using Cutadapt (Martin, 2011). At the end of this step (Fig. 1, step 4), the number of reads per amplicon is known: some amplicons could be over-sequenced and some others under-represented. For each amplicon, several different sequences co-existed due to the presence of viral variants and sequencing errors. At this step, we postulated that the most abundant read represents the dominant variant of the population for each amplicon. Our method is slightly different from the one usually used when the consensus sequence is generated from alignment with the majority voting strategy. Indeed, in consensus extraction method the columns of the alignment are considered as independent one to another and the majority base is selected whereas in our method the whole amplicon sequence is taking into account which could result in different calling sequence. Fig. 2 shows an example of different calling results between the two methods.
Fig. 2.
Difference observed between sequences generated from alignment combined with consensus extraction and full-amplicon based extraction.
When a dominant genome exists in population but with a proportion lower than 50%, the two methods can return different sequences. For example in Fig. 2, if the dominant genome, representing 1/3 in population, carries a G and all the other variants carry an A at this position, our method will return the G from the dominant variant whereas the consensus method will vote for A at this position which is the major base at this position. However, in the case where a dominant variant represents more than 50% in population, the two methods become equivalent and produce the same final sequence. To identify the most abundant variant sequence for each amplicon (Fig. 1, step 5), a clustering step at 100% identity to group and count identical reads is performed using CD-hit (Fu et al., 2012). In the final step of the protocol (Fig. 1, step 6), the most abundant read for each amplicon is used as the representative fragment for the assembly step performed using CAP3 (Huang and Madan, 1999) leading to the dominant genome sequence.
V-ASAP is freely available on gitub https://github.com/caboche/V-ASAP as a docker container. Parameters used for each program are detailed in the Material and Methods section.
2.2. Validation of V-ASAP on Zika virus
To validate our analytical method, we used an available public data on Zika virus (Quick et al., 2017). In this study, 35 primer pairs were designed and the PCR products were sequenced with Illumina overlapping paired-end reads leading to 35 amplicons of 400 bp covering the entire genome of Zika virus (around 10,300 bp). These data were comparable with the data produced in our protocol and the complete genome sequences obtained by the authors were available making this study a good validation dataset.
We applied V-ASAP to this dataset and compared the results with the sequences obtained from the classical method consisting in aligning reads against a reference sequence and extracting a consensus sequence from this alignment (see section “Material and methods - Validation on Zika virus” for more details about the selection of reference sequences and parameters for both approaches). We used four different reference sequences (KU853012.1, HQ224499.1, HQ234501.1, KF268948.1) for the alignment-based method in order to study the impact of the reference sequence selection. These four reference sequences were selected for presenting an increasing phylogenetic distance from the sequenced viruses (see Supplementary material part 1 for the phylogenetic tree). The results are shown in Table 1.
Table 1.
Output comparison of V-ASAP and alignment-based approach using different reference sequences.
| Method | V-ASAP | Alignment | Alignment | Alignment | Alignment |
|---|---|---|---|---|---|
| Reference sequence | None | KU853012.1 | HQ234499.1 | HQ234501.1 | KF268948.1 |
| SRR5121076 (KY317939 – 10608 bp) | |||||
| # contigs | 2 | 2 | 2 | 32 | 33 |
| Total length | 10162 | 10450 | 9546 | 2722 | 2889 |
| N50 | 5254 | 5275 | 4923 | 115 | 111 |
| # unaligned contigs | 0 | 0 | 0 | 13 | 14 |
| Genome fraction (%) | 95.796 | 98.482 | 89.989 | 21.380 | 20.249 |
| # Ns | 0 | 3 | 10 | 3 | 8 |
| # mismatches | 0 | 1 | 0 | 0 | 0 |
| # indels | 0 | 0 | 0 | 0 | 0 |
| SRR5121078 (KY317936 – 10389 bp) | |||||
| # contigs | 3 | 3 | 7 | 28 | 29 |
| Total length | 9965 | 10,061 | 8876 | 2340 | 2449 |
| N50 | 5254 | 5255 | 4584 | 115 | 111 |
| # unaligned contigs | 0 | 0 | 1 | 12 | 11 |
| Genome fraction (%) | 95.919 | 96.843 | 85.388 | 18.433 | 18.760 |
| # Ns | 0 | 0 | 0 | 1 | 23 |
| # mismatches | 0 | 0 | 0 | 0 | 0 |
| # indels | 0 | 0 | 0 | 0 | 0 |
| SRR5121079 (KY317937 – 10462 bp) | |||||
| # contigs | 2 | 2 | 4 | 29 | 33 |
| Total length | 10,162 | 10,209 | 9588 | 2569 | 2773 |
| N50 | 5254 | 2255 | 4924 | 111 | 107 |
| # unaligned contigs | 0 | 0 | 0 | 11 | 13 |
| Genome fraction (%) | 97.132 | 97.582 | 91.617 | 19.853 | 20.340 |
| # Ns | 0 | 0 | 13 | 27 | 9 |
| # mismatches | 0 | 0 | 1 | 0 | 0 |
| # indels | 0 | 0 | 0 | 0 | 0 |
As expected, the results showed that the consensus sequences obtained with alignment-based method varied a lot depending on the selected reference sequence: the more the reference and studied sequences were close the better were the results. With the closest reference sequence (KU853012.1), the results of the three assemblies were reliable: more than 96% of the sequence was covered in 2 or 3 contigs with very few errors (insertions, deletions or mismatches). Only one mismatch was observed for the consensus sequence generated with SRR5121076 dataset. This error in consensus calling was due to a misalignment of several reads in this region: the ends of a number of reads at this position did not align, probably due to an incorrect trimming of these reads in the original dataset. This example showed that the correct alignment of reads is necessary to call the correct consensus and that the alignment step has to be very carefully performed, an incorrect trimming or an incorrect parameters tuning could have a strong impact on results. When the reference sequence was divergent from the studied sequence, the quality of the obtained consensus sequence rapidly fall down. The coverage decreased (around 90% with HQ234499.1 and only around 20% with the more distant sequences HQ234501.1 and KF268948.1). Moreover the assembly became very fragmented with an increasing number of contigs. Another interesting feature was the number of unaligned contigs, i.e. contigs produced by the method but which did not align against the true sequence. This number increased dramatically when the reference sequence was more divergent. This was due to the well-known over-fitting bias of the alignment-based method (Archer et al., 2010): some bases of the reference sequence were selected in place of the true bases from reads because of incorrect alignment of reads. The reference-free method used in V-ASAP resulted in sequences close to the expected ones with metrics similar to the one obtained with the closest reference sequence. The assembly covered more than 95% of the sequence with only 2 or 3 contigs and without any errors for the three datasets. The sequence produced with alignment-based method with the closest reference was a little longer (between 0% and 2%) compared to the sequence obtained with V-ASAP. This difference was due to bad trimming of some reads which were longer than the amplicon sizes. This bad-trimmed reads were not considered in V-ASAP but were in the alignment-based approach leading to additional bases in the consensus sequence. All the results showed that V-ASAP avoid biases due to the alignment step, and even if a close reference sequence is not available, our pipeline is able to produce a relevant sequence which is not the case, as demonstrated here, with alignment-based methods.
2.3. Development and validation of the protocol for HCoV-OC43
The goal of this study was to directly sequence clinical samples from patients infected with HCoV-OC43 isolated at the University Hospital of Lille (France) from nasal swab (noted ‘MDSX’) or bronco-alveolar lavage (noted ‘PR2’) with a protocol that could be used in routine clinical context. HCoV-OC43 has a genome of more than 30 kb and some development, optimization and validation steps were necessary.
Choice of the amplicon length
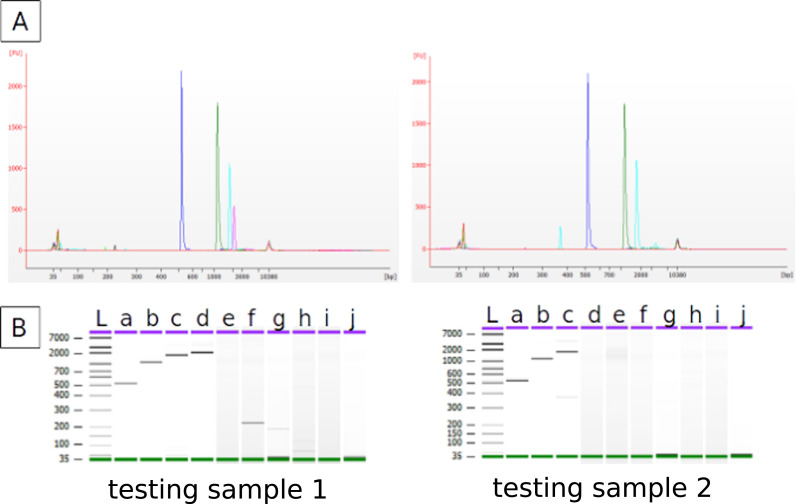
Several sequencing-based methods were used to obtain HCoV-OC43 full-length genomes. Sanger sequencing was the first method used with variable size of amplified regions (St-Jean et al., 2004, Vijgen et al., 2005). Targeted viral nucleic acid capture and RNA sequencing library was recently used to sequence four human HCoV-OC43 isolates (Dinwiddie et al., 2018). The most usual method is to divide the genome into 2.5 kb regions which are amplified, fragmented and sequenced with HTS technologies (Cotten et al., 2013, Taboada et al., 2016). However, depending on the viral load and on the RNA quality, the amplification of the 2.5 kb genome fragments could be troublesome. We tested the possibility to amplify long viral genome fragments from HCoV-OC43 infected samples by testing RT-PCR amplifications of different lengths, from ~500 bp to ~5000 bp with an increment-step of ~500 bp (see Supplementary material part 2 for more details). The Fig. 3 shows the amplification results on two testing clinical samples used for protocol setting which were chosen to be representative of the other clinical samples to be analyzed in this study. Specific amplified fragments were only obtained for short lengths (below ~2000 bp). The first testing sample gave only amplification for products between ~500 bp and ~2000 bp and the second sample only gave amplifications till the ~1500 bp fragment. The fact that no amplification was detected above ~2000 bp illustrates the disqualification of an approach which would start by a long range PCR. The 500 bp fragment size could be amplified for the two testing samples with a good efficiency showing that viral regions of 500 bp had a high probability to be amplified. In addition, fragment of 500 bp were of technical interest as they could be directly sequenced with 300 bp Illumina paired-end overlapping reads. We therefore decided to divide the genome into ~500 bp overlapping fragments.
Fig. 3.
Coronavirus genome amplification trials by RT-PCR for different fragment lengths. A: Capillary electrophoresis profiles of RT-PCR products of the obtained different fragments. B: Agarose gel-like profiles. L: ladder with the sizes alongside (in bp); a to j: RT-PCR product from ~500p to ~5000 bp with an increment-step of ~500 bp.
Primer design and grouping for PCR multiplex
To generate a reliable consensus sequence on which to design primers, 47 HCoV-OC43 genome sequences were aligned using CLC GenomicsWorkbench (Qiagen), the accession numbers of the 47 genomes used are available in Supplementary material part 3. The primers were designed in conserved regions to obtain amplicons of 500–550 bp with a minimum overlap of 16 bp between each amplicon (after trimming of primer sequences) leading to a list of 99 primer pairs (see Supplementary material for the list of primers). In order to decrease the number of PCR reactions, we pooled PCR primer pairs in 12 groups each containing between 3 and 12 PCR primer pairs. The primer pairs were grouped according to their positions on the genome avoiding too short distance between concomitant pairs. A first try of RT-PCR amplicon sequencing performed using a first grouping of primer pairs and amplification efficiency was assessed according to the number of reads obtained for each amplicon after quality filtering. Groups were reorganized to cluster strong amplifications in same groups and weaker amplifications in other groups always keeping the distance criteria in mind. This optimization of the protocol allowed to obtain an amplification for all amplicons and to minimize the difference of amplification efficiencies between amplicon within a same group and between groups. Despite this optimization, a difference in the number of reads per amplicon was observed from 1 to 50,000 reads between the most sequenced amplicon and the less one during the preliminary tests.
After multiplex PCR amplification, the PCR products from the 12 amplicon groups were mixed in equimolar proportions, a library was prepared for each clinical sample and then sequenced (see Material and methods section).
Distribution of variant sequences per amplicon
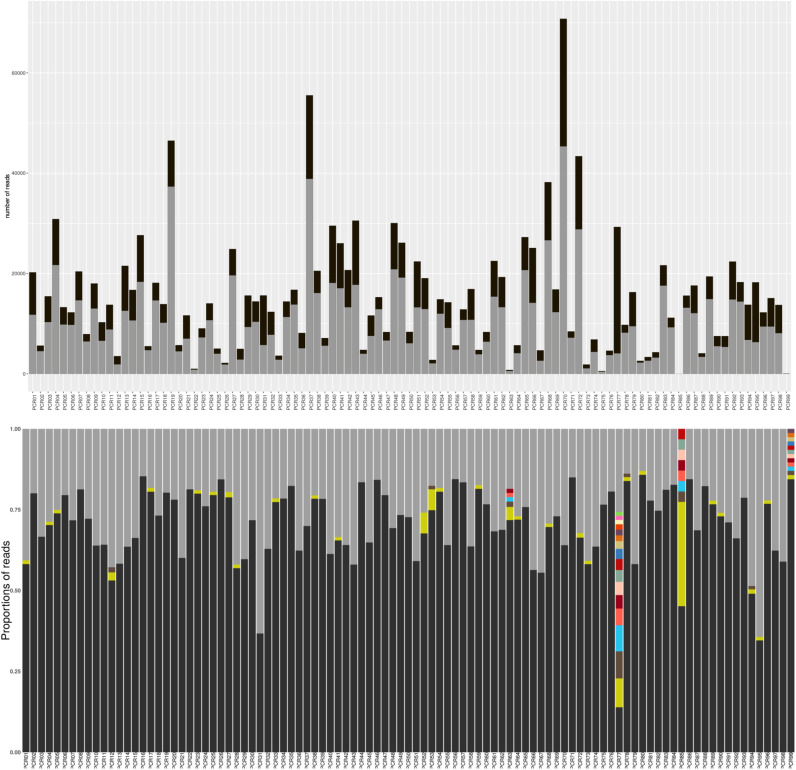
In order to validate that a dominant variant exists for each amplicon, we studied the cluster size distribution, i.e. the number of identical reads in each cluster, for each amplicon. Fig. 4 shows the distribution of cluster sizes for the MDS4 clinical sample.
Fig. 4.
Study of the cluster size distribution for the 99 amplicons (PCR01 to PCR99) from the MDS4 sample. The top panel shows the number of reads for each amplicon (in black) and the number of reads in the biggest cluster (in gray). The bottom panel shows the proportion of each clusters for each amplicon. The proportion of the biggest cluster is in black and the proportion of clusters containing less than 1% of reads were summed up and are in gray.
The cluster size distribution showed that the number of reads was very different between amplicons (Fig. 4 top panel). For example, more than 40,000 reads were obtained for amplicon 70 and only 65 reads for amplicon 99. However, the distribution of the reads in individual amplicons was relatively similar as a dominant sequence was systematically observed for each amplicon representing more than 50% of the reads for 94 of the 99 amplicons (Fig. 4 bottom panel). Only amplicon 77 showed a different profile with several sequences in relatively high proportions. The same experiment was performed for each clinical samples leading to the same conclusions (see Supplementary material part 4). We studied the amplicon 77 that showed a different distribution. We compared the reads from the clusters representing high proportions (more than 30 reads). Almost all reads showed differences in the last 8 nucleotides which contained a stretch of four Ts. This region was overlapping with the next amplicon (Amplicon 78) which did not show this variability problem. We first discarded analytical problem (e.g. bad primer trimming) and we concluded that this variability was due to an amplification or sequencing artifact. However, this artifact did not impact the final built sequence in our method as the region was also covered by another amplicon. Finally, the results showed that selecting the most abundant read as the representative fragment for each amplicon seemed to reflect the reality.
2.4. Sequencing of HCov-OC43 genomes from clinical samples
The complete protocol was applied to 11 clinical samples from patients infected with HCoV-OC43. The sequencing data were analyzed with V-ASAP. The sequencing results and metrics are shown in Table 2. In comparison with previous studies and despite a longer genome sequence, the sequencing of coronavirus genome was very specific. The number of reads that mapped against a HCoV-OC43 sequence was very high (more than 92% for 9 of the samples), showing that the primer design and the biological protocol led to few noise in sequencing data. Only MDS5 and PR2 samples showed very low percentages of mapped reads, 11.30% and 9.39% respectively, due to the fact that the major part of reads was locally aligned against human sequences. For the 11 samples, the number of merged reads was comprised between 1,811,280 and 2,309,880 providing enough sequencing data for reliable analysis.
Table 2.
Results and metrics from the sequencing of the 11 clinical samples.
| Sample | Viralload | Number of freezing/thawing | Library [c](nM)a | Mappedreads (%)b | Mergedreads | Amplicons <10readsc | Amplicons <100readsd | Contigse | Assemblysize (bp)f |
|---|---|---|---|---|---|---|---|---|---|
| MDS1 | + | 3 | ND (7.9) | 92.03 | 2,270,961 | 25 | 41 | 8 | 28.915 |
| MDS2 | + | 3 | 4.76 | 98.87 | 1,972,807 | 0 | 2 | 1 | 30.665 |
| MDS4 | + | 2 | 6.97 | 99.02 | 1,811,280 | 0 | 3 | 1 | 30.664 |
| MDS5 | + | 6 | 0.46 (2.10) | 11.30 | 2,309,880 | 70 | 95 | 17 (+19) | 16.516 (+8738) |
| MDS6 | + | 3 | 3.49 | 97.45 | 1,971,524 | 1 | 7 | 1 | 30.664 |
| MDS11 | +++ | 2 | 5.77 | 99.03 | 1,973,800 | 0 | 2 | 1 | 30.665 |
| MDS12 | ++ | 3 | 0.63 (14.10) | 98.79 | 2,130,557 | 9 | 17 | 1 | 30.419 |
| MDS14 | ++ | 4 | 5.34 | 92.51 | 2,164,740 | 0 | 7 | 1 | 30.664 |
| MDS15 | ++ | 4 | 2.98 (45.20) | 98.47 | 2,118,341 | 4 | 10 | 2 | 30.471 |
| MDS16 | ++ | 3 | 2.43 (37) | 95.81 | 2,180,964 | 4 | 10 | 1 | 30.668 |
| PR2 | + | 3 | 0.29 (1.90) | 9.39 | 2,182,806 | 68 | 81 | 17 (+18) | 19.045 (+8062) |
ND: not detected. Numbers in brackets correspond to the concentration obtained after library amplification.
The “mapped reads” column shows the number of reads mapped against the KX344031 sequence after filtering of PhiX control reads.
Number of amplicons having a sequencing depth lower than 10 reads after merging.
Number of amplicons having a sequencing depth lower than 100 reads after merging.
Number of contigs produced with CAP3. Number of singlets are in brackets.
Size of the final assembly. Numbers in brackets correspond to the cumulative size of singlets.
Some of our samples showed very low library quantity (MDS1, MDS5, MDS12, MDS15, MDS16 and PR2) and a library amplification was needed for these samples. Sequencing depth was not uniform between amplicons along the genome: some amplicons were over-sequenced (until 48,000×) and some others were only sequenced a few times or not at all. Semi-quantitative results of viral load in each sample showed that all samples with relatively high viral load (noted as ++ and +++ in Table 2) led to a complete or nearly-complete assembly (1 or 2 contigs covering more than 99% of the genome sequence). Two of the samples, MDS2 and MDS4, with lower viral load (noted as + in Table 2) also led to a complete assembly with none of their amplicons with a depth below 10 × . On the opposite, poor results were obtained for MDS5 and PR2, leading to a final assembly covering only 53% and 63%, respectively, of the genome sequence. The poor results obtained with MDS5 were probably due to a degradation of the extracted RNA. Indeed, contrary to the other samples, it was subjected to 6 freezing/thawing as for this patient other viruses were searched before searching for HcoV-OC43 presence. We can postulate that the repetitive freezing/thawing of the MDS5 sample caused the degradation of the viral RNA. The PR2 sample is the only sample obtained by broncho-alveolar lavage for which a lower efficiency of the extraction is usually observed, explaining the poor sequencing efficiency and so the poor resulting assembly.
Using this new protocol, full-length assembled genomes (in a single contig) were obtained for 7 out of the 11 clinical samples. For MDS15 sample, 2 contigs were obtained covering more than 99% of the entire genome. 8 contigs leading to a final assembly of 28,915 bp was obtained for MDS1 sample for which the library concentration was not detectable before amplification.
3. Conclusions
Here we introduced a complete protocol called V-ASAP for whole-genome virus sequencing based on multiplex PCR and amplicon sequencing, and a new reference-free analytical pipeline leading to the sequence of the dominant viral genome. We validated our analytical approach on published Zika data and applied the entire protocol to sequence 11 clinical samples infected with HCoV-OC43 virus collected at the University Hospital of Lille. V-ASAP led to a complete or a nearly complete assembly for 9 out of the 11 samples and this even for samples with low viral load. Contrary to classical analytical process, our method avoids alignment biases and is able to assemble virus genome from families presenting highly divergent sequences (only conserved regions are necessary for primer design). It was the first time that the multiplex PCR method was successfully used for amplification of a genome of more than 30 kb. This protocol opens possibilities in the field of molecular virology firstly because it can be easily applied to sequence and assemble other virus genomes. It is also of particular interest to explore over time the evolution of a virus genome during the infection process as one can follow which variants take over the others.
4. Material and methods
4.1. Samples collection and description
Samples were collected from patients hospitalized at the University Hospital of Lille. HCoV-OC43 RNA detection was obtained using Anyplex II RV16 (RV16) kit (Seegene, South Korea) according to the manufacturer's recommendations (Radko et al., 2017). Semi-quantitative results are expressed on a scale from + to +++. Ten of the samples come from nasal swab (noted ‘MDSX’) and 1 from broncho-alveolar lavage (PR2). Viral RNA were extracted using the QIAamp Viral RNA Mini kit (Qiagen, Courtaboeuf, France) according to the manufacturer's protocol. RNA were eluted in 80 µl of DNAse-RNAse free water.
4.2. Amplicon sequencing of HCoV-OC43
16 µl of extracted RNA were reversed transcribed into cDNA using random hexamers primers (Life Technologies) and 400U of SuperScript III reverse transcriptase enzyme (Life Technologies) during 2 h at 45 °C. Obtained cDNA were amplified with the 2X PCR Precision Master Mix (Applied Biological Materials Inc) and multiple primer pairs divided into 12 groups (see primers sequence and groups in Supplementary data). PCR conditions were 40 cycles of annealing at 60 °C for 20 s and extension at 72 °C for 60 s. Amplified products were purified with NucleoMag NGS Clean-up and Size Select (Macherey-Nagel, Hoerdt, France). Purified amplicons were quantified by fluorimetric method (using Qubit dsDNA HS, ThermoFisher Scientific). Amplicons from each group were normalized to have the same equimolar concentration and pooled together to obtain one pool per initial sample.
Sequencing libraries were prepared with 75 ng to 1 µg of pooled DNA, using the NxSeq AmpFREE Low DNA Library Kit and Illumina-compatible adaptors (Lucigen). Each library had a unique 6 bases index. Libraries were quantified with the KAPA Library Quantification Kit Illumina Platforms (Kapa Biosystems) and checked by microfluidic electrophoresis (Bioanalyzer High Sensitivity DNA Kit, Agilent Technologies).
Before sequencing, libraries were normalized at 2 nM and pooled (MDS2, MDS4, MDS6, MDS11 and MDS14). For samples with low library concentration (MDS1, MDS5, MDS12, MDS15, MDS16 and PR2), libraries were amplified by 10 PCR cycles with P5/P7 primers (which are specific of Illumina adaptors), and then normalized to 2 nM before pooling. Libraries were paired-end sequenced (5 or 6 libraries per sequencing run) with 2 × 300 cycles on the Illumina MiSeq.
4.3. V-ASAP
Several public tools are used in V-ASAP. CASPER was used for read merging (option -j). PhiX reads were filtered out with bowtie2 (parameter–local). Reads were then sorted by amplicon with cutadapt (option–discard-untrimmed) and primer sequences were trimmed. For each amplicon, reads were then clustered using CD-HIT (parameters -c 1 -d 0 -s 1). Finally the representative reads for each amplicon were assembled with CAP3 (-o 16 -s 251 -j 31 -p 66 -i 21).
V-ASAP is freely accessible on https://github.com/caboche/V-ASAP as a docker container.
4.4. Validation on Zika virus
Zika datasets (SRR5121076, SRR5121078, SRR5121079) were obtained from SRA (NCBI). Reads were already pre-processed: primers were trimmed and a quality control was probably performed. In our method, primer sequences are used to assign each read to its amplicon of origin, but in these datasets primer sequences were already trimmed and we had to adapt our method for this step. The 22 nucleotides following the true forward primers designed in the original study was considered as “fake forward primers” (the list of these 35 primers is given in Supplementary material part 5). The reverse primers could not be used because the read ends in SRA were trimmed for quality. The “fake forward primers” were used as input of V-ASAP analysis with a change in parameters of CASPER (-g 0.27 which is the threshold for mismatch ratio of best overlap region) and the minimum read size was fixed to 350 bp in the clustering step. All the others steps of V-ASAP remained unchanged.
To study the impact of the reference sequence selection in the alignment-based approach, we selected four sequences based on their increasing phylogenetic distance from the studied Zika sequences. From the phylogenic tree (see Supplementary material part 1), we selected a sequence close to the studied genomes (KU853012.1), a sequence a little further but still on the same branch of the phylogenetic tree (HQ234499.1) and two sequences more phylogenetically distant (HQ234501.1 and KF268948.1). These 4 reference genomes were used for alignment of reads with novoalign (-r Random -l 40 -g 40 -x 20 -t 502). The alignment BAM file was then converted into a pileup file with Samtools mpileup command. VarScan was used with the pileup file to generate the list of all SNPs and indels with a minimum coverage of 1 read. An home-made script was then used to call the consensus: at each position with a minimal coverage of 8 reads, the base with a frequency of more than 51% was called.
Acknowledgements
We thank Cécile Lecoeur for generation of some of the graphs of this article and Maï Zahran for reading the revised version of the manuscript. The work was supported by a Lille Hospital University grant for the project EPI-CoV.
Acknowledgments
Competing interests
The authors declare that they have no competing interests.
Data availability
Sequencing reads are available from BioProject PRJNA507154. Genome sequences were deposited in GenBank with accession numbers: MK303620 for MDS2, MK303621 for MDS4, MK303619 for MDS6, MK303622 for MDS11, MK303623 for MDS12, MK303624 for MDS14, MK327281 for MDS15 and MK303625 for MDS16.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2019.03.006.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- Archer J., Rambaut A., Taillon B.E., Harrigan P.R., Lewis M., Robertson D.L. The evolutionary analysis of emerging low frequency HIV-1 CXCR4 using variants through time--an ultra-deep approach. PLoS Comput. Biol. 2010;6:e1001022. doi: 10.1371/journal.pcbi.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caboche S., Audebert C., Lemoine Y., Hot D. Comparison of mapping algorithms used in high-throughput sequencing: application to Ion Torrent data. BMC Genom. 2014;15:264. doi: 10.1186/1471-2164-15-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Lam T.T., Watson S.J., Palser A.L., Petrova V., Grant P., Pybus O.G., Rambaut A., Guan Y., Pillay D., Kellam P., Nastouli E. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg. Infect. Dis. 2013;19 doi: 10.3201/eid1905.130057. (736–42B) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinwiddie D.L., Hardin O., Denson J.L., Kincaid J.C., Schwalm K.C., Stoner A.N., Abramo T.J., Thompson T.M., Putt C.M., Young S.A., Dehority W.N., Kennedy J.L. Complete genome sequences of four novel human coronavirus OC43 isolates associated with severe acute respiratory infection. Genome Announc. 2018;6 doi: 10.1128/genomeA.00452-18. (e00452-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey M.I., Bradley C.W., Holden K.L., Hewins P., Ngui S.-L., Tedder R., Jumaa P., Smit E. Use of genome sequencing to identify hepatitis C virus transmission in a renal healthcare setting. J. Hosp. Infect. 2017;96:157–162. doi: 10.1016/j.jhin.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Houldcroft C.J., Beale M.A., Breuer J. Clinical and biological insights from viral genome sequencing. Nat. Rev. Microbiol. 2017;15:183–192. doi: 10.1038/nrmicro.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlihan C.F., Frampton D., Ferns R.B., Raffle J., Grant P., Reidy M., Hail L., Thomson K., Mattes F., Kozlakidis Z., Pillay D., Hayward A., Nastouli E. Use of whole-genome sequencing in the investigation of a nosocomial influenza virus outbreak. J. Infect. Dis. 2018;218:1485–1489. doi: 10.1093/infdis/jiy335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S., Lee B., Yoon S. CASPER: context-aware scheme for paired-end reads from high-throughput amplicon sequencing. BMC Bioinforma. 2014;15(Suppl 9):S10. doi: 10.1186/1471-2105-15-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10. [Google Scholar]
- Posada-Cespedes S., Seifert D., Beerenwinkel N. Recent advances in inferring viral diversity from high-throughput sequencing data. Virus Res. 2017;239:17–32. doi: 10.1016/j.virusres.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K., Oliveira G., Robles-Sikisaka R., Rogers T.F., Beutler N.A., Burton D.R., Lewis-Ximenez L.L., de Jesus J.G., Giovanetti M., Hill S.C., Black A., Bedford T., Carroll M.W., Nunes M., Alcantara L.C.J., Sabino E.C., Baylis S.A., Faria N.R., Loose M., Simpson J.T., Pybus O.G., Andersen K.G., Loman N.J. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radko S., Ian Stuart J., Zahariadis G. Evaluation of three commercial multiplex assays for the detection of respiratory viral infections. J. Virol. Methods. 2017;248:39–43. doi: 10.1016/j.jviromet.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean J.R., Jacomy H., Desforges M., Vabret A., Freymuth F., Talbot P.J. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboada B.T., Isa P., Espinoza M.A., Aponte F.E., Arias-Ortiz M.A., Monge-Martínez J., Rodríguez-Vázquez R., Díaz-Hernández F., Zárate-Vidal F., Wong-Chew R.M., Firo-Reyes V., del Río-Almendárez C.N., Gaitán-Meza J., Villaseñor-Sierra A., Martínez-Aguilar G., García-Borjas M., Noyola D.E., Pérez-Gónzalez L.F., López S., Santos-Preciado J.I., Arias C.F. Complete genome sequence of human coronavirus OC43 isolated from Mexico. Genome Announc. 2016;4 doi: 10.1128/genomeA.01256-16. (e01256-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan G., Forbi J.C., Xia G.-L., Fonseca-Ford M., Vazquez R., Khudyakov Y.E., Montiel S., Waterman S., Alpuche C., Goncalves Rossi L.M., Luna N. Full-length genome characterization and genetic relatedness analysis of hepatitis A virus outbreak strains associated with acute liver failure among children. J. Med. Virol. 2014;86:202–208. doi: 10.1002/jmv.23843. [DOI] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moës E., Thoelen I., Wollants E., Lemey P., Vandamme A.-M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Data Availability Statement
Sequencing reads are available from BioProject PRJNA507154. Genome sequences were deposited in GenBank with accession numbers: MK303620 for MDS2, MK303621 for MDS4, MK303619 for MDS6, MK303622 for MDS11, MK303623 for MDS12, MK303624 for MDS14, MK327281 for MDS15 and MK303625 for MDS16.