Abstract
The recent discovery of Middle East Respiratory Coronavirus and another novel dromedary camel coronavirus UAE-HKU23 in dromedaries has boosted interest in search of novel viruses in dromedaries. In this study, fecal samples of 203 dromedaries in Dubai were pooled and deep sequenced. Among the 7330 assembled viral contigs, 1970 were assigned to mammalian viruses. The largest groups of these contigs matched to Picobirnaviridae, Circoviridae, Picornaviridae, Parvoviridae, Astroviridae and Hepeviridae. Many of these viral families were previously unknown to dromedaries. In addition to the high abundance of contigs from Circoviridae (n=598 with 14 complete genomes) and Picobirnaviridae (n=1236), a high diversity of contigs from these two families was found, with the 14 Circoviridae complete genomes forming at least five clusters and contigs from both genogroup I and genogroup II potentially novel picobirnaviruses. Further studies comparing the incidence of these viral families in healthy and sick dromedaries will reveal their pathogenic potential.
Keywords: Metagenomic analysis, Dromedary camel, Novel virus
Highlights
-
•
The viromes of fecal samples from dromedaries in the Middle East were analyzed.
-
•
Circoviridae and Picobirnaviridae are most abundant among the mammalian viruses.
-
•
A high diversity of contigs from Circoviridae and Picobirnaviridae was observed.
Introduction
Camels are one of the most unique mammals on earth. In particular, they have shown perfect adaptation to desert life where the daytime temperature is very high, diurnal temperature range is large and the supply of food and water is scarce. Such adaptations are made through their distinct anatomical and physiological properties, such as short but thick fur, long legs, water conservation and unique fat metabolism. Therefore, camels were used for transportation of people and goods as well as for military uses in the past. In addition, they also provide a good source of meat, milk and wool. They are also important recreational animals in the Middle East and are used for camel racing. Having been associated with humans for at least 5000 years, camels usually pose little physical danger to humans. Occasionally, infectious diseases, such as brucellosis, can be transmitted from camels to humans. Dromedary camels are one of the two surviving old world camel species, namely Camelus dromedarius (dromedary or one-humped camel), which inhabits the Middle East and North and Northeast Africa; and Camelus bactrianus (Bactrian or two-humped camel), which inhabits Central Asia. Among the 20 million camels on earth, 90% are dromedaries.
The recent emergence of Middle East Respiratory Coronavirus (MERS-CoV) from the Middle East and the presence of neutralizing antibodies against MERS-CoV from dromedaries in the Middle East have boosted interest in the search of novel viruses in dromedaries (de Groot et al., 2013, Lau et al., 2013, Perera et al., 2013, Reusken et al., 2013, Zaki et al., 2012). Viruses of at least eight families, including Paramyxoviridae, Flaviviridae, Herpesviridae, Papillomaviridae, Picornaviridae, Poxviridae, Reoviridae and Rhabdoviridae, have been found to infect camels (Al-Ruwaili et al., 2012, Intisar et al., 2009, Khalafalla et al., 2010, Ure et al., 2011, Wernery et al., 2014, Wernery et al., 2008, Wernery and Zachariah, 1999, Yousif et al., 2004). Recently, we have discovered a novel coronavirus, named dromedary camel coronavirus UAE-HKU23 (DcCoV UAE-HKU23), in dromedaries (Woo et al., 2014b). As camels are closely associated with humans, knowledge on the variety of viruses present in this hardy group of animals is important to understand their potential for emergence. In this study, we analyzed the viromes of fecal samples of dromedaries in the Middle East, which is the first metagenomic study on animals of the family Camelidae. The interestingly large number and high diversity of contigs from the Circoviridae and Picobirnaviridae families were also discussed.
Results
Metagenomic analysis of dromedary fecal samples
Fecal samples of 203 dromedaries were pooled and deep sequenced using the Illumina HiSeq 2500 instrument, generating 29,247,514 paired-end 151-bp sequence reads. De novo assembly of the metagenome was performed using IDBA-UD to confirm isolation of viral genomes using default parameter with minimum read length of 200. There were 159,388 contigs ranging in size from 200 to 14,611 bp with a mean contig length of 540 bp.
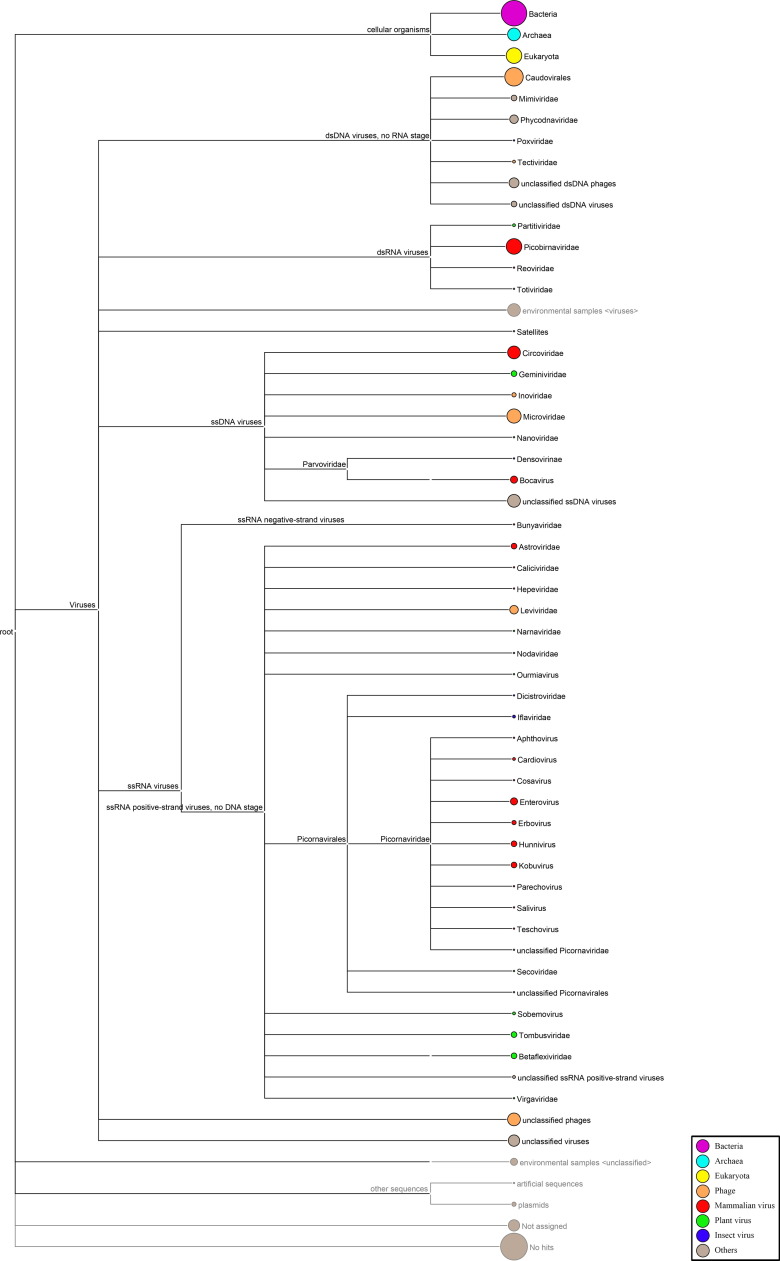
Among 159,388 contigs, 7330 were viral sequences. The most abundant fraction of viral contigs matched to the bacteriophages, including those of the order Caudovirales (n=3805), family Microviridae (n=509) and unclassified phages (n=319) ( Fig. 1). Viral contigs related to plant viruses included those of the families Geminiviridae (n=17), Betaflexiviridae (n=15), Totiviridae (n=2), Nanoviridae (n=1) and Partitiviridae (n=4); and those related to insect viruses included those of the families Iflaviridae (n=5), Dicistroviridae (n=3), Poxviridae (n=3), and Nodaviridae (n=2) and subfamily Densovirinae (n=2) (Fig. 1).
Fig. 1.
Taxonomic classification of sequences with similarity to viruses. Circles located next to taxa are logarithmically proportional to the total number of contigs with BLASTx E<10−5. The figure was generated using MEGAN version 4.70.4.
One thousand nine hundred and seventy (26.9%) of the 7330 viral contigs were assigned to mammalian viruses (Fig. 1). The largest group of the contigs matched to double-stranded RNA viruses in the family Picobirnaviridae (n=1236), followed by single-stranded DNA viruses in the family Circoviridae (n=598). The remaining contigs with homology to the most represented families of mammalian viruses were, in order of decreasing abundance, Picornaviridae [kobuviruses (n=17), enteroviruses (n=26), hunnivirus (n=14), encephalomyocarditis virus (n=4)]; Parvoviridae [porcine bocavirus (n=22), human bocavirus (n=5), feline bocavirus (n=1), gorilla bocavirus (n=1)]; Astroviridae [porcine astrovirus (n=8), feline astrovirus (n=7)]; Hepeviridae [HEV (n=3)]; Reoviridae [rotavirus (n=3)] and Caliciviridae [feline norovirus (n=2)]. These viral contigs showed a wide range of sequence identity to known viruses, suggesting some of these sequences might be derived from novel viruses.
Alignment of sequences of Circoviridae and Picobirnaviridae
The 598 and 1236 contigs that belonged to the families Circoviridae and Picobirnaviridae respectively were analyzed by BLASTx. In both families Circoviridae and Picobirnaviridae, contigs that encoded the corresponding RdRp, capsid proteins and hypothetical proteins were observed (Supplementary Fig. 1).
Phylogenetic analysis of complete genomes in Circoviridae
Repeated terminal sequences in the contigs indicated a circular genome. Fourteen contigs containing complete circular genomes of the Circoviridae family, ranging in size from 2516 to 2977 bp ( Fig. 2). Overall, nucleotide identities to known members of the Circoviridae family were less than 75% for all the genomes. Therefore, according to the ICTV criteria (www.ictvdb.org) which state that circoviruses of the same species should share >75% and >70% nucleotide identity in their complete genome and capsid protein sequences respectively, these viruses found in dromedaries should be novel species in the Circoviridae family.
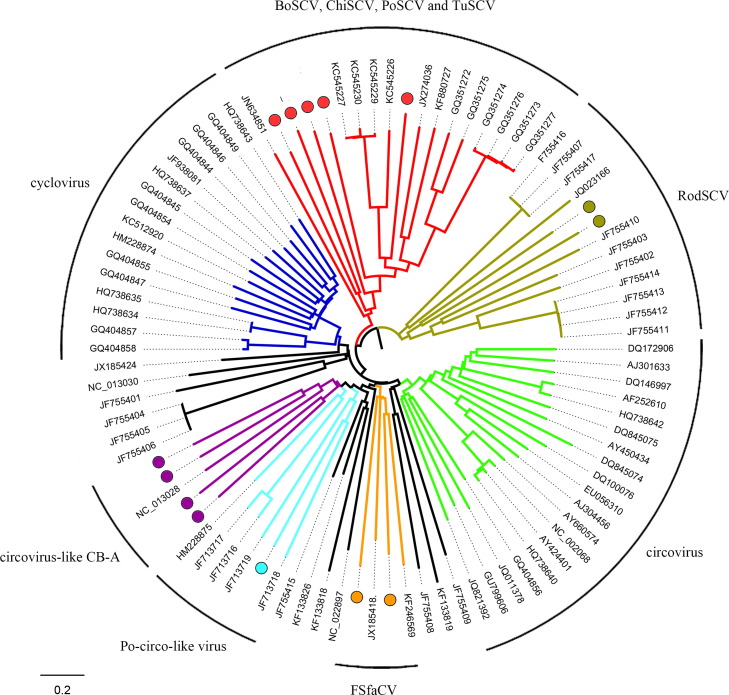
Fig. 2.
Genomic organization of the novel circular genomes identified in this study (A) and representative genomes in the Circoviridae family (B). The 2 major ORFs, encoding the putative replication-associated protein (rep) and the putative capsid protein (cap) and other ORFs are shown. The locations of the potential stem-loop at the origin of replication are marked. The junctions of alternative splicing sites (AS) were indicated with bridge lines. DcSCV, dromedary camel stool-associated circular DNA virus (KM573763–KM573776); FSfaCV, fur seal feces associated circular DNA virus (KF246569); Po-Circo-like virus 41, porcine circovirus-like virus 41 (JF713718); BoSCV, bovine stool-associated circular DNA virus (JN634851); PoSCV, porcine stool-associated circular DNA virus (KF193403); RodSCV, rodent stool-associated circular virus (JF755402); GoCV, goose circovirus (GU320569); PCV2, porcine circovirus 2 (FR823451).
Phylogenetic tree of these 14 complete genomes were constructed with representative complete genomes of circovirus, cyclovirus and circo-like virus sequences, starting at Rep ATG. These 14 complete genomes formed at least five clusters, including one related to porcine circovirus-like virus, five related to bovine stool-associated circular DNA virus (BoSCV), two related to fur seal feces-associated circular DNA virus (FSfaCV), two related to rodent stool associated circular genome virus (RodSCV) and four related to other cirovirus-like virus ( Fig. 3).
Fig. 3.
Phylogenetic analysis based on complete genome nucleotide sequences of Circoviridae. The scale bar indicates the number of nucleotide substitutions per residue. Solid circles mark the contigs determined in this study. Porcine circovirus-like virus, fur seal feces-associated circular DNA virus, bovine stool-associated circular DNA virus, circoviruses, cycloviruses, circovirus-like virus and rodent stool associated circular genome virus are represented by cyan, orange, red, green, blue, purple and citron branches respectively.
Phylogenetic analysis of sequences in Picobirnaviridae
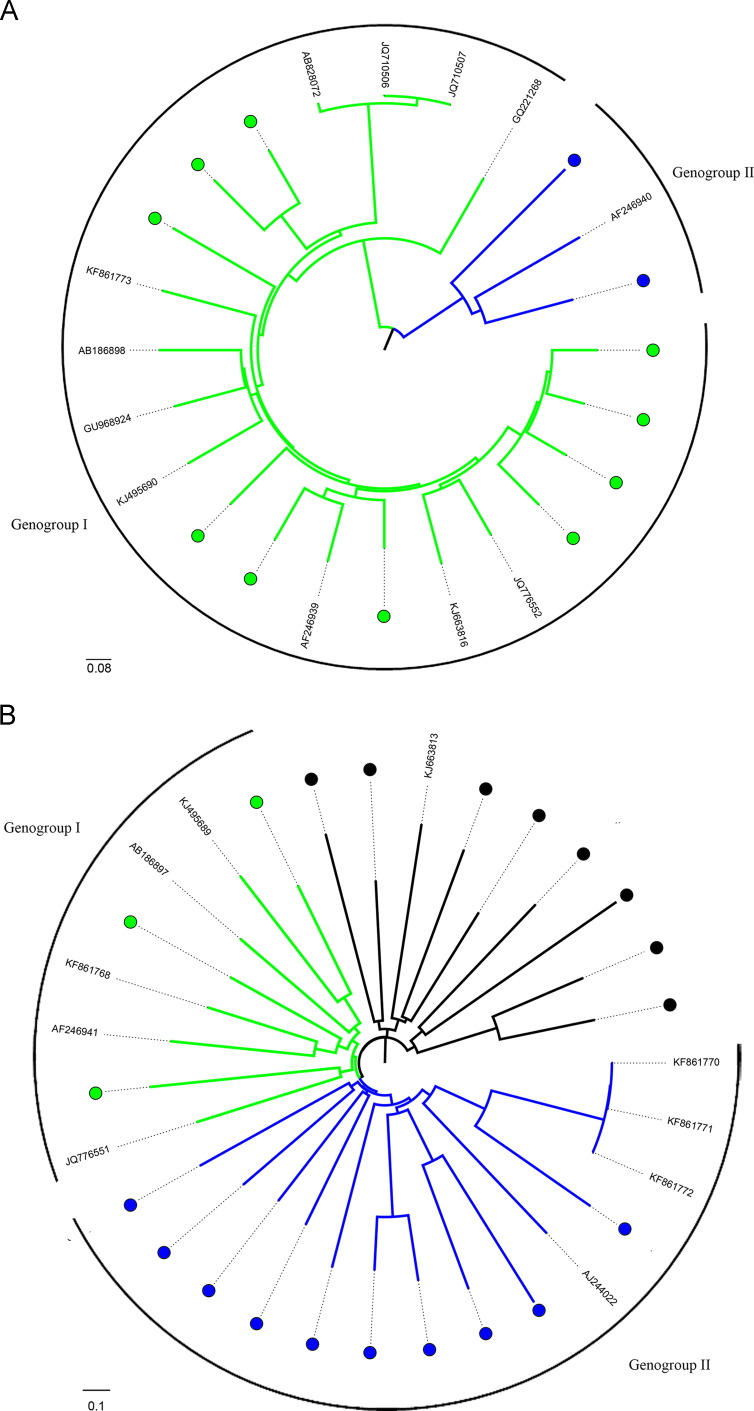
The 12 and 21 contigs that belonged to the family Picobirnaviridae with complete RdRp and capsid genes respectively were further aligned with all available complete RdRp and capsid genes of picobirnaviruses. Phylogenetic tree of the complete RdRp and capsid genes of the picobirnavirus genome is shown in Fig. 4. The contigs were highly diverse. Contigs that belonged to both genogroup I and genogroup II picobirnaviruses were observed. In addition, distinct branches that were not clustered with either genogroup I or genogroup II picobirnaviruses were also observed, suggesting that there may be one or more additional genogroups in picobirnaviruses.
Fig. 4.
Phylogenetic analysis based on complete Picobirnaviridae RNA dependent RNA polymerase (Panel A) and capsid (Panel B) nucleotide sequences. The scale bar indicates the number of nucleotide substitutions per residue. Solid circles mark the contigs determined in this study. Picobirnavirus genogroup I and genogroup II are represented by green and blue branches respectively.
Discussion
In this first metagenomic study on viromes in animals of the family Camelidae, more than 500 contigs (including 14 complete genomes) and around 25% of mammalian virus contigs observed in dromedary fecal samples belonged to the Circoviridae family. Members of the Circoviridae family are small non-enveloped circular single-stranded DNA viruses found in a wide variety of mammals and birds. Circovirus infections are very common and geographically widely distributed. Although subclinical infections are common, circovirus infections have been suggested to be associated with psittacine beak and feather disease, infectious chicken anemia, circovirus disease of pigeons, and the postweaning multisystemic wasting syndrome of pigs (Biagini et al., 2011). Among the metagenomic studies on fecal samples of other mammals, only one study on fecal samples of pigs showed a comparable high number of sequences from the Circoviridae family ( Table 1) (Sachsenroder et al., 2014). At least five metagenomic studies did not show any circovirus sequence (Lager et al., 2012, Li et al., 2011a, Li et al., 2011b, Smits et al., 2013, van den Brand et al., 2012). As for sequence diversity, the 14 complete genomes formed at least five clusters related to different known members of the Circoviridae family, including porcine circovirus-like virus, BoSCV, FSfaCV, RodSCV and other cirovirus-like virus, were observed in this study (Fig. 3). This high diversity of sequences from the Circoviridae family was rarely seen in other metagenomic studies.
Table 1.
Abundance of circoviruses- and picobirnaviruses-related reads in metagenomic analysis of fecal samples in various mammals.
| References | Animal | No. of fecal samples pooled | Next-generation sequencing platform | Abundance of circoviruses-related reads | Abundance of picobirnaviruses-related reads |
|---|---|---|---|---|---|
| Li et al. (2011b) | California sea lion | 47 | Genome sequencer FLX instrument | − | + |
| Li et al. (2010) | Bata | 480 | Genome sequencer FLX instrument | + | − |
| Wu et al. (2012) | Batb | 216 | Illumina senome snalyzer | + | − |
| Ge et al. (2012) | Batc | 6 | Illumina genome analyzer | + | − |
| Bodewes et al. (2013) | Red fox | 13 | Genome sequencer FLX instrument | + | + |
| Phan et al. (2011) | Wild rodentd | 105 | Genome sequencer FLX instrument | + | + |
| Shan et al. (2011) | Pig | 36 | Genome sequencer FLX instrument | + | − |
| Sachsenroder et al. (2014) | Pig | 8 | Genome sequencer FLX instrument | +++ | + |
| Lager et al. (2012) | Pig | 4 | Genome sequencer FLX instrument | − | + |
| Sachsenroder et al. (2014) | Pig | 5 | Genome sequencer FLX instrument | ++ | ++ |
| Cheung et al (2013) | Pig | 6 | Genome sequencer FLX instrument | + | + |
| van den Brand et al. (2012) | Pine marten | 4 | Genome sequencer FLX instrument | − | − |
| van den Brand et al. (2012) | European badger | 3 | Genome sequencer FLX instrument | + | − |
| Smits et al. (2013) | Ferret | 39 | GS Junior instrument | − | + |
| Li et al. (2011a) | Dog | 18 | Genome sequencer FLX instrument | − | − |
| Present study | Dromedary camel | 203 | Illumina senome Analyzer | +++ | +++ |
Include Myotis velifer, Nycticeius humeralis, Perimyotis subflavus, Antrozous pallidus and Tadarida brasiliensis.
Include Myotis ricketti (n=29), Rhinolophus ferrumequinum (n=15), Myotis myotis (n=28), Rhinolophus sinicus (n=19), Hipposideros armiger (n=15), Hipposideros pomona (n=23), Hipposideros cineraceus (n=20), Ia io (n=11), Tylonycteris robustula (n=10), Rhinolophus affinis (n=19) and Miniopterus schreibersii (n=27).
Include Myotis ricketti (n=2), Scotophilus kuhlii (n=2), Hipposideros armiger (n=1) and Myotis spp. (n=1).
Include Peromyscus crinitus (n=7), Peromyscus maniculatus (n=20), Peromyscus truei (n=1), Peromyscus boylii (n=4), Mus musculus (n=20), Neotoma cinerea, Microtus longicaudus (n=1) and Microtus pennsylvanicus (n=51).
The dromedary fecal samples also contain a large number (more than 1000 contigs and more than half of all mammalian virus contigs) and high diversity of picobirnavirus sequences. Picobirnaviruses are small non-enveloped bisegmented double-stranded RNA viruses found in human and a wide variety of mammals and birds. Since its first discovery in fecal samples of humans and rats in 1988 (Pereira et al., 1988a, Pereira et al., 1988b), picobirnaviruses have been reported in other mammals and birds (Browning et al., 1991, Gallimore et al., 1993, Ludert et al., 1991, Masachessi et al., 2007, Nates et al., 2011) and environmental water samples (Hamza et al., 2011). The pathogenicity of picobirnaviruses has not been established. Although picobirnaviruses have been detected in fecal samples from children with diarrhea and in immunocompromised patients, they have also been found in individuals without diarrhea (Delmas, 2011). Although >500 sequences (most are partial sequences) of picobirnaviruses are available in the GenBank database, phylogenetic and evolutionary studies have been hampered by the small number of complete genome sequences of picobirnaviruses available. So far, there are only four complete genome sequences of genogroup I picobirnaviruses, including one from a human picobirnavirus, another from a porcine picobirnavirus, the third from a turkey picobirnavirus and the fourth from a novel picobirnavirus, named otarine picobirnavirus, we discovered in the fecal sample of a California sea lion in an oceanarium in Hong Kong recently (Woo et al., 2012). For these four available genomes, the larger genome segment, segment 1, is 2.3–2.5 kb in length and encodes the capsid protein and one to two putative proteins of unknown function, while the smaller genome segment, segment 2, is 1.6–1.7 kb in length and encodes the viral RdRp. Moreover, no genogroup II picobirnavirus complete genome sequence is available. This large number of picobirnavirus sequences observed in dromedary fecal samples has never been found in metagenomic studies in other animals. For example, pigs are well-known to have a relatively high prevalence of picobirnavirus infections (Banyai et al., 2008, Chen et al., 2014, Ganesh et al., 2012, Giordano et al., 2011, Martinez et al., 2010, Smits et al., 2011), but in the various metagenomic studies on fecal samples of pigs, picobirnavirus sequences only constituted 0–7% of all mammalian virus sequences (Cheung et al., 2013, Lager et al., 2012, Sachsenroder et al., 2012, Sachsenroder et al., 2014, Shan et al., 2011). In fact, in some metagenomic studies, such as those on bats, pine martens, European badgers and dogs, no picobirnavirus sequence was observed (Table 1) (Ge et al., 2012, Li et al., 2011a, Li et al., 2010, van den Brand et al., 2012, Wu et al., 2012). As for the diversity, data from the present metagenomic study suggested the presence of large numbers of both genogroup I and genogroup II picobirnavirus contigs with high diversity in dromedaries (Fig. 4). This is different from results obtained from metagenomic studies for fecal samples of other animals, in which mainly genogroup I picobirnavirus sequences were observed (Bodewes et al., 2013, Cheung et al., 2013, Lager et al., 2012, Li et al., 2011b, Phan et al., 2011, Sachsenroder et al., 2012, Sachsenroder et al., 2014, Smits et al., 2013). Complete genome sequencing of these picobirnaviruses in dromedaries would be invaluable in understanding genome evolution in this understudied family of virus.
In addition to circoviruses and picobirnaviruses, the present metagenomic analysis also revealed other viral families in dromedaries, many of which are previously not known in dromedaries, such as hepatitis E virus, picornaviruses, parvoviruses and astroviruses. The use of centrifugation steps and filters in metagenomics studies may result in the loss of viruses, particularly those with large virions. Unlike three recent studies which detected the presence of MERS-CoV in nose swabs of dromedaries in Qatar, Saudi Arabia and Egypt (Alagaili et al., 2014, Chu et al., 2014, Haagmans et al., 2014), no MERS-CoV was found in the present metagenomic study on fecal samples. This is probably due to the inherent tissue tropism of MERS-CoV. Similarly, our recently discovered DcCoV UAE-HKU23 was also not found in the present metagenomic study on fecal samples collected from adult dromedaries (Woo et al., 2014b). This is probably because DcCoV UAE-HKU23 was mainly detected in dromedary calves, whereas its prevalence in adult dromedaries was very low (Woo et al., 2014b). The plant and insect viral sequences observed in fecal samples of dromedaries were probably results of ingestion of plants and insects by the dromedaries. As for the mammalian virus families, hepatitis E virus has never been reported in camels and its prevalence, genome sequence and phylogenetic analysis have been reported elsewhere (Woo et al., 2014a). As for picornaviruses, astroviruses and parvoviruses, they are also commonly observed in previous metagenomic studies using fecal samples of other animals (Bodewes et al., 2013, Phan et al., 2011, Shan et al., 2011). Parvoviruses and astroviruses are important pathogens in various groups of mammals and these two families of viruses are previously not described in dromedaries. Since some contigs were found to be most closely related to human bocavirus, complete genome sequencing and further phylogenetic analysis would be useful to further understand the relationship between these “dromedary bocaviruses” and human bocaviruses. Although some members of picornaviruses are able to infect Bactrians and cause diseases such as foot-and-mouth disease, dromedaries were known to be not susceptible to these agents and camels are not the reservoir of these picornaviruses (Wernery et al., 2014). Further studies comparing the incidence of these three important families of viruses in healthy and sick dromedaries will reveal their importance in this unique group of animals.
Materials and methods
Sample collection
All fecal samples of dromedaries (Camelus dromedarius) were left-over specimens submitted for coprological studies to Central Veterinary Research Laboratory in Dubai, United Arab Emirates from January to July 2013. The samples originated from different camel premises where dromedaries were kept for racing, including both dromedaries for routine check-up (n=200) and those with diarrhea (n=3). A total of 203 adult dromedaries have been tested in this study.
Sample preparation for Illumina sequencing
The viral transport medium containing the 203 fecal samples were pooled, 200 μl (~0.2 g feces per 2 ml viral transport medium) each, and centrifuged at 10,000g for 10 min. The supernatant was then filtered through a 0.45-μm filter (Millipore, Massachusetts, USA) to remove eukaryotic and bacterial cell-sized particles. The filtrate was treated with a cocktail of DNase enzymes consisting of 14 U of turbo DNase (Ambion, Austin, TX, USA), 20 U of benzonase (Novagen, Madison, WI, USA) and 20 U of RNase One (Promega, Wisconsin, USA) at 37 °C for 60 min in 1×DNase buffer (Ambion, Austin, TX, USA) to digest unprotected nucleic acids. Total RNA from the sample was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). Reverse transcription was performed using SuperScript III reverse transcriptase (Invitrogen, San Diego, CA, USA) and a random primer containing a 20-base arbitrary sequence at the 5′ end followed by a randomized octamer (8 N) at the 3′ end. A single round priming and extension was performed using Klenow fragment polymerase (New England Biolabs, Massachusetts, USA). PCR amplification with primer consisting of only the 20-based arbitrary sequence of the random primer was performed in 20 cycles of 94 °C for 15 s, 60 °C for 30 s and 68 °C for 1 min and a final extension at 68 °C for 7 min in an automated thermal cycler (Applied Biosystems, Foster City, CA, USA). Standard precautions were taken to avoid PCR contamination and no amplified PCR product was observed in negative control. The PCR product was purified using the MinElute PCR Purification Kit (Qiagen, Hilden, Germany) following the manufacturer׳s protocol with slight modification. The purified DNA was eluted in 15 μl of EB buffer and used as the template for library construction.
Library construction for Illumina sequencing
The metagenomic library was prepared using Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) and Nextera XT Index kit (Illumina, San Diego, CA, USA) according to the manufacturer׳s protocol. Briefly, 1 ng of input DNA was tagmented by the Nextera XT transposome at 55 °C for 5 min. This transposome simultaneously fragmented the input DNA and added adapter sequence to the ends, allowing amplification by PCR in subsequent steps. The sequencing library with tagmented DNA was then amplified in 12 cycles of 95 °C for 10 s, 55 °C for 30 s and 72 °C for 30 s and a final extension at 72 °C for 5 min in an automated thermal cycler (Applied Biosystems, Foster City, CA, USA). Amplified DNA library was purified using 1.8×AMPure XP beads (Beckman Coulter, Danvers, MA, USA) to remove very short library fragments from the population. The amplified DNA library was analyzed using the 2100 Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA, USA) to ensure that the sizes of the amplified products were from 250 bp to 1500 bp. Purified sequencing library was quantified using KAPA library quantification kit (Kapa Biosystems, Wilmington, MA, USA) and subsequently sequenced on Illumina HiSeq 2500 with 151 bp paired-end reads (Rapid Run Mode). Image analysis and base calling were performed with SCS2.8/RTA1.8 (Illumina, San Diego, CA, USA). FASTQ file generation and the removal of failed reads were performed using CASAVA ver.1.8.2 (Illumina, San Diego, CA, USA). The Illumina data are available from the NCBI SRA database (http://www.ncbi.nlm.nih.gov/sra) accession number SRP047227.
Analysis of sequence reads
Illumina sequence reads were adapter and quality trimmed using Trimmomatic with the Nextera adapter fasta sequences and following parameters: leading 3, trailing 3, sliding window 4:15, minimum length 36 bp (Lohse et al., 2012). Trimmed paired-end reads were de novo assembled in silico with IDBA-UD 1.1.1 with default parameters and fixed threshold for a minimum contig length of 200 bp (Peng et al., 2012). IDBA-UD algorithm is based on the de Bruijn graph approach for assembling reads. The contigs that aligned to rRNA sequences from the SILVA rRNA database were initially removed using Bowtie2 (Langmead and Salzberg, 2012). The remaining contigs were compared to non-redundant protein sequences (nr) database from NCBI (http://www.ncbi.nlm.nih.gov), which contains non-redundant sequences from GenBank translations (i.e. GenPept) and sequences from other databanks (Refseq, PDB, SwissProt, PIR and PRF), using BLASTx with an E-value cutoff of 10−5. BLAST results were parsed to save the best hits for each sequence. The best-hit sequences were individually annotated to note the sources of the matching sequences (virus, phage, bacteria, archaea and eukaryotes). Sequences were also analyzed using a metagenomic annotation tool, MEGAN version 4.70.4, to assign each sequence into different taxa present in the metagenomic sequences using the NCBI taxonomic database (Huson et al., 2011).
Phylogenetic analysis of sequences in Circoviridae and Picobirnaviridae
After de novo assembly, there were 598 and 1236 contigs belonging to the families Circoviridae and Picobirnaviridae respectively. Fourteen contigs containing complete circular genomes of the Circoviridae family were used for phylogenetic analysis. The 14 complete genome sequences have been submitted to GenBank with accession numbers KM573763–KM573776. Twelve and 21 contigs that covered complete RdRp and capsid genes respectively in the picobirnavirus genome were used for phylogenetic analysis. The 12 RdRp and 21 capsid sequences have been submitted to GenBank with accession numbers KM573798–KM573809 and KM573777–KM573797, respectively.
Phylogenetic analysis was performed by the neighbor-joining method, using Jukes–Cantor substitution model with gamma distributed rate variation and bootstrap values calculated from 1000 trees.
Acknowledgments
We thank Dr. Wing-Man Ko, Secretary for Food and Health and Dr. Constance Chan, Director of Department of Health, HKSAR, The Peoples׳ Republic of China; members of the Centre for Genomic Sciences, The University of Hong Kong, for their technical support; and Shanty Jose and Sweena Liddle from Central Veterinary Research Laboratory for their help in sample preparation.
This work is partly supported by the HKSAR Health and Medical Research Fund; Seed Funding for TRS and Strategic Research Theme Fund, The University of Hong Kong; Theme-based Research Scheme, Research Grant Council Grant, University Grant Council; and Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2014.09.020.
Contributor Information
Patrick C.Y. Woo, Email: pcywoo@hkucc.hku.hk.
Ulrich Wernery, Email: cvrl@cvrl.ae.
Appendix A. Supplementary materials
Supplementary Fig. 1. Sequence identity distribution analysis of contig sequences related to Circoviridae (Panel A) and Picobirnaviridae (Panel B). Each dot represents an assembled sequence contig with the corresponding amino acid identity (best BLASTx match with E<10−5). The figure was generated using Excel based on the blast results with manual annotation.
Supplementary Material
References
- Al-Ruwaili M.A., Khalil O.M., Selim S.A. Viral and bacterial infections associated with camel (Camelus dromedarius) calf diarrhea in North Province, Saudi Arabia. Saudi J. Biol. Sci. 2012;19:35–41. doi: 10.1016/j.sjbs.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagaili A.N., Briese T., Mishra N., Kapoor V., Sameroff S.C., de Wit E., Munster V.J., Hensley L.E., Zalmout I.S., Kapoor A., Epstein J.H., Karesh W.B., Daszak P., Mohammed O.B., Lipkin W.I. Middle East respiratory syndrome coronavirus infection in dromedary camels in saudi arabia. mBio. 2014;5:e00884–00814. doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyai K., Martella V., Bogdan A., Forgach P., Jakab F., Meleg E., Biro H., Melegh B., Szucs G. Genogroup I picobirnaviruses in pigs: evidence for genetic diversity and relatedness to human strains. J. Gen. Virol. 2008;89:534–539. doi: 10.1099/vir.0.83134-0. [DOI] [PubMed] [Google Scholar]
- Biagini P., Bendinelli M., Hino S., Kakkola L., Mankertz A., Niel C., Okamoto H., Raidal S., Teo C.G., Todd D. Circoviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy, Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses, International Union of Microbiological Societies, Virology Division. Elsevier Academic Press; USA: 2011. pp. 343–349. [Google Scholar]
- Bodewes R., van der Giessen J., Haagmans B.L., Osterhaus A.D., Smits S.L. Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J. Virol. 2013;87:7758–7764. doi: 10.1128/JVI.00568-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning G.F., Chalmers R.M., Snodgrass D.R., Batt R.M., Hart C.A., Ormarod S.E., Leadon D., Stoneham S.J., Rossdale P.D. The prevalence of enteric pathogens in diarrhoeic thoroughbred foals in Britain and Ireland. Equine Vet. J. 1991;23:405–409. doi: 10.1111/j.2042-3306.1991.tb03751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Sun H., Lan D., Hua X., Cui L., Yuan C., Yang Z. Molecular detection of genogroup I and II picobirnaviruses in pigs in China. Virus genes. 2014;48:553–556. doi: 10.1007/s11262-014-1058-8. [DOI] [PubMed] [Google Scholar]
- Cheung A.K., Ng T.F., Lager K.M., Bayles D.O., Alt D.P., Delwart E.L., Pogranichniy R.M., Kehrli M.E., Jr. A divergent clade of circular single-stranded DNA viruses from pig feces. Arch. Virol. 2013;158:2157–2162. doi: 10.1007/s00705-013-1701-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu K.W., Poon L.M., Gomaa M., Shehata M., Perera A.P.M., Zeid A., Rifay S., Siu Y., Guan Y., Webby J., Ali A., Peiris M., Kayali G. MERS coronaviruses in dromedary camels, Egypt. Emerg. Infect. Dis. 2014;20:1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Middle east respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B. Picobirnaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy, Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses, International Union of Microbiological Societies, Virology Division. Elsevier Academic Press; USA: 2011. pp. 535–539. [Google Scholar]
- Gallimore C., Lewis D., Brown D. Detection and characterization of a novel bisegmented double-stranded RNA virus (picobirnavirus) from rabbit faeces. Arch. Virol. 1993;133:63–73. doi: 10.1007/BF01309744. [DOI] [PubMed] [Google Scholar]
- Ganesh B., Banyai K., Kanungo S., Sur D., Malik Y.S., Kobayashi N. Detection and molecular characterization of porcine picobirnavirus in feces of domestic pigs from kolkata, India. Indian J. Virol.: Off. Organ of Indian Virol. Soc. 2012;23:387–391. doi: 10.1007/s13337-012-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li Y., Yang X., Zhang H., Zhou P., Zhang Y., Shi Z. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 2012;86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M.O., Martinez L.C., Masachessi G., Barril P.A., Ferreyra L.J., Isa M.B., Valle M.C., Massari P.U., Nates S.V. Evidence of closely related picobirnavirus strains circulating in humans and pigs in Argentina. J. Infect. 2011;62:45–51. doi: 10.1016/j.jinf.2010.09.031. [DOI] [PubMed] [Google Scholar]
- Haagmans B.L., Al Dhahiry S.H., Reusken C.B., Raj V.S., Galiano M., Myers R., Godeke G.J., Jonges M., Farag E., Diab A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., Al Romaihi H.E., Al Khal A., Bermingham A., Osterhaus A.D., Alhajri M.M., Koopmans M.P. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I.A., Jurzik L., Uberla K., Wilhelm M. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011;45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Huson D.H., Mitra S., Ruscheweyh H.J., Weber N., Schuster S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intisar K.S., Ali Y.H., Khalafalla A.I., Mahasin E.A., Amin A.S. Natural exposure of Dromedary camels in Sudan to infectious bovine rhinotracheitis virus (bovine herpes virus-1) Acta Trop. 2009;111:243–246. doi: 10.1016/j.actatropica.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Khalafalla A.I., Saeed I.K., Ali Y.H., Abdurrahman M.B., Kwiatek O., Libeau G., Obeida A.A., Abbas Z. An outbreak of peste des petits ruminants (PPR) in camels in the Sudan. Acta Trop. 2010;116:161–165. doi: 10.1016/j.actatropica.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Lager K.M., Ng T.F., Bayles D.O., Alt D.P., Delwart E.L., Cheung A.K. Diversity of viruses detected by deep sequencing in pigs from a common background. J. Vet. Diagn. Investig.: Off. Publ. Am. Assoc. Vet. Lab. Diagn. Inc. 2012;24:1177–1179. doi: 10.1177/1040638712463212. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Li K.S., Tsang A.K., Lam C.S., Ahmed S., Chen H., Chan K.H., Woo P.C., Yuen K.Y. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of Pipistrellus Bat Coronavirus HKU5 in Japanese Pipistrelle: implications for the origin of the novel middle east respiratory syndrome coronavirus. J. Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Pesavento P.A., Shan T., Leutenegger C.M., Wang C., Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 2011;92:2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Shan T., Wang C., Cote C., Kolman J., Onions D., Gulland F.M., Delwart E. The fecal viral flora of California sea lions. J. Virol. 2011;85:9909–9917. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Victoria J.G., Wang C., Jones M., Fellers G.M., Kunz T.H., Delwart E. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 2010;84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Bolger A.M., Nagel A., Fernie A.R., Lunn J.E., Stitt M., Usadel B. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludert J.E., Hidalgo M., Gil F., Liprandi F. Identification in porcine faeces of a novel virus with a bisegmented double stranded RNA genome. Arch. Virol. 1991;117:97–107. doi: 10.1007/BF01310495. [DOI] [PubMed] [Google Scholar]
- Martinez L.C., Masachessi G., Carruyo G., Ferreyra L.J., Barril P.A., Isa M.B., Giordano M.O., Ludert J.E., Nates S.V. Picobirnavirus causes persistent infection in pigs. Infect. Genet. Evol.: J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2010;10:984–988. doi: 10.1016/j.meegid.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Masachessi G., Martinez L.C., Giordano M.O., Barril P.A., Isa B.M., Ferreyra L., Villareal D., Carello M., Asis C., Nates S.V. Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch. Virol. 2007;152:989–998. doi: 10.1007/s00705-006-0900-2. [DOI] [PubMed] [Google Scholar]
- Nates S.V., Gatti M.S.V., Ludert J.E. The picobirnavirus: an integrated view on its biology, epidemiology and pathogenic potential. Future Virol. 2011;6:13. [Google Scholar]
- Peng Y., Leung H.C., Yiu S.M., Chin F.Y. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- Pereira H.G., Fialho A.M., Flewett T.H., Teixeira J.M., Andrade Z.P. Novel viruses in human faeces. Lancet. 1988;2:103–104. doi: 10.1016/s0140-6736(88)90032-3. [DOI] [PubMed] [Google Scholar]
- Pereira H.G., Flewett T.H., Candeias J.A., Barth O.M. A virus with a bisegmented double-stranded RNA genome in rat (Oryzomys nigripes) intestines. J. Gen. Virol. 1988;69(Pt 11):2749–2754. doi: 10.1099/0022-1317-69-11-2749. [DOI] [PubMed] [Google Scholar]
- Perera R., Wang P., Gomaa M., El-Shesheny R., Kandeil A., Bagato O., Siu L., Shehata M., Kayed A., Moatasim Y., Li M., Poon L., Guan Y., Webby R., Ali M., Peiris J., Kayali G. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveillance. 2013;18:20574. doi: 10.2807/1560-7917.es2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- Phan T.G., Kapusinszky B., Wang C., Rose R.K., Lipton H.L., Delwart E.L. The fecal viral flora of wild rodents. PLoS Pathog. 2011;7:e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Vries L.S., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortazar-Schmidt C., Drosten C., Koopmans M.P. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachsenroder J., Twardziok S., Hammerl J.A., Janczyk P., Wrede P., Hertwig S., Johne R. Simultaneous identification of DNA and RNA viruses present in pig faeces using process-controlled deep sequencing. PloS One. 2012;7:e34631. doi: 10.1371/journal.pone.0034631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachsenroder J., Twardziok S.O., Scheuch M., Johne R. The general composition of the faecal virome of pigs depends on age, but not on feeding with a probiotic bacterium. PloS One. 2014;9:e88888. doi: 10.1371/journal.pone.0088888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T., Li L., Simmonds P., Wang C., Moeser A., Delwart E. The fecal virome of pigs on a high-density farm. J. Virol. 2011;85:11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Poon L.L., van Leeuwen M., Lau P.N., Perera H.K., Peiris J.S., Simon J.H., Osterhaus A.D. Genogroup I and II picobirnaviruses in respiratory tracts of pigs. Emerg. Infect. Dis. 2011;17:2328–2330. doi: 10.3201/eid1712.110934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Raj V.S., Oduber M.D., Schapendonk C.M., Bodewes R., Provacia L., Stittelaar K.J., Osterhaus A.D., Haagmans B.L. Metagenomic analysis of the ferret fecal viral flora. PloS One. 2013;8:e71595. doi: 10.1371/journal.pone.0071595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ure A.E., Elfadl A.K., Khalafalla A.I., Gameel A.A., Dillner J., Forslund O. Characterization of the complete genomes of Camelus dromedarius papillomavirus types 1 and 2. J. Gen. Virol. 2011;92:1769–1777. doi: 10.1099/vir.0.031039-0. [DOI] [PubMed] [Google Scholar]
- van den Brand J.M., van Leeuwen M., Schapendonk C.M., Simon J.H., Haagmans B.L., Osterhaus A.D., Smits S.L. Metagenomic analysis of the viral flora of pine marten and European badger feces. J. Virol. 2012;86:2360–2365. doi: 10.1128/JVI.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernery U., Kinne J., Schuster R.F. OIE Book; Paris, France: 2014. Infectious disorders of camelids. [Google Scholar]
- Wernery U., Knowles N.J., Hamblin C., Wernery R., Joseph S., Kinne J., Nagy P. Abortions in dromedaries (Camelus dromedarius) caused by equine rhinitis A virus. J. Gen. Virol. 2008;89:660–666. doi: 10.1099/vir.0.82215-0. [DOI] [PubMed] [Google Scholar]
- Wernery U., Zachariah R. Experimental camelpox infection in vaccinated and unvaccinated dromedaries. Zent. Veterinar. Reihe B. J. Vet. Med. Ser. B. 1999;46:131–135. doi: 10.1111/j.0931-1793.1999.00250.x. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Bai R., Teng J.L., Lee P., Martelli P., Hui S.W., Yuen K.Y. Complete genome sequence of a novel picobirnavirus, otarine picobirnavirus, discovered in California sea lions. J. Virol. 2012;86:6377–6378. doi: 10.1128/JVI.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Teng J.L., Tsang A.K., Joseph M., Wong E.Y., Y., T., Sivakumar S., Xie J., Bai R., Wernery R., Wernery U., Yuen K.Y. New hepatitis E virus genotype in camels, the Middle East. Emerg. Infect. Dis. 2014;20:1044–1048. doi: 10.3201/eid2006.140140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Wernery U., Wong E.Y., Tsang A.K., Johnson B., Yip C.C., Lau C.C., Sivakumar S., Cai J.P., Fan R.Y., Chan K.H., Mareena R., Yuen K.Y. Novel betacoronavirus in dromedaries of the middle East, 2013. Emerg. Infect. Dis. 2014;20:560–572. doi: 10.3201/eid2004.131769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ren X., Yang L., Hu Y., Yang J., He G., Zhang J., Dong J., Sun L., Du J., Liu L., Xue Y., Wang J., Yang F., Zhang S., Jin Q. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 2012;86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousif A.A., Braun L.J., Saber M.S., Aboelleil T., Chase C.C.L. Cytopathic genotype 2 bovine viral diarrhea virus in dromedary camels. Arab J. Biotechnol. 2004;7:8. [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Sequence identity distribution analysis of contig sequences related to Circoviridae (Panel A) and Picobirnaviridae (Panel B). Each dot represents an assembled sequence contig with the corresponding amino acid identity (best BLASTx match with E<10−5). The figure was generated using Excel based on the blast results with manual annotation.
Supplementary Material